Abstract
Tremendous efforts have been made these last decades to increase our knowledge of intracellular degradative systems, especially in the field of autophagy. The role of autophagy in the maintenance of cell homeostasis is well documented and the existence of defects in the autophagic machinery has been largely described in diseases and aging. Determining the alterations occurring in the many forms of autophagy that coexist in cells and tissues remains complicated, as this cellular process is highly dynamic in nature and can vary from organ to organ in the same individual. Although autophagy is extensively studied, its functioning in different tissues and its links with other biological processes is still poorly understood. Several assays have been developed to monitor autophagy activity in vitro, ex vivo, and in vivo, based on different markers, the use of various inhibitors and activators, and distinct techniques. This review emphasizes the methods applied to measure (macro-)autophagy in tissue samples and in vivo via a protein, which centrally intervenes in the autophagy pathway, the microtubule-associated protein 1A/1B-light chain 3 (MAP1LC3), which is the most widely used marker and the first identified to associate with autophagosomal structures. These approaches are presented and discussed in terms of pros and cons. Some recommendations are provided to improve the reliability of the interpretation of results.
Keywords: autophagy, autophagic flux, MAP1LC3, lysosome, in vivo autophagy assays
1. Introduction
Autophagy (from the ancient Greek “αὐτόφαγος”, meaning “self-eating”) is a critical component of cell homeostasis. This pivotal process contributes to the cellular quality control and energetic regulation of tissues via the lysosomal processing and recycling of vital cellular components [1]. Three main forms of autophagy are classically described that are macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA), but in fact, there are many more forms and subtypes of each (as the different types of microautophagy, for example), working concomitantly in the cell to control its viability. Some of these processes are highly selective, some others are less, but all share a certain degree of selectivity to mediate the degradation of specific targets [2,3,4,5,6].
Much autophagic research has been originally done by exploiting organisms such as the versatile yeast and drosophila, and then followed by mammals. Over the past decades, significant advances have been made, leading to a better understanding of the molecular mechanisms driving autophagy in its many intricate physiological functions. These activities are thought to be particularly important in all aspects of cell life including differentiation and development, adaptation to metabolic stresses, degradation of dangerous cargo (e.g., misfolded or aggregated proteins, proteins produced daily in excess, damaged organelles, intracellular pathogens), and regulation of immune responses. It occurs, however, that these normally protective functions become harmful when autophagy processes are altered. Recent studies centered on the molecular biology of autophagy have especially addressed these issues in diseases as diverse as autoimmune and inflammatory syndromes, cancer, neurodegeneration, infectious diseases, cardiovascular disorders, metabolic diseases, intestinal bowel diseases, ischemia–reperfusion injury, and aging [5,7,8,9,10,11,12]. Mutations in numerous autophagy-related (ATG) genes have been linked to specific diseases, although rarely to one single pathology [4]. These mutations affect the proper functioning and regulation of autophagy and lysosomal activity, as well as cargo delivery. Their consequences also interfere in the crosstalk of autophagy with the many other forms of cellular circuits, such as apoptosis, necrosis, and ferroptosis [6].
These last years, the molecular elements of autophagy pathways characterizing macroautophagy, CMA, and more recently endosomal microautophagy (eMI), have been extensively studied. The cascade of events giving rise to the coordinated emergence of diversified organelles, the implication of multiple ATG genes, the identification of proteins complexes, for example, the ULK1 serine–threonine kinase (involving ULK1, FIP200, ATG13, and ATG101) and the VPS34/BECLIN-1 (BECN1) complex, which act sequentially in the autophagic processes, have been precisely examined [4,13,14,15,16]. These studies have permitted to pinpoint a number of markers that characterize different steps or forms of the autophagy process [17]. The combined use of drugs acting as activators or inhibitors on selected components and of genetically manipulated eukaryotic models have greatly contributed to the identification of successive steps and key components of the main autophagic pathways [14,18]. Particular attention should be given when certain drugs (e.g., bafilomycin A1) are used experimentally as their effect can critically depend on the time and concentration used as well as on the cell type. These drugs have also allowed demonstrating the potential interest of targeting autophagy for therapeutic applications [7,9,17]. Examples of such pharmacological agents and their main mechanism of action are listed in Table 1. Several comprehensive reviews extensively document their effects and some of their limitations [4,7,9,10,17,18,19,20,21,22].
Table 1.
Examples of autophagic and lysosomal activators and inhibitors (non-exhaustive list) 1.
| Autophagy Activators | Mechanism of Action |
| KU-0063794 | Inhibits MTORC1 and MTORC2 |
| Lithium, carbamezapine, sodium valproate | Reduces intracellular inositol levels by inhibiting their synthesis |
| Metformin | Activates AMPK |
| NVP-BEZ235 | Inhibits PI3K/MTOR |
| Rapamycin, everolimus, temsirolimus (CCI-779), deforolimus, AZD8055 | Interact wih FkBP-12 and the complex inhibits the activity of MTOR |
| Resveratrol | Activates SIRT1 |
| Spermidine | Inhibits several acetyltransferases, e.g., EP300, IKI3, and SAS3 |
| Statins (simvastatin) | Inhibits PKB/AKT |
| Sucrose | Impairs lysosomal function |
| Tamoxifen | Increases the intracellular level of ceramide. Abolishes the inhibitory effect of PI3K |
| TAT-Beclin-1 | Inhibits the activity of PI3KC3 through competing of the binding of BECN1 to its negative regulator GAPR-1 |
| Torkinib (PP242), torin 1 | Inhibits MTORC1/2 |
| Trehalose | Impairs lysosomal function |
| Tunicamycin | Induces ER stress in cells by inhibiting the first step of the biosynthesis of N-linked glycans in the proteins |
| Autophagy Inhibitors | Mechanism of Action |
| Azithromycin | Inhibits lysosomal v-ATPase and prevents the acidification process |
| Bafilomycin A1, concanamycin A | Both specifically bind to the V0 domain of v-ATPase and inhibit the intracellular pH gradients in endocytic and secretory organelles |
| CQ/HCQ | Increases the lysosomal pH (v-ATPase-independent) |
| Colchicine, Lys05, monensin, nigericine, nocodazole, SAHA (vorinostat), vinblastine | Inhibits autophagosome-lysosome fusion |
| E64d/pepstatin A | Inhibits cysteine and aspartic proteases |
| Leupeptin | Inhibits serine and cysteine proteases |
| NH4Cl | Increases the lysosomal pH (v-ATPase-independent) |
| P140 peptide | Inhibits the chaperone activity of HSPA8 |
| Wortmannin, 3-methyladenine, LY294002 | Inhibits class III PI3K |
Discovered in our team, a 21-mer phosphopeptide called P140, which primarily targets CMA in vitro and ex vivo and corrects altered macroautophagy occurring in inflammatory pathological contexts, showed its therapeutic effectiveness both in animal models of autoimmunity [26,27,28,29,30] and in patients with lupus [31,32]. Given to lupus mice with active disease, P140 reduced the overexpression level of lysosomal-associated membrane protein (LAMP)2A and HSPA8, two key players of CMA [33,34,35]. In vitro, P140 inhibited CMA in a cell line that stably expressed a CMA reporter [34,35]. In NOD.H-2h4 mice that develop a primary Sjögren’s-like syndrome, P140 rescued sick mice from some autophagy defects presumably associated with the ULK1 complex, and significantly reduced the formation of tertiary lymphoid structures in salivary glands, a hallmark characteristic of this disease in patients [30]. The expression of BECN1, which was raised in diseased NOD.H-2h4 mice, a feature that could explain, at least in part, the clinical and biological beneficial effect of P140 in this mouse model.
