Abstract
Background
Lung cancer (LC) is one of the leading causes of cancer-related mortality in China and worldwide. Despite the progress in diagnosis and treatment of LC, the prognosis of LC remains poor. Studies have demonstrated that long non-coding RNAs (lncRNAs) play a critical role in carcinogenesis and cancer development.
Methods
Here we examined the expression and potential function of lnc-RAB11B-AS1 in LC both in vitro and in vivo. All experiments in this study were conducted using A549 and PC-9 cell lines according to protocols described in this paper. The clinic characteristics were analyzed using logistic regression, cox model, log rank test, biochemical analysis using qRT-PCR, transfections, nude mice model, and cell biological analysis using Transwell assay, CCK-8 assay, flow cytometry, and rescue experiments, and immunohistochemistry.
Results
The results showed that lnc-RAB11B-AS1 was significantly overexpressed in LC tissues compared to the corresponding non-tumor tissues. Patients with a higher level of lnc-RAB11B-AS1 expression showed a poorer overall survival rate. Functionally, overexpression of lnc-RAB11B-AS1 promotes cell proliferation, migration and invasion abilities of LC cell lines, which suggests lnc-RAB11B-AS1 may play an oncogenic role in LC. lnc-RAB11B-AS1 was located in physical contiguity with RAB11B gene and found positively regulates the RAB11B expression, and the protein levels of RAB11B in LC tissues also found to positively correlated with the level of lnc-RAB11B-AS1 expression. RAB11B silencing partially abrogated lnc-RAB11B-AS1-induced proliferation of the LC cell lines used in this study.
Conclusions
This study provided a novel evidence into the function of lncRNA-driven carcinogenesis. Our findings highlighted the importance of lnc-RAB11B-AS1 and RAB11B in LC progression and indicated that lnc-RAB11B-AS1 may serve as a novel and valuable prognostic biomarker for LC.
Keywords: lncRNA, lung cancer (LC), lnc-RAB11B-AS1, RAB11B, survival
Introduction
Lung cancer (LC) is one of the most common malignant respiratory tumors and remains a serious health threat (1). The morbidity and mortality rates of LC rank first in the spectrum of cancers and have kept increasing in the past few decades in China (2-4). The 5-year overall survival rate of LC patients is 16.1% in China (5), while in rural areas is only 11.2% (6). The main reason for the poor survival rate of LC patients is that most LC patients were diagnosed at an advanced stage, and a large number of the LC patients were sustained malignant proliferation and extensive lymphatic metastasis. Although studies have identified tons of genes that are involved in LC tumorigenesis and tumor metastasis, the molecular mechanisms underlying the tumorigenesis and metastasis are not yet well understood. Therefore, a detailed understanding of the relevant mechanisms and molecular pathways of activated lncRNA in LC is crucial to discovery of new anti-cancer therapeutic targets.
Long non-coding RNAs (lncRNAs) are a sub-class of non-coding RNAs over 200 nucleotides in length and lack of protein coding capacity. LncRNAs played crucial roles in multiple biological processes through regulating genes expression of proteins involved in various processes, such as carcinogenesis, cell proliferation, migration and invasion (7). LncRNAs are divided into five types: (I) bidirectional; (II) intergenic; (III) intronic; (IV) sense; and (V) anti-sense (anti-sense lncRNAs, as-lncRNAs) (8). Previous studies have demonstrated that as-lncRNAs are functional because the special location with its natural antis-sense transcripts. In addition, recent studies have demonstrated a link between as-lncRNAs and cancer progression. For example, ZEB1-AS1 (7) was reported to promote tumor metastasis and its overexpression predicted poor prognosis in hepatocellular carcinoma (HCC). KRT7-AS (9) was shown to promote cancer cell progression in gastric cancer (GC), and another study showed that HNF1A-AS1 (10) functions as a competing endogenous RNA in colon cancer. These reports have demonstrated the involvement of as-lncRNAs in different cancers and their potential as biomarkers for the early detection, diagnosis and treatment of cancer. Recent studies showed that upstream anti-sense transcripts of as-lncRNAs played a critical rule in transcriptional regulation of corresponding gene expression (11). Sequence analysis showed that most as-lncRNAs originate from the promoters of the corresponding mRNAs in a head-to-head conformation. Thus, there seems to be an obvious potential to investigate these as-lncRNAs as an approach to study the well-known tumor-suppressors or oncogenes with a natural anti-sense transcript.
The RAS superfamily was first reported as oncogenes in mice by Jenifer Harvey in 1960s (12) and to date, over 150 genes of the RAS super-family have been identified. The RAS superfamily proteins are divided into five sub-classes: Ras, Rho, Ran, Arf and Rab (13). Approximately 60 of Rab proteins have been identified in the human genome (14). We previously found that RAB11B upregulated in osteosarcoma and negatively correlated with the expression level of the corresponding natural anti-sense transcript lnc-RAB11B-AS1 (15). We found that lnc-RAB11B-AS1 functioned as a tumor suppressor in osteosarcoma. However, another study conducted by Feng et al. (16) found that lnc-RAB11B-AS1 was upregulated in GC and the overexpression was correlated with clinical stage, metastasis and overall survival of the GC patients. A recent research reported that upregulation of lnc-RAB11B-AS1 could enhance the ability of cell migration and invasion in breast cancer cell lines both in vitro and in vivo via hypoxia-inducible factor 2 (HIF-2) (17) Lnc-RAB11B-AS1 is a 1022-bp transcript with 3 exons and located on human chromosome 19q13.2 (chr19: 8,439,260-8,455,575, hg19) on the reverse strand, a region that has been associated with high risk for several cancers, such as ovarian carcinoma (18), prostate cancer (19), pancreatic cancer (20), neuroblastoma (21) and chronic obstructive pulmonary disease (22,23). Although these studies have indicated a link between lnc-RAB11B-AS1 and cancer, the biological functions of lnc-RAB11B-AS1 in LC remained to be clarified. Furthermore, recent studies showed that the expression level of an mRNA correlated with the level of the corresponding anti-sense transcript (11). We therefore speculated whether lnc-RAB11B-AS1 regulates RAB11B expression, promotes LC progress and worsening LC prognosis.
In this study, we investigated the expression pattern and clinical significance of lnc-RAB11B-AS1 in LC patients and examined the functions of lnc-RAB11B-AS1 in LC cell lines. We also examined the potential function of lnc-RAB11B-AS1 in regulating RAB11B expression in LC.
Methods
Study subjects
All the LC patients involved in the present study were Han Chinese people from Southern and Eastern China. A total of 276 paired samples of LC tissues and paired normal tissues were used in the present study, 182 of which were collected from the Affiliated Hospitals of Guangzhou Medical University, the First Affiliated Hospital affiliated with Kunming University and Cancer Hospital affiliated with Kunming University between 2008 and 2015, and the rest of the samples were collected from the First Affiliate Hospital of Soochow University between 2007 and 2016. The LC patients in the study had no genetic connections with one another. The present study was approved by the Ethics Committee of Guangzhou Medical University (No. GMU201481473040) and we strictly followed the related clinical research guidelines. All study participants involved in the present study were provided written informed consent.
Cell culture
Human lung adenocarcinoma cell lines A549 and PC-9 and human embryonic kidney cell line 293 (HEK-293) were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Science (Shanghai Institute of Cell Biology, China). A549 and PC-9 cell lines were cultured in RPMI-1640 medium (Gibco, Life Technologies, USA) and HEK-293 cell line was cultured in DMEM medium (Gibco). All cell lines were grown in 10% (volume ratio) fetal bovine serum (FBS)-containing culture medium and all cell lines were cultured in a humidified atmosphere containing 5% CO2 at 37 °C.
qRT-PCR analysis
Total RNA was extracted from LC tissues and cell line samples using Trizol Reagent (Life Technologies) according to the manufacturer’s instructions. RIN (RNA integrity number) was determined to detect RNA integrity using an Agilent Bioanalyzer 2100 (Agilent Technologies, CA, USA). Lnc-RAB11B-AS1 and RAB11B expression levels were detected in all tissue cDNA (complementary DNA) samples using quantitative real-time polymerase chain reaction (qRT-PCR). ACTB expression level was used as an internal control of the qRT-PCR assay. All primers used are listed in Table S1. All qRT-PCR analyses were performed using ABI 7900HT system (Applied Biosystems, CA, USA), and the master mix of the qRT-PCR assay was also purchased from Applied Biosystems (ABI Power SYBR Green PCR Master Mix). The relative expression levels of the genes involved in this study were calculated using the comparative threshold cycle (Ct) (2-ΔΔCt) method.
Table S1. Primers used in this study.
| Gene | Forward primer (5'->3') | Reverse primer (5'->3') |
|---|---|---|
| lnc-RAB11B-AS1 | GGAACATGTTTACATGGACTTTGT | TCTTTGTTCTTGTTTGTTTTCTTTCT |
| RAB11B | CGTACTACCGTGGTGCAGTG | ATGACGATGTTGCTGTCTGC |
| RAB11A | TCTCAGGGCAGTTCCTACAGA | ACCACATTGTTGCTTGGAGAC |
| RAB25 | GACTGCTCTTCCTGGAGACCT | TCTGTCTCTGCTTGGACACCT |
| ACTB | GGCGGCACCACCATGTACCCT | AGGGGCCGGACTCGTCATACT |
Subcellular fractionation
Nuclear and cytosolic fractions used in this study were extracted from LC cell lines using a nuclear/cytoplasmic isolation kit (Biovision, San Francisco, CA, USA) according to the manufacturer’s protocol.
