Abstract
The ongoing novel coronavirus disease (COVID-19) pandemic makes us painfully perceive that our bullet shells are blank so far for fighting against severe human coronavirus (HCoV). In spite of vast research work, it is crystal clear that the evident does not warrant the commercial blossoming of anti-HCoV drugs. In this circumstance, drug repurposing and/or screening of databases are the only fastest option. This study is an initiative to recapitulate the medicinal chemistry of severe acute respiratory syndrome (SARS)-CoV-2 (SARS-CoV-2). The aim is to present an exquisite delineation of the current research from the perspective of a medicinal chemist to allow the rapid development of anti-SARS-CoV-2 agents.
Keywords: COVID-19, SARS-CoV-2, Anti-HCoV agent, Drug repurposing, Target based screening, Molecular modelling
Graphical abstract
1. Introduction
Coronaviruses (CoVs) typically cause mild to severe respiratory and intestinal infections in mammals, including humans [1,2]. It belongs to the family Coronaviridae which comes under the order Nidovirales [3,4]. CoVs are classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus and Deltacoronavirus. The first two genera (i.e., alpha- and betacoronavirus mainly infect mammals, whereas, gammacoronaviruses and deltacoronaviruses foul avian species.
More than 60 years have passed since the identification of first human CoV (HCoV) was documented as a respiratory tract modulator [5,6]. In December 2019, several clusters (epidemiologically associated with a seafood and animal market in Wuhan, China) of patients suffering fever, illness, severe respiratory tract infections and pneumonia of unknown origin were reported [[7], [8], [9], [10], [11], [12]]. This finally leaded to the isolation of a novel coronavirus (2019-nCoV) and the disease recently named as COVID-19. World Health Organization (WHO) already characterized COVID-19 as world pandemic [13]. This infection has spread over to 216 countries and territories [14].
Before COVID-19 outbreak, there were six species of HCoVs that were reported for their association with respiratory tract infections (Table 1 ).
Table 1.
Different types of human coronavirus (HCoVs).
| Discovery | HCoV genera | Coronaviruses | Natural Host | Cellular receptor |
|---|---|---|---|---|
| 1966 | α-CoV | HCoV-229E | Bats | Human aminopeptidase N (CD13) |
| 1967 | β-CoV | HCoV-OC43 | Cattle | 9-O-Acetylated sialic acid |
| 2003 | β-CoV | SARS-CoV | Palm Civets | ACE2 |
| 2004 | α-CoV | HCoV-NL63 | Palm Civets, Bats | ACE2 |
| 2005 | β-CoV | HCoV-HKU1 | Mice | 9-O-Acetylated sialic acid |
| 2012 | β-CoV | MERS-CoV | Bats, Camels | DPP4 |
| 2019 | β-CoV | SARS-CoV-2 | Bats, ? | ACE2 |
These six HCoVs strains are - (a) HCoV-229E, (b) HCoV-OC43, (c) HCoV- Hong Kong University 1 (HCoV-HKU1), (d) HCoV-NL63, (e) severe acute respiratory syndrome (SARS)-CoV (SARS-CoV) and (f) Middle East respiratory syndrome (MERS)-CoV (MERS-CoV) [15]. The seventh strain of HCoV is novel coronavirus (2019-nCOV aka SARS-CoV-2) which is taxonomically belongs to the Betacoronavirus genre and possesses high nucleotide sequence similarity with SARS-CoV and MERS-CoV [[16], [17], [18], [19], [20]].
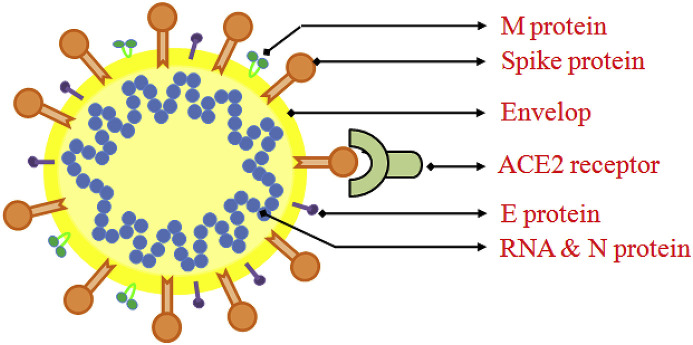
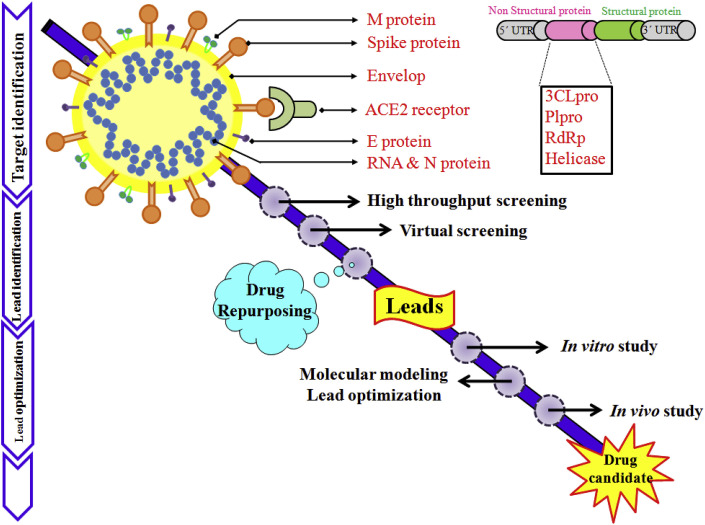
SARS-CoV-2 is a positive-sense single-stranded RNA viruses surrounded by an envelope (Fig. 1 ).
Fig. 1.
Schematic representation of coronavirus structure showing M (membrane) protein, S (Spike) protein, E (envelope) protein, N (nucleocapsid) protein & RNA along with the receptor ACE2.
SARS-CoV-2, about 30,000 bp single-stranded RNA virus, utilizes host cellular components to accomplish its physiological affairs such as viral entry, the assembly as well as budding of virions, genomic replication, and protein synthesis, subsequently executes pathological damage to the host [[21], [22], [23]]. Thus, punctuating any juncture of viral life cycle by small molecules, peptides, vaccines or physical elements may potentially gain therapeutic benefit to host. Depending on several viral targets (Fig. 1) related to the stages of viral life cycle, novel anti-viral agents may be designed and discovered. Nonetheless, different structure-based modeling techniques and numerous ligand-based computational techniques may be fruitful strategy to design newer inhibitors against SARS-CoV-2 [[24], [25], [26]].
Meanwhile, the hefty menace posed by current outbreak of COVID-19, it is obvious that the scientific community is looking for effective drugs within plausible time. The coherent development and well organised strategies remains the only hope to triumph the battle against partially known SARS-CoV-2. Now, repurposing of existing anti-viral drugs based on previous ground work of closely related coronavirus and rapid screening of drug databases is one of the strategic and economic ways to eradicate COVID-19 pandemic [[27], [28], [29]]. The traditional bioinformatics and chemo-informatics approaches readily generated new data into SARS-CoV-2 research at an explosive pace.
Considering the severity of the spread of COVID-19, this study is in-line with the concept of finding the chemo-types to expedite the process of anti-HCoV drug discovery. Here, an exquisite picture of the recent research including target-based and biological screening is provided. We includes virtual (in silico) as well as experimental (in vitro) screening approaches in response to SARS-CoV-2 reported until April 2020. The main aim is to provide the scientific community with an overview of the medicinal chemistry of SARS-CoV-2 to allow the rapid development of anti-viral agents. This work, a part of our rational drug design and discovery [[30], [31], [32], [33], [34], [35], [36]], is an initiative to pave the way of anti-SARS-CoV-2 drug discovery paradigm that could help to facilitate the global efforts to fight against COVID-19.
2. Targets for therapeutic intervention
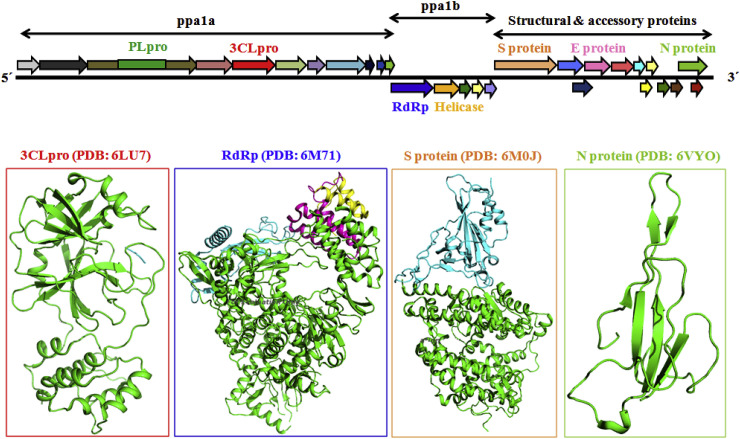
The details structural biology of the SARS-CoV-2 virus is yet to be discovered. It contains a 30,000 bp, single-stranded positive sense RNA genome which is encapsulated within a membrane envelope (Fig. 1) [[37], [38], [39]]. It recruits multi-subunit replication machinery [40]. The genome of SARS-CoV-2 comprises about 30,000 nucleotides with ten Open Reading Frames (ORFs). The 3′ terminal regions encodes for several structural proteins including spike (S), membrane envelope (E) and nucleo-capsid (N) proteins (Fig. 1) whereas, the 5′ terminal ORF1ab responsible for two viral replicase polyproteins namely pp1a and pp1b. Upon proteolytic cleavage of these two viral replicase polyproteins fabricate sixteen non-structural proteins (nsp) (Fig. 2 ) [37].
