Abstract
Heavy metals are environmental pollutants affect the integrity and distribution of living organisms in the ecosystem and also humans across the food chain. The study targeted the removal of copper (Cu2+) from aqueous solutions, depending on the biosorption process. The bacterial candidate was identified using 16S rRNA sequencing and phylogenetic analysis, in addition to morphological and cultural properties as Azotobacter nigricans NEWG-1. The Box-Behnken design was applied to optimize copper removal by Azotobacter nigricans NEWG-1 and to study possible interactive effects between incubation periods, pH and initial CuSO4 concentration. The data obtained showed that the maximum copper removal percentage of 80.56% was reached at run no. 12, under the conditions of 200 mg/L CuSO4, 4 days’ incubation period, pH, 8.5. Whereas, the lowest Cu2+ removal (12.12%) was obtained at run no.1. Cells of Azotobacter nigricans NEWG-1 before and after copper biosorption were analyzed using FTIR, EDS and SEM. FTIR analysis indicates that several functional groups have participated in the biosorption of metal ions including hydroxyl, methylene, carbonyl, carboxylate groups. Moreover, the immobilized bacterial cells in sodium alginate-beads removed 82.35 ± 2.81% of copper from the aqueous solution, containing an initial concentration of 200 mg/L after 6 h. Azotobacter nigricans NEWG-1 proved to be an efficient biosorbent in the elimination of copper ions from environmental effluents, with advantages of feasibility, reliability and eco-friendly.
Subject terms: Metals, Applied microbiology
Introduction
The world population is continuously increasing, with anticipation to reach six billion by the end of 2050. Otherwise, agricultural productivity facing water shortage, depleting soil fertility and various abiotic stresses. Furthermore, the estimates revealed annual losses in agricultural land, mainly occurred through industrialization, pollution, desertification and urban development1. Pollution of soil and water with heavy metals is one of the major stresses that affect agricultural production, local population health, natural resources and the balance of the ecosystem2,3. In addition, industrial pollution from heavy metals leads to contamination of sites in a large area, resulting in significant changes in the structure and/or biosphere of the soil as a result of transition activities4. Heavy metals are metallic chemical elements that have relatively high density and are toxic at low concentrations. Copper is one of these metals, which is necessary as trace element for growth and metabolic processes in living organisms, serving as an essential micronutrient ion that is involved in some metabolic processes as being a co-factor of many metalloenzymes where it plays a role in the active sites of these enzymes, but it is toxic at high concentrations5. Metals, including copper is present in many wastewater sources and industries including, fertilizer, wood preservatives, printing operations, paint manufacturing, copper polishing, wire drawing, electronics plating, printed circuit board manufacturing, paper and pulp, metal cleaning and refineries6–8. Plants are highly sensitive to the toxicity of Cu2+, exhibiting metabolic disorders and inhibition of growth when the amount of Cu2+ in the tissues increases slightly above the normal levels9. Otherwise, the excessive human consumption of copper in the drinking water causes gastrointestinal irritation, headaches, central nervous disorders accompanied by depression, hepatic and kidney damage, extensive capillary damage, strong mucosal irritation, stomach cramps, diarrhea, vomiting, nausea and probable liver and kidney necrotic damages10–12. Therefore, it is mandatory to remove copper ions to be in allowable limits without virulence levels to organisms.
Previously, the traditional procedures for removal of heavy metals from the ecosystem may include chemical and precipitation treatment, adsorption techniques, ion exchange and electrochemical and oxidation processes. These previous techniques were found to generate toxic products, too expensive and energy requirements. Generally, the bioremediation process is an eco-friendly technique that has emerged as a cost-effective alternative to traditional technologies. Additionally, the bioremediation process is a biological technique that relies on the use of biological materials like microorganisms in biosorption and/or the bioaccumulation of such heavy metal pollutants13–15. It could effectively minimize the risk of contaminants across the removal of toxic heavy metals from soil or groundwater3. The bioremediation process has many advantages e.g. low waste production, the safety of biological processes to the environment, low energy demand and self-sustainability16,17. Soil microorganisms are a new applicable strategy to remediate soil contaminant with heavy metals, since discovering the role of microorganisms in metal mobility and availability to the plants18. Microorganisms have a variety of mechanisms for handling high heavy metals concentrations and often are limited to one or a few metals19. In addition, the kinetics of microbial process for heavy metals removal are depending upon the complexation, metal ions reduction, efflux or as acting as electron acceptors in anaerobic respiration16,20. For example, previous studies showed the role of mycorrhizal fungi, N-fixing rhizobacteria and free-living rhizosphere bacteria in removing heavy metals either through direct microbial action and/or encouraging plant growth21,22. Further, rhizospheric bacteria increased the uptake of Cd2+ in Brassica napus23. The role of Azotobacter chroococcum with zeolite as a carrier for removing heavy metals from contaminated soil has been reported20,24. Another study referred to the capacity of actinobacteria in the removal of heavy metals25. Otherwise, El-Naggar et al.26 the efficacy of algal biomass of Gelidium amansii in the biosorption of the lead element from aqueous solution. Similarly, the biosorption of some heavy metals from aqueous solution has been carried out by the algal biomass27,28.
Azotobacter species are saprophytic, nitrogen-fixing bacteria that are widely distributed in soil, water and in combination with some plants. Therefore, in the present study Azotobacter nigricans NEWG-1 as an ecofriendly bacterium was used for the biosorption of copper ions from the medium under different growth conditions. Statistical modeling-approach was used for optimization of the biosorption process. In addition, characterization of the bacterial biomass and application of the immobilized bacterial cells for copper removal were studied.
Results and discussion
Heavy metals represent the main pollutants in land and water resources, threatening the picture and distribution of organisms. Anthropogenic sources of metal contamination included five categories (i) metal mining, (ii) atmospheric deposition, (iii) agricultural practice (iv), industrial processes, and (v) waste disposal29,30. The risk factor of heavy metals can be due to accumulation in the human body across food chain31. The impact of heavy metals could extend to change the microbial communities and activities32. However, the most traditional remediation methods such as landfill, thermal treatment, acid leaching and electro-reclamation do not provide reasonable solutions due to low efficiency, costs and time consuming33. Remediation of heavy metals from contaminated water is a global challenge and requires new technology to enhance the process. Biological treatment was found to be more efficient and low cost34. One effective and promising process is phytoremediation, which is the use of plants to detoxify pollutants28,35. The use of the microbial approach in heavy metals leaching is another type of biological treatment, it is a promising approach since discovering the role of soil microorganisms in leaching the heavy metals4,36.
The present study is designed to leach copper ion by Azotobacter nigricans NEWG-1 using Box-Benken design as affected by the independent variables, i.e. incubation periods, pH values and Cu2+ concentrations.
Cultural and morphological features of Azotobacter sp. strain NEWG-1
Azotobacter genus produces flat, slimy, paste-like colonies with a diameter of 5–10 mm. The growth is favored at a temperature of 20–30 °C. Morphological characteristics of Azotobacter sp. isolate NEWG-1 was observed by Gram-stained slides after incubation on nitrogen-free Asby’s medium at 30 °C for 28 h. Microscopic observation of Azotobacter sp. isolate NEWG-1 showed oval or spherical large bacteria (1–2 μm in diameter) that form thick-walled cysts and may produce large quantities of capsular slime. Morphological examination revealed that the Azotobacter sp. isolate NEWG-1 is Gram-negative, aerobic, free-living, heterotrophic bacteria, and fix atmospheric nitrogen so it can grow in the absence of nitrogen such as the modified nitrogen-free Asby’s medium.
rRNA gene sequence analysis and phylogenetic analysis
The obtained 16S rRNA sequence of Azotobacter sp. isolate NEWG-1 was determined and the amplified 16S rRNA fragment gave sequence with 1500 bp (Fig. 1). The obtained 16S rRNA sequence was subjected to the BLAST search37 of the GenBank database. The phylogenetic tree (Fig. 2) was constructed using MEGA 3.0 software38. Accordingly, Azotobacter sp. isolate NEWG-1 was identified as Azotobacter nigricans NEWG-1 and the 16S rRNA sequence had been deposited in the DDBJ/EMBL-Bank/GenBank database under the accession number LC485953.
Figure 1.
