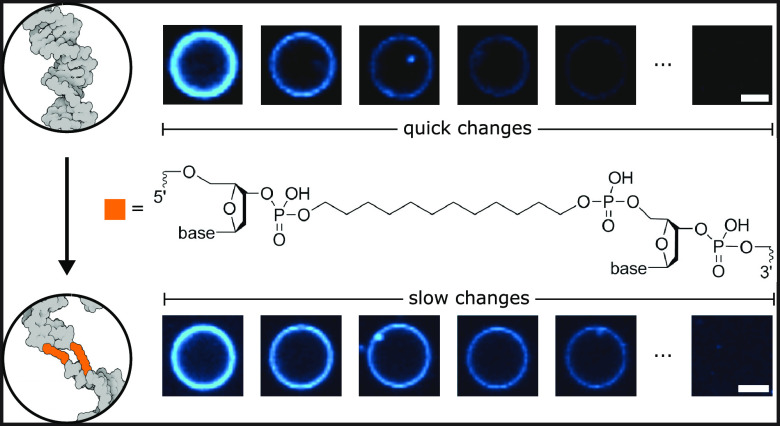
Abstract
Lipid membranes, enveloping all living systems, are of crucial importance, and control over their structure and composition is a highly desirable functionality of artificial structures. However, the rational design of protein-inspired systems is still challenging. Here we have developed a highly functional nucleic acid construct that self-assembles and inserts into membranes, enabling lipid transfer between inner and outer leaflets. By designing the structure to account for interactions between the DNA, its hydrophobic modifications, and the lipids, we successfully exerted control over the rate of interleaflet lipid transfer induced by our DNA-based enzyme. Furthermore, we can regulate the level of lipid transfer by altering the concentration of divalent ions, similar to stimuli-responsive lipid-flipping proteins.
Keywords: DNA nanotechnology, lipid membranes, synthetic ion channel, dodecane, lipid flipping, molecular dynamics
The phospholipid membrane envelopes every cell and is an important signaling platform for all organisms. Embedded within it, transmembrane proteins are molecular machines responsible for the membrane’s properties.1 Their relatively complex structure allows for a high level of control over the bilayer shape2 and the lipid composition.3 Such a degree of control makes protein-inspired transmembrane structures a desirable tool and the development of these structures a vibrant field of study.4−13
One of the materials currently explored as a prospective protein-mimicking building block is DNA due to the ease with which it may be modified, designed, and assembled using complementary base pairing. Starting with a synthetic DNA–origami membrane channel,14 a whole variety of structures were developed,15−21 all designed to insert into the membrane via hydrophobic anchors; the nucleic acid scaffold is rarely modified.22 Because of the DNA’s charge, the hydrophilic headgroups, rather than the hydrophobic tails of the lipids, face toward the membrane-spanning DNA. This results in the formation of a toroidal pore, shaped by the merged bilayer leaflets. Such arrangement of lipids has many implications, namely, the ion flow on the DNA–lipid interface21 or lipid transfer between bilayer leaflets.23 Here we report the dependency of both of these effects on the hydrophilicity of the DNA-based constructs’ membrane-spanning domain and present an artificial, protein-inspired structure exhibiting control over the rate and the level of interleaflet lipid transfer.
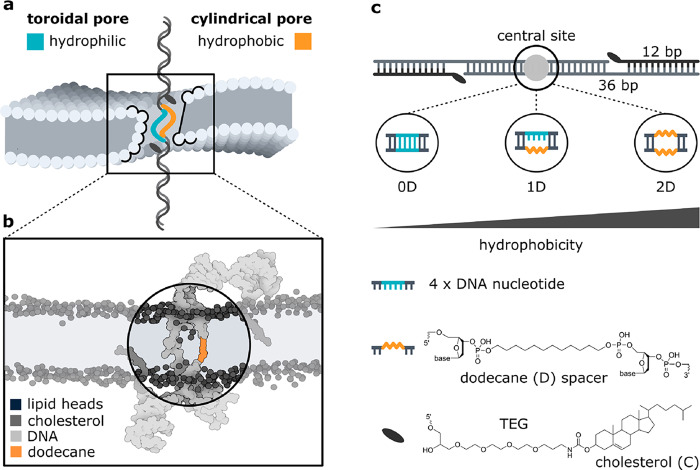
Knowing that leaflet merging results from DNA–lipid interactions, we hypothesized that the shape of the pore formed by transmembrane nanostructures depends on the hydrophilicity of the membrane-spanning domain. Indeed, the effect has been attributed to the mechanisms of naturally occurring lipid-flipping proteins.24 To confirm this idea, we have designed a membrane-inserting DNA duplex with four nucleotides in the central part of one of its strands replaced by a 12-carbon chain (dodecane). The membrane-spanning part of the structure is therefore half hydrophilic and half hydrophobic. An illustration of this construct embedded in a bilayer is shown in Figure 1a. The schematic was created based on the results of all-atom molecular dynamics (MD) simulations, shown in Figure 1b. Upon 1 μs of simulation, lipids were found with their headgroups predominantly pointing toward the unmodified strand, whereas no lipids were present within the pore on the side of the hydrophobic domain.
