Abstract
The COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is a current global threat for which there is an urgent need to search for an effective therapy. The transmembrane spike (S) glycoprotein of SARS-CoV-2 directly binds to the host angiotensin-converting enzyme 2 (ACE2) and mediates viral entrance, which is therefore considered as a promising drug target. Considering that new drug development is a time-consuming process, drug repositioning may facilitate rapid drug discovery dealing with sudden infectious diseases. Here, we compared the differences between the virtual structural proteins of SARS-CoV-2 and SARS-CoV, and selected a pocket mainly localizing in the fusion cores of S2 domain for drug screening. A virtual drug design algorithm screened the Food and Drug Administration-approved drug library of 1234 compounds, and 13 top scored compounds were obtained through manual screening. Through in vitro molecular interaction experiments, eltrombopag was further verified to possess a high binding affinity to S protein plus human ACE2 and could potentially affect the stability of the ACE2-S protein complex. Hence, it is worth further exploring eltrombopag as a potential drug for the treatment of SARS-CoV-2 infection.
Keywords: SARS-CoV-2, Virtual drug design, Homology Modeling, Surface plasmon resonance
Abbreviations: ACE2, angiotensin-converting enzyme 2; kon, association rate; koff, dissociation rate; E, envelope protein; KD, equilibrium dissociation constant; M, membrane protein; N, nucleocapsid protein; RBD, receptor-binding domain; RU, resonance unit; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; S, spike; SPR, surface plasmon resonance; TSA, thermo shift assay
Graphical abstract
1. Introduction
Beginning in early December 2019, cases of suspected novel viral pneumonia were reported in Wuhan, Hubei Province, China. The infectious disease, now termed COVID-19 by the World Health Organization, manifests clinically as fever, cough, shortness of breath, muscle ache, and low leucocyte count (Chen et al., 2020). Human-to-human transmission has been indicated in epidemiological studies (Chan et al., 2020; Li et al., 2020).
COVID-19 has now spread globally (Bogoch et al., 2020; Holshue et al., 2020; Yoo and Hong, 2020). Epidemiological data provided by the World Health Organization indicates that, as of May 25, 2020, 5,304,772 patients have been infected by COVID-19, with 342,029 related deaths in over 230 countries, areas, or territories. Meanwhile, the spread of the pandemic is still accelerating.
Respiratory tract samples were collected from patients and a novel coronavirus was identified as the pathogenic agent by the Chinese Center for Disease Control and Prevention (Chinese CDC) on January 07, 2020 (Chinese CDC, 2020). This virus is a member of the Coronaviridae family, with the third highest pathogenic potential. Other members of the family include the highly pathogenic SARS-CoV and Middle East respiratory syndrome coronavirus, identified in 2003 (Zhong et al., 2003) and 2012 (Hilgenfeld and Peiris, 2013), respectively. The novel causative virus, designated severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) by the Coronavirus Study Group, is an enveloped, positive-sense, single-stranded RNA beta-coronavirus that has high similarity with SARS-CoV genome (Wu et al., 2020), yet the virulence is not exactly identical. SARS-CoV-2 tends to have a longer latent period and less lethality, compared to SARS-CoV. Alterations in coronavirus virulence may be attributable to the differences in the spike (S) gene. SARS-CoV-2 genome sequencing revealed that the S protein receptor-binding domain directly binds the host angiotensin-converting enzyme 2 (ACE2), thereby acting as a receptor for viral entry (Wan et al., 2020). Computational modeling of the S protein structure has also confirmed this result (Xu et al., 2020), indicating the possibility for drug screening using computer simulation.
The ongoing pandemic is regarded as a “Public Health Emergency of International Concern.” However, there are still no specific antiviral drugs and vaccines available.
S proteins comprise a large class of glycoproteins with N-terminal (S1) and C-terminal (S2) domains, which have distinct functions. It is generally believed that the S1 hypervariable region is closely related to coronavirus tropism, while S2 is necessary for mediating membrane fusion. The S1 fragment has pronounced variability, while the S2 fragment also changes in some variants. Therefore, a complete understanding of the pathogenicity of the coronavirus requires detailed insights into the processes of receptor recognition and membrane fusion (Gui et al., 2017). Because of the indispensability of S protein in viral entrance to host cells, it is a hot target for drug screening to prevent the entrance of SARS-CoV-2 into the host cells.
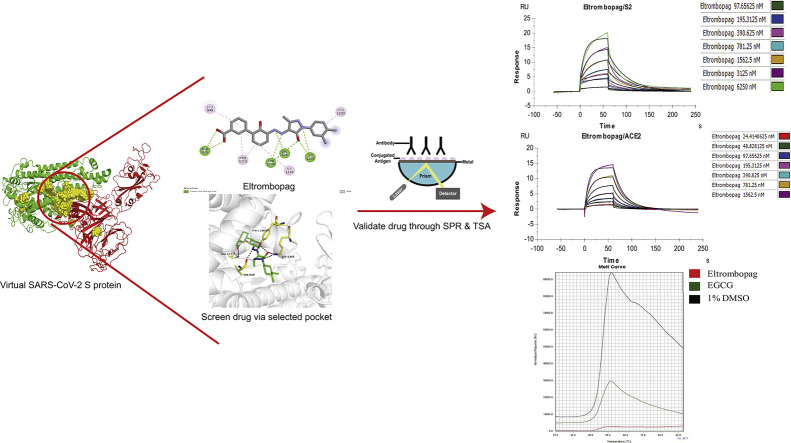
In this study, we describe a structure-based virtual screening model and subsequent high throughput screening methodology, based on the S protein structure of SARS-CoV-2 (Fig. S1). The data will provide insights into the development of potentially efficacious drugs. More importantly, the virtual screening model also offers a reasonable, economic, and rapid method to screen drugs and find out-of-guide application of approved drugs in other diseases and in future possible epidemic or pandemic situations.
2. Materials and methods
2.1. S protein structure simulation and virtual screening
The virtualized S protein, reported by Zhang et al., and cryo-EM S protein structure reported by McLellan et al., were used for the virtualization of the S protein, assessment, characteristic analysis, and drug screening (Wrapp et al., 2020; Zhang et al., 2020). The SARS-CoV-2 S protein sequence was downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/nuccore/NC_045512). SARS-CoV-2 S protein structure was generated using information of the SARS-CoV and SARS-CoV-2 spike glycoprotein structure, downloaded from the PDB database (https://www.rcsb.org/, PDB ID: 5X58, 5WRG, 6U7H, 6CRV, 6LZG). The SARS-CoV-2 spike protein binding pocket was generated using PyMOL software (https://pymol.org/2/). Docking platform, powered with autodock-vina at Beijing Beike Deyuan Bio-Pharm Technology Co. Ltd.(http://www.vslead.com/), was used against the FDA drug database to generate the hit list, and the parameter setup at (center for x = −2.075, y = 16.878, z = 573.956; size for x = 26, y = 26, z = 26. num_modes = 9). The top 100 drugs were selected for further visual inspection. Of these, 13 with highest docking scores were used for more assays.
2.2. TM-align
TM-align is an algorithm for the comparison of sequence independent protein structure, which generates optimized residue-to-residue alignment based on structural alignment, using heuristic dynamic programming iterations. The TM-score is generated to scale the structural similarity of proteins, with values in the range of 0–1. A perfect match between two structures is indicated by a value of 1, values below 0.2 indicates two randomly chosen unrelated proteins, and values higher than 0.5 generally assume the same fold in SCOP/CATH. The similarity between spike protein model (QHD43416.pdb), reported by the Zhang lab (Zhang et al., 2020) and the reported SARS-CoV-2 spike protein structure (6LZG.pdb) was analyzed by TM-align (Version 20,190,822). For comparison, chain 1 (A316309, 1273 residues) and chain 2 (B316309, 959 residues) of QHD4316 were used.
