Abstract
Background
Lung cancer is a heterogeneous malignant tumor involving more than 50 histological subtypes. Currently, molecularly targeted drugs have been shown to have promising applications in the clinical treatment of lung cancer. This study aims to explore the expression patterns and prognostic potential of enolase 2 (ENO2) in lung cancer.
Methods
Differential expressions of ENO2 in lung cancer cases were analyzed using the Oncomine database. Meanwhile, the prognostic potentials of ENO2 in lung cancer were assessed by deploying the Kaplan-Meier plotter database.
Results
Forty-one studies reported a significant difference in ENO2 expression between tumors and the normal healthy control tissues. Among all the studies, there was an upregulation of ENO2 in 29 studies, and downregulation in 12 studies. 9/41 studies revealed upregulated ENO2 in distinct types of tumor tissues, including cervical cancer, esophageal cancer, kidney cancer, leukemia, melanoma, pancreatic cancer, sarcoma, and lung cancer. Furthermore, upregulated ENO2 was identified in 365 cases of lung cancer (P<0.05). By analyzing the Kaplan-Meier Plotter database, the ENO2 level was negatively correlated to the overall survival of lung cancer patients (P<0.05). Subsequently, subgroup analysis revealed that the prognostic potential of ENO2 was much more pronounced in lung adenocarcinoma patients (P<0.05).
Conclusions
ENO2 is upregulated in lung cancer tissues and linked to the prognosis. It can be used as a therapeutic target for developing lung cancer drugs.
Keywords: Lung cancer, enolase 2 (ENO2), Oncomine
Introduction
Lung cancer is a heterogeneous malignant tumor involving more than 50 histological subtypes (1-3). The incidence and mortality rates of lung cancer rank first place in China and worldwide (4,5). The GLOBOCAN database has estimated that there were 2.09 million new cases of lung cancer and 1.76 million deaths worldwide in 2018.
The two main subtypes of lung cancer are small-cell lung carcinoma (SCLC) and non-small-cell lung carcinoma (NSCLC), and the latter can be further categorized into adenocarcinoma, squamous-cell carcinoma (SCC) and large-cell lung carcinoma (LCLC). Radical surgery is preferred for early-stage NSCLC and some cases of locally advanced NSCLC. However, the long-term effects of radical surgery are unsatisfactory (6). Significant efforts have been made for exploring molecular biology and genomics of lung cancer, and therapeutic strategies for lung cancer have been improved as well.
Currently, molecularly targeted drugs have been shown to have promising applications in the clinical treatment of lung cancer (7). Enolase 2 (ENO2), also known as neuron-specific enolase (NSE), promotes the conversion of β-glycerol phosphate to acetone dihydroxyphosphate, which is of significance in glycolysis. Glycolysis can trigger tumor cell proliferation by supplying the energy required by the tumor (8,9). Therefore, we have speculated that ENO2 plays a role in tumor progression. In recent research, cancer-associated databases have been extensively used. At present, there are few reports on the relationship between lung cancer and ENO2. Therefore, this study aims to explore the expression pattern of ENO2 in lung cancer and its prognostic significance using the Oncomine database and Kaplan-Meier Plotter database, respectively. Our results may supply theoretical evidence for clarifying the potential mechanisms of lung cancer progression.
Methods
Data acquisition from the Oncomine database
The available data was assessed using the Oncomine database (https://www.oncomine.org//resource//login.html) according to the following criteria: (I) gene: ENO2; (II) cancer type: lung cancer; (III) data type: all; (IV) analysis type: cancer vs. normal analysis; (V) thresholds: P value <1E−4, fold change >2 and gene rank = top 10%. A significant difference was set at a P value <0.05.
Kaplan-Meier plotter database analysis on survival in lung cancer
Survival analysis data of lung cancer patients were searched in the Kaplan-Meier Plotter Database (http://kmplot.com/analysis/) according to the following criteria: (I) cancer: lung cancer; (II) gene: ENO2; (III) split patients by auto select best cutoff; (IV) survival: OS. A significant difference was set at P value <0.05.
