Abstract
Background
Carbapenem-resistant Klebsiella pneumoniae (CRKP) has become an urgent global public health issue, but its distribution has obvious regional differences. The purpose of this study was to investigate the patient-based disease burden and molecular epidemiology of CRKP infections in a tertiary hospital in northern Jiangsu Province in China.
Methods
A retrospective, epidemiological survey of CRKP infections in our hospital from January to December 2016 was conducted to collect clinical and epidemiologic data. Non-duplicated clinical CRKP isolates were collected for the resistance-associated genes and clonal correlation analysis by PCR, sequencing and multilocus sequence typing (MLST).
Results
252 CRKP infection cases were collected, and the annual CRKP infection incidence of the hospital during 2016 was 14.64 per 10,000 hospital discharges (252/172,112*10,000) and 13.78 per 100,000 patient days (252/1,829,190*100,000). The patient-based disease burden concentrated on antimicrobial exposure history (133/224, 59.37%)—the most dominant STs. KPC-2 (120/128, 93.8%) was the predominant carbapenemase and ST11 (98/128, 76.5%) was the dominant STs. One isolate was detected with harboring blaKPC-2 and blaMCR-1 simultaneously.
Conclusions
Patient-based disease burden and KPC-2-producing ST11 Klebsiella pneumonia caused in higher CRKP incidence in the hospital. The emergence of CRKP with blaKPC-2 and blaMCR-1 should be of concern.
Keywords: Carbapenem-resistant Klebsiella pneumonia, ST11, infection
Introduction
Klebsiella pneumonia is a common opportunistic Enterobacteriaceae bacterium, which mostly affects patients with compromised immune systems, and causes various system infections. In the past decade, abusing antibiotics, especially third-generation cephalosporins and carbapenems, for Klebsiella pneumonia infections led to a rapid increase in drug resistance (1). In 2007, a carbapenem-resistant Klebsiella pneumonia (CRKP) was reported firstly in Zhejiang Province (2). Given CRKP’s receptivity to horizontal gene transfer and recombination with mobile genetic elements, it had been reported in every province in China and many parts of the world (3). CRKP was resistant to most antimicrobials and caused high rates of morbidity and mortality and had become an emerging threat to public health worldwide (4). China Antimicrobial Surveillance Network (CHINET) showed the rate of carbapenem resistance to the Enterobacteriaceae had risen from 0.7% in 2000 to 14% in 2015, and the carbapenem resistance rates to K. pneumoniae had increased from 3% in 2005 to 18.9% in 2016 (5,6).
In a multicenter retrospective research in China, the incidence rate of Carbapenem-resistant Enterobacteriaceae (CRE) infection was average 4.0 per 10,000 discharges (7), which was slightly higher than the incidence of European countries and the United States, 1.3 per 10,000 hospital and 2.93 per 100,000 population admissions, respectively (8,9), but the detail material displayed in one hospital from northern Jiangsu Province, the incidence was a staggering 14.9 per 10,000 discharges in this study, which was higher than the national average (4.0 per 10,000 discharges) (7). The reason caused such a high incidence of CRE was unknown, and most previous studies focused on the analysis of risk factors and molecular epidemiology of CRKP and rarely involved the burden of disease in CRKP patients. So, the epidemiological information survey was necessary for the prevention and control of CRKP.
Previous research showed CRKP isolates accounted for the absolute advantage of clinical CRE strains in China (10), so in the current study, we aimed to collect CRKP isolates to investigate such a high CRE infection incidence, by examining the patient’s disease burden and molecular epidemiology of antibiotic resistance genes, and the relationship between each other to obtain the measures to control and treat infection of such strains.
Methods
Ethical approval
The Hospital Ethics Clerk approved the study. The medical records and biological specimens used in this retrospective study were obtained from earlier clinical diagnosis and treatment, and the committee exempted the patient’s informed consent.