2.3. Surface plasmon resonance assay
Equilibrium dissociation constant (KD) values were measured by surface plasmon resonance (SPR) method using, BIACore™ —S200 at 25 °C and a fixed Protein-tag CM5 chip (GE health care) at a certain resonance unit (RU). The details of each chip (human ACE2, SARS-CoV-2 S1 + S2 ECD, SARS-CoV2 S2 ECD) are shown in Supplementary Table S1. Briefly, the methylated dextran biosensor chips were activated by hydrochloric acid N-ethyl-N′-(3-dimethylaminopropyl) carbodiimide and N-hydroxy-succinimide, according to the supplier's instructions. All proteins were acquired from Sino Biological Inc. Human ACE2 or SARS-CoV-2 spike protein (S1 + S2,S2) was diluted with 10 mM sodium acetate pH (4.5 or 5.5) to 50 μg/mL, and then injected at a flow rate of 5 μL/min, until approximately 12,000 or 17,000 RU of the coupled protein were obtained. After injection of human ACE2 or SARS-CoV-2 S protein, 1 M ethanolamine was injected to block unreacted groups. For kinetic measurements, eltrombopag (97.7 nM to 1562.5 nM, or 97.7 nM to 6250 nM) was serially diluted twice at 25 °C, at a flow rate of approximately 30 μL/min in PBS (containing 0.05% Tween 20 and 3% dimethylsulfoxide). A simple one-to-one Langmuir combination model (BIAcore S200 Evaluation Software version 1.1) was used to calculate the association rate (kon) and dissociation rate (koff) by simultaneously fitting the association and dissociation sensorgrams. KD was calculated as the ratio of koff/kon.
2.4. Thermo shift assay
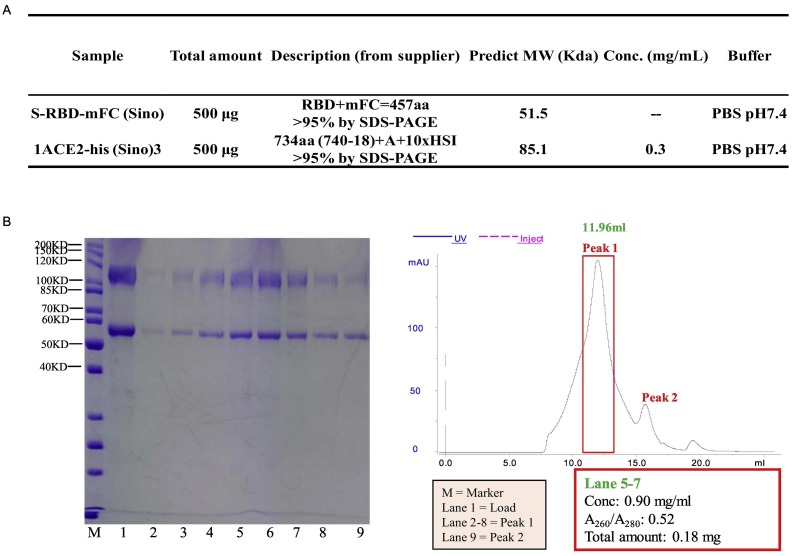
S-RBD (S protein receptor binding domain) and ACE2 protein complex was prepared by mixing S-RBD and ACE2 (Fig. S9-A) in a 1:1 ratio and incubating at 4 °C for 3 h. Then, the sample was concentrated to 0.2 mL for thermo shift assay (TSA) (Fig. S9-B). To test the Tm (melting temperature) of the RBD-ACE2 complex, 5 μL metal-ion working solution was added to 96-well RE-PCR plate (AXYGEN, USA), and then 10 μL RBD-ACE2 complex was added to each well and the mixture incubated at 4 °C for 1 h. Subsequently, 5 μL dye SYPRO orange working solution was added to each well. The plate was run in an ABI7500 fast real-time PCR system with a ramp rate of 1 °C per minute from 25 °C and ending at 99 °C. Next, different compounds were assessed to determine their effect on RBD-ACE2 protein stability, using the same procedure.
Fig. S9.
Experiment details of thermo shift assay (TSA). (A) Prepared S-RBD-mFB (sino) and ACE-2-His protein sample. (B) Concentrate S-RBD-ACE2 complex. S-RBD (mFc tag): 51.5 kD, ACE2: 85.1 kD. Collect lane 5– 7 as concentrated protein complex, based on SDS-PAGE result.
3. Results
3.1. Model of the SARS-CoV-2 structural proteins
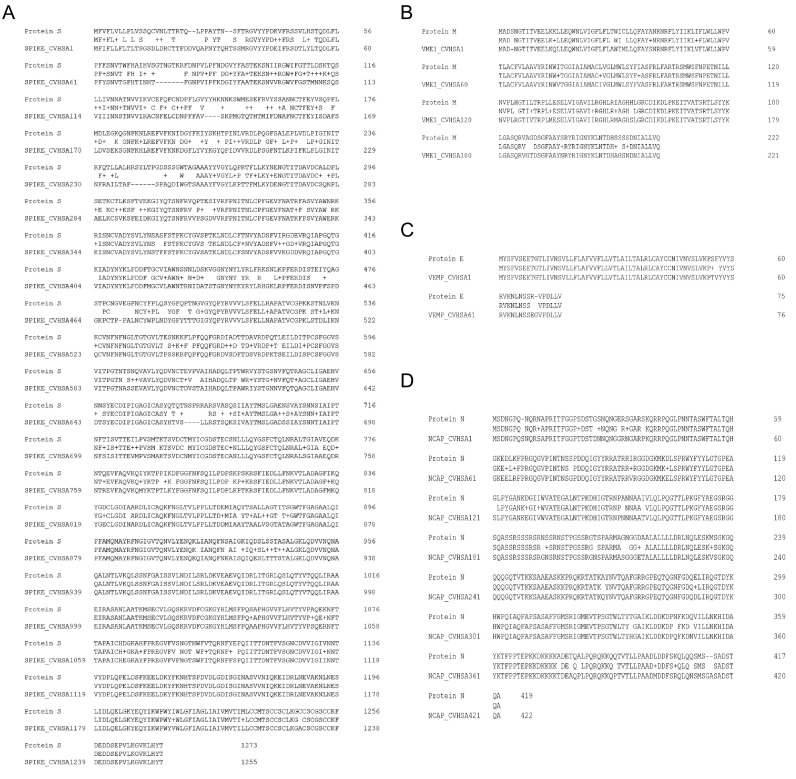
We compared the amino acid sequences of SARS-CoV-2 structural proteins, including the S protein, membrane protein (M), envelope protein (E), and nucleocapsid protein (N) with that of SARS-CoV. Although high similarity was observed, alterations in part of the amino acid sequences were also evident (Fig. S2).
Fig. S2.
Amino acid sequence differences between SARS-CoV-2 and SARS-CoV. Amino acid sequences of SARS-CoV-2 or SARS-CoV S protein (A), M protein (B), E protein, (C) and N protein (N) were downloaded and compared.
To screen effective drugs for COVID-19 therapy, it is important to elucidate the structures of proteins involved in the pathogenesis of the disease. By simulating the binding affinity between candidate drugs and protein, the drugs showing high binding affinity can be screened and further evaluated for efficacy. However, during “Public Health Emergency” such as a pandemic, structure-based drug screening of drugs is not effective in addressing the urgency for the search of effective drugs. By virtualizing the protein structure, it is possible to obtain a reliable structure that can be used to perform rapid and economic drug screening. Considering that the invasion of SARS-CoV-2 into host cells is mediated by S protein, a virtualized S protein was used for our drug screening.