Another peptide, the cell-penetrating peptide Tat-BECN1, rather acts as an activator of autophagy [36]. It was found to exert protection against viral and bacterial infection both in vitro and in vivo, and some effectiveness in other pathological conditions such as heart failure in response to cardiac hypertrophy, for example [4]. The activity of some rationally-designed analogs of Tat-BECN1 peptide, like Tat-D11 (Novus Biologicals), prove significantly more potent than the original sequence in different assays and indications. A peptide named humanin initially identified from surviving neurons in patients with Alzheimer’s disease (AD), was described as a direct enhancer of CMA that acts by increasing substrate binding and translocation into lysosomes [37]. It interacts with HSP90 and stabilizes the binding of this chaperone to CMA cargos as they bind to the lysosomal membrane. It was shown that humanin exerts cardioprotective and neuroprotective properties in diseases such as AD, cardiovascular diseases, stroke, myocardial infarction, diabetes, and cancer. These peptides and other molecules that target specific autophagy elements represent potential therapeutics that might be successfully applied to treat patients affected by pathologies for which current drugs are clearly insufficient with severe secondary effects, or to individuals who are refractory to the existing medications.
When developing new chemical strategies of intervention that target autophagy, an important step is to demonstrate that in vivo, and not only in vitro or ex vivo, the molecule is effective on the expected autophagic target that has been identified in the upstream investigations. Indeed, in vivo, it may well be that the molecule induces its effect as thought, i.e., directly on the expected autophagy component but it might also act via an autophagy-independent mechanism or on an alternative metabolic system that in fine impact autophagy. It is effectively well known that numerous autophagy-related markers are also involved in other metabolic pathways. It is the case of BECN1, HSPA8, MTOR, and many others [4,23]. Yet, measuring autophagy in vivo or even in tissue samples (biopsies) collected from patients, for example, remains a challenge. Assessment of autophagic flux in whole organisms is even more challenging. Specific assays are limited and often remain non validated given the variety of tissues and organisms that are studied. The aim of this review is to gather useful information compiled from the existing literature and generated by our own experience, to analyze the validity of proposed methods and to discuss their potential and limitations. An emphasis is placed on microtubule-associated protein 1A/1B-light chain 3 (MAP1LC3) that is central in the autophagy process and especially in macroautophagy. Present at low and variable levels of expression in a normal situation, its distribution is widespread, for example in the bone marrow, brain, heart, placenta, thyroid, bladder, and several other organs and tissues.
2. The Microtubule-Associated Protein 1A/1B-Light Chain 3 (MAP1LC3)
Together with sequestosome (SQSTM1)/p62, MAP1LC3 is the most widely used marker that is followed experimentally to evaluate the extent of autophagy in cells. MAP1LC3 is a member of the highly conserved ATG8 protein family. It was initially described in the yeast (S. cerevisiae) and later identified in mammals [38,39]. MAP1LC3 belongs to a large family of seven members composed of MAP1LC3A, 3B, 3B2, 3C, and the γ-aminobutyric acid (GABA)-receptor-associated proteins (GABARAP, GABARAPL1, GABARAPL2) proteins. The NMR structure of MAP1LC3B (residues 1-120) has been determined (Figure 1) [40]. The members of this protein family share a common ubiquitin-like (Ubl) structures present at the C-terminus and two α-helices at their N-terminus, which are likely involved in protein–protein and lipid–protein interactions and post-translational modifications that occur during the autophagy process [41,42,43,44]. In the cytosol, newly synthesized MAP1LC3B is cleaved by the autophagy-related 4B cysteine peptidase (ATG4B) at the C-terminal glycine residues120 leading to MAP1LC3-I (Figure 1). Phosphatidylethanolamine (PE)-conjugation of MAP1LC3-I resulting in the lipidated MAP1LC3-II form requires ATG7 and ATG3, as well as the ATG12–ATG5–ATG16L1 complex [45,46,47]. Formation of the latter involves conjugation of ATG12 to ATG5, in a process mediated by ATG7 and ATG10, and further binding of the ATG12–ATG5 conjugate to an ATG16L1 dimer. In this process, ATG7 plays a central role as this E1-like activating enzyme enables both the lipidation of ATG8 proteins (MAP1LC3) and conjugation of ATG12 to ATG5. It is the only enzyme common to both conjugation pathways. At the isolation membrane and autophagosomes, MAP1LC3-II displays significant functions in the autophagy process, such as elongation, sealing isolation membranes, recognizing cargos, etc. [47]. The Ubl structures encompass β-strands with hydrophobic pockets, which are implicated in protein interactions. MAPL1LC3 is also present in the nucleus of a variety of cell types. In response to starvation, nuclear MAP1LC3 is deacetylated and trafficked out of the nucleus into the cytoplasm where it exerts its functions in autophagy. The protein is also enriched in nucleoli where it binds nuclear and nucleolar constituents, such as microtubule-associated protein 1B, tubulin, and ribosomal proteins.
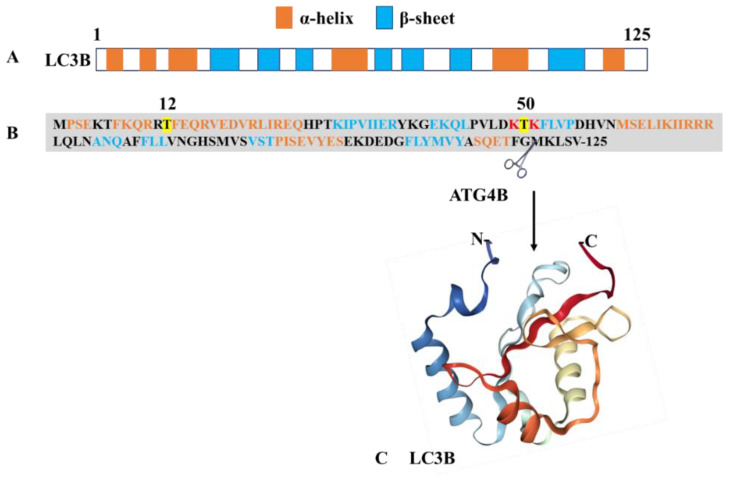
Figure 1.
Structure of MAP1LC3B. The protein presents as a monomer with a molar mass of 13.6 kDa. (A) Pictorial illustration showing the alignment of α-helices and β-sheets in LC3B. (B) Primary structure of MAP1LC3B (NCBI reference ID: NP_073729.1) showing the positions of α-helixes and β-sheets. The sites of phosphorylation and acetylation are highlighted in yellow and red, respectively. The UbI domain is located at the C-terminus of the protein. (C) Three-dimensional (3D)-structure (PDB ID: 1V49) of the MAP1LC3B fragment 1–120. Figure generated using https://www.rcsb.org/, ref. doi:10.1093/bioinformatics/bty419.
MAP1LC3 proteins interact with many cofactors and ligands. All interacting proteins display a common ubiquitin-binding domain (UBD) and a short hydrophobic LC3-interacting region (LIR) motif that contains an N-terminal sequence W (tryptophan)xxL (leucine). The W residue is surrounded by other aromatic residues, tyrosine (Y) and phenylalanine (F), acidic residues, and sometimes serine (S) and threonine (T) residues to compose a consensus W/F/YXXL/I/V motif. These residues interact with the basic residues present in the Ubl domain of MAP1LC3 via electrostatic bridges (Figure 1). LIR motif-containing proteins encompass two hydrophobic residues, which are accommodated into the MAP1LC3 binding pockets [48]. The LIR motifs are post-translationally regulated and contribute to the different functions displayed by MAP1LC3 proteins, including selective autophagy process [48,49,50,51].