Fig. 2.
Schematic plot of the SARS-CoV-2 genome and proteome showing different polyproteins (pp1a and pp1b) along with the structural and accessory proteins. Abbreviations used are: PL2-Pro, papain-like protease; 3CLpro, virus main protease; RdRp, RNA-dependent RNA polymerase; Helicase, Zn2+-dependent helicase; S protein, spike protein; E, envelope glycoprotein; M, matrix; N, nucleocapsid; PDB, protein data bank.
Currently there are no broad spectrum inhibitors of SARS-CoV-2 viral proteins are reported. Mainly, five SARS-CoV-2 targets are being investigated including receptor binding domain (RBD)/Spike protein, N protein, E protein, 3CLpro and RdRp (Fig. 2) [[41], [42], [43]]. However, most of the viral proteins would be potential future targets and detailed understanding of their role is required to stimulate chemotherapeutic arbitration against HCoVs. A comparative list of reported crystal structures of 2019-nCoV is depicted in Table 2 as available from Protein Data Bank [44].
Table 2.
List of reported crystal structures of SARS-CoV-2 as available from Protein Data Bank (April, 2020).
| Target | PDB | Date | Method |
|---|---|---|---|
| Chimeric receptor-binding domain complexed with its receptor human ACE2 | 6VW1 | Deposited: 2020-02-18 Released: 2020-03-04 |
X-RAY DIFFRACTION Resolution: 2.68 Å |
| Spike receptor-binding domain bound with ACE2 | 6M0J | Deposited: 2020-02-21 Released: 2020-03-18 |
X-RAY DIFFRACTION Resolution: 2.45 Å |
| Receptor binding domain in complex with human antibody CR3022 | 6W41 | Deposited: 2020-03-09 Released: 2020-03-25 |
X-RAY DIFFRACTION Resolution: 3.08 Å |
| receptor binding domain in complex with CR3022 Fab | 6YLA | Deposited: 2020-04-06 Released: 2020-04-15 |
X-RAY DIFFRACTION Resolution: 2.42 Å |
| SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | 6M71 | Deposited: 2020-03-16 Released: 2020-04-01 |
ELECTRON MICROSCOPY Resolution: 2.90 Å |
| Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with MES | 6WCF | Deposited: 2020-03-30 Released: 2020-04-15 |
X-RAY DIFFRACTION Resolution: 1.06 Å |
| SARS-CoV-2 3CL protease (3CL pro) in complex with a novel inhibitor | 6M2N | Deposited: 2020-02-28 Released: 2020-04-15 |
X-RAY DIFFRACTION Resolution: 2.20 Å |
| Peptide-bound SARS-CoV-2 Nsp9 RNA-replicase | 6W9Q | Deposited: 2020-03-23 Released: 2020-04-08 |
X-RAY DIFFRACTION Resolution: 2.05 Å |
| The N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein | 6YI3 | Deposited: 2020-03-31 Released: 2020-04-08 |
SOLUTION NMR |
| The crystal structure of papain-like protease of SARS-CoV-2 | 6W9C | Deposited: 2020-03-22 Released: 2020-04-01 |
X-RAY DIFFRACTION Resolution: 2.70 Å |
| The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | 6LU7 | Deposited: 2020-01-26 Released: 2020-02-05 |
X-RAY DIFFRACTION Resolution: 2.16 Å |
| Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in complex with AMP | 6W6Y | Deposited: 2020-03-18 Released: 2020-03-25 |
X-RAY DIFFRACTION Resolution: 1.45 Å |
| The 1.9 A Crystal Structure of NSP15 Endoribonuclease from SARS-CoV-2 in the Complex with a Citrate | 6W01 | Deposited: 2020-02-28 Released: 2020-03-11 |
X-RAY DIFFRACTION |
| Crystal structure of RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2 | 6VYO | Deposited: 2020-02-27 Released: 2020-03-11 |
X-RAY DIFFRACTION Resolution: 1.70 Å |
| SARS-CoV-2 spike ectodomain structure (open state) | 6VYB | Deposited: 2020-02-25 Released: 2020-03-11 |
ELECTRON MICROSCOPY Resolution: 3.20 Å |
| The 2019-nCoV RBD/ACE2-B0AT1 complex | 6M17 | Deposited: 2020-02-24 Released: 2020-03-11 |
ELECTRON MICROSCOPY Resolution: 2.90 Å |
| Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 | 6VXS | Deposited: 2020-02-24 Released: 2020-03-04 |
X-RAY DIFFRACTION Resolution: 2.03 Å |
| Crystal Structure of NSP15 Endoribonuclease from SARS-CoV-2. | 6VWW | Deposited: 2020-02-20 Released: 2020-03-04 |
X-RAY DIFFRACTION Resolution: 2.20 Å |
| The crystal structure of papain-like protease of SARS-CoV-2 | 6W9C | Deposited: 2020-03-22 Released: 2020-04-01 |
X-RAY DIFFRACTION Resolution: 2.70 Å |
The spike glycoprotein of coronavirus is the main conciliator of entry into the host cells [[45], [46], [47], [48], [49], [50], [51], [52]]. This spike glycoprotein contains (i) a large ecto-domain, (ii) a single-pass transmembrane anchor, and (iii) a short C-terminal intracellular tail [44]. The ecto-domain consists of a receptor-binding unit S1 and a membrane-fusion S2 stalk. Basically, the receptor-binding unit S1 binds to a specific cell surface receptor via its receptor binding domain (RBD), whereas the trimeric S2 fuses the viral membranes and host cell to enable the entry of viral genomes into host cells (Fig. 2). Researchers already identified angiotensin converting enzyme 2 (ACE2) as a functional receptor for SARS-CoV [[48], [49], [50], [51]]. The crowned shaped spike glycoprotein of CoVs binds directly to ACE2 on the host cells surface and plays critically in virus infection. ACE2 is expressed widely with conserved primary structures throughout the animal kingdom. ACE2 from fish, amphibians, reptiles, birds, to mammals can potentially interact with RBD of SARS-CoV-2 [44]. Therefore, blocking of the RBD and ACE2 interaction is an obvious therapeutic intervention to treat diseases caused by CoVs. Specific antibodies or small molecular inhibitors can disrupt the interaction of RBD with ACE2.
Among the set of sixteen non-structural proteins, nsp5 is identified to play a pivotal role in the life cycle of SARS-CoV-2 replication as well as maturation (Fig. 2). Being a key component, the nsp5 is termed as main protease (Mpro). Like other RNA viruses, the functional significance of this Mpro or chymotrypsin-like protease (3CLpro) of SARS-CoV-2 emerges as a fascinating drug target for the development of anti-viral agents.
Recently, the 3D structure of 3CLpro of SARS-CoV-2 was reported [1]. Like other coronaviruses Mpro, it also consists of three domains. The domain I (comprising of 8–101 amino acids) and II (102–184 amino acids) are anti-parallel β-barrels resembling the chymotrypsin. Besides, the domain III (201–306 amino acids) essentially comprehended of five α-helices and arrayed into a antiparallel globular cluster. Interestingly, a long loop region (185–200 residues) bridged domain II and III. The substrate-binding pocket of SARS-CoV-2 virus Mpro is located in a cleft between domain I and II [1]. The binding site possesses a Cys-His catalytic dyad with other crucial amino acid residues including F140, L141, N142, G143, S144, M165, E166, Q189 and T190.