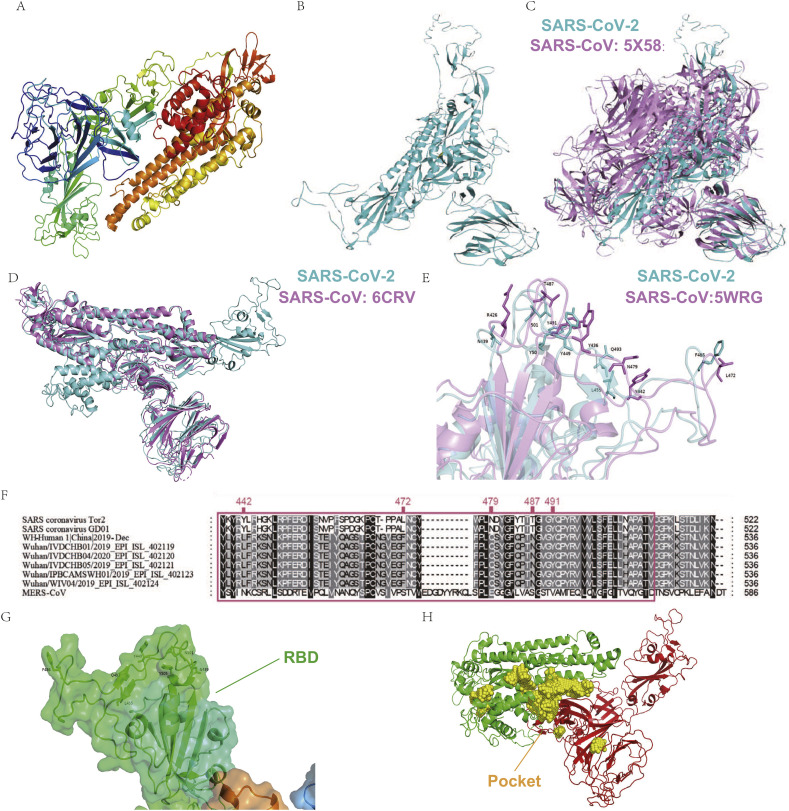
As recently reported by Zhang et al., the virtualized SARS-CoV-2 S protein tertiary structure was successfully established (Zhang et al., 2020). Since the structure of SARS-CoV-2 S protein was also reported recently (Wrapp et al., 2020), we compared the virtualized S protein from Zhang lab (QHD43416.pdb) (Zhang et al., 2020) with the reported S protein (6LZG.pdb) via TM-align by two different chains and obtained TM scores of 0.646 and 0.849, respectively; suggesting a high structural similarity between the two proteins (Fig. S3). The characteristic analysis and drug screening were completed by the S protein from Zhang lab. The structure of SARS-CoV-2 S protein was obtained by multi-template modeling (Fig. 1-A; template protein ID: 5 × 58, 5WRG, 6U7H, 6CRV, and 6LZG), or by ab initio model through I-TASSER (Fig. 1-B). The comparison between the ab initio model of SARS-CoV-2 and SARS-CoV S protein (5 × 58) showed high similarity (Fig. 1-C). In addition, to assess the accuracy of the virtualization, the structure of SARS-CoV-2 M, E, and N proteins were also generated via ab initio model through I-TASSER (Fig. S4) and assessed via Procheck (Fig. S5, Fig. S6).
Fig. S3.
TM score of modeling and reported S protein structure of SARS-CoV-2. The structure of spike model (QHD43416.pdb) was compared to the reported S protein (6LZG.pdb) via TM-align. Two different chains, chain 1 (A316309) and chain 2 (B316309), were used, respectively.
Fig. 1.
The tertiary structure of SARS-CoV-2 Spike (S) protein and the pocket for drug screening.
The tertiary structure of SARS-CoV-2 S protein was acquired via homology modeling (A) or ab initio model (B, cyan). (C) Comparison of SARS-CoV-2 S protein (cyan) and SARS-CoV S protein 5 × 58 (mauve). (D) Identification of SARS-CoV-2 S protein RBD (cyan) by comparing to SARS-CoV S protein 6CRV (mauve). (E) Comparison of SARS-CoV-2 S protein RBD (cyan) with SARS-CoV 5WRG RBD (mauve). (F) Comparison of RBD amino acid sequences between different strains of SARS-CoV and SARS-CoV-2. (G) The RBD of S protein (green area) show insufficient stability for future drug screening. (H) The selected binding pocket (yellow area) for drug screening,
Fig. S4.
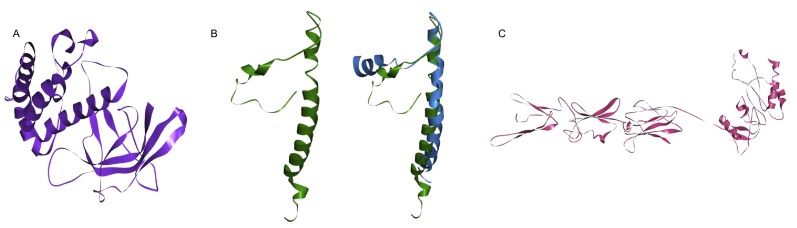
Tertiary structures of SARS-CoV-2 and SARS-COV M, E, N protein. M, E, N protein structure of SARS-CoV-2 were acquired through ab initio modeling. (A) M protein. (B) E protein (Green: SARS-CoV-2, Blue: SARS-CoV 2MM4). (C) N protein.
Fig. S5.
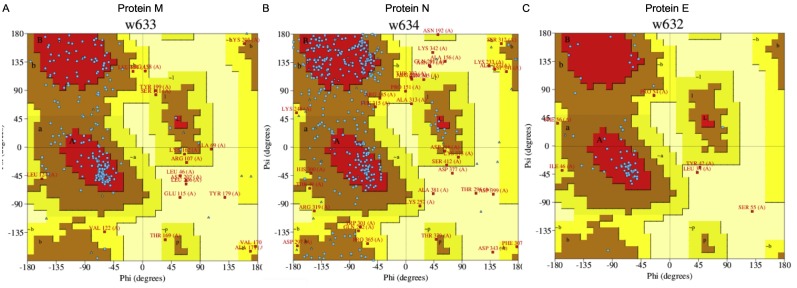
Ramachandran plot of SARS-CoV-2 proteins. The protein structure M protein (A), N protein (B), and E protein (C) were assessed by Procheck and depicted using a Ramachandran plot.
Fig. S6.
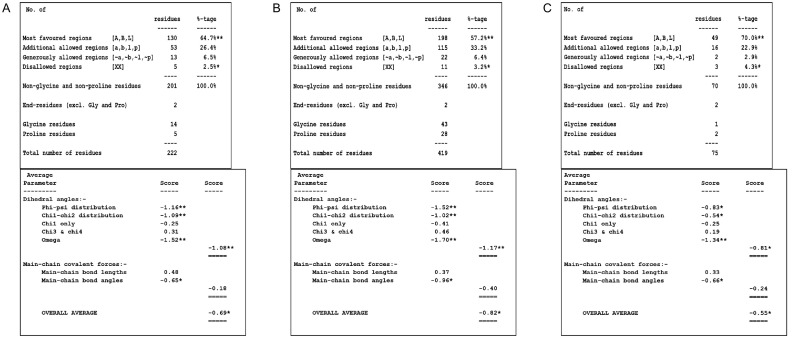
Ramachandran plot statistics. The Ramachandran plot statistics of M protein (A), N protein (B), and E protein (C) were listed. The percentage or residues in the most favored regions indicated the quality of model (>90% is thought to possess great quality), while the value of G factors provides information on unusual property (<−0.5 represents unusual, and <−1.0 represents high unusual).
The homology model of SARS-CoV-2 S protein structure was further compared with the full-length S protein structure (6CRV) of SARS-CoV to identify the receptor-binding domain (RBD) (Fig. 1-D), the domain responsible for binding to the receptor. Comparison of amino acid sequence and RBD structure showed that SARS-CoV-2 RBD had a highly similar structure to SARS-CoV RBD (Fig. 1-E), despite alterations in four of the five key amino acids: Y442L, L472F, N479Q, T487N, Y491 (Fig. 1-E, F).
3.2. Design of the pocket for drug screening
S protein RBD of both SARS-CoV-2 and SARS-CoV binds to ACE2 to mediate further viral entrance and is therefore an important target for drug screening (Gui et al., 2017; Walls et al., 2020). However, the RBD of SARS-CoV-2 S protein is not suitable for determining the protein-binding pocket to screen drugs (Fig. 1-G). The RBD of the SARS-CoV-2 S protein was mainly composed of loops, with only a small portion of β-sheets, and this kind of structure is not stable enough to allow the binding of small molecular drugs. After calculation, the pocket with enough stability for future protein-drug interaction was selected for drug screening (Fig. 1-H).
The selected pocket comprised multiple parts of S protein, including: S1: Val 42-Ser50, Glu 281-Val289, Lys300-Ile312, Gly601-Ser605; S2: Pro812-Glu868, Ser937-Val976, and Asn1192-Ala1222. Most of the binding sites were identified in the S2 domain. Notably, the two binding sites Ser937-Val976 and Asn1192-Ala1222 showed high overlaps with the HR1 and HR2 fusion core of S2 domain, respectively (Xia et al., 2020), thus suggesting that the selected pocket possessed a high probability to affect the S2 domain-mediated fusion of SARS-CoV-2.
