Abstract
Background
Antibiotic resistance is a serious public health challenge exacerbated by the widespread use of β-lactam and glycopeptide antibiotics. The identification of resistances is crucial, and CHROMID ESBL medium has been developed to detect enterobacteria with extended-spectrum β-lactamases (ESBL). The objective of this study was to evaluate the potential of this medium to detect other types of resistant bacteria.
Methods
Vancomycin, cefoxitin, imipenem, and cefepime disks were used to measure growth on CHROMID ESBL medium of β-lactam-resistant Gram-negative (83 with ESBL, 57 with carbapenemases, 35 with AmpC and 3 Stenotrophomonas maltophilia) and Gram-positive [37 vancomycin-susceptible (vancoS) microorganisms and 21 vancomycin-resistant (vancoR) Enterococcus faecium] clinical isolates (retrospective study) and colonization by the aforementioned bacteria (prospective study), using 649 rectal swabs, 314 pharyngeal swabs, and 44 swabs from other localizations.
Results
Retrospective study: species grown on the medium exhibited different colors. Growth on the medium was observed for: all ESBL enterobacteria, which were susceptible to imipenem and cefoxitin; 95% of isolates with carbapenemases, mostly resistant to imipenem; 80% of those with AmpC; 86% of vancoR E. faecium isolates; and 42% of vancoS E. faecalis isolates, with large growth inhibition halos around the vancomycin disk. Prospective study: vancoR E. faecium, ESBL Klebsiella, Pseudomonas with carbapenemases, A. baumannii (mostly from rectal swabs), S. maltophilia, Achromobacter xylosoxidans, and Burkholderia cenocepacia (mostly from pharyngeal swabs) were isolated from the 246 positive samples.
Conclusions
CHROMID ESBL medium permitted the differential growth of Gram-negative bacteria, many with ESBL and carbapenemases. ESBL enterobacteria were susceptible to imipenem, carbapenemase-producing microorganisms grew around the imipenem disk, and vancoR E. faecium was isolated on the medium. Results of the prospective study demonstrate the potential clinical relevance of this medium. S. maltophilia was more frequently detected with pharyngeal swabs and ESBL Klebsiella, A. baumannii, and Pseudomonas with rectal swabs.
Keywords: Multiresistant bacteria, chromogenic media, CHROMID ESBL medium
Introduction
Numerous clinical isolates develop resistance to β-lactam and glycopeptide antibiotics. According to the World Health Organization (WHO), bacteria currently posing a worldwide global threat include: carbapenemase-producing Acinetobacter baumannii, Pseudomonas aeruginosa (CPP), and Enterobacteriaceae (CPE) as well as extended-spectrum β-lactamases (ESBLs), methicillin-resistant (metiR) Staphylococcus aureus, and vancomycin-resistant (vancoR) Enterococcus faecium (1). A. baumannii is an opportunistic pathogen that can cause pneumonia, bacteremia, meningitis, urinary tract infections, and peritonitis, among other infections. A. baumannii has been prioritized by the WHO for developing novel antibiotic therapies because of the increasing prevalence of resistant strains (2-4). P. aeruginosa is a highly ubiquitous bacteria and an opportunistic pathogen responsible for bacteremias, urinary tract infections, and pneumonia, among other infections, and it is the main cause of morbidity and mortality in patients with cystic fibrosis (5-7). The Enterobacteriaceae family is part of normal human microbiota, but Klebsiella, Escherichia, and Enterobacter genera can cause pneumonia and bloodstream and urinary tract infections, among other severe diseases. It has become necessary to use carbapenems against ESBL enterobacteria, favoring the development of carbapenem-resistant carbapenemase-producing bacteria (8-10). Finally, E. faecium and E. faecalis can produce opportunistic infections. VancoR isolates have largely been recorded in developing countries, although increasing globalization is spreading this phenomenon worldwide (11,12). After the diagnosis of infection by these pathogens, it is crucial to explore their presence in the digestive tract of patients and their contacts using inexpensive, simple, comprehensive, rapid, and effective methods (13-15). Techniques developed to detect ESBL enterobacteria, frequently encountered in the hospital setting, include utilization of the transparent medium CHROMID ESBL (bioMérieux, France). It contains cefpodoxime, allowing the detection of ESBL enterobacteria colonies (16-18), along with substances that inhibit Gram-positive bacteria growth, and chromogenic substrates that are used to presumptively identify different genera and species by their color, as follows: pink/burgundy for Escherichia coli; blue/green for Klebsiella, Enterobacter, Serratia, or Citrobacter; and light to dark brown for Proteae (16-19). Other chromogenic media are also available to detect vancoR enterococci, presumptively differentiating vancoR E. faecium from E. faecalis (20,21).
The incorporation of cefoxitin (FOX), cefepime (FEP), and imipenem (IMP) disks on a solid culture medium may reveal microorganisms resistant to these antibiotics, which all have well-documented antibiotic activity. In this way, CHROMID ESBL medium, designed to recover only ESBL-producing enterobacteria from rectal swabs, may hypothetically detect β-lactam-resistant Gram-negative and vancoR Gram-positive bacteria through the addition of standard antibiotic disks. To our best knowledge, this is the first published study on the possible clinical usefulness of CHROMID medium to detect resistant non-ESLB-producing bacteria.
The objective of this study was to measure the growth of Gram-negative bacteria with different types of β-lactam-resistance on CHROMID ESBL culture plates and to determine the selection of Gram-positive bacteria on these media.
Methods
A retrospective study was conducted on the growth of β-lactam-resistant Gram-negative clinical isolates and vancoR Gram-positive clinical isolates on CHROMID ESBL medium with different antibiotic disks. In addition, a prospective study was performed to detect colonization by the same bacteria.