The nsp12 of SARS-CoV-2 encoded viral RNA-dependent RNA polymerase (RdRp) together with co-factors nsp7 and nsp8 boasts polymerase activity (Fig. 2). RdRp is a pivotal enzyme in the life cycle of all RNA viruses including CoV, human immune deficiency virus (HIV), Hepatitis C Virus (HCV) and Zika Virus (ZIKV). Gao and co-workers [40] reported the cryo-EM structure of full-length nsp12 of SARS-CoV in complex with cofactors nsp7 and nsp8. The active site domain of the SARS-CoV-2 RdRp is set up by conserved polymerase motifs A-G in the palm domain [40]. RdRp active site is appointing two successive aspartate residues projected from a beta-turn structure making them surface accessible through the nucleotide channel. As configured to SARS-CoV, the RdRp of SARS-CoV-2 contains a larger N-terminal extension and polymerase domain. Multiple sequence alignment (MSA) of SARS-CoV-2 RdRp disclosed percent sequence identity against different HCoV strains such as the Alpha-coronavirus (229E: 48.55% and NL63: 48.79%) and the Beta-coronavirus (OC43: 55.07%, HKU1: 48.16%, MERS-CoV: 56.76% and SARS-CoV: 90.18%). Thus, the SARS-CoV RdRp is the closest strain to the RdRp of SARS-CoV-2 [53]. This structural information may furnish a basis for the design of new anti-COVID-19 agents or drug repurposing against viral proteins.
3. Molecular modeling and in silico virtual screening against SARS-CoV-2
Novel coronavirus (COVID-19) is hardly 180 days old. Scanty knowledge about the molecular mechanisms of the disease is obstructing the attempts to develop successful anti-viral agents. In consequence, animal models capable of mimicking the human physiological responses to SARS-CoV-2 infections are sketchy so far.
Until precise molecular and structural biology underlying SARS-CoV-2 replication and each of the proteins’ details functions are available, bioinformatics and molecular modelling techniques are the only handy strategy. In silico virtual screening techniques are proficient to identifying leads against putative targets. Five viral targets are currently being considered such as receptor binding domain (RBD)/Spike protein, N protein, E protein, 3CLpro and RdRp.
3.1. Targeting receptor binding domain (RBD)/spike protein
Chen and co-workers [44] suggested unique structural features of the spike glycoprotein RBD of SARS-CoV-2. However, the RBDs of SARS-CoV and SARS-CoV-2 share 72% amino acid sequences identity and disclose highly similar ternary structures. On the contrary, SARS-CoV-2 exhibits a distinct loop with flexible glycyl replacing rigid prolyl residues in SARS-CoV. Molecular modeling study unfolded that a phenylalanine moiety (Phe486) in the flexible loop involves critically in its penetration into a deep hydrophobic pocket of ACE2. This observation may partially confer a stronger interaction between SARS-CoV-2 and ACE2 of the host cell. Moreover, spike glycoprotein of SARS-CoV-2 exhibits higher binding affinity for its receptor in comparison to other CoVs [44]. These suggest the aggressiveness of SARS-CoV-2 than SARS-CoV-1.
This observation was in line with the analysis of Xu et al. [44] where the computational model of RBD domain of the SARS-CoV-2 S protein established strong interaction with human ACE2.
Robson [54] performed a preliminary bioinformatics studies to propose a synthetic vaccine and peptidomimetic antagonist against the Spike glycoprotein of SARS-CoV-2. The author employed Q-UEL language to perform the bioinformatics approach. KRSFIEDLLFNKV was identified as a well conserved sequence motif that corresponds to the known cleavage sites of the SARS virus. This sequence motif formed the basis for design of specific synthetic vaccine epitope and peptidomimetic agent [54].
A group of scientists from the Cairo University, Egypt predicted COVID-19 spike binding site to a cell-surface receptor namely Glucose Regulated Protein 78 (GRP78) by employing structural bioinformatics in combination with protein-protein docking [55]. Firstly, a homology model of SARS-CoV-2 spike protein was built by using SARS-CoV spike (PDB: 6ACD, chain C) as a template. Further sequence and structural alignments with the Pep42 cyclic peptide hypothesized four cyclic region (starting and ending with cysteine residues connected by a disulfide bond) of SARS-CoV-2 spike protein as possible binding sites to GRP78. The protein-protein docking by HADDOCK software revealed the involvement of the Substrate Binding Domain β (SBD β) of GRP78 (PDB: 5E84) and the receptor-binding domain of the SARS-CoV-2 spike protein in recognition of the host cell receptor. Moreover, the favourable binding was observed between regions III (C391-C525) and IV (C480- C488) of SARS-CoV-2 spike protein model and GRP78. Notably, the region IV was supposed to be a pivotal driving force for GRP78 binding (predicted binding affinity: 9.8 kcal/mol). Prediction of this binding site sheds light on the mode of envelope protein recognition by the GRP78 substrate-binding domain for future endeavours [55].
3.2. Targeting N protein
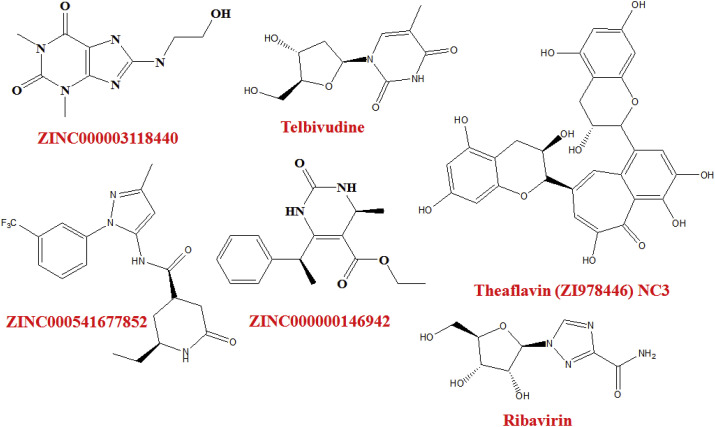
In a molecular modeling study to explore potential inhibitors of RNA binding to N terminal domain (NTD) of Nucleocapsid protein (N protein), Sarma et al. [56] pointed out two potential hits namely ZINC000003118440 and ZINC000000146942 (Fig. 3 ).
Fig. 3.
Structure of the virtual hits.
The authors employed two NTD structures of N proteins namely 2OFZ and 1SSK. Firstly, a set of diverse compounds from Asinex and Maybridge library were docked. Then 15 compounds for each of the targets were prioritized with significant docking scores. Further MM-GBSA binding free energy, pharmacokinetic properties (QikProp) and drug-likeness (SwissADME) as well as molecular dynamics (MD) studies were performed to screen the compounds. Out of these two potential hits, one compound was a theophylline derivative. Since theophylline derivative is commonly used as a bronchodilator, hence, the author further screened approved bronchodilators against the putative N protein RNA binding site of CoVID-19 [56]. The approved bronchodilators showed MM-GBSA binding affinity in the following order:
Formeterol > Terbutaline > Ipratropium bromide > Tiotropium Bromide > Theophylline > Salbutamol.
Recently, native or non-native protein-protein interactions (PPIs) are emerged as a target for structure-based screening of small molecule. It may be an alternative drug design paradigm which could accelerate anti-viral drug discovery against various pathogens [[57], [58], [59]]. Since the orthostatic/allosteric stabilization of non-native PPIs of SARS-CoV-2 nucleocapsid protein results abnormal protein oligomerization and finally leading to loss of viral activity. Scientists from National Chung Hsing University, Taiwan acclaimed non-native PPIs of N-terminal domain of the MERS-CoV nucleocapsid protein (MERS-CoV N-NTD) [60]. They reported a crystal structure of MERS-CoV N-NTD in a non-native dimeric configuration which turned a target for virtual screening of orthosteric stabilizers from Acros and ZINC drug databases. This finding provides valuable insight and further motivations into the design of new anti-virals based on stabilizing a non-native protein interaction interface of N protein [60].
3.3. Targeting E protein
Gupta and co-workers [61] employed computational techniques to explore the best possible structure of the SARS-CoV-2 E protein present in the PDB database. The author reported that E protein of SARS-CoV-2 is a pentameric protein comprised of 35 α-helices and 40 loops. Near about 8000 compounds were docked whereas 700 compounds were prioritized with significant affinities or docking scores. The docking study with E protein of SARS-CoV-2 and phytochemicals like Belachinal, Macaflavanone E &Vibsanol divulged that amino acids such as V25 and F26 play crucially in the binding interactions. The functional behaviour of E protein of SARS-CoV-2 after 200 ns molecular dynamics studies revealed that α-helix and loops of E protein escapades random movement and modulates normal ion channel activity to succour the pathogenesis in human and other vertebrates. Unlikely the random manoeuvre of the E protein of SARS-CoV-2 gets reduced after binding with inhibitors [61].
3.4. Targeting chymotrypsin-like protease (3CLpro)
Khan et al. [62] pinpointed three FDA-approved drugs such as Remdesivir, Saquinavir and Darunavir) and two small molecules (flavone and coumarin derivatives) as possible inhibitors of 3CLpro by target-based virtual screening. A number of 8000 compounds were docked and top 700 primary hits were distinguished by significant affinities or docking scores. Then the binding interaction between the active site residue of 3CLpro and the selected compounds were extensively scrutinized using the PLIF module in MOE. Further, MD simulation and binding free energy calculations were recorded to evaluate the dynamic behaviour, stability of protein-ligand contact, and binding affinity of the hits [62].