Plasmids, cell transfection and stable cell lines
The full-length complimentary DNA of human lnc-RAB11B-AS1 and small hairpin RNA (shRNA) targeting lnc-RAB11B-AS1 were both synthesized by GeneCopoeia (MD, USA). The sequence of the full-length lnc-RAB11B-AS1 complimentary DNA was cloned into lentivirus expression vector pEZ-Lv206, and the shRNA targeting lnc-RAB11B-AS1 was cloned in to the lentivirus expression vector psi-LVRH1MP. Both vectors contained a gene encoding red fluorescence protein, which could be detected by an inverted fluorescence microscope to determine transfection efficiency. The resulting constructs were verified by sequencing. Stably transfected LC cell lines were established according to the protocol of GeneCopoeia, and the transfection reagents (Lenti-Pac HIV Expression Packaging Kit) were also purchased from GeneCopoeia. All stable cell lines were verified by both fluorescence and qRT-PCR.
Cell proliferation assay
The A549 and PC-9 cell lines with stably overexpressed and down-expressed lnc-RAB11B-AS1 were seeded in 96-well plates (200 cells per well), and the proliferation of the seeded cells were examined using the Cell Counting Kit-8 (CCK-8) (Engreen Biosystem Co. Ltd., China) according to the manufacturer’s protocol. The absorbance of the cell lines was measured at 450 nm at a series of time points (0, 24, 48, 72 and 96 hours after the cells were seeded).
Flow cytometry assays
For cell cytometry assays, A549 and PC-9 cell lines with stably overexpressed and down-expressed lnc-RAB11B-AS1 were labeled with propidium iodide (PI) (7seaharmtech, Shanghai, China) and analyzed using flow cytometry. For apoptosis assays, Annexin V-fluorescein isothiocyanate (FITC)/PI (Multisciences, Hangzhou, China) staining was performed and the apoptosis of the cells were examined by flow cytometry according to the manufacturer’s protocol.
Clonogenic assay
Stably transfected cell lines were plated into 6-well plates in triplicate at a density of 100 cells/plate and regularly cultured in routine conditions for 2 weeks. The colonies were stained with Giemsa (Beijing Solarbio Science & Technology Co. Ltd., China) and then counted (the colonies contain more than 50 cells) and imaged. The clonogenic assay was conducted in triplicate.
Cell migration and invasion assays
Transwell assays were conducted in an 8 µm Trans-well chamber (Costar, Corning Incorporated, NY, USA). A549 and PC-9 cell lines with stably overexpressed and down-expressed lnc-RAB11B-AS1 were trypsinized and re-suspended by FBS-free culture medium with 0.1% bovine serum albumin (BSA). Cells (2×104) were seeded into each chamber, which was placed into a 24-well plate containing culture medium with 20% FBS (Gibco). After 48 hours culture, non-migrated cells in the upper section of the chamber were removed lightly. Migrated cells number were manually counted in 10 random fields by a microscope, and the migrated cells numbers were averaged.
Cell invasion assays were conducted using chambers with Matrigel (Corning), and assays were performed as described for migration assays.
All experiments were conducted in triplicate.
Xenograft model in nude mice
BALB/c-nu nude mice (6-week-old) were purchased from Nanjing Biomedical Research Institute of Nanjing University (Nanjing, China). Stably transfected A549 and PC-9 cell lines were re-suspended at a concentration of 1×107 cells/mL in phosphate buffer (PBS). A total of 54 mice were randomly divided into 9 groups (three male mice and three female mice in each group) as follows: A549 lnc-RAB11B-AS1 overexpression & Control, A549 lnc-RAB11B-AS1 silenced & Control, PC-9 lnc-RAB11B-AS1 overexpression & Control, PC-9 lnc-RAB11B-AS1 silenced & Control, and a group of mice were injected with PBS only as a control for the whole experiments (data of this group was not shown). Each mouse was subcutaneously injected in the back flank with 0.2 mL of the indicated cell suspension (a total of 2×106 cells). Tumor growth was monitored over 21 days, and we measured the tumor length and width and obtained imaging every 3 days. The volume of the tumor was calculated as follows: Vtumor = length × width × width ×0.5. This study was approved by the Ethics Committee of Guangzhou Medical University (No. GZYDW201503512). All animal experiments were conducted according to the relevant regulations.
Immunohistochemistry (IHC)
Immunohistochemistry assay was used to detect the RAB11B protein level in the cancer patients’ samples, we selected 2 pairs of tissues with a high expression level of lnc-RAB11B-AS1, which were confirmed by qRT-PCR experiment. The experiment was conducted at a routine procedure. The tissue samples used was formalin-fixed, were embedded in paraffin and sliced. The sections were then incubated with monoclonal antibodies against RAB11B (1:500, ab228954, abcam, USA) overnight at 4 °C, and then incubated with a secondary antibody (1:500, ab150117, abcam), the 3,3’-diaminobenzidine (DAB) was used to be the chromogen. Hematoxylin was used to be the dying material. Finally, the RAB11B protein level was evaluated.
Luciferase assays
pGL3-RAB11B vector and pGL3-Control vector were co-transfected with lnc-RAB11B-AS1 overexpression and lnc-RAB11B-AS1 downregulation using Lipofectamine 3000 (Invitrogen) into A549 and PC-9 cell lines. The relative luciferase activity of each group was normalized by Renilla luciferase activity 48 h after transfection.
Statistical analysis
Statistical analyses were performed using SPSS 19.0. The differences between groups was calculated by χ2 test. One-way analysis of variance test was used to examine the influence of lnc-RAB11B-AS1 expression on RAB11B. Overall survival rates of different groups were calculated by the Kaplan-Meier survival analysis and the log-rank test was applied for comparison. P values less than 0.05 were considered significant.
Results
Expression pattern of lnc-RAB11B-AS1 in LC
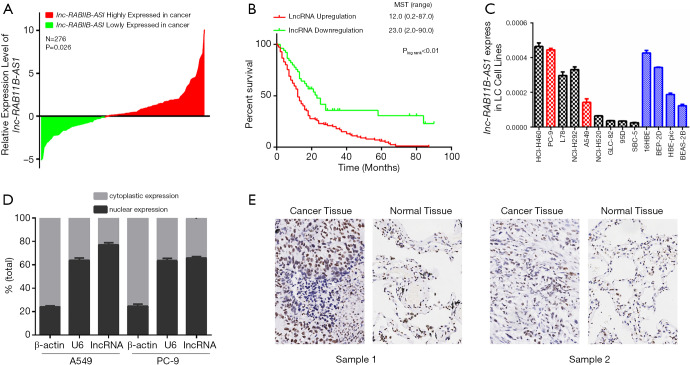
The clinical characteristics of the 276 patients with LC included in this study are shown in Table S2 [this data was published in one of our previous studies which was a study based on the same study subjects (24)]. As shown in Figure 1A, lnc-RAB11B-AS1 was significantly upregulated in cancerous tissues from the LC patients compared with corresponding normal tissues, with an average 1.47-fold increased expression (P=0.026).
Table S2. Demographics and clinical characteristics of studied samples.
| Characteristics | Southern samples, N (%) | Eastern samples, N (%) | Pearson χ2 | P |
|---|---|---|---|---|
| Total | 182 (65.9) | 94 (34.1) | ||
| Age(years) | ||||
| <60 | 105 (57.7) | 51 (54.3) | 0.298 | 0.585 |
| ≥60 | 77 (42.3) | 43 (45.7) | ||
| Sex | ||||
| Female | 52 (28.6) | 28 (29.8) | 0.045 | 0.8329 |
| Male | 130 (71.4) | 66 (70.2) | ||
| Family history of cancer | ||||
| No | 161 (88.5) | 84 (89.4) | 0.050 | 0.822 |
| Yes | 21 (11.5) | 10 (10.6) | ||
| Smoking | ||||
| No | 67 (36.8) | 30 (31.9) | 0.653 | 0.419 |
| Yes | 115 (63.2) | 64 (68.1) | ||
| Drinking | ||||
| No | 137 (75.3) | 73 (77.7) | 0.194 | 0.660 |
| Yes | 45 (24.7) | 21 (23.3) | ||
| Gold stage | ||||
| I + II | 71 (39.0) | 29 (30.1) | 2.287 | 0.515 |
| III + IV | 111 (61.0) | 65 (69.9) | ||
| Histological types | ||||
| Adenocarcinoma | 86 (47.3) | 41 (43.6) | 1.681 | 0.794 |
| Squamous cell carcinoma | 51 (28.0) | 31 (33.0) | ||
| Large cell carcinoma | 6 (3.3) | 5 (5.3) | ||
| Small cell lung cancer | 19 (10.4) | 8 (8.5) | ||
| Other carcinomas* | 20 (11.0) | 9 (9.6) |
*, mixed-cell or undifferentiated carcinoma.
Figure 1.