Study design and Bacterial strains
A retrospective, epidemiologic surveillance survey of CRKP infection was conducted in a tertiary hospital with over 4,000 beds in northern Jiangsu province, China from January to December 2016. Inpatients with CRKP positive cultures from either blood, cerebrospinal fluid (CSF), lower respiratory secretion, urine or wound secretion were selected for clinical characteristics investigation. Scheduling of all surveillance survey was designed to gather clinical and epidemiologic information, containing specimen tested time, type of culture specimen, sample origin, vital statistics, hospital-acquired or community-acquired infection, the tourist history, health care history in the past year, medical conditions experienced, invasive operation and history of antibiotic use within 30 days before culture, types of infection, clinical signs, therapy, and outcomes.
Non-duplicate CRKP strains of partial patients were collected to study the molecular epidemiology features. All the strains were identified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Bruker Daltonics, Germany).
MLST analysis
Seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB and tonB) for multilocus sequence typing (MLST) were amplified according to protocols provided on the K. pneumonia MLST website (http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html). The sequences of the polymerase chain reaction (PCR) products were performed by comparing it with the international K. pneumoniae MLST database to get the sequence type (ST).
Antimicrobial susceptibility test
The susceptibility of CRKP isolates to 17 antibiotics consist of ertapenem, meropenem, imipenem, ceftazidime, ceftriaxone, cefotaxime, aztreonam, cefepime, piperacillin/Tazobactam, ciprofloxacin, levofloxacin, gentamicin, tobramycin, amikacin, trimethoprim/Sulfamethoxazole, tigecycline and colistin were obtained using the automated VITEK-2 compact system. Susceptibility breakpoints were interpreted asper the recommendations of the Clinical and Laboratory Standards Institute (http://www.clsi.org/). For colistin and tigecycline, the breakpoints were defined according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/). Escherichia coli ATCC 25922 and K. pneumoniae ATCC700603 were used as a quality control strain for the antimicrobial susceptibility tests.
Identification of antibiotic resistance genes
Carbapenemase genes (blaKPC, blaNDM, blaIMP, and blaVIM); extended-spectrum β-lactamases (ESBLs) (blaCTX-M, blaTEM, blaSHV), and AmpC β-lactamases (blaDHA, and blaCMY) and colistin resistance gene (blaMCR-1) were detected by PCR amplification, and the primer sequences and reaction conditions refer to previous studies. The PCR amplicons of resistance genes were sequenced, and the gene sequences were compared on the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/blast/).
Definitions
If a Klebsiella pneumoniae strain were resistant to at least one of the carbapenem agents (imipenem, ertapenem and meropenem), it would be defined as a CRKP according to the recommendation of the Center for Disease Control and Prevention.
Statistical analyses
All the statistical analyses were performed by WHONET (version 5.6) and SPSS software package (version 18.0). The annual incidence rate for CRKP infections was calculated as the counts of CRKP infection cases divided by the counts of hospital discharges in 2016. The incidence of CRKP infection was also calculated as the counts of CRKP infection cases per 100,000 inpatient-days. Descriptive statistics summarized the clinical and epidemiological characteristics of CRKP infections. The score of the Charlson complication index was calculated. Continuous variables were represented as the median of the median range. For categorical variables, the percentages of patients or isolates in each category were calculated.
Results
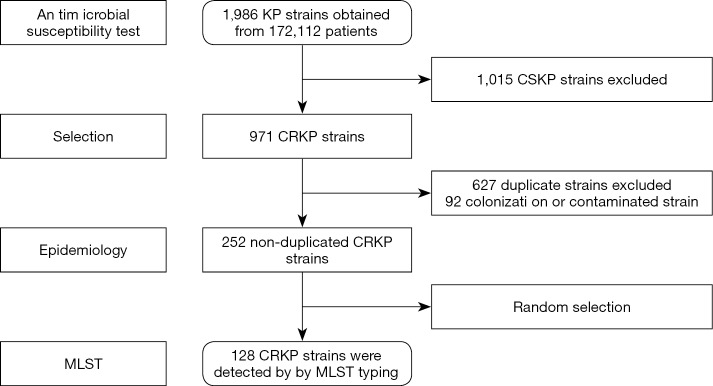
Between January 1 and December 31, 2016, a total of 252 cases with CRKP infection were included for the investigation according to the exclusion criteria (Figure 1). The CRKP burden showed that the annual CRKP infection incidence of the hospital during 2016 was 14.64 per 10,000 hospital discharges (252/172, 112*10,000) and 13.78 per 100,000 patient days (252/1,829, 190*100,000).