Results
Differential expressions of ENO2 in tumors
By searching the Oncomine database, we obtained 447 studies involving ENO2 expressions in different tumor types. Among them, 41 studies reported a significant difference in ENO2 expression between the tumors and normal tissues, which were recruited for further analysis. Among them, there was an upregulation of ENO2 in 29 studies, and downregulation in 12 studies. As shown in Figure 1, downregulated ENO2 was reported in the brain and central nervous system tumors (n=9), leukemia (n=2), and prostate cancer (n=1). Upregulated ENO2 was reported in the cervical cancer (n=3), esophageal cancer (n=2), kidney cancer (n=7), leukemia (n=2), melanoma (n=1), pancreatic cancer (n=2), sarcoma (n=4), lung cancer (n=4) and others (n=4).
Figure 1.
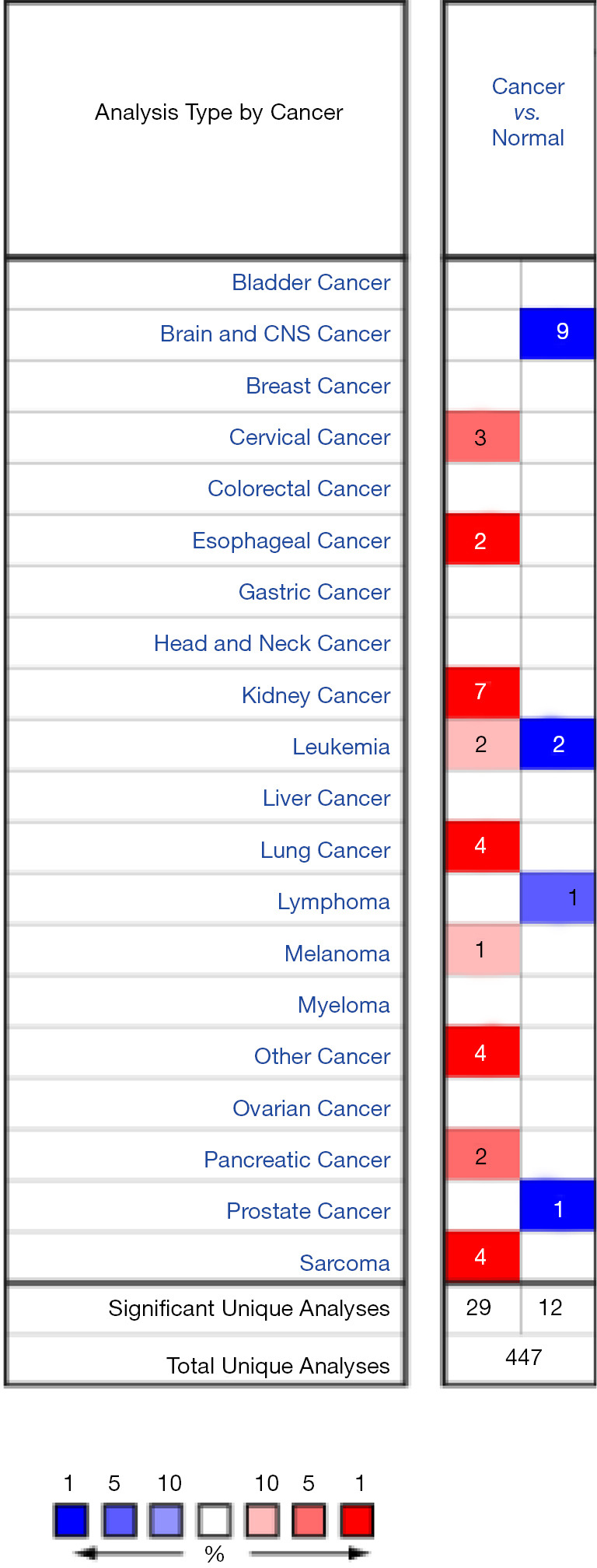
Differential expressions of ENO2 in tumors. ENO2, enolase 2.