Kandeel and Al-Nazawi [63] reported statistics of pairwise sequence comparison matrix among the main protease (Mpro) of CoVs. A high pairwise sequence alignment identity (96.08%) was found between SARS-CoV-2 and SARS-CoV-1 Mpro, whereas only 51.61% identity was exposed for SARS-CoV-2 and MERS-CoV Mpro. In consequence, the number of amino acid differences between SARS-CoV-2 and SARS-CoV-1 Mpro was only 12, while it was 153 between SARS-CoV-2 and MERS-CoV Mpro. An early virtual screening (VS) study of FDA approved drugs (retrieved from Selleckchem Inc.) against the first resolved SARS-CoV-2 Mpro crystal structure (PDB: 6LU7) was performed. That helps the repurposing of already approved drugs to eradicate COVID-19 [63]. FDA approved drugs were de-slated and optimized using OPLS2005 force field by the aid of Ligprep software. Similarly, the protein was optimized by protein preparation module in Maestro software package (Schrodinger LLC, NY, USA). The standard precision (SP docking) of Schrodinger glide docking module was employed for VS. Depending on the highest docking score, top 20 FDA approved drugs were highlighted including Chromocarb (a vasoprotective), Ribavirin (Anti-viral agent), Telbivudine (anti-hepatitis B virus), Vitamin B12 and Nicotinamide (Vitamin), Aminophylline (Bronchodilator), Triflusal (Cardiovascular drug), Bemegride (CNS stimulant), Aminosalicylate Sodium and Pyrazinamide (Antituberculosis agents), Temozolomide (Anticancer), Methazolamide (used in Glaucoma), Tioxolone (Anti-acne agent), Propylthiouracil (Antithyroid agent), Cysteamine HCl (Nephropathiccystinosis), Methoxamine hydrochloride (Alpha-adrenergic agonist), Zonisamide (Anticonvulsant), (+,-)-Octopamine HCl (Adrenergic agonist), Amiloride hydrochloride (Diuretic).
Detailed scanning of the binding mode of these drugs with SARS-CoV-2 Mpro conferred that hydrophobic and hydrogen bonding interactions were the main imperators for binding. Interestingly, Telbivudine (Fig. 3), an anti-hepatitis B virus agent, bind with the SARS-CoV-2 Mpro through hydrogen bond interactions with amino acid residues S49 and Q189. Moreover, a broad spectrum antiviral agent, Ribavirin (Fig. 3), interacted with the SARS-CoV-2 Mpro by forming hydrogen bonds with side chain amino acid residue Q189 and the backbone amino acid residue T25. Ribavirin is officially approved against respiratory syncytial virus (RSV) infection. It is also used in combination with interferon α2b against hepatitis C virus. Moreover, it also exhibited potency against SARS-CoV infection [63].
In a study to distinguish potential active herbs against 2019-nCoV, Ma et al. [64] performed molecular docking based virtual screening of traditional Chinese medicine database (TCMD) 2009 by CDOCKER program against 3CLpro and papain-like protease (PLpro) of SARS-CoV-2. The VS study against Mpro screened 12,322 active components. On the other hand, VS approach against PLpro screened 11,294 potential ingredients and representative active ingredients such as ginger ketophenol, ginkgol alcohol, ferulic acid, etc. [64].
Deep docking (DD) methodology that enable fast prediction of docking scores and expand the repertoire of structure-based VS of billions of compound in a very short time. A group of scientists from the University of British Columbia, Canada [65] applied DD method to screen 1.3 billion compounds from ZINC15 library and recognized top 1000 hits against Mpro of SARS-CoV-2 (PDB: 6LU7). Upon careful observation of the interaction between ZINC000541677852 (Fig. 3) and SARS-CoV-2 Mpro conferred that the P1, P2 and P3 groups occupied the binding pocket by forming hydrophobic interactions. In addition, it formed hydrogen bond interactions with three backbone amino acid residues C145, L141, H164 and one side chain amino acid residue Q192 [65].
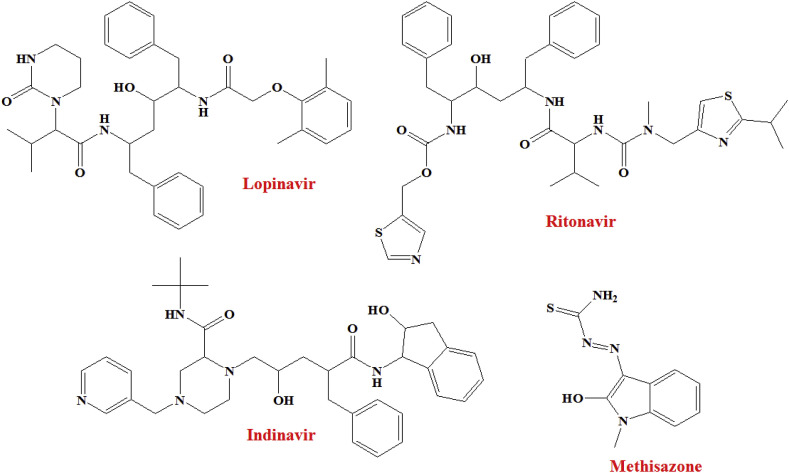
Shah and co-workers [66] selected SARS-CoV-2 Mpro proteins (PDB: 5R7Y, 5R7Z, 5R80, 5R81, 5R82) having resolution < 2 Å, R-Value Free < 0.30, R-Value Work < 0.25 to perform molecular docking study of 61 anti-viral agents. The Maestro interface (Schrödinger Suite, LLC, NY) was used to perform the molecular docking study. Compounds exhibited dock score of −6.5 or less were taken as promising molecules. This comparative analysis suggested that Asunaprevir, Indinavir, Galidesivir, Lopinavir, Remdesivir, Ritonavir, ABT450, CGP42112A and Marboran (Methisazone) interacted with >2 SARS-CoV-2 Mpro structures. Moreover, an anti-HIV drug Lopinavir showed binding affinities for all five structures (with a dock score of less than −6.5), whereas, Indinavir and Ritonavir (Fig. 4 ) interacted with 4 out of 5 proteins.
Fig. 4.
Structure of Lopinavir, Indinavir, Ritonavir and Methisazone.
In addition, Methisazone (an inhibitor of protein synthesis), CGP42112A (an angiotensin AT2 receptor agonist) and ABT450 (an inhibitor of the non-structural protein 3–4A) may be emerged as a new hits against SARS-CoV-2 [66].
3.5. Targeting RNA dependent RNA polymerase (RdRp)
A scientist from Cairo University, Egypt performed homology modelling of SARS-CoV-2 RdRp by the aid of Swiss Model, a automated homology modelling web server, using SARS-CoV-1 RdRp (PDB: 6NUR, chain A) as a template [52]. The resulted SARS-CoV-2 RdRp homology model expressed 97.08% sequence identity to the template. The model was validated based on the Ramachandran plot which showed 100% of the residues in the allowed regions, 97.5% in the most favoured region. Further, the molecular docking was employed by utilizing AutoDock Vina to test some direct-acting antiviral (DAA) drugs including Sofosbuvir, Ribavirin, Remidisvir, IDX-184 against COVID-19 RdRp. A grid box was chosen at the SARS-CoV-2 RdRp by utilizing the AutoDock tools using x, y, z coordinates of 142.1, 138.7, 150.0, respectively in 30 × 30 × 30 Å dimensions. The active site aspartates (D255 and D256) were kept flexible during the docking study. The optimization of the ligands were performed using MM3 and PM6 force field after that further optimization was carried out through B3LYP functional of Density Functional Theory (DFT) based quantum mechanics. The binding energies and interaction complexes formed after docking conferred that IDX-184, Sofosbuvir, and Ribavirin can tightly bind to the RdRp of SARS-CoV-2. Interestingly, Guanosine derivative IDX-184, Sofosbuvir and Ribavirin interacted with RdRp through multiple hydrogen bonding. Additionally, Sofosbuvir and IDX-184 form metal interaction through the Mg+2 with E702 and D652, respectively. Sofosbuvir exhibit two hydrophobic interactions with D651and Y510 whereas, IDX-184 form a salt bridge D514, which referred for the increased stabilization of the interactions. Hence, strong binding with SARS-CoV-2 RdRp, these agents can contradict the function of RdRp in the life cycle of 2019-nCoV viruses leading to viral eradication [52].
In another study, Elfiky [67] reported SARS-CoV-2 RdRp targeted molecular docking study of some anti-polymerase drugs which have been approved for use against various viruses. Not surprisingly, Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir exhibited promising binding affinity against SARS-CoV-2 RdRp. These results were also consistence with previous one [52]. Guanosine derivative (IDX-184), Setrobuvir, YAK has potential to block the SARSCoV-2 strain. Since these drugs have already passed the ADME and toxicity measurements these may be used as a new therapeutic drug candidate against SARS-CoV-2.