MAP1LC3 plays a key role in the autophagosome biogenesis machinery from the isolation membrane to the lysosomal stage. The recruitment of MAP1LC3 and other ATG proteins triggers vesicle expansion in a concerted manner and intervenes in the initial steps of membrane curvature. During this complex sequential process, MAP1LC3 is finely regulated by diverse post-translational modifications, which favor or on the contrary alter its activity. Thus, phosphorylation of MAP1LC3B at Thr50 by serine/threonine-protein kinases STK3/STK4 is essential for the autophagosome–lysosomal fusion process [52]. Phosphorylation at residue Ser12 by phosphokinase A (PKA) keeps MAP1LC3B unavailable for conjugation to PE and inhibits the autophagy process [53]. This modification is essential for phagophore expansion; its failure leads to defects in autophagosome formation. During stress or starvation conditions, PKA activity is inhibited and MAP1LC3B remains available for conjugation. On the other hand, acetylation regulates the MAP1LC3B translocation from the nucleus to the cytosol. The reason for the presence of LC3B in the nucleus remains an unanswered question. Acetylation at residues Lys49 and Lys51 makes the MAP1LC3B protein confined to the nucleus and not available for cytosolic ligases [54]. Of note, Lys49 and Lys51 form ionic interactions with ligands, especially with the LIR motifs. In stress conditions, especially starvation, deacetylation of MAP1LC3B by sirtuin 1 results in the availability of MAP1LC3B for ligases [54].
3. Current MAP1LC3-Based Methods Designed for Autophagy Research
MAP1LC3-based assays are widely used in current autophagy research [22,55,56,57,58]. In general, they offer the possibility to monitor the cellular autophagic activity in a routine manner using conventional laboratory equipment. A number of advantageous criteria, such as sensitivity, performance, specimen throughput, volume requirements, the limit of detection, ease of execution, instrument workspace, and the costs of equipment and disposal, are met. Commonly, the detection of processed MAP1LC3B-II by Western blot (Figure 2) or fluorescence studies, together with electron microscopy for autophagosome formation (Figure 3), is sufficient to qualify autophagy in cells.
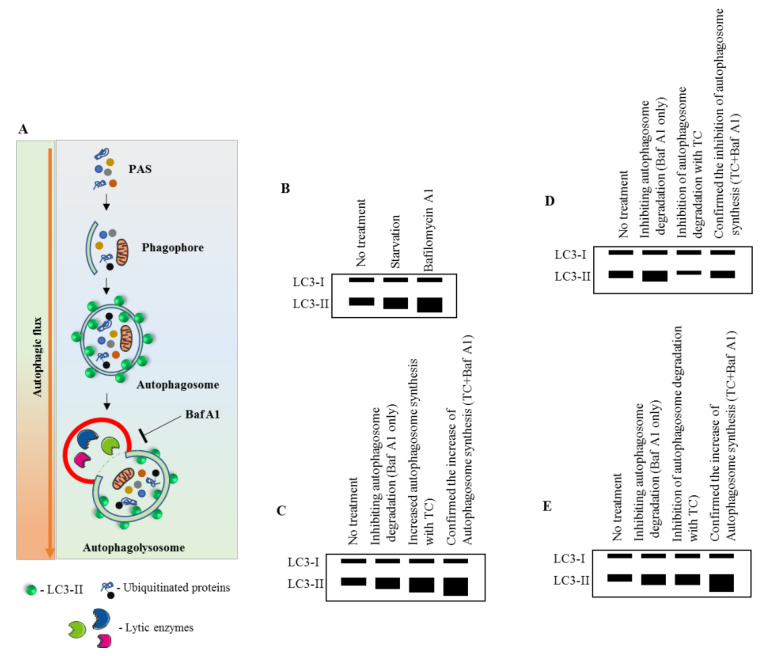
Figure 2.
Immunoblot measurement of MAP1LC3B turnover and its dynamics. (A) Pictorial representation of the autophagic flux process. Baf A1 inhibits the autophagy process by blocking the autophagosome–lysosomal fusion. (B–E) Effect of a test compound on MAP1LC3-II expression. Without the use of Baf A1, it would be unpredictable to assess whether the compound is increasing the autophagosome synthesis or inhibiting the autophagosome degradation. Modified from [59]. See abbreviations in the abbreviations section.
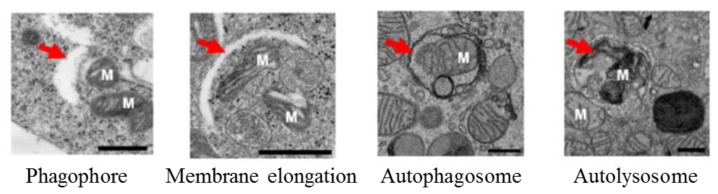
Figure 3.
Macroautophagy process under electron microscopy. Red arrows are indicating the phagophore, membrane elongation, autophagosome, and autolysosome. M: mitochondria, scale bar: 500 nm. Adapted from [60].
These basic methods, applied to primary cells and cell lines, are often adequate for the first series of screening tests, but they may appear insufficient to further develop knowledge for therapeutic purposes. Indeed, they suffer each from intrinsic limitations that can introduce errors in the data analyses and survey results (Table 2). Furthermore, and most importantly, they often apply to cells and not to tissues and organs and are routinely performed in vitro/ex vivo and not in vivo.
Table 2.
MAP1LC31-based standard assay for assessing autophagy and their limitations 1.
| Techniques | Special Attention, Limitations and Pitfalls |
|---|---|
| Western blot assay (Figure 2) |
|
| Fluorescence microscopy for detecting endogenous MAP1LC3 |
|
| Fluorescence microscopy for detecting reporters (e.g., GFP-MAP1LC3, mRFP-GFP-MAP1LC3, …) |
|
| Flow cytometry |
|
| Multispectral imaging flow cytometry | |
| Bioluminescence |
|
| MAP1LC3B time-resolved fluorescence transfer (TR-FRET) assay |
|
| Electron microscopy (Figure 3) |
|
| Long-lived protein degradation |
|
| LDH sequestration assay |
|
| Dextran sequestration assay |
1 A few other conventional methods and/or nonspecific methods, such as isotope release from long-lived protein degradation (radiolabeling long-lived proteins with radioactive amino acid residues, such as [14C]-leucine, [3H]-leucine, [14C]-valine, or [35S]-methionine) and LDH assay (using LDH as a cargo) are not reviewed here as they are not routinely used. 2We are well aware that determining the ATG protein levels or the number of autophagosomes alone does not provide the overall estimation of autophagic activity since the process is very dynamic, and MAP1LC3-II can quickly degrade within the lysosomes. In fact, the two opposite scenarios, namely induction of autophagy or blockade in the downstream steps of autophagy leading to defective degradation, can result in an increased number of autophagosomes. Therefore, it is necessary to evaluate the autophagic flux by measuring the level of expression of MAP1LC3-I and -II in the presence of lysosomal inhibitors such as bafilomycin A1 or others. See abbreviations in the abbreviations section.
In general, these methods have the capability for a high sample throughput and are used to sift large numbers of such samples for further application. As indicated, however, they also have some disadvantages and intrinsic limitations. The results that are generated to support possible changes in autophagy processes have also to be distinguished from effects induced by the many other forms of cell death [22,83].
4. MAP1LC3-Based Methods Designed for Studying Autophagy in Tissues
Several methods have been described to detect MAP1LC3 directly in fresh and fixed tissues collected from patients and experimental animal models. They are listed in Table 3 with their respective advantages and limitations (see also the Box 1).
Table 3.
MAP1LC3-based methods to measure autophagy in biopsies.
| Method | Advantages | Limitations |
|---|---|---|
| Immunohistochemistry |
|
|
| Western blot analysis (from FFPE tissue) [84] |
|
|
| In-situ hybridization [85] |
|
|
See abbreviations in the abbreviations section.
Box 1. Central aspects to consider while monitoring autophagy.
General considerations
-
-
Autophagy is a dynamic mechanism that requires flux measurements to be monitored.
-
-
An increase of autophagy markers or factors detected at an early stage of the process may not give a clear idea on the entire process of autophagy as the blockade may appear at later stages. -Selection of mouse strain, while transfecting MAP1LC3, is a prominent factor in autophagy research, since for example, basal autophagy markedly differs between mouse strains.