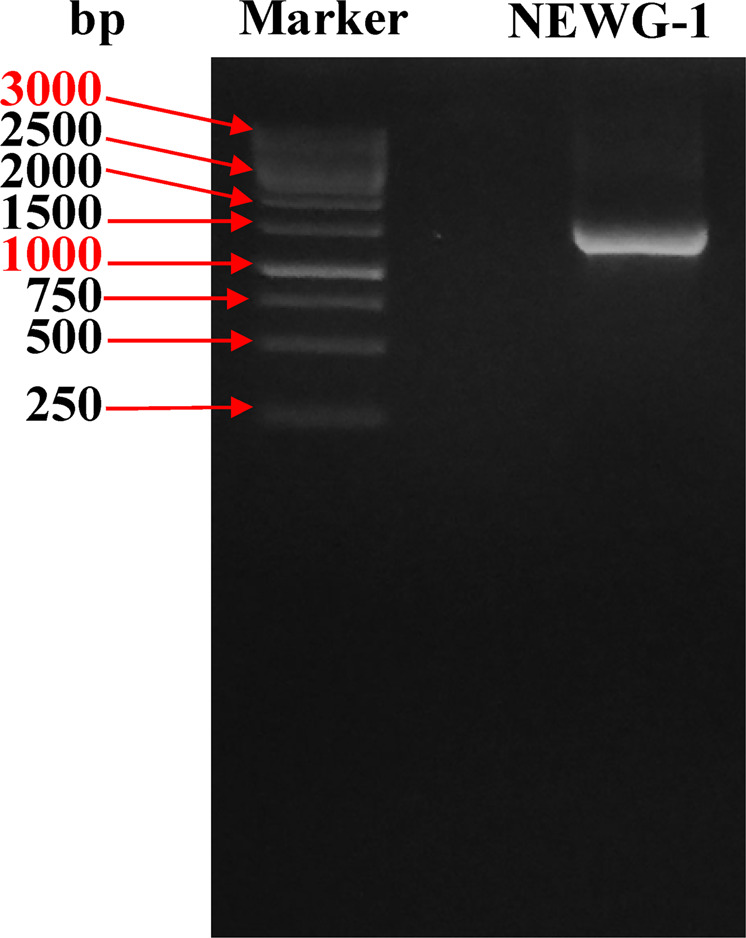
Agarose gel electrophoresis showed the PCR product of the amplified Azotobacter nigricans NEWG-1 16S rRNA fragment.
Figure 2.
Phylogenetic tree obtained by neighbor-joining analysis showing Azotobacter nigricans NEWG-1 position within the genus Azotobacter.
Optimization of medium condition using Box-Benken
The design matrix of Box-Behnken was employed to determine the interactive effects of the independent variables, i.e. incubation periods, initial culture pH, and initial concentration of Cu2+ in culture medium growth to obtain the maximum copper removal% by Azotobacter nigricans NEWG-1. The experimental design matrix of Box-Behnken with 15 runs and the corresponding actual and predicted values of Cu2+ removal % were introduced in Table 1. Data obtained illustrate the variations in Cu2+ removal %, in which it ranged from 12.12 to 80.56%. These variations indicated that the process of optimization was important for the maximization of the removal efficiency of copper by Azotobacter nigricans NEWG-1. Data also show the highest value of Cu2+ removal (80.56%) occurred in run no.12. Whereas, the lowest value of Cu2+ removal was in run no.1, being 12.12%. As can be seen, the predicted values of Cu2+ removal are very close to the corresponding actual values, reflecting the accuracy of such model in the prediction of the response.
Table 1.
Experimental fermentation conditions of the tested variables based on Box-Behnken design and the corresponding observed and fitted Cu2+ removal % by Azotobacter nigricans NEWG-1.
| Std | Run | Tested variables | Cu2+ removal (%) | Residuals | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Incubation time (X1) | pH (X2) | CuSO4 concentration (X3) | ||||||||
| Coded | Actual (days) | Coded | Actual | Coded | Actual (mg/L) | Experimental | Predicted | |||
| 9 | 1 | 0 | 4 | −1 | 5.5 | −1 | 50 | 14.73 | 15.16 | −0.43 |
| 3 | 2 | −1 | 2 | 1 | 8.5 | 0 | 125 | 52.58 | 55.69 | −3.11 |
| 15 | 3 | 0 | 4 | 0 | 7 | 0 | 125 | 60.31 | 58.89 | 1.42 |
| 6 | 4 | 1 | 6 | 0 | 7 | −1 | 50 | 50.19 | 52.87 | −2.68 |
| 13 | 5 | 0 | 4 | 0 | 7 | 0 | 125 | 59.57 | 58.89 | 0.68 |
| 7 | 6 | −1 | 2 | 0 | 7 | 1 | 200 | 70.88 | 68.20 | 2.68 |
| 14 | 7 | 0 | 4 | 0 | 7 | 0 | 125 | 56.8 | 58.89 | −2.09 |
| 1 | 8 | −1 | 2 | −1 | 5.5 | 0 | 125 | 12.12 | 14.97 | −2.85 |
| 4 | 9 | 1 | 6 | 1 | 8.5 | 0 | 125 | 67.47 | 64.62 | 2.85 |
| 8 | 10 | 1 | 6 | 0 | 7 | 1 | 200 | 60.32 | 63.60 | −3.28 |
| 12 | 11 | 0 | 4 | 1 | 8.5 | 1 | 200 | 80.56 | 80.13 | 0.43 |
| 2 | 12 | 1 | 6 | −1 | 5.5 | 0 | 125 | 18.68 | 15.57 | 3.11 |
| 5 | 13 | −1 | 2 | 0 | 7 | −1 | 50 | 42.02 | 38.74 | 3.28 |
| 10 | 14 | 0 | 4 | 1 | 8.5 | −1 | 50 | 72.37 | 72.54 | −0.17 |
| 11 | 15 | 0 | 4 | −1 | 5.5 | 1 | 200 | 47.9 | 47.74 | 0.16 |
Evaluation of Box-Behnken results
The experimental data of Box-Behnken results was subjected to the statistical analysis and ANOVA. As shown in Table 2, the overall model F-value=43.5. Model F-value is calculated as the ratio of mean square regression and mean square residual, the higher the F-value, the higher the significance of the model. The P-value of the overall model is very low (0.0003), this implies that the model is significant. The lower P-value (≤0.05) means the higher significance of the model. Another indication of the adequacy of the model is the non-significant lack of fit (P-value = 0.128), which is a prerequisite for the fit of the overall model.
Table 2.
Analysis of variance for the response surface of Cu2+ removal % by Azotobacter nigricans NEWG-1 obtained by the Box-Behnken design.
| Source of variance | Degrees of freedom | Contribution, % | Sum of square | Mean of square | F-value | P-value | Coefficient estimate | |
|---|---|---|---|---|---|---|---|---|
| Overall model | 9 | 98.7 | 6163.84 | 684.87 | 43.49 | 0.0003* | 58.89 | |
| Linear effect | X1 | 1 | 0.7 | 45.41 | 45.41 | 2.88 | 0.1502 | 2.38 |
| X2 | 1 | 64.6 | 4029.78 | 4029.78 | 255.92 | <0.0001* | 22.44 | |
| X3 | 1 | 12.9 | 807.02 | 807.02 | 51.25 | 0.0008* | 10.04 | |
| Total linear | 3 | 78.2 | 4882.2 | 1627.4 | 103.4 | 0.000* | ||
| Square effect | X12 | 1 | 5.1 | 340.93 | 340.93 | 21.65 | 0.0056* | −9.61 |
| X22 | 1 | 8.7 | 494.41 | 494.41 | 31.40 | 0.0025* | −11.57 | |
| X32 | 1 | 2.6 | 159.30 | 159.30 | 10.12 | 0.0245* | 6.57 | |
| Total square | 3 | 16.4 | 1020.6 | 340.2 | 21.6 | 0.003* | ||
| Interaction effect | X1X2 | 1 | 0.3 | 17.35 | 17.35 | 1.10 | 0.3420 | 2.08 |
| X1X3 | 1 | 1.4 | 87.70 | 87.70 | 5.57 | 0.0647 | −4.68 | |
| X2X3 | 1 | 2.5 | 156.00 | 156.00 | 9.91 | 0.0254* | −6.25 | |
| Total interaction | 3 | 4.2 | 261.1 | 87.0 | 5.5 | 0.048* | ||
| Error effect | Pure Error | 2 | 0.1 | 6.85 | 3.42 | |||
| Lack-of-Fit | 3 | 1.2 | 71.88 | 23.96 | 7.0 | 0.1276 | ||
| Total error | 5 | 1.3 | 78.7 | 15.8 | ||||
| Overall total | 14 | 100.0 | ||||||
| R2 | 98.74% | Std. Dev. | 3.97 | |||||
| Adj R2 | 96.47% | Mean | 51.10 | |||||
| Pred R2 | 81.33% | C.V.% | 7.77 | |||||
| Adeq Precision | 20.1125 | PRESS | 1165.54 | |||||
*Indicates significant effect. “Std. Dev. is the standard deviation, the coefficient of determination (R2), Adj R2 is the adjusted-R2, Pred R2 is the predicted-R2 and PRESS is the prediction error sum of squares, Adeq Precision is adequate precision, C.V is the Coefficient of variation”.