Figure 1.
A flowchart for the CRKP choice process in this study. CRKP, carbapenem-resistant Klebsiella pneumoniae.
Of the 252 individual cases, there were 85 female patients (33.7%) and 167 male patients (62.3%), respectively, and the median age was 59 years (interquartile range, 50 to 73; Table 1). The largest proportion is between 65 and 69 years old (91/252, 36.11%). 49.21% (124/252) of the patients presented with more than one underlying disease, and the median Charlson Comorbidity Index was 1 (interquartile range: 0–2). Co-morbidities include hypertension (108/252, 42.86%), followed by ICU stay (75/252, 29.76%), neurological diseases (66/252, 26.19%) and pulmonary diseases (59/252, 23.64%). The rate of use of urinary catheterization and central venous catheters was 89.29% (225/252) and 50.79% (128/252), respectively. Eighty-eight point eighty-nine percent (224/252) of patients had a history of antimicrobial using in 30 days before the culture, and most of the cases had the use of multiple antibiotics (133/224, 59.37%).
Table 1. Demographic characteristics of patients infected with CRKP.
| Characteristics | No./Total (%) |
|---|---|
| Female | 85/252 (33.73) |
| Age, median (interquartile range), years | 59 [50~73] |
| 0–4 | 5/252 (1.98) |
| 5–17 | 4/252 (1.59) |
| 18–49 | 55/252 (21.83) |
| 50–64 | 72/252 (28.57) |
| 65–79 | 91/252 (36.11) |
| ≥80 | 25/252 (9.92) |
| Travel history to Southeast Asia | 1/252 (0.4) |
| Health care exposure during the prior year | |
| Acute care hospitalization | 17/252 (6.75) |
| A resident of a long-term care facility | 3/252 (1.20) |
| The long-term acute care facility | 2/252 (0.79) |
| Underlying conditions | |
| Charlson Comorbidity Index, median (interquartile range) | 1 (0~2) |
| None | 128/252 (50.79) |
| Cancer | 10/252 (4.0) |
| Diabetes mellitus | 43/252 (17.06) |
| Heart diseases | 18/252 (7.14) |
| Hypertension | 108/252 (42.86) |
| Immunodeficiency | 0/252 (0.00) |
| ICU stay | 75/252 (29.76) |
| Liver diseases | 6/252 (2.38) |
| Neurological diseases | 66/252 (26.19) |
| Pulmonary diseases | 59/252 (23.41) |
| Renal diseases | 7/252 (2.78) |
| Receipt of corticosteroids | 6/252 (2.38) |
| Receipt of immunosuppressor | 0/252 (0.00) |
| Smoking history | 30/252 (11.90) |
| Indwelling devices prior to culture | |
| Arterial cannula | 6/252 (2.38) |
| Central venous catheter | 128/252 (50.79) |
| Gastric tube | 11/252 (4.36) |
| Tracheal cannula | 112/252 (44.44) |
| Tracheotomy | 77/252 (30.56) |
| Urinary catheter | 225/252 (89.29) |
| Antimicrobials use prior to the culture within 30 days | |
| None | 28/252 (11.11) |
| Single antimicrobial use | 91/224 (40.63) |
| Multiple antimicrobials use | 133/224 (59.37) |
| 3rd or 4th generation cephalosporins use | 112/224 (50.00) |
| Carbapenems use | 55/224 (24.55) |
CRKP, carbapenem-resistant Klebsiella pneumoniae.
The clinical features and outcomes of the cases were presented in Table 2. Eighty-two point ninety-four percent (209/252) of the patients presented lower respiratory tract infection, followed by urinary tract infection (11.90%, 151/252). The most obvious increase in inflammatory markers was C-reactive protein (252/320, 78.8%), followed by white blood cell count (151/252, 59.92%) and procalcitonin (118/252, 46.82%). In terms of discharge and outcome, there were 135 (135/252, 53.57%) patients returned home, followed by 15 patients discharged to subsequent outpatient (15/252, 5.95%) or other hospitals (11/252, 4.37%), 3 patients discharged to another long-term acute facility (3/252, 1.19%). The overall mortality rate was 38.09% (96/252) with in-hospital mortality of 23.41% (59/252) and 30 day’s mortality 14.68% (37/252).
