Abstract
Background
The modification and regulation of N6-methyladenosine (m6A) at mRNA level can affect the development and progression in various tumors. ALKBH5, as an m6A demethylase, plays different roles in tumors by regulating the m6A modification of mRNA. However, its role in renal cell carcinoma (RCC) remains unclear.
Methods
First, levels of ALKBH5 in RCC tissues and cell lines were verified by qRT-PCR and western blot. We analyzed the relationship between ALKBH5 and the clinicopathological characteristics of RCC patients and the influence of ALKBH5 on the prognosis of patients. Then we generated ALBKH5-overexpression, ALBKH5-knockdown stable RCC cell lines and their control cell lines. Through cell proliferation assay, colony formation assay, cell invasion and tumor migration assay, cell cycle assay and xenograft studies, we studied the ALKBH5 roles in RCC cell lines. AURKB was predicted to be its potential target based on TCGA database analysis and verified by western blot. The role of AURKB in RCC was verified by TCGA database and Kaplan-Meier analysis with TMA immunohistochemical analysis. Finally, the specific molecular mechanism of ALKBH5 targeting AURKB was explored by dual-luciferase reporter assay, RNA immunoprecipitation (RIP), m6A dot-blot assay, m6A RNA Immunoprecipitation (MeRIP) assay, and mRNA stability assay.
Results
We found that ALKBH5 was highly expressed in both RCC tumor tissues and cell lines. Clinicopathological analysis showed that high ALKBH5 expression was associated with larger tumor volume (P=0.017) and higher TNM staging (P=0.006), and worse prognosis (log rank: P=0.0199). The cellular functional assays showed that stably overexpression ALKBH5 could promote the cell proliferation, colony formation, cell migration and cell invasion of renal cell carcinoma cells in vitro and promote tumor growth in vivo. In contrast, ALKBH5 knocked down inhibited cell proliferation, colony formation, migration and invasion of renal cell carcinoma cells in vitro. Based on TCGA database analysis, AURKB was predicted highly expressed in RCC and a potential target of ALKBH5. Both database prediction and TMA immunohistochemical analysis supported that AURKB could affect the prognosis of RCC patients (P values of 5.5e-08 and 0.0004, respectively) and was regulated by ALKBH5 expression level. Subsequent mechanism experiments showed that ALKBH5 regulated the expression of AURKB by regulating the stability of AURKB mRNA in the m6A-dependent manner, and finally promoted cell proliferation. Furthermore, we found that hypoxia-induced HIF could up-regulate both expressions of AURKB and ALKBH5.
Conclusions
Our findings suggest that ALKBH5 may play a carcinogenic role in renal cell carcinoma by stabilizing AURKB mRNA in a m6A-dependent manner. These data suggest that ALKBH5 may play a key role in RCC and targeting the ALKBH5 signaling pathway may be a promising strategy for the treatment of RCC.
Keywords: ALKBH5, renal cell cancer, N6-methyladenosine (m6A), AURKB
Introduction
Renal cell carcinoma (RCC) is one of the most common tumors in both men and women, accounting for 5% or 3% of malignant neoplasms respectively (1). Approximately 75% of patients diagnosed with RCC have localized disease. RCC is considered to be highly resistant to chemotherapy and radiotherapy, and surgery is still the current standard for the treatment of localized RCC (2,3). However, about 30–50% of intermediate- and high-risk RCC patients will develop metastatic RCC after surgical removal of the primary tumor (4). At the time of diagnosis, about 30% of cases are locally advanced or develop metastases. Recently, molecular targeted therapy has improved the overall survival of patients with metastatic RCC (5). However, due resistance to targeted drugs, the long-term prognosis remains poor. The identification of adjuvant therapy is an unmet need for RCC. Thus, thoroughly revealing the molecular mechanism in cancer is vital for the development of effective RCC therapies.
The post-transcriptional regulation of mRNA is the primary gene regulatory mechanism in eukaryotes, with N6-methyladenosine (m6A) being the most abundant and prevalent mRNA modifier in RNAs (6,7). The m6A modification of RNA is dynamic, and includes installation, erasure, and recognition, along with regulation by methyltransferase, demethylases, or readers (8,9). M6A regulates a series of RNA biological functions, such as the RNA splicing, translation efficiency, and mRNA stability (10). In recent years, scientists have reported that m6A-dependent RNA regulation could affect tumor initiation and progression in various cancers, including acute myeloid leukemia (AML) (11), breast cancer (12), bladder cancer (13), glioblastoma (14), pancreas cancer (15), colorectal cancer (16), and hepatic carcinoma (17). In RCC, we found that 1 methyltransferase-like protein in the methyltransferase complex, METTL3, acted as a tumor suppressor (18). Meanwhile, another component of the complex, Wilms tumor 1-associated protein (WTAP) promoted RCC progression by regulating stability of CDK2 mRNA (19). Zhuang et al. found that the fat mass and obesity-associated protein (FTO), another m6A demethylase, could suppresses clear cell RCC via FTO-PGC-1α signaling pathway (20). However, the role of the other components involved in m6A methylation regulation for RCC, along with the underlying mechanisms, is still not fully elucidated.
The m6A demethylase AlkB homolog 5 (ALKBH5) is localized in the nucleus and expressed in most tissues (21,22). It is known that ALKBH5 can influence gene expression, nuclear RNA transfer, and RNA metabolism (22). Recently, ALKBH5 was found to be involved in the progression of cancers and regulated through hypoxia-inducible factor (HIF) 1 in cancer cells (23). In breast cancer cells, ALKBH5 was shown to be directly targeted by HIF-1α and regulated by HIF-2α, and induce the phenotype of cancer stem cells by mediating NANOG mRNA m6A-demethylation, suggesting that ALKBH5 may play an important tumorigenic role (24). Furthermore, Zhang et al. demonstrated that ALKBH5 induced lower m6A level which helped to promote tumor progression in glioblastoma (25). Further study showed that ALKBH5 played a key role for breast cancer initiation (26) and gastric metastasis (27). ALKBH5 was also found to promote cell proliferation through interacting with DDX3 and AGO2 by regulating m6A levels (28). Moreover, in a study of epithelial ovarian cancer, ALKBH5 could reduce the autophagy and promote tumor growth and invasion through regulating the mRNA stability of Bcl-2 (29). However, it was also found that ALKBH5 could inhibit pancreatic tumor development by mediating the m6A-demethylation of lncRNA (30). Taken together, the literature suggests that ALKBH5 participates in the development of cancers by regulating m6A level and manifests variably in different cancer types. Still, the function and related mechanisms of ALKBH5 in RCC remain unclear.
In this study, the roles of ALKBH5 and related mechanisms in RCC were explored resulting in the following observations: (I) upregulated ALKBH5 was detected in RCC cell lines and tissues and correlated with poor outcomes; (II) ALKBH5 accelerated the cell growth in vitro and in vivo in RCC; (III) ALKBH5 promoted cell proliferation of RCC via regulating mRNA stability of AURKB in an m6A-dependant manner; (IV) HIF-induced hypoxia could upregulate the expression of AURKB by activating ALKBH5. Therefore, ALKBH5 may function as an oncogene in RCC and serve as a prognostic biomarker and therapeutic strategy in clinic.
Methods
Clinical specimens
RCC and matched adjacent normal tissue were collected from patients admitted to the Department of Urology of the First Affiliated Hospital of Nanjing Medical University from January 2008 to February 2010. These patients were undergoing radical nephrectomy and none had received chemotherapy, radiotherapy, or targeting therapy before surgical operation. All cases were individually categorized by independent pathologists. This study was ethically authorized by the Local Ethics Committees of the First Affiliated Hospital of Nanjing Medical University. We obtained informed consent from all the patients to use their data for research purposes.