Retrospective study
Clinical isolates
The study included 178 strains of Gram-negative bacteria with different resistance mechanisms (CLSI 2018 criteria) isolated from clinical samples in the Microbiology Department of our hospital in Granada, Spain: 83 with ESBL (43 K. pneumonie, 38 E. coli, 2 Proteus mirabilis); 57 with carbapenemases: Klebsiella pneumoniae carbapenemases (KPC) (8 K. pneumoniae), carbapenemase type IMP (IMPase) (16 CPP, 1 K. pneumoniae), Verona integron-encoded metallo-beta-lactamase (VIM) (4 Enterobacter cloacae, 3 K. pneumonie, 3 Klebsiella oxytoca, 1 P. aeruginosa, 1 E. coli), and oxacillinases (OXA) (15 A. baumannii, 4 K. pneumonie, 1 K. oxytoca); 35 with AmpC and FOX-resistance (16 E. cloacae, 7 E. coli, 2 Citrobacter freundii, 2 Klebsiella aerogenes, 2 P. mirabilis, 2 K. pneumonie, 1 K. oxytoca, 1 Raoutella ornithinolytica, 1 Serratia marcescens, 1 Citrobacter amalonaticus); and 3 Stenotrophomonas maltophilia. The study also included 58 Gram-positive isolates: 37 vancomycin-susceptible (vancoS) [12 E. faecalis, 9 metiR Staphylococcus haemolitycus, 9 S. aureus (6 metiR), 6 E. faecium, and 1 metiR Staphylococcus epidermidis], and 21 vancoR E. faecium. Isolates were identified with the MicroScan system (Beckman Coulter, USA) and mass spectrometry (Maldi-Tof®, Bruker Daltonik GmbH, Germany). Resistance was characterized using the MicroScan system with subsequent carbapenemase determination, if applicable, by means of the Rapidec® Carba NP colorimetric test (BioMerieux, France), immunochromatography (NG-Test Carba, NG Biotech, France; and OXA-23 K-Set, CorisBioConcept, Belgium), or the E-test for vancomycin (VA) resistance (MIC Strip, Liofilchem). Carbapenemase production and resistance to VA were confirmed by the Andalusian Molecular Typing Laboratory of the Spanish PIRASOA Program using appropriate molecular biology techniques. AmpC production was defined by resistance to FOX and synergy with cloxacillin. Carbapenemase production was ruled out with the Rapidec® Carba NPcolorimetric test (BioMerieux, France).
Investigation of bacteria growth on CHROMID ESBL medium
An 0.5 McFarland suspension of each isolate was prepared from colonies grown on lamb blood agar (Becton Dikinson, USA). Next, a sterilized swab was soaked with the homogenized suspension, excess liquid was eliminated, and it was seeded in a uniform manner on half of the plate, streaking the bacterial load on the other half with a calibrated inoculation loop. FEP (30 µg, Becton Dickinson), FOX (30 µg, Becton Dickinson), and IMP (10 µg, Becton Dickinson) disks were then placed equidistantly on the seeded area. For Gram-positive bacteria, a VA disk (30 µg, Becton Dickinson) was also added at a distance from the IMP disk. The medium was then incubated for 48 h at 37 °C, with readings at 24 and 48 h. The isolate was considered susceptible if the inhibition halo was ≥1.5 cm at 24 h and resistant if it was <1.5 cm.
Prospective study
Clinical samples
These were samples from individuals aged >14 years studied by the Microbiology Department of Virgen de las Nieves University Hospital of Granada, which covers a population of around 440,000 inhabitants. Between November 1 2018 and April 30 2019, 1,007 samples from 441 patients (some with multiple episodes) were studied for suspicion of colonization by metiR S. aureus and ESBL Klebsiella; they comprised 649 rectal swabs, 314 pharyngeal swabs, and 44 swabs from other sites. Samples were seeded on MRSAII (BIORAD, France), selective for metiR S. aureus, and on CHROMID ESBL. The presence of vancoR enterococcus, CPE, carbapenemase-producing Pseudomonas (CPP), A. baumannii, and other non-glucose-fermenting Gram-negative bacilli was also studied on CHROMID ESBL.
Microorganism growth on CHROMID ESBL medium
The sample was inoculated on the medium by spreading the swab content on one half of the plate and streaking the bacterial load on the other half with a calibrated inoculation loop. Next, FEP, FOX, and IMP disks were placed for growth/inhibition measurement. All plates were incubated for 48 h at 37 °C and were read at 24 and 48 h. The result was then reported as: (I) absence of bacterial growth at 48 h of incubation; (II) growth of one or more morphotypes at 24 h but with insufficient bacterial load for reading halos; in this case, each colony morphotype of clinical interest was tested on the medium with the three antibiotic disks, again incubating the plates at 37 °C for 24 h for halo reading; or (III) growth with sufficient bacterial load for analysis after 24 h of incubation. The morphology, coloring, and disk halo results were recorded. The plate was re-incubated, acting as in the previous step when necessary, and isolates were identified by mass spectrometry (Maldi-Tof®, Bruker Daltonik GmbH, Germany). Susceptibility studies were conducted using the MicroScan system, with subsequent carbapenemase determination, when applicable, by means of the Rapidec® Carba NP colorimetric test (BioMerieux), immunochromatography (NG-Test Carba, NG Biotech; and OXA-23 K-Set, CorisBioConcept), or the E-test for VA resistance (MIC Strip, Liofilchem). Carbapenemase production results were confirmed by the Andalusian Molecular Typing Laboratory of the Spanish PIRASOA Program using the appropriate molecular biology tests.
Microorganism growth on MRSAII medium (BIORAD)
Plates were inoculated as described above; with incubation for 48 h at 37 °C and readings at 24 and 48 h. Colonies suspected of being metiR S. aureus (red color) were examined by mass spectrometry (Maldi-Tof®).
Microsoft Excel 2010 was used to conduct a descriptive analysis of the data, calculating absolute and relative frequencies for categorical variables.
Results
Retrospective study
Behavior of Gram-negative isolates on CHROMID ESBL culture medium
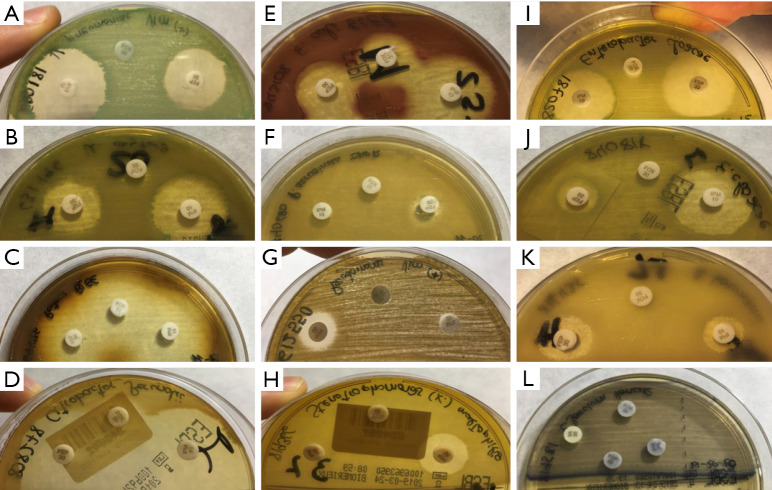
Plate readings were similar between 24 and 48 h in all cases. Colors varied among the different species (Figure 1). E. coli strains grown on CHROMID ESBL medium appeared pink/burgundy in color, Klebsiella, Enterobacter, Serratia, and Citrobacter genera appeared blue/green, and Proteus, Providencia, and Morganella appeared light to dark brown, consistent with the manufacturer’s indications. The manufacturer’s list of bacteria that grow with specific colors does not include Pseudomonas or Acinetobacter, which were whitish/transparent, or Stenotrophomonas, which were light brown/green, similar to the color of K. oxytoca but less intense and closer to light brown/yellow.