Table 2. Clinical characteristics and outcomes of patients infected with CRKP.
| Characteristics | No./total (%) |
|---|---|
| Infection types | |
| Intra-abdominal infection | 18/252 (7.14) |
| Urinary tract infection | 30/252 (11.90) |
| Lower respiratory tract infection | 209/252 (82.94) |
| Bacteremia | 23/252 (9.13) |
| Meningitis | 25/252 (9.92) |
| Other infections | 3/252 (1.19) |
| APACHE II Score | 20 (4~48) |
| Septic shock | 3/252 (1.19) |
| Laboratory findings | |
| White blood cell, /mm3 | |
| Median (Interquartile range) | 13.82 (3.6~88.8) |
| Subgroup | |
| <4,000 (/mm3) | 30/252 (11.90) |
| >10,000 (/mm3) | 151/252 (59.92) |
| C-reactive protein >10 mg/L | 118/252 (46.82) |
| Procalcitonin | |
| >2 ng/mL | 40/252 (15.87) |
| Clinical outcomes | |
| Length of stay, median (interquartile range), days | 37.11 (3~730) |
| ICU admission | 204/252 (80.95) |
| Length of ICU stay, median (interquartile range), days | 14.44 (1~112) |
| Discharge disposition | |
| Home | 127/252 (50.540) |
| Long-term acute facility | 3/252 (1.19) |
| Follow-up outpatient | 15/252 (5.95) |
| Transfer to other hospitals | 11/252 (4.37) |
| Mortality | |
| In-hospital mortality | 59/252 (23.41) |
| 30 day’s mortality | 37/252 (14.68) |
CRKP, carbapenem-resistant Klebsiella pneumoniae.
The antimicrobial resistance profiles of 252 CRKP strains are summarized in Table 3. All CRKP strains showed multidrug resistance, i.e., they showed resistance to 3 or more classes of drugs. Of these 252 CRKP strains, they showed an exceptionally low sensitivity rate (6/252, 2.4%) to meropenem, while displayed nearly complete resistance to ertapenem and imipenem except for 2 isolates showed intermediate to imipenem. And all isolates showed resistant rates of more than 95% to ceftazidime, ceftriaxone, cefotaxime, cefepime, aztreonam, piperacillin/tazobactam. Amikacin, Trimethoprim/Sulfamethoxazole, gentamicin and levofloxacin showed activities against CRKP with resistant rates of 58.7%, 78.3%, 84.5% and 88.5%, respectively. Polymyxin and tigecycline remained excellent susceptibility (>90%).
Table 3. Antimicrobial susceptibility testing results of 252 CRKP isolates based on local clinical laboratories (%, n).
| Antimicrobial agents | S [n] | I [n] | R [n] |
|---|---|---|---|
| Ertapenem | 0 [0] | 0 [0] | 100 [252] |
| Meropenem | 2.4 [6] | 4.4 [11] | 93.3 [235] |
| Imipenem | 0 [0] | 0.8 [2] | 99.2 [250] |
| Ceftazidime | 1.2 [3] | 0 [0] | 98.8 [249] |
| Ceftriaxone | 0 [0] | 0 [0] | 100 [252] |
| Cefotaxime | 0.8 [2] | 0 [0] | 99.2 [250] |
| Aztreonam | 2.8 [7] | 0 [0] | 97.2 [245] |
| Cefepime | 2.4 [6] | 0.8 [2] | 96.8 [244] |
| Piperacillin/Tazobactam | 3.6 [9] | 0.4 [1] | 96.0 [242] |
| Ciprofloxacin | 6.4 [16] | 1.6 [4] | 92.1 [232] |
| Levofloxacin | 8.7 [22] | 2.8 [7] | 88.5 [223] |
| Gentamicin | 11.5 [29] | 4.0 [10] | 84.5 [252] |
| Tobramycin | 7.5 [19] | 2.4 [6] | 90.1 [227] |
| Amikacin | 36.9 [93] | 4.4 [11] | 58.7 [105] |
| Trimethoprim/sulfamethoxazole | 23.0 [58] | 3.2 [8] | 73.8 [186] |
| Tigecycline | 92.5 [233] | 3.2 [8] | 4.4 [11] |
| Colistin | 99.6 [251] | 0 [0] | 0.4 [1] |
CRKP, carbapenem-resistant Klebsiella pneumoniae.