Tissue microarray (TMA) and immunohistochemistry (IHC)
TMA was made from 96 formalin-fixed and paraffin-embedded RCC tumors samples. We performed IHC to assess ALKBH5 and AURKB protein level on TMA. These samples were stained with primary antibodies in the following manner: anti-ALKBH5 antibody (1:200, Sigma, USA) or anti-AURKB antibody (1:200, Abcam, USA). Standard staining protocols were used (19). The stained tissues were graded by staining intensity (SI) and percentage of positive cells (PP). The SI score ranged from 0 to 3 points (0, negative staining; 1, weak staining; 2, moderate dyeing; 3, strong staining), while PP was divided into 5 types: 0 (0% positive cells), 1 (<10%), 2 (11–50%), 3 (51–80%), 4 (>80%). SI and PP scores were multiplied to obtain the final staining scores, with a rank from 0 to 12. Two urologists rated the positive level of immunohistochemical staining. The low-staining [0–7] and high-staining [8–12] groups were grouped according to different scores.
Cell culture and transfection
Human RCC cell lines (Caki-1, Caki-2, 769P, 786-0, and ACHN) and a normal renal tubular epithelial cell line (HK-2) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cell lines Caki-1 and Caki-2 were cultured using McCoy’s 5A medium (Gibco, USA). 769P and 786-0 cells were maintained in RPMI-1640 medium (Gibco, USA), while AHCN and HK-2 cells were grown in DMEM media (Gibco, USA). Next, 1% penicillin/streptomycin solution (Gibco, USA) and 10% fetal bovine serum (BI, Israel) were added into all media at 37 °C in an incubator with 5% CO2.
For the inhibitor experiment, AZD1152-HQPZ and hesperidin (MCE, USA) were treated after cell attachment at concentrations of 10 and 120 µM. Colony formation and CCK8 assay were used to evaluate the inhibitory effect after 48 h of inhibitor treatment.
Lentiviruses of ALKBH5 overexpression and knockdown were constructed by Novobio (Novobio, China). In 6-well plates, 50% confluence of cells were seeded and infected with the ALKBH5 overexpression lentivirus (ALKBH5), a negative control (NC), ALKBH5 knockdown lentivirus (shALKBH5-1, shALKBH5-2), and a scramble control (SCR). Meanwhile, to enhance infection efficiency, polybrene (5 µg/mL) was added. After selection for 2 weeks with puromycin (4 µg/mL), the stably transfected cells were generated.
AURKB siRNA (5'-GCAGAGATCGAAATCCAGG-3') and the negative controls were purchased from GenePharma (GenePharma, China). Lipofectamine 2000 kit (Invitrogen, USA) was used for transfections, and cells were collected 48 h later. The role of siRNA was evaluated by CCK8 and colony formation.
RNA isolation and real-time quantitative polymerase chain reaction (qRT-PCR)
Total RNA was extracted from clinical specimens or cultured cell lines using Trizol reagent (Invitrogen, USA) and cDNA was synthesized using Primescript RT Reagent (TaKaRa, Japan). qRT-PCR was performed using SYBR® Premix Ex Taq™ Reagent (TaKaRa, Japan) and StepOne Plus Real-Time PCR system (Applied Biosystems, USA). The primers were as follows:
ALKBH5, Forward: 5'- CGCAAGGCAGACCCTGAT -3'
Reverse: 5'- CGGTTCTCTTCCTTGTCCATCT -3'
AURKB, Forward: 5'-CAGTGGGACACCCGACAT-3'
Reverse: 5'- GTACACGTTTCCAAACTTGCC -3'
β-actin, Forward: 5'-CCTGGCACCCAGCACAAT-3'
Reverse: 5'-GCTGATCCACATCTGCTGGAA-3'.
The ABI Step One Software version 2.1, 2−ΔΔCt method was used to calculate fold changes in mRNA expression, and normalization was based on β-actin.
Protein isolation and western blot
Total cellular proteins were lysed by RIPA buffer with protease and phosphatase inhibitors (Pierce, USA). BCA kit (Beyotime, China) was used to determine the protein concentration. Samples were separated by 10% or 12% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF, Millipore, USA) membrane. After incubation of the primary antibodies anti-ALKBH5 antibody (1:1,000; Sigma, USA), anti-HIF-1α antibody (1:1,000, CST, USA), anti-HIF-2α antibody (1:1,000, CST, USA), rabbit anti-AURKB antibody (1:1,000; Abcam, USA), or mouse anti-GAPDH (1:1,000; Cell Signal Technology, USA) at 4 °C for overnight, the membranes were then incubated with horseradish peroxidase (HRP)-labeled secondary antibodies (CST, USA) for 60 min at 37 °C. After washing 3 times, a chemiluminescence system (Bio-Rad, USA) and Image Lab Software (Bio-Rad, USA) were used to detect and analyze signals.
Cell proliferation assay
Cell proliferation was detected by Cell Counting Kit 8 (CCK8) assay (Dojindo, Japan). For each well 3×103 cells were seeded into 96-well plates. The cells were cultured for 24, 48, 72, and 96 h, incubated with CCK8 at 37 °C for 3 h; the absorbance at 450 nm was measured with a microplate reader.
Colony formation assay
The pretreated cells were seeded into 6-well plates with 600 cells per well. After cells were attached, they were cultured for 2 weeks continuously. The colonies were fixed with paraformaldehyde and dyed by 0.1% crystal violet. Colonies were counted under a microscope in each well, and all cell colonies contained 50 or more cells.
Cell cycle assay
For the cell cycle assay, 1×106 cells were collected, washed in phosphate-buffered saline (PBS), and fixed in 75% precooled ethanol at −20 °C for 24 h. Then, the cells were washed twice with PBS, and stained using propidium iodide (BD, USA) at 15 °C for 0.5 h. Next, the cells were detected by flow cytometry (Becton Dickinson, USA), and the cell cycle was analyzed with Cell Quest Modfit software.
Xenograft studies
For the xenograft study, 5- to 6-week-old BALB/c nude mice were obtained from the Nanjing University Animal Center. The in vivo experiments were ethically authorized by the Ethics Committee of Nanjing Medical University. The nude mice were randomly grouped into 2 groups, of 5 mice each; 786-0 cells (7×106 in 100 µL PBS) were stabilized with ALKBH5 knockdown lentiviral transfection vector (shALKBH5) or scramble vector (SCR) via subcutaneous injection into the left armpit of each mouse. The volumes and weights of the resulting tumors were measured every 5 days. The tumor volume was calculated as follows: volume = length × width2 × 0.52. After 30 days, the mice were sacrificed, and the tumors were harvested.
Luciferase assay
The dual-luciferase reporter assay was performed in triplicate according to Promega’s instructions. The full-length AURKB 3'-UTR and the mutated AURKB 3'-UTR were inserted into the Pezx-FR02 vector (Promega, USA). Lipofectamine 3000 reagent (Invitrogen, USA) was used to transfect RCC cell (Caki-1 and 786-0) ALKBH5 knockdown cells (shALKBH5), and the scramble cells (SCR) for 48 h. Next, luciferase assays were conducted with a luciferase assay system (Promega, USA). The relative luciferase activity was considered to be the ratio between the luciferase activity induced by ALKBH5 and that induced by SCR.
RNA immunoprecipitation (RIP)
RIP experiments were completed with Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA). Briefly, RCC cells (2×107) were lysed with RIP lysis buffer, co-incubated with 5 µg of magnetic beads with anti-ALKBH5 antibody, and nonimmunized with IgG overnight at 4 °C. After RNA was purified, RT-PCR and qRT-PCR were performed to assess AURKB transcription in ALKBH5 or IgG immunocomplexes.