3.3. In silico screening identified 13 FDA-approved drugs as potential binders
High-throughput virtual screening is widely used in the process of drug discovery. Since ACE2 works as the receptor of S protein, a series of drugs or small-molecule peptides could be designed to prevent the virus from fusing further with the membrane of the target host cell. Total 1234 small molecules in the FDA-approved drug library were docked into the selected binding pocket of S2 domain. The top 100 computationally scored molecules were identified. Finally, 13 potentially therapeutic compounds were selected through manual screening of relevant mechanisms of action, drug toxicity, and binding power (range of −9.3 to −12.3 kcal/mol).
The in silico studies showed that all the selected 13 FDA drugs had relatively good interaction with the S2 subunits and were ranked according to their binding affinities (Table 1). The ligand-amino acid interactions are summarized in Table 2 and all structure and drug-protein interactions are shown in Fig. 2 and Fig. S7, respectively.
Table 1.
Top 13 drugs screened after docking.
| Compounds | Affinity (kcal/mol) | Drug Bank Id | PubChem CID | FDA Approved Application | Targets | References |
|---|---|---|---|---|---|---|
| Dactinomycin | −12.3 | DB00970 | 457,193 | Antitumor drug | Inhibition of RNA and protein synthesis to induce p53-independent apoptosis | (Koba and Konopa, 2005, Karpinski and Adamczak, 2018) |
| Bictegravir | −10.4 | DB11799 | 129,626,368 | Anti-AIDS drug | Prevention of HIV integrase (IN)-mediated replication | (Sax et al., 2017, Gallant et al., 2017) |
| Temsirolimus | −10.3 | DB06287 | 134,812,825 | Antitumor drug | Inhibition of mTORC1 | (Pópulo et al., 2012, Yang et al., 2013) |
| Dolutegravir | −9.8 | DB08930 | 54,726,191 | Anti-AIDS drug | Impairing the function of the HIV integrase-DNA complex | (Zamek-Gliszczynski et al., 2019) |
| Elbasvir | −9.7 | DB11574 | 71,661,251 | Anti-HCV drug | Inhibiting non-structural protein 5A (NS5A) to suppress RNA replication and assembly of HCV | (Bell et al., 2016) |
| Irbesartan | −9.6 | DB01029 | 3749 | Hypertension drug | ARB,inhibition of the classical RAS pathway, upregulation of ACE2 | (Lewis et al., 2001, Yisireyili et al., 2018) |
| Gliquidone | −9.6 | DB01251 | 91,610 | Diabetes drug | Blocking ATP-dependent K+ channels and promoting the release of insulin | (Bonora et al., 1992, Ocana et al., 1993, Bachmann et al., 1979) |
| Lanreotide | −9.6 | DB06791 | 71,349 | Anti-acromegaly drugs | Inhibiting the somatostatin receptors (SSTs), activating MAPK pathway and regulating nitric oxide | (Giustina et al., 2014, Narayanan and Kunz, 2016) |
| Glycyrrhizic acid | −9.6 | DB13751 | 14,982 | Hepatoprotective drug | Regulation of the immune system and inhibition of the translocation of nuclear factor-kappa B and viral replication | (Ming and Yin, 2013, Li et al., 2014, Pastorino et al., 2018) |
| Tasosartan | −9.5 | DB01349 | 60,919 | Antihypertensive drug | Long-acting ARB and interacting with both AT1 and AT2 receptors | (Dina and Jafari, 2000) |
| Velpatasvir | −9.5 | DB11613 | 67,683,363 | Anti-HCV drug | Similar to Elbasvir | (Targett-Adams et al., 2011, Miller, 2017) |
| Eltrombopag | −9.4 | DB06210 | 135,449,332 | Treatment of ITP | Non-peptide thrombopoietin (TPO) receptor agonist | (Erickson-Miller et al., 2009, Kim et al., 2018) |
| Azilsartan medoxomil | −9.3 | DB08822 | 25,210,270 | Hypertension drug | ARB,blocking the AT1 receptor without affecting the AT2 receptor | (Hjermitslev et al., 2017) |
The name, Drug Bank ID, PubChem CID, FDA approved application and Targets are listed. Abbreviations: mTOR1: mammalian target of rapamycin complex 1; RAS: renin-angiotensin system; MAPK: mitogen-activated protein kinase; ARB: angiotensin receptor blocker; AT1/2: angiotensin type 1/2 receptor.
Table 2.
Summary of ligand-protein interaction sites in the pocket.
| Drug Name | Hydrogen Bonds | Hydrophobic Interactions | Carbon-hydrogen interactions | Other interactions |
|---|---|---|---|---|
| Dactinomycin | TYR1209,ASN953 | PRO123, LYS278, LEU48, TRP1217, ILE1210 | SER46, ASN280 | ILE1210 (π-sigma) |
| Glycyrrhizic Acid | LEU303, ASN824, THR827, GLN949, LEU1205, TYR1209, TRP1217 | ALA956, LYS1205 | VAL826 | TRP-1217 (π-donor) |
| Eltrombopag | VAL826, ALA829, LYS1205, TYR1209 | LEU849, LEU1203, ILE1210, PRO1213 | ||
| Azilsartan medoxomil | THR827, GLN949, TRP1217 | LEU945, ILE1210, PRO1213 | ASN824, TRY1209 | |
| Bictegravir | ASN824, LYS825, LYS1205, TYR1209,TRP1217 | LEU849, LEU945, LEU1203, TRP1212, PRO1213 | THR827 | ASN1199 (π-anion) |
| Temsirolimus | PHE823, ASN824, LYS825, THR827, ASN 953, TYR 1209, TRP1217 | ALA956, ILE1210, PRO1213, TRP1217, TYR 1209 | VAL826, THR307 | |
| Dolutegravir | ASN824, LYS825, LYS1205, TYR1209, TRP1217 | LEU849, LEU945, LEU1203, PRO1213 | THR827 | ASN1199 (π-anion) |
| Elbasvir | ASN824, GLN949, ASN953, GLN957 | LEU303, TYR313, LEU828, LEU849, TRP1212, PRO1213, TRP1217 | THR827 | TYR1209 (π-donor) LYS825 (amide-π) |
| Irbesartan | THR827, LYS1205, TYR1209, TRP1217 | LYS1205, ILE1210, PRO1213 | ||
| Gliquidone | LYS1205, TYR1209, TRP1217 | LEU849, LEU945, TRP1212, PRO1213 | LEU828, TYR1209 | TYR1209 (π‑sulfur) |
| Lanreotide | ASN960, LYS964, ASP1199, LYS1205, TYR1209 | LEU303, LYS304, LEU828, LEU1203 | LYS1205 | ALA952, ALA956 (π-sigma) GLU309 (π-anion) PHE833 (π-π) |
| Tasosartan | GLU1195, TYR1209 | LYS825, TYR1209, PRO1213, TRP1217 | ASP1199 (π-anion) LYS1205 (π-cation) |
|
| Velpatasvir | ASN824, VAL826, GLN949, ASN960, LYS1205, TYR1209 | LEU48, LYS304, LEU945, ILE1210 | LEU303 | LYS964 (π-cation) |
The amino acids interacting with each drug and the interaction types are summarized.
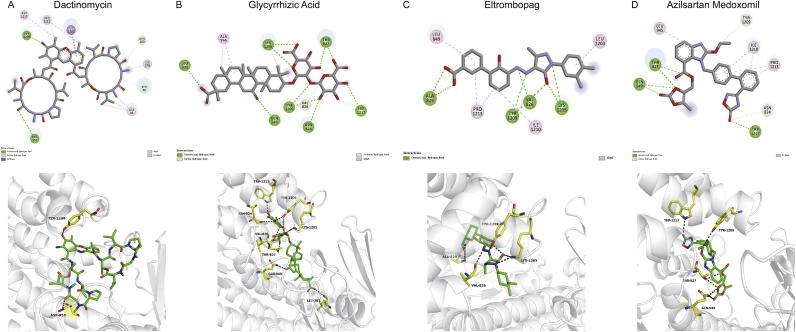
Fig. 2.
Interaction sites between drugs and SARS-CoV-2 S protein.