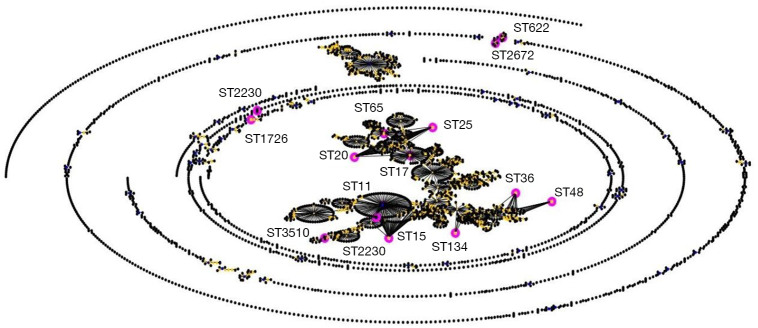
The clinical outcomes showed that the ICU admission rate of patients with CRKP infection was astonishing 80.95% (204/252). It suggested that CRKP isolates were collected from the ICU ward. A total of 128 isolates were collected in this experiment. And 15 STs were found within the 128 strains by MLST typing (Figure 1). The major type was ST11 (98/128, 76.6%), followed by ST48 (12/128, 9.4%), ST134 (3/128, 2.3%), ST2230 (33/128, 2.3%). Other STs included 2 or fewer isolates in each ST, which were be comprised of, ST25 (2/128, 1.6%), ST2270 (1/128, 0.78%), ST15 (1/128, 0.78%) and so on (Table 4). All 15 STs were clustered into 7 CCs (CC11, CC15, CC17, CC20, CC25, CC36, CC48) and eight singletons according to the eBURST analysis algorithm (Figure 2), showing the genetic diversity of CRKP in this study.
Table 4. Drug resistance gene distribution among the molecular types of 128 CRKP isolates from pediatric patients.
| Sequence type [15] | Isolates [128] | Carbapenemase [125] | AmpC β-lactamase [66] | Penicllinases and Extended-spectrum β-lactamase [128] |
|---|---|---|---|---|
| ST11 | 98 | KPC-2 [97], NDM-5 [1] | DHA-1 [59] | TEM-1 [89], SHV-12 [64], SHV-11 [17], CTX-M-65 [65], CTX-M-3 [1], CTX-M-14 [3], CTX-M-27 [1] |
| ST48 | 12 | KPC-2 [11] | TEM-1 [12], SHV-12 [8], SHV-11 [3] | |
| ST134 | 3 | KPC-2 [3] | DHA-1 [3] | TEM-1 [2], SHV-12 [2], CTX-M-65 [3] |
| ST2230 | 3 | KPC-2 [2] | TEM-1 [3], SHV-12 [3], CTX-M-65 [2] | |
| ST25 | 2 | KPC-2 [2] | TEM-1 [2], SHV-12 [2], CTX-M-3 [2] | |
| ST2270 | 1 | DHA-1 [1] | TEM-1 [1], SHV-12 [1], CTX-M-27 [1] | |
| ST15 | 1 | KPC-2 [1] | TEM-1 [1], SHV-11 [1], CTX-M-65 [1] | |
| ST65 | 1 | IMP-4 [1] | TEM-1 [1], SHV-12 [1], CTX-M-65 [1] | |
| ST3501 | 1 | KPC-2 [1] | DHA-1 [1] | TEM-1 [1], SHV-12 [1], CTX-M-65 [1] |
| ST1726 | 1 | KPC-2 [1] | TEM-1 [1], SHV-12 [1], CTX-M-65 [1] | |
| ST36 | 1 | NDM-1 [1] | DHA-1 [1] | TEM-1 [1], SHV-12 [1], CTX-M-15 [1] |
| ST2672 | 1 | NDM-5 [1] | TEM-1 [1], SHV-12 [1], CTX-M-55 [1] | |
| ST622 | 1 | NDM-5 [1] | TEM-1 [1], SHV-12 [1], CTX-M-15 [1] | |
| ST20 | 1 | KPC-2 [1] | DHA-1 [1] | TEM-1 [1], SHV-11 [1], CTX-M-65 [1] |
| ST17 | 1 | KPC-2 [1] | TEM-1 [1], SHV-12 [1], CTX-M-65 [1] |
CRKP, carbapenem-resistant Klebsiella pneumoniae.