-
-
Detecting an increasing number of lysosomes may not reflect an increase in autophagic activity. LysoSensor may help to differentiate between the effects of autophagosome to lysosomes.
-
-
It can be difficult to propose conclusions in some instances, especially in infections, in which the life cycle of pathogens affects the basal autophagic pathway.
Technical considerations
-
-
Lack of highly specific autophagy inhibitors and activators remains a limiting factor.
-
-
Autophagy-inducing agents, either for autophagy flux measurement or therapy, behave differently upon time and dosage.
-
-
To evaluate novel molecules on inhibition or activation of autophagy, it is strongly recommended to use standard controls such as starved conditions and knockdown/knockout of key ATG proteins (e.g., ATG3, 5, 7, 9a, 16L1, FIP200, AMBRA1, BECN1) as a starting point.
-
-
Autophagy modulator screening assays using GFP-MAP1LC3 benefit from secondary probes (GFP-MAP1LC3-RFP-LC3∆G, mRFP-GFP-MAP1LC3, mCherry-GFP-MAP1LC3) to clearly discriminate the effects on autophagosomes from effects on lysosomes or autolysosomes.
Specific considerations that apply when organs/tissue are studied
-
-
The autophagic process (type, regulation, intensity) can differ from cell to cell, organ to organ/tissue in the same individual.
-
-
Many fluorescence methods used in autophagy monitoring require specialized fixation methods, fresh tissue preparation, and rapid visualization. Most of the classical methods are not applicable in the case of the brain.
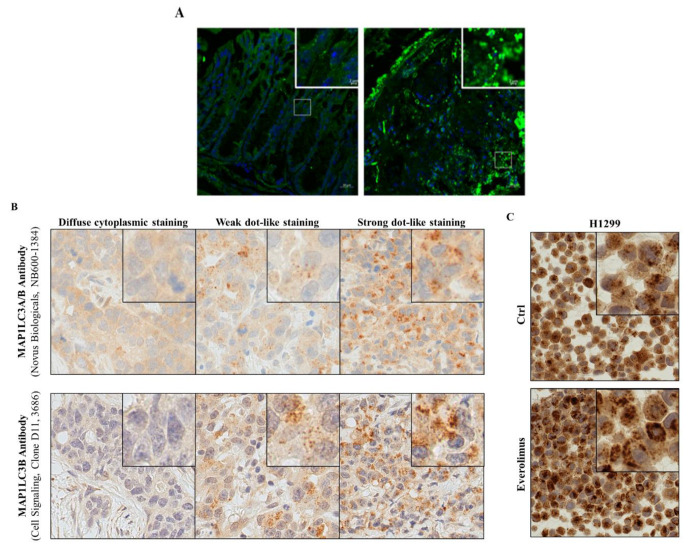
Examples are highlighted in Figure 4 using mouse colonic tissues, and human lung cancer tissues and cell lines.
Figure 4.
Immunofluorescence for MAP1LC3B. (A) Colon tissue sections of a control mouse and a mouse treated by TNBS to induce acute colitis. Staining for DNA with DAPI (blue) and for MAP1LC3 with Alexa-fluor labeled specific antibodies; (B) MAP1LC3A/B immunohistochemistry staining of three archived FFPE samples from patients with an adenocarcinoma of the lung. Different MAP1LC3A/B expression levels were visualized using two different antibodies, as indicated. (C) MAP1LC3A/B immunohistochemistry staining of FFPE-H1299 NSCLC cells that were either left untreated (Ctrl) or treated with everolimus, a selective inhibitor of MTOR activity used to stimulate autophagy. Detailed instructions on how the quantification of LC3B dot formation is done can be found in [84,86,87]. See abbreviations in the abbreviations section.
A variety of suppliers provide adequate antibodies that should be evaluated and finely calibrated in each cell/tissue and methodological condition to gain an optimal signal/background ratio. A non-exhaustive list is given in Table 4.
Table 4.
Non-exhaustive list of MAP1LC3 antibodies.
| Antibodies | Applications, Conditions 1 | Limitations |
|---|---|---|
| Polyclonal anti-MAP1LC3B antibody (#NB600-1384, Novus Biologicals) | Western blot 1:1000 in 5% (w/v) non-fat milk, 3h, room temperature. For example, ref [84,86,87] |
MAP1LC3A cross-reaction |
| Polyclonal affinity-purified rabbit anti-MAP1LC3B antibody (#2775; Cell signaling Technology) | Western blot, 1:1000 Used with human primary cells (PBMCs, dendritic cells, monocytes, macrophages), cell lines. For example ref [12] |
|
| Rabbit anti-MAP1LC3B antibody (#3868,Cell Signaling) | Immunofluorescence 1:200 in PBS-1% (w/v) BSA. | |
| Anti-MAP1LC3B antibody (#M186-3, MBL Inc.) | Western blot 0.5 µg/mL and immunofluorescence 5 µg/mL in TBS containing 1% non-fat milk. Used for example with sciatic nerves sections from Lewis rats; with malignant glioblastoma-derived U-251MG cells; with mouse spleen and salivary glands cells; with colonic cells from mice with colitis (Figure 4). For example, ref. [29,30,57] |
|
| Monoclonal rabbit anti-MAP1LC3B antibody, clone D11 (#3686, Cell Signaling) | Immunofluorescence. For example ref [84,86,87] |
See the legend of Figure 4B |
| Polyclonal affinity-purified rabbit anti-MAP1LC3A/B antibody (#4108; Cell signaling Technology) | Immunofluorescence 1:200 dilution. Used with cell lines. For example, ref [12] |
1 These conditions are routinely used in our respective laboratories. They have to be adapted to each type of substrate and technique. See abbreviations in the abbreviations section.
As indicated, although MAP1LC3-II has been widely used as a marker to detect autophagy in various tissue samples from patients (biopsies or extraction of tissues at autopsy) or animal models, these assays cannot provide information on the dynamics of autophagy process and flux.
Detecting co-localization of key autophagy proteins or post-translational modifications associated with an active autophagy state might represent new approaches to better describe autophagic activity in tissue. In addition, the quality of the tissue materials from patients and animals, e.g., the time from removing the specimen to tissue fixation, is key to adequately assess autophagy. Clinical studies sometimes rely on quantifying MAP1LC3 staining intensity rather than analyzing MAP1LC3 dot formation to evaluate autophagic activity in archived patient samples. This is clearly less conclusive and should be avoided. Analyzing MAP1LC3 punctae formation, although not measuring the autophagic flux per se, better represents increased autophagic activity (Figure 4B,C) [87]. Improved and new techniques are critical to enabling autophagy flux analyses in the human tissues.
5. MAP1LC3-Based Methods Designed for Autophagy In Vivo
A number of approaches have been described to detect MAP1LC3 in living experimental animals. They are listed in Table 5 with respective performances and merits.
Table 5.
Methods to measure autophagy in vivo 1,2.
| Method | Advantages | Limitations |
|---|---|---|
| Transgenic mice expressing GFP-MAP1LC3 and fluorescence microscopy | Allow the formation of autophagosomes to be studied. Used to study macroautophagy and mitophagy. | Do not permit the formation of autolysosome to be studied as GFP loses its fluorescence at acidic pH in lysosomes. Other transgenic mice are required [88] No measurement of autophagic flux. Cells possess auto-fluorescent punctate structures such as lipofuscin that is detectable in the green spectra. Always compare to non-transgenic control littermates [66]. |
| mCherry-GFP-MAP1LC3 and mRFP-GFP-MAP1LC3 mouse and fluorescence microscopy | High time resolution. | Technical difficulty in distinguishing RFP/GFP double-positive and single positive punctae. Lack of performance to measure the basal autophagic flux. |
| GFP-MAP1LC3-RFP-LC3∆G mRNA (injected in animal eggs) and fluorescence microscopy | Measure the basal (low) and induced autophagy flux in embryos and tissues of zebrafish and mice [70]. |
1 SBI-0206965 (Adooq Bioscience), a potent and selective inhibitor of ULK1, can be used to inhibit autophagy in vivo; is given intraperitoneally into mice at 2 mg/kg body weight in DMSO, once per day for 7 days [89]; other autophagy blockers can be used in vivo as control, e.g., CQ/HCQ, NH4Cl, bafilomycin A1 (in certain strict conditions), colchicine, vinblastine, and the inhibitor of lysosomal enzymes leupeptide [66]. 2 Measurement of chaperone-mediated autophagy (CMA) activity in vivo has been described [90]. See abbreviations in the abbreviations section.