The interaction between different drugs and SARS-CoV-2 S protein were virtualized to calculate binding affinity. The structure of each drug and the corresponding amino acids interacting with each drug are shown: (A) Dactinomycin (DB00970, PubChem CID: 457193); (B) Glycyrrhizic acid (DB13751, PubChem CID: 14982); (C) Eltrombopag (DB06210, PubChem CID: 135449332); (D) Azilsartan medoxomil (DB08822, PubChem CID: 25210270).
Fig. S7.
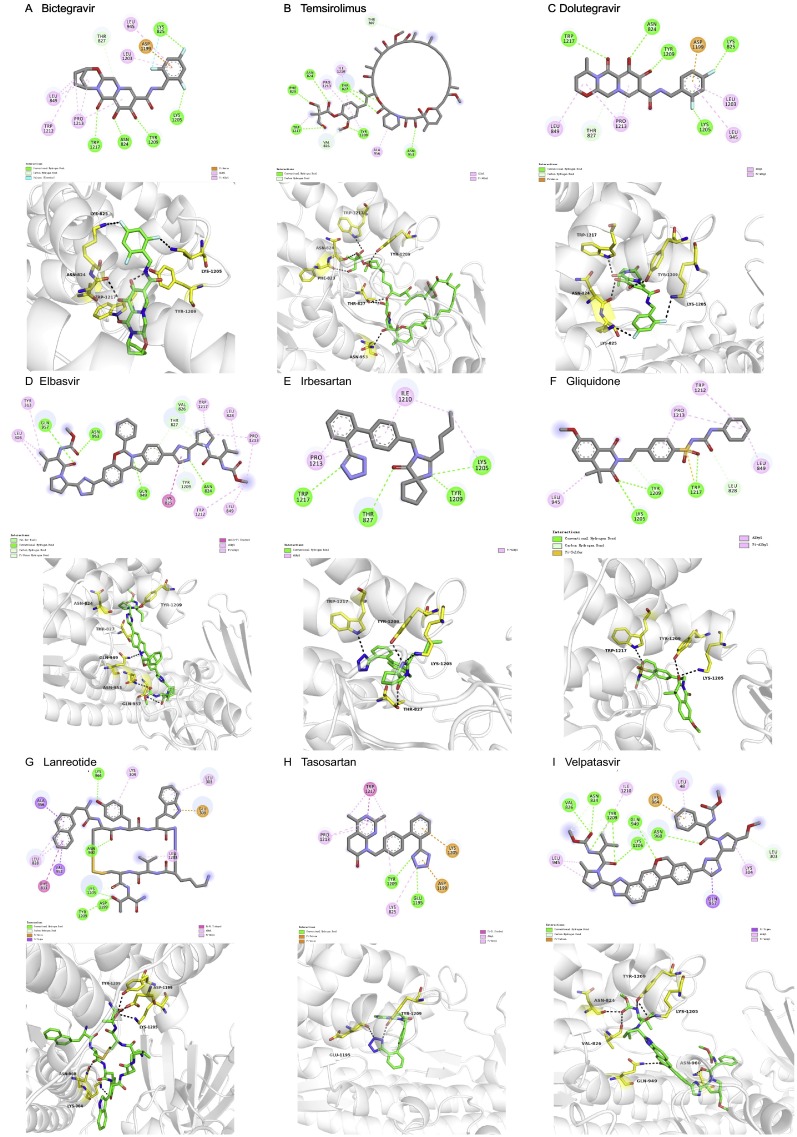
Interaction models of different drugs and SARS-CoV-2 S protein. The interaction between different drugs and SARS-CoV-2 S protein were virtualized to calculate binding affinity. The structure and interacting spots of each drug are shown. (A) Bictegravir (DB11799, PubChem CID: 129626368); (B) Temsirolimus (DB06287, PubChem CID: 134812825); (C) Dolutegravi (DB08930, PubChem CID: 54726191); (D) Elbasvir (DB11574, PubChem CID: 71661251); (E) Irbesartan (DB01029, PubChem CID: 3749); (F) Gliquidone (DB01251, PubChem CID: 91610); (G) Lanreotide (DB06791, PubChem CID: 71349); (H) Tasosartan (DB01349, PubChem CID: 60919); (I) Velpatasvir (DB11613, PubChem CID: 67683363).
The screening results (Table 1) showed that a series of anti-tumor drugs (dactinomycin, temsirolimus),anti-AIDS drugs (bictegravir, dolutegravir, velpatasvir), anti-HCV drugs (elbasvir, velpatasvir), hypertension drugs (irbesartan, tasosartan, azilsartan medoxomil), anti-diabetic drugs (gliquidone), anti-acromegaly drug (lanreotide), hepatoprotective drug (glycyrrhizic acid), and a drug for thrombocytopenia (eltrombopag) may have a high binding affinity to S protein.
Dactinomycin (Fig. 2-A, Table 1, Table 2), had the highest docking score of −12.3 kcal/mol with the S protein. Dactinomycin formed hydrogen bonds with TYR1209 and ASN953 residues of the viral proteins. π-sigma interactions with ILE1210; hydrophobic interactions with PRO1213, LYS278, LEU48, TRP1217, and ILE1210; and hydrocarbon interactions with SER46 and ASN280 were evident (Fig. 3 ). Dactinomycin belongs to the actinomycin group of antibiotics extracted from Actinomyces antibioticus and is widely used as an antitumor drug and functions by inhibiting RNA and protein synthesis to induce p53-independent apoptosis. The possible mechanisms of action include interaction with DNA, stabilization of cleavable complexes of topoisomerase I and II, and promotion of free radical formation (Koba and Konopa, 2005). Dactinomycin is mostly used to treat Wilms tumor, Ewing sarcoma, neuroblastomas, and trophoblastic tumors (Karpinski and Adamczak, 2018).
Fig. 3.
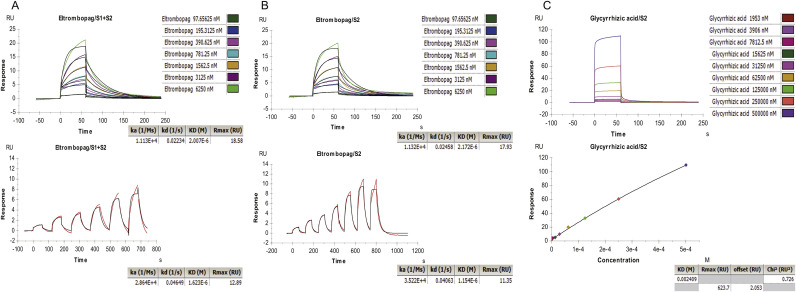
SPR analysis of interactions of different drugs and SARS-CoV-2 S protein.
(A, B) Representative SPR data for various concentrations of eltrombopag binding to S1 + S2 domain or S2 domain at pH 5.5. (C) Representative SPR data for various concentrations of glycyrrhizic acid binding to S2 domain at pH 5 0.5. RU, resonance units.
Glycyrrhizic acid (Fig. 2-B, Table 1, Table 2), had a docking score of −9.6 kcal/mol with viral proteins. Glycyrrhizic acid formed hydrogen bonds with LEU303, ASN824, THR827, GLN949, LEU1205, TYR1209, and TRP1217. It showed carbon‑hydrogen interactions with VAL826, π-donor interactions with TRP1217, and hydrophobic interactions with ALA956 and LYS1205. It is extracted from the root of the licorice plant, Glycyrrhizia glabra, a member of the Fabaceae family. Different extracts from this family have a wide range of pharmacological activities, including antibacterial, anti-inflammatory, antiviral, antioxidant, and antidiabetic activities (Pastorino et al., 2018). The mechanism of action of glycyrrhizic acid is complex. The compound exists in α and β forms. The α form is predominantly distributed in the liver and displays anti-inflammatory activity by inhibiting the translocation of nuclear factor-kappa B, suppressing the function of tumor necrosis factor-alpha, and inhibiting CD4+ T cell proliferation via phosphoinositide 3-kinase/Akt (Ming and Yin, 2013). Glycyrrhizic acid is widely used in China and Japan as a hepatoprotectant (Li et al., 2014). Furthermore, glycyrrhizic acid displays broad-spectrum antiviral activity against viruses, including vaccinia virus, herpes simplex virus, vesicular stomatitis virus, and hepatitis B virus (Ming and Yin, 2013; Sun et al., 2019). The possible mechanisms of action are the inhibition of viral replication and regulation of the immune system.