The P-values were also used as a measure to verify the significance of each variable which, in fact, is necessary to understand mutual interaction patterns between the test variables. Among the test variables, the P-values of X2 (initial culture pH) and X3 (initial concentration of Cu2+) were significant, whereas, X1 (incubation period) was not significant. Regarding the square effect, all the variables were significant. Finally, X2X3 was the only significant interaction. The total effects of linear, square and interaction were found to be significant, being 0.000, 0.003 and 0.048; respectively.
These data were confirmed by the Pareto chart (Fig. 3), which was used to determine the contribution of each variable to the variability in the Cu2+ removal. The absolute values of the effects were plotted in descending order. The significance of the individual effect of each of the tested variables at the level of 0.05 was designated using the reference line on the chart. In these results, the incubation period had no significant effect on Cu2+ removal, on the other hand, pH followed by initial Cu2+ were the most efficient variables.
Figure 3.
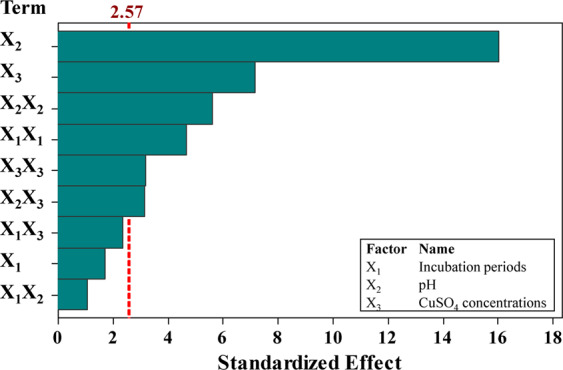
Pareto chart of the standardized effects of the tested parameters on Cu2+ removal % by Azotobacter nigricans NEWG-1.
In order to assign the fitness of the model based on the experimental data, further statistics parameters were calculated. Data standard deviation measured a low value of 3.97. The lower the value of the standard deviation the better the model describes the response. The value of the prediction error sum of squares (PRESS) is 1165.54. A smaller value of PRESS indicates the model’s has better predictive efficiency.
Regarding the coefficient of determination (R2), it was found to be 98.74%. R2 is the variation that occurs in the response when applying the model. It is one of the important measures that usually used to determine how well the model fits the data (copper removal). The higher R2 value means that the model better explains the data. The adjusted-R2 explains the variance in the response as affected by the independent variables. However, the present adjusted-R2 value was 96.47%, this is high enough to explain the response as a result of the model. The predicted-R2 was calculated in order to estimate how well the model predicts the response for new experiments. The predicted-R2 of the current model was estimated to be 81.33%. Models with higher values of predicted-R2 have greater ability to predict the response.
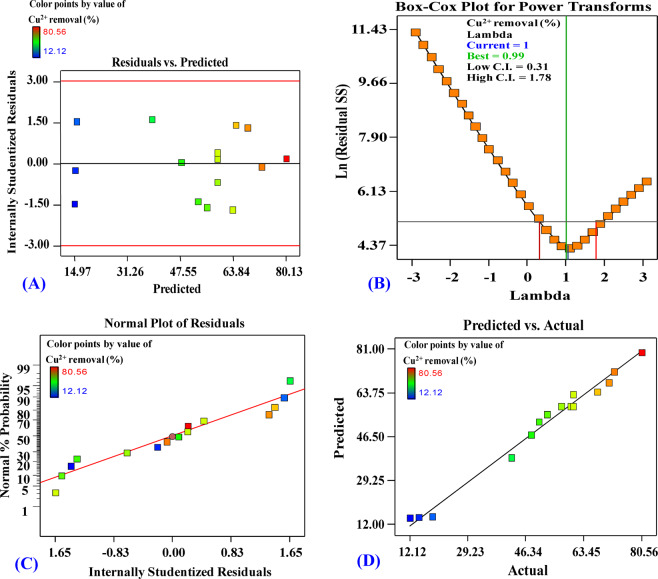
To verify that the model is adequate to meet the assumptions of the analysis, some analytical statics were performed (Fig. 4). The residuals were plotted against fitted values (Fig. 4A), the residuals on the plot fall randomly around the center line and no trends or patterns could be observed when displayed against the fitted values of Cu2+ biosorption. The residuals not correlated and scattered randomly and independently on both sides of the center line, indicating that residuals have constant variance and the model is adequate and meets the assumptions of the analysis.
Figure 4.
(A) Correlation between the residual and predicted values, (B) Box-Cox plot, (C) The normal probability plot of the residuals, (D) Correlation between the experimental and predicted values for Cu2+ removal by Azotobacter nigricans NEWG-1 determined by the second-order polynomial equation.
In Box-Cox plot (Fig. 4B), the current lambda (λ) was equal to 1.0, the best λ was 0.99, whereas the confidence interval (C.I.) was between low (0.31) and high (1.78), since the best value of lambda lies between the points of low and high C.I. so, there was no recommended transform of the data. The normal probability plot (Fig. 4C) indicates that the residuals follow a straight line, meaning that the data were normally distributed. In consent with the Box-Cox plot, no needs for data transform. Plotting of the actual versus the predicted response values (Fig. 4D) showed that the data points split evenly along the 45-degree line. This can help detect the value(s) that are not easily predicted by the model.
According to the previous ANOVA, the second-order polynomial equation for Cu2+ biosorption by Azotobacter nigricans NEWG-1 was calculated as coded units as follows;
| 1 |
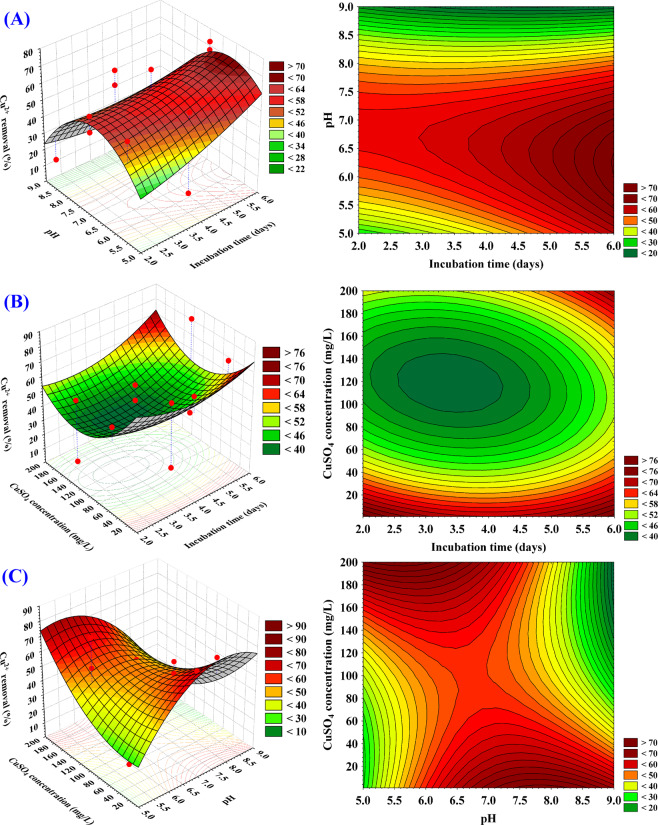
where X1 is the coded value of incubation time, X2 is the coded value of initial culture pH and X3 is the coded value of initial concentration of Cu2+ Three-dimensional (3D) surface and contour plots. To understand the interactive effects of the three tested variables and the optimum values of each variable required for the maximum Cu2+ removal %, the 3D surface and contour plots were established by plotting Cu2+ removal % on the Z-axis against two of the variables and the third variable is holed at the center point (shown in Fig. 5A–C). Figure 5A represents the Cu2+ removal % as the simultaneous effect of incubation time (X1), pH (X2) while the initial concentration of CuSO4 was kept at the central point (125 mg/L). The Cu2+ removal % increased with the increment of incubation time and with an increase in initial pH, the Cu2+ removal % increased beyond pH 7. A further rise in the initial pH leads to a gradual reduction in Cu2+ removal %. By solving the Eq. (1), the highest Cu2+ removal % of 70.28% could be reached using the optimal predicted levels of incubation time of 4.43 days, initial pH of 8.5 when the initial concentration of CuSO4 was kept at 125 mg/L.
Figure 5.
Three-dimensional surface plot of Cu2+ removal % by Azotobacter nigricans NEWG-1, showing the interactive effects of two variables at a time of the three tested variables factors, holding the third factor at the center point.
