Abstract
Background
The coronavirus disease 2019 (COVID-19) has rapidly become a pandemic worldwide. The value of chest computed tomography (CT) is debatable during the treatment of COVID-19 patients. Compared with traditional chest X-ray radiography, quantitative CT may supply more information, but its value on COVID-19 patients was still not proven.
Methods
An automatic quantitative analysis model based on a deep network called VB-Net for infection region segmentation was developed. A quantitative analysis was performed for patients diagnosed as severe COVID 19. The quantitative assessment included volume and density among the infectious area. The primary clinical outcome was the existence of acute respiratory distress syndrome (ARDS). A univariable and multivariable logistic analysis was done to explore the relationship between the quantitative results and ARDS existence.
Results
The VB-Ne model was sensitive and stable for pulmonary lesion segmentation, and quantitative analysis indicated that the total volume and average density of the lung lesions were not related to ARDS. However, lesions with specific density changes showed some influence on the risk of ARDS. The proportion of lesion density from −549 to −450 Hounsfield unit (HU) was associated with increased risk of ARDS, while the density was ranging from −149 to −50 HU was related to a lowered risk of ARDS.
Conclusions
The automatic quantitative model based on VB-Ne can supply useful information for ARDS risk stratification in COVID-19 patients during treatment.
Keywords: Coronavirus disease 2019 (COVID-19), quantitative computed tomography assessment (quantitative CT assessment), acute respiratory distress syndrome (ARDS), artificial intelligence (AI)
Introduction
The coronavirus disease 2019 (COVID-19), which is caused by a novel coronavirus SARS-COV-2 (1,2), has rapidly developed into a pandemic and arisen public attention. Although a large number of studies has demonstrated the benefit of chest computed tomography (CT) in the diagnostic workups of COVID-19 (3-5), some expert consensus or guidelines still suggest that chest CT is only more favorably indicated than chest x-ray radiography for making the early diagnosis of COVID-19 and helping the discharge decision of patients (6). The usefulness of chest CT in severe COVID-19 patients is still in controversy, for reasons of availability, cost, and increased risk of cross-infection and radiation dosage.
In routine clinical practice, CT can only supply limited information about respiratory status using qualitative assessment. However, with the development of artificial intelligence (AI) technology, CT is far beyond a qualitative and basic quantitative examination. Quantitative CT has been widely used in many filed, including the respiratory system (7). As for COVID-19, some previous studies investigated the differential diagnosis, severity rating, and prognosis prediction using a qualitative CT combined with AI technology in COVID-19 patients (8,9). For instance, Li et al. (10) established a total severity score to classify the mild and severe type COVID-19 patients as well as Shen et al. (11) used some quantitative metrics, such as lesion volume, mean lesion density to stratify the severity of COVID-19. However, little is known about the potential of quantitative CT in monitoring the acute respiratory distress syndrome (ARDS) status in patients during treatment as which is the primary cause of ventilation use in COVID-19 patients and attention also should be paid, especially in some early stage asymptomatic patients.
Some studies suggested that density composition might also influence patients’ respiratory function and clinical outcomes besides the total volume and location of the infectious lesion (12). However, the existing assessment method, whether qualitative or quantitative, could not evaluate the density composition of infectious lesions thoroughly. Thus, the purpose of this study was to develop a quantitative method based on deep-learning (DL) segmentation to check the ARDS status in COVID-19 patients during treatment. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3554).
Methods
DL-based segmentation network: VB-Net
Due to the significant variations of both shape and position across different COVID-19 patients, traditional segmentation methods often fail and need manual interaction. We developed a deep network called VB-Net for infection region segmentation (13). This model a light-weighted extension of a 3-D convolutional neural network that combines V-Net (14) by using the bottle-neck structure (15). The speed of VB-Net is much faster than V-Net because of the bottle-neck structure (15,16).
In the bottle-net design of VB-Net, three layers using the 1×1×1, 3×3×3 and 1×1×1 convolution kernel was used to replace the original kernels. The first 1×1×1 kernel layer reduces the number of channels, and the 3×3×3 kernel layer processes the data, and the original channels of feature maps are restored by another 1×1×1 kernel layer. In this way, we not only reduce the model size and inference time but also use the cross-channel features, and VB-Net is more applicable to 3D volumetric data. The network structure of VB-Net is shown in Figure 1.
Figure 1.