Figure 2.
An eBURST analysis of the K. pneumonia MLST database (accessed on August 10, 2019). The sequence types found in this study were marked with pink circles.
Among the 128 studied isolates, all strains were detected as positive for ESBL genes and 66 strains co-carried ESBL and AmpC genes (Table 4). The stains often carried 3 or 4 different types of broad-spectrum β-lactamase genes, and the most frequently one was blaTEM-1 (92.3%, 118/128), followed by blaSHV (88.3%, 113/128), blaCTX-M-65 (56.3%, 72/128), blaCTX-M-14 (2.3%, 3/128), blaCTX-M-15 (1.6%, 2/128) and others (2.3%, 3/128). For the targeted carbapenemase genes, blaKPC-2 (93.8%, 120/128), blaNDM-5 (2.3%, 3/128), blaNDM-1 (0.8%, 1/128) and blaIMP-4 (0.8%, 1/128) were detected among the isolates. One colistin-resistant isolate co-carried blaKPC-2 and blamcr-1 was identified in this study.
Discussion
Infections caused by CRKP, with increasing resistance rates and mortality, are a pressing issue for public health (11). Previous multicenter or multinational studies showed the CRE incidence in the European countries, the United States and China, which was 1.3 per 10,000 hospital admissions, 2.93 per 100,000 population and 4.0 per 10,000 discharges, respectively (7-9). But the details of the report show that CRE incidence a hospital in Jiangsu, China, shows an amazing 14.97% per 10,000 hospital discharges (7). Given the absolute advantage of CRKP strains in the proportion of CRE strains, our study combined the patient-based disease burden and the epidemiological characteristics of clinical CRKP infection cases to reveal the cause of high incidence in this hospital. The current study showed the annual CRKP infection incidence of the hospital during 2016 was 14.64 per 10,000 hospital discharges and 13.78 per 100,000 patient days. The level of CRKP infection incidence was consistent with previous reports, which was far higher than the level of European and American countries and the average level of China (7,8).
Patient’s disease burden
As previously reported, elderly patients, hypertension, diabetes, ICU stay, neurological disease, lung disease, and various aggressive procedures are important risk factors for CRKP infections (12,13). Demographic data in our study showed that the incidence of most risk factors was almost the same as the national level, but the incidence of ICU stay and various aggressive procedures was significantly higher than the national average level (7). To our best knowledge, the patient’s disease burden, higher urinary tube use (89.29%), tracheotomy (30.56%) and ICU stay (29.76%) may be significant contributors to CRKP with high incidence in this hospital.
History of antimicrobial use, especially for the use of 3rd or 4th generation cephalosporins and carbapenems before infection, was confirmed as risk factors correlated with CRKP infections in previous studies (14). In our study, 88.89% of patients received antibiotic treatment within 30 days before a positive culture. Of these patients, 91 received single antibiotic treatment, and 133 received multiple antibiotic treatments. A total of 112 (112/224, 50.00%) and 55 (55/224, 24.55%) of 224 patients received 3rd or 4th generation cephalosporin or carbapenems. Excessive exposure to multiple antimicrobials uses and 3rd or 4th generation cephalosporins were the cause of higher CRKP incidence similar to previous reports (15).
Molecular epidemiology
Several mechanisms are leading to carbapenem resistance. Carbapenemases, which are largely responsible for resistance, include KPC, NDM-1, VIM, IMP, OXA-48 and so on (16). These enzymes can be encoded not only by chromosomal genes but also by mobile elements obtained, such as plasmids and transposons, which is the main reason for the rapid spread of the epidemic. Producing KPC is the most frequent mechanism of resistance to carbapenems in K. pneumonia. KPC-producing K. pneumoniae has spread worldwide in the past decades. In China, KPC-2 is the main cause of carbapenem resistance. In China, KPC-producing is the most common mechanism for carbapenemase resistance in K. pneumoniae (17). Following the earlier previous report, we detected 120 (93.8%) stains harbored the KPC-2 carbapenemases. Two class B metallobeta-lactamases, NDM (NDM-1, NDM-5) and IMP-4 were detected in 4 isolates. This suggested that KPC-2 producing was the main cause of carbapenem resistance in this hospital.
