Abstract
Background
Coronavirus disease 2019 (COVID-19) has spread rapidly worldwide from Wuhan. An easy-to-use index capable of the early identification of inpatients who are at risk of becoming critically ill is urgently needed in clinical practice. Hence, the aim of this study was to explore an easy-to-use nomogram and a model to triage patients into risk categories to determine the likelihood of developing a critical illness.
Methods
A retrospective cohort study was conducted. We extracted data from 84 patients with laboratory-confirmed COVID-19 from one designated hospital. The primary endpoint was the development of severe/critical illness within 7 days after admission. Predictive factors of this endpoint were selected by LASSO Cox regression model. A nomogram was developed based on selected variables. The predictive performance of the derived nomogram was evaluated by calibration curves and decision curves. Additionally, the predictive performances of individual and combined variables under study were evaluated by receiver operating characteristic curves. The developed model was also tested in a separate validation set with 71 laboratory-confirmed COVID-19 patients.
Results
None of the 84 inpatients were lost to follow-up in this retrospective study. The primary endpoint occurred in 23 inpatients (27.4%). The neutrophil-to-lymphocyte ratio (NLR) and C-reactive protein (CRP) were selected as the final prognostic factors. A nomogram was developed based on the NLR and CRP. The calibration curve and decision curve indicated that the constructed nomogram model was clinically useful. The AUCs for the NLR, CRP and Combined Index in both training set and validation sets were 0.685 (95% CI: 0.574–0.783), 0.764 (95% CI: 0.659–0.850), 0.804 (95% CI: 0.702–0.883), and 0.881 (95% CI: 0.782–0.946), respectively.
Conclusions
Our results demonstrated that the nomogram and Combined Index calculated from the NLR and CRP are potential and reliable predictors of COVID-19 prognosis and can triage patients at the time of admission.
Keywords: Coronavirus disease 2019 (COVID-19), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), prognostic, neutrophil-to-lymphocyte ratio (NLR), C-reactive protein (CRP)
Introduction
In December 2019, several cases of pneumonia of unknown etiology, now known as coronavirus disease 2019 (COVID-19), occurred in Wuhan, China. The disease has rapidly spread from Wuhan to other provinces in China and around the world. As of March 4, 2020, a total of 84,022 cases in China have been laboratory confirmed. Globally, confirmed cases have been documented in 77 countries and 3,198 deaths caused by COVID-19 have been reported (1).
According to the Novel Coronavirus Pneumonia Prevention and Control Program (5th edition) published by the National Health Commission of China, COVID-19 patients were categorized into four types: mild, common, severe and critical (2). The clinical classifications are as follows: (I) mild: patients present mild symptoms with no imaging sign of pneumonia; (II) common: patients have fever and respiratory tract symptom with imaging sign of pneumonia; (III) severe: patient who meets any of the following conditions: patients present respiratory distress with respiratory rate ≥30 breath/min, SpO2 (oxygen saturation) ≤93% on room air, and PaO2 (arterial blood oxygen partial pressure)/FiO2 (fraction of inspired oxygen) ≤300 mmHg (1 mmHg =0.133 kPa); (IV) critical: patient meets any of the following conditions: patient presents respiratory failure and requires mechanical ventilation support, patient presents shock, and patient presents multiple organ dysfunction syndrome and requires ICU admission (3). Most mild/common patients have a good prognosis, while the mortality of severe or critical type patients with COVID-19 is considerable (4). Hence, preventing mild or common cases from progressing into severe/critical cases is of paramount value to reduce mortality. However, factors dictating the severity of this newly emerged disease are not well defined. In addition, dyspnea was one of the major characteristics of severe- and critical-type COVID patients. The median duration from illness onset to dyspnea was 5–8 days (5,6). Hence, the establishment of a simple and effective model to predict the progression of COVID-19 in the early phase is urgently needed. The data from such research are scarce but are of paramount importance.
This study aims to develop a useful model based on routine laboratory assessment results along with demographic information to predict the 7-day severity of COVID-19 among patients, which will be of considerable value for the early identification of admitted patients who are at risk of becoming critically ill. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-2372).