M6A dot blot assay
The m6A dot-blot was carried on a Bio-Dot instrument (Bio-Rad Laboratories Inc., USA). Briefly, the RNA samples were denatured under vacuum conditions and were found to be nitrocellulose. The membranes were stained by methylene blue to examine RNA loading after UV cross-linking. To detect m6A levels, membranes were incubated with rabbit anti-m6A antibody (1:1,000, Abcam, USA) overnight at 4 °C. After this, the membranes with HRP-labeled rabbit IgG secondary antibody was washed by a large quantity of 0.1% TBST at room temperature for 60 min. Then, washing by TBST was performed again, and the ECL Western Blotting Detection Kit (Millipore, USA) was used to analyze the results. Dots were detected by Bio-Rad chemiluminescence system.
M6A RNA immunoprecipitation (MeRIP) assay
To measure the m6A level in AURKB mRNA, MeRIP was performed. One µg of divisible m6A antibody was bound to protein A-agarose slurry (Millipore) overnight at 4 °C. Next, 100 µg of divisible total RNA was incubated with the antibody in RNase inhibitor-supplemented immunoprecipitation buffer (750 mM NaCl, 50 mM Tris·HCl, and 0.5% Igepal CA-630) at 4 °C for 3 h. Then divisible total RNA was incubated with 4.2 µL of proteinase K in 300 µL of elution buffer (1 mM EDTA, 5 mM Tris·HCl, and 0.05% SDS) at 50 °C for 90 min to elute RNA from the beads. m6A+ RNA was then purified by phenol/chloroform extraction and analyzed with RT-qPCR. Nucleotide sequences of the m6A AURKB mRNA primers were as follows: 5'-CAGTGGGACACCCGACAT-3' and 5'- GTACACGTTTCCAAACTTGCC -3'.
mRNA stability
Cells transfected with ALKBH5 lentivirus, ALKBH5 knockdown lentivirus, or the control lentivirus were treated with 5 µg/mL actinomycin D (Act D) for 0, 1, 2, 4, 6, and 8 h. Total RNAs were extracted, and then analyzed by qRT-PCR. The transcript level of mRNA was calculated as the relative half-life of mRNA and controlled based on standards of β-actin.
Statistical analysis
STATA 22.0 software (StataCorp) was used for statistical analysis. Data were expressed as mean ± standard deviation (SD). The chi-squared test and the Student’s t-test were applied to determine the diversity in the treatment groups. The survival curve was plotted by Kaplan-Meier method and compared by log-rank test. P value <0.05 was considered statistically significant.
Results
ALKBH5 was significantly upregulated in RCC tissues and correlated with RCC patient prognosis
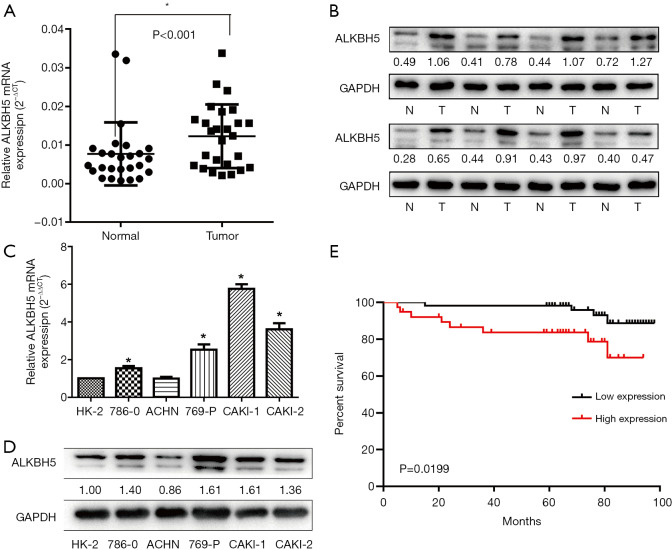
We first examined the mRNA and protein level of ALKBH5 in 26 paired RCC samples by western blot and qRT-PCR. The results showed that ALKBH5 was significantly upregulated in RCC tissues compared to the adjacent normal tissues (Figure 1A,B). We also examined ALKBH5 expression in RCC cell lines and found that ALKBH5 was upregulated in RCC cell lines (Caki-1, Caki-2, ACHN, 786-0, and 769P) compared to HK2, a normal renal tubule epithelium cell (Figure 1C,D).
Figure 1.
ALKBH5 was upregulated in RCC tissues and cell lines and acted as a prognostic factor for RCC patients. (A) ALKBH5 mRNA expression in 26 pairs of RCC tissues and matched adjacent tissues. (B) ALKBH5 protein expression in RCC tissues and matched adjacent tissue. Eight cases are shown as examples. The fold change of ALKBH5 is shown below each lane. (C,D) ALKBH5 mRNA and protein expression in RCC cell lines and normal epithelium cells of renal tubule HK2. (E) Kaplan-Meier survival curves depicting overall survival of patients with RCC based on ALKBH5 expression conditions. Data are presented as mean ± SD. *. P<0.05. RCC, renal cell carcinoma.
We performed IHC on the TMA of 96 RCC cases with long-term follow-up in order to explore the relationship between clinicopathologic features and ALKBH5 expression. We found that ALKBH5 expression was significantly correlated with TNM stage and tumor size (Table 1). The high expression group was associated with higher tumor-node-metastasis (TNM) stage and larger tumor size. No significant correlation was observed between ALKBH5 expression and age, gender or histological grade. Furthermore, patients with high ALKBH5 expression had a worse prognosis and worse overall survival rate (log rank: P=0.0199) compared to those with low ALKBH5 expression based on Kaplan–Meier survival curves (Figure 1E). Thus, these results suggest that ALKBH5 might be fundamentally involved in RCC progression and has potential as a prognostic marker.
Table 1. Association of ALKBH5 expression with the clinicopathologic characteristics of renal cancer patients (chi-squared test).
| Parameters | Number of cases | ALKBH5 expression | P value | |
|---|---|---|---|---|
| Low (%) | High (%) | |||
| Age (years) | 0.507 | |||
| <60 | 58 (60.4) | 36 (62.0) | 22 (37.9) | |
| ≥60 | 38 (39.6) | 21 (55.3) | 17 (44.7) | |
| Gender | 0.308 | |||
| Male | 60 (62.5) | 38 (63.3) | 22 (36.7) | |
| Female | 36 (37.5) | 19 (52.8) | 17 (47.2) | |
| Tumor size (cm) | 0.017* | |||
| ≤4 | 51 (53.1) | 36 (70.6) | 15 (29.4) | |
| >4 | 45 (46.9) | 21 (46.7) | 24 (53.3) | |
| Histological grade | 0.152 | |||
| I–II | 78 (81.3) | 49 (62.8) | 29 (37.2) | |
| III–IV | 18 (18.7) | 8 (44.4) | 10 (55.6) | |
| TNM stage | 0.006** | |||
| I | 73 (76.0) | 51 (69.9) | 22 (30.1) | |
| II–IV | 23 (24.0) | 10 (43.5) | 13 (56.5) | |
| AURKB | 0.034* | |||
| Negative | 59 (61.5) | 40 (67.8) | 19 (32.2) | |
| Positive | 37 (38.5) | 17 (46.0) | 20 (54.0) | |
*, P value <0.05 represents statistical significance; **, P value <0.01 represents statistical significance.
ALKBH5 promoted RCC tumorigenesis in vitro and in vivo
To clarify the function of ALKBH5 in vitro, we selected Caki-1 and 786-0 stable transfection cells with ALKBH5 overexpression lentivirus, knockdown lentivirus, or control lentivirus. The ALKBH5 expression levels were detected by qRT-PCR and western blot (Figure S1A,B).