Expression pattern of ENO2 in lung cancer tissues
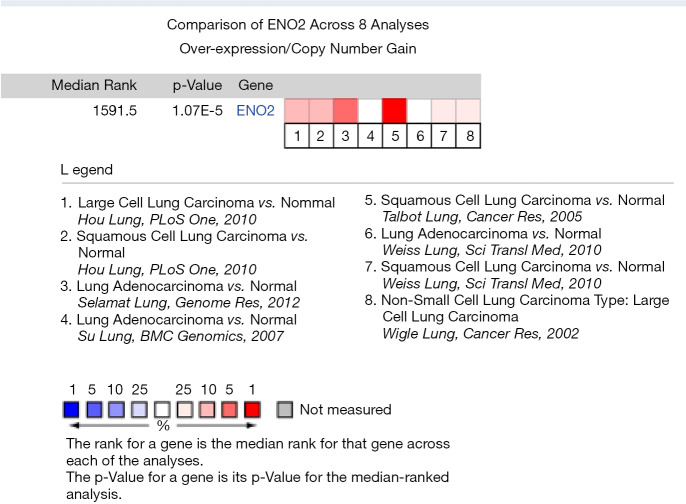
Differential expressions of ENO2 between lung cancer tissues (adenocarcinoma, SCC, and LCLC) and adjacent normal ones were reported in 4 studies (3 datasets and 4 studies), involving 365 tissue samples. The 4 studies, including Talbot (10), Selamat (11), Hou et al. (12), and others, were published in the PLoS One, Genome Res, BMC Genomics, Cancer Res and Sci Transl Med journal datasets. Based on the meta-analysis findings, ENO2 was upregulated in lung cancer tissues compared to that in normal tissues (the median level of ENO2 in all differentially expressed genes was 1591.5, P=1.07E−5) (Figure 2).
Figure 2.
Expression pattern of ENO2 in lung cancer tissues in the Oncomine database. ENO2, enolase 2.
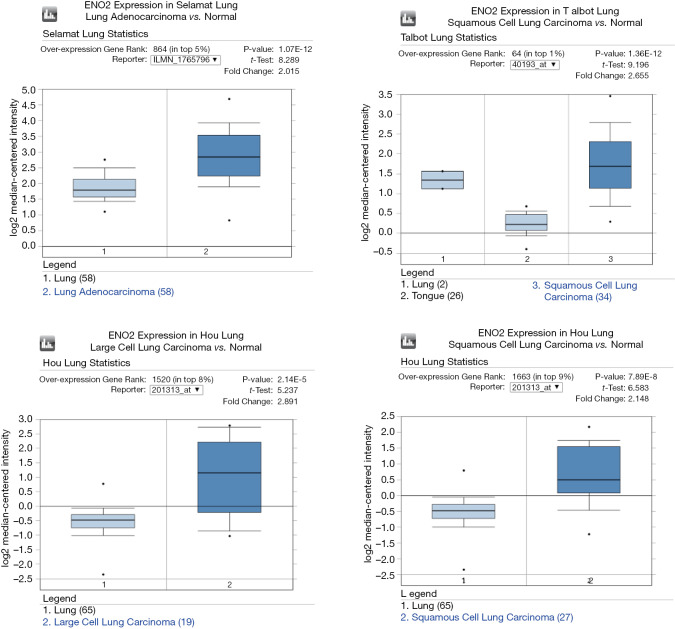
Differential expressions of ENO2 in lung cancer microarrays
In a group of 4 microarray datasets, including Talbot (10), Selamat (11), Hou et al. (12), and others, ENOS was significantly upregulated in lung cancer tissues than in normal tissues (P<0.05) (Figure 3).
Figure 3.
Differential expressions of ENO2 in lung cancer microarrays. ENO2, enolase 2.
Prognostic potential of ENO2 in lung cancer
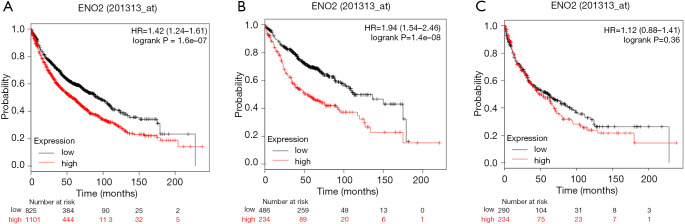
Survival analysis was conducted using the Kaplan-Meier Plotter database to address the prognostic potential of ENO2 in lung cancer. It is shown that the ENO2 level was negatively correlated to the overall survival of lung cancer patients. Compared with lung cancer patients expressing a low level of ENO2, those with an elevated level of ENO2 had worse survival (P<0.01). Subgroup analyses further identified that ENO2 was related to the overall survival of adenocarcinoma patients (P<0.01). Furthermore, it was unrelated to that in SCC patients (P=0.36) (Figure 4).