Methods
Study design and participants
For this retrospective, cohort, single-center study, we recruited patients from January 8, 2020 to February 14, 2020 from the General Hospital of Central Theater Command, which was a designated hospital for COVID-19 patients. According to the policy put in place by the local government, both laboratory-confirmed and suspected cases were admitted to this hospital. Only laboratory-confirmed COVID-19 patients with mild/common types on admission were enrolled in this study. A confirmed case of COVID-19 was defined as a positive result on high-throughput sequencing or real-time reverse-transcriptase-polymerase-chain-reaction (RT-PCR) assay using nasal or pharyngeal swab specimens (6-8). Laboratory assessments consisted of a complete blood count, blood chemical analysis, myoglobin (Myo), coagulation testing, assessment of liver and renal function, and measures of electrolytes, C-reactive protein, procalcitonin, lactate dehydrogenase, and creatine kinase. All the data of enrolled cases, including demographic information, clinical symptoms or signs, clinical outcomes and laboratory findings on admission, were extracted from electronic medical records. For cases receiving multiple laboratory tastings during their hospitalization, only results on the day of admission were used for analysis. If multiple laboratory tastings were tested on the day of admission, the results which were the closet to admission time were used for analysis. The primary endpoint or event of interest in this study was the development of severe/critical disease within 7 days of admission. Hence, the final follow-up day for each enrolled patient was the seventh day after admission.
This study was approved by the Ethics Commission of General Hospital of Central Theatre Command {[2020]017-1}. The data are anonymous, and the requirement for informed consent was therefore waived.
Statistical analysis
IBM SPSS Statistics 22.0 and R 3.6.2 were used for statistical analyses. All tests were two-tailed, and P less than 0.05 was considered statistically significant. The chi-square test or Fisher’s exact probability test was used to compare count data. A Kolmogorov-Smirnov test or Shapiro-Wilk test was used to test the normality of the continuous variables. For continuous data that met the criteria of the normality test, an independent t-test was used for the comparison. Otherwise, the Mann-Whitney U test was used. The penalized Cox regression model with Least absolute shrinkage and selection operator (LASSO) was used to identify the most important factors of 7-day critical or severity transition in COVID-19 patients (9,10). The Cox proportional hazard model was used for univariate and multivariate analysis of prognosis, and a nomogram was developed for variables that were statistically significant (11). The concordance index (C-index) was calculated to evaluate the predictive accuracy. Additionally, time-dependent receiver operating characteristic (ROC) curves, including the area under the curve (AUC) and its 95% confidence interval (95% CI), were analyzed by time ROC package to evaluate the performance of prognostic prediction (12). AUCs were compared according to the method described by DeLong et al. (13). The optimal threshold for each selected factor and combined index was determined when the Youden index achieved the highest value. On the basis of the optimal cut-off given by the ROC curve analysis, all of the factors under study were transformed to binary variables, and their corresponding no severe illness survival rates were calculated by the Kaplan-Meier method.
Results
Demographic profiles and laboratory findings of the participants on admission
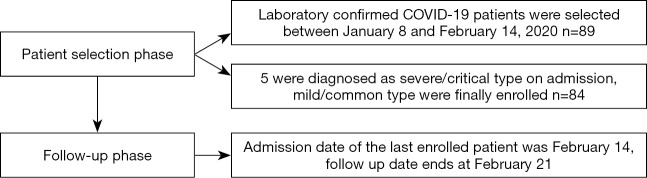
Eighty-four inpatients with COVID-19 were enrolled in this study (Figure 1). The clinical characteristics and laboratory findings are shown in Table 1. The median age of these participants was 53 years (range, 20–91 years). A total of 27.4% of patients were younger than 39 years, while 32.1% were above the age of 60 years. A total of 47% were males. A total of 37.0% had at least one underlying disease (e.g., hypertension and coronary heart disease). The most common underlying disease in this study population was hypertension (16/84, 19.0%). The second most common underlying disease was coronary heart disease (8/84, 9.5%). The status of patients on admission was judged by doctors according to the American Thoracic Society guideline (14) and was categorized as non-severe in 41 patients and severe in 43 patients. The definition of severe here was different from that of severe COVID-19 according to the National Health Commission of China. As of February 21, 73 patients (86.9%) remained in the hospital, 8 were discharged from the hospital, and 3 died.
Figure 1.
Flowchart of patient selection and follow up.
Table 1. Clinical characteristics and laboratory findings of 84 enrolled in patients with COVID-19.
