Abstract
Background
N6-methyladenosine (m6A), the most internal mRNA modification, is involved in various cancers. However, the function of m6A in hepatocellular carcinoma (HCC) is still not well explored. Here, we aimed to develop a prognostic model consist of m6A associated genes in HCC.
Methods
The mRNA expression profiles and the corresponding clinical information from the patients with HCC were obtained from The Cancer Genome Atlas (TCGA) database, m6A differentially expressed genes (DEGs) were screened by univariate, Lasso and multivariate Cox regression analyses to develop the prognostic model. The differentially expressed m6A genes in HCC tissues were verified by quantitative reverse transcription PCR (qRT-PCR).
Results
The DEGs were obtained between the HCC (n=374) and normal tissues (n=50). Nine m6A genes were correlated with the prognosis, and five of them (KIAA1429, METTL3, YTHDF1, YTHDF2, ZC3H13) were included in the prognostic model by Lasso regression analyses. Four genes (KIAA1429, METTL3, YTHDF1, YTHDF2) were proved significantly high expression in our ten pairs of matched HCC and normal tissues by PCR. The HCC patients were divided into two groups (high- and low-risk) according to the risk score. The high-risk group associated with a significant poor prognosis (P=1.72×10–4). The time-dependent receiver operating characteristic (ROC) curve analysis confirmed the good performance of this prognostic model (AUC =0.617). After univariate [P<0.001, 1.213 (1.136−1.295)] and multivariate Cox regression analyses [P<0.001, 1.198 (1.115−1.288)] by combined with other clinical factors, this prognostic model was identified as an independent prognostic factor of HCC patients.
Conclusions
The m6A genes were differentially expressed between HCC and normal tissues, and associated with the prognosis of HCC.
Keywords: m6A, mRNA, hepatocellular carcinoma (HCC), prognosis
Introduction
Hepatocellular carcinoma (HCC) accounts for the majority of primary liver cancers. Because of hepatitis B or C virus (HBV or HCV) infection or alcohol abuse, the most of HCC patients have a background of liver disease (1-3). The prognosis of HCC is very poor. M6A has been shown a reversible modification in messenger RNA (mRNA) and can regulate processing, translation and decay of mRNA. Dysregulation of m6A genes have an effect on oncogene expression, indicating the m6A and tumorigenesis are related (4-6). Emerging evidence suggests that m6A modification is associated with the tumor proliferation, differentiation, invasion and metastasis in malignant tumors (7,8). We aimed to basing on the m6A genes to establish a prognostic model of HCC.
The traditional prognostic predictors of HCC mainly depend on the tumor size, clinical stage, AFP and the pathological findings (9-11). Currently, there are some prognostic model based on the differentially expressed genes (DEGs) between HCC and para-carcinoma tissue. However, the predictive sensitivity and specificity of these above factors are not high.
The m6A associated genes are being studied extensively in various tumors (12). The modification of m6A methylation including three types of protein factors: “writers” (methyltransferases, METTL3, METTL14, and KIAA1429), “erasers” (demethylases, ALKBH5 and FTO), and “readers” (YTH domain containing RNA binding proteins, YTHDF1, YTHDF2 and YTHDF3) (13,14). Although the m6A has been studied in many other tumors, the role of m6A in HCC is still not well defined. Furthermore, the prognostic function of m6A genes still not been explored in HCC.
This project designed to build a new prognostic model of HCC patients. Basing on the clinical and transcriptome sequencing data of 377 HCC patients from TCGA database, the m6A DEGs between HCC and para-carcinoma tissue were analyzed. This prognostic model will provide a more accurate and effective prediction of HCC patients’ prognosis, and it’s also the first time to combine m6A associated genes to predict the prognosis of HCC.
Methods
Data source
The mRNA expression profiles and the clinical data from the patients with HCC were downloaded from the TCGA, which containing 374 HCC tissues and 50 normal tissues. As the data from the TCGA are publicly available and open-access, our ethics committees did not need to approve the study. The current research follows the TCGA data access policies and publication guidelines. The ten pairs of matched HCC and para-carcinoma tissue, which were used to verify the expression of the m6A genes, were obtained from the liver surgery department of Peking Union Medical College Hospital, and approved by the Medical Ethics Committee of Peking Union Medical College Hospital of the Chinese Academy of Medical Sciences (CAMS) & PUMC.