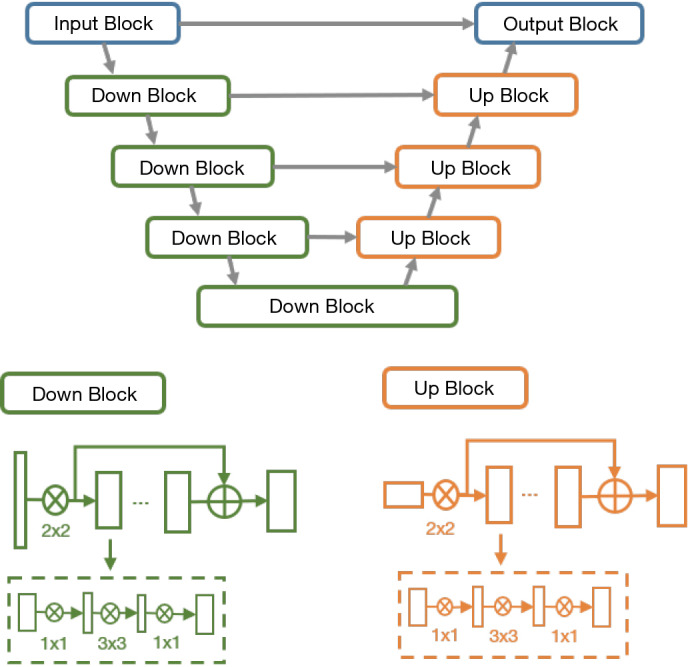
The network structure for COVID-19 infection segmentation. The dashed boxes show the bottle-neck structures inside the V-shaped network (14). COVID-19, coronavirus disease 2019.
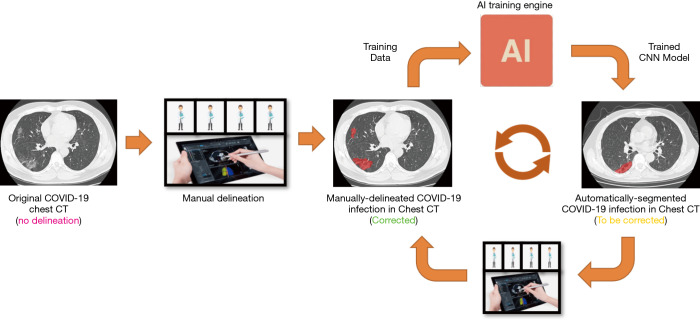
In addition to using the new model, the human-in-the-loop (HITL) strategy was also adopted to update the DL model iteratively. The aim is to minimize the efforts of delineating the training data. The basic idea was to annotate a group of images to train the model manually and then apply the model to new data for further manual correction. Gradually increased training data can be lineated in this HITL strategy. Depending on the number of training samples, the HITL training strategy converged after 3–4 iterations. Figure 2 illustrates the process of the proposed HITL training strategy.
Figure 2.
The human-in-the-loop workflow. COVID-19, coronavirus disease 2019; CT, computed tomography.
Training dataset and image acquisition parameters
The model used in this paper was trained by 2,565 COVID-19 cases, and 2,785 negative cases, where COVID-19 was diagnosed by RT-PCR of nasal and pharyngeal swab specimens (17), and negative cases were composed of pulmonary nodules cases, other pneumonia cases, and healthy cases. Training cases were from Shanghai, Jiangsu and Wuhan Province of China. In this paper, we applied the VB-Net model for segmenting our cohort of patients whose primary outcome was ARDS. Our goal is to quantitatively analyze the image data during the treatment of these patients.
Model application for application patient cohort
This retrospective study was approved by the proper ethics review board. The requirement for informed consent was waived because the anonymized study did not alter any diagnosis and treatment of the patients.
All patients diagnosed with COVID-19 by positive RT-PCR results using nasal and pharyngeal swab specimens in our hospital from Jan to Feb 2020 were screened in the study. After screening the clinical and CT records, only patients who were classified into severe cases in their medical records were included for further analysis. Patients with a missing medical record or aged <18 years were excluded.
The non-contrast chest CT examinations were performed for each patient when their competent doctors thought it was necessary to assess their respiratory status. CT examination was prohibited when the patient could not get rid of the ventilator. The detail of chest CT scan parameters was showed in Supplementary file.