Figure 1.
Coloring of the different species studied on the CHROMID ESBL plate. (A) K. pneumoniae; (B) K. oxytoca; (C) P. mirabilis; (D) Citrobacter spp.; (E) E. coli; (F,G) P. aeruginosa; (H) S. maltophilia; (I,J) E. cloacae; (K) A. baumannii; (L) vancoR E. faecium.
Table 1 lists the results for growth on the medium, which was observed in all ESBL-producing K. pneumoniae, E. coli, and P. mirabilis isolates. Among the 57 carbapenemase-producing isolates, no growth was observed in 1 (6.3%) out of 16 CPP with IMPase, 1 (33.3%) out of 3 K. pneumoniae with VIM, or in the sole K. oxytoca isolate with OXA.
Table 1. Number of isolates grown on CHROMID ESBL medium, percentage by microorganism, and mean percentage by type of resistance.
| Resistance mechanisms | Microorganisms | No. isolates | Positive growth (%) | Mean % growth |
|---|---|---|---|---|
| ESBL | Klebsiella pneumoniae | 43 | 43 (100.0) | 100.0 |
| Proteus mirabilis | 2 | 2 (100.0) | ||
| Escherichia coli | 38 | 38 (100.0) | ||
| Carbapenemases | 94.7 | |||
| KPC | Klebsiella pneumoniae | 8 | 8 (100.0) | 100.0 |
| IMPase | Pseudomonas aeruginosa | 16 | 15 (93.8) | 94.1 |
| Klebsiella pneumoniae | 1 | 1 (100.0) | ||
| VIM | Enterobacter cloacae | 4 | 4 (100.0) | 91.7 |
| Klebsiella pneumoniae | 3 | 2 (66.7) | ||
| Klebsiella oxytoca | 3 | 3 (100.0) | ||
| Pseudomonas aeruginosa | 1 | 1 (100.0) | ||
| Escherichia coli | 1 | 1 (100.0) | ||
| OXA | Acinetobacter baumannii | 15 | 15 (100.0) | 95.0 |
| Klebsiella pneumoniae | 4 | 4 (100.0) | ||
| Klebsiella oxytoca | 1 | 0 (0) | ||
| AmpC | Citrobacter freundii | 2 | 1 (50.0) | 80.0 |
| Citrobacter amalonaticus | 1 | 1 (100.0) | ||
| Escherichia coli | 7 | 4 (57.1) | ||
| Enterobacter cloacae | 16 | 16 (100.0) | ||
| Klebsiella aerogenes | 2 | 2 (100.0) | ||
| Proteus mirabilis | 2 | 1 (50.0) | ||
| Klebsiella pneumoniae | 2 | 2 (100.0) | ||
| Klebsiella oxytoca | 1 | 1 (100.0) | ||
| Raoutella ornithinolytica | 1 | 0 (0) | ||
| Serratia marcescens | 1 | 0 (0) | ||
| Other mechanisms | Stenotrophomonas maltophilia | 3 | 3 (100.0) | 100.0 |
ESBL, extended-spectrum β-lactamases; KPC, Klebsiella pneumoniae carbapenemases; IMPase, carbapenemases type IMP; VIM, Verona integron-encoded metallo-beta-lactamase; OXA, oxacillinases.
With regard to the isolates with AmpC, no growth was observed for R. ornithinolytica or S. marcescens, whereas a recovery of 50.0% was observed for C. freundii, 57.1% for E. coli, and 50.0% for P. mirabilis. For the remaining microorganisms, 100.0% of isolates were recovered.
Table 2 reports the presence or absence of halos ≥1.5 cm around IMP, FEP, and FOX disks. All ESBL enterobacteria were susceptible to IMP and FOX, while the susceptibility to FEP was variable.
Table 2. Number of isolates showing halos ≥1.5 cm (susceptibility, S) or not (resistance, R) around cefoxitin (FOX), cefepime (FEP), and imipenem (IMP) disks according to the type of resistance.
| Mechanisms | Microorganisms | IMP/FEP/FOX | Total | |||||
|---|---|---|---|---|---|---|---|---|
| S/S/S | S/R/S | R/R/R | S/S/R | R/S/R | S/R/R | |||
| Other mechanisms | S. maltophilia | – | – | – | – | 3 | – | 3 |
| AmpC | K. oxytoca | 1 | – | – | – | – | – | 1 |
| K. pneumoniae | 2 | – | – | – | – | – | 2 | |
| P. mirabilis | – | – | 1 | – | – | – | 1 | |
| K. aerogenes | – | – | – | 2 | – | – | 2 | |
| E. cloacae | 2 | 1 | – | 12 | – | 1 | 16 | |
| E. coli | 4 | – | – | – | – | – | 4 | |
| C. amalonaticus | 1 | – | – | – | – | – | 1 | |
| C. freundii | 1 | – | – | – | – | – | 1 | |
| CBP | ||||||||
| OXA | K. pneumoniae | – | – | – | – | – | – | 4 |
| A. baumannii | – | – | 1 | 14 | – | – | 15 | |
| VIM | E. coli | 1 | – | – | – | – | – | 1 |
| P. aeruginosa | – | – | 1 | – | – | – | 1 | |
| K. oxytoca | 2 | – | – | 1 | – | – | 3 | |
| K. pneumoniae | – | – | – | 2 | – | – | 2 | |
| E. cloacae | 1 | – | – | 3 | – | – | 4 | |
| IMPase | K. pneumoniae | – | – | – | 1 | – | – | 1 |
| P. aeruginosa | – | – | 5 | – | 10 | – | 15 | |
| KPC | K. pneumoniae | 4 | – | 3 | 1 | – | – | 8 |
| ESBL | P. mirabilis | 2 | – | – | – | – | – | 2 |
| E. coli | 38 | – | – | – | – | – | 38 | |
| K. pneumoniae | 36 | 7 | – | – | – | – | 43 | |
CBP, carbapenemases; ESBL, extended spectrum β-lactamases; KPC, Klebsiella pneumoniae carbapenemases; IMPase, carbapenemases type IMP; VIM, Verona integron-encoded metallo-beta-lactamase; OXA, oxacillinases.
Behavior of Gram-positive isolates on CHROMID ESBL culture medium
Among enterococci, growth was observed as bluish punctiform colonies (Figure 1) and was recorded in 18 (86%) of vancoR E. faecium isolates close to the VA disk and in 5 (42%) of vancoS E. faecalis isolates with halos ≥1.5 cm. No staphylococci grew on the medium.