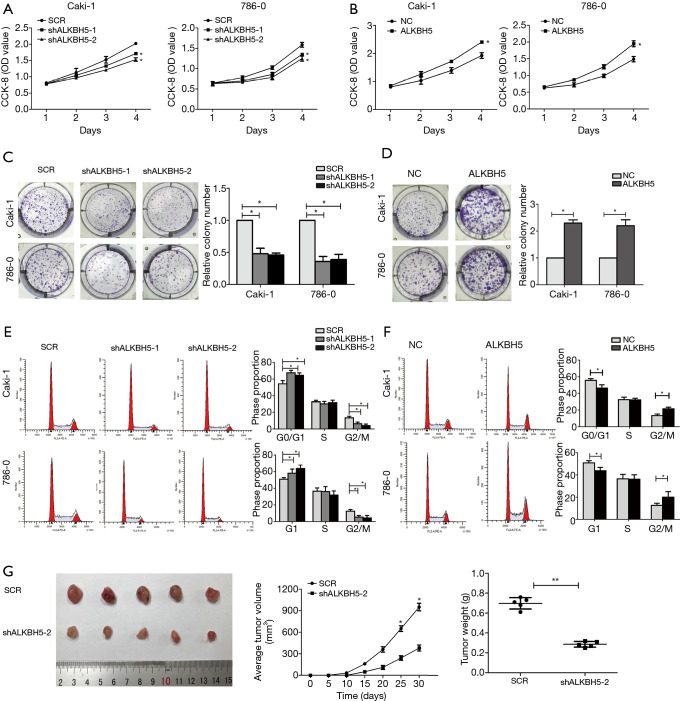
CCK-8 assay results indicated that ALKBH5 knockdown decreased cell proliferation (Figure 2A), while ALKBH5 overexpression increased cell proliferation (Figure 2B). Colony formation assay results revealed that ALKBH5 knockdown inhibited cell colony formation efficiency (Figure 2C), whereas cell colony formation efficiency was enhanced after ALKBH5 overexpression compared to the control group (Figure 2D). Furthermore, cell cycle results demonstrated that the percentage of G1 phase was increased while the percentage of G2/M phase was decreased in ALKBH5 knockdown cells (Figure 2E), while the opposite effects were found in ALKBH5 overexpression cells (Figure 2F). The effects of ALKBH5 in vivo were further studied. We used ALKBH5 knockdown or the control cells to generate a subcutaneous xenograft tumor mouse model. As expected, ALKBH5 knockdown cells (shALKBH5-2) showed smaller tumor volume and weight compared to the control cells (SCR) (Figure 2G).
Figure 2.
ALKBH5 promoted RCC tumorigenesis in vitro and in vivo. (A,B) Knockdown of ALKBH5 inhibited Caki-1 and 786-0 cell proliferation (A), whereas overexpression of ALKBH5 enhanced cell proliferation (B). (C,D) Colony formation assay showed that ALKBH5 knockdown significantly decreased the cloning number of Caki-1 and 786-0 cells compared with the control group, whereas overexpression of ALKBH5 enhanced the cloning number. (E,F) Cell cycle of Caki-1 and 786-0 cells analyzed by flow cytometry after the knockdown or overexpression of ALKBH5. The histogram indicates the percentage of cells. (G) Subcutaneous tumor model of 786-0 cells with ALKBH5 knockdown. Tumor volume and weight were measured at the indicated weeks after mice were transplanted. Data are presented as mean ± SD. *, P<0.05; **, P<0.01. RCC, renal cell carcinoma.
In addition to this, Transwell migration demonstrated that knockdown of ALKBH5 could decrease the cells crossing the membrane (Figure S2A), yet cell migration capability was increased in ALKBH5 overexpression cells (Figure S2B). Similarly, the invasion capability was inhibited in ALKBH5 knockdown cells (Figure S2C). On the contrary, the cell invasion capability was enhanced after ALKBH5 overexpression in 786-0 and Caki-1 cell lines (Figure S2D). The combined data indicate that ALKBH5 may act as a positive regulator in tumor proliferation, invasion, and migration in RCC cells,.
ALKBH5 regulated AURKB expression and correlated with AURKB expression
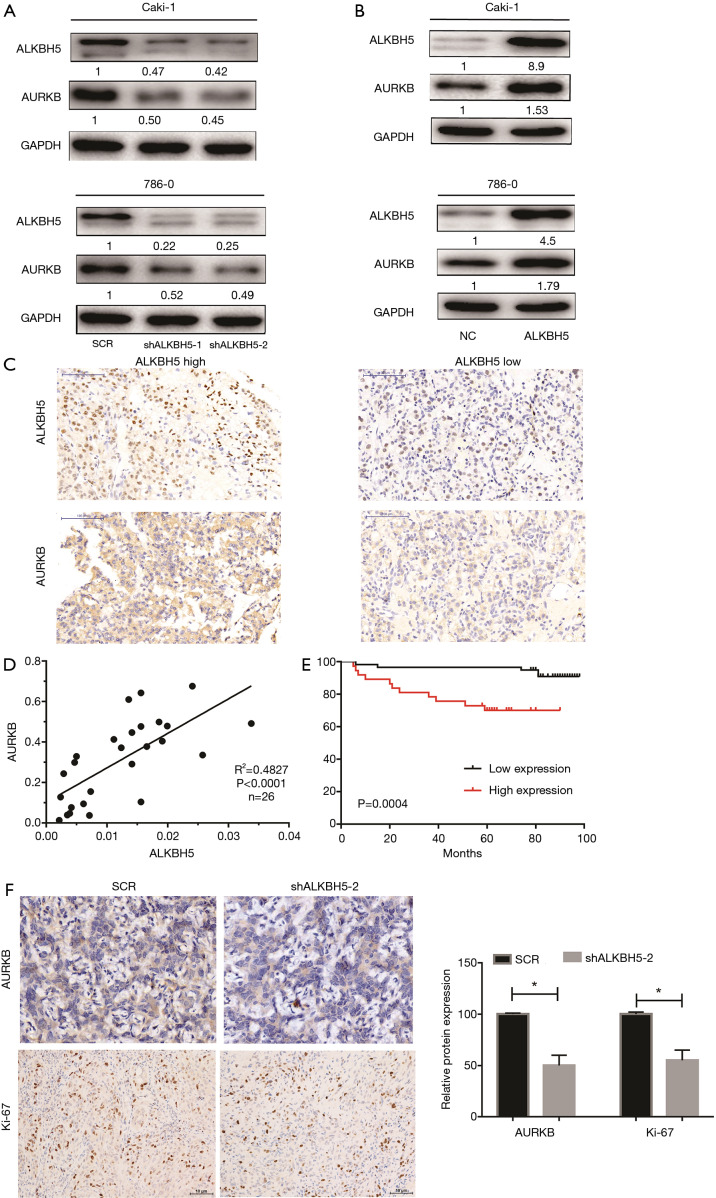
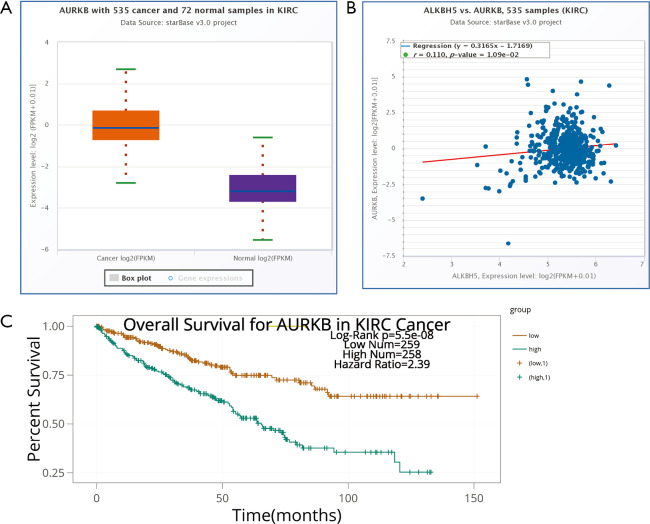
To delineate the ALKBH5-mediated molecular function in RCC cells, bioinformatic analysis was conducted to explore potential mRNAs regulated by ALKBH5. The Cancer Genome Atlas (TCGA) database (http://starbase.sysu.edu.cn/panCancer.php) analysis revealed that AURKB was upregulated in RCC (Figure S3A) and positively correlated with ALKBH5 expression (Figure S3B). Western blot assays confirmed that ALKBH5 knockdown reduced the expression of AURKB (Figure 3A), while ALKBH5 overexpression upregulated the expression of AURKB in RCC cells (Figure 3B). Expression of AURKB was also positively correlated with expression of ALKBH5 in RCC tissues by IHC (Figure 3C) and qRT-PCR (Figure 3D). Kaplan-Meier survival curves showed a significant correlation between high AURKB expression and overall poor survival in RCC patients (Figure 3E), consistent with the results obtained from TCGA database (http://starbase.sysu.edu.cn/panCancer.php) (Figure S3C). Finally, we found that Ki67 (a proliferation marker of tumors) and AURKB expression were significantly decreased after ALKBH5 knockdown of the subcutaneous xenograft tumor model by IHC analysis (Figure 3F).