| Clinical characteristics | All patients (n=84) | Primary endpoint | ||
|---|---|---|---|---|
| Event (n=23) | Free-of-event (n=61) | P | ||
| Age (years), median (range) | 53 (20.0–91.0) | 67 (31.0–91.0) | 51 (20.0–83.0) | 0.125 |
| Age group, n (%) | ||||
| ≤39 | 23 (27.4) | 2 (8.7) | 21 (34.4) | 0.026 |
| 40–59 | 34 (40.5) | 6 (26.1) | 28 (45.9) | 0.321 |
| ≥60 | 27 (32.1) | 15 (65.2) | 12 (19.7) | <0.001 |
| Male sex, n (%) | 47 (56.0) | 16 (69.6) | 31 (50.8) | 0.125 |
| Body mass index (kg/m2), median (range) | 24.31 (15.24–36.13) | 24.80 (15.24–36.13) | 22.73 (17.80–27.43) | 0.245 |
| Underlying disease, n (%) | ||||
| Any | 31 (37.0) | 14 (60.9) | 17 (27.9) | 0.010 |
| Hypertension | 16 (19.0) | 10 (43.5) | 6 (9.8) | 0.001 |
| COPD | 2 (2.4) | 0 (0.0) | 2 (3.3) | – |
| Diabetes | 7 (8.3) | 3 (13.0) | 4 (6.6) | 0.378 |
| Coronary heart disease | 8 (9.5) | 6 (26.1) | 2 (3.3) | 0.004 |
| Cancer§ | 3 (3.6) | 1 (4.3) | 2 (3.3) | 0.793 |
| Chronic renal disease | 5 (6.0) | 1 (4.3) | 4 (6.6) | 0.726 |
| Hepatitis B infection | 3 (3.6) | 1 (4.3) | 2 (3.3) | 0.793 |
| Cerebrovascular disease | 5 (6.0) | 3 (12.9) | 2 (3.3) | 0.116 |
| Status on admission (severe), n (%)& | 43 (51.2) | 11 (47.8) | 32 (52.5) | 0.808 |
| qSOFA score | ||||
| 0 | 82.1% (69/84) | 56.5% (13/23) | 91.8% (56/61) | <0.001 |
| 1 | 17.9% (15/84) | 43.5% (10/23) | 8.2* (5/61) | <0.001 |
| Time (days)*, median (range) | NA | 2 (1.0–7.0) | NA | NA |
| Outcomes (as of Feb 21), n (%) | ||||
| Still in hospital | 73 (86.9) | 18 (78.3) | 55 (90.1) | 0.320 |
| Discharge from hospital | 8 (9.5) | 2 (8.7) | 6 (9.8) | 0.874 |
| Died | 3 (3.6) | 3 (13.0) | 0 | – |
| Laboratory findings, median (range) | ||||
| White blood cells | 4.70 (1.70–14.90) | 4.60 (2.0–11.4) | 5.4 (1.7–14.9) | 0.274 |
| Neutrophil | 3.05 (0.86–14.15) | 2.80 (0.96–10.07) | 3.44 (0.86–14.15) | 0.203 |
| Lymphocyte | 1.05 (0.21–3.74) | 1.16 (0.33–3.74) | 0.78 (0.21–3.74) | <0.001 |
| NLR | 2.82 (0.67–67.38) | 2.61 (0.67–11.31) | 5.17 (1.28–67.38) | <0.001 |
| RBC | 4.28 (2.29–5.58) | 4.29 (3.46–5.44) | 4.18 (2.99–5.58) | 0.818 |
| Hemoglobin | 128 (75.0–158.0) | 127 (75.0–158.0) | 129 (90.0–156.0) | 0.944 |
| RDW | 12.2 (10.8–17.4) | 12.15 (10.8–17.4) | 12.5 (11.2–16.6) | 0.047 |
| Platelets | 170 (70.0–399.0) | 177 (102.0–370.0) | 156 (70.0–399.0) | 0.019 |
| ESR | 25 (3.0–139.0) | 24 (3.0–80.0) | 29 (6.0–138.0) | 0.144 |
| CRP | 15.2 (0.5–126.0) | 12.90 (0.5–74.60) | 37.69 (1.2–126.0) | 0.009 |
| Interleukin-6 | 16.75 (1.5–200.8) | 13.1 (1.5–187.8) | 27.70 (13.20–200.8) | 0.007 |
| MYO | 24.59 (0.1–763.0) | 21.72 (21.0–763.0) | 59.97 (0.3–290) | 0.006 |
| pH | 7.42 (7.38–7.47) | 7.41 (7.38–7.45) | 7.42 (7.39–7.47) | 0.155 |
| PCO2 | 4.57 (3.39–6.05) | 4.82 (3.74–6.05) | 4.46 (3.39–5.45) | 0.325 |
| PO2 | 12.90 (5.30–28.80) | 10.00 (5.30–18.40) | 16.40 (10.30–28.80) | 0.002 |
According to the presence or absence of the primary endpoint, the patients were categorized into the event group (23 patients) and free-of-event group (61 patients). §, cancers referred to any malignancy. All cases were characterized by stable disease. &, the condition of patients on admission was judged by doctors according to the American Thoracic Society guidelines. Hence, they were divided into severe and non-severe groups. *, time here means from admission to developing severe/critical disease. NLR, neutrophil-to-lymphocyte ratio; RBC, red blood cell; RDW, red blood cell distribution width; ESR, erythrocyte sedimentation rate; qSOFA, quick sequential organ failure assessment.