Despite the availability of various approaches for analyzing the autophagy in vivo in experimental models, and their potential use to screen therapeutic molecules and study the fundamental process of autophagy, their translation into the clinic remains limited. Further research is necessary to develop suitable probes that are safer and can be used in humans to analyze autophagy in vivo.
6. Conclusions
This short survey clearly identifies the crucial need for developing more performant assays permitting to screen autophagy regulator molecules in vivo. In view of the lack of suitable techniques to analyze autophagy flux in vivo in humans, the best possible solution would be to investigate autophagy flux by using cells derived from the affected organs/tissues of the patients with aberrations in the autophagy. Alternatively, induced pluripotent stem cell technology (3D tissue cultures or organoids) could be exploited in order to reprogram human somatic cells to obtain disease-relevant cell types for the investigations. Genome-wide studies have reported several disease-associated loci and genes that affect autophagy [4,91]. Though ex vivo, these cells provide an opportunity to investigate the role of autophagy in the pathogenesis of diseases and to get a clue on if therapeutic molecules that are deemed beneficial in the animal models could be used in the patients.
It is also important that MAP1LC3-based analyses need to be complemented with lysosomal function to confirm the intact autophagy process. Though MAP1LC3 analyses indicate the autophagy process, the functional intactness of lysosomes is critical to complete the process via degradation of macromolecules and damaged cellular components. In fact, several muscular disorders such as X-linked myopathy with excessive autophagy, inclusion body myositis (IBM), IBM-associated with Paget’s disease of the bone, frontotemporal dementia, and amyotrophic lateral sclerosis are associated with reduced proteolytic activity of lysosomes through enhanced autophagosome biogenesis was observed [92,93]. This feature has also been observed in lupus settings (Wang and Muller, unpublished data).
We deliberately focused this article on MAP1LC3-based analyses and macroautophagy. We listed the different ways of MAP1LC3 quantification in different settings and the arguments pro and against these measurements (see also Box 1 for recommendation). We emphasized here the importance of complementing MAP1LC3 autophagy measurements with nonMAP1LC3-based techniques. Other markers should be systematically followed such as SQSTM1, BECLIN-1, WIPI-1, ATG5/12, ATG14, ATG16L1, VPS34, LAMP2A, and others if possible, to support the results obtained with MAP1LC3. It is also important to distinguish the noncanonical autophagy process known as LC3-associated phagocytosis (LAP) from the canonical autophagy process. LAP and canonical autophagy processes do not require hierarchical intervention of all of the ATG proteins [94]. Both pathways do not play the same functions in cellular homeostasis and physiology, and therefore differentiating both processes can be highly relevant [95]. Recent studies on monocyte/macrophages revealed that LAP has a significant role in phenotype differentiation, particularly on the anti-inflammatory phenotype [96]. In addition to its role in regulating autoimmune response [97], LAP-mediated protective response against hepatic and systemic inflammation are also documented [98].
As reviewed in this article, biochemical assays, electron microscopy, and light microscopy are useful in assessing the autophagic process, however, these assays are cell type-, context-, and time-dependent. Immunologists or cell biologists who have equipped the laboratory with many techniques, should consider using as many markers possible and also, at least, two independent techniques to confirm the autophagic process. Autophagic flux analyses are especially recommended. Despite advancements in the methods to measure autophagy, there is still a lack of completeness, which should be answered with future developments. Novel methods, both in vitro and especially in vivo, to detect changes in the autophagy process in living tissues, are particularly awaited. They are decisive to discover and develop new, more effective drugs that target the autophagy machinery for patient care.
Acknowledgments
We thank Sruthi Retnakumar (Strasbourg), Sabina Berezowska (Bern), and Anna M. Schläfli (Bern) for giving access to the results shown in Figure 4.
Glossary
| Amphisomes | are single-membrane compartment issue from the fusion of autophagosomes and endosomes |
| Autolysosomes | are single-membered acidic vesicle formed by the fusion of autophagosome and lysosome |
| Autophagosomes | are well discriminated double-membrane vesicles in which intracellular material (incl. organelles and fragments of organelles) are sequestered and delivered to the lysosome for degradation |
| Endosomes | are single-membrane compartment formed by the process of endocytosis |
| Lysosomes | are digestive organelles of the cell, which bear an acidic environment (pH 4.5 to 5.0) to degrade material via different processes |
| v-ATPases | are membrane-associated proton pumps, which maintain the acidic environment in organelles, such as the endosomes, lysosomes, trans-Golgi network, and secretory granules. |
| Abbreviations: | |
| AMPK | adenosine monophosphate-activated protein kinase |
| ATG | autophagy-related |
| ATP | adenosine triphosphate |
| Baf A1 | bafilomycin A1 |
| BSA | bovine serum albumin |
| LC3-I/-II | MAP1LC3-I/-II |
| CQ/HCQ | chloroquine/hydroxychloroquine |
| Ctrl | control nonsmall |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DMSO | dimethyl sulfoxide |
| ER | endoplasmic reticulum |
| FFPE | formalin-fixed paraffin-embedded |
| FkBP-12 | FK506 binding protein-12 kDa |
| GAPR-1 | Golgi-associated pathogenesis related-1 protein |
| GFP | green fluorescent protein |
| GFP-MAP1LC3-RFP-LC3∆G | construct in which the C-terminus of GFP-LC3 is fused to RFP-LC3 protein deleted from its C-terminal glycine |
| HSP | heat shock protein |
| LDH | lactate dehydrogenase |
| MAP1LC3 | microtubule-associated protein 1A/1B-light chain 3 |
| mRFP | monomeric red fluorescent protein |
| MTOR | mechanistic target of rapamycin |
| MTORC1 | mammalian target of rapamycin complex 1 |
| NSCLC | nonsmall cell lung cancer |
| PBMCs | peripheral blood mononuclear cells |
| PBS | phosphate-buffered saline |
| PI3K | phosphatidylinositol 3-kinase |
| PKB/AKT | protein kinase B |
| RFP | red fluorescent protein |
| SAHA | suberanilohydroxamic acid |
| SIRT1 | sirtuin 1 |
| TBS | Tris-buffered saline |
| TC | trial compound |
| TNBS | 2,4,6 trinitrobenzene sulfonic acid |
| v-ATPase | vacuolar-type H+-ATPase |
Author Contributions
All authors wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the French Centre National de la Recherche Scientifique, Région Grand-Est and the FHU ARRIMAGE, the Laboratory of Excellence Medalis (ANR-10-LABX-0034), Initiative of Excellence (IdEx), Strasbourg University, the University of Strasbourg Institute for Advanced Study (USIAS), and the Swiss National Science Foundation (31003A_173219 to M.P.T.). M.P.T. and S.M. acknowledge the support of the TRANSAUTOPHAGY COST Action (CA15138). S.M. also thanks the Club francophone de l’autophagie (CFATG) and the European Regional Development Fund of the European Union in the framework of the INTERREG V Upper Rhine program.