A simple qualitative assessment was done by two independent radiologists after each CT examination, including the change of volume, density, location lesions. The respiratory assessment was estimated by arterial blood gas analysis after each CT examination.
Quantification assessment of COVID-19 infection
The entire pipeline for quantitative COVID-19 assessment includes the following steps:
For each patient, a given chest CT is first fed into the DL-based segmentation system, which segments the infection regions, the whole lung, the lung lobes, and all the bronchopulmonary segments;
-
The following quantitative metrics are then calculated to quantify infectious regions of the image of that patient:
❖ Volumes of infection in the whole lung, and volumes of infection in each lobe and each bronchopulmonary segment.
❖ Percentage of infection (POI) in the whole lung, each lobe, and each bronchopulmonary segment. They are used to measure the severity of COVID-19 and the distribution of infection within the lung.
❖ Hounsfield unit (HU) histograms within different infection regions. Different HU range zones or components are used, including (zone 1: <−750), (zone 2: from −750 to −650), (zone 3: −649 to −550), (zone 4: −549 to −450), (zone 5: −449 to −350), (zone 6: −349 to −250), (zone 7: −249 to −150), (zone 8: −149 to −50), (zone 9: >−49) inside the infection region.
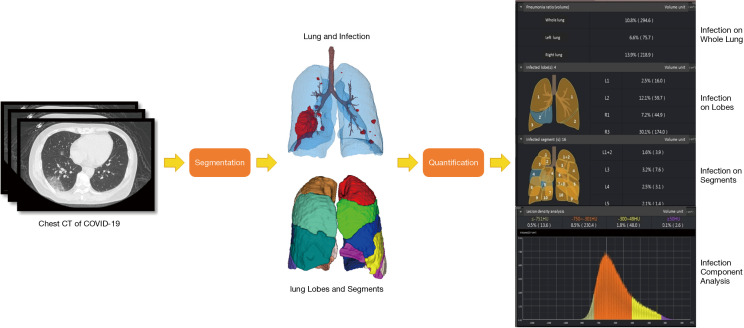
❖ Figure 3 shows the entire pipeline for the quantitative COVID-19 assessment. A chest CT scan is first fed to the DL-based segmentation system, which generates infection areas, the whole lung, lung lobes, and all the bronchopulmonary segments, respectively. Then, the quantitative metrics are calculated to quantify infection regions of the patient. The quantification supplies the basis for measuring the severity of COVID-19 from the CT perspective and for tracking longitudinal changes during the treatment course.
❖ The primary outcome of this study is the existence of ARDS. It was measured by the arterial partial pressure of oxygen(PaO2)/oxygen fraction ratio of inhalation gas(FiO2) <300. Considered age was described as a risk factor in many previous study, we used age as an effect modifier in our study (1,4,5).
Figure 3.
Pipeline for quantifying COVID-19 infection. A chest CT scan is first fed into the DL-based segmentation system. Then, quantitative metrics are calculated to characterize infection regions in the CT scan, such as infection volumes and POIs in the whole lung, lung lobes, and bronchopulmonary segments. COVID-19, coronavirus disease 2019; CT, computed tomography; POI, percentage of infection.
Statistical analysis
Statistical and correlation analysis methods are used to analyze the data and to supply the basis for measuring the severity of COVID-19 from the CT perspective and for tracking longitudinal changes during the treatment course.
Continuous variables were reported as the mean (standard deviation or std) or median [interquartile range (IQR)]. Student’s t-test or Mann-Whitney U test was used to compare between-group differences (presence and non-presence of primary composite endpoints) based on distributions. Categorical variables were presented as n (%) and compared using χ2 or Fisher exact test. Then, a two-step, multivariable logistic regression models was used to find the risk factors of CT quantitative data. First, all significant factors in χ2 or Fisher’s exact test will be put into univariable logistic regression. Then, a second multivariable logistic regression was done with the significant factors in the first regression. Statistical significance was set at 2-tailed P<0.05. All analyses were performed on the R software (Version 3.6.3).
Results
Training of the model
Based on the segmentation model was evaluated by 300 separate image datasets. It turned out that the average Dice similarity coefficient is 91.6%±10.0% (median 92.2%, IQR 89.0–94.6%; range, 9.6–98.1%). The average volume error is 10.7±16.7 cm3 (median 5.9 cm3, IQR 1.8–13.9 cm3; range, 0.0–89.6 cm3). The mean POI estimation errors are 0.3% for the whole lung, 0.5% for lung lobes, and 0.8% for bronchopulmonary segments. Eighty-six point seven percent of lung-lobe POIs and 81.6% of bronchopulmonary-segment POIs are accurately estimated with differences equal or less than 1%. The result of the full evaluation was showed in Table S1.