According to the presence or absence of the primary endpoint, the patients were categorized into the event group (23 patients) and the free-of-event (FOE) group (61 patients). The differences in the status of enrolled patients on admission were not significant (P=0.808). The participants aged above 60 years in the event group were more likely to develop severe/critical disease within 7 days after admission than those in the FOE group (65.2% vs. 19.7%, P<0.001). The presence of any underlying disease, hypertension, and coronary heart disease was more common in the event group (P=0.010, 0.001, and 0.004, respectively). The median days from admission to the primary endpoint was 2 (range, 1–7). The three non-survivors were all from the event group. No deaths occurred in the FOE group as of February 21.
Absolute lymphocyte count, the neutrophil-to-lymphocyte ratio (NLR), red blood cell distribution width (RDW), platelets (PLTs), C-reactive protein (CRP), interleukin-6 (IL-6), and MYO in the event group were significantly different from those in the FOE group (P<0.05). The event group was characterized by dyspnea; hence, the PO2 concentration in the event group was obviously lower than that in the FOE group (P=0.002).
Selection of predictive indicators for the 7-day endpoint
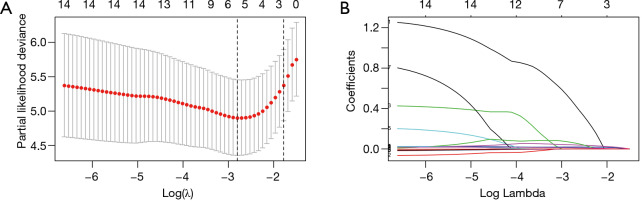
A total of 14 variables, including age, sex, underlying disease [treated as a categorical variable (yes vs. no)], complete blood count, MYO, and IL-6, were included in the LASSO regression. The results showed that underlying disease, age, NLR, RDW, PLT and CRP were predictive indicators for the 7-day endpoint (Figure 2). The abovementioned indicators were included in the Cox multivariate regression analysis. After adjustment, the results showed that NLR and CRP were independent predictive indicators with hazard ratios above 1.
Figure 2.
Selection of prognostic factors using the LASSO regression model. (A) LASSO regression used 10-fold cross-validation to determine the optimal tuning parameter (λ). The partial likelihood deviance was plotted vs. log(λ). (B) LASSO coefficient profiles of the 14 factors of COVID-19.
Nomogram construction
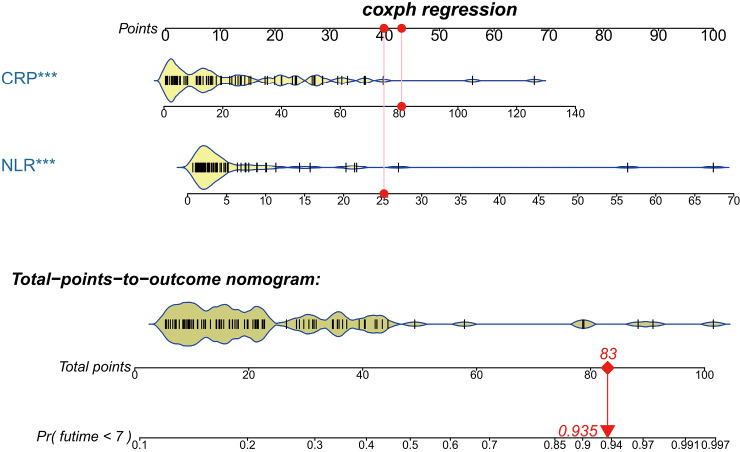
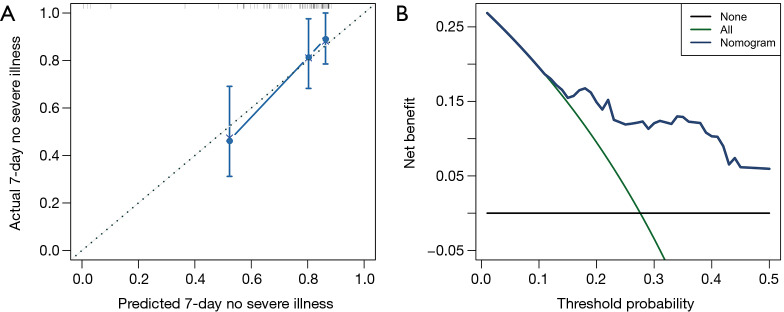
The nomogram was developed based on the NLR and CRP and was used to quantitatively predict the free-of-event 7-day survival rates (Figure 3). The C-index of the nomogram was 0.784 (95% CI: 0.680–0.888). The calibration curve depicted in Figure 4 shows that the derived nomogram performed well when compared to the actual results. Findings from the decision curve analysis showed that using the developed nomogram to predict the severe illness probability added more net benefit than either the treat-all or treat-none scheme, indicating that the constructed nomogram model was clinically useful.