Conflicts of Interest
S.M. is co-inventor of CNRS-ImmuPharma patents on P140 peptide; has received research funds (paid to institution). The other authors declare no conflicts of interest. All declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Bento C.F., Renna M., Ghislat G., Puri C., Ashkenazi A., Vicinanza M., Menzies F.M., Rubinsztein D.C. Mammalian Autophagy: How Does It Work? Annu. Rev. Biochem. 2016;85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 2.Gatica D., Lahiri V., Klionsky D.J. Cargo recognition and degradation by selective autophagy. Nature. 2018;20:233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirkin V., Rogov V.V. A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Mol. Cell. 2019;76:268–285. doi: 10.1016/j.molcel.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Levine B., Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias E., Cuervo A.M. Pros and Cons of Chaperone-Mediated Autophagy in Cancer Biology. Trends Endocrinol. Metab. 2019;31:53–66. doi: 10.1016/j.tem.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J., Cao F., Yin H.-L., Huang Z.-J., Lin Z.-T., Mao N., Sun B., Wang G. Ferroptosis: Past, present and future. Cell Death Dis. 2020;11:1–13. doi: 10.1038/s41419-020-2298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonam S.R., Wang F., Muller S. Autophagy: A new concept in autoimmunity regulation and a novel therapeutic option. J. Autoimmun. 2018;94:16–32. doi: 10.1016/j.jaut.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Levy J.M.M., Thorburn A. Autophagy in cancer: Moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 2019;27:843–857. doi: 10.1038/s41418-019-0474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonam S.R., Wang F., Muller S. Lysosomes as a therapeutic target. Nat. Rev. Drug Discov. 2019;18:923–948. doi: 10.1038/s41573-019-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Retnakumar S.V., Muller S. Pharmacological Autophagy Regulators as Therapeutic Agents for Inflammatory Bowel Diseases. Trends Mol. Med. 2019;25:516–537. doi: 10.1016/j.molmed.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Hazari Y., Pedro J.M.B.-S., Hetz C., Galluzzi L., Kroemer G. Autophagy in hepatic adaptation to stress. J. Hepatol. 2020;72:183–196. doi: 10.1016/j.jhep.2019.08.026. [DOI] [PubMed] [Google Scholar]
- 12.Das M., Karnam A., Stephen-Victor E., Gilardin L., Bhatt B., Sharma V.K., Rambabu N., Patil V., Lecerf M., Käsermann F., et al. Intravenous immunoglobulin mediates anti-inflammatory effects in peripheral blood mononuclear cells by inducing autophagy. Cell Death Dis. 2020;11:50. doi: 10.1038/s41419-020-2249-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klionsky D.J., Eskelinen E.L., Deretic V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes. wait, I’m confused. Autophagy. 2014;10:549–551. doi: 10.4161/auto.28448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuma A., Komatsu M., Mizushima N. Autophagy-monitoring and autophagy-deficient mice. Autophagy. 2017;13:1619–1628. doi: 10.1080/15548627.2017.1343770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tekirdag K., Cuervo A.M. Chaperone-mediated autophagy and endosomal microautophagy: Joint by a chaperone. J. Biol. Chem. 2017;293:5414–5424. doi: 10.1074/jbc.R117.818237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker S.A., Ktistakis N.T. Autophagosome Biogenesis Machinery. J. Mol. Biol. 2020;432:2449–2461. doi: 10.1016/j.jmb.2019.10.027. [DOI] [PubMed] [Google Scholar]
- 17.Galluzzi L., Pedro J.M.B.-S., Levine B., Green D.R., Kroemer G. Pharmacological modulation of autophagy: Therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2017;16:487–511. doi: 10.1038/nrd.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubinsztein D.C., Codogno P., Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Y., Ren X., Hait W.N., Yang J.-M. Therapeutic targeting of autophagy in disease: Biology and pharmacology. Pharmacol. Rev. 2013;65:1162–1197. doi: 10.1124/pr.112.007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gros F., Muller S. Pharmacological regulators of autophagy and their link with modulators of lupus disease. Br. J. Pharmacol. 2014;171:4337–4359. doi: 10.1111/bph.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vakifahmetoglu-Norberg H., Xia H.-G., Yuan J. Pharmacologic agents targeting autophagy. J. Clin. Investig. 2015;125:5–13. doi: 10.1172/JCI73937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klionsky D.J., Abdelmohsen K., Abe A., Abedin J., Abeliovich H., Acevedo-Arozena A., Adachi H., Adams C.M., Adams P.D., Adeli K., et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonam S.R., Ruff M., Muller S. Hspa8/Hsc70 in Immune Disorders: A Molecular Rheostat that Adjusts Chaperone-Mediated Autophagy Substrates. Cells. 2019;8:849. doi: 10.3390/cells8080849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.-J., Coppes R.P., Engedal N., Mari M., Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sobota J.A., Back N., Eipper B.A., Mains R.E. Inhibitors of the V0 subunit of the vacuolar H + ATPase prevent segregation of lysosomal and secretory-pathway proteins. J. Cell Sci. 2009;122:3542–3553. doi: 10.1242/jcs.034298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schall N., Page N., Macri C., Chaloin O., Briand J.-P., Muller S. Peptide-based approaches to treat lupus and other autoimmune diseases. J. Autoimmun. 2012;39:143–153. doi: 10.1016/j.jaut.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Muller S., Brun S., Rene F., de Seze J., Loeffler J.-P., Jeltsch-David H. Autophagy in neuroinflammatory diseases. Autoimmun. Rev. 2017;16:856–874. doi: 10.1016/j.autrev.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 28.Brun S., Schall N., Bonam S.R., Bigaut K., Mensah-Nyagan A.-G., de Seze J., Muller S. An autophagy-targeting peptide to treat chronic inflammatory demyelinating polyneuropathies. J. Autoimmun. 2018;92:114–125. doi: 10.1016/j.jaut.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Li B., Wang F., Schall N., Muller S. Rescue of autophagy and lysosome defects in salivary glands of Mrl/lpr mice by a therapeutic phosphopeptide. J. Autoimmun. 2018;90:132–145. doi: 10.1016/j.jaut.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Voynova E., Lefebvre F., Qadri A., Muller S. Correction of autophagy impairment inhibits pathology in the Nod.H-2h4 mouse model of primary Sjögren’s syndrome. J. Autoimmun. 2020;108:102418. doi: 10.1016/j.jaut.2020.102418. [DOI] [PubMed] [Google Scholar]
- 31.Zimmer R., Scherbarth H.R., Rillo O.L., Gomez-Reino J.J., Muller S. Lupuzor/P140 peptide in patients with systemic lupus erythematosus: A randomised, double-blind, placebo-controlled phase IIb clinical trial. Ann. Rheum. Dis. 2012;72:1830–1835. doi: 10.1136/annrheumdis-2012-202460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilhelm M., Wang F., Schall N., Kleinmann J.-F., Faludi M., Nashi E.P., Sibilia J., Martin T., Schaeffer E., Muller S. Lupus Regulator Peptide P140 Represses B Cell Differentiation by Reducing HLA Class II Molecule Overexpression. Arthritis Rheumatol. 2018;70:1077–1088. doi: 10.1002/art.40470. [DOI] [PubMed] [Google Scholar]