M6A genes and the prognosis of HCC
We searched the current literatures about m6A, and list all the m6A genes. Then download the raw counts of the HCC mRNA expression profiles from the TCGA databases. The differentially expressed m6A genes were calculated using R package with an adjusted P value of <0.05 were considered significant difference. We performed a Consensus Cluster Plus analysis based on the expression level of mRNAs, and divided the HCC patients into two clusters. Then a Principal Component Analysis (PCA) were conducted between these two clusters. We also performed a survival analysis in the two clusters based on the clinical data from the TCGA, to identify the relationship between the expression level of m6A genes and the prognosis of HCC.
The m6A gene-related prognostic model
We analyzed the correlation between the expression level of each m6A gene and the overall survival (OS) of HCC patients by univariate analysis. The gene was considered significant when the P value was <0.05. Then, a Lasso-penalized Cox regression analysis was employed to identify the prognostic model. The HCC patients were divided into high and low risk group according to the risk score, and a Kaplan-Meier (K-M) survival analysis were conducted between the low- and high-risk group. ROC curve analyses were performed to assess the predictive power of the prediction model. Univariate and multivariate Cox regression analyses were performed to determine whether the prognostic model could be independent of other clinical variables (including age, gender, histologic grade and tumor stage) for patients with HCC. All reported P values were two-sided. The hazard ratio (HR) and 95% confidence intervals were calculated.
PCR validation of the expression of the m6A genes included in the prognostic model
According to the results of bioinformatics analysis, the expression level of m6A genes, which included in the prognostic model, were validated by PCR using ten pairs of matched HCC and para-carcinoma tissue obtained from our center. Total RNA of tissue and cells was extracted with Trizol (Thermo Fisher Scientific) and all reagents used in the experiment were RNase-free. Actin was used as internal reference in PCR.