Expression pattern of lnc-RAB11B-AS1 and its association with lung cancer survival. (A) Expression pattern of lnc-RAB11B-AS1 in lung cancer tissues. The expression level of lnc-RAB11B-AS1 was significantly higher in lung cancerous tissues compared with adjacent normal lung tissues. Results are shown as Log (tumor/normal). P was calculated by the paired t-test. (B) Effect of lnc-RAB11B-AS1 expression on lung cancer survival. Patients with high expression of lnc-RAB11B-AS1 showed a significantly poorer survival than those with low expression of lnc-RAB11B-AS1. P was calculated by the log-rank test. (C) Relative expression of lnc-RAB11B-AS1 in nine lung cancer cell lines and three normal pulmonary epithelial cell lines. (D) Expression of lnc-RAB11B-AS1 in nuclear and cytoplasmic fractions of LC cells. (E) Expression of RAB11B protein in lung cancer tissues.
Associations between lnc-RAB11B-AS1 expression and LC progression
We next categorized LC patients based on lnc-RAB11B-AS1 expression using the medium level in LC tissues. Patients with expression levels equal to or greater than the cut-off value were categorized in the high lnc-RAB11B-AS1 expression group, and those with expression levels lower than the cut-off value were defined as the low lnc-RAB11B-AS1 expression group. The expression status of lnc-RAB11B-AS1 was statistically different in patients at different clinic stages (P<0.001), with lymph node metastasis (N stage) (P<0.001) and with distant metastasis (M stage) (P<0.001) (Table 1). These results showed that the LC patients with a higher level of lnc-RAB11B-AS1 expression had a higher risk for advanced clinic stage, N stage and M stage.
Table 1. Association between the expression level of lnc-RAB11B-AS1 and the clinic characteristics of the patients.
| Clinic characteristics | Southern samples, N (%) | Pa | Eastern samples, N (%) | Pa | Merged samples, N (%) | Pa | |||
|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | Low | High | ||||
| Age | |||||||||
| <60 | 38 (36.2) | 67 (63.8) | 0.462 | 21 (41.2) | 30 (58.8) | 0.233 | 59 (37.8) | 97 (62.2) | 0.180 |
| ≥60 | 32 (41.6) | 45 (58.4) | 23 (53.5) | 20 (46.5) | 55 (45.8) | 65 (54.2) | |||
| Sex | |||||||||
| Female | 23 (44.2) | 29 (55.8) | 0.312 | 15 (53.6) | 13 (46.4) | 0.392 | 38 (47.5) | 42 (52.5) | 0.182 |
| Male | 47 (36.2) | 83 (63.8) | 29 (43.9) | 37 (56.1) | 76 (38.8) | 120 (61.2) | |||
| Family history of cancer | |||||||||
| No | 92 (38.5) | 99 (61.5) | 0.971 | 40 (47.6) | 44 (52.4) | 0.648 | 102 (41.6) | 143 (58.4) | 0.755 |
| Yes | 8 (38.1) | 13 (61.9) | 4 (40.0) | 6 (60.0) | 12 (38.7) | 19 (61.3) | |||
| Family history of lung cancer | |||||||||
| No | 64 (37.9) | 105 (62.1) | 0.554 | 44 (48.4) | 47 (51.6) | 0.099 | 108 (41.5) | 152 (58.5) | 0.750 |
| Yes | 6 (46.2) | 7 (53.8) | 0 (0) | 3 (100.0) | 6 (37.5) | 10 (62.5) | |||
| Smoking | |||||||||
| No | 32 (47.8) | 35 (52.2) | 0.049 | 15 (50.0) | 15 (50.0) | 0.671 | 47 (48.5) | 50 (51.5) | 0.076 |
| Yes | 38 (33.0) | 77 (67.0) | 29 (45.3) | 35 (54.7) | 67 (37.4) | 112 (62.6) | |||
| Drinking | |||||||||
| No | 53 (38.7) | 84 (61.3) | 0.913 | 38 (52.1) | 35 (47.9) | 0.057 | 91 (43.3) | 119 (56.7) | 0.222 |
| Yes | 17 (37.8) | 28 (62.2) | 6 (28.6) | 15 (71.4) | 23 (34.8) | 43 (65.2) | |||
| Surgery | |||||||||
| No | 46 (38.0) | 75 (62.0) | 0.862 | 28 (48.3) | 30 (51.7) | 0.717 | 74 (41.3) | 105 (58.7) | 0.987 |
| Yes | 24 (39.3) | 37 (60.7) | 16 (44.4) | 20 (55.6) | 40 (41.2) | 57 (58.8) | |||
| Chemotherapy | |||||||||
| No | 21 (33.3) | 42 (66.7) | 0.301 | 10 (34.5) | 19 (65.5) | 0.110 | 31 (33.7) | 61 (66.3) | 0.069 |
| Yes | 49 (41.2) | 70 (58.8) | 34 (52.3) | 31 (47.7) | 83 (45.1) | 101 (54.9) | |||
| Radiotherapy | |||||||||
| No | 43 (41.3) | 61 (58.7) | 0.356 | 18 (37.5) | 30 (62.5) | 0.065 | 61 (40.1) | 91 (59.9) | 0.661 |
| Yes | 27 (34.6) | 51 (65.4) | 26 (56.5) | 20 (43.5) | 53 (42.7) | 71 (57.3) | |||
| Clinical stages | |||||||||
| I + II | 44 (62.0) | 27 (38.0) | <0.001 | 20 (69.0) | 9 (31.0) | 0.004 | 64 (64.0) | 36 (36.0) | <0.001 |
| III + IV | 26 (23.4) | 85 (61.5) | 24 (36.9) | 41 (63.1) | 60 (34.1) | 116 (65.9) | |||
| T | |||||||||
| 1+2 | 43 (41.4) | 61 (58.6) | 0.356 | 23 (51.1) | 22 (48.9) | 0.423 | 66 (44.3) | 83 (55.7) | 0.274 |
| 3+4 | 27 (34.6) | 51 (65.4) | 21 (42.9) | 28 (57.1) | 48 (37.8) | 79 (62.2) | |||
| N | |||||||||
| 0 | 34 (51.5) | 32 (48.5) | 0.006 | 27 (62.8) | 16 (37.2) | 0.004 | 61 (56.0) | 48 (44.0) | <0.001 |
| 1+2+3 | 36 (31.0) | 80 (69.0) | 17 (33.3) | 34 (66.7) | 53 (31.7) | 114 (68.3) | |||
| M | |||||||||
| 0 | 62 (48.1) | 67 (51.9) | <0.001 | 33 (55.0) | 27 (45.0) | 0.034 | 95 (50.3) | 94 (49.7) | <0.001 |
| 1 | 8 (15.1) | 45 (84.9) | 11 (32.4) | 23 (67.6) | 19 (21.8) | 68 (78.2) | |||
| Histological types | |||||||||
| Adenocarcinoma | 37 (43.0) | 49 (57.0) | 0.325 | 20 (48.8) | 21 (51.2) | 0.471 | 57 (44.9) | 70 (55.1) | 0.747 |
| Squamous cell carcinoma | 21 (41.2) | 30 (58.8) | 12 (38.7) | 19 (61.3) | 33 (40.2) | 49 (59.8) | |||
| Large cell carcinoma | 1 (16.7) | 5 (83.3) | 2 (40.0) | 3 (60.0) | 3 (27.3) | 8 (72.7) | |||
| Small cell lung cancer | 4 (21.1) | 15 (78.9) | 6 (75.0) | 2 (25.0) | 10 (37.0) | 17 (63.0) | |||
| Other carcinomasb | 7 (35.0) | 13 (65.0) | 4 (44.4) | 5 (55.6) | 11 (37.9) | 18 (62.1) | |||
a, Pearson χ2 test P value; b, mixed-cell or undifferentiated carcinoma.
Associations between the lnc-RAB11B-AS1 expression and LC prognosis
We next analyzed the effect of lnc-RAB11B-AS1 expression on LC survival. We obtained complete follow-up data from 163 patients (110, 60.4% in southern population, 53, 56.4% in eastern population) [listed in Table S3, and this data was published in one of our previous studies which was a study based on the same study subjects (24)]. We conducted Kaplan-Meier survival analysis and log-rank test, and found that the LC patients with a higher level of lnc-RAB11B-AS1 expression experienced a significantly shorter medium survival time than the ones with a lower lnc-RAB11B-AS1 expression level (12.0 vs. 23.0 months, Plog-rank <0.01; Figure 1B).