Clinical application
Eighty-six patients were enrolled in our study. After screening and exclusion, 27 patients were finally included in the analysis. Three, 55 and 4 patients were excluded because of age, not severe type and incomplete medical records. The mean age of patients was 60.3 [37–87], the median age of patients was 59 (IQR =33.5), and the male proportion was 66.67% (18/27). The median time from the first symptom to admission was six days (IQR =3.5), while the median time from the admission to classification into the severe patient was one day (IQR =2.5). The most common first symptom of these patients was fever (24/27, 88.89%) and cough (21/27, 77.78%), the most common comorbidity of these patients was hypertension (13/27, 48.15%). One hundred twenty-three CT scans were performed on these patients. The median time from admission to the first CT scan was three days (IQR =5.5), while the median of times of CT scans for each patient was four times (IQR =2). The median time of the gap between the two CT scans was five days (IQR =3).
Qualitative assessment results and the quantitative results of each CT scan were matched with its corresponding respiratory record. Among the 117 CT scans, 29.06% (34/117) of the clinical records of the day showed patients had ARDS. Eighty-seven point two percent (102/117) of CT scans were done with oxygen therapy. Notably, there were two times when patients finished the CT without oxygen therapy, which means patients were asymptomatic, but the arterial blood gas analysis showed that patients had ARDS.
The median area proportion of the infectious area was 10.2% (IQR =14.8%), while the mean density of the infectious area was −557.83±150.38 HU. The highest proportion of different density interval was zone 1, which composed 57.54%±30.58% area of the whole infectious region on average.
We analyzed the relationship between the age of patients, the area proportion of total infectious area (V), the average density of the whole infection (ρ), the area proportion of infection area in inner lung zone (Vi) and in the peripheral lung zone (Vp), the area proportion of specific region among the whole infection region of zone 1 (n1), zone 2 (n2), zone 3 (n3), zone 4 (n4), zone 5 (n5), zone 6 (n6), zone 7 (n7), zone 8 (n8) and zone 9 (n9) with the ARDS existence risk respectively. The single logistics regression result showed that age, V (estimate =0.035, P<0.001), ρ (estimate =0.004, P<0.001), Vi (estimate =0.087, P<0.001), Vp (estimate =0.044, P<0.001), zones 1–9 (zone 1: estimate =−2.111, P=0.038, zone 2: estimate =−1.848, P=0.035, zone 3: estimate =1.326, P=0.048, zone 4: estimate =7.114, P=0.008, zone 5: estimate =13.585, P=0.014, zone 6: estimate =19.066, P=0.004, zone 7: estimate =18.1540, P=0.005, zone 8: estimate =12.637, P=0.013, zone 9: estimate =5.236, P=0.033) were related to ARDS existence. Then, a multivariate logistics regression was performed with all the above positive factors (P<0.05) and age, which was reported as a risk factor for ARDS existence in COVID-19 in many previous studies (1,4,5). The multivariate logistics showed that only age (P=0.0118), the area proportions of zone 4 (P=0.003), zone 8 (P=0.021) were significantly related to ARDS existence independently (Table 1).
Table 1. Multivariate logistic regression result of the quantitative variates on the ARDS existence.
| Variables | Estimate Std. | Std. Error | Z value | Pr (>|z|) |
|---|---|---|---|---|
| Intercept | 2.03 | 7.433 | 0.274 | 0.783 |
| Age | 0.04 | 0.016 | 2.781 | 0.005 |
| V | −0.07 | 0.074 | −1.014 | 0.310 |
| Vi | 0.10 | 0.066 | 1.644 | 0.100 |
| Vp | 0.03 | 0.035 | 0.883 | 0.377 |
| ρ | 0.01 | 0.010 | 0.942 | 0.346 |
| n1 | 0.24 | 3.240 | 0.076 | 0.393 |
| n2 | 16.95 | 15.990 | 1.060 | 0.288 |
| n3 | −95.00 | 51.710 | −1.837 | 0.066 |
| n4 | 237.79 | 109.410 | 2.173 | 0.029 |
| n5 | −250.25 | 142.200 | −1.760 | 0.078 |
| n6 | 1.99 | 124.580 | 0.016 | 0.987 |
| n7 | 195.11 | 116.940 | 1.668 | 0.095 |
| n8 | −164.73 | 71.400 | −2.307 | 0.021 |
| n9 | 27.08 | 19.160 | 1.413 | 0.157 |
ρ, the average density of whole infection; V, area proportion of total infectious area; Vi, he area proportion of infection area in inner lung zone; Vp, the area proportion of specific region among the whole infection region; nx, the area proportion of Zx among the whole infection region.