Figure 3.
CRP and the NLR were used to develop a nomogram to predict the probability of noncritical illness on the seventh day. For example (indicated by a solid red circle and arrow), the CRP and NLR results of an admitted patient were 80 mg/L and 25, respectively. The points for his/her CRP and NLR were approximately 43 and 40, respectively. Hence, the total number of points for this patient was 83, which indicated a probability of 0.935 for developing severe/critical illness after admission. CRP, C-reactive protein; NLR, neutrophil-to-lymphocyte ratio.
Figure 4.
Validation and clinical usefulness of the nomogram. (A) Calibration curves of nomograms in terms of agreement between the predicted risk and actual observed outcomes; (B) decision curve analysis of the nomogram for 7-day survival with no severe illness.
Predictive performance of the NLR, CRP and Combined Index
The coefficients of the NLR and CRP from the Cox regression model were 0.051 and 0.020, respectively. Survival ROC curve analysis was used to analyze the predictive performance of the NLR, CRP and Combined Index (Figure 5). The optimal cut-offs and corresponding sensitivity and specificity are listed in Table 2. As expected, Combined Index in training set yielded an increased AUC of 0.804 (95% CI: 0.702–0.883; P=0.000) with 73.91% sensitivity and 76.67% specificity, which indicated that there was an additive effect in the predictive value of the two biomarkers. To confirm the reliability of the developed model, we calculated the Combined Index in a separate validation set with 71 laboratory-confirmed COVID-19 patients. An additive effect with AUC of 0.881 (95% CI: 0.782–0.946, P<0.001) was also obtained in the validation set, suggesting that the prognostic model had good sensitivity and specificity in predicting patients 7-day no severe illness survival.
Figure 5.
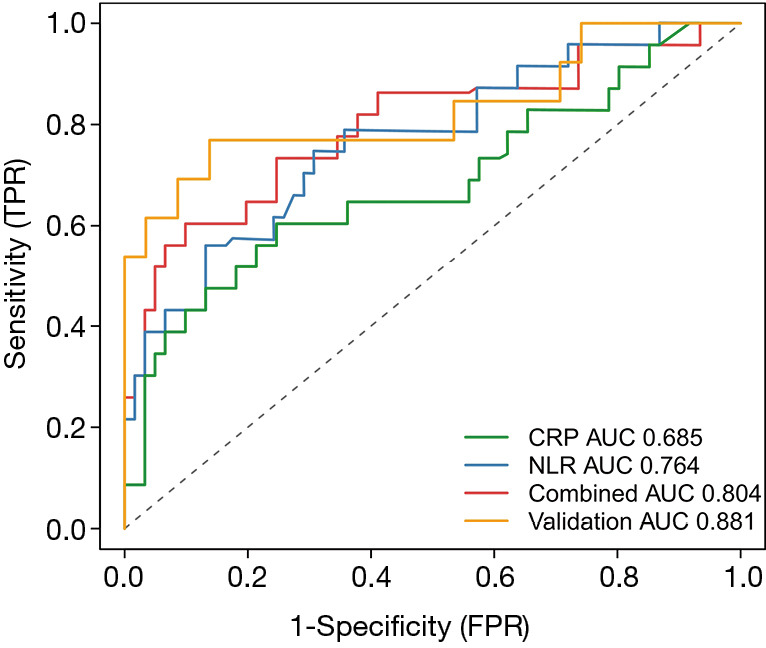
ROC curve analysis using the NLR, CRP and Combined Index for predicting the 7-day endpoint. CRP yielded an AUC of 0.685 (95% CI: 0.574–0.783), while NLR yielded an AUC of 0.764 (95% CI: 0.659–0.850). The combined index in both training and validation sets yielded increased AUCs of 0.804 (95% CI: 0.702–0.883) and 0.881 (95% CI: 0.782–0.946), respectively. neutrophil-to-lymphocyte ratio; CRP, C-reactive protein; ROC, receiver operating characteristic; AUC, area under the curve.
Table 2. Predictive performance of CRP, the NLR and Combined Index for the 7-day endpoint.
| Variables | Cut-off | AUC (95% CI) | Sensitivity | Specificity | P* |
|---|---|---|---|---|---|
| CRP | 25.96 | 0.685 (0.574–0.783) | 60.87% | 75.00% | <0.001 |
| NLR | 4.87 | 0.764 (0.659–0.850) | 56.52% | 86.89% | <0.001 |
| Combined Index (training) | 0.66 | 0.804 (0.702–0.883) | 73.91% | 76.67% | <0.001 |
| Combined Index (validation) | 0.66 | 0.881 (0.782–0.946) | 76.92% | 91.38% | <0.001 |
*, P value for AUC. NLR, neutrophil-to-lymphocyte ratio; CRP, C-reactive protein; AUC, area under the curve.