Table S3. Association between the clinic characteristics and the prognosis of the lung cancer patients.
| Variables | Southern samples | Eastern samples | Merged samples | MST (month) | log-rank P value | Crude HR (95% CI) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case, N | Death, N (%) | Case, N | Death, N (%) | Case, N | Death, N (%) | ||||||
| Total | 110 | 88 (80%) | 53 | 45 (84.9) | 163 | 133 (81.6) | |||||
| Age | |||||||||||
| <60 | 64 | 50 (78.1) | 34 | 27 (79.4) | 98 | 77 (78.6) | 24.5 | 1.00 (Ref.) | |||
| ≥60 | 46 | 38 (82.6) | 19 | 18 (94.7) | 65 | 56 (86.2) | 22.5 | 0.575 | 1.10 (0.78–1.56) | ||
| Sex | |||||||||||
| Female | 38 | 29 (76.3) | 16 | 13 (81.3) | 54 | 42 (77.8) | 26.8 | 1.00 (Ref.) | |||
| Male | 72 | 59 (81.9) | 37 | 32 (86.5) | 109 | 91 (83.5) | 22.2 | 0.345 | 0.84 (0.58–1.21) | ||
| Family history of cancer | |||||||||||
| No | 100 | 79 (79.0) | 45 | 38 (84.4) | 145 | 117 (80.7) | 24.4 | 1.00 (Ref.) | |||
| Yes | 10 | 9 (90.0) | 8 | 7 (87.5) | 18 | 16 (88.9) | 19.0 | 0.353 | 1.28 (0.76–2.17) | ||
| Smoking | |||||||||||
| No | 44 | 32 (72.7) | 20 | 15 (75.0) | 64 | 47 (73.5) | 29.4 | 1.00 (Ref.) | |||
| Yes | 66 | 56 (84.9) | 33 | 30 (90.9) | 99 | 86 (86.9) | 20.0 | 0.015 | 1.56 (1.09–2.22) | ||
| Drinking | |||||||||||
| No | 83 | 67 (80.7) | 40 | 34 (85.0) | 123 | 101 (82.1) | 24.2 | 1.00 (Ref.) | |||
| Yes | 27 | 21 (77.8) | 13 | 11 (84.6) | 40 | 32 (80.0) | 21.3 | 0.600 | 1.11 (0.75–1.66) | ||
| Stage | |||||||||||
| I + II | 42 | 31 (73.8) | 13 | 10 (76.9) | 55 | 41 (74.5) | 32.2 | 1.00 (Ref.) | |||
| III + IV | 68 | 57 (83.8) | 40 | 35 (87.5) | 108 | 92 (85.2) | 19.1 | 0.004 | 1.73 (1.19–2.52) | ||
| Histological types | |||||||||||
| Adenocarcinoma | 56 | 44 (78.6) | 20 | 16 (80.0) | 76 | 60 (78.9) | 29.0 | 1.00 (Ref.) | |||
| Squamous cell carcinoma | 30 | 25 (83.3) | 18 | 20 (90.0) | 48 | 45 (93.8) | 19.7 | 0.114 | 1.38 (0.93–2.04) | ||
| Large cell carcinoma | 2 | 2 (100.0) | 3 | 3 (100.0) | 5 | 5 (100.0) | 12.6 | 0.070 | 2.34 (0.93–5.86) | ||
| Small cell lung cancer | 11 | 9 (81.8) | 4 | 2 (50.0) | 15 | 11 (73.3) | 12.9 | 0.095 | 1.76 (0.91–3.40) | ||
| Other carcinomasa | 11 | 8 (72.7) | 6 | 6 (100.0) | 17 | 14 (93.3) | 15.6 | 0.083 | 1.68 (0.93–3.00) | ||
a, mixed-cell or undifferentiated carcinoma. MST, median survival time; HR, hazard ratio; ref, reference.
We conducted a COX regression for multivariate analysis and found that the chemotherapy (HR =0.67; 95% CI, 0.45–0.99, P=0.048) and high lnc-RAB11B-AS1 expression (HR =1.76; 95% CI, 1.09–2.85, P=0.020) were statistically associated with the overall survival of the patients (Table 2). We further analyzed the merged samples in a stratification way; the results are shown in Table 3.
Table 2. COX model of the significant variables.
| Variables | HR (95% CI) | Cox model P value |
|---|---|---|
| Age | 1.43 (1.00–2.04) | 0.053 |
| Sex | 1.28 (0.80–2.04) | 0.311 |
| Smoking | 1.58 (1.00–2.50) | 0.051 |
| surgery | 0.71 (0.47–1.08) | 0.108 |
| Chemotherapy | 0.67 (0.45–0.99) | 0.048 |
| Radiotherapy | 0.70 (0.49–1.02) | 0.060 |
| Stage | 1.17 (0.95–1.44) | 0.136 |
| lnc-RAB11B-AS1 over-expression | 1.76 (1.09–2.85) | 0.020 |
Table 3. Association between the expression level of lnc-RAB11B-AS1 and the prognosis of the lung cancer patients.
| Variables | lnc-RAB11B-AS1 lowly expressed | lnc-RAB11B-AS1 highly expressed | Adjusted HR (95% CI)a | Phomob | Pinterc | |||
|---|---|---|---|---|---|---|---|---|
| Case N | Death, N (%) | Case, N | Death, N (%) | |||||
| Age | ||||||||
| <60 | 28 | 13 (46.4) | 70 | 64 (91.4) | 2.58 (1.42–4.71) | 0.361 | 0.362 | |
| ≥60 | 23 | 16 (69.6) | 42 | 40 (95.2) | 1.74 (0.96–3.16) | |||
| Sex | ||||||||
| Female | 25 | 16 (64.0) | 29 | 26 (89.7) | 2.02 (1.12–3.65) | 0.742 | 0.712 | |
| Male | 26 | 13 (50.0) | 83 | 78 (94.0) | 2.34 (1.23–4.46) | |||
| Family history of cancer | ||||||||
| No | 45 | 24 (53.3) | 100 | 93 (93.0) | 2.40 (1.52–3.78) | 0.227 | 0.112 | |
| Yes | 6 | 5 (83.3) | 12 | 11 (91.7) | 1.15 (0.38–3.46) | |||
| Family history of lung cancer | ||||||||
| No | 49 | 27 (55.1) | 108 | 100 (92.3) | 2.20 (1.43–3.38) | 0.417 | 0.760 | |
| Yes | 2 | 2 (100.0) | 4 | 4 (100.0) | 65.32 (0.02-251377) | |||
| Smoking | ||||||||
| No | 30 | 17 (56.7) | 34 | 30 (88.2) | 2.45 (1.32–4.54) | 0.373 | 0.381 | |
| Yes | 21 | 12 (59.8) | 78 | 74 (94.9) | 1.63 (0.82–3.01) | |||
| Drinking | ||||||||
| No | 43 | 28 (65.1) | 80 | 73 (91.3) | 1.80 (1.16–2.80) | 0.092 | 0.083 | |
| Yes | 8 | 1 (12.5) | 32 | 31 (96.9) | 10.44 (1.42–76.80) | |||
| Stage | ||||||||
| I + II | 32 | 18 (56.3) | 23 | 23 (100.0) | 1.91 (0.86–4.27) | 0.805 | 0.776 | |
| III + IV | 19 | 11 (57.9) | 89 | 81 (91.0) | 2.15 (1.31–3.53) | |||
| Surgery | ||||||||
| No | 35 | 21 (60.0) | 75 | 70 (93.3) | 1.88 (1.15–3.08) | 0.327 | 0.416 | |
| Yes | 16 | 8 (50.0) | 37 | 34 (91.9) | 3.05 (1.33–7.02) | |||
| Chemotherapy | ||||||||
| No | 12 | 10 (83.3) | 40 | 36 (90.0) | 1.20 (0.59–2.47) | 0.094 | 0.100 | |
| Yes | 39 | 19 (48.7) | 72 | 68 (94.4) | 2.55 (1.52–4.27) | |||
| Radiotherapy | ||||||||
| No | 28 | 18 (64.3) | 64 | 62 (96.9) | 2.10 (1.23–3.57) | 0.797 | 0.826 | |
| Yes | 23 | 11 (47.8) | 48 | 42 (87.5) | 2.35 (1.20–4.61) | |||
| Histological types | ||||||||
| Adenocarcinoma | 26 | 14 (53.8) | 50 | 46 (92.0) | 1.98 (1.08–3.62) | 0.967 | 0.411 | |
| Squamous cell carcinoma | 16 | 10 (62.5) | 34 | 33 (97.1) | 2.20 (1.08–4.50) | |||
| Large cell carcinoma | 1 | 1 (100.0) | 4 | 4 (100.0) | – | |||
| Small cell lung cancer | 4 | 2 (50.0) | 11 | 9 (81.2) | 2.80 (0.59–13.31) | |||
| Other carcinomasd | 4 | 2 (50.0) | 13 | 12 (92.3) | 2.66 (0.58–12.15) | |||
a, HRs were adjusted for age, sex, stage and metastasis in a Cox regression model; b, P value of homogeneity test between strata for the related HRs of the lowly expressed and highly expressed lnc-RAB11B-AS1; c, P value of test for the multiplicative interaction between the expression level of lnc-RAB11B-AS1 and selected variables on cancer death in Cox regression models; d, mixed-cell or undifferentiated carcinoma.
Expression levels of lnc-RAB11B-AS1 in LC cell lines
We next examined lnc-RAB11B-AS1 expression patterns in 9 LC cell lines and 4 normal lung epithelial cell lines and found varying levels of expression (Figure 1C). PC-9 and A549 cells showed relatively high lnc-RAB11B-AS1 expression and thus we selected these cell lines for further evaluation. In both cell lines. lnc-RAB11B-AS1 was mostly localized in the nucleus (Figure 1D), which supports the possibility of its role as a transcriptional regulator in LC development.