Prospective study
No microorganisms were isolated in 761 (75.6%) of the 1,007 samples. Table 3 exhibits the distribution of microorganisms in the remaining 246 samples by sample type. The only bacterium showing a similar detection rate between pharyngeal and rectal samples was S. maltophilia. Among the 40 episodes in which both samples were studied, the same bacterium was detected in both swabs in 16 cases, in rectal swab alone in 13 cases, and in pharyngeal swab alone in 11 cases. Isolates are listed in Table 4. The only cases of B. cenocepacia and A. xylosoxidans were detected in pharyngeal swabs.
Table 3. Number of positive episodes of each bacterium by type of sample.
| Microorganisms | Positive samples (%) | ||||
|---|---|---|---|---|---|
| Total | Rectal swabs | Pharyngeal swabs | Other | Isolate description | |
| A. baumanii | 55 | 45 (81.8) | 6 (10.9) | 4 (7.3) | 6 OXA-23 |
| VancoR enterococci | 20 | 20 (100.0) | 0 (0) | 0 (0) | 15 E. faecium; 4 E. faecalis; 1 E. casseliflavus |
| ESBL K. oxytoca | 5 | 5 (100.0) | 0 (0) | 0 (0) | – |
| ESBL K. pneumoniae | 68 | 63 (92.6) | 3 (4.4) | 2 (2.9) | – |
| P. aeruginosa | 23 | 21 (91.3) | 1 (4.3) | 1 (4.3) | 22 IMPase; 1 VIM |
| P. putida | 2 | 2 (100.0) | 0 (0) | 0 (0) | 2 VIM |
| MetiR S. aureus | 48 | 12 (25.0) | 36 (75.0) | 0 (0) | – |
| S. maltophilia | 23 | 12 (52.2) | 11 (47.8) | 0 (0) | – |
| A. xylosoxidans | 1 | 0 (0) | 1 (100.0) | 0 (0) | – |
| B. cenocepacia | 1 | 0 (0) | 1 (100.0) | 0 (0) | – |
OXA, oxacillinases; VancoR, vancomycin-resistant; ESBL, extended-spectrum β-lactamases; IMPase, carbapenemases type IMP; VIM, Verona integron-encoded metallo-beta-lactamase; MetiR, methicillin-resistant.
Table 4. Comparison of episodes in which both types of sample were available.
| Rectal swabs | Pharyngeal swabs | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MetiR S. aureus | S. maltophilia | ESBL K. pneumoniae | A. baumannii | VancoR Enterococci | P. aeruginosa | ||||||||||||
| Yes | No | Yes | No | Yes | No | Yes | No | Yes | No | Yes | No | ||||||
| Yes | 0 | 0 | 5 | 1 | 5 | 3 | 6 | 3 | 0 | 1 | 0 | 5 | |||||
| No | 2 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |||||
MetiR, methicillin-resistant; ESBL, extended-spectrum β-lactamases; VancoR, vancomycin-resistant.
Discussion
Retrospective study
Behavior on the culture medium with disks of Gram-negative isolates
This color system offers a simple technique to detect the growth of different microorganisms and their presumptive species or genus, although the color is not always easy to distinguish, hampering the discrimination of species. For this reason, the presumptive identification of bacteria on the manufacturer’s list (22) should be confirmed with another technique such as mass spectrometry. Clear morphological differences were observed between non-fermenting Gram-negative enterobacteria and bacilli.
K. pneumoniae, E. coli, and P. mirabilis, acknowledged as potential ESBL producers (23), all grew on the medium and were all susceptible to IMP and FOX. ESBL production is known to generate resistance against extended-spectrum cephalosporins but not against cephamycins (e.g., FOX) or carbapenems (e.g., IMP). The growth of seven K. pneumoniae was not affected by FEP. ESBL development can confer resistance against 3rd and 4th generation cephalosporins but this is much less frequent, as reflected in the markedly lower number of FEP-resistant than FEP-susceptible strains that grew on the medium. Although resistance to FEP is less common, it is not recommended as monotherapy against infection by ESBL enterobacteria to avoid spreading resistance, and it should only ever be used in combination with aminoglycosides or fluoroquinolones (24-26).
Three of the carbapenemase-producing isolates (5.3% of the total with carbapenemases) did not grow on CHROMID ESBL medium, despite previously demonstrating resistance to carbapenems. The strains were revived and inoculated on both blood plates and CHROMID ESBL medium to rule out inoculation error or death of the strain. Their lack of growth may be attributable to the presence of multiple antibiotics on the medium, i.e., IMP or FEP with cefpodoxime on CHROMID ESBL, a combination that has proven to be active against resistant bacteria (27-30). It may also be related to the theory of development by loss of unnecessary genes, given that thawed isolates were seeded on blood agar plates before their study, with the potential loss of resistance genes not required in this medium, which does not select for this characteristic (31). Finally, errors in the placing of samples on the medium cannot be ruled out. One study limitation is that genetic analysis was not carried out to elucidate the reasons for non-growth.
With regard to the isolates that showed growth, CPP with IMPase and VIM were resistant, as expected, because IMPase and VIM are metallo-β-lactamases, which confer resistance to almost all β-lactams, IMP, and FOX, with variable susceptibility to FEP (32,33). Three of the eight KPC-producing isolates were resistant to all three antibiotics, consistent with the known resistance of KPC to carbapenems and cephalosporins, with occasional susceptibility to some 3rd- and 4th-generation cephalosporins (32,34), while five of the remaining isolates were susceptible to FEP and IMP, with variable resistance to FOX. As noted above, the absence of resistance to IMP may be due to the loss of resistance genes, such as those that produce KPC. In vitro studies have observed an IMP-susceptible phenotype among KPC-producing strains (35,36). The remaining IMPase- and VIM-producing isolates that grew on the medium also exhibited phenotypes of susceptibility to FEP and IMP with variable resistance to FOX. Although these findings do not agree with reported data on metallo-β-lactamase enzymes, account should be taken of a possible synergic effect between the disk and antibiotic on the medium (37). The degree of resistance of isolates with VIM and IMPase has been found to vary, and these resistance genes are generally observed in plasmid integrons, favoring their loss (37,38). Finally, only one of the OXA-producing A. baumannii isolates showed the expected phenotypical resistance to IMP (and to FEP and FOX). In contrast, the 14 other cases of A. baumannii were resistant to FOX and susceptible to FEP and IMP. Various authors have proposed that OXA has a low hydrolytic activity against carbapenems (39,40), which may explain the observation of IMP susceptibility halos on the medium and the susceptibility of OXA-producing K. pneumoniae isolates.