The variations in Cu2+ removal (%) efficiency by Azotobacter nigricans NEWG-1 may be influenced by independent variables, which effect on biosorption process. Park et al.39 reported that the physicochemical factors have an influence on the biosorption performance. Moreover, Wang40 studying the biosorption process of copper ion by Saccharomyces cerevisiae. The efficacy of Cu2+ removal enhanced from 1 to 20 mg/L, while it was decreased in the concentrations range from 25 to 50 mg/L. The pH value was found to have a positive effect on the biosorption process. Another suggested mechanism for copper removal throughout the process of adsorption is the ion-exchange. Additionally, the cell surface and mucus layer also play critical role in both absorption and adsorption processes of metal-ions through which, the functional-groups that occurred onto cell surface make complexes with the metal-ions to act as organization atom of leaching process41. Besides, negative charges of the carboxyl anionic groups and phosphoric acid on the microbial cell wall surface, allows the metal ions to attach and/or pass across the cell membrane42. Moreover, the pH values influence the sites of sorbate43 through the competition that occurs among metal ions and protons.
Rohini and Jayalakshmi44 reported a bioremediation process of copper ions using Bacillus cereus with a maximum tolerable capacity of 600 mg/L copper under optimal growth conditions. Whereas, Pugazhendhi et al.45 reported the efficacy of Ralstonia solanacearum for biosorption of lead from aqueous solution.
The 3D surface and contour plots in Fig. 5B illustrate Cu2+ removal % as a function of incubation time (X1) and initial concentration of CuSO4 (X3) while pH (X2) was fixed at 7. Figure 5B indicates that low Cu2+ removal % is supported by lower incubation time (X1), the increase in incubation time, the Cu2+ removal % increases and the maximum Cu2+ removal % obviously obtained at near 4 days of incubation. On the other side, the initial concentration of CuSO4 (X3) up to 200 mg/L had a steady positive effect on the Cu2+ biosorption by Azotobacter nigricans NEWG-1. By analysis of Fig. 5B and solving the Eq. (1), the maximum predicted Cu2+ removal % of 75.64% could be reached using pH 7, 200 mg/L of the initial concentration of CuSO4 and incubation period of 3.8 days.
However, the increases of metal ions could affect the quantity of the bio-absorbed heavy metal per unit weight of biomass26. Another study suggesting that the leaching process falls into two categories (i) biosorption (passive) using non-living cells (ii) bioaccumulation using living cells46,47. However, another investigation pointed out that the leaching process depends upon the covalent interaction of metal ion on the cell surface or inside the cell by different methods48. The adsorption ability of species of Azotobacter, Pseudomonas, Serratia and Klebsiella to heavy metals has been reported49. Further, the copper ions were found to be utilized in the metabolic process of bacteria, e.g. cytochrome c oxidase50. Moreover, leaching of Cr(VI) using fungal biomass has been investigated51.
Figure 5C shows Cu2+ removal % as influenced by pH (X2) and initial concentration of CuSO4 (X3) by maintaining the incubation time (X1) at 4 days. With an increased concentration of CuSO4; Cu2+ removal % by Azotobacter nigricans NEWG-1 was increased and the maximum Cu2+ removal % has been obtained at a high level of CuSO4. The Cu2+ removal % increases with the increment of pH and the maximum Cu2+ removal % obviously obtained at pH 8 and the further increase in pH led to a decrease in Cu2+ removal %. By analysis of Fig. 5C and solving the Eq. (1), the maximal predicted Cu2+ removal % of 81.19% could be reached using 4 days of incubation time and the optimal predicted levels of pH 8 and initial concentration of CuSO4 of 200 mg/L.
Model verification
In order to determine the optimal combination of the tested variables, that maximize the Cu2+ biosorption, the response optimization was carried out to increase the efficacy of Azotobacter nigricans NEWG-1 in Cu2+ biosorption. The optimal predicted levels of 4 days of the incubation time, pH 8 and 200 mg/L of initial CuSO4 to maximize the efficacy of Cu2+ biosorption in the fermentation medium were estimated. The theoretical estimated Cu2+ biosorption based on the equation of the quadratic model at such condition was 81.19%. In order to confirm the optimization results, the theoretical calculations from the regression equation were laboratory validated using the estimated levels of the tested variables, the laboratory value of Cu2+ biosorption was found to be 80.56%. This value is closely related to the theoretical value, confirming the accuracy of the proposed model.
In this study, the maximum Cu2+ removal by Azotobacter nigricans strain NEWG-1 was 80.56% under the conditions of 200 mg/L CuSO4, 4 days incubation period and pH 8.
Narasimhulu52 used response surface methodology to optimize the process variables for batch biosorption of Cu2+ by Bacillus subtilis. The optimum levels of process variables for the highest biosorption of Cu2+ (78.4%) by Bacillus subtilis were determined to be contact time of 30 min, biomass concentration of 2 mg/mL, pH of 4 and temperature of 32 °C using CuSO4 at an initial concentration of 10 mg/L. On the other hand, Rajeshkumar and Kartic53 used one factor at a time optimization method to determine the optimal values of various physicochemical variables on the biosorption of Cu2+ by Bacillus sp., the physicochemical variables for maximum biosorption of Cu2+ (88%) were pH (8), temperature 35°C when the initial Cu2+ concentration was 100 mg/L. Moreover, Choińska-Pulit et al.54 used Box-Behnken design to optimize the biosorption process of Cu2+ by Pseudomonas azotoformans JAW1 in an aqueous medium. The maximum biosorption of Cu2+ by Pseudomonas azotoformans JAW1 (63.32%) was achieved at the optimum levels of process variables (concentration of the biosorbent of 2 g/L, the initial metal concentration of 25 mg/L and pH 6). Also, El-Ahwany55 used Plackett-Burman experimental design in 12 experimental runs to evaluate the significance of 11 process variables on Cu2+ biosorption by Oenococcus oeni PSU1. The most significant variables were mixing speed, immobilization and initial copper concentration. These variables were selected and further optimized using the three-level Box–Behnken design to determine the optimal level each. The estimated optimal levels of these three variables for maximum Cu2+ biosorption by Oenococcus oeni PSU1 (85%) were immobilization (2.27%), mixing speed (136 rpm) and metal concentration (8.6 mg/mL).
Manohari and Yogalakshmi56 studied the Cu2+ ions removal process by genera of Bacillus and Arthrobacter, using Box-Behnken design. Three variables (pH, temperature and Cu2+ concentration) were selected to find out the optimal levels and to evaluate the combined effects of these variables on Cu2+ removal efficiency by isolated bacterial consortium2. The optimal values of the selected variables for the highest Cu2+ bioremoval of 82.8% were 168 h of incubation at a temperature of 32.5°C, pH 5 and 600 mg/L copper concentration2. On the other hand, Ghosh and Saha57 used central composite design to optimize the bioremediation process variables and to study the effects and interactions of these variables on Cu2+ removal from aqueous solution by Stenotrophomonas maltophilia PD2. Initial pH, initial copper concentration and contact time on removal % of Cu2+ was studied using central composite design. The maximum experimental Cu2+ removal % by copper-resistant Stenotrophomonas maltophilia PD2 (90%) was obtained at the optimal levels of initial copper concentration (50 mg/L), contact time (26 h) and pH (5.5). The efficiency of leaching was 63.32%. As well as, the bacterial organism, Bacillus licheniformis has the ability to remove 80% copper ion under pH 6.5 and contact time of 216 h58. In the context, the studies of Karthik et al.59,60 reported the removal of Cr(VI) by using Cellulosimicrobium funkei strain AR8.
Fourier transform infrared (FTIR) analysis
Dried cells of Azotobacter nigricans NEWG-1 were analyzed before and after copper biosorption (Fig. 6) to investigate the interaction of copper ion with functional groups of the cells wall of the bacterium. Several functional groups have the ability to bind with metal ions, such as amino, phosphate, carboxylate and hydroxyl groups61. Biosorption of metal ions takes place via the ion exchange process on the cell surface. The adsorbent spectra were measured in the range between 400 and 4000 cm−1 wave number62. The chart of analysis indicates that the spectra of 3452 and 3439 cm−1 O-H stretching vibrations. 2923 and 2927 cm−1 (asymmetric CH2 stretch), it is probable that the band observed at 2856 cm−1 is the difference between the hydroxyl stretching vibrational frequency and the hydroxyl translation frequency. 1741, 1730 cm−1 is the wavenumber for C=O in the carboxyl group (-COOH) or carbonyl ester 1638, 1639 cm−1 is most likely due to the presence of a C=O stretch in an amide bond. A band appeared at 1634 cm−1 was assigned to C=N stretching vibrational band.1449, 1444 cm−1 (CH2 bending vibration). 1409 cm−1 (COO-) carboxylate group. 1410 cm−1 (C-O). The bands at 1155 and 1158 cm−1 are due to the C-O stretching, whereas bands at 1077 and 1041 cm−1 are due to strong C-O stretching (primary alcohol). 670 cm−1 strong cis-disubstituted alkenes. Bands at 670, 672 cm−1 and around 447 cm−1 indicate the bidentate ligand, while the absence of the band at 670-630 cm−1 reveals monodenticity. 558 cm−1 was assigned to CuO stretching vibrations. Copper –oxygen (Cu-O) stretching bands have been notable at 447 cm−1. The lower frequency regions of IR spectra of all complexes recorded weak bands around 447-558 cm−1 is attributed to Cu-N bonds63.