Figure 3.
ALKBH5 regulated AURKB expression in RCC cells and correlated with AURKB expression in human RCC tissues. (A,B) AURKB expression was obviously decreased in ALKBH5-knockdown cells and increased in ALKBH5 overexpression cells as revealed by western blotting. (C) IHC analysis of ALKBH5 and AURKB in RCC at 200× magnification. Scale bars indicate 100 µm. Yellow staining or brown staining indicates positive. (D) A moderate positive correlation between the expression of ALKBH5 and AURKB mRNAs was found in 26 paired RCC tissues. (E) Kaplan-Meier survival curves showed that AURKB upregulation was significantly associated with poor overall survival in RCC patients. (F) IHC analysis of AURKB and Ki67 in xenografts. The histogram indicates the AURKB and Ki67-positive cells from the panel. Yellow staining or brown staining indicates positive. *, P<0.05. RCC, renal cell carcinoma; IHC, immunohistochemistry.
Taken together, we can infer that ALKBH5 regulates expression of AURKB and is associated with the expression of AURKB in human RCC.
ALKBH5 promoted AURKB mRNA demethylation and enhanced its stability in an m6A-dependant manner
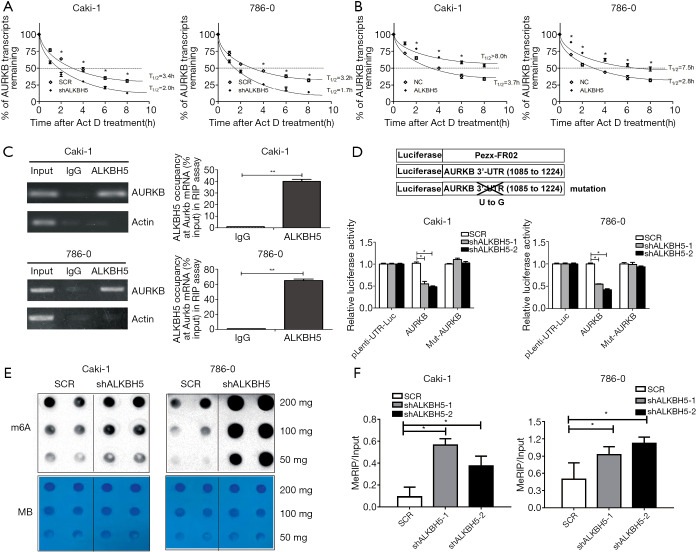
First, we used actinomycin D (Act D) to treat the ALKBH5 knockdown or overexpression cells. The results suggested the relative half-life of AURKB was significantly decreased by ALKBH5 knockdown (Figure 4A). Moreover, ALKBH5 overexpression significantly increased the relative half-life of AURKB mRNA (Figure 4B). These results demonstrated that ALKBH5 increased the mRNA stability of AURKB.
Figure 4.
ALKBH5 enhanced the stability of the AURKB transcript by direct binding to AURKB mRNA in an m6A-dependent manner. (A) The half-life of AURKB transcript was decreased after ALKBH5 knockdown. (B) The half-life of AURKB transcript was enhanced by ALKBH5 overexpression. (C) RIP assays showed that ALKBH5 interacted with AURKB mRNAs in 786-0 and Caki-1 cells. The agarose electrophoresis results of the PCR products are shown in the left panel. The qRT-PCR results of RIP assays are shown in the right panel. (D) Relative luciferase activities of the reporter carrying AURKB 3'-UTR with either wild-type or mutation after transfection into ALKBH5-knocked down or the control cells were measured. Relative luciferase activities were normalized to Renilla luciferase activity. (E) m6A-modified RNAs were measured by dot blot using m6A antibody in 786-0 and Caki-1 cells with or without knockdown of ALKBH5. ALKBH5 knockdown resulted in significantly increased m6A levels in a dose-dependent manner. MB, methylene blue (as loading control). (F) MeRIP assays showed that ALKBH5 knockdown caused a significant increase in the m6A levels of AURKB in RCC cells. Data are presented as mean ± SD. *, P<0.05; **, P<0.01.
Next, RIP assays indicated that the anti-ALKBH5 antibody could significantly enrich AURKB mRNAs, compared with the anti-IgG antibody (Figure 4C). We also performed a dual-luciferase assay which confirmed that AURKB 3'-UTR was needed to decrease AURKB expression by ALKBH5 knockdown. Caki-1 and 786-0 ALKBH5 knockdown cells (shALKBH5) and the control cells (SCR) were transfected with Pezx-FR02 vector reporters which carried AURKB 3'-UTR, mutation of AURKB 3'-UTR, or empty vector. Knockdown of ALKBH5 could significantly decrease luciferase activities of AURKB 3'-UTR reporter vector, but mutated AURKB 3'-UTR or the empty vector (Figure 4D).
Finally, we investigated whether levels of m6A RNA in RCC cells are modified after a decrease of ALKBH5 expression. Compared to the control cells, knockdown of ALKBH5 markedly increased the m6A levels in RCC cells (Figure 4E). MeRIP assays showed that ALKBH5 knockdown caused a significant increase in the m6A levels of the 3'-UTR of AURKB mRNAs in RCC cells (Figure 4F).
Taken together, these results suggest that ALKBH5 can promote AURKB mRNA demethylation and enhance its stability in an m6A-dependent manner.