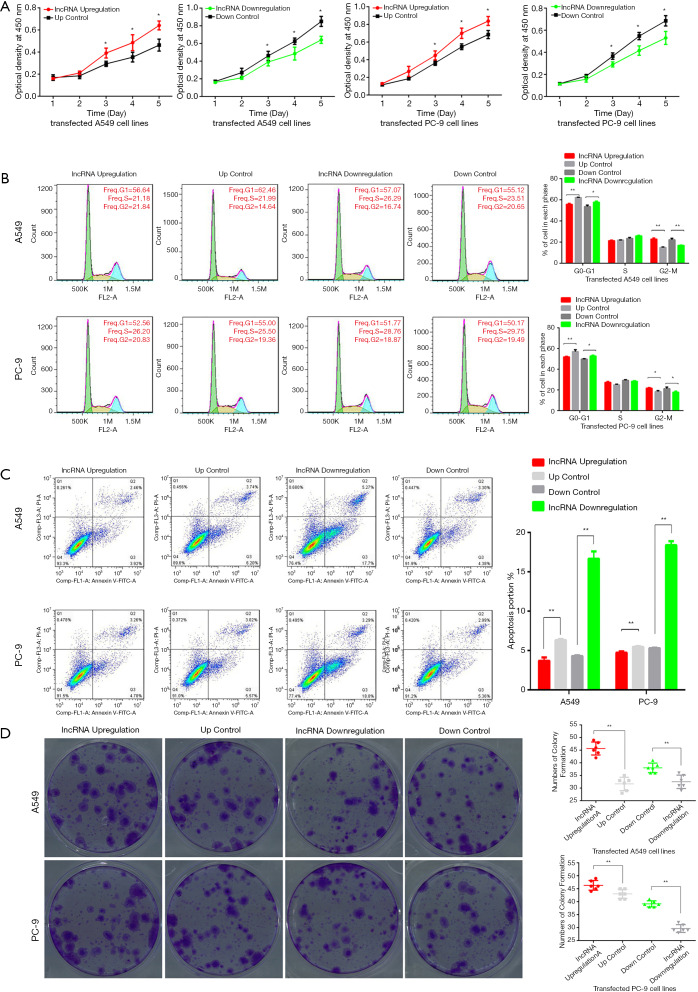
lnc-RAB11B-AS1 overexpression promotes proliferation ability and decreases apoptosis of LC cells in vitro
We next conducted CCK-8 assays to assess the influence of lnc-RAB11B-AS1 on LC cell proliferation. We generated PC-9 and A549 cell lines stably expressing a lnc-RAB11B-AS1 overexpression construct or shRNA targeting lnc-RAB-AS1 as described in Methods. CCK-8 results showed that lnc-RAB11B-AS1 upregulation significantly increased cell proliferation rates in vitro, while lnc-RAB11B-AS1 silencing resulted in reduced cell proliferation rates in both cell lines (P<0.05; Figure 2A). We further examined the effect of lnc-RAB11B-AS1 expression on the cell cycle and found that overexpression of lnc-RAB11B-AS1 resulted in fewer cells in G1 and more cells in M phase, while cells with stably down-expressed lnc-RAB11B-AS1 showed increased cells in G1 phase and fewer cells in M phase (P<0.05; Figure 2B). Similar results were found in both PC-9 and A549 cell lines. In addition, overexpression of lnc-RAB11B-AS1 caused an increase in apoptosis, while stably down-expressed lnc-RAB11B-AS1 led to a decrease in apoptosis in both PC-9 and A549 cell lines (P<0.05; Figure 2C).
Figure 2.
Lnc-RAB11B-AS1 promotes cancer cell proliferation, apoptosis, colony formation and cell cycle progression in LC cells in vitro. (A) Lnc-RAB11B-AS1 over-expression significantly enhanced lung cancer cell viability, while silencing of lnc-RAB11B-AS1 reduced cell viability of both A549 and PC-9 cell lines; (B) over-expression of lnc-RAB11B-AS1 decreased the numbers of G1 phase cells and increased the numbers of cells in M phase, while silencing of lnc-RAB11B-AS1 had the opposite effects; (C) Lnc-RAB11B-AS1 over-expression caused a decrease in apoptosis and silencing increased the apoptosis rate in both A549 and PC-9 cell lines; (D) the tablet clone formation efficiency of the transformed cell lines showed significant increases in lnc-RAB11B-AS1 overexpressed cell lines and decreases in lnc-RAB11B-AS1 silenced cell lines. *, P value <0.05; **, P value <0.01.
lnc-RAB11B-AS1 overexpression increases colony formation ability of LC cells
We next evaluated whether lnc-RAB11B-AS1 expression impacted the clonogenic ability of LC cell lines. The results showed that the colony formation counts in LC cell lines were statistically increased when lnc-RAB11B-AS1 was upregulated compared with control groups, while silencing lnc-RAB11B-AS1 significantly reduced the colony numbers both in PC-9 and A549 cell lines (P<0.05; Figure 2D).
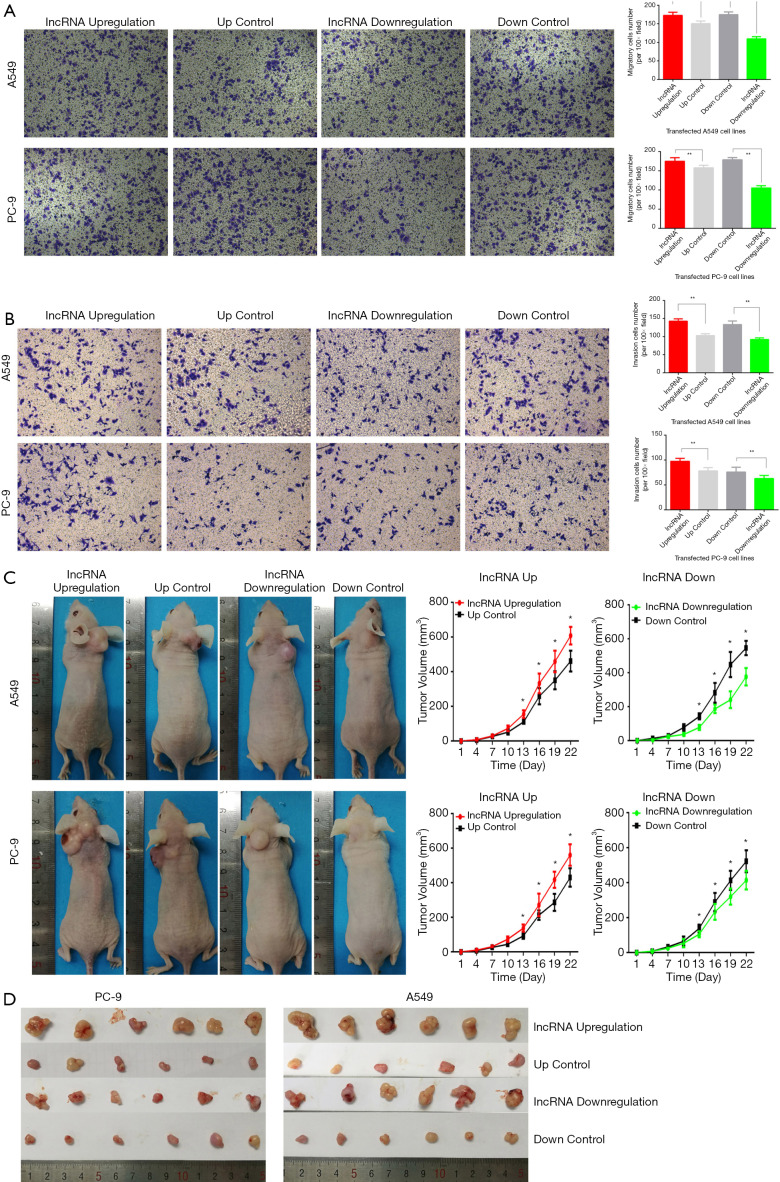
lnc-RAB11B-AS1 overexpression promotes LC cell migration and invasion abilities in vitro
As epithelial-mesenchymal transition (EMT) is an important property of tumor metastasis, we next performed TRANSWELL assays to explore the influence of lnc-RAB11B-AS1 on cell migration and invasion abilities. Cell migration assay results showed that upregulation of lnc-RAB11B-AS1 statistically increased the migratory ability of the LC cell lines, while lnc-RAB11B-AS1 knockdown significantly reduced this ability (P<0.05; Figure 3A). Similar effects were also observed in invasion assays (P<0.05; Figure 3B).
Figure 3.
Lnc-RAB11B-AS1 promotes cancer cell migration and invasion activities in LC cells in vitro and enhances tumor growth in vivo. (A,B) Cell lines with overexpressed lnc-RAB11B-AS1 showed significantly higher migration and invasion activities compared with controls, while cell lines silenced for lnc-RAB11B-AS1 exerted significantly lower abilities; (C,D) tumor growth was increased in xenograft mice implanted with stable transfected in lnc-RAB11B-AS1 overexpressed cells and attenuated in xenograft mice implanted with lnc-RAB11B-AS1 silenced cells compared with controls. All results are shown as mean ± SD. P<0.05, calculated by the one-way ANOVA test. *, P value <0.05; **, P value <0.01.
lnc-RAB11B-AS1 promotes tumor growth in vivo
To study the influence of lnc-RAB11B-AS1 on the proliferation abilities of LC cells in vivo, we generated a BALB/c-nu nude mice xenograft model using stably transfected PC-9 and A549 cell lines. A statistically increase in tumor volume and weight was observed in the lnc-RAB11B-AS1 upregulation groups (both PC-9 and A549 cell lines), while the lnc-RAB11B-AS1 downregulation groups showed a statistically decrease in tumor volume and weight (P<0.05; Figure 3C,D).