The lowest growth rates were observed in AmpC-producing isolates (80%), which may be explained by the inability of Gram-negative bacteria to grow on CHROMID ESBL medium. However, the genes encoding AmpC resistance to bacteria are often integrated within the bacterial chromosome, reducing the likelihood of their loss, although there have been reports of plasmid-determined AmpC-type β-lactamases that are more readily lost by the bacterium (41). The lack of genetic analysis to explore this issue is a study limitation, as acknowledged above. It should also be taken into account that AmpC is less effective against 3rd and 4th generation cephalosporins, and the cefpodoxime on the medium may have inhibited their growth. Nevertheless, this lack of growth is useful in clinical practice because it signifies that these microorganisms are, in principle, not relevant for the diagnosis. E. cloacae and K. aerogenes were resistant to FOX and susceptible to both FEP and IMP, in line with previous studies (32,42-45). Some isolates grown on the medium exhibited susceptibility to FOX and IMP with variable resistance to FEP, a similar phenotype to that of microorganisms with ESBL resistance, which may also have been present. One isolate of E. cloacae was resistant to FOX and FEP but susceptible to IMP, indicating a mixture of AmpC- and ESBL-type resistance, which are not mutually exclusive. The phenotype of Citrobater genus was similar to that of ESBL-producing isolates, possibly due to the loss of expression of genes encoding AmpC resistance through the freezing and thawing of isolates on blood agar. Escherichia presented the same phenotype, which may also be due to very low ampC expression levels, without showing resistance (46). The absence of AmpC-related resistance in K. pneumoniae and K. oxytoca isolates may be attributable to the localization of the gene in plasmids in this genus, facilitating its loss (41). The sole isolate of P. mirabilis was resistant to FOX, FEP, and IMP, which is not characteristic of AmpC-type resistance and is more similar to a combination of ESBL and AmpC production, given that IMP resistance is not related to carbapenemases in this species. S. maltophilia was resistant to FOX and IMP, indicating a different resistance mechanism, such as a reduced number of porins (32,41,47).
In general, despite the possible effect of thawing strains on a non-selective medium, the results show a pattern for each resistance type (ESBL, carbapenemase, and AmpC) on CHROMID ESBL medium. Most strains with ESBL and carbapenemases exhibited growth, strains with ESBL were susceptible to IMP and FOX when the three antibiotic disks were added, while strains with carbapenemases were not susceptible to FOX. The lack of resistance to IMP may be due to the handling of the strain or may point to the need for a higher halo cutoff point than 1.5 cm. Growth was less frequent in isolates with AmpC, while the growth of three strains of K. pneumoniae and K. oxytoca with susceptibility to the three disks also suggests that the halo cutoff point should be >1.5 cm.
Behavior on culture medium with disks of Gram-positive isolates
VancoR enterococci colonies grown on CHROMID ESBL acquired a bluish color similar to the blue-green color produced by K. pneumoniae or K. aerogenes, although the two genera are readily differentiated by the morphology of their colonies (small and punctiform in the case of enterococci). Given that the manufacturer of CHROMID ESBL only describes characteristic colorings for Gram-negative bacteria, there is no reference for the Gram-positive bacteria and non-fermenting bacilli obtained in this study.
None of the isolates of the Staphylococcus genus grew on CHROMID ESBL medium in the presence of VA, because they all had a phenotype susceptible to this antibiotic. They also showed no growth beyond the range of VA disks, although Staphylococcus possesses a high degree of resistance to β-lactam antibiotics due to the presence of β-lactamases, which would favor the development of these isolates on the medium (48). It can therefore be deduced that the medium also contains another type of substrate not reported by the manufacturer that does not permit the development of vancoS isolates. The study included both VA-resistant and VA-susceptible strains of the Enterococcus genus. There has been a major increase in the resistance of this genus to VA over the past few years, mainly in E. faecium (49-51), enabling the growth of the majority of the present vancoR isolates on the medium. The vancoR E. faecium isolates grew with and without VA disks. In contrast, the vancoS E. faecium isolates showed no growth, as in the case of the Staphylococcus genus. Seven isolates of E. faecalis did not grow on the medium, being vancoS isolates, although five vancoS isolates showed growth beyond the range of VA activity, which may indicate their resistance to a possible factor in the medium that usually inhibits vancoS isolates.
The data obtained in this study indicate that the CHROMID ESBL medium can be used to detect Gram-negative bacteria and may also detect vancoR enterococci, even without the utilization of VA disks.
Prospective study
All vancoR enterococci isolates were obtained from rectal swabs, as expected, given that this type of bacteria colonizes the gastrointestinal tract (52-54). Klebsiella genus isolates were mainly obtained from rectal swabs, because the Enterobacteriaceae family is characterized by colonizing the digestive tract (55,56), although cases of K. pneumoniae were also found in pharyngeal samples. Authors have recently studied the colonization of this species in respiratory tract areas (57). Likewise, A. baumannii was much more frequently detected in rectal swabs than in pharyngeal swabs, because it usually colonizes the gastrointestinal tract, although respiratory tract isolates have occasionally been reported (58,59). The situation for Pseudomonas genus is similar except that it initially colonizes the respiratory tract and is implicated in respiratory infections (60-62). This does not agree with the data obtained in the prospective study, although there was a higher likelihood of obtaining isolates from rectal swabs because of their larger number (n=649) in comparison to pharyngeal samples (n=314). In contrast, all A. xylosoxidans and B. cenocepacia isolates were from swabs of the pharynx, the site usually colonized by these species (63,64), and most metiR S. aureus grew from pharyngeal swabs for the same reason (65-67), although a small proportion was found in swabs of the rectum, which can also be colonized by this species (68). Finally, almost the same number of S. maltophilia isolates was found in each type of swab, although S. maltophilia more frequently colonizes the respiratory tract (69,70). The larger number of rectal swabs should again be taken into account. The results obtained for Pseudomonas may be explained by the long-term treatment of these patients with extended-spectrum respiratory antibiotics, with the intestine becoming a reservoir for this pathogen.
Clinical implications
This study makes a timely methodological contribution, given the increasing number of patients colonized by bacteria that are resistant to β-lactam antibiotics via different genetic mechanisms and that must be detected. The application of molecular biology tests for these patients would carry elevated hospital costs, because they are highly sensitive and detect all possible resistant microorganisms and resistances, generating uncertainty in the interpretation of results and leading to the unnecessary isolation of patients within the hospital.
Novel findings
Utilization of the CHROMID ESBL medium as in the present study allows the detection of resistant and viable (living) microorganisms and provides simple, rapid, economic, and approximate information on the resistance mechanisms of the bacteria colonizing the patient. Further studies should explore the utilization of other antibiotic disks and/or different halo cutoff points to improve the sensitivity and specificity of results.