AURKB interference reversed the proliferation promoting effects of ALKBH5 on RCC cells
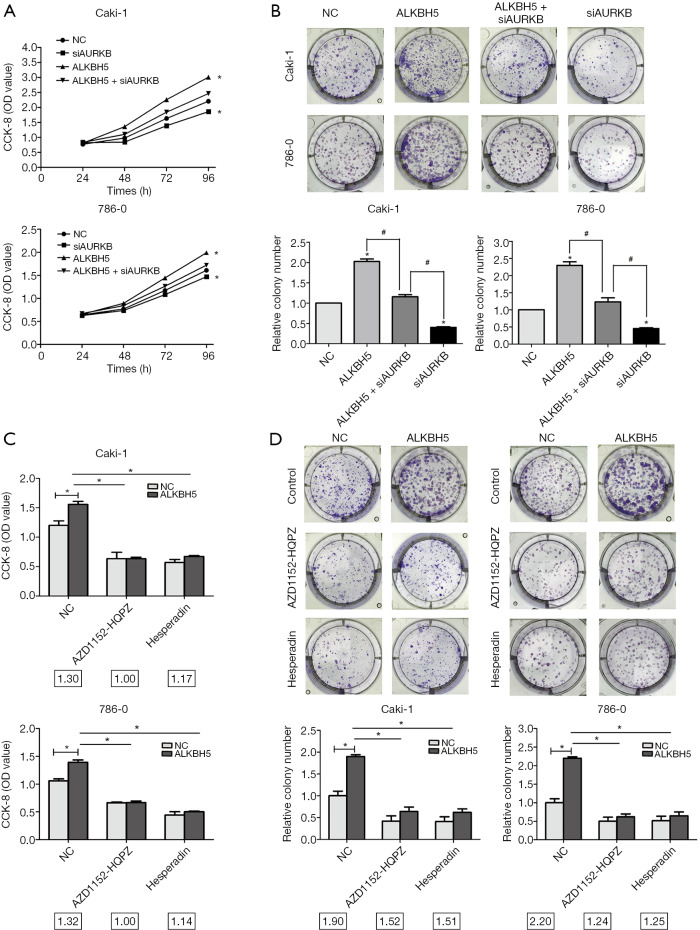
To confirm AURKB’s influence on the proliferation of ALKBH5-regulated cells, ALKBH5 overexpressed (ALKBH5) and control cells (NC) were selected and transfected with AURKB small interference RNA (siAURKB) or a control (CTRi). Our data revealed that AURKB knockdown decreased cell growth in the CCK8 assay, and it reversed the effect of ALKBH5-induced cell growth (Figure 5A). The results of colony formation assays were similar (Figure 5B).
Figure 5.
Reduced AURKB expression suppressed the proliferation induced by ALKBH5 in RCC cells. (A,B) CCK8 and clone formation assays were used to measure the effect of AURKB small interference RNA (siAURKB) on Caki-1 and 786-0 cells with ALKBH5 overexpression. (C,D) The effect of AURKB inhibitor (AZD1152-HQPZ and hesperidin) on Caki-1 and 786-0 cells with ALKBH5 overexpression was measured by CCK8 and clone formation assays. The colony fold (NC/ALKBH5) is stated under the histogram. Data are presented as mean ± SD. *, P<0.05; #, P<0.05. RCC, renal cell carcinoma.
To investigate whether ALKBH5 promotes cell proliferation by an AURKB-dependent mechanism, we selected 2 commonly used selective AURKB inhibitors AZD1152-HQPZ and hesperidin. We found that both AZD1152-HQPZ and hesperidin decreased the OD value fold (NC/ALKBH5) and the colony fold (NC/ALKBH5) in the CCK8 experiment (Figure 5C) and in colony formation (Figure 5D), respectively. The results showed that proliferation ability was markedly decreased when these inhibitors inhibited AURKB function in 786-0 and Caki-1 cells. These results confirmed that proliferation was promoted by a decrease of ALKBH5 when AURKB function was inhibited.
HIF induced by hypoxia up-regulated the ALKBH5 and AURKB expression in RCC cells
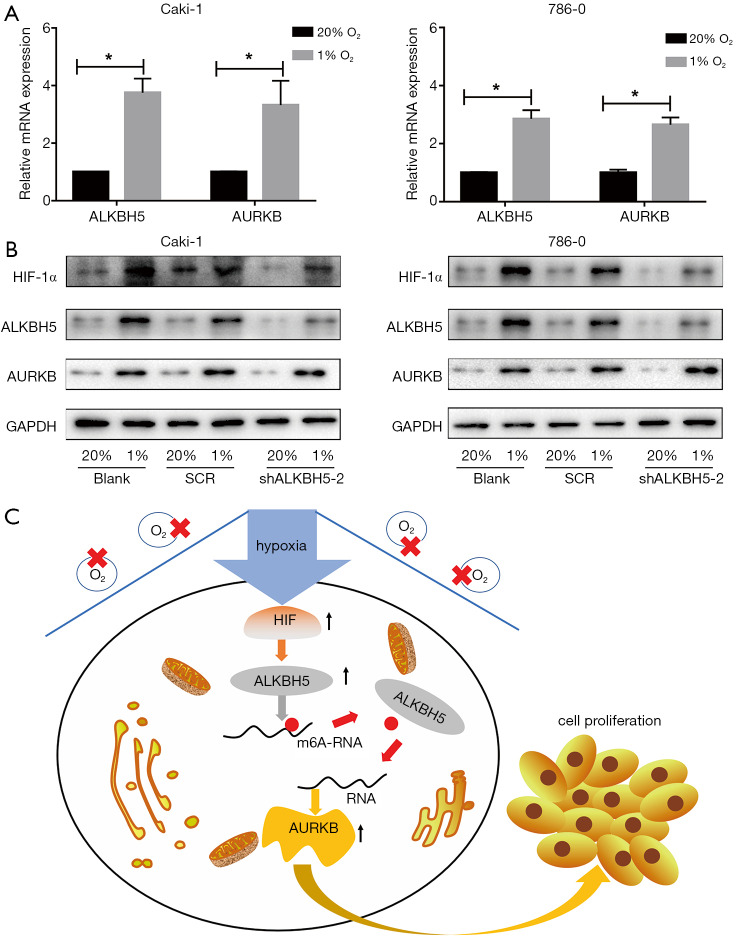
To explore whether ALKBH5 expression is regulated by HIF induced by hypoxia on the expression of ALKBH5 in RCC cells, we exposed RCC cells to 20% or 1% O2 for 24 h/48 h. The results showed that the expression of ALKBH5 and AURKB remarkably increased in RCC cells both at the mRNA (Figure 6A) and protein levels (Figure 6B), with HIF-1α or HIF-2α expression increased (Figure 6B).
Figure 6.
HIF induced by hypoxia upregulated ALKBH5 expressions in RCC. (A) ALKBH5 and AURKB mRNAs were upregulated in the 1% O2 group as compared with 20% O2 group as revealed by qRT-PCR. Caki-1 and 786-0 cells were exposed to 20% or 1% O2. RNA was extracted at 24 h. Data are presented as mean ± SD from 3 independent experiments; *, P<0.05. (B) HIF-1α. HIF-2α, ALKBH5, and AURKB proteins were up-regulated in the 1% O2 group as compared to the 20% O2 group as revealed by western blot. Caki-1 and 786-0 cells were exposed to 20% or 1% O2. Protein was extracted at 48 h. (C) Working model: ALKBH5 promoted RCC cell proliferation by stabilizing AURKB mRNAs in an m6A-dependent manner. RCC, renal cell carcinoma; qRT-PCR, real-time quantitative polymerase chain reaction.
Discussion
In our study, we confirmed that ALKBH5 plays a key regulatory role in the progression of RCC. We found that ALKBH5 could promote the proliferation of RCC by regulating AURKB in an m6A-dependent manner, which supports ALKBH5 signaling as a promising therapeutic strategy in RCC.
Panneerdoss et al. reported that ALKBH5 knockdown inhibited cell proliferation, invasion, and migration in cancers, such as breast cancer, cervical cancer, liver cancer and prostate cancer (31). We also found that ALKBH5 was significantly upregulated in RCC cell lines. ALKBH5 knockdown remarkably inhibited proliferation, invasion, and migration in RCC cell lines, whereas ALKBH5 overexpression produced the opposite phenomenon. Similarly, the in vivo assay in nude mice showed that 786-0 cells with ALKBH5 knockdown formed smaller tumors than those of the control cells. These results confirm that ALKBH5 acts as an oncogene in RCC. Additionally, we found that ALKBH5 was also upregulated in RCC tissues and was associated with TMN stage and tumor size in patients. Furthermore, the prognosis of RCC patients with high ALKBH5 expression was worse, which highlights its potential prognostic value for RCC patients. Zhang et al. also reported that ALKBH5 promoted tumor progression in glioblastoma (25), while Fukumoto et al. reported that downregulation of FTO and ALKBH5 upregulates Wnt signaling by increasing m6A modification in FZD10 mRNA, leading to PARPi resistance (32). This means that the regulatory network of ALKBH5 may be involved in carcinogenesis. All these results strongly support the carcinogenic role of ALKBH5 in RCC cancer. Therefore, it is of great significance to explore the downstream targets of ALKBH5.