Lung and co-workers [68] identified Theaflavin, a polyphenolic constituent present in black tea, against RdRp after screening eighty-three chemical structures from traditional Chinese medicinal compounds along with their similar structures retrieved from ZINC15 database. The three-dimensional RdRp structures of SARS-CoV-2 (NCBI: YP_009725307.1), SARS-CoV (NP_828869.1) and MERS-CoV (YP_009047223.1) were modelled using Modeller 7. After molecular docking with idock (https://github.com/HongjianLi/idock), the theaflavin showed promising binding energy against RdRp of SARS-CoV-2 (idock score: 9.11 kcal/mol), MERS-CoV (idock score: 8.26 kcal/mol) and SARS-CoV (idock score: 8.03 kcal/mol). Hydrophobic interactions were found to be the driving force in binding. Theaflavin formed hydrogen bonds with D452, R553 and R624 of SARS-CoV-2 RdRp. Additionally, a π-cation interaction was found with R553 [68].
3.6. Targeting multiple proteins
Calligari et al. [69] cashed in the molecular docking technique to examine the affinity of several inhibitors (previously developed for another viral pathogen) to SARS-CoV-2 viral proteins (the main 3C-like protease, S protein, RdRp, Nucleocapsid protein). HIV inhibitors Saquinavir (DB01232: Autodock Vina Scoring −9.3 kcal/mol), Indinavir (DB00224: Vina Scoring −8.7 kcal/mol), Tipranavir (DB00932: Vina Scoring −8.6 kcal/mol), Ritonavir (DB00503: Vina Scoring −8.1 kcal/mol), Lopinavir (DB01601: Vina Scoring −8.1 kcal/mol), Atazanavir (DB01072: Vina Scoring −8.0 kcal/mol), Nelfinavir (DB00220: Vina Scoring −7.9 kcal/mol), Amprenavir (DB00701: Vina Scoring −7.7 kcal/mol), Darunavir (DB01264: Vina Scoring −7.6 kcal/mol), Fosamprenavir (DB01319: Vina Scoring −7.2 kcal/mol) were identified as promising molecules against SARS-CoV-2 Mpro. This results were also consistence with hepatitis C virus (HCV) inhibitors Faldaprevir (DB11808: Vina Scoring −8.4 kcal/mol) and Asunaprevir (DB11586: Vina Scoring −8.1 kcal/mol). Surprisingly, HCV NS3/4A protease inhibitor Simeprevir (DB06290: AutodockVina Scoring −10.0 kcal/mol) was recognized as the best Autodock Vina scoring drug. In spite of HCV main protease measure very low identity with the SARS-CoV-2 homolog (only 7.5%), thus this finding was thunderbolt [69]. Therefore, it may be inferred that the similar topology of active site of both proteases was the main driving force in binding of Simeprevir. It fitted well into the two hydrophobic pockets fringed the catalytic dyad H41-C145 of SARS-CoV-2 Mpro. It was also fitted into the hydrophobic loop F181-F185. Moreover, the binding of Simeprevir to SARS-CoV-2 protease was anchored by three hydrogen bonding interactions with E166, G143 and N142 [69]. In the same article, the homology modelling of SARS-CoV-2 S protein by the aid of iTasser server was done by using SARS-CoV spike (PDB: 5WRG, 5 × 58, 5XLR) as a template. Global pairwise sequence alignment measured that SARS-CoV-2 S protein shares about 76% of its primary sequence with its homolog in addition an overall similarity of 87%. Autodock Vina molecular docking of the homology modelled structure of S protein predicted Umifenovir (DB13609), Enfuvirtide (DB00109), and Pleconaril (DB05105) as potential inhibitors [69].
In a study to repurposing existing drugs against current pandemic COVID-19, Wu et al. [70] predicted some potential drugs acting on a certain target or multiple targets of SARS-CoV-2. Bioinformatics based homology modelling was utilised to build possible targets such as viral Mpro, PLpro, RdRp, helicase, Spike, etc. Next, these modelled proteins and human relative proteins including human ACE2 and type-II transmembrane serine protease enzymes were forwarded to systematically analyse and screen ZINC drug database (ZDD) along with database of traditional Chinese medicine and natural products and the database of commonly used 78 anti-viral drugs [70]. This study seems to be very interesting due to its deep discussions and vast target predictability. The lead candidates emerged from this study required in vitro and in vivo studies for further conformations.
4. What efforts are taken to identify anti-SARS-CoV-2 agents?
The new drug discovery and development takes more than five to ten years of investigations as well as cost billions of dollars. Thus, drug repositioning is the only cheap strategy to respond immediately. FDA-approved drugs justify safe alternatives if at least modest anti-SARS-CoV-2 activity can be achieved. Currently, Academia and industry personnel are involved in the testing of – (i) approved drugs and/or (ii) drug candidates in clinical trials. The in vitro screenings of FDA approved drugs as well as the compounds which are currently under clinical trials phases were well documented [[71], [72], [73], [74], [75], [76], [77]] and the need for further in vivo testing to facilitate drug discovery efforts against SARS-CoV-2.
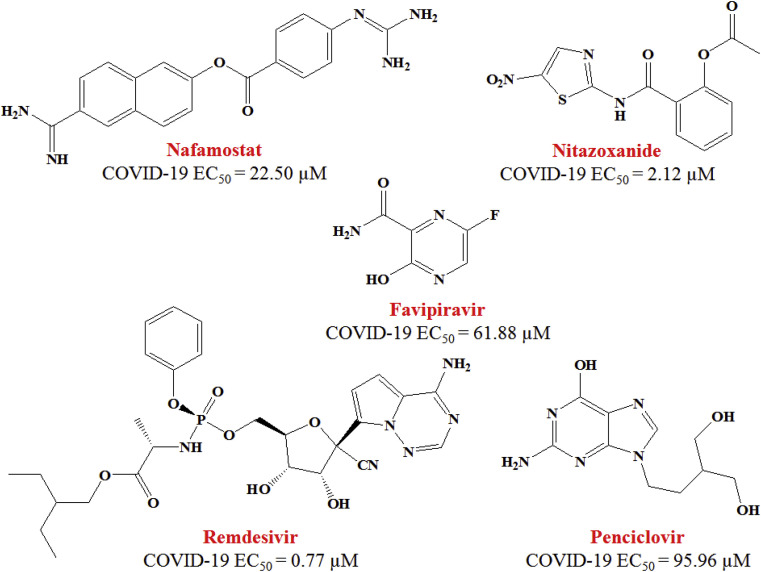
Nafamostat (Fig. 5 ) is a potent membrane fusion inhibitor of MERS-CoV [78]. Now, it has been found to inhibit the 2019-nCoV infection (EC50 = 22.50 μM, CC50 > 100 μM, SI > 4.44).
Fig. 5.
Structure and EC50 values of Nafamostat, Nitazoxanide, Favipiravir, Remdesivir and Penciclovir against SARS-CoV-2 in Vero E6 cells.
In comparison with Nafamostat, Nitazoxanide (Fig. 5) exhibited 10-fold more potency to inhibit the SARS-CoV-2 infection with a half-maximal effective concentration (EC50) = 2.12 μM, half-maximal cytotoxic concentration (CC50) > 35.53 μM and selectivity index (SI) > 16.76) [72]. Besides, this is agent marketed as an anti-protozoal drug [79] and also reported to possess anti-viral action against a broad range of viruses [80].
In the same article, Wang et al. [72] reported anti-COVID-19 property of three nucleoside analogs such as Favipiravir (EC50 = 61.88 μM; CC50 > 400 μM; SI > 6.46), Ribavirin (EC50 = 109.50 μM; CC50 > 400 μM; SI > 3.65) and Penciclovir (EC50 = 95.96 μM; CC50 > 400 μM; SI > 4.17). In near future, in vivo experiment is needed which will further justify the property of these above mentioned inhibitors of SARS-CoV-2.
An adenosine analogue Remdesivir (Fig. 5) showed a wide array of anti-viral property from various cultured cells to nonhuman primate (NHP) models. This agent (GS-5734) is currently under clinical observation against Ebola virus infection. Broad spectrum antiviral properties of Remdesivir have been illustrated few years ago [81]. Now, it has been found to effectively block SARS-CoV-2 infection at low-micromolar concentration [72]. It exhibited EC50 of 0.77 μM against 2019-nCoV in Vero E6 cells with CC50 > 100 μM; selectivity index (SI) > 129.87). Remdesivir also inhibited virus infection in SARS-CoV-2 sensitive human liver cancer Huh-7 cells [72].