Figure 1.
Schematic representation of the designed DNA nanostructure. (a) Hydrophilicity of the membrane-spanning domain determines the structure of the DNA–lipid interface. (b) Snapshot from an all-atom MD simulation of the 1D construct. No lipid headgroups are present in the proximity of the dodecane spacer. (c) Schematic representation of the double-stranded DNA construct, highlighting its membrane-anchoring (cholesterol) and internal (dodecane) modifications. Three different designs varying in hydrophobicity were used, with zero (0D), one (1D), or two (2D) dodecane spacers placed in the structure’s central site.
Realizing that the toroidality of the pore can be disrupted by modifying the membrane-spanning part, we investigated three versions of the aforementioned DNA duplex, featuring zero (0D), one (1D), or two (2D) dodecane spacers placed in the central site of the structure. All tested duplexes were additionally modified with two cholesterol moieties (2C) (Figure 1c, Supplementary Table 1), which facilitate their attachment and insertion into the bilayer (Supplementary Figure 1). In the absence of cholesterol modifications, no attachment caused by the presence of the C12 chain (0C 2D) was observed (Supplementary Figure 2). The three structures were physically folded using a commercially available C12-modified strand. The structures were characterized and tested experimentally via ionic current recordings, whereas an additional fluorescent tag allowed optical measurements further described in this work. Additionally, all-atom MD simulations enabled a thorough understanding of the details of the systems.
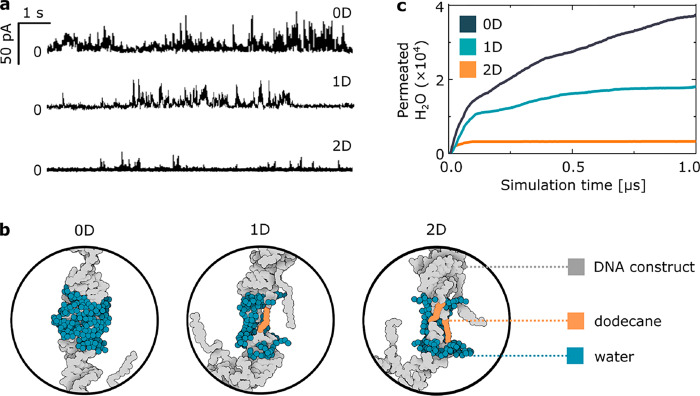
The design of the duplexes, especially the long separation of the hydrophobic anchors (24 bp ≈ 8 nm), causes transient insertions of the structure, which has been previously reported for a similar construct.21 The mode of insertion can be studied by recording the ionic current across a lipid membrane in the presence of structures. Figure 2a shows three representative current traces, one for each of the studied designs. The results indicate that (I) the structures do not form a stable pore but induce current bursts while spanning the bilayer and (II) with an increasing number of internal dodecane modifications, the total number of ions transported across the membrane is reduced. The experimental results suggest quantitative differences in the sizes of water channels formed during the insertion, controlled by the construct’s hydrophobicity, as expected. (See also Supplementary Figure 3.)
Figure 2.
Experiments and simulations reveal the DNA-induced transient water channel in a lipid bilayer. (a) Representative current trace for each of the three designs. (b) Snapshots highlighting the number of water molecules in the channel after 0.8 μs of MD simulations of DNA constructs in a lipid bilayer. Lipids and ions are not shown for clarity. (c) Results of all-atom MD simulations, showing the number of water molecules permeated through the membrane as a function of simulation time.