Aurora B, as a member of the chromosomal passenger complex, is a key regulator in cell mitosis (33), and is involved in the processes of chromosome condensation, chromosome-microtubule interaction, sister chromatid cohesion, cytokinesis, and the spindle assembly checkpoint (34). Aurora B expression has also been shown to play a key role in tumorigenesis (35), progression (36), and chemotherapy response (37). At present, Aurora B is a well-known molecular target, and selective inhibitors have been investigated in clinical trials (38,39). Our study conducted bioinformatic analysis to explore the potential mRNAs regulated by ALKBH5 and found that AURKB was elevated in RCC tissues and positively related with the expression of ALKBH5. We then verified the expression of AURKB in the ALKBH5-knocked down and overexpressed RCC cells and found that overexpressed ALKBH5 significantly increased AURKB the expression. We also found that AURKB expression was upregulated in RCC tissues and positively correlated with ALKBH5 expression in tissues of RCC patients. Kaplan-Meier survival curves showed a significant correlation between high AURKB expression and poor overall survival in RCC cases. Consistent with our results, Li et al. reported that high AURKB expression was positively corelated with worse prognosis in RCC cases (40), while Li et al. also reported that silencing AURKB with siRNA or inhibitors inhibited cell proliferation in RCC (41). We also found that AURKB siRNA and AURKB inhibitors could reduce the proliferation induced by ALKBH5 in RCC overexpression cells. These results suggest that AURKB interference reverses proliferation by promoting the effects of ALKBH5 on RCC cells.
As an m6A demethylase, ALKBH5 was found to regulate the targeted gene expression in an m6A-dependant manner. The results of dual-luciferase reporter assay revealed that ALKBH5 could recognize 3'-UTR in AURKB mRNA. The results of RIP assay revealed that ALKBH5 could enrich AURKB mRNA. The results of MeRIP assay revealed that ALKBH5 could increase the stability of AURKB mRNA by increasing m6A level. Then protein levels of AURKB were increased eventually. Overall, these findings indicate that ALKBH5 may promote proliferation by regulating AURKB in an m6A-dependent manner.
The hypoxia of tumors is related to invasive tumor phenotypes, therapeutic resistance, and poor outcomes (42). The response of cells to hypoxia is primarily mediated by the transcription factors from the HIF family, which regulates multiple genes in progression of cancer cells (43,44). Previous studies have indicated that under hypoxia BCSCs have a higher ALBKH5 expression in an HIF-1α- and HIF-2α-dependent mode, ultimately leading to breast cancer stem cell (BCSC) enrichment in tumors (24). We consistently found that ALKBH5 expression could be upregulated by HIF-1α in Caki-1 cells and regulated by HIF-2α in 786-0 cells, which are HIF-1α defective cells (45). The present work showed that ALKBH5 may act as an oncogene in RCC by stabilizing AURKB mRNA in an m6A-dependent manner. A working model is displayed in Figure 6C. These data suggest that ALKBH5 may figure critically in targeting ALKBH5 signaling, and it is hoped that these findings will contribute to a potential therapeutic strategy in RCC.
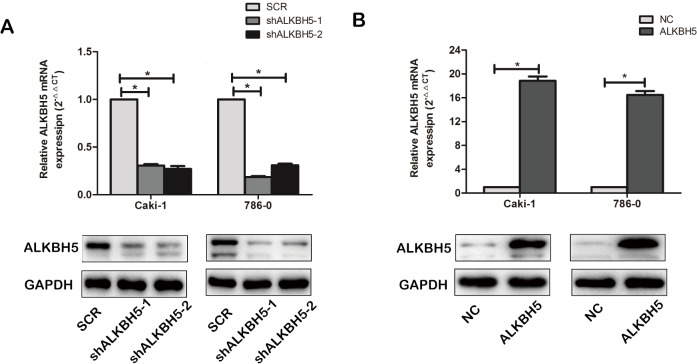
Figure S1.
The efficiency of ALKBH5 knockdown and overexpression in 786-0 and Caki-1 cell lines. 786-0 and Caik-1 cell lines were infected with ALKBH5-overexpressed lentivirus (termed as ALKBH5), a negative control (termed as NC), ALKBH5 knockdown lentivirus (termed as shALKBH5-1, sh ALKBH5-2), and a scramble control (termed as SCR). The efficiency of ALKBH5 knockdown (A) and overexpression (B) in 786-0 and Caki-1 cell lines was screened by real-time quantitative polymerase chain reaction (qRT-PCR) and western blot. Data are presented as mean ± SD. *, P<0.05.
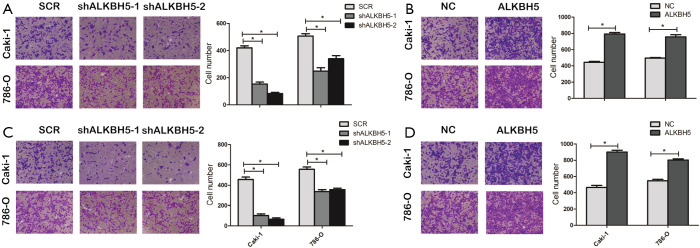
Figure S2.
ALKBH5 regulated RCC cell migration and invasion. (A,B) Effects of ALKBH5 knockdown or overexpression on cell migration in 786-0 and Caki-1 cell lines by Transwell migration assays. (C,D) Effects of ALKBH5 knockdown or overexpression on cell invasion in 786-0 and Caki-1 cell lines by Transwell invasion assay. The results are expressed as the number of invaded cells per field compared with respective control (magnification, ×100). Data are presented as mean ± SD. *, P<0.05. RCC, renal cell carcinoma.
Figure S3.
AURKB was positively correlated with ALKBH5 expression and predicted poor survival in RCC patients from TCGA database. AURKB was upregulated in RCC tissues samples downloaded from TCGA database. (B) A scatter plot of ALKBH5 and AURKB relative expression in tumor samples downloaded from TCGA database. (2-tailed Spearman’s correction, R =0.110). (C) Kaplan-Meier survival curves revealed that AURKB was a prognostic factor for RCC. RCC, renal cell carcinoma; TCGA, The Cancer Genome Atlas.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was financially sponsored by the Jiangsu Province “333” project (LGY2018055), the Jiangsu Province Six Talent Peaks Project (2015-WSW-025), and the National Natural Science Foundation of China (no. 81602235, 81772711).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was ethically authorized by the Local Ethics Committees of the First Affiliated Hospital of Nanjing Medical University (No. NJMU-EA-2017-319). We obtained informed consent from all the patients to use their data for research purposes.