The patients with CRKP infection in the hospital were mainly concentrated in the ICU (204/252, 80.95%) ward, and the lower respiratory tract infections (209/252, 82.94%) were the main type of infections. It has been reported the major MLST type of CRKP was ST11 in China (18). The MLST analysis in our study showed that the most prevalent clone was ST11 (98/128, 76.6%), which were also mainly from the lower respiratory tract infections and ICU ward. In this KPC-2 type producing CRKP, the drug-resistant genes, which are mediated by plasmids, are extremely easy to spread horizontally among bacteria and are likely to cause outbreaks of nosocomial infections (18). This highly aggregated KPC-2 producing CRKP with lower respiratory tract infections from the ICU ward suggested some possible local outbreak in this hospital. And outbreaks in some specific wards possibly played a major role in higher CRKP incidence in this hospital and caused a high mortality rate (38.09%), which ranged from approximately 20% to 70% in previous data (7,19). The possible outbreaks need to be brought to the attention of the infection control department, and some effective measures should be implemented to reduce the higher CRKP incidence of this hospital.
Colistin belongs to the family of cationic polypeptide antibiotics, with broad-spectrum ability against multiple resistant bacterial infections, which is commonly supposed to the last line of defense to the CRE infections, but the description of colistin resistance by a plasmid-mediated resistance gene, blaMCR-1, in 2015 raised concerns in CRKP (20). To our best knowledge, one isolate was resistant to meropenem, ertapenem, imipenem, and colistin except for tigecycline in this study, and both MCR-1 and KPC-2 genes were detected. Both genes harbored in one strain were reported in Brazil, which was plasmid-mediated and detected in K. pneumonia ST437 (21). This is the first report of K. pneumoniae ST11 co-harboring the blaKPC-2 and blaMCR-1 in China, suggesting that the difficult situation of K. pneumoniae resistance in this hospital.
Conclusions
We have to admit that our research has some limitations, such as the sample size is not large enough, and some clinical data cannot be fully obtained to reflect the actual infection situation. However, we did find some unique reasons for the high incidence of CRKP. Patient’s disease burden, contained higher urinary tube use, tracheotomy, ICU stay, generation cephalosporins and carbapenems exposure history, and some possible outbreaks of KPC-2-producing ST11 CRKP in specific wards played major roles in higher CRKP incidence in the hospital. These results suggest that targeted measures to reduce patients’ disease burden and control cluster outbreaks should be implemented in this hospital to control the high incidence of CRKP.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This research was supported by the National Natural Science Foundation of China (81871734), Jiangsu Provincial Medical Talent (ZDRCA2016053), Six Talent Peaks Project of Jiangsu Province (WSN-091), Advanced Health Talent of the Six-One Project of Jiangsu Province (LGY2016042), Xuzhou Science and Technology Planning Project (KC19160).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Ethics Committee approved the study of the Affiliated Hospital of Xuzhou Medical University (IRB: XYFY2015-JS005-01).
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editors (Zhi-De Hu, Bing Gu) for the series “Advances in laboratory tests for infectious diseases” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.03.122). The series “Advances in Laboratory Tests for Infectious Diseases” was commissioned by the editorial office without any funding or sponsorship. BG served as the unpaid Guest Editor of the series and serves as an unpaid Managing Editor of Annals of Translational Medicine. The other authors have no other conflicts of interest to declare.