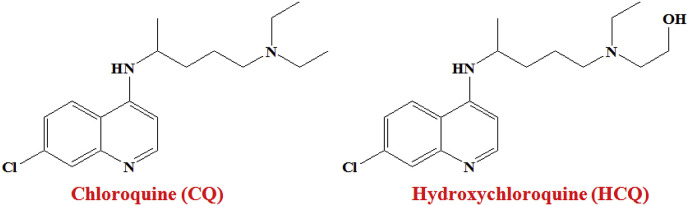
A widely-used anti-malarial drug Chloroquine (CQ, Fig. 6 ) has been used for more than 70 years [82], now, found to shows clinical potency against COVID-19.
Fig. 6.
Structure of Chloroquine (CQ) and Hydroxychloroquine (HCQ).
The molecular mechanism of Chloroquine against SARS-CoV is known [83,84]. Very recently, Wang et al. [72] reported time-of-addition assay that explained the function of CQ (EC50 = 1.13 μM; CC50 > 100 μM; SI > 88.50) at both entry as well as at post-entry stages of the novel corona virus infection in Vero E6 cells.
The same group of researches further evaluated the in vitro anti-SARS-CoV-2 effect of Hydroxychloroquine (HCQ, Fig. 6) sulphate, a derivative of CQ, in comparison to CQ [73]. Hydroxychloroquine sulphate [85] was introduced long before, first synthesized in 1946 [73]. Upon introduction of a hydroxyl group into CQ resulted in about 40% less toxic agent than CQ in animals. Both CQ and HCQ are weak bases and restrict the virus infection by- (i) triggering endosomal pH which is essential for virus/cell fusion and (ii) interfering with the glycosylation of ACE2 receptor and spike protein of coronavirus [73]. Additionally, both CQ and HCQ obstructed the SARS-CoV-2 transport from early endosomes (EEs) to endolysosomes (ELs).
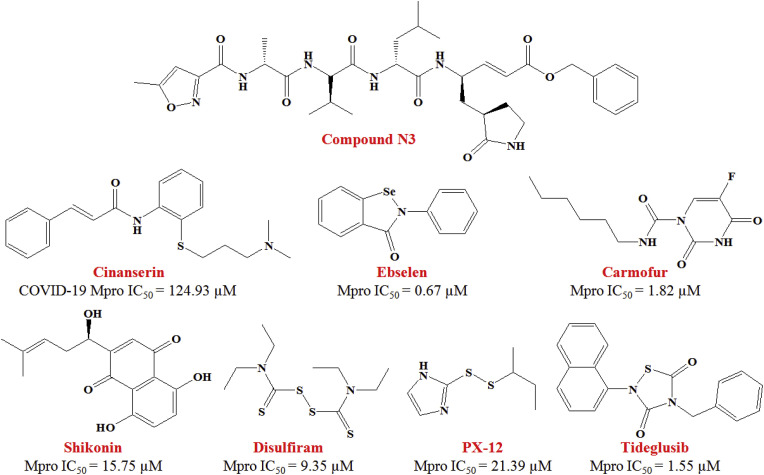
Jin and co-workers [1] executed structure-assisted drug design, molecular docking, VS as well as high-throughput screening to pinpoint new leads by targeting the 3CLpro of SARS-CoV-2. The same study first time reported the crystal structure of SARS-CoV-2 Mpro (PDB: 6LU7) in complex with a mechanism-based inhibitor, N3 (Fig. 7 ).
Fig. 7.
Small-molecular inhibitors of SARS-CoV-2 main protease (Mpro).
A molecular docking study with PDB: 6LU7 suggested that the ligand (i.e., Cinanserin, a serotonin antagonist, Fig. 7) adequately fitted into the substrate-binding site and formed two cation-π interactions with His41 and Glu166 of SARS-CoV-2 Mpro. This in silico observations was found in agreement with the in vitro SARS-CoV-2 inhibition assay where Cinanserin exhibited IC50 value of 124.93 μM. Further, a combination of SARS-CoV-2 Mpro structure-based VS and high-throughput screening of more than 10,000 compounds (containing approved drugs, clinical trials compounds, and other active compounds) picked six lead molecules as promising SARS-CoV-2 Mpro inhibitors (IC50 range: 0.67–21.4 μM). Among these identified inhibitors, Disulfiram and Carmofur are FDA-approved drugs, whereas Ebselen, Tideglusib, Shikonin, TDZD-8 and PX-12 are undergoing currently in preclinical studies (Fig. 7). Besides, Ebselen also possessed effective anti-viral activity in cell-based assays. This interesting study done by Jin et al. [1] not only demonstrated a robust screening strategy but also open a new panorama to rapid discovery of lead molecules targeting SARS-CoV-2 Mpro.
An attempt was made by Su and collaborators to introduce the first SARS-CoV-2 MLpro crystal structure complied with a non-covalent inhibitor named Baicalein [43]. This compound exhibited promising SARS-CoV-2 Mpro inhibitory activity (IC50 = 0.94 μM) along with a dose-dependent inhibition on the replication of SARS-CoV-2 (EC50 = 1.69 μM). At the receptor binding site, Baicalein forms interaction with catalytic Glu166 and oxyanion loop residues 138–145. More interestingly, it exhibited very distinct binding mode than the other covalent or peptidomimetic 3CLpro inhibitors [43].
In vitro screening of 48 drugs was screened against HCoVs infection [71]. Immunofluorescence analysis with an antibody specific for the novel HCoV viral N protein was scored for each drug treated cells. The dose-response curve (DRC) was developed after analysing the confocal microscope images of both viral N protein and cell nuclei. Remdesivir (SARS-CoV-2 IC50 = 11.41 μM), Lopinavir (SARS-CoV-2 IC50 = 9.12 μM) and Chloroquine (SARS-CoV-2 IC50 = 7.28 μM) were used as reference drugs. Consequently, 24 drugs showed good activities with IC50 ranges of 0.1–10 μM. An anti-helminthic drug, Niclosamide, and a corticosteroid used to treat asthma and allergic rhinitis, Ciclesonide, emerged as SARS-CoV-2 inhibitors with IC50 of 0.28 μM and 4.33 μM, respectively. Notably, Niclosamide reduces MERS-CoV replication by inhibiting SKP2 activity leading to enhancement in autophagy [86]. Thus, a similar mechanism may be introduced by Niclosamide to hamper SARS-CoV-2 infection [71].
Zhang et al. [87] reported structure-based design of α-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication. These showed good inhibitory properties against the isolated proteases, viral replicons, virus-infected Huh7 cells. Near-equipotency against the enteroviruses, alphacoronaviruses, and betacoronaviruses was observed upon optimization of the P2 substituent of the α-ketoamides. The cyclopentylmethyl and cyclohexylmethyl at the P2 substituent disposed low-micromolar EC50 values against the three virus genera in cell cultures. Compound 11r was found promising against MERS-CoV in virus-infected Huh7 cells [87]. By dint of high similarity among the 3CLproteases of coronavirus, it is awaited that compound 11r is expected to display good anti-viral activity against COVID-19 in near future.
Meanwhile, the clinical trial studies of some molecules have been started against COVID-19 mainly through drug repurposing [88]; those are depicted in Table 3 .
Table 3.
List of effective molecules targeting SARS-CoV-2.
| Molecules | SARS-CoV-2 Target | Target disease |
|---|---|---|
| Remdesivir (GS-5734) | RNA-dependent RNA polymerase | Anti-Ebola |
| Favipiravir | RdRp | Anti-influenza |
| Ivermectin | Viral Protease | Anti-parasitic agent, anti-HIV |
| Lopinavir/Ritonavir | Viral Protease | Anti-HIV |
| APN01 | Blocking Virus–Cell Membrane Fusion | undergone phase II trial for ARDS |
| Hydroxychloroquine | Blocking Virus–Cell Membrane Fusion | Antimalarial and anti-autoimmune agent |
| Arbidol Hydrochloride (Umifenovir) | Blocking Virus–Cell Membrane Fusion | Inhibitor of influenza and arboviruses |
| Pegylated interferon with ribavirin | Replication inhibitor | Anti-HCV, anti-HIV |
5. Concluding remarks
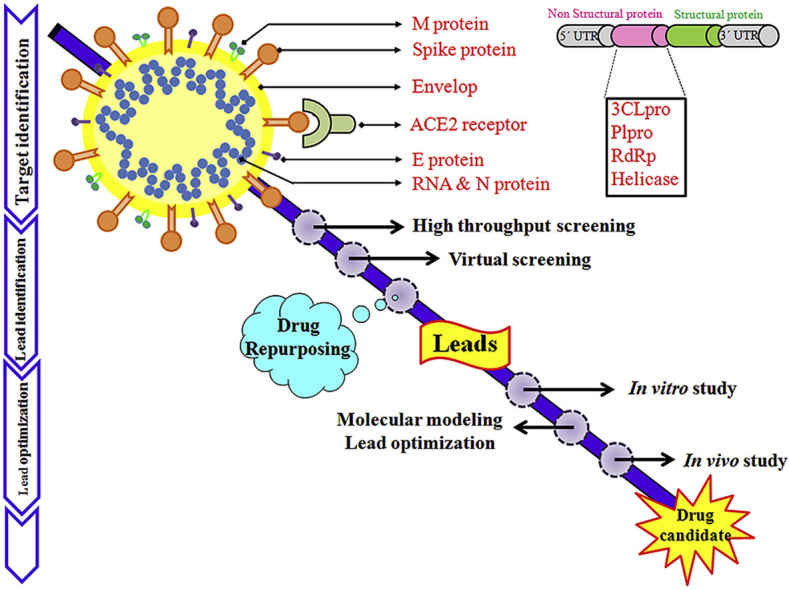
The medicinal chemistry of SARS-CoV-2 infection is still in its infancy, with target specific lead molecules are yet to identify. This study deals with the information currently available on potential targets for therapeutic invention and screening of new compounds or drug repurposing against SARS-CoV-2 (Fig. 8 ).