To further investigate the interactions between the inserted structures and the surrounding lipids, we performed all-atom MD simulations. An initial examination of a 2D construct in the absence of a lipid membrane showed that when in an aqueous solution, the dodecane modifications adopt a contracted conformation rather than appearing fully stretched (Supplementary Figure 4). However, after being placed in a membrane, the C12 chains extend to span through the hydrophobic core, whereas DNA moves out from the hydrophobic region. Therefore, even though a toroidal pore was formed around each construct, the induced water channels noticeably differed between each system (Figure 2b); the number of water molecules in the pore decreased with the increasing hydrophobicity of the central site, agreeing with the observed differences between experimentally obtained conductance traces. After 1 μs of simulations, dodecane was fully stretched, which caused closure of the pore and the subsequent cessation of water and lipid transfer (Supplementary Figure 5). The presence of a stretched dodecane spacer affected the molecular arrangements of the created pore, resulting in a smaller number of permeated water molecules (Figure 2c) as well as fewer lipids transferred between merged leaflets (Supplementary Figure 6).
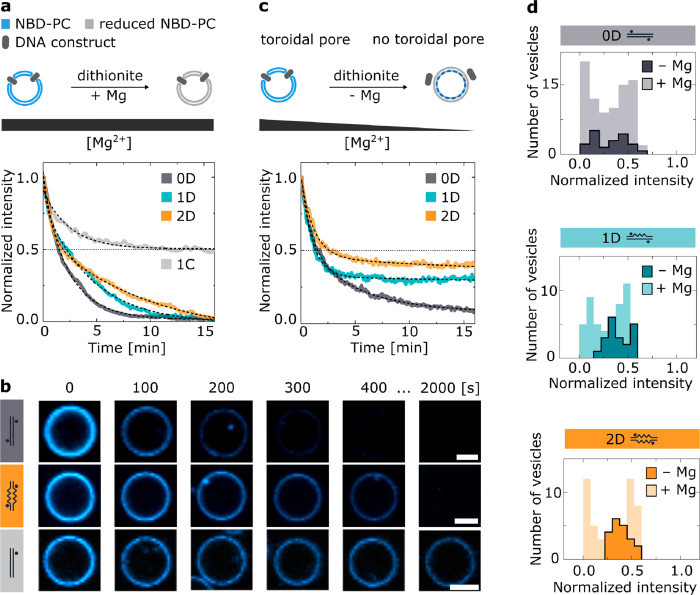
To further observe the disruption of the toroidal pore and its effects on the rate of lipid transfer, we experimentally examined the movement of lipids between the inner and outer leaflets of the bilayer. This movement is intrinsically rare due to a high energy barrier.25 However, when the toroidal pore is formed upon DNA duplex insertion, the lipids can move unhindered between both—now merged—leaflets.23 The change in the bilayer’s composition is studied via an optical assay based on a redox reaction. The system under study consisted of giant unilamellar vesicles (GUVs) made of 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) lipids containing a 0.5 wt % addition of nitrobenzoxadiazole (NBD)-labeled PC lipids. Upon the addition of a reducing agent, the fluorescent NBD molecule turns into its nonfluorescent derivative.25 When no toroidal pore is formed, in the presence of the membrane-impermeable reductant, the fluorescence intensity of the vesicle decreases to ∼50% because only the outer leaflet labels are bleached. Once the toroidal pore is formed, NBD lipids from the inner leaflet diffuse to the outside of the vesicle, where they, too, are reduced. Consequently, a complete loss of the fluorescence signal is observed (Figure 3a). In these studies, dithionite ([S2O4]2–) was chosen as the reductant due to its negative charge, ensuring membrane impermeability.23,26 The charge also prevents its translocation through the DNA-induced pore, as barely any transport of negative ions has been observed during MD simulations (Supplementary Figure 7). The fluorescence stability of the occasionally appearing internal vesicles is an additional confirmation of the dithionite membrane impermeability in the presence of the inserting DNA (Supplementary Figure 8). Furthermore, in a previous study by our group, dithionite leakage through pores induced by a much larger DNA construct was shown to be negligible on the time scale of the experiment.23 We studied the effect of the designed duplexes on the composition of a lipid bilayer by incubating vesicles with each of the structures and recording their fluorescence upon the addition of dithionite. After the experiment, two populations of vesicles can be distinguished, with optical signals decreased to either 50 or 0% (Supplementary Figure 9). The first group was assigned to the vesicles with no toroidal pore, indicating the absence of DNA insertions. These results can be compared with control experiments performed for noninserting 0C and 1C structures (Supplementary Figure 10). The population of vesicles that exhibited a complete loss of fluorescence represents the membranes with merged leaflets and therefore with a DNA duplex spanning through the bilayer.