On the basis of the optimal cutoff based on the ROC curve analysis, all the indicators under study were transformed into binary variables, and risk ratios were calculated. The results of risk ratios demonstrated that increased CRP, NLR, and Combined Index in training set and validation sets were associated with a higher incidence of 7-day endpoint occurrence, with risk ratios of 4.667 (95% CI: 1.681–12.956), 8.458 (95% CI: 1.962–36.471), 7.792 (95% CI: 2.613–23.233) and 10.400 (95% CI: 2.224–48.624), respectively (Table 3).
Table 3. Risk ratios of CRP, the NLR and Combined Index.
| Variables | RR (95% CI) | P |
|---|---|---|
| CRP ≥25.93 | 4.667 (1.681–12.956) | <0.001 |
| NLR ≥4.87 | 8.458 (1.962–36.471) | <0.001 |
| Combined Index (training) ≥0.66 | 7.792 (2.613–23.233) | <0.001 |
| Combined Index (validation) ≥0.66 | 10.400 (2.224–48.624) | <0.001 |
CRP, C-reactive protein; NLR, neutrophil-to-lymphocyte ratio.
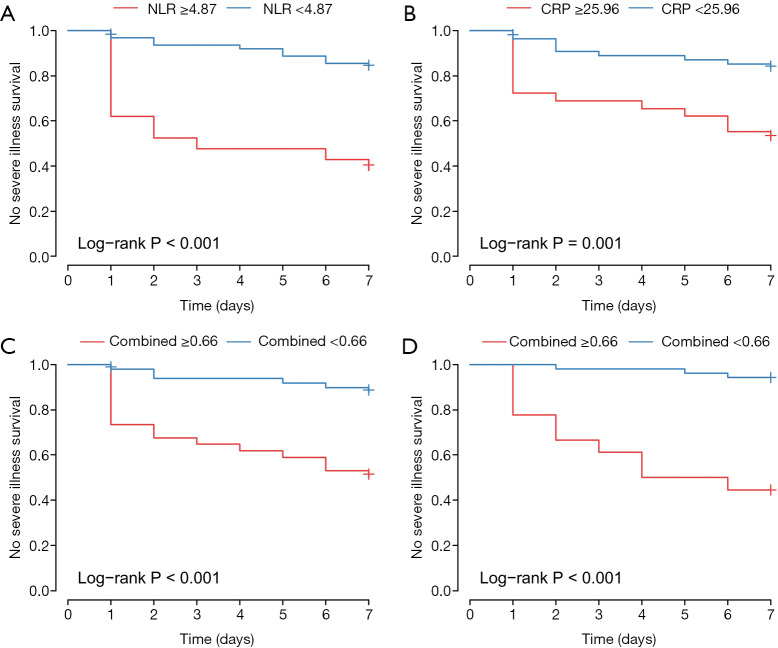
Kaplan-Meier curves of subgroups stratified by the NLR, CRP and Combined Index
None of the 84 inpatients were lost to follow-up in this retrospective study. The primary endpoint occurred in 23 inpatients (27.4%), including 3.6% (3/84) who died. Participants were categorized into two subsets according to the optimal cut-off value of CRP, the NLR and Combined Index in the training set, which is shown in Table 3. Kaplan-Meier curves showed that the difference between the two subgroups was statistically significant. Validation results further showed that patients in the validation model with high Combined Index had poor outcome compared to those with low Index (Figure 6).
Figure 6.
Kaplan-Meier curves of survival in patients without severe illness according to binary risk group stratification by (A) NLR (low risk: <4.87; high risk: ≥4.87), (B) CRP (low risk: <25.96; high risk: ≥25.96), (C) Combined Index in the training set (low risk: <0.66; high risk: ≥0.66) and (D) Combined Index in the validation set (low risk: <0.66; high risk: ≥0.66). The mean survival time according to Kaplan-Meier curves is shown in Table S1. CRP, C-reactive protein; NLR, neutrophil-to-lymphocyte ratio.
Discussion
COVID-19 has recently been declared a public health emergency of international concern by the World Health Organization (WHO); the responsible pathogen has been identified as a novel enveloped RNA beta-coronavirus that has currently been named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), previously named 2019 novel coronavirus (15,16). Similarities in demographic information and clinical features between SARS-CoV-2 and previous beta-coronavirus infections have been noted. Participants aged older than 60 years, especially those with underlying disease, were more likely to develop the severe/critical type within 7 days after admission, which is in agreement with previous studies (17,18).