Fig. 8.
Drug discovery approaches against novel coronavirus.
As the 3D structure of the SARS-CoV-2 3C-like protease bears 96% identity with its ortholog from (SARS-CoV). Interestingly, the residues involved in the catalysis, substrate binding and dimerization of 3CLpro are 100% conserved. In addition, the polyprotein pp1ab sequences are highly similar (86% identity). Depending upon the alike substrate specificities and high identities, we are of the opinion that the previous progress of specific SARS-CoV inhibitors development can undertaking a course of action on the design and discovery of inhibitors against SARS-CoV-2. Our group have already explored the structural properties important for SARS-CoV viral 3Clike protease inhibitors [30]. Recently in a collaborative work, our research team suggested the implications of naphthyl derivatives against SARS-CoV-2 PLpro enzyme though in-depth ligand-receptor interaction analysis [89]. We have already predicted some in-house glutamine-based molecules to use as a seed for drug design and optimization against PLpro of SARS-CoV-2 [90].
In fact, some other anti-viral drugs can also be taken into consideration. In this regards, target-based VS is one of the most important approaches used for drug repurposing. The computational analyses are not subordinate but are a right choice to enrich the basal knowledge during the long process leading to drug development (Fig. 8). Until any clear-cut treatment approach is prescribed for COVID-19, the use of already approved drugs is only alternative strategy. Howbeit, relatively limited computational resource or biased in silico screening may scattered the linearity of drug discovery of novel coronavirus. In the near future, the virtual hits may serve as a promising drug like molecule against SARS-CoV-2 after details in vitro and in vivo laboratory investigations.
Moreover, the availability of X-ray crystal structures of the important viral proteins will trigger more exhaustive docking calculation of diverse chemotypes. It is crystal clear that the prevention of COVID-19 requires strong and sustainable global collaborative work [91]. Data sharing is exigent to fill the knowledge gaps on this global pandemic. Further progress of the scientific understanding regarding the structural and molecular biology of SARS-CoV-2 will legitimate the shape of lead compounds to achieve therapeutic goals. The development of medicinal chemistry through bioinformatics and chemo-informatics studies remains indispensable with a bit of savoir faire.
Declaration of competing interest
The authors have no conflict of interests.
Acknowledgment
Sk. Abdul Amin sincerely acknowledges Council of Scientific and Industrial Research (CSIR), New Delhi, India for awarding the Senior Research Fellowship (SRF) [FILE NO.: 09/096(0967)/2019-EMR-I, Dated: 01-04-2019]. Tarun Jha is also thankful for the financial support from RUSA 2.0 of UGC, New Delhi, India to Jadavpur University, Kolkata, India. Dr. Shovanlal Gayen of Dr. Harisingh Gour University, Sagar, India is gratefully acknowledged for his critical discussion. Sk. Abdul Amin thankfully acknowledge Mr. Subhabrata Ghose, Mrs. Tisha Mukherjee Sarkar for their critical reading of the manuscript. We are very much thankful to the Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India for providing the research facilities.
Biographies
Sk. Abdul Amin is a Senior Research Fellow at Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India. He is working under the guidance of Tarun Jha. His research area includes design and synthesis of small molecules with anti-cancer and anti-viral properties, computational chemical biology, and large-scale structure-activity relationship analysis. He has published sixty two research/review articles in different reputed peer-reviewed journals and three book chapters. He enjoys a good conversation on science, regional history, contemporary art and books.
Tarun Jha, a Professor at Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India, has supervised 16 Ph.D. students and guided nine research projects funded by different organizations. He has published more than 150 research articles in different reputed peer-reviewed journals. His research area includes design and synthesis of anti-cancer small molecules. Prof. Jha is a member of the Academic Advisory Committee of National Board of Accreditation (NBA), New Delhi, India.
References
- 1.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020 doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 2.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 6.McIntosh K., Dees J.H., Becker W.B., Kapikian A.Z., Chanock R.M. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. U.S.A. 1967;57:933–940. doi: 10.1073/pnas.57.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wuhan Municipal Health Commission 2019. http://wjw.wuhan.gov.cn/front/web/showDetail/2019123108989
- 8.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.She J., Jiang J., Ye L., Hu L., Bai C., Song Y. 2019 novel coronavirus of pneumonia in Wuhan, China: emerging attack and management strategies. Clin. Transl. Med. 2020;9:19. doi: 10.1186/s40169-020-00271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benvenuto D., Giovanetti M., Salemi M., Prosperi M., De Flora C., Junior Alcantara L.C., Angeletti S., Ciccozzi M. The global spread of 2019-nCoV: a molecular evolutionary analysis. Pathog. Glob. Health. 2020;114:64–67. doi: 10.1080/20477724.2020.1725339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Q., Lin Q., Jin S., You L. Coronavirus 2019-nCoV: a brief perspective from the front line. J. Infect. 2020;80:373–377. doi: 10.1016/j.jinf.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020
- 14.https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 15.Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov. Today. 2020 doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Habibzadeh P., Stoneman E.K. The novel coronavirus: a bird’s eye view. Int. J. Occup. Environ. Med. 2020;11:65–71. doi: 10.15171/ijoem.2020.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. eCollection 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan Y., Zhu H.L., Zhou C. Advance of promising targets and agents against COVID-19 in China. Drug Discov. Today. 2020 doi: 10.1016/j.drudis.2020.02.011. pii: S1359–6446(20)30098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekins S., Mottin M., Ramos P.R.P.S., Sousa B.K.P., Neves B.J., Foil D.H., Zorn K.M., Braga R.C., Coffee M., Southan C., Puhl A.C., Andrade C.H. Déjà vu: stimulating open drug discovery for SARS-CoV-2. Drug Discov. Today. 2020 doi: 10.1016/j.drudis.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang W., Yang Y., Rao Y., Rao X. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vaccines. 2020;5:18. doi: 10.1038/s41541-020-0170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets. Antivir. Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J. Med. Virol. 2020;92:479–490. doi: 10.1002/jmv.25707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiessling L., Chen P., Wang J., Li J.P. Fighting the coronavirus outbreak. ACS Chem. Biol. 2020;15:799–801. doi: 10.1021/acschembio.0c00175. [DOI] [PubMed] [Google Scholar]
- 27.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 28.Wang J. Fast identification of possible drug treatment of coronavirus disease -19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020 doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odhar H.A., Ahjel S.W., Albeer A.A.M.A., Hashim A.F., Rayshan A.M., Humadi S.S. Molecular docking and dynamics simulation of FDA approved drugs with the main protease from 2019 novel coronavirus. Bioinformation. 2020;16:236–244. doi: 10.6026/97320630016236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adhikari N., Baidya S.K., Saha A., Jha T. Structural insight into the viral 3Clike protease inhibitors: comparative SAR/QSAR approaches. In: Gupta S.P., editor. Viral Proteases and Their Inhibitors. Academic Press; USA: 2017. pp. 317–402. (Chapter 11) [Google Scholar]
- 31.Amin S.A., Adhikari N., Jha T. Design of aminopeptidase N inhibitors as anti-cancer agents. J. Med. Chem. 2018;61:6468–6490. doi: 10.1021/acs.jmedchem.7b00782. [DOI] [PubMed] [Google Scholar]
- 32.Halder A.K., Saha A., Jha T. Exploration of structural and physicochemical requirements and search of virtual hits for aminopeptidase N inhibitors. Mol. Divers. 2013;17:123–137. doi: 10.1007/s11030-013-9422-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutta S., Halder A.K., Adhikari N., Amin S.A., Das S., Saha A., Jha T. Synthesis, anticancer activity, SAR and binding mode of interaction studies of substituted pentanoic acids. Future Med. Chem. 2019;11:1679–1702. doi: 10.4155/fmc-2018-0361. [DOI] [PubMed] [Google Scholar]
- 34.Banerjee S., Amin S.A., Baidya S.K., Adhikari N., Jha T. Exploring the structural aspects of ureido-amino acid-based APN inhibitors: a validated comparative multi-QSAR modelling study. SAR QSAR Environ. Res. 2020;31:325–345. doi: 10.1080/1062936X.2020.1734080. [DOI] [PubMed] [Google Scholar]