Footnotes
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3079
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3079). The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med 2005;353:2477-90. 10.1056/NEJMra043172 [DOI] [PubMed] [Google Scholar]
- 3.Di Pierro G, Mini E, Roviello G. Cabozantinib in advanced non-clear-cell renal cell carcinoma: is it the way clearer now? Ann Transl Med 2019;7:S229. 10.21037/atm.2019.07.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rydzanicz M, Wrzesinski T, Bluyssen HA, et al. Genomics and epigenomics of clear cell renal cell carcinoma: recent developments and potential applications. Cancer Lett 2013;341:111-26. 10.1016/j.canlet.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 5.Patel DN, Figlin RA, Kim HL. Adjuvant treatment for renal cell carcinoma: do we finally have a major breakthrough? Clin Adv Hematol Oncol 2016;14:907-14. [PubMed] [Google Scholar]
- 6.Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 2012;149:1635-46. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu W, Wang JZ, Xu Z, et al. Detection of N6methyladenosine modification residues (Review). Int J Mol Med 2019;43:2267-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ke S, Alemu EA, Mertens C, et al. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev 2015;29:2037-53. 10.1101/gad.269415.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji L, Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res 2012;22:624-36. 10.1038/cr.2012.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu N, Parisien M, Dai Q, et al. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 2013;19:1848-56. 10.1261/rna.041178.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vu LP, Pickering BF, Cheng Y, et al. The N 6-methyladenosine (M 6 A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 2017;23:1369-76. 10.1038/nm.4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niu Y, Lin Z, Wan A, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer 2019;18:46. 10.1186/s12943-019-1004-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han J, Wang JZ, Yang X, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer 2019;18:110. 10.1186/s12943-019-1036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visvanathan A, Patil V, Arora A, et al. Essential Role of METTL3-mediated M 6 A Modification in Glioma Stem-Like Cells Maintenance and Radioresistance. Oncogene 2018;37:522-33. 10.1038/onc.2017.351 [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Bai R, Li M, et al. Excessive miR-25-3p maturation via N 6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun 2019;10:1858. 10.1038/s41467-019-09712-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Yang X, Chen Z, et al. M 6 A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer 2019;18:87. 10.1186/s12943-019-1014-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N 6 -methyladenosine-dependent primary MicroRNA processing. Hepatology 2017;65:529-43. 10.1002/hep.28885 [DOI] [PubMed] [Google Scholar]
- 18.Li X, Tang J, Huang W, et al. The M6A methyltransferase METTL3: acting as a tumor suppressor in renal cell carcinoma. Oncotarget 2017;8:96103-16. 10.18632/oncotarget.21726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang J, Wang F, Cheng G, et al. Wilms' tumor 1-associating protein promotes renal cell carcinoma proliferation by regulating CDK2 mRNA stability. J Exp Clin Cancer Res 2018;37:40. 10.1186/s13046-018-0706-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang C, Zhuang C, Luo X, et al. N6-methyladenosine demethylase FTO suppresses clear cell renal cell carcinoma through a novel FTO-PGC-1alpha signalling axis. J Cell Mol Med 2019;23:2163-73. 10.1111/jcmm.14128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013;49:18-29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Zhao JC. Update: Mechanisms Underlying N 6-Methyladenosine Modification of Eukaryotic mRNA. Trends Genet 2016;32:763-73. 10.1016/j.tig.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thalhammer A, Bencokova Z, Poole R, et al. Human AlkB Homologue 5 Is a Nuclear 2-oxoglutarate Dependent Oxygenase and a Direct Target of Hypoxia-Inducible Factor 1α (HIF-1α). PLoS One 2011;6:e16210. 10.1371/journal.pone.0016210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C, Samanta D, Lu H, et al. Hypoxia Induces the Breast Cancer Stem Cell Phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A 2016;113:E2047-56. 10.1073/pnas.1602883113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Zhao BS, Zhou A, et al. M 6 A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017;31:591-606 e6. [DOI] [PMC free article] [PubMed]
- 26.Aguilo F, Zhang F, Sancho A, et al. Coordination of m(6)A mRNA Methylation and Gene Transcription by ZFP217 Regulates Pluripotency and Reprogramming. Cell Stem Cell 2015;17:689-704. 10.1016/j.stem.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Guo S, Piao HY, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem 2019;75:379-89. 10.1007/s13105-019-00690-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah A, Rashid F, Awan HM, et al. The DEAD-Box RNA Helicase DDX3 Interacts With M 6 A RNA Demethylase ALKBH5. Stem Cells Int 2017;2017:8596135. 10.1155/2017/8596135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu H, Gan X, Jiang X, et al. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J Exp Clin Cancer Res 2019;38:163. 10.1186/s13046-019-1159-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y, Hu H, Wang Y, et al. ALKBH5 Inhibits Pancreatic Cancer Motility by Decreasing Long Non-Coding RNA KCNK15-AS1 Methylation. Cell Physiol Biochem 2018;48:838-46. 10.1159/000491915 [DOI] [PubMed] [Google Scholar]
- 31.Panneerdoss S, Eedunuri VK, Yadav P, et al. Cross-talk among writers, readers, and erasers of M 6 A regulates cancer growth and progression. Sci Adv 2018;4:eaar8263. [DOI] [PMC free article] [PubMed]
- 32.Fukumoto T, Zhu H, Nacarelli T, et al. N 6-Methylation of Adenosine of FZD10 mRNA Contributes to PARP Inhibitor Resistance. Cancer Res 2019;79:2812-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta 2008;1786:60-72. [DOI] [PubMed] [Google Scholar]
- 34.Ma HT, Poon RY. How protein kinases co-ordinate mitosis in animal cells. Biochem J 2011;435:17-31. 10.1042/BJ20100284 [DOI] [PubMed] [Google Scholar]
- 35.Katayama H, Brinkley WR, Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev 2003;22:451-64. 10.1023/A:1023789416385 [DOI] [PubMed] [Google Scholar]
- 36.Bertran-Alamillo J, Cattan V, Schoumacher M, et al. AURKB as a target in non-small cell lung cancer with acquired resistance to anti-EGFR therapy. Nat Commun 2019;10:1812. 10.1038/s41467-019-09734-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beussel S, Hasenburg A, Bogatyreva L, et al. Aurora-B protein expression is linked to initial response to taxane-based first-line chemotherapy in stage III ovarian carcinoma. J Clin Pathol 2012;65:29-35. 10.1136/jclinpath-2011-200212 [DOI] [PubMed] [Google Scholar]
- 38.Dar AA, Goff LW, Majid S, et al. Aurora kinase inhibitors--rising stars in cancer therapeutics? Mol Cancer Ther 2010;9:268-78. 10.1158/1535-7163.MCT-09-0765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katayama H, Sen S. Aurora kinase inhibitors as anticancer molecules. Biochim Biophys Acta 2010;1799:829-39. 10.1016/j.bbagrm.2010.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Zhang ZF, Chen J, et al. VX680/MK-0457, a potent and selective Aurora kinase inhibitor, targets both tumor and endothelial cells in clear cell renal cell carcinoma. Am J Transl Res 2010;2:296-308. [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Zhou W, Wei L, et al. The effect of Aurora kinases on cell proliferation, cell cycle regulation and metastasis in renal cell carcinoma. Int J Oncol 2012;41:2139-49. 10.3892/ijo.2012.1633 [DOI] [PubMed] [Google Scholar]
- 42.Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol 2009;10:992-1000. 10.1016/S1470-2045(09)70240-2 [DOI] [PubMed] [Google Scholar]
- 43.Rini BI. New strategies in kidney cancer: therapeutic advances through understanding the molecular basis of response and resistance. Clin Cancer Res 2010;16:1348-54. 10.1158/1078-0432.CCR-09-2273 [DOI] [PubMed] [Google Scholar]
- 44.Wigerup C, Pahlman S, Bexell D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol Ther 2016;164:152-69. 10.1016/j.pharmthera.2016.04.009 [DOI] [PubMed] [Google Scholar]
- 45.Liu W, Chen H, Wong N, et al. Pseudohypoxia induced by miR-126 deactivation promotes migration and therapeutic resistance in renal cell carcinoma. Cancer Lett 2017;394:65-75. 10.1016/j.canlet.2017.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as