Discussion
We presented a new deep learning-based model to assess the density and location of the infectious region precisely and comprehensively. The result of the primary clinical application also showed that our model was effective and stable in monitoring COVID-19 patients with severe infection.
In this work, we explored deep learning to segment COVID-19 infectious regions within lung fields on CT scans. With the proposed algorithm integrated, our system showed advantages from two aspects. First, our system supplies fast and correct segmentation for COVID-19 infectious regions, compared to manual contouring. Second, with exact segmentation, our system supplies quantitative information that is necessary to track disease progression and analyze longitude changes of COVID-19 during the entire treatment period. Besides, compared with traditional qualitative method, our system could provide a more objective assessment of the infectious lesions because it was not dependent on the experience of radiologist. Thus, we believe that this deep learning system for COVID-19 quantification will open many new research directions of interest in this community. The first potential research application of this system is to quantify longitudinal changes in the follow-up CT scans of COVID-19 patients. Another research application of our system is to explore the quantitative lesion distribution specifically related to COVID-19.
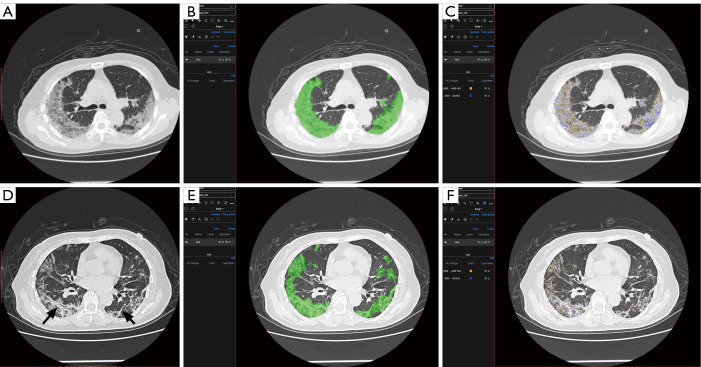
The clinical application provided evidence of some hypotheses on COVID-19 treatment from physicians. The regression result showed that after adjusting the age, which was a definite risk factor of a bad outcome in COVID-19 patients, the total volume and average density of the whole infectious region were not related to the ARDS existence. This result explained the poor efficiency of manual assessment to predict the disease progression, where the V and ρ was the main content of observation. However, a relationship between the area of specific densities and the location of the infectious segments with the risk of ARDS existence was observed. For example, our results showed with the increase of the proportion of the zone 4 area in the infectious region, the risk of ARDS existence becomes higher, while the increase of zone 8 was related to the lower risk of ARDS existence. From our experience, the possible mechanism of such a result was that the regions with CT values located in zone 4 intervals were considered as the tissue between ground-glass opacity (GGO) and consolidation (as shown in Figure 4A,B,C). The increase of such regions’ area usually means the GGO region in the infection was turned into consolidation rapidly, which was related to the deterioration of the disease. While the region with CT values located in the zone 8 interval was considered as consolidation tissues that were turned into fibrous tissue, which could indicate the disease was on the mend (as shown in Figure 4D,E,F).
Figure 4.
A 79-year-old female was diagnosed with severe COVID-19. After seven days treatment, her symptoms relieved. (A,B,C) However, the chest CT showed that the area of infection increased rapidly. The quantitative analysis result showed several zone 4 area interval density in her infectious area. Her symptoms deteriorated the next day, and the gas arterial blood gas analysis showed ARDS. (D,E,F) After 12 days’ treatment, another Quantitative CT analysis result showed the area of Zone4 decrease while the area of zone 8 increase (the zone 8 density mainly distributed in fibrous tissue area, black arrow in D), which could indicate the disease was on the mend. COVID-19, coronavirus disease 2019; CT, computed tomography.