In summary, the CHROMID ESBL medium permitted the differential growth of Gram-negative bacteria with different types of β-lactam resistance, and the inhibition halos around FOX, IMP, and FEP disks revealed a pattern in which most isolates with ESBL and carbapenemases grew on the medium and ESBL enterobacteria were susceptible to IMP. The opposite was observed for carbapenemase producers, which were resistant to IMP. With regard to Gram-positive bacteria, the medium detected VA-resistant E. faecium with no need for the addition of a VA disk. However, despite this advantage, the application of other techniques to precisely determine resistances should not be ruled out. The prospective study demonstrated the potential clinical relevance of this technique, allowing the detection of colonization by Gram-negative bacteria designated by the WHO as the most important multiresistant pathogens. Finally, the combined utilization of CHROMID ESBL and MRSAII media revealed a more frequent detection of metiR S. aureus and S. maltophilia in pharyngeal swabs and of ESBL K. pneumoniae, A. baumannii, and CPP in rectal swabs, although these observations should be tested in larger patient series.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors are grateful to the Andalusian Molecular Typing Laboratory of the Spanish PIRASOA Program for the genetic studies of the clinical isolates.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editors (Zhi-De Hu, Bing Gu) for the series “Advances in laboratory tests for infectious diseases” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.02.158). The series “Advances in Laboratory Tests for Infectious Diseases” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
References
- 1.La OMS publica la lista de las bacterias para las que se necesitan urgentemente nuevos antibióticos. Available online: https://www.who.int/es/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed
- 2.Jiménez-Guerra G, Heras-Cañas V, Gutiérrez-Soto M, et al. Urinary tract infection by Acinetobacter baumannii and Pseudomonas aeruginosa: evolution of antimicrobial resistance and therapeutic alternatives. J Med Microbiol 2018;67:790-7. 10.1099/jmm.0.000742 [DOI] [PubMed] [Google Scholar]
- 3.Diomedi PA. Infecciones por Acinetobacter baumannii pan-resistente: Consideraciones epidemiológicas y de manejo antimicrobiano actualizado. Rev Chil Infectología 2005;22:298-320. 10.4067/S0716-10182005000600003 [DOI] [PubMed] [Google Scholar]
- 4.Pinzon JO, Mantilla JR, Valenzuela E, et al. Caracterización molecular de aislamientos de Acinetobacter baumannii provenientes de la unidad de quemados de un hospital de tercer nivel de Bogotá. Infectio 2006;10:71-8. [Google Scholar]
- 5.Cafora M, Deflorian G, Forti F, et al. Phage therapy against Pseudomonas aeruginosa infections in a cystic fibrosis zebrafish model. Sci Rep 2019;9:1527. 10.1038/s41598-018-37636-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stover CK, Pham XQ, Erwin AL, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000;406:959-64. 10.1038/35023079 [DOI] [PubMed] [Google Scholar]
- 7.Wang T, Hou Y, Wang R. A case report of community-acquired Pseudomonas aeruginosa pneumonia complicated with MODS in a previously healthy patient and related literature review. BMC Infect Dis 2019;19:130. 10.1186/s12879-019-3765-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soria-Segarra C, Soria-Segarra C, Catagua-González A, et al. Carbapenemase producing Enterobacteriaceae in intensive care units in Ecuador: Results from a multicenter study. J Infect Public Health 2020;13:80-8. 10.1016/j.jiph.2019.06.013 [DOI] [PubMed] [Google Scholar]
- 9.Sheu CC, Chang YT, Lin SY, et al. Infections caused by carbapenem-resistant Enterobacteriaceae: An update on therapeutic options. Front Microbiol 2019;10:80. 10.3389/fmicb.2019.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watkins RR, Van Duin D. Current trends in the treatment of pneumonia due to multidrug-resistant Gram-negative bacteria. Version 2. F1000Res 2019. doi: . 10.12688/f1000research.16517.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thu WP, Sinwat N, Bitrus AA, et al. Prevalence, antimicrobial resistance, virulence genes and class 1 integrons of Enterococcus faecium and Enterococcus faecalis from pigs, pork and humans in Thai-Laos border provinces. J Glob Antimicrob Resist 2019;18:130-8. 10.1016/j.jgar.2019.05.032 [DOI] [PubMed] [Google Scholar]
- 12.O’Dea M, Sahibzada S, Jordan D, et al. Genomic, antimicrobial resistance and public health insights into Enterococcus spp. from Australian chickens. J Clin Microbiol 2019. doi: . 10.1128/JCM.00319-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bush K, Courvalin P, Dantas G, et al. Tackling antibiotic resistance. Nat Rev Microbiol 2011;9:894-6. 10.1038/nrmicro2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fariñas MC, Martínez-Martínez L. Infecciones causadas por bacterias gramnegativas multirresistentes: enterobacterias, Pseudomonas aeruginosa, Acinetobacter baumannii y otros bacilos gramnegativos no fermentadores. Enferm Infecc Microbiol Clin 2013;31:402-9. 10.1016/j.eimc.2013.03.016 [DOI] [PubMed] [Google Scholar]
- 15.Oteo J, Bou G, Chaves F, et al. Métodos microbiológicos para la vigilancia del estado de portador de bacterias multirresistentes. Enferm Infec Microbiol Clin 2017;35:667-75. 10.1016/j.eimc.2015.12.013 [DOI] [PubMed] [Google Scholar]
- 16.Glupczynski Y, Berhin C, Bauraing C, et al. Evaluation of a new selective chromogenic agar medium for detection of extended-spectrum β-lactamase-producing Enterobacteriaceae. J Clin Microbiol 2007;45:501-5. 10.1128/JCM.02221-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paniagua R, Valverde A, Coque TM, et al. Assessment of prevalence and changing epidemiology of extended-spectrum β-lactamase-producing Enterobacteriaceae fecal carriers using a chromogenic medium. Diagn Microbiol Infect Dis 2010;67:376-9. 10.1016/j.diagmicrobio.2010.03.012 [DOI] [PubMed] [Google Scholar]
- 18.Réglier-Poupet H, Naas T, Carrer A, et al. Performance of chromID ESBL, a chromogenic medium for detection of Enterobacteriaceae producing extended-spectrum β-lactamases. J Med Microbiol 2008;57:310-5. 10.1099/jmm.0.47625-0 [DOI] [PubMed] [Google Scholar]