The expression of lnc-RAB11B-AS1 was positively correlated with its sense-cognate gene RAB11B in LC
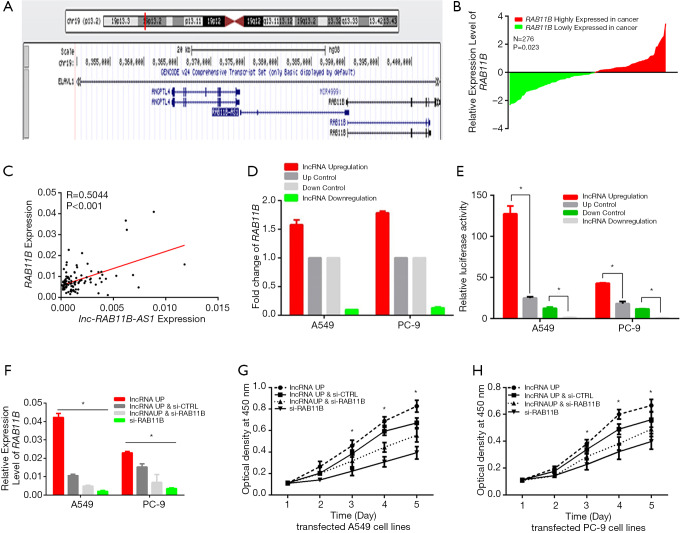
Sequence analysis showed that lnc-RAB11B-AS1 is located in physical contiguity with RAB11B (Figure 4A) and forms a ‘head-to-tail’ pairing pattern with 448 bp of full complementarity sequence. We next detected the expression levels of RAB11B by qRT-PCR using the same groups of LC tissue samples used to detect lnc-RAB11B-AS1 levels and conducted a correlation analysis. A statistical increase of RAB11B expression level was observed in LC tissues when compared with the paired normal tissues (P=0.023; Figure 4B). The correlation analysis demonstrated a positive correlation between the expression levels of lnc-RAB11B-AS1 and RAB11B (R =0.5044, P<0.001; Figure 4C).
Figure 4.
Correlation between lnc-RAB11B-AS1 and RAB11B expression. (A) The relative position of lnc-RAB11B-AS1 and RAB11B in human genome; (B) relative expression level of RAB11B in LC patients (N=276). P<0.01; (C) correlation between lnc-RAB11B-AS1 and RAB11B; (D) high expression of lnc-RAB11B-AS1 resulted in high RAB11B expression, while low expression of lnc-RAB11B-AS1 caused low RAB11B expression in stably transfected A549 and PC-9 cell lines; (E) upregulation of lnc-RAB11B-AS1 induced a significant increase in the luciferase activity of pGL3-Promoter-RAB11B in LC cell lines; (F) relative expression of RAB11B was measured by qPCR in A549 and PC-9 cells stably silenced for lnc-RAB11B-AS1 and transfected with si-RAB11B or si-CTRL; (G,H) CCK-8 assays were conducted to detect the effect of si-RAB11B on stably overexpression lnc-RAB11B-AS1 cell lines. *, P value <0.05; **, P value <0.01.
To confirm these findings, we also examined the levels of RAB11B expression in the lnc-RAB11B-AS1 stable transfected cell lines. Our results showed that RAB11B mRNA level was statistically increased when lnc-RAB11B-AS1 was upregulated, and decreased in those cells with lnc-RAB11B-AS1 silencing (Figure 4D).
lnc-RAB11B-AS1 promotes LC progress via correlated with RAB11B
We next performed luciferase reporter assay to more closely examine the correlation between lnc-RAB11B-AS1 and RAB11B gene. Reporter assays were performed in LC cell lines using a pGL3-RAB11B. Transfection of the reporter in cells with lnc-RAB11B-AS1 upregulation resulted in a significant increase in the luciferase activity in both LC cell lines, while lnc-RAB11B-AS1 silenced cell lines showed decreased luciferase activity (Figure 4E). The above results showed a positive relationship between lnc-RAB11B-AS1 and the promoter of RAB11B in LC cell lines.
lnc-RAB11B-AS1 promotes LC progression via upregulating RAB11B
We next analyzed the associations between the expression levels of RAB11B and LC progression. We stratified the LC patients into high and low expression groups using the medium level in LC tissues. Patients with expression levels equal to or greater than the cut-off value were categorized into the high RAB11B expression group, and those with expression levels lower than the cut-off value were defined as the low RAB11B expression group. The status of RAB11B expression was statistically different in patients at different clinic stages (P<0.001), N stage (P<0.001) and M stage (P<0.001) (Table 4). These results indicate that patients with a higher level of RAB11B expression would have a higher risk for advanced clinic stages, N stage and M stage.
Table 4. Association between the expression level of RAB11B and the clinic characteristics of the patients.
| Clinic characteristics | Southern samples, N (%) | Pa | Eastern samples, N (%) | Pa | Merged samples, N (%) | Pa | |||
|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | Low | High | ||||
| Age | |||||||||
| <60 | 46 (43.8) | 59 (56.2) | 0.762 | 22 (43.1) | 29 (56.9) | 0.919 | 68 (43.6) | 88 (56.4) | 0.856 |
| ≥60 | 32 (41.6) | 45 (58.4) | 19 (44.2) | 24 (55.8) | 51 (42.5) | 69 (57.5) | |||
| Sex | |||||||||
| Female | 24 (46.2) | 28 (53.8) | 0.570 | 12 (42.9) | 16 (57.1) | 0.923 | 36 (45.0) | 44 (55.0) | 0.686 |
| Male | 54 (41.5) | 76 (58.5) | 29 (43.9) | 37 (56.1) | 83 (42.3) | 113 (57.7) | |||
| Family history of cancer | |||||||||
| No | 67 (41.6) | 94 (58.4) | 0.348 | 37 (44.0) | 47 (56.0) | 0.807 | 104 (42.4) | 141 (57.6) | 0.529 |
| Yes | 11 (52.4) | 10 (47.6) | 4 (40.0) | 6 (60.0) | 15 (45.2) | 16 (54.8) | |||
| Family history of lung cancer | |||||||||
| No | 73 (43.2) | 96 (56.8) | 0.740 | 40 (44.0) | 51 (56.0) | 0.715 | 113 (43.5) | 147 (56.5) | 0.640 |
| Yes | 5 (38.5) | 8 (61.5) | 1 (33.3) | 2 (66.7) | 6 (37.5) | 10 (62.5) | |||
| Smoking | |||||||||
| No | 31 (46.3) | 36 (53.7) | 0.478 | 14 (46.7) | 16 (53.3) | 0.683 | 45 (46.4) | 52 (53.6) | 0.419 |
| Yes | 47 (40.9) | 68 (59.1) | 27 (42.2) | 37 (57.8) | 74 (41.9) | 105 (58.1) | |||
| Drinking | |||||||||
| No | 60 (43.8) | 77 (56.2) | 0.655 | 31 (42.5) | 42 (57.5) | 0.675 | 91 (43.3) | 119 (56.7) | 0.897 |
| Yes | 18 (40.0) | 27 (60.0) | 10 (47.6) | 11 (52.4) | 28 (42.4) | 38 (57.6) | |||
| Surgery | |||||||||
| No | 54 (44.6) | 67 (55.4) | 0.497 | 25 (43.1) | 33 (56.9) | 0.899 | 79 (44.1) | 100 (55.9) | 0.643 |
| Yes | 24 (39.3) | 37 (60.7) | 16 (44.4) | 20 (55.6) | 40 (41.2) | 57 (58.8) | |||
| Chemotherapy | |||||||||
| No | 24 (38.1) | 39 (61.9) | 0.345 | 12 (41.4) | 17 (58.6) | 0.770 | 36 (39.1) | 56 (60.9) | 0.344 |
| Yes | 54 (45.4) | 65 (54.6) | 29 (44.6) | 36 (55.4) | 83 (45.1) | 101 (54.9) | |||
| Radiotherapy | |||||||||
| No | 47 (45.2) | 57 (54.8) | 0.462 | 22 (45.8) | 26 (54.2) | 0.658 | 69 (45.4) | 83 (54.6) | 0.397 |
| Yes | 31 (39.7) | 47 (60.3) | 19 (41.3) | 27 (58.7) | 50 (40.3) | 74 (59.7) | |||
| Clinical stages | |||||||||
| I + II | 40 (56.3) | 31 (43.7) | 0.003 | 18 (62.1) | 11 (37.9) | 0.016 | 58 (58.0) | 42 (42.0) | <0.001 |
| III + IV | 38 (34.2) | 73 (65.8) | 23 (35.4) | 42 (64.6) | 61 (34.7) | 115 (65.3) | |||
| T | |||||||||
| 1+2 | 50 (48.1) | 54 (51.9) | 0.100 | 19 (42.2) | 26 (57.8) | 0.794 | 69 (46.3) | 80 (53.7) | 0.246 |
| 3+4 | 28 (35.9) | 50 (64.1) | 22 (44.9) | 27 (55.1) | 50 (39.4) | 77 (60.6) | |||
| N | |||||||||
| 0 | 35 (53.0) | 31 (37.0) | 0.036 | 26 (60.5) | 17 (39.5) | 0.002 | 61 (56.0) | 48 (44.0) | <0.001 |
| 1+2+3 | 43 (37.1) | 73 (62.9) | 15 (29.4) | 36 (70.6) | 58 (34.7) | 109 (65.3) | |||
| M | |||||||||
| 0 | 63 (48.8) | 66 (51.2) | 0.011 | 32 (53.3) | 28 (46.7) | 0.012 | 95 (50.3) | 94 (49.7) | <0.001 |
| 1 | 15 (28.3) | 38 (71.7) | 9 (26.5) | 25 (73.5) | 24 (27.6) | 63 (72.4) | |||
| Histological types | |||||||||
| Adenocarcinoma | 39 (45.3) | 47 (54.7) | 0.693 | 20 (48.8) | 21 (51.2) | 0.600 | 59 (46.5) | 68 (53.4) | 0.612 |
| Squamous cell carcinoma | 20 (39.2) | 31 (60.8) | 10 (32.3) | 21 (67.7) | 30 (36.6) | 52 (63.4) | |||
| Large cell carcinoma | 4 (66.7) | 2 (33.3) | 2 (40.0) | 3 (60.0) | 6 (54.5) | 5 (45.5) | |||
| Small cell lung cancer | 7 (36.8) | 12 (63.2) | 4 (50.0) | 4 (50.0) | 11 (40.7) | 16 (59.3) | |||
| Other carcinomasb | 8 (40.0) | 12 (60.0) | 5 (55.6) | 4 (44.4) | 13 (44.8) | 16 (55.2) | |||
a, Pearson χ2 test P value; b, mixed-cell or undifferentiated carcinoma.