Figure 4.
Prognostic potential of ENO2 in lung cancer. (A) Lung cancer; (B) lung adenocarcinoma; (C) lung squamous cell carcinoma. ENO2, enolase 2.
Discussion
As a strain of long-chain acidic dimer protein, ENO2 has two enolase isoenzymes (γγ and αγ) or 433 amino acids. ENO2 is expressed in nerve cells and neuroendocrine cells. Tumor cells are derived from these amino acids. Furthermore, it is also expressed in red blood cells, platelets, breast tissue, prostate, and uterus tissues (13,14). ENO2 has been reported to be upregulated in glioma, gastric cancer, prostate cancer, and many other types of malignant tumors (15-17). Yan et al. (17) suggested that ENO2 was significantly upregulated in hypoxic and serum-starved glioma cells. The silence of ENO2 inhibited glioma cell growth, showing that glycolysis triggers tumor cell growth under stress. Their research also proposed that the C-terminal peptide of ENO2 could regulate the cytoskeleton structure of actin through RhoA kinase, thus influencing glioma cell metastasis. It is suggested that ENO2 is also correlated to the metastasis of malignant tumors.
Recent studies have focused on the potential of ENO2 as a tumor biomarker (18). ENO2 can be utilized as a useful biomarker for diagnosing, predicting the prognosis and recurrence, and treating malignant tumors (e.g., seminoma, medullary thyroid carcinoma, and neuroblastoma). Liu et al. (19) showed that serum level of ENO2 was correlated with the immune types and risk stratification of acute lymphoblastic leukemia (ALL), as well as a serum level of lactate dehydrogenase (LDH). The overexpression of ENO2 mRNA induces cell proliferation, glycolysis, and glucocorticoid tolerance in ALL cases via upregulating glycolysis-associated genes and Akt activity. Hence, ENO2 can serve as a biomarker for chemotherapy efficacy and recurrence in ALL patients. The potential of ENO2 in diagnosing and predicting the prognosis and therapeutic efficacy in lung cancer has been previously reported (20,21).
In this study, we analyzed differential expressions of ENO2 in different tumor tissues using the Oncomine database. In 41 studies reporting the significant difference in ENO2 expression, nine studies revealed upregulated ENO2 in different types of tumor tissues, including cervical cancer, esophageal cancer, kidney cancer, leukemia, melanoma, pancreatic cancer, sarcoma, and lung cancer. Furthermore, upregulated ENO2 was identified in 365 cases of lung cancer, including adenocarcinoma, SCC, and LCLC. We after that analyzed the prognostic potential of ENO2 in lung cancer using the Kaplan-Meier Plotter database. Kaplan-Meier Plotter database, an extensively applied online database to predict the correlation between gene expressions and tumor prognosis, harbors the reliable information of 3,452 lung cancer samples. Our findings uncovered that the ENO2 level was negatively correlated to the overall survival of lung cancer. That is, an elevated level of ENO2 predicted poor prognosis of patients. The later subgroup analysis yielded the conclusion that the prognostic potential of ENO2 was much more pronounced in lung adenocarcinoma patients.