Based on this result, we recommend a quantitative evaluation of some specific density infectious segment rather than the assessment of the whole volume or density of the infectious region. However, it was difficult for radiologists to distinguish the adjacent interval manually. Thus, an automatic density segmentation was necessary. Besides, although not significantly, our result showed that the increase of infection is found in the inner zone of the lung could lead to a higher risk of ARDS existence. The current experience showed that the initiation of COVID-19 infection mostly found in the peripheral zone of the lung. In other words, when the inner zone gets infected, it means the infection is progressing rapidly.
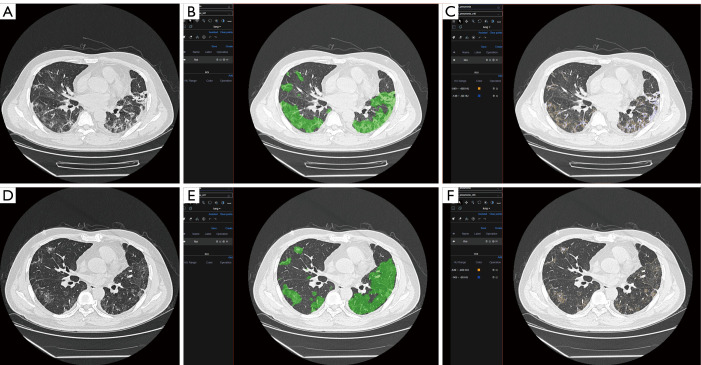
The most important benefit of quantitative assessment is the physicians could be conscious of the disease progression earlier than the onset of related symptoms. For instance, a 79-year-old female was diagnosed with severe COVID-19. After seven days of treatment, her symptoms relieved. The quantitative assessment showed that the area of infection increased rapidly and obvious zone 4 interval density in her infectious area (Figure 4A,B,C). Her symptoms deteriorated the next day, and the gas arterial blood gas analysis showed ARDS. There were many studies indicated that patients could have a large area of infection before symptom onset (18,19), but only some of them needed ventilation. For example, Figure 5 showed another case with extensive infection in the bilateral lung. Interestingly, his respiratory status stayed fine during the whole treatment. After a quantitative assessment, we found the density and location of his infection distributed in the minimal risk intervals of our analysis. The above two cases showed that the application of automatic CT assessment could help physicians discriminate those who were at higher risk better. In this study, we explored the relationship of chest CT imaging findings with the risk of ARDS in COVID-19 patients using an automatic quantitative assessment model, and the preliminary observations were quite encouraging. However, there are several limitations to our study. Due to the relatively small number of included patients and chest CT data, the power of statistical analysis may be influenced, and its stability might be a concern. Further multicenter prospective validation study with a large size cohort is needed. The second limitation is that the novelty of the algorism of the quantitative assessment model may not be very extraordinary compared to previously reported ones. However, the functionalities of this model are diverse and suited well for the application in various clinical scenarios of COVID-19, especially in supporting treatment decisions. Thirdly, the imaging data acquired by CT is quantitatively analyzable, although there are still some disputes about the role of chest CT over X-ray radiography in the diagnostic and therapeutic workups for COVID-19 patients. Further, the quantified imaging biomarkers can provide us with unique insights into the pathological changes and progression of this cunning viral pneumonia, helping both the diagnosis and treatment.
Figure 5.
A 56-year-old male was diagnosed with serve COVID-19, which had an extensive infection in the bilateral lung. Interestingly, his respiratory status stayed fine during the whole treatment. (A,B,C) was the first CT scans after admission. (D,E,F) was the CT scans after seven days treatment. In both CT scans’ quantitative analysis, zone 4 only composed of a limited proportion of the whole infection area. COVID-19, coronavirus disease 2019; CT, computed tomography.
Conclusions
In this study, we explored the relationship of chest CT imaging findings with the existence of ARDS in COVID-19 patients using an automatic quantitative assessment model, and our results found that the model was sensitive and stable for the lesion segmentation. The preliminary application of the model showed that the quantitative analysis of the density and location of infectious regions was related to the risk of ARDS existence in severe COVID 19 patients. Our assessment model supplies an excellent prospect of the quantitative CT application during the COVID-19 treatment, but a further prospective study was needed to draw a definite conclusion.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Supplementary
GE Healthcare CT scanner protocol: tube voltage, 120 kV (automatic adjustment); tube current, 200–500 mAs; rotation time, 0.5 second; section thickness, 0.625 mm; collimation, 0.625 mm; pitch, 1; matrix, 512×512; and inspiration breath hold. Reconstruction was performed with a bone algorithm with a thickness of 1 mm and an interval of 1 mm.