- 33.Page N., Gros F., Schall N., Décossas M., Bagnard D., Briand J.-P., Muller S. Hsc70 blockade by the therapeutic peptide P140 affects autophagic processes and endogenous MHII presentation in murine lupus. Ann. Rheum. Dis. 2010;70:837–843. doi: 10.1136/ard.2010.139832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macri C., Wang F., Tasset I., Schall N., Page N., Briand J.-P., Cuervo A.M., Muller S. Modulation of deregulated chaperone-mediated autophagy by a phosphopeptide. Autophagy. 2015;11:472–486. doi: 10.1080/15548627.2015.1017179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang F., Muller S. Manipulating Autophagic Processes in Autoimmune Diseases: A Special Focus on Modulating Chaperone-Mediated Autophagy, an Emerging Therapeutic Target. Front. Immunol. 2015;6:252. doi: 10.3389/fimmu.2015.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shoji-Kawata S., Sumpter R., Leveno M., Campbell G.R., Zou Z., Kinch L., Wilkins A.D., Sun Q., Pallauf K., MacDuff N., et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong Z., Tasset I., Diaz A., Anguiano J., Tas E., Cui L., Kuliawat R., Liu H., Kühn B., Cuervo A.M., et al. Humanin is an endogenous activator of chaperone-mediated autophagy. J. Cell Biol. 2017;217:635–647. doi: 10.1083/jcb.201606095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 39.Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 40.Kwon D.H., Kim L., Kim B.-W., Kim J.H., Roh K.-H., Choi E.-J., Song H.K. A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between Lc3B and RavZ. Biochem. Biophys. Res. Commun. 2017;490:1093–1099. doi: 10.1016/j.bbrc.2017.06.173. [DOI] [PubMed] [Google Scholar]
- 41.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. Lc3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shvets E., Fass E., Scherz-Shouval R., Elazar Z. The N-terminus and Phe52 residue of Lc3 recruit p62/Sqatm1 into autophagosomes. J. Cell Sci. 2008;121:2685–2695. doi: 10.1242/jcs.026005. [DOI] [PubMed] [Google Scholar]
- 43.Le Grand J.N., Chakrama F.Z., Seguin-Py S., Fraichard A., Delage-Mourroux R., Jouvenot M., Boyer-Guittaut M. Gabarapl1 (GEC1): Original or copycat? Autophagy. 2011;7:1098–1107. doi: 10.4161/auto.7.10.15904. [DOI] [PubMed] [Google Scholar]
- 44.Jacomin A.-C., Samavedam S., Promponas V.J., Nezis I. iLIR database: A web resource for Lir motif-containing proteins in eukaryotes. Autophagy. 2016;12:1945–1953. doi: 10.1080/15548627.2016.1207016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakatogawa H. Two ubiquitin-like conjugation systems that mediate membrane formation during autophagy. Essays Biochem. 2013;55:39–50. doi: 10.1042/bse0550039. [DOI] [PubMed] [Google Scholar]
- 46.Martens S., Fracchiolla D. Activation and targeting of ATG8 protein lipidation. Cell Discov. 2020;6:23. doi: 10.1038/s41421-020-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maruyama T., Noda N.N. Autophagy-regulating protease Atg4: Structure, function, regulation and inhibition. J. Antibiot. 2017;71:72–78. doi: 10.1038/ja.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Popelka H., Klionsky D.J. Analysis of the native conformation of the Lir/Aim motif in the Atg8/Lc3/Gabarap-binding proteins. Autophagy. 2015;11:2153–2159. doi: 10.1080/15548627.2015.1111503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Birgisdottir A.B., Lamark T., Johansen T. The Lir motif–crucial for selective autophagy. J. Cell Sci. 2013;126:3237–3247. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 50.Wild P., McEwan D.G., Dikic I. The LC3 interactome at a glance. J. Cell Sci. 2013;127:3–9. doi: 10.1242/jcs.140426. [DOI] [PubMed] [Google Scholar]
- 51.Lee Y.-K., Lee J.-A. Role of the mammalian Atg8/Lc3 family in autophagy: Differential and compensatory roles in the spatiotemporal regulation of autophagy. BMB Rep. 2016;49:424. doi: 10.5483/BMBRep.2016.49.8.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkinson D.S., Jariwala J.S., Anderson E., Mitra K., Meisenhelder J., Chang J.T., Ideker T., Hunter T., Nizet V., Dillin A. Phosphorylation of Lc3 by the Hippo kinases Stk3/Stk4 is essential for autophagy. Mol. Cell. 2015;57:55–68. doi: 10.1016/j.molcel.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cherra S.J., Kulich S.M., Uechi G., Balasubramani M., Mountzouris J., Day B.W., Chu C.T. Regulation of the autophagy protein LC3 by phosphorylation. J. Cell Biol. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang R., Xu Y., Wan W., Shou X., Qian J., You Z., Liu B., Chang C., Zhou T., Lippincott-Schwartz J., et al. Deacetylation of Nuclear Lc3 Drives Autophagy Initiation under Starvation. Mol. Cell. 2015;57:456–466. doi: 10.1016/j.molcel.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 55.Barth S., Glick D., MacLeod K. Autophagy: Assays and artifacts. J. Pathol. 2010;221:117–124. doi: 10.1002/path.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mizushima N., Yoshimori T., Levine B., Yoshimorim T. Methods in Mammalian Autophagy Research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang F., Li B., Schall N., Wilhelm M., Muller S. Assessing Autophagy in Mouse Models and Patients with Systemic Autoimmune Diseases. Cells. 2017;6:16. doi: 10.3390/cells6030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Runwal G., Stamatakou E., Siddiqi F.H., Puri C., Zhu Y., Rubinsztein D.C. Lc3-positive structures are prominent in autophagy-deficient cells. Sci. Rep. 2019;9:10147. doi: 10.1038/s41598-019-46657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubinsztein D.C., Cuervo A.M., Ravikumar B., Sarkar S., Korolchuk V.I., Kaushik S., Klionsky D.J. In search of an “autophagomometer”. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 60.Jung M., Choi H., Mun J.Y. The autophagy research in electron microscopy. Appl. Microsc. 2019;49:1–7. doi: 10.1186/s42649-019-0012-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D.M. Mtrc1 Senses Lysosomal Amino Acids Through an Inside-Out Mechanism That Requires the Vacuolar H+ ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Juhász G. Interpretation of bafilomycin, pH neutralizing or protease inhibitor treatments in autophagic flux experiments: Novel considerations. Autophagy. 2012;8:1875–1876. doi: 10.4161/auto.21544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Florey O., Gammoh N., Kim S.E., Jiang X., Overholtzer M. V-ATPase and osmotic imbalances activate endolysosomal LC3 lipidation. Autophagy. 2014;11:88–99. doi: 10.4161/15548627.2014.984277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zois C.E., Giatromanolaki A., Sivridis E., Papaiakovou M., Kainulainen H., Koukourakis M.I. “Autophagic flux” in normal mouse tissues: Focus on endogenous Lc3A processing. Autophagy. 2011;7:1371–1378. doi: 10.4161/auto.7.11.16664. [DOI] [PubMed] [Google Scholar]
- 65.Koukourakis M., Giatromanolaki A., Zois C., Sivridis E. Lc3 immunostaining pitfalls. Histopathology. 2013;62:962–963. doi: 10.1111/his.12080. [DOI] [PubMed] [Google Scholar]
- 66.Moulis M., Vindis C. Methods for Measuring Autophagy in Mice. Cells. 2017;6:14. doi: 10.3390/cells6020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mizushima N. Chapter 2 Methods for Monitoring Autophagy Using Gfp-Lc3 Transgenic Mice. Methods Enzymol. 2009;452:13–23. doi: 10.1016/s0076-6879(08)03602-1. [DOI] [PubMed] [Google Scholar]
- 68.Shvets E., Fass E., Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008;4:621–628. doi: 10.4161/auto.5939. [DOI] [PubMed] [Google Scholar]