Results
The m6A genes and the prognosis of HCC
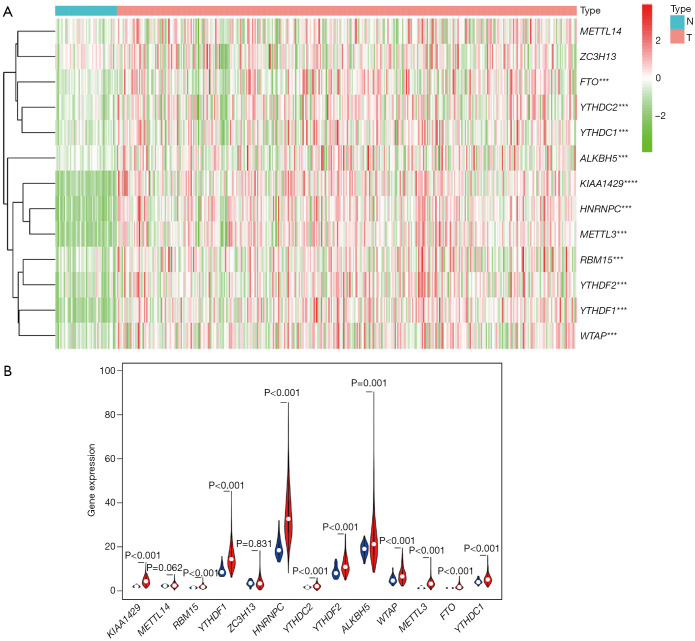
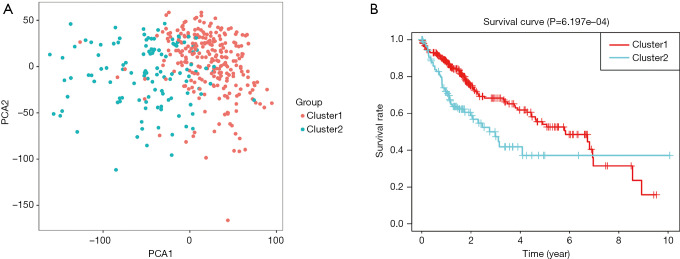
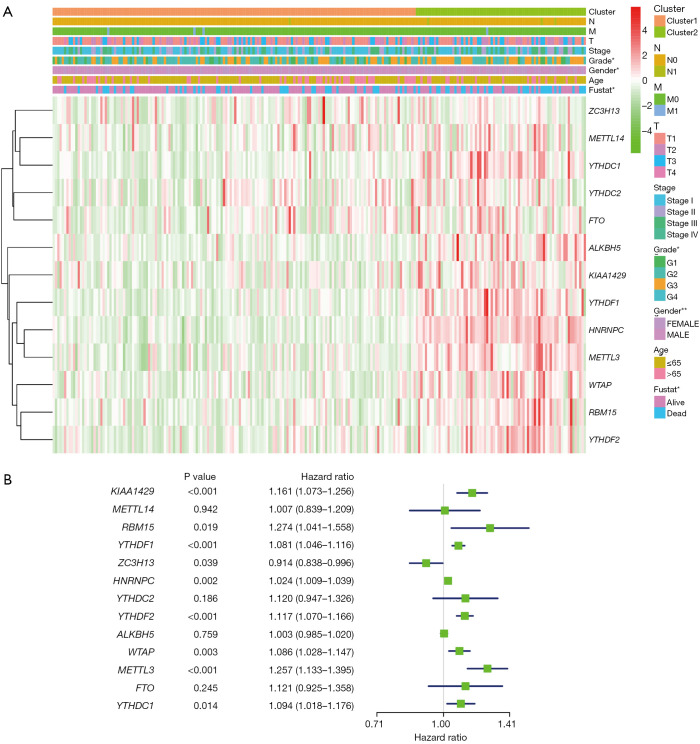
According to the current literature reports, we confirmed 13 m6A genes. A total of 11 differentially expressed m6A mRNAs (P<0.05) were identified in the mRNA expression profiles of HCC tissues (n=374) compared with normal tissues (n=50), as shown in the heat map and violin plot (Figure 1). Further, the correlation between all the m6A genes’ expression level in HCC were identified. Based on the consensus clustering approach, we divided the 374 HCC patients into two clusters, according to the expression level of mRNAs in HCC. The result of PCA indicated that these two clusters were independently divided (Figure 2A). The survival analysis Cluster 2 showed a poorer prognosis (P=6.197×10-4) compared with cluster 1 (Figure 2B). And we verified that the m6A genes’ expression level in cluster 2 were higher than cluster 1 by heat map analyzing (Figure 3A).
Figure 1.
(A) Heat map of the differential expressed m6A genes between hepatocellular carcinoma and normal tissues, N indicates normal tissues, T indicates hepatocellular carcinoma tissues, “***” indicates the P value of this differential expressed gene is less than 0.001. (B) The violin plot of each differential expressed m6A genes between hepatocellular carcinoma and normal tissues.
Figure 2.
Tumor sub-type analysis. (A) Two clusters of HCC patients divided by PCA. (B) Survival analysis of these two clusters, and the cluster 2 showed a poorer prognosis (P=6.197×10–4). HCC, hepatocellular carcinoma; PCA, Principal Component Analysis.
Figure 3.
The prognostic value of m6A genes in HCC patients. (A) The heat map indicates the higher expression level of m6A genes in cluster 2. (B) Univariate analysis of the relationship between the expression level of each m6A gene and the prognosis of HCC, and 9 m6A genes were proven to be related to prognosis of HCC. HCC, hepatocellular carcinoma.