Figure 6.
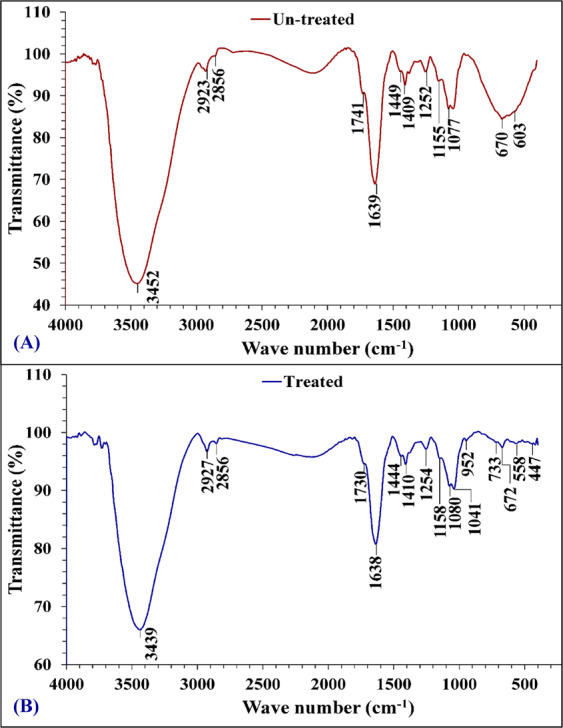
FTIR analysis of Azotobacter nigricans NEWG-1 cells: (A) before and (B) after biosorption of Cu2+ ions from aqueous solution.
Scanning electron microscopy (SEM)
Figure 7 shows a micrograph of Azotobacter nigricans NEWG-1 cells before and after the adsorption of copper ions. It is obvious that the normal, non-treated cells have a regular shape. They are oval or spherical large cells with a thick-wall and large slime capsular (Fig. 7A). On the other side, after the bacterial adsorption of copper ions, Fig. 7B showed dispersed cells with irregular shapes, probably due to the toxicity of the copper ions on the cell wall. In contrast to vegetative cells, cyst formation could be also observed, which enables bacterial cells to resist adverse environmental factors such as the concentration of nutrients.
Figure 7.
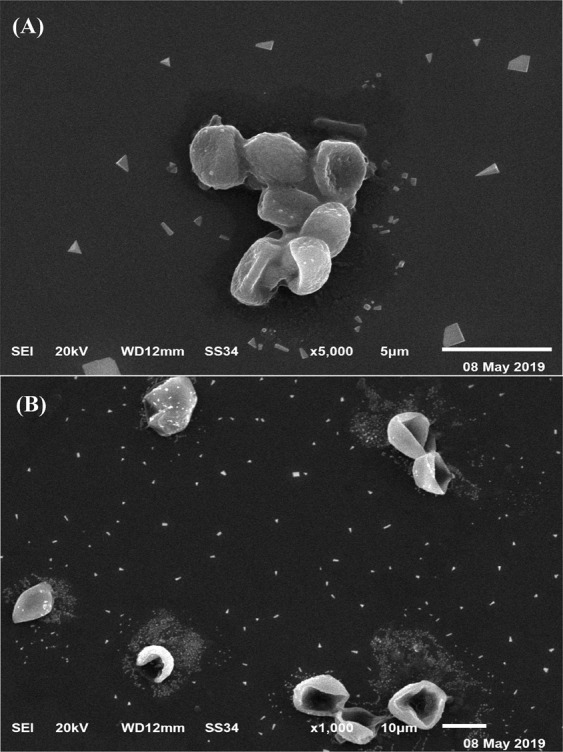
SEM micrograph of Azotobacter nigricans NEWG-1 cells: (A) before and (B) after biosorption of Cu2+ ions from aqueous solution.
Electron dispersive spectroscopy (EDS)
The EDS study was conducted in order to obtain the elementary data (Fig. 8). The EDS spectrum reveals that the binding of Cu2+ onto the cells surface of the tested bacterium is evident in Fig. 8B.
Figure 8.
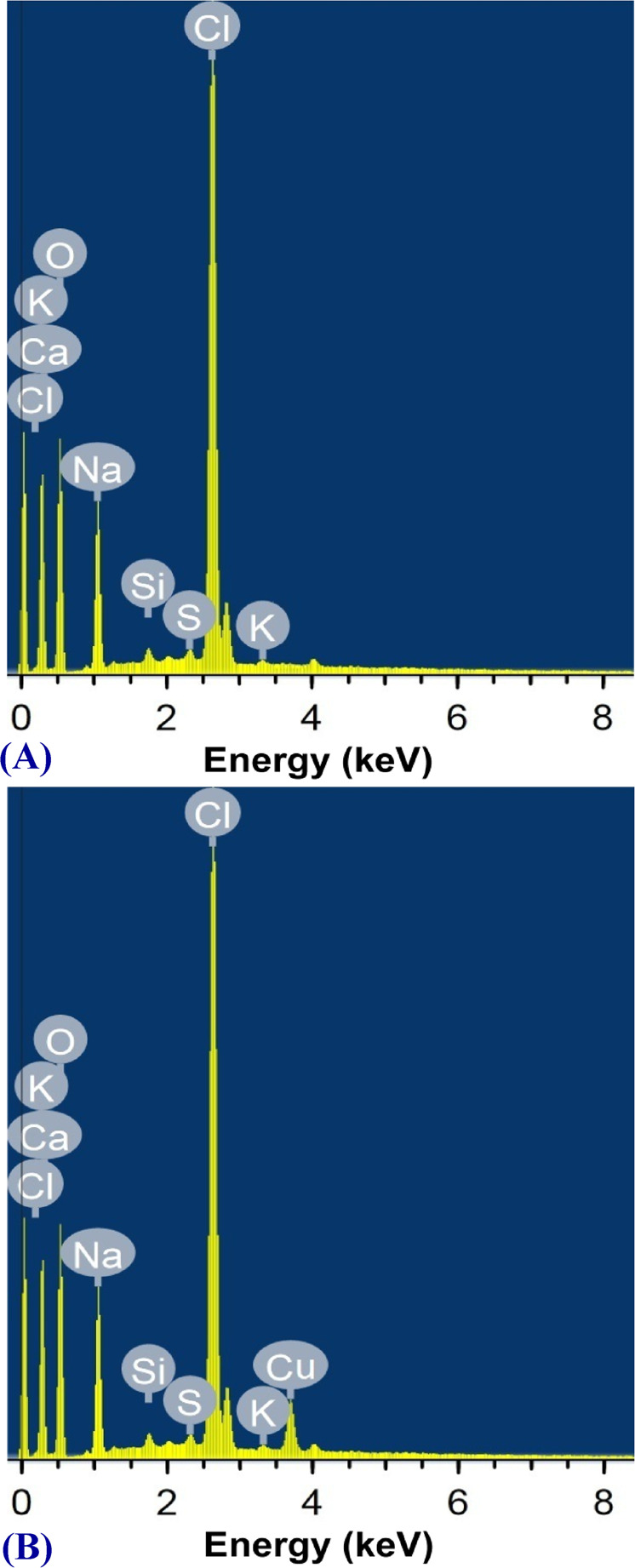
EDS analysis of Azotobacter nigricans NEWG-1 cells: (A) before and (B) after biosorption of Cu2+ ions from aqueous solution.
Copper removal by the alginate-immobilized cells
The ability of the immobilized Azotobacter nigricans NEWG-1 to remove copper ions from aqueous solution was studied, and the findings are shown in Figs. 9 and 10. The results show that the treatment of aqueous copper solution (200 mg/L) with immobilized Azotobacter nigricans NEWG-1 cells for 6 h resulted in the Cu2+ removal percentage of 82.35 ± 2.81%, which is significantly higher than those of using sodium alginate beads as a control (78%) (Fig. 10). Immobilization of Azotobacter nigricans NEWG-1 cells in polysaccharide beads, made of sodium alginate, could be more freely suspended cells to remove copper ions from wastewater as an alternative to current physical and chemical treatment techniques. Azotobacter nigricans strain NEWG-1, is an efficient and safe Cu2+ biosorbent, which makes possible to use the bacterium in the treatment of wastewater. The immobilized cells are more efficient in removing metals than the free cells15,64 and the size of the beads that are used in biomass immobilization is also a significant factor65.
Figure 9.