Eltrombopag (Fig. 2-C, Table 1, Table 2) was found to have a docking score of −9.4 kcal/mol with viral proteins. DB06210 formed hydrogen bonds with VAL826, ALA829, LYS1205, and TYR1209 and had hydrophobic interactions with LEU849, LEU1203, ILE1210, and PRO1213. Eltrombopag is a small-molecule, non-peptide thrombopoietin (TPO) receptor agonist and is used to upregulate platelet counts in idiopathic thrombocytopenia patients. Eltrombopag binds to the juxtamembrane domain of the TPO receptor and activates downstream Janus kinase/signal transducer and activator of transcription, AKT, and MAPK pathways (Kim et al., 2018). The signal cascade promotes the proliferation and differentiation of megakaryocytes to increase the production of platelets (Kim et al., 2018). Notably, eltrombopag interacts with the TPO receptor at a distinct site from endogenous TPO (Erickson-Miller et al., 2009).
Azilsartan medoxomil (Fig. 2-D, Table 1, Table 2) displayed a docking score of −9.3 kcal/mol with viral proteins. Azilsartan medoxomil formed hydrogen bonds with THR827, GLN949, and TRP1217. It had carbon‑hydrogen interactions with ASN824 and TYR1209, and had hydrophobic interactions with LEU945, ILE1210, and PRO1213. Azilsartan medoxomil is an angiotensin receptor blocker (ARB) used for the treatment of hypertension. Azilsartan medoxomil is a prodrug and is further hydrolyzed in the intestine to the active component azilsartan (Hjermitslev et al., 2017). Azilsartan blocks the AT1 receptor without affecting the AT2 receptor, thus mediating vasodilatation, reducing aldosterone release, and reducing sympathetic stimulus of blood vessels and the kidney to lower blood pressure (Hjermitslev et al., 2017).
Similarly, the other 9 drugs also showed high affinity with the designed S2 pocket. The information for the drugs is briefly summarized in Table 1 and Table 2, and the interaction model was shown in Fig. S7.
3.4. Verification of the protein-drug interaction via surface plasmon resonance and thermo shift assay
Immediately after the simulation, we used SPR to verify ten potential compounds. Considering that the pocket used for screening comprised mainly the S2 domain and partially the S1 domain, the chips were prepared via different S protein domains (S1 + S2, S2) of SARS-CoV-2. Based on the binding force and non-specific binding, we found that eltrombopag may be a potentially useful drug. With SPR, we showed that at pH 5.5, eltrombopag can not only bind efficiently to S2 domain as expected (KD = 2.172 × 10−6 M), but also bind to S1 + S2 domain (KD = 2.007 × 10−6 M) (Fig. 3-A, B, Table 3 ), without much difference. Meanwhile, the Rmax in both assays are lower than 20RU (resonance units), indicating the specificity of the interactions tested. This result confirmed that the S2 domain works as the major drug-binding site, and the existence of S1 domain does not disturb the interaction between eltrombopag and the pocket, thus suggesting the potential of eltrombopag binding to the intact SARS-CoV-2 S protein.
Table 3.
The binding affinity between 10 drugs and SARS-CoV-2 spike protein S2/(S1 + S2) domain tested through SPR.
| Sample Name | M.W (Da) | KD (M) | Rmax (RU) |
|---|---|---|---|
| Eltrombopag (S1 + S2) | 442.47 | 2.007E-6 | 18.58 |
| Eltrombopag | 442.47 | 2.172E-6 | 17.93 |
| Bictegravir | 449.38 | ND | ND |
| Dolutegravir | 419.38 | ND | ND |
| Temsirolimus | 1030.29 | ND | ND |
| Elbasvir | 882.02 | ND | ND |
| Irbesartan | 428.54 | ND | ND |
| Gliquidone | 527.63 | ND | ND |
| Glycyrrhizic acid | 822.93 | ND | ND |
| Velpatasvir | 883.02 | ND | ND |
| Azilsartan Medoxomil | 568.53 | ND | ND |
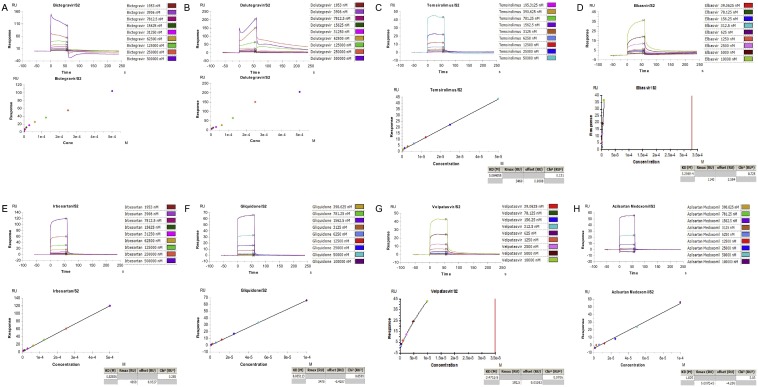
In contrast, glycyrrhizic acid, the herb reported to possess wide-spectrum anti-viral activity which showed high binding affinity in the virtual screening, did not provide good results in the SPR assay. The calculated KD is 0.0024 M, with the Rmax of 623.7RU, showing a very weak and unspecific binding (Fig. 3-C) and is therefore marked as ND (non-detectable) in Table 3, which suggests a poor interaction between glycyrrhizic acid and S2 domain. In addition, the SPR assay results of other drugs also showed high KD and Rmax value, suggesting similar poor interactions (Fig. S8).
Fig. S8.
SPR analysis of interactions of other drugs and SARS-CoV-2 S protein. SPR data for various concentrations of different drugs: (A) Bictegravir, (B) Dolutegravir, (C) Temsirolimus, (D) Elbasvir, (E) Irbesartan, (F) Gliquidone, (G) Velpatasvir, (H) Azilsartan Medoxomil binding to SARS-CoV-2 S2 domain at pH 5.5.
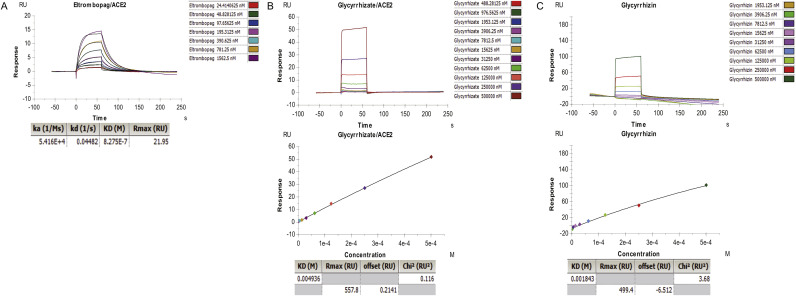
Taking into account that ACE2 functions as an indispensable receptor of viral S protein, an assay was performed to further test the binding affinity between drugs and ACE2. Again, eltrombopag showed an excellent binding affinity to human ACE2 extracellular domain with a KD = 8.275 × 10−7 M and Rmax = 21.95 RU (resonance units, RU) (Fig. 4 -A). In contrast, the high KD (0.0049 M and 0.0018 M)and Rmax values (557.8 RU and 499.4 RU) of glycyrrhizate and glycyrrhizin showed poor binding affinity to human ACE2 (Fig. 4-B, C). These results showed the potential of eltrombopag binding to both SARS-CoV-2 S protein and human ACE2 and reveals the possibility of inhibition of viral entrance.
Fig. 4.
SPR analysis of interactions of different drugs and human ACE2.
(A) Representative SPR data for various concentrations of eltrombopag binding to human ACE2 at pH 4.5. (B, C) Representative SPR data for various concentrations of glycyrrhizate, glycyrrhizin binding to human ACE2 at pH 4.5.