- 35.Sarkar R., Banerjee S., Amin S.A., Adhikari N., Jha T. Histone deacetylase 3 (HDAC3) inhibitors as anticancer agents: a review. Eur. J. Med. Chem. 2020;192:112171. doi: 10.1016/j.ejmech.2020.112171. [DOI] [PubMed] [Google Scholar]
- 36.Mondal S., Adhikari N., Banerjee S., Amin S.A., Jha T. Matrix metalloproteinase-9 (MMP-9) and its inhibitors in cancer: a minireview. Eur. J. Med. Chem. 2020;194:112260. doi: 10.1016/j.ejmech.2020.112260. [DOI] [PubMed] [Google Scholar]
- 37.Angeletti S, Benvenuto D, Bianchi M, Giovanetti M, Pascarella S, Ciccozzi M. COVID-2019: the role of the nsp2 and nsp3 in its pathogenesis. J. Med. Virol.. doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed]
- 38.Saikatendu K.S., Joseph J.S., Subramanian V., Clayton T., Griffith M., Moy K., Velasquez J., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1’’-phosphate dephosphorylation by a conserved domain of nsP3. Structure. 2005;13:1665–1675. doi: 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020 doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.https://swissmodel.expasy.org/repository/species/2697049
- 42.https://zhanglab.ccmb.med.umich.edu/COVID-19/
- 43.Su H. Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. bioRxiv. 2020 doi: 10.1101/2020.04.13.038687. oi: [DOI] [Google Scholar]
- 44.RCSB Protein Data Bank https://www.rcsb.org/
- 45.Chen Y., Guo Y., Pan Y., Zhao Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020;525:135–140. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;16:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020 doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 52.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robson B. Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput. Biol. Med. 2020;119:103670. doi: 10.1016/j.compbiomed.2020.103670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020;80:554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarma P., Sekhar M., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D.P., Medhi B. In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain) J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1753580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ni D., Lu S., Zhang J. Emerging roles of allosteric modulators in the regulation of protein-protein interactions (PPIs): a new paradigm for PPI drug discovery. Med. Res. Rev. 2019;39:2314–2342. doi: 10.1002/med.21585. [DOI] [PubMed] [Google Scholar]
- 58.Bosch J. PPI inhibitor and stabilizer development in human diseases. Drug Discov. Today Technol. 2017;24:3–9. doi: 10.1016/j.ddtec.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 59.Sijbesma E., Hallenbeck K.K., Leysen S., de Vink P.J., Skóra L., Jahnke W., Brunsveld L., Arkin M.R., Ottmann C. Site-directed fragment-based screening for the discovery of protein−protein interaction stabilizers. J. Am. Chem. Soc. 2019;141:3524–3531. doi: 10.1021/jacs.8b11658. [DOI] [PubMed] [Google Scholar]
- 60.Lin Shan-Meng, Lin Shih-Chao, Hsu Jia-Ning, Chang Chung-ke, Chien Ching-Ming, Wang Yong-Sheng, Wu Hung-Yi, Jeng U-Ser, Kehn-Hall Kylene, Hou Ming-Hon. Structure-based stabilization of non-native Protein−Protein interactions of coronavirus nucleocapsid proteins in antiviral drug design. J. Med. Chem. 2020;63:3131–3141. doi: 10.1021/acs.jmedchem.9b01913. [DOI] [PubMed] [Google Scholar]
- 61.Gupta M.K., Vemula S., Donde R., Gouda G., Behera L., Vadde R. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khan S.A., Zia K., Ashraf S., Uddin R., Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1751298. [DOI] [PubMed] [Google Scholar]
- 63.Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020;251:117627. doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma J., Huo X.Q., Chen X., Zhu W.X., Yao M.C., Qiao Y.J., Zhang Y.L. Study on screening potential traditional Chinese medicines against 2019-nCoV based on Mpro and PLP. Zhongguo Zhongyao Zazhi. 2020;45:1219–1224. doi: 10.19540/j.cnki.cjcmm.20200216.401. [DOI] [PubMed] [Google Scholar]
- 65.Ton A.-T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol. Inf. 2020;39:2000028. doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shah B., Modia P., Sagar S.R. Insilico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci. 2020;252:117652. doi: 10.1016/j.lfs.2020.117652. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 rna dependent rna polymerase (RdRp): a molecular docking study. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lung J., Lin Y.-S., Yang Y.-H., Chou Y.-L., Shu L.-H., Cheng Y.-C., Liu H.T., Wu C.-Y. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 2020;92:693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Calligari P., Bobone S., Ricci G., Bocedi A. Molecular investigation of SARS–CoV-2 proteins and their interactions with antiviral drugs. Viruses. 2020;12:445. doi: 10.3390/v12040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. BioRxiv. 2020 doi: 10.1101/2020.03.20.999730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang M., Cao R.2, Zhang L.1, Yang X.1, Liu J.1, Xu M.1, Shi Z.1, Hu Z.3, Zhong W.4, Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weston S., Haupt R., Logue J., Matthews K., Frieman M.B. FDA approved drugs with broad anti-coronaviral activity inhibit SARS-CoV-2 in vitro. BioRxiv. 2020 doi: 10.1101/2020.03.25.008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA- approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J., Ma X., Yu F., Liu J., Zou F., Pan T., Zhang H. BioRxiv; 2020. Teicoplanin Potently Blocks the Cell Entry of 2019-nCoV. [DOI] [Google Scholar]
- 77.Touret F., Gilles M., Barral K., Nougairede A., Decroly E., Lamballerie X.D., Coutard B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. BioRxiv. 2020 doi: 10.1101/2020.04.03.023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamamoto M., Matsuyama S., Li X., Takeda M., Kawaguchi Y., Inoue J.I., Matsuda Z. Identification of Nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob. Agents Chemother. 2016;60:6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fox L.M., Saravolatz L.D. Nitazoxanide: a new thiazolide antiparasitic agent. Clin. Infect. Dis. 2005;40:1173–1180. doi: 10.1086/428839. [DOI] [PubMed] [Google Scholar]
- 80.Rossignol J.-F. Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antivir. Res. 2014;110:94–103. doi: 10.1016/j.antiviral.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Amirian E.S., Levy J.K. Current knowledge about the antivirals remdesivir (GS-5734) and GS-441524 as therapeutic options for coronaviruses. One Health. 2020;9:100128. doi: 10.1016/j.onehlt.2020.100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Slater A.F. Chloroquine: mechanism of drug action and resistance in Plasmodium falciparum. Pharmacol. Ther. 1993;57:203–235. doi: 10.1016/0163-7258(93)90056-j. [DOI] [PubMed] [Google Scholar]
- 83.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;12:105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Plantone D., Koudriavtseva T. Current and future use of chloroquine and hydroxychloroquine in infectious, immune, neoplastic, and neurological diseases: a mini-review. Clin. Drug Invest. 2018;38:653–671. doi: 10.1007/s40261-018-0656-y. [DOI] [PubMed] [Google Scholar]
- 86.Gassen N.C., Niemeyer D., Muth D., Corman V.M., Martinelli, Gassen A., Hafner K., Papies J., Mösbauer K., Zellner A.5, Zannas A.S., Herrmann A., Holsboer F., Brack-Werner R., Boshart M., Müller-Myhsok B., Drosten C., Müller M.A., Rein T. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019;10:5770. doi: 10.1038/s41467-019-13659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang L., Lin D., Kusov Y., Nian Y., Ma Q., Wang J., von Brunn A., Leyssen P., Lanko K., Neyts J., de Wilde A., Snijder E.J., Liu H., Hilgenfeld R. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 88.Tu Y.F., Chien C.S., Yarmishyn A.A., Lin Y.Y., Luo Y.H., Lin Y.T., Lai W.Y., Yang D.M., Chou S.J., Yang Y.P., Wang M.L., Chiou S.H. A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020;21:2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Amin S.A., Ghosh K., Jha T., Gayen S. 2020. Exploring Naphthyl Derivatives as Corona Virus Papain-like Protease (PLpro) Inhibitors: A Hope in Anti-SARS-CoV-2 Drug Discovery. (under review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amin S.A., Ghosh K., Gayen S., Jha T. Chemical-informatics approach to COVID-19 drug discovery: Monte Carlo based QSAR, virtual screening and molecular docking study of some in-house molecules as papain-like protease (PLpro) inhibitors. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1780946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qu G., Li X., Hu L., Jiang G. Animperative need for research on the role of environmental factors in transmission of novel coronavirus (COVID-19) Environ. Sci. Technol. 2020;54:3730–3732. doi: 10.1021/acs.est.0c01102. [DOI] [PubMed] [Google Scholar]