Immobilization of Azotobacter nigricans NEWG-1in alginate beads during Cu2+ ions removal from aqueous solution. (A) Separating funnel packed with alginate beads, (B) Separating funnel packed with alginate- Azotobacter nigricans NEWG-1 beads.
Figure 10.
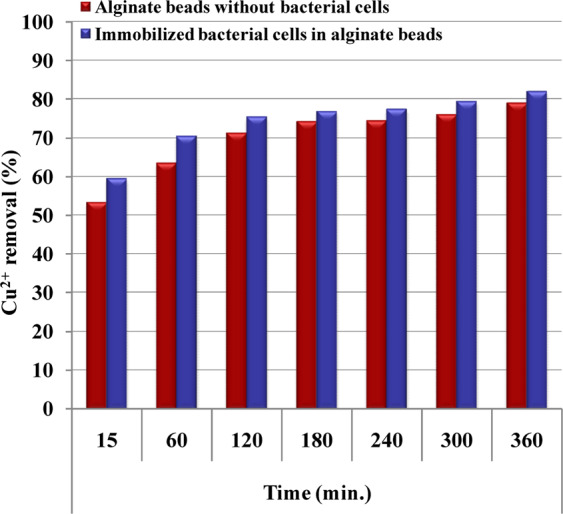
Application of immobilized Azotobacter nigricans NEWG-1 cells in Cu2+ ions removal from aqueous solution.
Materials and Methods
Microorganism and culture maintenance
Azotobacter sp. was kindly obtained from the Department of Microbiology, Soils, Water and Environment Research Institute, Agricultural Research Center (Affiliation ID: 60019332), Giza, Egypt. Nitrogen-free Ashby medium consisting of (g/L) 5 glucose, 5 mannitol, 0.1 CaCl2·2H2O, 0.1 MgSO4·7H2O, 0.005 Na2MoO4·2H2O, 0.9 K2HPO4, 0.1 KH2PO4, 0.01 FeSO4·7H2O, 5 CaCO3, 15 g agar, pH 7.3 was used for the maintenance for the bacterial strain66. The bacterium was grown on slants of the medium supported with 15 g for 72 h, and sub-cultured periodically66. The sterilization was carried out at 121 °C for 20 min.
Identification of Azotobacter sp. NEWG-1
rRNA sequencing
DNA extraction for the bacterial sample was performed using the thermo Gene JET Genomic DNA Purification Kit (#K0721). The PCR reaction and sequencing were performed according to the method of El-Naggar et al.67.
Inoculum preparation
For inoculum preparation, the bacterium was grown on the Asby’s medium under shaking at 30 °C for 72 h, the cell bacterial count was adjusted to obtain 108 CFU/mL, 5% (v/w) inoculum was used to inject 50 mL of fermentation medium in 250 mL Erlenmeyer flasks.
Fermentation medium
Nitrogen-free Ashby medium, consisting of (g/L) 5 glucose, 5 mannitol, 0.1 CaCl2·2H2O, 0.1 MgSO4·7H2O, 0.005 Na2MoO4·2H2O, 0.9 K2HPO4, 0.1 KH2PO4, 0.01 FeSO4·7H2O, 5 CaCO3, 15 g agar, pH 7.3 was used as fermentation medium for the bacterial strain66.
Optimization of Cu2+ biosorption
The experiment was performed to find out a suitable mathematical model to be used to optimize the Cu2+ removal by Azotobacter nigricans NEWG-1 under liquid state fermentation conditions. The effect and interaction of the three factors; incubation time (X1), initial culture pH (X2) and initial concentration of Cu2+ (X3) in the medium were investigated using Box-Behnken experimental design. To address the best combination of the three factors, each was investigated at three levels during the fermentation, i.e. X1 at 2, 4 and 6 days, X2 at 5.5, 7.0 and 8.5 and X3 at 50, 125 and 200 mg/L. After performing the Box-Behnken experimental design, the residual Cu2+ in the fermentation medium was determined and the removal percent of Cu2+ was calculated and fitted to the second-order polynomial quadratic model equation:
| 2 |
where; Y is the Cu2+ removal %, Xi, and Xj are independent variables; β0 model constant, βi, is linear Coefficients; βii is the quadratic coefficients and βij, is cross-product coefficients. The model was laboratory validated to ensure the fitness of the theoretically calculated value of each factor, using the previous equation.
Determination of Cu2+ bioremoval %
The solutions of Cu2+ have been made to reach the required concentrations in mg/L by dissolving copper (II) sulfate in 100 mL distilled water. At the end of the trials the residual copper was analyzed according to AOAC68 by Atomic Absorption Spectrophotometer (AAS) (Buck scientific 210 VGP, Inc.). The characteristic wavelengths were element-specific and accurate to 0.01- 0.1 nm. The apparatus has digital absorbance capable of operating at a wavelength of 324.8 nm for copper and has a detection limit of 0.005 mg/L. The line sources used in AAS is the single hollow cathode lamp. Analysis of copper was conducted by air acetylene flow FAAS. The sample absorbance displayed on a digital terminal screen. AAS was used for quantitative determination by using air acetylene flow rate of 1.0 L/min flame atomic absorption spectrophotometer for copper.
Experimental design and statistical analysis
The design of the Box-Behnken matrix and statistical analysis of variance (ANOVA) were performed using the statistical software packages Minitab (version 18, Minitab Inc., U.S.A.) and “Design-Expert software version 7 for Windows. The STATISTICA software (Version 8.0, StatSoft Inc., Tulsa, USA) was used to plot the 3D surface plots”.
FTIR spectroscopy
Before and after Cu2+ removal, the cells of Azotobacter nigricans NEWG-1 were analyzed using Fourier transform infrared spectroscopy to detect the functional groups present in the cells. With KBr pellets, the bacterial cells were implemented. The FTIR spectra of Azotobacter nigricans NEWG-1 were measured in the range between 400 and 4000 cm−1 using Thermo Fisher Nicolete IS10, “USA spectrophotometer at Spectral Analyses Unit, Faculty of Science, Mansoura University, Mansoura, Egypt”.
SEM investigation
To evaluate removal of the Cu2+ and to examine the surface of bacterial cells, dry cells of Azotobacter nigricans NEWG-1 (before and after removal of the Cu2+) were gold-coated and examined at various magnifications using SEM “scanning electron microscope, JEOL TEM-2100 attached to a CCD camera at an accelerating voltage of 200 kV at Central Laboratory, Electron Microscope Unit, Faculty of Agriculture, Mansoura University, Mansoura, Egypt”.
EDS evaluation
Energy-dispersive X-ray analysis was performed by means of JEOL TEM-2100 connected to a CCD camera at an accelerating voltage of 200 kV at Central Laboratory, Electron Microscope Unit, Faculty of Agriculture, Mansoura University, Mansoura, Egypt.
Cells immobilization
4 g sodium alginate dissolved into 100 mL of distilled water with continuous stirring for 30 min at 60 °C to prepare the solution of 4 percent sodium alginate69. Following cooling, the sterile sodium alginate gel was supplied with Azotobacter nigricans NEWG-1 cells of 105 CFU/mL from 72 h grown vegetative cells with stirring for 5 min at room temperature. The beads with a diameter of 1.5 ± 0.2 mm were produced by adding drop-wise of the alginate-bacterial biomass combination into a cold sterile solution of 2.5% CaCl2 through 3 mL syringe with gentle stirring at room temperature to make spheres. The resulting beads were washed several times with sterilized distilled water in order to remove any residues of CaCl2 from the surface of the beads and then stored at 4 °C overnight in distilled water for stabilization and hardness of the beads. Similarly, sodium alginate beads were prepared without incorporating bacterial biomass and applied as control. Separating funnel experiment was conducted to determine the efficiency of the alginate-bacterial beads to remove the metal ions. The Simax glass separating funnel was packed with the alginate-bacterial beads, a metal solution has been added (200 mg/L). Samples (5 mL) were collected regularly (every 30 min) from the separating funnel effluent at a flow rate of 3 mL/min. the collected fractions were analyzed with the Atomic Absorption Spectrophotometer (AAS) (Buck scientific 210 VGP, Inc.). The difference of the metal solution concentration prior and after adsorption determined the biosorption capacity of metal ions by the bacterial cells.
Conclusion
Azotobacter nigricans NEWG-1 showed high efficacy in copper removal (80.56%) after 6 h, encouraging the application of such bacterium according the proposed procedure to remove copper ions from similar natural aqueous polluted-solutions. Ultimately, offering an effective and eco-friendly procedure for the eradication of copper ions from wastewater.