We also examined the effect of RAB11B on the proliferation ability of LC cell lines using CCK-8 assays. We generated PC-9 and A549 cell lines expressing a siRNA targeting RAB11B as described in Methods. The above results showed that LC cell lines with silenced RAB11B showed reduced proliferation compared with control cells (Figure 4F,G,H). These results indicate that RAB11B functions in regulating LC cell proliferation and imply a role for RAB11B in promoting LC progression.
Discussion
Our previous studies found that lnc-RAB11B-AS1 was involved in cell cycle and metastasis regulation in osteosarcoma (15), and was reported to correlate with the advanced clinic stage, metastasis, and overall survival rate in gastric cancer patients (16) and promote hypoxia-mediated angiogenesis and metastasis in breast cancer patients (17). However, its role in LC development remains to be clarified. In this study, we identified a novel mechanism by which lnc-RAB11B-AS1 functions to promote LC development through regulating RAB11B expression. lnc-RAB11B-AS1 was overexpressed in LC tissues compared with the paired normal tissues. High expression levels of lnc-RAB11B-AS1 correlated with an advanced cancer stage, N, M status and poor overall survival rate in LC patients. The above data suggest that lnc-RAB11B-AS1 could act as a potential prognostic biomarker for the survival of LC patients and may be a novel potential therapeutic target in treating LC patients.
Our previous studies found that lnc-RAB11B-AS1 was down-regulated in osteosarcoma (15). However, Feng et al. (16) and Niu et al. (17) reported that lnc-RAB11B-AS1 was over-expressed in gastric cancer and breast cancer respectively. This is the first study to describe the expression pattern and function of lnc-RAB11B-AS1 in LC. We observed significantly higher expression levels of lnc-RAB11B-AS1 in LC tissues compared with corresponding non-cancerous tissues and had a positive correlation between the expression levels of lnc-RAB11B-AS1 and RAB11B. The expression level of lnc-RAB11B-AS1 was statistically correlated with clinic stage, N stage and M stage in LC patients. LC patients with upregulated lnc-RAB11B-AS1 expression showed a poorer prognosis and worse overall survival rate compared with those with downregulated lnc-RAB11B-AS1 expression, indicating that lnc-RAB11B-AS1 may function as a prognostic biomarker for LC. Gain-of-function and loss-of-function experiments conducted in this study showed that lnc-RAB11B-AS1 promoted cell cycle changes and induced apoptosis, proliferation, migration and invasion abilities of LC cells in vitro. Furthermore, xenograft experiments showed that overexpression of lnc-RAB11B-AS1 increased tumor growth in vivo. These results indicate that lnc-RAB11B-AS1 may play an oncogenic role in LC development and progression.
Increasing studies have shown that as-lncRNAs play a crucial role in cancer development and some may function as cancer biomarkers. For example, lnc-MUC5B-AS1 promotes metastasis through regulating its natural antisense transcript MUC5B in lung adenocarcinoma (25). Lnc-ZEB1-AS1 was reported to promote tumor metastasis and predicts poor prognosis in liver cancer (7). Lnc-ZEB2-AS1 was reported to promote proliferation and inhibit apoptosis in human lung cancer cells (26). Thus, in addition to these lncRNAs, lnc-RAB11B-AS1 may be another novel preventive and therapeutic biomarker for LC. The xenograft model showed that lnc-RAB11B-AS1 plays a crucial role in controlling LC cell proliferation in vivo, which indicates that lnc-RAB11B-AS1 might be a therapeutic target of LC.
Although our results indicate that lnc-RAB11B-AS1 might function as an oncogene in LC, the underlying molecular mechanisms remain to be clarified. Recent studies showed that methylation of the promoter of as-lncRNAs could be a potential strategy to dysregulate the expression of as-lncRNA, and thus affect the corresponding mRNA expression. For example, a previous report revealed a CpG island spanning the transcription start site of lnc-ZEB1-AS1, and lnc-ZEB1-AS1 methylation was negatively correlated with the lnc-ZEB1-AS1 expression (7). We observed a similar mechanism in lnc-RAB11B-AS1 in osteosarcoma in our previous study (15). We found that DNA hypomethylation of the promoter could result in an increase in the expression level of lnc-RAB11B-AS1. Our data showed that lnc-RAB11B-AS1 distributes mainly in the nucleus and partly in the cytoplasm. Literatures showed that nuclear lncRNAs involved in transcriptional regulation both in cis and in trans, and played a crucial role in the modulation of chromosomal interactions, chromatin looping, transcription factor trapping, gene methylation, recruitment of transcription factors, and chromatin modification (27,28). Thus, we speculate that hypomethylation in the promoter region may cause the upregulation of lnc-RAB11B-AS1 in LC. Canzio et al. (29) reported a new mechanism of the antisense lncRNAs functioned in distance-independent enhancer and promotor demethylation, thus could be a potential mechanism of how lnc-RAB11B-AS1 regulate RAB11B.
Sequence analysis showed that lnc-RAB11B-AS1 is located in physical contiguity with RAB11B, and formed a ‘head-to-tail’ pairing pattern with 448 bp full complementarity sequence. Therefore, we can suggest that lnc-RAB11B-AS1 may function as a super-enhancer or co-factor in the transcriptional regulation way (30) to regulate the target gene RAB11B. Luciferase reporter assays showed that lnc-RAB11B-AS1 overexpression promotes RAB11B expression, which supports the super-enhancer hypothesis. Furthermore, the qPCR results showed a positive correlation between the expression of lnc-RAB11B-AS1 and the expression of RAB11B, which demonstrated the positive regulation of lnc-RAB1B-AS1 on RAB11B in LC.
RAB11B is a member of the RAB11 family, which includes RAB11A, RAB11B and RAB25 (or RAB11C) (31). Multiple diseases have been correlated with RAB11 family proteins, such as Alzheimer’s disease, Huntington’s disease and type 2 diabetes, and RAB11 proteins have been associated with carcinogenesis and cancer development (32). Gebhardt et al. (33) found RAB11 proteins were functional in human skin cancer. Yoon et al. (34) demonstrated that RAB11 enhances the invasiveness and migration capabilities of breast cancer cells. RAB11B was first reported by Lai et al. (35) in mice in 1994 and has been associated with various diseases (35), such as breast cancer (36) and non-small cell lung cancer (37). Dysregulated 19p13.2 genomes were also correlated with pancreatic cancer, smoking associated chronic obstructive pulmonary disease (22,23,38), colon cancer (39) and ovarian mucinous carcinoma (18).
In this study, we found that overexpression of lnc-RAB11B-AS1 positively correlated with RAB11B expression level in LC tissues as well as stably transfected LC cell lines. We also examined whether lnc-RAB11B-AS1 regulates the RAB11B promoter activity, and indeed, lnc-RAB11B-AS1 exhibited a significant effect on promoting RAB11B promoter activity. Furthermore, RAB11B inhibition partially abrogated lnc-RAB11B-AS1-induced LC cell lines invasion and cancer metastasis, while silencing of lnc-RAB11B-AS1 suppressed RAB11B expression and decreased cell invasion and cancer metastasis. Although we found that lnc-RAB11B-AS1 had a positive regulation on RAB11B in LC on RNA level, we did not detect if the RAB11B protein was influenced by lnc-RAB11B-AS1, the mechanism of how lnc-RAB11B-AS1 was upregulated in LC remains to be clarified, and that’s what we plan to explore next.
In conclusion, our results provide the first evidence that lnc-RAB11B-AS1 may function as an oncogene to facilitate cell proliferation, invasion and migration ability, which possibly through the regulation on RAB11B in LC. We found that upregulation of lnc-RAB11B-AS1 in LC cell lines statistically promoted tumor growth, metastasis, cell proliferation and inhibit apoptosis both in vivo and in vitro, which increased the possibility that lnc-RAB11B-AS1 might be a promising new biomarker and therapeutic target for LC.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank Dr. Yuan Guo, Dr. Lin Liu, Dr. Dongsheng Huang and Dr. Yumin Zhou for their assistance on subjects’ recruitment.