Prognostic model establishing and survival analysis
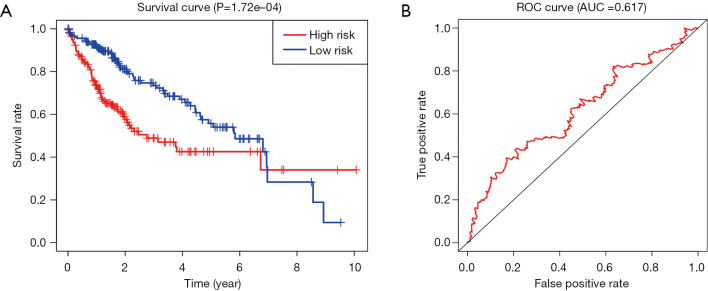
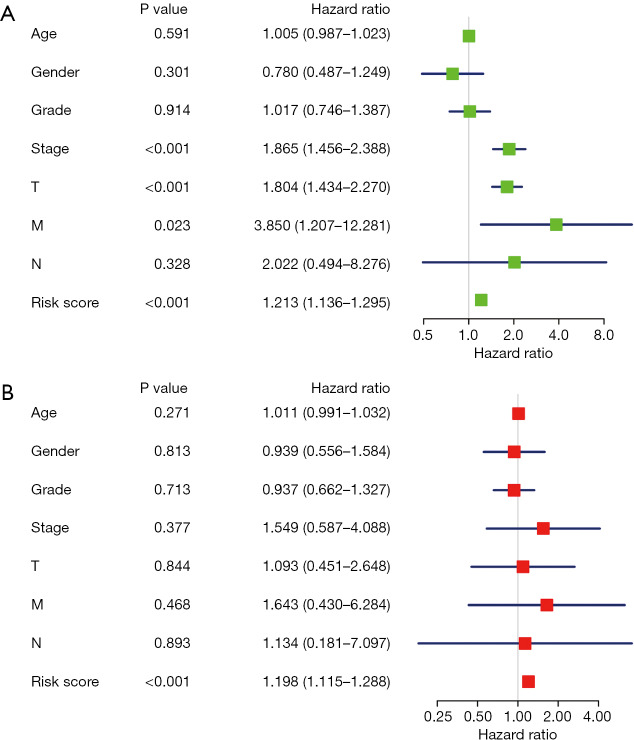
We confirmed 9 m6A genes were associated with poor prognosis in HCC by univariate analysis (Figure 3B). After Lasso-penalized Cox analysis, 5 m6A genes (KIAA1429, METTL3, YTHDF1, YTHDF2, ZC3H13) were included in the risk model. Based on the risk score, the 374 HCC patients were classified into two groups, high- and low-risk group. The high-risk group with a poorer prognosis (P=1.72×10-4) compared with low-risk group by survival analysis (Figure 4A). ROC analysis revealed that the AUC of this model was 0.617 (Figure 4B). The results of univariate [P<0.001, 1.213 (1.136–1.295)] and multivariate Cox regression analysis [P<0.001, 1.198 (1.115−1.288)] indicated that this prognostic model was an independent prognostic factor for HCC patients (Figure 5).
Figure 4.
The test of the prognostic model. (A) Survival analysis revealed that the high-risk group with a poorer prognosis compared with the low-risk group (P=1.72×10–4). (B) The ROC analysis indicated that the AUC of this prognostic model was 0.617. ROC, receiver operating characteristic.
Figure 5.
Identify the independence of this prognostic model as a predict factor of HCC. (A) Univariate analysis and (B) multivariate Cox regression analysis confirmed the independence of this prognostic model. HCC, hepatocellular carcinoma.
The PCR results
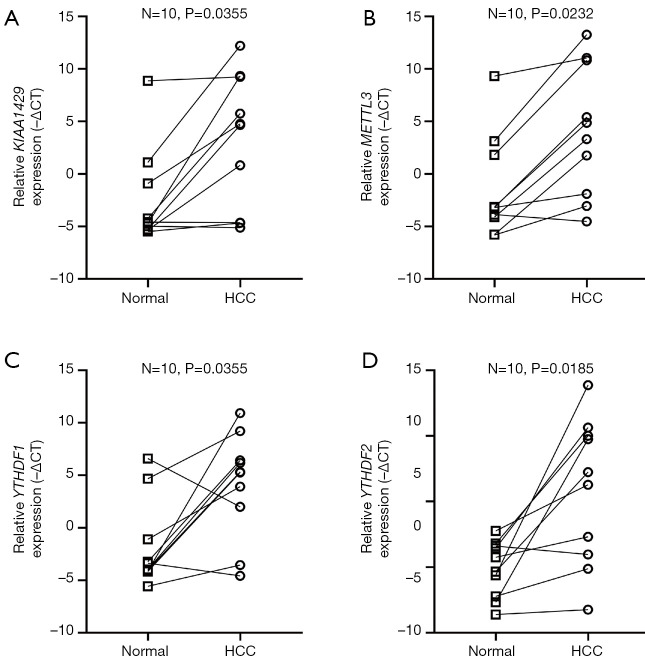
As the difference of the expression level of gene ZC3H13 was not significant between HCC and para-carcinoma tissue, we only validated the rest four m6A genes (KIAA1429, METTL3, YTHDF1, YTHDF2) included in the prognostic model. These four genes were all significantly high expressed in HCC (P=0.0355, P=0.0232, P=0.0355 and P=0.0185 respectively) (Figure 6) in accordance with the result of the TCGA data.
Figure 6.