Author contributions
A.A.G. proposed the research concept, providing some necessary tools for experiments, carried out some of the experiments, collected the data and contributed substantially to the writing of the manuscript. N.E.E. providing some necessary tools for experiments, experimental instructions, collected the data, participated in the data statistical analysis, contributed to the interpretation of the results, contributed substantially to the writing of the manuscript, reviewing and revising of the manuscript. W.I.S. proposed the topics, performed some of the experiments, experimental instructions, participated in the statistical analysis and coordinated and contributed to writing and critical reviewing of the final manuscript. M.S.E. proposed the research concept, performed some of the experiments, experimental instructions, contributed to the interpretation of the results, contributed substantially to the writing of the manuscript, reviewing and revising of the manuscript. A.Y.E. performed some of the experiments, experimental instructions, contributed to the interpretation of the results, contributed substantially to the writing of the manuscript, participated in the manuscript revision. All authors read and approved the final manuscript.
Data availability
The Sanger sequencing data of 16S rRNA sequence had been deposited in the DDBJ/EMBL-Bank/GenBank database under the accession number LC485953.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dόόs BR. Population growth and loss of arable land. Global Environ Change. 2002;12:303–311. [Google Scholar]
- 2.Alqadami AA, et al. Efficient removal of toxic metal ions from wastewater using a recyclable nanocomposite: a study of adsorption parameters and interaction mechanism. Journal of Cleaner Production. 2017;156:426–436. [Google Scholar]
- 3.Siebielec S, et al. Long term insight into biodiversity of a smelter wasteland reclaimed with biosolids and by-product lime. Sci Total Environ. 2018;636:1048–1057. doi: 10.1016/j.scitotenv.2018.04.372. [DOI] [PubMed] [Google Scholar]
- 4.Ettler V. Soil contamination near non-ferrous metal smelters: a review. Appl Geochem. 2016;64:56–74. [Google Scholar]
- 5.Harris ED. Cellular copper transport and metabolism. Ann Rev Nutrition. 2000;20:291–310. doi: 10.1146/annurev.nutr.20.1.291. [DOI] [PubMed] [Google Scholar]
- 6.Agarwal M, Singh K. Heavy metal removal from wastewater using various adsorbents: a review. Journal of Water Reuse and Desalination. 2017;7(4):387–419. [Google Scholar]
- 7.Han W, Fu F, Cheng Z, Tang B, Wu S. Studies on the optimum conditions using acid-washed zero-valent iron/aluminum mixtures in permeable reactive barriers for the removal of different heavy metal ions from wastewater. Journal of hazardous materials. 2016;302:437–446. doi: 10.1016/j.jhazmat.2015.09.041. [DOI] [PubMed] [Google Scholar]
- 8.Wang F, Lu X, Li X. Selective removals of heavy metals (Pb2+, Cu2+nd Cd2+) from wastewater by gelation with alginate for effective metal recovery. J Hazard Mater. 2016;308:75–83. doi: 10.1016/j.jhazmat.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 9.Fernandes JC, Henriques FS. Biochemical, physiological, and structural effects of copper in plants. Bot Rev. 1991;57:246–273. [Google Scholar]
- 10.Kalavathy MH, Karthikeyan T, Rajgopal S, Miranda LR. Kinetic and isotherm studies of Cu (II) adsorption onto H3PO4- activated rubber wood sawdust. J Colloid Interface Sci. 2005;292:354–362. doi: 10.1016/j.jcis.2005.05.087. [DOI] [PubMed] [Google Scholar]
- 11.Benvenuti T, Rodrigues MAS, Bernardes AM, Zoppas-Ferreira J. Closing the loop in the electroplating industry by electrodialysis. J Clean Prod. 2017;155:130–13825. [Google Scholar]
- 12.Cardoso SL, Costa CSD, Nishikawa E, da Silva MGC, Vieira MGA. Biosorption of toxic metals using the alginate extraction residue from the brown algae Sargassum filipendula as a natural ion-exchanger. J Clean Prod. 2017;165:491–499. [Google Scholar]
- 13.Samuel MS, Chidambaram R. Hexavalent chromium biosorption studies using Penicillium griseofulvum MSR1 a novel isolate from tannery effluent site: Box–Behnken optismization, equilibrium, kinetics and thermodynamic studies. Journal of the Taiwan Institute of Chemical Engineers. 2015;49:156–164. [Google Scholar]
- 14.Annadurai ST, Arivalagan P, Sundaram R, Mariappan R, Munusamy AP. Batch and column approach on biosorption of fluoride from aqueous medium using live, dead and various pretreated Aspergillus niger (FS18) biomass. Surfaces and Interfaces. 2019;15:60–69. [Google Scholar]
- 15.Barquilha CER, Cossich ES, Tavares CR, Silva EA. Biosorption of nickel (II) and copper(II) ions in batch and fixed bed columns by free and immobilized marine algae Sargassum sp. J Clean Prod. 2017;150:58–64. [Google Scholar]
- 16.Haferburg G, Kothe E. Metallomics: lessons for metal liferous soil remediation. Appl Microbiol Biotechnol. 2010;87:1271–1280. doi: 10.1007/s00253-010-2695-z. [DOI] [PubMed] [Google Scholar]
- 17.Jacob, J. M. et al. Biological approaches to tackle heavy metal pollution: a survey of literature. Journal of Environmental Management217, 56–70 (2018). [DOI] [PubMed]
- 18.Idris R, Trifonova R, Puschenreiter M, Wenzel WW, Sessitsch A. Bacterial communities associated with flowering plants of the Ni hyper accumulator Thaspi goesingense. Appl Environ Microbiol. 2004;70:2667–2677. doi: 10.1128/AEM.70.5.2667-2677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piddock LJ. Multi-drug-resistance efflux pumps-not just for resistance. Nat Rev Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 20.Baars O, et al. Crochelin, siderophores with a novel iron-chelating moiety from the nitrogen-fixing bacterium Azotobacter chroococcum. Angew Chem Int Ed. 2018;57:536–541. doi: 10.1002/anie.201709720. [DOI] [PubMed] [Google Scholar]
- 21.Khan MS, Zaidi A, Aamil M. Biocontrol of fungal pathogen by the use of plant growth promoting rhizobacteria and nitrogen fixing microorganisms. Ind J Bot Soc. 2002;81:255–263. [Google Scholar]
- 22.Wang, F. Y., Lin, X. G. & Yin, R. Role of microbial inoculation and chitosan in phytoextraction of Cu, Zn, Pb and Cd by Elsholtzia splendens a field case. Environ Pollut147, 248–255 (2007). [DOI] [PubMed]
- 23.Sheng XF, Xia JJ. Improvement of rape (Brassicanapus) plant growth and cadmium uptake by cadmium-resistant bacteria. Chemosphere. 2006;64:1036–1042. doi: 10.1016/j.chemosphere.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 24.Juwarkar AA, Prachi J, Singh SK, Rayalu S. Remediation of metal contaminated soil using a novel biotechnological approach. Environ Sci. 2006;1:4–6. [Google Scholar]
- 25.El-Baz, S. Bioremediation of heavy metals by antibacterial: Review. Amr J Innov Res App Sci 359–369 (2017).
- 26.El-Naggar NE, Hamouda RA, Mousa TE, Abdel-Hamid MS, Rabei NH. Satistical optimization for cadmium removal using Ulva fasciata biomass: characterization, immobilization and application for almost complete cadmium removal from aqueous solutions. Sci Rep. 2018;8:12456. doi: 10.1038/s41598-018-30855-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Naggar NE, Hamouda RA, Mousa IE, Abdel-Hamid MS, Rabei NH. Biosorption optimization, characterization and application of Gelidium amansii biomass for complete Pb2+ removal from aqueous solutions. Sci Rep. 2018;8:13456. doi: 10.1038/s41598-018-31660-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shanmugaprakash, M., Venkatachalam, S., Rajendran, K. & Pugazhendhi, A. Biosorptive removal of Zn (II) ions by Pongamia oil cake (Pongamia pinnata) in batch and fixed-bed column studies using response surface methodology and artificial neural network. Journal of Environmental Management227, 216–228 (2018). [DOI] [PubMed]
- 29.Haferburg G, Kothe E. Microbes and metals: interactions in the environment. J Basic Micrbiol. 2007;47(6):453–467. doi: 10.1002/jobm.200700275. [DOI] [PubMed] [Google Scholar]
- 30.Dulias, R. The impact of mining on the landscape. A study of the upper Silesian coal basin in Poland. Environmental Science and Engineering. Springer P. 209 (2016).