- 19.del Castillo MC, López-Cerezo L, Casal M, et al. Evaluación del medio chromID ESBL para la detección de portadores de enterobacterias productoras de betalactamasas de espectro extendido. Enferm Infecc Microbiol Clin 2011;29:471-2. 10.1016/j.eimc.2011.01.011 [DOI] [PubMed] [Google Scholar]
- 20.Matheeussen V, Loens K, Scott C, et al. Quality of molecular detection of vancomycin resistance in enterococci: results of 6 consecutive years of Quality Control for Molecular Diagnostics (QCMD) external quality assessment. Eur J Clin Microbiol Infect Dis 2019;38:1633-41. 10.1007/s10096-019-03591-2 [DOI] [PubMed] [Google Scholar]
- 21.Jahansepas A, Ahangarzadeh Rezaee M, Hasani A, et al. Molecular epidemiology of vancomycin-resistant Enterococcus faecalis and Enterococcus faecium isolated from clinical specimens in the northwest of Iran. Microb Drug Resist 2018;24:1165-73. 10.1089/mdr.2017.0380 [DOI] [PubMed] [Google Scholar]
- 22.Bou G, Fernández-Olmos A, García C, et al. Métodos de identificación bacteriana en el laboratorio de microbiología. Enferm Infecc Microbiol Clin 2011;29:601-8. 10.1016/j.eimc.2011.03.012 [DOI] [PubMed] [Google Scholar]
- 23.Seral García C, Pardos De La Gándara M, Castillo García FJ. Betalactamasas de espectro extendido en enterobacterias distintas de Escherichia coli y Klebsiella. Enferm Infecc Microbiol Clin 2010;28:12-8. 10.1016/S0213-005X(10)70003-3 [DOI] [PubMed] [Google Scholar]
- 24.Jiménez-Guerra G, Hoyos-Mallecot Y, Rodríguez-Granger J, et al. Rapid test for detection of susceptibility to cefotaxime in Enterobacteriaceae. Rev Argent Microbiol 2016;48:320-4. [DOI] [PubMed] [Google Scholar]
- 25.Jiménez-Guerra G, Heras-Cañas V, Béjar Molina LDC, et al. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae from urinary tract infections: Evolution of antimicrobial resistance and treatment options. Med Clin (Barc) 2018;150:262-5. 10.1016/j.medcli.2017.07.023 [DOI] [PubMed] [Google Scholar]
- 26.Sánchez-García JM, Sorlózano-Puerto A, Navarro-Marí JM, et al. Evolution of the antibiotic-resistance of microorganisms causing urinary tract infections: A 4-year epidemiological surveillance study in a hospital population. Rev Clin Esp 2019;219:116-123. 10.1016/j.rce.2018.07.005 [DOI] [PubMed] [Google Scholar]
- 27.Baddour LM, Yu VL, Klugman KP, et al. Combination antibiotic therapy lowers mortality among severely ill patients with pneumococcal bacteremia. Am J Respir Crit Care Med 2004;170:440-4. 10.1164/rccm.200311-1578OC [DOI] [PubMed] [Google Scholar]
- 28.Band VI, Hufnagel DA, Jaggavarapu S, et al. Antibiotic combinations that exploit heteroresistance to multiple drugs effectively control infection. Nat Microbiol 2019;4:1627-35. 10.1038/s41564-019-0480-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chow JW, Yu VL. Combination antibiotic therapy versus monotherapy for gram-negative bacteraemia: a commentary. Int J Antimicrob Agents 1999;11:7-12. 10.1016/S0924-8579(98)00060-0 [DOI] [PubMed] [Google Scholar]
- 30.Gupta V, Datta P. Next-generation strategy for treating drug resistant bacteria: Antibiotic hybrids. Indian J Med Res 2019;149:97-106. 10.4103/ijmr.IJMR_755_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albalat R, Cañestro C. Evolution by gene loss. Nat Rev Genet 2016;17:379-91. 10.1038/nrg.2016.39 [DOI] [PubMed] [Google Scholar]
- 32.Araújo M, Santos C, Lages D. Carbapenem resistant Enterobacteriaceae – the basics for every medical specialty. Port J Nephrol Hypert 2019;33:176-81. [Google Scholar]
- 33.Gómez Álvarez CA, Leal Castro AL, Pérez de Gonzalez MJ, et al. Mecanismos de resistencia en "Pseudomonas aeruginosa": entendiendo a un peligroso enemigo. Rev Fac Med 2005;53:27-34. [Google Scholar]
- 34.Malathi K, Anbarasu A, Ramaiah S. Identification of potential inhibitors for Klebsiella pneumoniae carbapenemase-3: a molecular docking and dynamics study. J Biomol Struct Dyn 2019;37:4601-13. 10.1080/07391102.2018.1556737 [DOI] [PubMed] [Google Scholar]
- 35.Arnold RS, Thom KA, Sharma S, et al. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South Med J 2011;104:40-5. 10.1097/SMJ.0b013e3181fd7d5a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weisenberg SA, Morgan DJ, Espinal-Witter R, et al. Clinical outcomes of patients with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae after treatment with imipenem or meropenem. Diagn Microbiol Infect Dis 2009;64:233-5. 10.1016/j.diagmicrobio.2009.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suárez CJ, Kattan JN, Guzman AM, et al. Mecanismos de resistencia a carbapenems en P. aeruginosa, Acinetobacter y Enterobacteriaceae y estrategias para su prevención y control. Infectio 2006;10:85-93. [Google Scholar]
- 38.Rodríguez MC, Ruiz del Castillo B, Rodríguez-Mirones C, et al. Caracterización molecular de aislados clínicos de Pseudomonas aeruginosa productores de la metalobetalactamasa VIM-2 en Cantabria, España. Enferm Infecc Microbiol Clin 2010;28:99-103. 10.1016/j.eimc.2009.01.013 [DOI] [PubMed] [Google Scholar]
- 39.Shu L, Dong N, Lu J, et al. Emergence of OXA-232 carbapenemase-producing Klebsiella pneumoniae that carries a pLVPK-Like virulence plasmid among elderly patients in China. Antimicrob Agents Chemother 2019. doi: . 10.1128/AAC.02246-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cano ME, Domínguez MA, Ezpeleta C, et al. Cultivos de vigilancia epidemiológica de bacterias resistentes a los antimicrobianos de interés nosocomial. Enferm Infecc Microbiol Clin 2008;26:220-9. 10.1016/S0213-005X(08)72694-6 [DOI] [PubMed] [Google Scholar]
- 41.Tafur JD, Torres JA, Villegas MV. Mechanisms of antibiotic resistance in Gram negative bacteria. Infectio 2008;12:217-26. [Google Scholar]
- 42.Jiménez-Guerra G, Borrego-Jiménez J, Gutiérrez-Soto B, et al. Susceptibility evolution to antibiotics of Enterobacter cloacae, Morganella morganii, Klebsiella aerogenes and Citrobacter freundii involved in urinary tract infections: an 11-year epidemiological surveillance study. Enferm Infecc Microbiol Clin 2019. doi: . 10.1016/j.eimc.2019.07.010 [DOI] [PubMed] [Google Scholar]