The PCR validation of four m6A genes included in the prognostic model. KIAA1429, METTL3, YTHDF1 and YTHDF2 were all significantly high expressed in HCC tissues, which in accordance with the results of TCGA. HCC, hepatocellular carcinoma.
Discussion
The current prognostic predictors of HCC are lack of specificity and sensitivity (15,16). The function of m6A associated genes in tumorigenesis and progression has been intensively explored. But the prognostic value of m6A genes in cancers still not well verified. Our study designed to analysis the differentially expressed m6A genes and establish a prognostic model of HCC. Firstly, we confirmed 11 m6A genes were high expressed in HCC compared with para-carcinoma tissue, and the high expression level of m6A genes associated with a poor prognosis of HCC. Then we developed a prognostic model consist of m6A genes. According to the risk score, HCC patients were divided into high- and low-risk group, and the high-risk group have a significant poor prognosis after survival analysis. After univariate and multivariate Cox regression analyses, this prognostic model be confirmed as an independent prognostic factor of HCC.
KIAA1429 is an important methyltransferase, which categorized as the components of ‘writers’ in m6A modification. There are studies revealed that reduction of KIAA1429 can significantly inhibited tumor proliferation, migration, and invasion by regulating mRNA methylation levels of ID2 in HepG2 cells. KIAA1429 increased the m6A level of ID2 mRNA, and this will decreased the ID2 expression and facilitated cell invasion and migration in HCC (17). It has been reported that GATA3 was the downstream target of KIAA1429, and the KIAA1429 produced m6A methylation on the 3'-UTR of GATA3 pre-mRNA, leading to GATA3 pre-mRNA degradation. Overexpression of KIAA1429 was correlated with poor prognosis in HCC (18).
METTL3 is an RNA methyltransferase involved in mRNA decay, translation and biogenesis control via m6A modification (19,20). METTL3 was reported to be related with gastrointestinal tumors, such as gastric cancer (21-23), colorectal cancer (24,25) and pancreatic cancer (26). As to HCC, METTL3 can inhibits SOCS2 expression, which could promote HCC cell proliferation, migration and colony formation (27). Meanwhile, METTL3 was observed to be upregulated in liver cancer and was found to promote its progression. In addition, METTL3 upregulation contributes to HCC metastasis and associated with a poor prognosis in HCC patients.
YT521-B homology domain family (YTHDF) proteins are the ‘readers’ of m6A, which affect the stability and translation efficiency of m6A-containing RNAs. YTHDF1 was reported to enhance translational efficiency of m6A-modifed mRNAs and significantly upregulated in HCC. Furthermore, survival analysis showed that lower YTHDF1expression level was associated with better prognosis of HCC patients (28). Bioinformatics enrichment analysis of YTHDF1 co-expressed genes revealed that YTHDF1 played a key role in regulating HCC cell cycle progression and metabolism. It’s a potential target in treatment and prognosis for HCC.
YTHDF2 also be proven associated with many tumors like colon cancer (29). Based on the current evidence, the high expression of YTHDF2 can suppress cell proliferation and tumor growth in HCC cells. Mechanistically, YTHDF2 via bound the m6A modification site of EGFR 3’-UTR to enhance the degradation of EGFR mRNA, which may repress cell proliferation and growth in HCC (30). There is another study indicated that YTHDF2 reduction in HCC cells promoted vascular reconstruction, inflammation and metastatic progression, which associated with the prognosis of HCC patients (31).
However, there are some limitations in this study. First, the population in the TCGA database are mainly White people and Black people, and extrapolating the results to other ethnic groups needs to be carefully substantiated. Second, this study mainly focused on bioinformatics analysis. Although the high expression of the four m6A genes in HCC are identified by PCR, this study still lacks of mechanism verification. Our next work will continue to focusing on the specific mechanisms of these genes in liver cancer. Third, the test performance of this model is still relatively low, and we will also explore the possibility of containing more prognostic variables to further improve the performance.