- 31.Zafar S, Aqil F, Ahmad I. Metal tolerance and biosorption potential of filamentous fungi isolated from metal contaminated agricultural soil. Bioresour Technol. 2007;98(13):2557–2561. doi: 10.1016/j.biortech.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 32.Matyar F, Kaya A, Dincer S. Antibacterial agents and heavy metal resistance in Gram-negative bacteria isolated from sea water, shrimp and sediment in Iskenderun Bay, Turkey. Sci Total Env. 2008;407(1):279–285. doi: 10.1016/j.scitotenv.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 33.Jing YD, He ZL, Yang XE. Role of soil rhizobacteria in phytoremediation of heavy metal contaminated soils. J Zhejiang Uni Sci. 2007;8:192–207. doi: 10.1631/jzus.2007.B0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samuel MS. EA Abigail, M., & Ramalingam, C. Biosorption of Cr (VI) by Ceratocystis paradoxa MSR2 using isotherm modelling, kinetic study and optimization of batch parameters using response surface methodology. PloS one. 2015;10:e0118999. doi: 10.1371/journal.pone.0118999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hadi F, Bano A. Effect of diazotrophs (Rhizobium and Azatebactor) on growth of maize (Zea mays L.) and accumulation of lead (Pb) in different plant parts. Pak J Bot. 2010;42:4363–4370. [Google Scholar]
- 36.Pugazhendhi A, Ranganathan K, Kaliannan T. Biosorptive removal of copper (II) by Bacillus cereus isolated from contaminated soil of electroplating industry in India. Water, Air, & Soil Pollution. 2018;229:76. [Google Scholar]
- 37.Altschul, S. F. & Koonin, E. V. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem Sci23(11), 444–447 (1998). [DOI] [PubMed]
- 38.Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biolevolution28(10), 2731–2739 (2011). [DOI] [PMC free article] [PubMed]
- 39.Park D, Yun YS, Park JM. The past, present, and future trends of biosorption. Biotechnol Bioproc Eng. 2010;15(1):86–102. [Google Scholar]
- 40.Wang Y. Optimization of cadmium, zinc and copper biosorption in an aqueous solution by Saccharomyces cerevisiae. Int J Chem. 2012;1:1–13. [Google Scholar]
- 41.Jin Y, Luan Y, Ning Y, Wang L. Effects and mechanisms of microbial remediation of heavy metals in soil: A critical review. Appl Sci. 2018;8(8):1336. [Google Scholar]
- 42.Brady D, Duncan JR. Cations loss during accumulation of heavy metal cations by Saccharomyces cerevisiae. Biotechnol Lett. 1994;16:543–548. [Google Scholar]
- 43.Vijayaraghavan K. Bacterial biosorbents and biosorption. Biotechnol Adv. 2008;26:266–291. doi: 10.1016/j.biotechadv.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 44.Rohini B, Jayalakshmi S. Bioremediation potential of Bacillus cereus against copper and other heavy metals. Int J Adv Res Biol Sci. 2015;2(2):200–209. [Google Scholar]
- 45.Pugazhendhi A, Boovaragamoorthy GM, Ranganathan K, Naushad M, Kaliannan T. New insight into effective biosorption of lead from aqueous solution using Ralstonia solanacearum: characterization and mechanism studies. Journal of Cleaner Production. 2018;174:1234–1239. [Google Scholar]
- 46.Donmez G, Aksu Z. Bioaccumulation of copper (ii) and nickel (ii) by the non-adapted and adapted growing Candida sp. Water Research. 2001;35(6):1425–1434. doi: 10.1016/s0043-1354(00)00394-8. [DOI] [PubMed] [Google Scholar]
- 47.Arivalagan, P., Singaraj, D., Haridass, V. & Kaliannan, T. Removal of cadmium from aqueous solution by batch studies using Bacillus cereus. Ecological Engineering71, 728–735 (2014).
- 48.Bhanoori M, Venkateswerlu G. In vivo chitin-cadmium complexation in cell wall of Neurospora crassa. Biochim Biophys Acta. 2000;1523(1):21–8. doi: 10.1016/s0304-4165(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 49.Kloepper JW, Lifshitz R, Zablotowicz RM. Free living bacterial inocula for enhancing crop productivity. Trends Biotechnol. 1989;7:39–43. [Google Scholar]
- 50.Bhateria R. & Snehlata A review: Role of rhizospheric bacteria in phytoremediation of heavy metal contaminated soil. Inte J of Curr Research. 2013;5(12):3897–3907. [Google Scholar]
- 51.Samuel MS. Isotherm modelling, kinetic study and optimization of batch parameters using response surface methodology for effective removal of Cr (VI) using fungal biomass. PloS one. 2015;10(3):1–15. doi: 10.1371/journal.pone.0116884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narasimhulu K. Optimization of Batch Biosorption of Cr (VI) and Cu (II) Ions from Wastewater using Bacillus subtilis. SF J Material Res Let. 2017;1:3. [Google Scholar]
- 53.Rajeshkumar R, Kartic N. Removal of Cu2+ ions from aqueous solutions using copper resistant bacteria. Our Nature. 2011;9:49–54. [Google Scholar]
- 54.Choińska-Pulit A, Sobolczyk-Bednarek J, Łaba W. Optimization of copper, lead and cadmium biosorption onto newly isolated bacterium using a Box-Behnken design. Ecotoxicol Environ Safety. 2018;149:275–283. doi: 10.1016/j.ecoenv.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 55.El-Ahwany, A. M. D. Statistical analysis and optimization of copper biosorption capability by Oenococcus oeni PSU-1 In. Afr J Biotechnol11(18), 4225–4233 (2012).
- 56.Manohari R, Yogalakshmi KN. Optimization of copper (II) removal by response surface methodology using root nodule endophytic bacteria isolated from Vigna unguiculata. Water, Air, & Soil Pollu. 2016;227(8):285. [Google Scholar]
- 57.Ghosh A, Saha P. Optimization of copper bioremediation by Stenotrophomonas maltophilia PD2. J Environ Chem Eng. 2013;1:159–163. [Google Scholar]
- 58.Akshatha J, Udayashankara N, Kslokesh TH, Sudarshan BL. Bioremediation of lead, nickel and copper by metal resistant Bacillus licheniformis isolated from mining site: Optimization of operating parameters under laboratory conditions. Int J res Eng Technol. 2017;5(5):13–32. [Google Scholar]
- 59.Karthik, C., et al. Evaluation of Cr (VI) reduction mechanism and removal by Cellulosimicrobium funkei strain AR8, a novel haloalkaliphilic bacterium. Journal of hazardous Materials333, 42–53 (2017a). [DOI] [PubMed]
- 60.Karthik, C., Ramkumar, V. S., Pugazhendhi, A., Gopalakrishnan, K., & Arulselvi, P. I. Biosorption and biotransformation of Cr (VI) by novel Cellulosimicrobium funkei strain AR6. Journal of the Taiwan Institute of Chemical Engineers70, 282–290 (2017b).
- 61.Hussein, H., Krull, R., Abou El-Ela, S. I., & Hempel, D. C. Interaction of the different heavy metal ions with immobilized bacterial culture degrading xenobiotic wastewater compounds. In Proceedings of the second international water association world water conference, Berlin, Germany1519, p.1519 (2001).
- 62.Garg U, Kaur MP, Jawa GK, Sud D, Garg VK. Removal of cadmium (II) from aqueous solutions by adsorption on agricultural waste biomass. J Hazard Mater. 2008;154:1149–1157. doi: 10.1016/j.jhazmat.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 63.Khalil SM. Synthesis, spectroscopic and magnetic studies on metal complexes of 5-methyl-3-(2-hydroxyphenyl) pyrazole. J Coordination Chem. 2003;56(12):1013–1024. [Google Scholar]
- 64.Valdez C, Perengüez Y, Mátyás B, Guevara MF. Analysis of removal of cadmium by action of immobilized Chlorella sp. micro-algae in alginate beads. F1000 Res. 2018;1:7–54. doi: 10.12688/f1000research.13527.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mehta SK, Gaur JP. Use of algae for removing heavy metal ions from wastewater: progress and prospects. Cit Rev Biotechnol. 2005;25:113–152. doi: 10.1080/07388550500248571. [DOI] [PubMed] [Google Scholar]
- 66.Kızılkaya R. Yield response and nitrogen concentrations of spring wheat (Triticum aestivum) inoculated with Azotobacter chroococcum strains. Ecological Eng. 2008;33(2):150–156. [Google Scholar]
- 67.El-Naggar NE. Extracellular production of the oncolytic enzyme, L-asparaginase, by newly isolated Streptomyces sp. strain NEAE-95 as potential microbial cell factories: Optimization of culture conditions using response surface methodology. Curr Pharm Biotechnol. 2015;16(2):162–178. doi: 10.2174/1389201015666141113123910. [DOI] [PubMed] [Google Scholar]
- 68.AOAC. Official Methods of Analysis. 20th ed, Association of Official Analytical Chemists. Washington, DC, USA (2016).
- 69.Kumar SS, Saramma AV. Nitrate and phosphate uptake by immobilized cells of Gloeocapsa gelatinosa. J Mar Biol Ass India. 2012;54:119–122. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Sanger sequencing data of 16S rRNA sequence had been deposited in the DDBJ/EMBL-Bank/GenBank database under the accession number LC485953.