Siemens CT protocol: tube voltage, 120 kV; tube current, 110 mAs (automatic adjustment); rotation time, 0.5 second; section thickness, 0.75 mm; collimation, 0.6 mm; pitch, 1; matrix, 512×512; and inspiration breath hold. Reconstruction was performed with a bone algorithm with a thickness of 1 mm and an interval of 1 mm.
Table S1. Quantitative evaluation of the deep learning segmentation system on the validation dataset. The Dice coefficients, volume estimation error and POI estimation error in the whole lung, lung lobes, and bronchopulmonary segments were calculated to assess the automatic segmentation accuracy.
| Accuracy metrics | Mean | Standard deviation | Median | 25% IQR | 75% IQR | Number of infected samples |
|---|---|---|---|---|---|---|
| Dice similarity coefficient | 91.6% | 10.0% | 92.2% | 89.0% | 94.6% | 300 |
| Volume estimation error (cm3) | 10.7 | 16.7 | 5.9 | 1.8 | 13.9 | 300 |
| POI (the whole lung) | 0.3% | 0.4% | 0.1% | 0.0% | 0.4% | 300 |
| POI (left upper lobe) | 0.4% | 1.0% | 0.1% | 0.0% | 0.4% | 233 |
| POI (left lower lobe) | 0.7% | 1.6% | 0.3% | 0.1% | 1.0% | 267 |
| POI (right upper lobe) | 0.3% | 0.7% | 0.1% | 0.0% | 0.5% | 213 |
| POI (right middle lobe) | 0.3% | 0.7% | 0.1% | 0.0% | 0.5% | 204 |
| POI (right lower lobe) | 0.6% | 1.1% | 0.3% | 0.1% | 0.9% | 275 |
| POI (left upper lobe/posterior tip) | 0.5% | 1.0% | 0.1% | 0.0% | 0.5% | 189 |
| POI (left upper lobe/anterior) | 0.5% | 1.2% | 0.2% | 0.0% | 0.5% | 158 |
| POI (left upper lobe/upper tongue) | 0.7% | 1.7% | 0.2% | 0.0% | 0.9% | 192 |
| POI (left upper lobe/lower tongue) | 0.7% | 1.8% | 0.2% | 0.0% | 0.8% | 175 |
| POI (left lower lobe/dorsal) | 0.9% | 2.1% | 0.4% | 0.1% | 1.2% | 224 |
| POI (left lower lobe/anterior medial basal) | 0.6% | 1.4% | 0.2% | 0.0% | 0.8% | 209 |
| POI (left lower lobe/outer basal) | 1.1% | 2.5% | 0.5% | 0.1% | 1.7% | 228 |
| POI (left lower lobe/posterior basal) | 1.1% | 2.4% | 0.5% | 0.1% | 1.6% | 233 |
| POI (right upper lobe/apical) | 0.4% | 1.1% | 0.1% | 0.0% | 0.5% | 142 |
| POI (right upper lobe/back) | 0.7% | 1.7% | 0.2% | 0.0% | 0.8% | 186 |
| POI (right upper lobe/anterior) | 0.4% | 1.1% | 0.1% | 0.0% | 0.9% | 151 |
| POI (right middle lobe/lateral) | 0.6% | 1.5% | 0.1% | 0.0% | 0.6% | 183 |
| POI (right middle lobe/medial) | 0.3% | 0.8% | 0.1% | 0.0% | 0.4% | 167 |
| POI (right lower lobe/dorsal) | 0.9% | 1.9% | 0.4% | 0.1% | 1.4% | 233 |
| POI (right lower lobe/inner basal) | 0.6% | 1.4% | 0.3% | 0.1% | 0.9% | 162 |
| POI (right lower lobe/anterior basal) | 0.6% | 1.4% | 0.1% | 0.0% | 0.9% | 210 |
| POI (right lower lobe/outer basal) | 0.9% | 1.8% | 0.4% | 0.1% | 1.2% | 236 |
| POI (right lower lobe/posterior basal) | 1.0% | 2.0% | 0.5% | 0.1% | 1.6% | 249 |
IQR, interquartile range; POI, percentage of infection.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective study was approved by the West China Hospital Sichuan University Biomedical ethics review board (No. 2020140). The requirement for informed consent was waived because the anonymized study did not alter any diagnosis and treatment of the patients.