This prognostic model can not only provide a prediction of HCC patients’ prognosis, as for the m6A genes, it will also point one direction for further research of HCC mechanism, and the potential value in diagnosis and treatment of HCC still to be explored.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by the International Science and Technology Cooperation Projects (2016YFE0107100); and the CAMS Innovation Fund for Medical Science (CIFMS) (2017-I2M-4-003 and 2018-I2M-3-001).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by our institutional ethics committee. Written informed consent was obtained from the patient for publication of this manuscript and any accompanying images.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2894). The authors have no conflicts of interest to declare.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Xie DY, Ren ZG, Zhou J, et al. Critical appraisal of Chinese 2017 guideline on the management of hepatocellular carcinoma. Hepatobiliary Surg Nutr 2017;6:387-96. 10.21037/hbsn.2017.11.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villanueva A. Hepatocellular Carcinoma. N Engl J Med 2019;380:1450-62. 10.1056/NEJMra1713263 [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Weng H, Su R, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)- Methyladenosine RNA Demethylase. Cancer Cell 2017;31:127-41. 10.1016/j.ccell.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Eckert MA, Harada BT, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol 2018;20:1074-83. 10.1038/s41556-018-0174-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vu LP, Pickering BF, Cheng Y, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 2017;23:1369-76. 10.1038/nm.4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui Q, Shi H, Ye P, et al. m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep 2017;18:2622-34. 10.1016/j.celrep.2017.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwok CT, Marshall AD, Rasko JE, et al. Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol 2017;10:39. 10.1186/s13045-017-0410-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SH, Moon DB, Kim WJ, et al. Preoperative prognostic values of alpha-fetoprotein (AFP) and protein induced by vitamin K absence or antagonist-II (PIVKA-II) in patients with hepatocellular carcinoma for living donor liver transplantation. Hepatobiliary Surg Nutr 2016;5:461-9. 10.21037/hbsn.2016.11.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li ZL, Han J, Liu K, et al. Association of family history with long-term prognosis in patients undergoing liver resection of HBV-related hepatocellular carcinoma. Hepatobiliary Surg Nutr 2019;8:88-100. 10.21037/hbsn.2018.11.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eiro N, Vizoso FJ. Importance of tumor/stroma interactions in prognosis of hepatocellular carcinoma. Hepatobiliary Surg Nutr 2014;3:98-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer 2019;18:103. 10.1186/s12943-019-1033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet 2014;15:293-306. 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015;161:1388-99. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhan P, Ji YN. Prognostic significance of TP53 expression for patients with hepatocellular carcinoma: a meta-analysis. Hepatobiliary Surg Nutr 2014;3:11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu B, Yang XR, Xu Y, et al. Systemic immune-inflammation index predicts prognosis of patients after curative resection for hepatocellular carcinoma. Clin Cancer Res 2014;20:6212-22. 10.1158/1078-0432.CCR-14-0442 [DOI] [PubMed] [Google Scholar]
- 17.Cheng X, Li M, Rao X, et al. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. Onco Targets Ther 2019;12:3421-8. 10.2147/OTT.S180954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan T, Li H, Zhang D, et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer 2019;18:186. 10.1186/s12943-019-1106-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbieri I, Tzelepis K, Pandolfini L, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 2017;552:126-31. 10.1038/nature24678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin S, Choe J, Du P, et al. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 2016;62:335-45. 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue B, Song C, Yang L, et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer 2019;18:142. 10.1186/s12943-019-1065-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Chen C, Ding Q, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2019. [Epub ahead of print]. 10.1136/gutjnl-2019-319639 [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Zhang M, Ge S, et al. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med 2019;8:4766-81. 10.1002/cam4.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng R, Cheng Y, Ye S, et al. m6A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther 2019;12:4391-402. 10.2147/OTT.S201052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T, Hu PS, Zuo Z, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer 2019;18:112. 10.1186/s12943-019-1038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taketo K, Konno M, Asai A, et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol 2018;52:621-9. [DOI] [PubMed] [Google Scholar]
- 27.Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018;67:2254-70. 10.1002/hep.29683 [DOI] [PubMed] [Google Scholar]
- 28.Zhao X, Chen Y, Mao Q, et al. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomarkers 2018;21:859-68. 10.3233/CBM-170791 [DOI] [PubMed] [Google Scholar]
- 29.Tanabe A, Tanikawa K, Tsunetomi M, et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1alpha mRNA is translated. Cancer Lett 2016;376:34-42. 10.1016/j.canlet.2016.02.022 [DOI] [PubMed] [Google Scholar]
- 30.Zhong L, Liao D, Zhang M, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett 2019;442:252-61. 10.1016/j.canlet.2018.11.006 [DOI] [PubMed] [Google Scholar]
- 31.Hou J, Zhang H, Liu J, et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer 2019;18:163. 10.1186/s12943-019-1082-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as