Collectively, this study analyzed the expression pattern and prognostic potential of ENO2 in lung cancer cases using Oncomine and Kaplan-Meier Plotter databases. It is uncovered that ENO2 is upregulated in lung cancer patients, and highly expressed ENO2 does not favor the overall survival in lung cancer. Our conclusions may supply a new path for exploring the mechanisms of lung cancer and developing diagnostic or prognostic biomarkers, as well as target drugs. Notably, some limitations of this study should be noteworthy. First, the heterogeneity between studies should not be neglected. Secondly, the small sample size in some studies may lead to biases. Thirdly, our conclusion can be limited because all data were solely acquired from the Oncomine database and we will further confirm the role of ENO2 in lung cancer through cytology experiments and animal model experiments in the near future. We will also test the expression of ENO2 in lung cancer tissues and prove the clinicopathological correlation analysis between the expression of ENO2 and lung cancer. In future explorations, the potential role of ENO2 in lung cancer progression should be confirmed with experimental and clinical studies.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of Jiangsu Province (grant BK20191174) and National Natural Science Foundation of China (grant 81672281).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3354). The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7-30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Van Schil PE, Rami-Porta R, Asamura H. The 8th TNM edition for lung cancer: a critical analysis. Ann Transl Med 2018;6:87. 10.21037/atm.2017.06.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. 10.1097/JTO.0000000000000630 [DOI] [PubMed] [Google Scholar]
- 4.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 6.Pisters KM, Le Chevalier T. Adjuvant chemotherapy in completely resected non-small-cell lung cancer. J Clin Oncol 2005;23:3270-8. 10.1200/JCO.2005.11.478 [DOI] [PubMed] [Google Scholar]
- 7.Ai X, Guo X, Wang J, et al. The rapies for advanced non-small cell lung cancer. Oncotarget 2018;9:37589-607. 10.18632/oncotarget.26428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroemer G, Pouyssegur J. Tumor cell metabolism:cancer’s Achilles’heel. Cancer Cell 2008;13:472-82. 10.1016/j.ccr.2008.05.005 [DOI] [PubMed] [Google Scholar]
- 9.Wang SY, Zha XJ, Zhu XY, et al. Metabolic syndrome and its components with neuron-specific enolase: a cross-sectional study in large health check-up population in China. BMJ Open 2018;8:e020899. 10.1136/bmjopen-2017-020899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talbot SG, Estilo C, Maghami E, et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res 2005;65:3063-71. 10.1158/0008-5472.CAN-04-1985 [DOI] [PubMed] [Google Scholar]
- 11.Selamat SA, Chung BS, Girard L, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res 2012;22:1197-211. 10.1101/gr.132662.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One 2010;5:e10312. 10.1371/journal.pone.0010312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isgrò MA, Bottoni P, Scatena R. Neuron-specific enolase as a biomarker: Biochemical and clinical aspects. Adv Exp Med Biol 2015;867:125-43. 10.1007/978-94-017-7215-0_9 [DOI] [PubMed] [Google Scholar]
- 14.Vizin T, Kos J. Gamma-enolase:a well-known tumour marker, with a less-known role in cancer. Radiol Oncol 2015;49:217-26. 10.1515/raon-2015-0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park T, Lee YJ, Jeong SH, et al. Overexpression of Neuron-Specific Enolase as a Prognostic Factor in Patients with Gastric Cancer. J Gastric Cancer 2017;17:228-36. 10.5230/jgc.2017.17.e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muoio B, Pascale M, Roggero E. The role of serum neuron-specific enolase in patients with prostate cancer: a systematic review of the recent literature. Int J Biol Markers 2018;33:10-21. 10.5301/ijbm.5000286 [DOI] [PubMed] [Google Scholar]
- 17.Yan T, Skaftnesmo KO, Leiss L, et al. Neuronal markers are expressed in human gliomas and NSE knockdown sensitizes glioblastoma cells to radiotherapy and temozolomide. BMC Cancer 2011;11:524. 10.1186/1471-2407-11-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isgrò MA, Bottoni P, Scatena R. Neuron-specific enolase as a biomarker: Biochemical and clinical aspects. Adv Exp Med Biol 2015;867:125-43. 10.1007/978-94-017-7215-0_9 [DOI] [PubMed] [Google Scholar]
- 19.Liu CC, Wang H, Wang WD, et al. ENO2 promotes cell proliferation, glycolysis, and glucocorticoid-resistance in acute lymphoblastic Leukemia. Cell Physiol Biochem 2018;46:1525-35. 10.1159/000489196 [DOI] [PubMed] [Google Scholar]
- 20.Wang B, He YJ, Tian YX, et al. Clinical utility of haptoglobin in combination with CEA, NSE and CYFRA21-l for diagnosis of lung cancer. Asian Pac J Cancer Prev 2014;15:9611-4. 10.7314/APJCP.2014.15.22.9611 [DOI] [PubMed] [Google Scholar]
- 21.Ma R, Xu H, Wu J, et al. Identification of serum proteins and multivariate models for diagnosis and therapeutic monitoring of lung cancer. Oncotarget 2017;8:18901-13. 10.18632/oncotarget.14782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as