A notable laboratory finding in regard to COVID-19 was reduced absolute lymphocyte count with a normal or slightly decreased white blood cell count, which was different from the findings in patients with other beta-coronavirus infections, such as Middle East respiratory syndrome (19). In clinical practice, it has been observed that in some non-survivors, the absolute value of lymphocytes decreases progressively while the white blood cell count gradually increases over time, resulting in a “divergence” between the absolute value of neutrophils and lymphocytes, which gave us the idea that the neutrophil-to-lymphocyte ratio, namely, the NLR, may be correlated with the progression and prognosis of this newly emerged disease. In fact, numerous observational studies and meta-analyses have documented that increased NLR is associated with poor prognosis in various kinds of diseases, such as pancreatic cancer, some solid cancers, sepsis and bacterial infection (20). Our results showed that the NLR alone yielded a relatively high AUC (0.764, 95% CI: 0.659–0.850) to predict the 7-day endpoint, but the sensitivity was just 56.52%. This sensitivity is inadequate for a predictive indicator because sensitivity is the ability to diagnose those with the disease correctly among those identified as positive by the test. The lower the sensitivity, the more false-negative cases. The combined use of CRP and the NLR not only yielded an elevated AUC of 0.804 (95% CI: 0.702–0.883; P=0.000) but also resulted in a greatly increased sensitivity from 56.52% to 73.92% at the expense of a decrease in specificity. Moreover, the newly developed model was tested in a separate validation set with 71 laboratory-confirmed COVID-19 cases. Results showed that AUC in validation set was 0.881 (95% CI: 0.782–0.946) with 76.92% sensitivity and 91.38% specificity. A previous preprint also showed that the NLR was an early-identification risk factor for COVID-19 severe illness; however, in that study, the NLR was the only predictor of illness severity (2). In addition, the sample size and study design differ.
Our study further demonstrated that patients with an increased NLR (≥4.87) were as much as 8.458-times (95% CI: 1.962–36.471) more likely to develop the severe/critical type than those with a low NLR (<4.87). Patients with an increased Combined Index (≥0.66) in both training and validation sets had as much as 7.792-times (95% CI: 2.613–23.233) and 10.400-times (95% CI: 2.224–48.624) higher risks of developing the severe/critical type than those with a low Combined Index (<0.66). The results of Kaplan-Meier curves of subgroups stratified by binary NLR, CRP and Combined Index also demonstrated that the mean survival time in each group was significantly different (P<0.001 for all). The findings collectively suggested that the Combined Index developed from the NLR and CRP is a reliable predictive indicator for the prognosis of this still fairly new disease.
It is well known that qSOFA score was recommended by the Third International Consensus Definitions for Sepsis as a simple prompt to identify infected patients outside the ICU who are likely to be septic (21). The qSOFA score ranges from 0 to 3. The presence of 2 or more qSOFA points near the onset of infection indicated high risks of death or prolonged ICU stay. However, in the present study, the qSOFA scores of 69 patients (82.1%) were 0 point, while those of the rest were 1 point. None scored 2 or more points. This was in agreement with a recent published research in Chest, their results showed that patients with COVID-19 induced ARDS had much lower SOFA scores (22). Our newly developed model may be more suitable as a predictive index than qSOFA score in this case cohort under study.
Nomograms are commonly used graphic calculating devices to predict prognosis in medical disciplines and some other fields and have the ability to allow for the approximate graphic computation of the likelihood of a clinical event by integrating diverse prognostic and determinant variables (23). The nomogram developed in our study based on NLR and CRP is an easy-to-use graphic tool for triaging patients into risky categories to determine the likelihood of developing severe/critical types and can fulfil our desire to triage a large number of patients in a short time.
However, some limitations must be noted in this research. First, the time (in days) was calculated from admission to the endpoint, not from the onset of illness. In reality, it was very difficult to obtain laboratory assessment results of a patient if he or she did not go to the hospital. Second, because retrospective cohort design properties and small sample sizes are major limitations of the present study, our results should be interpreted with caution and validated in large-scale studies. Last but not the least, because of the limited time and emergency of outbreak of COVID-19, we did not carry out a thorough analysis of all vital signs and mental status, more researches and development of models based on these easily obtainable vital signs and mental status are needed to be done later.
Conclusions
Early detection, early reporting, early isolation, early diagnosis and early management have collectively contributed to the reduction in both daily new confirmed cases and mortality in critically ill patients in China, which has led to great progress in the battle against COVID-19. However, there still remain significant challenges for clinical physicians to stratify risk at the time of patient admission. Our results demonstrated that the nomogram and Combined Index, derived from the NLR and CRP, is a potential, reliable and easy-to-use predictor for COVID-19 prognosis and triage patients at the time of admission. However, our results should be interpreted with caution and validated in longitudinal studies in a larger cohort.