Footnotes
Reporting Checklist: The authors have completed the STROEB reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3554
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3554
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3554). The authors have no conflicts of interest to declare.
References
- 1.Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 2020;382:727-33. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phan LT, Nguyen TV, Luong QC, et al. Importation and Human-to-Human Transmission of a Novel Coronavirus in Vietnam. N Engl J Med 2020;382:872-4. 10.1056/NEJMc2001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei J, Xu H, Xiong J, et al. 2019 Novel Coronavirus (COVID-19) Pneumonia: Serial Computed Tomography Findings. Korean J Radiol 2020;21:501-4. 10.3348/kjr.2020.0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Dong C, Hu Y, et al. Temporal Changes of CT Findings in 90 Patients with COVID-19 Pneumonia: A Longitudinal Study. Radiology 2020. doi: . 10.1148/radiol.2020200843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis 2020;20:425-34. 10.1016/S1473-3099(20)30086-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubin GD, Ryerson CJ, Haramati LB, et al. The Role of Chest Imaging in Patient Management during the COVID-19 Pandemic: A Multinational Consensus Statement from the Fleischner Society. Radiology 2020. doi: . 10.1148/radiol.2020201365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kermany DS, Goldbaum M, Cai W, et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018;172:1122-31.e9. 10.1016/j.cell.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 8.Li L, Qin L, Xu Z, et al. Artificial Intelligence Distinguishes COVID-19 from Community Acquired Pneumonia on Chest CT. Radiology 2020. doi: . 10.1148/radiol.2020200905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang L, Han R, Ai T, et al. Serial Quantitative Chest CT Assessment of COVID-19: Deep-Learning Approach. Radiology 2020;2:e200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li K, Fang Y, Li W, et al. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19). Eur Radiol 2020. doi: . 10.1007/s00330-020-06817-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen C, Yu N, Cai S, et al. Quantitative computed tomography analysis for stratifying the severity of Coronavirus Disease 2019. J Pharm Anal 2020;10:123-9. 10.1016/j.jpha.2020.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bak SH, Lee HY, Kim JH, et al. Quantitative CT Scanning Analysis of Pure Ground-Glass Opacity Nodules Predicts Further CT Scanning Change. Chest 2016;149:180-91. 10.1378/chest.15-0034 [DOI] [PubMed] [Google Scholar]
- 13.Milletari F, Navab N, Ahmadi SA. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition 2016:770-8. [Google Scholar]
- 14.He K, Zhang X, Ren S, et al., editors. Deep Residual Learning for Image Recognition. IEEE Conference on Computer Vision & Pattern Recognition; 2016. [Google Scholar]
- 15.Han M, Zhang Y, Zhou Q, et al. Large-scale evaluation of V-Net for organ segmentation in image guided radiation therapy. Medical Imaging 2019: Image-Guided Procedures, Robotic Interventions, and Modeling; 2019: International Society for Optics and Photonics; 2019:109510O. [Google Scholar]
- 16.Mu G, Ma Y, Han M, et al. Automatic MR kidney segmentation for autosomal dominant polycystic kidney disease. Medical Imaging 2019: Computer-Aided Diagnosis; 2019: International Society for Optics and Photonics; 2019:109500X. [Google Scholar]
- 17.World Health Organization. Clinical management of severe acute respiratory infection when novel coronavirus (2019-nCoV) infection is suspected: interim guidance, 28 January 2020 . World Health Organization 2020. [cited 2020 April 14]. Available online: https://apps.who.int/iris/handle/10665/330893
- 18.Pan F, Ye T, Sun P, et al. Time Course of Lung Changes On Chest CT During Recovery From 2019 Novel Coronavirus (COVID-19) Pneumonia. Radiology 2020. doi: . 10.1148/radiol.2020200370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernheim A, Mei X, Huang M, et al. Chest CT Findings in Coronavirus Disease-19 (COVID-19): Relationship to Duration of Infection. Radiology 2020. doi: . 10.1148/radiol.2020200463 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as