References
- 1.Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011;17:1791-8. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei ZQ, Du XX, Yu YS, et al. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother 2007;51:763-5. 10.1128/AAC.01053-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcoleta AE, Berrios-Pasten C, Nunez G, et al. Klebsiella pneumoniae Asparagine tDNAs Are Integration Hotspots for Different Genomic Islands Encoding Microcin E492 Production Determinants and Other Putative Virulence Factors Present in Hypervirulent Strains. Front Microbiol 2016;7:849. 10.3389/fmicb.2016.00849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamma PD, Goodman KE, Harris AD, et al. Comparing the Outcomes of Patients With Carbapenemase-Producing and Non-Carbapenemase-Producing Carbapenem-Resistant Enterobacteriaceae Bacteremia. Clin Infect Dis 2017;64:257-64. 10.1093/cid/ciw741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu FP, Guo Y, Zhu DM, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005-2014. Clin Microbiol Infect 2016;22 Suppl 1:S9-14. 10.1016/j.cmi.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 6.Xiao YH, Wang J, Li Y, et al. Bacterial resistance surveillance in China: a report from Mohnarin 2004-2005. Eur J Clin Microbiol Infect Dis 2008;27:697-708. 10.1007/s10096-008-0494-6 [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Wang Q, Yin Y, et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae Infections: Report from the China CRE Network. Antimicrob Agents Chemother 2018. doi: . 10.1128/AAC.01882-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guh AY, Bulens SN, Mu Y, et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae in 7 US Communities, 2012-2013. JAMA 2015;314:1479-87. 10.1001/jama.2015.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grundmann H, Glasner C, Albiger B, et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis 2017;17:153-63. 10.1016/S1473-3099(16)30257-2 [DOI] [PubMed] [Google Scholar]
- 10.Zhang R, Liu L, Zhou H, et al. Nationwide Surveillance of Clinical Carbapenem-resistant Enterobacteriaceae (CRE) Strains in China. EBioMedicine 2017;19:98-106. 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 2015;112:E3574-81. 10.1073/pnas.1501049112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pang F, Jia XQ, Zhao QG, et al. Factors associated to prevalence and treatment of carbapenem-resistant Enterobacteriaceae infections: a seven years retrospective study in three tertiary care hospitals. Ann Clin Microbiol Antimicrob 2018;17:13. 10.1186/s12941-018-0267-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoxha A, Karki T, Giambi C, et al. Attributable mortality of carbapenem-resistant Klebsiella pneumoniae infections in a prospective matched cohort study in Italy, 2012-2013. J Hosp Infect 2016;92:61-6. 10.1016/j.jhin.2015.06.018 [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, Zhang Y, Yao X, et al. Risk factors and clinical outcomes for carbapenem-resistant Enterobacteriaceae nosocomial infections. Eur J Clin Microbiol Infect Dis 2016;35:1679-89. 10.1007/s10096-016-2710-0 [DOI] [PubMed] [Google Scholar]
- 15.Turner P, Pol S, Soeng S, et al. High Prevalence of Antimicrobial-resistant Gram-negative Colonization in Hospitalized Cambodian Infants. Pediatr Infect Dis J 2016;35:856-61. 10.1097/INF.0000000000001187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pitout JD, Nordmann P, Poirel L. Carbapenemase-Producing Klebsiella pneumoniae, a Key Pathogen Set for Global Nosocomial Dominance. Antimicrob Agents Chemother 2015;59:5873-84. 10.1128/AAC.01019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi Y, Wei Z, Ji S, et al. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 2011;66:307-12. 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- 18.Fu P, Tang Y, Li G, et al. Pandemic spread of blaKPC-2 among Klebsiella pneumoniae ST11 in China is associated with horizontal transfer mediated by IncFII-like plasmids. Int J Antimicrob Agents 2019;54:117-24. 10.1016/j.ijantimicag.2019.03.014 [DOI] [PubMed] [Google Scholar]
- 19.Fraenkel-Wandel Y, Raveh-Brawer D, Wiener-Well Y, et al. Mortality due to blaKPC Klebsiella pneumoniae bacteraemia. J Antimicrob Chemother 2016;71:1083-7. 10.1093/jac/dkv414 [DOI] [PubMed] [Google Scholar]
- 20.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016;16:161-8. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 21.Dalmolin TV, Martins AF, Zavascki AP, et al. Acquisition of the mcr-1 gene by a high-risk clone of KPC-2-producing Klebsiella pneumoniae ST437/CC258, Brazil. Diagn Microbiol Infect Dis 2018;90:132-3. 10.1016/j.diagmicrobio.2017.09.016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as