- 69.Morishita H., Kaizuka T., Hama Y., Mizushima N. A new probe to measure autophagic flux in vitro and in vivo. Autophagy. 2017;13:757–758. doi: 10.1080/15548627.2016.1278094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaizuka T., Morishita H., Hama Y., Tsukamoto S., Matsui T., Toyota Y., Kodama A., Ishihara T., Mizushima T., Mizushima N. An Autophagic Flux Probe that Releases an Internal Control. Mol. Cell. 2016;64:835–849. doi: 10.1016/j.molcel.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 71.Yoshii S., Mizushima N. Monitoring and Measuring Autophagy. Int. J. Mol. Sci. 2017;18:1865. doi: 10.3390/ijms18091865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eng K.E., Panas M.D., Hedestam G.B.K., McInerney G.M. A novel quantitative flow cytometry-based assay for autophagy. Autophagy. 2010;6:634–641. doi: 10.4161/auto.6.5.12112. [DOI] [PubMed] [Google Scholar]
- 73.Demishtein A., Porat Z., Elazar Z., Shvets E. Applications of flow cytometry for measurement of autophagy. Methods. 2015;75:87–95. doi: 10.1016/j.ymeth.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 74.Phadwal K., Alegre-Abarrategui J., Watson A.S., Pike L., Anbalagan S., Hammond E.M., Wade-Martins R., McMichael A., Klenerman P., Simon A.K. A novel method for autophagy detection in primary cells. Autophagy. 2012;8:677–689. doi: 10.4161/auto.18935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pugsley H.R. Quantifying autophagy: Measuring Lc3 puncta and autolysosome formation in cells using multispectral imaging flow cytometry. Methods. 2017;112:147–156. doi: 10.1016/j.ymeth.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 76.Schwinn M.K., Machleidt T., Zimmerman K., Eggers C.T., Dixon A.S., Hurst R., Hall M.P., Encell L.P., Binkowski B.F., Wood K.V. Crispr-Mediated Tagging of Endogenous Proteins with a Luminescent Peptide. ACS Chem. Biol. 2017;13:467–474. doi: 10.1021/acschembio.7b00549. [DOI] [PubMed] [Google Scholar]
- 77.Bresciani A., Spiezia M.C., Boggio R., Cariulo C., Nordheim A., Altobelli R., Kuhlbrodt K., Dominguez C., Muñoz-Sanjuan I., Wityak J., et al. Quantifying autophagy using novel Lc3B and p62 Tr-Fret assays. PLoS ONE. 2018;13:e0194423. doi: 10.1371/journal.pone.0194423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eskelinen E.-L., Reggiori F., Baba M., Kovács A.L., Seglen P.O. Seeing is believing: The impact of electron microscopy on autophagy research. Autophagy. 2011;7:935–956. doi: 10.4161/auto.7.9.15760. [DOI] [PubMed] [Google Scholar]
- 79.Dupont N., Leroy C., Hamaï A., Codogno P. Long-Lived Protein Degradation During Autophagy. Methods Enzymol. 2017;588:31–40. doi: 10.1016/bs.mie.2016.09.074. [DOI] [PubMed] [Google Scholar]
- 80.Seglen P.O., Overbye A., Saetre F. Methods in Enzymology. Elsevier BV; Amsterdam, The Netherlands: 2009. Chapter 5 Sequestration Assays for Mammalian Autophagy; pp. 63–83. [DOI] [PubMed] [Google Scholar]
- 81.Donohue E., Tovey A., Vogl A.W., Arns S., Sternberg E., Young R.N., Roberge M. Inhibition of Autophagosome Formation by the Benzoporphyrin Derivative Verteporfin. J. Biol. Chem. 2010;286:7290–7300. doi: 10.1074/jbc.M110.139915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Munafó D.B., I Colombo M. A novel assay to study autophagy: Regulation of autophagosome vacuole size by amino acid deprivation. J. Cell Sci. 2001;114:3619–3629. doi: 10.1242/jcs.114.20.3619. [DOI] [PubMed] [Google Scholar]
- 83.Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., Alnemri E.S., Altucci L., Amelio I., Andrews D.W., et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schläfli A.M., Adams O., Galván J.A., Gugger M., Savic S., Bubendorf L., Schmid R.A., Becker K.-F., Tschan M.P., Langer R., et al. Prognostic value of the autophagy markers Lc3 and p62/Sqstm1 in early-stage non-small cell lung cancer. Oncotarget. 2016;7:39544–39555. doi: 10.18632/oncotarget.9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dhingra A., Alexander D., Reyes-Reveles J., Sharp R., Boesze-Battaglia K. Microtubule-Associated Protein 1 Light Chain 3 (LC3) Isoforms in RPE and Retina. Springer Science and Business Media LLC; Cham, Switzerland: 2018. pp. 609–616. Retinal Degenerative Diseases. [DOI] [PubMed] [Google Scholar]
- 86.Langer R., Neppl C., Keller M.D., Schmid R., Tschan M.P., Berezowska S. Expression Analysis of Autophagy Related Markers Lc3B, p62 and Hmgb1 Indicate an Autophagy-Independent Negative Prognostic Impact of High p62 Expression in Pulmonary Squamous Cell Carcinomas. Cancers. 2018;10:281. doi: 10.3390/cancers10090281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schläfli A., Berezowska S., Adams O., Langer R., Tschan M.P. Reliable Lc3 and p62 Autophagy Marker Detection in Formalin Fixed Paraffin Embedded Human Tissue by Immunohistochemistry. Eur. J. Histochem. EJH. 2015;59 doi: 10.4081/ejh.2015.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li L., Wang Z.V., Hill J., Lin F. New Autophagy Reporter Mice Reveal Dynamics of Proximal Tubular Autophagy. J. Am. Soc. Nephrol. 2013;25:305–315. doi: 10.1681/ASN.2013040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rocchi A., Yamamoto S., Ting T.C., Fan Y., Sadleir K., Wang Y., Zhang W., Huang S., Levine B., Vassar R., et al. A Becn1 mutation mediates hyperactive autophagic sequestration of amyloid oligomers and improved cognition in Alzheimer’s disease. PLoS Genet. 2017;13:e1006962. doi: 10.1371/journal.pgen.1006962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Juste Y.R., Cuervo A.M. Methods in Molecular Biology. Springer Science and Business Media LLC; New York, NY, USA: 2019. Analysis of Chaperone-Mediated Autophagy; pp. 703–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang P., Mizushima N. Autophagy and human diseases. Cell Res. 2013;24:69–79. doi: 10.1038/cr.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ching J.K., Elizabeth S.V., Ju J.-S., Lusk C., Pittman S.K., Weihl C.C. Mtor dysfunction contributes to vacuolar pathology and weakness in valosin-containing protein associated inclusion body myopathy. Hum. Mol. Genet. 2012;22:1167–1179. doi: 10.1093/hmg/dds524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ramachandran N., Munteanu I., Wang P., Ruggieri A., Rilstone J.J., Israelian N., Naranian T., Paroutis P., Guo R., Ren Z.-P., et al. Vma21 deficiency prevents vacuolar ATPase assembly and causes autophagic vacuolar myopathy. Acta Neuropathol. 2013;125:439–457. doi: 10.1007/s00401-012-1073-6. [DOI] [PubMed] [Google Scholar]
- 94.Galluzzi L., Green D.R. Autophagy-Independent Functions of the Autophagy Machinery. Cell. 2019;177:1682–1699. doi: 10.1016/j.cell.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oikonomou V., Renga G., de Luca A., Borghi M., Pariano M., Puccetti M., Paolicelli G., Stincardini C., Costantini C., Bartoli A., et al. Autophagy and Lap in the Fight against Fungal Infections: Regulation and Therapeutics. Mediat. Inflamm. 2018:1–7. doi: 10.1155/2018/6195958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cunha L.D., Yang M., Carter R., Guy C., Harris L., Crawford J.C., Quarato G., Romero E.B., Kalkavan H., Johnson M.D.L., et al. LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell. 2018;175:429–441.e16. doi: 10.1016/j.cell.2018.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Heckmann B.L., Romero E.B., Cunha L.D., Magne J., Green D.R. Lc3-Associated Phagocytosis and Inflammation. J. Mol. Biol. 2017;429:3561–3576. doi: 10.1016/j.jmb.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wan J., Weiss E., ben Mkaddem S., Mabire M., Choinier P.-M., Picq O., Thibault-Sogorb T., Hegde P., Pishvaie D., Bens M., et al. Lc3-associated phagocytosis protects against inflammation and liver fibrosis via immunoreceptor inhibitory signaling. Sci. Transl. Med. 2020;12:eaaw8523. doi: 10.1126/scitranslmed.aaw8523. [DOI] [PubMed] [Google Scholar]