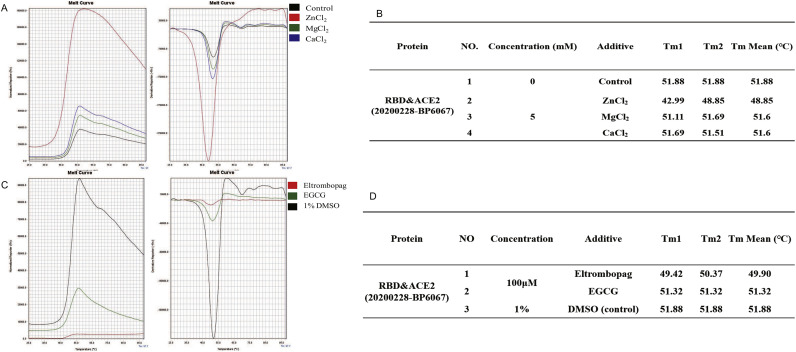
The pharmacological effect of eltrombopag on S-RBD-ACE2 protein complex was further verified by TSA. Results showed that metal ion solutions (MgCl2 and CaCl2) had no effect on the Tm of the RBD-ACE2 complex (Fig. 5 -A, B, 51.88 °C versus 51.6 °C). Notably, protein precipitation was observed after the addition of ZnCl2. Hence, the single curve of ZnCl2 was unreliable. The addition of compound eltrombopag had a slight effect on the RBD-ACE2 protein complex, which reduced the Tm from 51.88 °C to 49.9 °C (Fig. 5-C, D). These results confirmed that eltrombopag lowered the stability of S-RBD-ACE2 protein complex.
Fig. 5.
Eltrombopag affects the stability of RBD-ACE2 complex.
(A, B) The Tm of RBD-ACE2 complex with different metal ions (Black: Control; Red: 5 mM ZnCl2; Green: 5 mM MgCl2; 5 mM CaCl2) as control were tested by TSA. (C, D) The Tm of RBD-ACE2 complex with different compounds treatment (Red: 100 μM eltrombopag; Green: 100 μM EGCG (epigallocatechin gallate); Black: 1% DMSO) were tested by TSA. A decreased Tm value can be observed after eltrombopag treatment.
4. Discussion
Under the premise that the structure is not completely resolved, homology modeling and de novo modeling are considered to be reliable and rapid methods for drug screening. We combined sequence analysis with homology modeling to quickly obtain the SARS-CoV-2 S protein. The sequence analysis and homology modeling results showed that the S protein of SARS-CoV and SARS-CoV-2 share high similarity. Importantly, consistent with previous report, 4 of the 5 key amino acids of SARS-CoV-2 RBD changed when compared to SARS-CoV; yet the structure was completely preserved and showed similar characteristics with SARS-CoV (Wan et al., 2020). Moreover, the modeling S protein structure also matched well to the recently published SARS-CoV-2 S protein tertiary structure, according to the TM score, thus validating the accuracy of our homology modeling (Wrapp et al., 2020).The structure of this protein and the structures of other related structural proteins were ascertained through virtual drug design, rapid screening, and manual screening of FDA-approved drugs.
Both SARS-CoV and SARS-CoV-2 recognize human ACE2 via the RBD of S protein, which makes the domain a hot target for drug screening. Nevertheless, our virtual structure indicated that the RBD of SARS-CoV-2 is mainly composed of loops and is not stable enough for drug screening. As a replacement, we selected a stable pocket whose binding sites mainly localize in the S2 domain, more specifically, the HR1 and HR2 fusion core. Therefore, it could be inferred that drugs screened through this pocket may have a high probability to affect the S2 domain-mediated membrane fusion.
Based on the pocket selected, the 13 molecules identified in this study may be potential therapeutic drugs. These include a series of drugs that are used to treat viral infection, tumors, diabetes, hypertension, and other diseases.
We also discovered a Chinese herbal medicine glycyrrhizic acid that had a combination with S protein. Glycyrrhizic acid has been reported to be efficacious for the treatment of SARS infections (Hoever et al., 2005).
Notably, multiple ACEI/ARB drugs were screened in this study, showing their potential interaction with the pocket of SARS-CoV-2 protein. However, these drugs should be used with caution in COVID-19 patients. Much attention has been paid to ACEI/ARB drugs for their regulation of the level of ACE2, the specific receptor of SARS-CoV-2. However, the exact role of ACEI/ARB drugs in SARS-CoV-2 infection is not clear yet. ACEI/ARB drugs can indirectly upregulate ACE2 by blocking ARS, which is believed to be a risk factor. However, ACE2 possesses anti-inflammatory effect and many other protective functions, which means that upregulated ACE2 can protect humans from damages caused by SARS-CoV-2. Furthermore, it has been reported, that for patients suffering from both hypertension diseases and COVID-19, ACEI may protect them from SARS-CoV-2 infection (Meng et al., 2020). In summary, risky or protective, there is still much work to be done to figure out the relationship between ACEI/ARB drugs and COVID-19.
Eventually, 10 compounds were selected to undergo SPR test. The SPR results confirmed that eltrombopag possesses good binding affinity to both SARS-CoV-2 S protein and human ACE2, indicating its potential to affect both sides of viral entrance. Considering the domain participating in the formation of the pocket, eltrombopag could affect the S2 domain-mediated membrane fusion, thus preventing the viral entrance. This requires further verification in the future.
However, how eltrombopag functions through ACE2 remains to be explored. TSA verified that eltrombopag could lower RBD-ACE2 complex stability. This result showed that eltrombopag acts by binding to the S-RBD-ACE2 complex and lowering the protein complex stability. Nevertheless, it is still hard to draw a conclusion, since eltrombopag is mainly used for increasing the platelet count but not for blood pressure regulation. Although increased blood pressure presents as an adverse event in some patients, the mechanism is not clear yet (Ptushkin et al., 2018). It is worth exploring how eltrombopag functions after binding to ACE2, which can help us further understand the potential anti-SARS-CoV-2 mechanism of eltrombopag and its adverse effect of increasing the blood pressure.
Although the results from this virtual drug screening provided many drug options, there are few shortcomings in the study. Cell and animal experimental studies still need to be carried out for these drug candidates. Additionally, the accuracy of homology modeling is limited, compared to the structure analyzed by Cryo-EM, since the precision is affected by both parameters and experience of operators. For example, discrepancy between virtual screening and SPR test is observed, which further confirms that the screened drug requires more verification experiments to illustrate its characteristics. We used SPR assay in this research to show the binding affinity, and together with the virtual screening, we showed the enormous potential of eltrombopag as an anti SARS-CoV-2 treatment. The discrepancy between virtual screening and SPR test, such as in the case of glycyrrhizic acid, could have been caused by unspecific binding, which suggests the need for further verification experiments in the future.
This study utilized homology modeling to analyze the SARS-CoV-2 S protein structure; besides, virtual drug screening was also performed. Moreover, the study describes the high possibility of eltrombopag as a candidate for anti SARS-CoV-2 therapy. These findings will help guide future efforts to screen for other SARS-CoV-2 structural proteins and potential therapeutic compounds. In addition, this method to screen drugs based on virtualized protein structure is also a reasonable, economic, and fast method to screen drugs and reveal out-of-guide application of approved drugs, which can be used against many other diseases and in future possible epidemic or pandemic situations.
The following are the supplementary data related to this article.
Workflow chart of the structure-based virtual screening and subsequent high throughput screening methodology. The amino acid sequence of SARS-CoV-2 was downloaded from NCBI and analyzed. The structure of S, M, E, N protein was acquired by ab initio modeling or homology modeling (S protein), and the S protein docking pocket for drug screening was analyzed by comparing with SARS-CoV. By scoring the 1234 compounds from the FDA drug database compound library via the drug screening pocket, the top 100 compounds were selected. Among these, 13 compounds were further selected through visual inspection, which underwent surface plasmon resonance test and Thermo Shift Assay to verify the efficacy.
Information about the protein chips used in SPR test.
Funding
This work was supported by CAMS Innovation Fund for Medical Sciences (CIFMS) [No. 2016-I2M-3-011, 2016-I2M-1-002]; CAMS Innovation Fund for Medical Sciences No. (2020-I2M-CoV19-001); “13th Five-Year” National Science and Technology Major Project for New Drugs [No: 2019ZX09734001-002]; Tsinghua University-Peking University Center for Life Sciences [No. 045-160321001).
Declaration of Competing Interest
None.
Acknowledgments
The authors thank Beijing Beike Deyuan Bio-Pharm Technology Co. Ltd. and Beijing Novogene Co. Ltd. We would like to thank Editage (www.editage.cn) for English language editing.