Funding: This study was supported by the National Natural Science Foundation of China Grants 81803325 (DW); 81473040, 81673267, 81872694 (JL); 81402753, 81672303, 81871876 (LY); 81602289, 81872127 (FQ); Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program 2017BT01S155 (JL); Science Foundation for Distinguished Young Scholars in Jiangsu BK20160008 (YZ); Guangdong Provincial Major Projects Grants 2014KZDXM046 (JL); Guangdong Education Bureau Characteristic Innovation Project Grants 2015KTSCX116 and Guangzhou Science and Technology Program Pearl River Nova Projects Grants 201710010049 (LY). National Natural Science Foundation of Guangdong 2016A030313567 (LZ); Science and Technology Program of Guangzhou 201607010035 (LZ). Medical Science and Technology Project of Guangzhou 20191A011064 (DW). Guangdong Medical Science and Technology Research Project Grant A2019379 (DW). National Natural Science Foundation of Guangdong Province 2019A1515011407 (TL). Guangdong High School Young Innovative Talents Project Grant 2015KQNCX136 (FQ). Guangzhou Science Research Program General Project 201707010123 (FQ). Guangzhou Municipal Scientific Research Project 1201630073 (FQ). Natural Science Foundation of Guangdong Province 2019A1515011407 (TL).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The present study was approved by the Ethics Committee of Guangzhou Medical University (No. GMU201481473040) and we strictly followed the related clinical research guidelines. All study participants involved in the present study were provided written informed consent.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.04.52). The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zeng H, et al. Epidemiology of lung cancer in China. Thorac Cancer 2015;6:209-15. 10.1111/1759-7714.12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Zeng H, et al. The updated incidences and mortalities of major cancers in China, 2011. Chin J Cancer 2015;34:502-7. 10.1186/s40880-015-0042-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China, 2013. Cancer Lett 2017;401:63-71. 10.1016/j.canlet.2017.04.024 [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 6.Zeng H, Zheng R, Guo Y, et al. Cancer survival in China, 2003-2005: a population-based study. Int J Cancer 2015;136:1921-30. 10.1002/ijc.29227 [DOI] [PubMed] [Google Scholar]
- 7.Li T, Xie J, Shen C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene 2016;35:1575-84. 10.1038/onc.2015.223 [DOI] [PubMed] [Google Scholar]
- 8.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol 2013;26:155-65. 10.1038/modpathol.2012.160 [DOI] [PubMed] [Google Scholar]
- 9.Huang B, Song JH, Cheng Y, et al. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene 2016;35:4927-36. 10.1038/onc.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang C, Qiu S, Sun F, et al. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett 2017;410:50-62. 10.1016/j.canlet.2017.09.012 [DOI] [PubMed] [Google Scholar]
- 11.Wei W, Pelechano V, Jarvelin AI, et al. Functional consequences of bidirectional promoters. Trends Genet 2011;27:267-76. 10.1016/j.tig.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harvey JJ. An unidentified virus which causes the rapid production of tumors in mice. Nature 1964;204:1104-5. 10.1038/2041104b0 [DOI] [PubMed] [Google Scholar]
- 13.Goitre L, Trapani E, Trabalzini L, et al. The Ras superfamily of small GTPases: the unlocked secrets. Methods Mol Biol 2014;1120:1-18. 10.1007/978-1-62703-791-4_1 [DOI] [PubMed] [Google Scholar]
- 14.Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol 2010;22:461-70. 10.1016/j.ceb.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, Liu Z, Yang Y, et al. Long non-coding RNA RAB11B-AS1 prevents osteosarcoma development and progression via its natural antisense transcript RAB11B. Oncotarget 2018;9:26770-86. 10.18632/oncotarget.24247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng L, Yu Y, Li Y. Expression and clinical significance of lnc-RAB11B-AS1 in gastric cancer (in Chinese). Chin Clin Oncol 2017;22:592-7. [Google Scholar]
- 17.Niu Y, Bao L, Chen Y, et al. HIF-2-induced long non-coding RNA RAB11B-AS1 promotes hypoxia-mediated angiogenesis and breast cancer metastasis. Cancer Res 2020;80:964-75. 10.1158/0008-5472.CAN-19-1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelemen LE, Lawrenson K, Tyrer J, et al. Genome-wide significant risk associations for mucinous ovarian carcinoma. Nat Genet 2015;47:888-97. 10.1038/ng.3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet 2009;41:1122-6. 10.1038/ng.448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang JW, Shi ZZ, Shen TY, et al. Identification of genomic alterations in pancreatic cancer using array-based comparative genomic hybridization. PLoS One 2014;9:e114616. 10.1371/journal.pone.0114616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fransson S, Östensson M, Djos A, et al. Estimation of copy number aberrations: Comparison of exome sequencing data with SNP microarrays identifies homozygous deletions of 19q13.2 and CIC in neuroblastoma. Int J Oncol 2016;48:1103-16. 10.3892/ijo.2016.3349 [DOI] [PubMed] [Google Scholar]
- 22.Nedeljkovic I, Lahousse L, Carnero-Montoro E, et al. COPD GWAS variant at 19q13.2 in relation with DNA methylation and gene expression. Hum Mol Genet 2018;27:396-405. 10.1093/hmg/ddx390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan DM, Vincent TL, Salit J, et al. Smoking dysregulates the human airway basal cell transcriptome at COPD risk locus 19q13.2. PLoS One 2014;9:e88051. 10.1371/journal.pone.0088051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D, Yang B, Chen J, et al. Upregulation of long non-coding RNA RAB1A-2 induces FGF1 expression worsening lung cancer prognosis. Cancer Lett 2018;438:116-25. 10.1016/j.canlet.2018.09.016 [DOI] [PubMed] [Google Scholar]
- 25.Yuan S, Liu Q, Hu Z, et al. Long non-coding RNA MUC5B-AS1 promotes metastasis through mutually regulating MUC5B expression in lung adenocarcinoma. Cell Death Dis 2018;9:450. 10.1038/s41419-018-0472-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y, Hu Y, Hu M, et al. Long non-coding RNA ZEB2-AS1 promotes proliferation and inhibits apoptosis in human lung cancer cells. Oncol Lett 2018;15:5220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 2016;17:756-70. 10.1038/nrm.2016.126 [DOI] [PubMed] [Google Scholar]
- 28.Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016;539:452-5. 10.1038/nature20149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canzio D, Nwakeze CL, Horta A, et al. Antisense lncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin alpha Promoter Choice. Cell 2019;177:639-53.e15. 10.1016/j.cell.2019.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie JJ, Jiang YY, Jiang Y, et al. Increased Expression of the Long Non-coding RNA LINC01503, Regulated by TP63, in Squamous Cell Carcinoma and Effects on Oncogenic Activities of Cancer Cell Lines. Gastroenterology 2018;154:2137-51.e1. 10.1053/j.gastro.2018.02.018 [DOI] [PubMed] [Google Scholar]
- 31.Kelly EE, Horgan CP, McCaffrey1 MW. Rab11 proteins in health and disease. Biochem Soc Trans 2012;40:1360-7. 10.1042/BST20120157 [DOI] [PubMed] [Google Scholar]
- 32.Greenfield JP, Leung LW, Cai D, et al. Estrogen lowers Alzheimer beta-amyloid generation by stimulating trans-Golgi network vesicle biogenesis. J Biol Chem 2002;277:12128-36. 10.1074/jbc.M110009200 [DOI] [PubMed] [Google Scholar]
- 33.Gebhardt C, Breitenbach U, Richter KH, et al. c-Fos-dependent induction of the small ras-related GTPase Rab11a in skin carcinogenesis. Am J Pathol 2005;167:243-53. 10.1016/S0002-9440(10)62969-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon SO, Shin S, Mercurio AM. Hypoxia stimulates carcinoma invasion by stabilizing microtubules and promoting the Rab11 trafficking of the alpha6beta4 integrin. Cancer Res 2005;65:2761-9. 10.1158/0008-5472.CAN-04-4122 [DOI] [PubMed] [Google Scholar]
- 35.Lai F, Stubbs L, Artzt K. Molecular analysis of mouse Rab11b: a new type of mammalian YPT/Rab protein. Genomics 1994;22:610-6. 10.1006/geno.1994.1434 [DOI] [PubMed] [Google Scholar]
- 36.Yang TL, Su YR, Huang CS, et al. High-Resolution 19p13.2-13.3 Allelotyping of Breast Carcinomas Demonstrates Frequent Loss of Heterozygosity. Genes Chromosomes Cancer 2004;41:250-6. 10.1002/gcc.20080 [DOI] [PubMed] [Google Scholar]
- 37.Singh A, Misra V, Thimmulappa RK, et al. Dysfunctional KEAP1-NRF2 interaction in Non-Small-Cell Lung Cancer. PLoS Med 2006;3:e420. 10.1371/journal.pmed.0030420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Peng S, Cheng H, et al. Genetic Pleiotropy between Nicotine Dependence and Respiratory Outcomes. Sci Rep 2017;7:16907. 10.1038/s41598-017-16964-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang B, Jia WH, Matsuda K, et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet 2014;46:533-42. 10.1038/ng.2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as