Figure 3.
Controlling the rate and the level of lipid flipping through the DNA nanostructure’s architecture and the concentration of divalent cations. (a) Experimental results from the schematically illustrated bleaching assay, with magnesium concentration constant (4 mM, +Mg) throughout the experiment. The plot shows the fluorescence intensity time traces collected for the three constructs and the noninserting (1C) control upon dithionite addition at t = 0. Each plot is an average of at least three traces, indicative of the leaflet merging (Supplementary Figure 11). The black dashed lines represent the biexponential fit (Supplementary Table 2). (b) Representative confocal microscopy image sequences under +Mg conditions, showing the difference in the fluorescence decay rates of 0D and 2D structures, alongside the noninserting 1C structure. The scale bars indicate 5 μm. (c) Experimental results analogous to panel a, with the magnesium concentration decreasing by 1.5 mM (−Mg) throughout the experiment. Each plot is an average of at least three traces, indicated by the respective peaks in the histograms of the final intensities presented in panel d (Supplementary Figure 12). (d) Histograms of the final intensity values collected from three experiments for each DNA construct, for +Mg (N0D = 82, N1D = 49, N2D = 50) and −Mg (N0D = 24, N1D = N2D = 20) conditions (Supplementary Figure 9).
The fluorescence decay traces, averaged for the vesicles affected by the DNA insertion, are shown in Figure 3a. The observed rates of bleaching (0.25 ± 0.01, 0.21 ± 0.01, and 0.09 ± 0.01 min–1 for 0D, 1D, and 2D structures, respectively) correlate with the hydrophobicity of the central site (Supplementary Table 2). We hypothesize that the presence of the C12 spacer impedes the formation of a pathway for the lipids’ interleaflet movement, with its effect being especially striking when comparing the vesicles incubated with 0D and 2D constructs, as presented in Figure 3b. Even the structures with an entirely hydrophobic central site (2D) cause lipid transfer across the membrane, which suggests the transient merging of the leaflets during the insertion, agreeing with the ionic current measurements (Figure 2a).
The experiments involving DNA constructs are most commonly performed in the presence of magnesium ions to ensure the stability of DNA-based nanostructures.27,28 The stability of internally modified constructs shows higher sensitivity to changes in magnesium concentration, which we confirmed by spectroscopic measurements (Supplementary Figures 13 and 14, Supplementary Table 3) and polyacrylamide gel electrophoresis (Supplementary Figure 15, Supplementary Table 4). We concluded that the presence of Mg2+ may provide an additional handle to control DNA-induced lipid movements. Further experiments were conducted, as presented in Figure 3c, where the magnesium concentration was rapidly decreased upon the addition of dithionite. Therefore, the observed effects resulted from a combination of: (I) reducing agent bleaching the fluorophores, (II) DNA-induced pores influencing the accessibility of fluorophores to the bleaching factor, and (III) changes in ionic concentration affecting the stability of the DNA structure.
We compared the final intensities of the traced vesicles from the previously described experiments with constant magnesium concentration (+Mg) with the ones that were exposed to decreasing magnesium concentration (−Mg) (Figure 3d). When there are no changes in the divalent cation concentration, all three systems exhibit similar distributions of final fluorescence intensities. 50, 46.9, and 40% of vesicles studied in the presence of 0D, 1D, and 2D structures, respectively, had their final fluorescence intensity indicating the presence of lipid flipping (>70% bleaching; see Supplementary Figure 9 for details). However, when the concentration of Mg2+ is reduced, both 1D and 2D constructs exhibit only partial bleaching of the inner-leaflet lipids, suggesting that the DNA-induced lipid transfer was stopped during the experiment. We attribute this to the decreased stability of these structures in the reduced Mg2+ concentration (Supplementary Figures 13–15) and the subsequent disruption of lipid flipping occurring during the assay. As a result, only a fraction of lipids change their interleaflet position, which is reminiscent of the effects of natural scramblases, which exhibit control over the amount of transferred lipids.29 The level at which the lipid movement is terminated depends on the number of dodecane modifications, with 0D showing the lowest level of residual fluorescence. The change in cation concentration by 1.5 mM resulted in bleaching of around 92, 70, and 61% of lipids caused by the 0D, 1D, and 2D structures, respectively (Supplementary Figure 12, Supplementary Table 2). Our measurements indicate that we can not only control the rate of lipid flip-flop movements using hydrophobic modifications but also use an external stimulus to vary the level of interleaflet transfer.