References
- Bachmann W., et al. Extra-pancreatic action of sulfonylureas - effect of gliquidone on insulin and glucagon binding to rat-liver plasma-membranes. Eur. J. Clin. Investig. 1979;9(6):411–415. doi: 10.1111/j.1365-2362.1979.tb00905.x. [DOI] [PubMed] [Google Scholar]
- Bell A.M., et al. Elbasvir/Grazoprevir: a review of the latest agent in the fight against hepatitis C. Int. J. Hepatol. 2016 doi: 10.1155/2016/3852126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoch I.I., et al. Potential for global spread of a novel coronavirus from China. J. Travel Med. 2020 doi: 10.1093/jtm/taaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonora E., et al. Studies on the mechanism of action of sulfonylureas in type-ii diabetic subjects - gliquidone. J. Endocrinol. Investig. 1992;15(1):1–11. doi: 10.1007/BF03348644. [DOI] [PubMed] [Google Scholar]
- Chan J.F.-W., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet (London, England) 2020 doi: 10.1016/s0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet (London, England) 2020 doi: 10.1016/s0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina R., Jafari M. Angiotensin II-receptor antagonists: an overview. Am. J. Health Syst. Pharm. 2000;57(13):1231–1241. doi: 10.1093/ajhp/57.13.1231. [DOI] [PubMed] [Google Scholar]
- Erickson-Miller C.L., et al. Preclinical activity of Eltrombopag (SB-497115), an Oral, nonpeptide Thrombopoietin receptor agonist. Stem Cells. 2009;27(2):424–430. doi: 10.1634/stemcells.2008-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J., et al. Bictegravir, emtricitabine, and tenofovir alafenamide versus dolutegravir, abacavir, and lamivudine for initial treatment of HIV-1 infection (GS-US-380-1489): a double-blind, multicentre, phase 3, randomised controlled non-inferiority trial. Lancet. 2017;390(10107):2063–2072. doi: 10.1016/s0140-6736(17)32299-7. [DOI] [PubMed] [Google Scholar]
- Giustina A., et al. Investigational drugs targeting somatostatin receptors for treatment of acromegaly and neuroendocrine tumors. Expert Opin. Investig. Drugs. 2014;23(12):1619–1635. doi: 10.1517/13543784.2014.942728. [DOI] [PubMed] [Google Scholar]
- Gui M., et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27(1):119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir. Res. 2013;100(1):286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjermitslev M., et al. Azilsartan Medoxomil, an angiotensin II receptor antagonist for the treatment of hypertension. Basic Clin. Pharmacol. Toxicol. 2017;121(4):225–233. doi: 10.1111/bcpt.12800. [DOI] [PubMed] [Google Scholar]
- Hoever G., et al. Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus. J. Med. Chem. 2005;48(4):1256–1259. doi: 10.1021/jm0493008. [DOI] [PubMed] [Google Scholar]
- Holshue M.L., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski T.M., Adamczak A. Anticancer activity of bacterial proteins and peptides. Pharmaceutics. 2018;10(2) doi: 10.3390/pharmaceutics10020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.O., et al. Eltrombopag for use in children with immune thrombocytopenia. Blood Adv. 2018;2(4):454–461. doi: 10.1182/bloodadvances.2017010660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koba M., Konopa J. Actinomycin D and its mechanisms of action. Postepy higieny i medycyny doswiadczalnej (Online) 2005;59:290–298. [PubMed] [Google Scholar]
- Lewis E.J., et al. Renoprotective effect of the angiotensin-receptor antagonist irbesartan in patients with nephropathy due to type 2 diabetes. N. Engl. J. Med. 2001;345(12):851–860. doi: 10.1056/NEJMoa011303. [DOI] [PubMed] [Google Scholar]
- Li J.-y., et al. Glycyrrhizic acid in the treatment of liver diseases: literature review. Biomed. Res. Int. 2014 doi: 10.1155/2014/872139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J., et al. Renin-angiotensin system inhibitors improve the clinical outcomes of COVID-19 patients with hypertension. Emerg. Microb. Inf. 2020;9(1):757–760. doi: 10.1080/22221751.2020.1746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.M. Sofosbuvir-velpatasvir: a single-tablet treatment for hepatitis C infection of all genotypes. Am. J. Health Syst. Pharm. 2017;74(14):1045–1052. doi: 10.2146/ajhp60632. [DOI] [PubMed] [Google Scholar]
- Ming L.J., Yin A.C.Y. Therapeutic effects of Glycyrrhizic acid. Nat. Prod. Commun. 2013;8(3):415–418. [PubMed] [Google Scholar]
- Narayanan S., Kunz P.L. Role of Somatostatin analogues in the treatment of neuroendocrine Tumors. Hematol. Oncol. Clin. North Am. 2016;30(1):163–177. doi: 10.1016/j.hoc.2015.09.008. [DOI] [PubMed] [Google Scholar]
- Ocana M., et al. GLIQUIDONE, an ATP-dependent k+ channel antagonist, antagonizes morphine-induced HYPERMOTILITY. Eur. J. Pharmacol. 1993;239(1–3):253–255. doi: 10.1016/0014-2999(93)91006-9. [DOI] [PubMed] [Google Scholar]
- Pastorino G., et al. Liquorice (Glycyrrhiza glabra): a phytochemical and pharmacological review. Phytother. Res. 2018;32(12):2323–2339. doi: 10.1002/ptr.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pópulo H., et al. The mTOR signalling pathway in human cancer. Int. J. Mol. Sci. 2012;13(2):1886–1918. doi: 10.3390/ijms13021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptushkin V.V., et al. Thrombopoietin receptor agonists in the treatment of chronic resistant primary immune thrombocytopenia: efficacy and safety data in real clinical practice. Ter. Arkh. 2018;90(7):70–76. doi: 10.26442/terarkh201890770-76. [DOI] [PubMed] [Google Scholar]
- Sax P.E., et al. Coformulated bictegravir, emtricitabine, and tenofovir alafenamide versus dolutegravir with emtricitabine and tenofovir alafenamide, for initial treatment of HIV-1 infection (GS-US-380–1490): a randomised, double-blind, multicentre, phase 3, non-inferiority trial. Lancet. 2017;390(10107):2073–2082. doi: 10.1016/s0140-6736(17)32340-1. [DOI] [PubMed] [Google Scholar]
- Sun Z.-G., et al. Research Progress of Glycyrrhizic acid on antiviral activity. Mini-Rev. Med. Chem. 2019;19(10):826–832. doi: 10.2174/1389557519666190119111125. [DOI] [PubMed] [Google Scholar]
- Targett-Adams P., et al. Small molecules targeting hepatitis C virus-encoded NS5A cause subcellular redistribution of their target: insights into compound modes of action. J. Virol. 2011;85(13):6353–6368. doi: 10.1128/jvi.00215-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., et al. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.) 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020 doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., et al. mTOR kinase structure, mechanism and regulation. Nature. 2013;497(7448):217–223. doi: 10.1038/nature12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yisireyili M., et al. Angiotensin receptor blocker irbesartan reduces stress-induced intestinal inflammation via AT1a signaling and ACE2-dependent mechanism in mice. Brain Behav. Immun. 2018;69:167–179. doi: 10.1016/j.bbi.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Yoo J.H., Hong S.T. The outbreak cases with the novel coronavirus suggest upgraded quarantine and isolation in Korea. J. Korean Med. Sci. 2020;35(5):e62. doi: 10.3346/jkms.2020.35.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamek-Gliszczynski M.J., et al. Clinical extrapolation of the effects of Dolutegravir and other HIV Integrase inhibitors on folate transport pathways. Drug Metab. Dispos. 2019;47(8):890–898. doi: 10.1124/dmd.119.087635. [DOI] [PubMed] [Google Scholar]
- Zhang C., et al. Protein structure and sequence reanalysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. J. Proteome Res. 2020 doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N.S., et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362(9393):1353–1358. doi: 10.1016/s0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Workflow chart of the structure-based virtual screening and subsequent high throughput screening methodology. The amino acid sequence of SARS-CoV-2 was downloaded from NCBI and analyzed. The structure of S, M, E, N protein was acquired by ab initio modeling or homology modeling (S protein), and the S protein docking pocket for drug screening was analyzed by comparing with SARS-CoV. By scoring the 1234 compounds from the FDA drug database compound library via the drug screening pocket, the top 100 compounds were selected. Among these, 13 compounds were further selected through visual inspection, which underwent surface plasmon resonance test and Thermo Shift Assay to verify the efficacy.
Information about the protein chips used in SPR test.