Table S1. Mean survival time according to Kaplan-Meier curves.
| Variables | Low risk* (days) | High risk (days) | P |
|---|---|---|---|
| CRP | 6.4 (5.8–6.8) | 4.9 (3.9–5.9) | <0.001 |
| NLR | 6.5 (6.1–6.9) | 4.0 (2.7–5.3) | <0.001 |
| Combined Index in training set | 6.6 (6.2–7.0) | 4.7 (3.8–5.7) | <0.001 |
| Combined Index in validation set | 6.9 (6.6–7.0) | 4.2 (2.9–5.4) | <0.001 |
*, low risk for CRP, NLR and Combined Index are defined as <25.93, <4.87 and <0.66, respectively. Vice versa. CRP, C-reactive protein; NLR, neutrophil-to-lymphocyte ratio.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank Dr. Mengxue Yuan for her kind and professional assistance in calculating qSOFA scores during our revision.
Funding: This project was supported by the National Natural Science Foundation of China (81573254, 81872716). The funders played no role in the study design, data collection, data analysis, data interpretation, or writing of this manuscript.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Commission of General Hospital of Central Theatre Command {[2020]017-1}. The data are anonymous, and the requirement for informed consent was therefore waived.
Footnotes
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-2372
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-2372
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2372). The authors have no conflicts of interest to declare.
References
- 1.World Health Organization. Coronavirus disease (COVID-19) outbreak. Available online: https://www.who.int
- 2.Liu J, Liu Y, Xiang P, et al. Neutrophil-to-Lymphocyte Ratio Predicts Severe Illness Patients with 2019 Novel Coronavirus in the Early Stage. doi: https://doi.org/ 10.1101/2020.02.10.20021584 [DOI] [Google Scholar]
- 3.Cheng ZJ, Shan J. 2019 Novel coronavirus: where we are and what we know. Infection 2020;48:155-63. 10.1007/s15010-020-01401-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 2020;8:475-81. 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D, Hu B, Hu C, et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020;323:1061-9. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu DKW, Pan Y, Cheng SMS, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem 2020;66:549-55. 10.1093/clinchem/hvaa029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan L, Xu D, Ye G, Xia C, Wang S, Li Y, et al. Positive RT-PCR Test Results in Patients Recovered from COVID-19. JAMA 2020;323:1502-3. 10.1001/jama.2020.2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med 1997;16:385-95. [DOI] [PubMed] [Google Scholar]
- 10.Simon N, Friedman J, Hastie T, et al. Regularization Paths for Cox's Proportional Hazards Model via Coordinate Descent. J Stat Softw 2011;39:1-13. 10.18637/jss.v039.i05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Kattan MW. Drawing Nomograms with R: applications to categorical outcome and survival data. Ann Transl Med 2017;5:211. 10.21037/atm.2017.04.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med 2013;32:5381-97. 10.1002/sim.5958 [DOI] [PubMed] [Google Scholar]
- 13.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837-45. 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 14.Mandell LA, Wunderink RG, Anzueto A, et al. Infectious Diseases Society of America/American Thoracic Society Consensus Guidelines on the Management of Community-Acquired Pneumonia in Adults. Clin Infect Dis 2007;44:S27-72. 10.1086/511159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benvenuto D, Giovanetti M, Ciccozzi A, et al. The 2019-new coronavirus epidemic: evidence for virus evolution. J Med Virol 2020;92:455-9. 10.1002/jmv.25688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan W, Ni Z, Hu Y, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med 2020. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A Novel Coronavirus from Patients with Pneumonia in China 2019. N Engl J Med 2020. [Epub ahead of print]. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020;395:507-13. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park GE, Kang CI, Ko JH, et al. Differential Cell Count and CRP Level in Blood as Predictors for Middle East Respiratory Syndrome Coronavirus Infection in Acute Febrile Patients during Nosocomial Outbreak. J Korean Med Sci 2017;32:151-4. 10.3346/jkms.2017.32.1.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mei Z, Shi L, Wang B, et al. Prognostic role of pretreatment blood neutrophil-to-lymphocyte ratio in advanced cancer survivors: A systematic review and meta-analysis of 66 cohort studies. Cancer Treat Rev 2017;58:1-13. 10.1016/j.ctrv.2017.05.005 [DOI] [PubMed] [Google Scholar]
- 21.Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:801-10. 10.1001/jama.2016.0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang X, Du R, Wang R, et al. Comparison of Hospitalized Patients with Acute Respiratory Distress Syndrome Caused by COVID-19 and H1N1. Chest 2020. [Epub ahead of print]. 10.1016/j.chest.2020.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balachandran VPD, Gonen MP, Smith JJM, et al. Nomograms in oncology: more than meets the eye. Lancet Oncol 2015;16:e173-80. 10.1016/S1470-2045(14)71116-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as