We report synthetic DNA-based nanostructures that insert into membranes and exhibit a control over their interaction with surrounding lipids, mimicking natural membrane-spanning molecules. By introducing a protein-inspired hydrophobic domain, we changed the rate of lipid flipping induced by the DNA. Additionally, we demonstrated a fundamental connection between the design of the DNA structure and its ability to flip lipids in response to external stimuli like divalent salts. The removal of Mg2+ allowed for lipid flipping to be stopped, which, until now, was the ability of only specialized transmembrane proteins. The results of our experiments and simulations emphasize the importance of contextual design when creating new synthetic constructs. We show that the architecture of the membrane-targeting structure should not be treated in isolation but rather in the context of its interactions with the surrounding environment and, in a broader sense, its prospective applications,30 which is essential for the creation of a new generation of complex protein-mimicking molecular machineries.
Acknowledgments
We acknowledge support from the ERC consolidator grant (DesignerPores 647144) (U.F.K.), Winton Programme for the Physics of Sustainability (D.S.), Cambridge Trust Vice Chancellor’s Award (A.O.), the Engineering and Physical Sciences Research Council (EPSRC) (D.S. and A.O.), the National Science Foundation (USA) (DMR-1827346); and the National Institutes of Health (P41-GM104601). We acknowledge the supercomputer time provided to A.A. and H.J. through the XSEDE allocation grant (MCA05S028) and the Blue Waters petascale supercomputer system (UIUC).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.0c00990.
DNA sequences, experimental details, and supporting figures and tables (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Alberts B.; Johnson A.; Lewis J.; Raff M., Roberts K.; Walter P.. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, 2002. [Google Scholar]
- Kozlov M. M.; et al. Mechanisms shaping cell membranes. Curr. Opin. Cell Biol. 2014, 29, 53–60. 10.1016/j.ceb.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachowski A. Phospholipids in animal eukaryotic membranes: Transverse asymmetry and movement. Biochem. J. 1993, 294 (1), 1–14. 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabushi I.; Kuroda Y.; Yokota K. A,B,D,F-tetrasubstituted β-cyclodextrin as artificial channel compound. Tetrahedron Lett. 1982, 23 (44), 4601–4604. 10.1016/S0040-4039(00)85664-6. [DOI] [Google Scholar]
- Kobuke Y.; Ueda K.; Sokabe M. Artificial Non-Peptide Single Ion Channels. J. Am. Chem. Soc. 1992, 114 (20), 7618–7622. 10.1021/ja00046a002. [DOI] [Google Scholar]
- Carmichael V. E.; et al. Biomimetic Ion Transport: A Functional Model of a Unimolecular Ion Channel. J. Am. Chem. Soc. 1989, 111 (2), 767–769. 10.1021/ja00184a075. [DOI] [Google Scholar]
- Nakano A.; Xie Q.; Mallen J. V.; Echegoyen L.; Gokel G. W. Synthesis of a Membrane-Insertable, Sodium Cation Conducting Channel: Kinetic Analysis by Dynamic 23Na NMR. J. Am. Chem. Soc. 1990, 112 (3), 1287–1289. 10.1021/ja00159a083. [DOI] [Google Scholar]
- Voyer N.; Robitaille M. A Novel Functional Artificial Ion Channel. J. Am. Chem. Soc. 1995, 117 (24), 6599–6600. 10.1021/ja00129a027. [DOI] [Google Scholar]
- Tanaka Y.; Kobuke Y.; Sokabe M. A Non-Peptidic Ion Channel with K+ Selectivity. Angew. Chem., Int. Ed. Engl. 1995, 34 (6), 693–694. 10.1002/anie.199506931. [DOI] [Google Scholar]
- Ralston G. B.; Kuchel P. W.. Schaum’s Outline of Theory and Problems of Biochemistry; McGraw-Hill Education - Europe: New York, 1987. [Google Scholar]
- Lear J. D.; Schneider J. P.; Kienker P. K.; DeGrado W. F. Electrostatic effects on ion selectivity and rectification in designed ion channel peptides. J. Am. Chem. Soc. 1997, 119 (14), 3212–3217. 10.1021/ja9629672. [DOI] [Google Scholar]