- 43.Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev 2009;22:161-82. 10.1128/CMR.00036-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmieri M, Schicklin S, Pelegrin AC, et al. Phenotypic and genomic characterization of Ampc-producing Klebsiella pneumoniae from Korea. Ann Lab Med 2018;38:367. 10.3343/alm.2018.38.4.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lan NPH, Hien NH, Le Thi Phuong T, et al. Phenotypic and genotypic characteristics of ESBL and AmpC producing organisms associated with bacteraemia in Ho Chi Minh City, Vietnam. Antimicrob Resist Infect Control 2017;6:105. 10.1186/s13756-017-0265-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martínez Rojas DV. AmpC type betalactamases: Generalities and phenotype detection methods. Rev Soc Ven Microbiol 2009;29:78-83. [Google Scholar]
- 47.Pena Viña I. Enterobacterias productoras de carbapenemasas: tipos, epidemiología molecular y alternativas terapéuticas. Doctoral Thesis. Universidad Complutense de Madrid. 2016. Available online: https://eprints.ucm.es/38513/1/T37533.pdf
- 48.Lacueva Arnedo M. Resistencia a antibióticos en Staphylococcus aureus. Evolución y perspectiva actual. Grade Thesis. Universidad Complutense de Madrid. 2017. Available online: http://147.96.70.122/Web/TFG/TFG/Memoria/MANUEL%20LACUEVA%20ARNEDO.pdf
- 49.Ahumada SE, Otero RM, Noriega ER. Resistencia de los enterococos a los antimicrobianos: implicaciones clínicas. Enferm Infecc Microbiol 1999;19:227-35. [Google Scholar]
- 50.Casal MM, Causse M, Solís F, et al. Investigation of antimicrobial resistance to Enterococcus faecium. Rev Esp Quimioter 2012;25:180-2. [PubMed] [Google Scholar]
- 51.Cercenado E. Enterococcus: resistencias fenotípicas y genotípicas y epidemiología en España. Enferm Infecc Microbiol Clin 2011;29:59-65. 10.1016/S0213-005X(11)70045-3 [DOI] [PubMed] [Google Scholar]
- 52.Nguyen AH, Miller WR, Arias CA. Shape follows function: gastrointestinal signals for enterococcal colonization. Trends Mol Med 2019;25:464-6. 10.1016/j.molmed.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ponessa A, Gambandé T, All L, et al. Enterococos vancomicina resistentes: colonización en pacientes hospitalizados, en Rosario, Argentina. Acta Bioquímica Clínica Latinoamericana 2006;40:499-502. [Google Scholar]
- 54.Bayjanov JR, Baan J, Rogers MRC, et al. Enterococcus faecium genome dynamics during long-term asymptomatic patient gut colonization. Microb Genomics 2019. doi: 10.1099/mgen.0.000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chi C, Xue Y, Lv N, et al. Longitudinal gut bacterial colonization and its influencing factors of low birth weight infants during the first 3 months of life. Front Microbiol 2019;10:1105. 10.3389/fmicb.2019.01105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soria-Segarra C, González-Bustos P, López-Cerero L, et al. Tracking KPC-3-producing ST-258 Klebsiella pneumoniae outbreak in a third-level hospital in Granada (Andalusia, Spain) by risk factors and molecular characteristics. Mol Biol Rep 2020;47:1089-97. 10.1007/s11033-019-05203-w [DOI] [PubMed] [Google Scholar]
- 57.Dimitriou V, Biehl LM, Hamprecht A, et al. Controlling intestinal colonization of high-risk haematology patients with ESBL-producing Enterobacteriaceae: a randomized, placebo-controlled, multicentre, Phase II trial (CLEAR). J Antimicrob Chemother 2019;74:2065-74. 10.1093/jac/dkz124 [DOI] [PubMed] [Google Scholar]
- 58.Thom KA, Hsiao WWL, Harris AD, et al. Patients with Acinetobacter baumannii bloodstream infections are colonized in the gastrointestinal tract with identical strains. Am J Infect Control 2010;38:751-3. 10.1016/j.ajic.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gómez-Carcassés L, Pérez-Hernández L, Pujol-Enseñat Y, et al. Characterization of patients with Acinetobacter baumannii ventilator-associated pneumonia in progressive care units. Medisur 2016;14:[aprox. 14 p.]. [Google Scholar]
- 60.Vander H, Prabha V. Colonization of mouse vagina with Pseudomonas aeruginosa: A plausible explanation for infertility. Microb Pathog 2019;134:103602. 10.1016/j.micpath.2019.103602 [DOI] [PubMed] [Google Scholar]
- 61.Montero MM. Pseudomonas aeruginosa multiresistente: aspectos epidemiológicos, clínicos y terapéuticos. Doctoral Thesis. Universidad Autónoma de Barcelona; 2012. Available online: https://www.tdx.cat/handle/10803/107902
- 62.Garrós Garay J, Ruiz de Gordejuela E, García Cebrián F, et al. Pseudomonas aeruginosa infection-colonization in patients with bronchiectasias or COPD. Clinical features, microbiology and outcome. Gac Méd Bilbao 2002;99:63-8. [Google Scholar]
- 63.Lambiase A, Catania MR, del Pezzo M, et al. Achromobacter xylosoxidans respiratory tract infection in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis 2011;30:973-80. 10.1007/s10096-011-1182-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gartner S, Salcedo Posadas A, García Hernández G. Enfermedad respiratoria en la fibrosis quística. Protoc Diagn Ter Pediatr 2017;1:299-319. [Google Scholar]
- 65.Villasusa Páez I, Martínez Motas I, Álvarez García N, et al. Prevalencia de bacterias potencialmente patógenas en la nasofaringe de niños sanos de un círculo infantil de Ciudad de La Habana. Rev Cubana Med Trop 2006;58:181-9. [PubMed] [Google Scholar]
- 66.Fortaleza CR, Melo EC, Fortaleza CMCB. Nasopharyngeal colonization with methicillin-resistant Staphylococcus aureus and mortality among patients in an intensive care unit. Rev Lat Am Enfermagem 2009;17:677-82. 10.1590/S0104-11692009000500013 [DOI] [PubMed] [Google Scholar]
- 67.Fuentes Páez Y, Martínez Motas I, Sierra González G, et al. Nasopharyngeal colonization by potentially pathogenic bacteria found in healthy children from an elementary school. Rev Cubana Med Trop 2009;61:50-6. [Google Scholar]
- 68.Cervantes-García E, García-González R, María Salazar-Schettino P. Características generales del Staphylococcus aureus. Rev Mex Patol Clin Med Lab Lab 2014;61:28-40. [Google Scholar]
- 69.Apisarnthanarak A, Fraser VJ, Dunne WM, et al. Stenotrophomonas maltophilia intestinal colonization in hospitalized oncology patients with diarrhea. Clin Infect Dis 2003;37:1131-5. 10.1086/378297 [DOI] [PubMed] [Google Scholar]
- 70.Juliet LC, Fernández VA. Stenotrophomonas maltophilia. Rev Chilena Infectol 2006;23:247-8. 10.4067/S0716-10182006000300009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as