- Ghadiri M. R.; Granja J. R.; Buehler L. K. Artificial transmembrane ion channels from self-assembling peptide nanotubes. Nature 1994, 369, 301–304. 10.1038/369301a0. [DOI] [PubMed] [Google Scholar]
- Sakai N.; et al. Dendritic folate rosettes as ion channels in lipid bilayers. J. Am. Chem. Soc. 2006, 128 (7), 2218–2219. 10.1021/ja058157k. [DOI] [PubMed] [Google Scholar]
- Langecker M.; et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 2012, 338, 932–936. 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J. R.; Seifert A.; Fertig N.; Howorka S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 2016, 11 (2), 152–156. 10.1038/nnano.2015.279. [DOI] [PubMed] [Google Scholar]
- Arnott P. M.; Howorka S. A Temperature-Gated Nanovalve Self-Assembled from DNA to Control Molecular Transport across Membranes. ACS Nano 2019, 13 (3), 3334–3340. 10.1021/acsnano.8b09200. [DOI] [PubMed] [Google Scholar]
- Göpfrich K.; et al. DNA-tile structures induce ionic currents through lipid membranes. Nano Lett. 2015, 15 (5), 3134–3138. 10.1021/acs.nanolett.5b00189. [DOI] [PubMed] [Google Scholar]
- Göpfrich K.; et al. Large-Conductance Transmembrane Porin Made from DNA Origami. ACS Nano 2016, 10 (9), 8207–8214. 10.1021/acsnano.6b03759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franquelim H. G.; Khmelinskaia A.; Sobczak J. P.; Dietz H.; Schwille P. Membrane sculpting by curved DNA origami scaffolds. Nat. Commun. 2018, 9, 811. 10.1038/s41467-018-03198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y.; Endo M.; Sugiyama H. Mimicking membrane-related biological events by DNA origami manotechnology. ACS Nano 2015, 9 (4), 3418–3420. 10.1021/acsnano.5b01723. [DOI] [PubMed] [Google Scholar]
- Göpfrich K.; et al. Ion channels made from a single membrane-spanning DNA duplex. Nano Lett. 2016, 16 (7), 4665–4669. 10.1021/acs.nanolett.6b02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J. R.; Stulz E.; Howorka S. Self-assembled DNA nanopores that span lipid bilayers. Nano Lett. 2013, 13 (6), 2351–2356. 10.1021/nl304147f. [DOI] [PubMed] [Google Scholar]
- Ohmann A.; et al. A synthetic enzyme built from DNA flips 107 lipids per second in biological membranes. Nat. Commun. 2018, 9, 2426. 10.1038/s41467-018-04821-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T.; Yu K.; Hartzell H. C.; Tajkhorshid E. Lipids and ions traverse the membrane by the same physical pathway in the nhTMEM16 scramblase. eLife 2017, 6, e28671. 10.7554/eLife.28671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D.; Mcconnell H. M. Inside-Outside Transitions of Phospholipids in Vesicle Membranes. Biochemistry 1971, 10, 1111–1120. 10.1021/bi00783a003. [DOI] [PubMed] [Google Scholar]
- Alakoskela J. M. I.; Kinnunen P. K. J. Control of a redox reaction on lipid bilayer surfaces by membrane dipole potential. Biophys. J. 2001, 80 (1), 294–304. 10.1016/S0006-3495(01)76014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owczarzy R.; Moreira B. G.; You Y.; Behlke M. A.; Wälder J. A. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry 2008, 47 (19), 5336–5353. 10.1021/bi702363u. [DOI] [PubMed] [Google Scholar]
- Kielar C.; et al. On the Stability of DNA Origami Nanostructures in Low-Magnesium Buffers. Angew. Chem., Int. Ed. 2018, 57 (3), 9470–9474. 10.1002/anie.201802890. [DOI] [PubMed] [Google Scholar]
- Sahu S. K.; Gummadi S. N.; Manoj N.; Aradhyam G. K. Phospholipid scramblases: An overview. Arch. Biochem. Biophys. 2007, 462 (1), 103–114. 10.1016/j.abb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan S.; Ijäs H.; Linko V.; Keller A. Structural stability of DNA origami nanostructures under application-specific conditions. Comput. Struct. Biotechnol. J. 2018, 16, 342–349. 10.1016/j.csbj.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.