Abstract
Gene delivery within hydrogel matrices can potentially direct mesenchymal stem cells (MSCs) towards a chondrogenic fate to promote regeneration of cartilage. Here, we investigated whether the mechanical properties of the hydrogel containing the gene delivery systems could enhance transfection and chondrogenic programming of primary human bone marrow-derived MSCs. We developed collagen-I-alginate interpenetrating polymer network hydrogels with tunable stiffness and adhesion properties. The hydrogels were activated with nanocomplexed SOX9 polynucleotides to direct chondrogenic differentiation of MSCs. MSCs transfected within the hydrogels showed higher expression of chondrogenic markers compared to MSCs transfected in 2D prior to encapsulation. The nanocomplex uptake and resulting expression of transfected SOX9 were jointly enhanced by increased stiffness and cell-adhesion ligand density in the hydrogels. Further, transfection of SOX9 effectively induced MSCs chondrogenesis and reduced markers of hypertrophy compared to control matrices. These findings highlight the importance of matrix stiffness and adhesion as design parameters in gene-activated matrices for regenerative medicine.
Keywords: GAMs, IPNs, 3D transfection, SOX9, tissue engineering
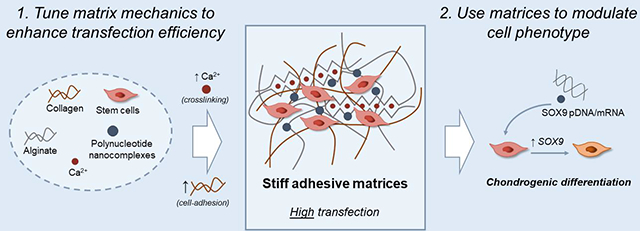
Graphical Abstract
1. Introduction
Directing differentiation of stem cells is a promising approach in regenerative medicine,(Merrell and Stanger 2016; Jopling, Boue, and Belmonte 2011) in order to generate specified cells to restore missing, damaged, or diseased tissue.(Murry and Keller 2008; Yamamizu et al. 2013) Several autologous and allogeneic stem cell products are available (i. e. Holoclar, Tigenix,, Temcell, Allocord) but existing therapies on the market mainly utilize growth factors delivered from polymer matrices to promote differentiation of tissue-resident stem cells.(Yannas et al. 1989; Lo et al. 2012) Limitations of these devices include the necessity of using high doses of growth factors together with a poor control over their release kinetics, which potentially can lead to side effects and complications.(Kang, Hsu, and Lehman 2017; Epstein 2013; DeVine et al. 2012) Gene therapy approaches based on viral vectors have been recently commercialized and could address problems of protein stability and dosing, but they pose concerns in terms of immunogenicity and integration-mediated genotoxicity.(D. Wang, Tai, and Gao 2019)
Hydrogels may aid in overcoming the limitations of protein-based regenerative strategies and viral vector approaches by delivering polynucleotides in a tissue-conductive three-dimensional (3D) environment, while inducing prolonged expression of growth factors and transcription factors that regulate tissue development.(Im, Kim, and Lee 2011; Raisin, Belamie, and Morille 2016) Known as Gene-Activated Matrices (GAMs), the first and more studied examples were based on the delivery of plasmid DNA (pDNA) or viral vectors. Recently, mRNA has been used for GAM activation providing enhanced transfection efficiency.(Elangovan et al. 2015; Balmayor et al. 2016; Khorsand et al. 2017) In either case, the effects of the hydrogel matrix itself remain to be considered. Previous studies on gene transfer on two-dimensional (2D) substrates have demonstrated that both material stiffness(Kong et al. 2005) and adhesion properties,(Y. A. Wang et al. 2007) as well as the presence of ECM proteins can modulate transfection.(Dhaliwal et al. 2010; Perlstein et al. 2003) Yet, conclusions obtained from 2D studies do not always translate to 3D systems(Chu and Kong 2012; Shepard et al. 2010; Dhaliwal, Oshita, and Segura 2013; Lei, Padmashali, and Andreadis 2009) due to the higher complexity of cell transfection within these environments.(Zhang et al. 2012, 2010) Moreover, the mechanical properties of GAMs may modulate the biological effect of the induced transgene, because cell fate can also be regulated intrinsically by stiffness, elasticity, and stress-relaxation.(Engler et al. 2006; Guilak, Cohen, and Estes 2009; Chaudhuri, Gu, Darnell, et al. 2015; Huebsch et al. 2010; Vining and Mooney 2017)
Here, we investigated whether imparting specific mechanical properties to a GAM environment can enhance 3D gene transfer and support chondrogenesis. To this end, we developed a set of interpenetrating polymer networks (IPNs) of collagen-I and alginate. With these IPNs, it is possible to independently tune the stiffness of the matrix, without significantly affecting its architecture, polymer concentration and adhesion ligand density.(Branco da Cunha et al. 2014) In addition, their composition resembles that of the native cartilaginous tissue(Nathaniel, Mow, and Foster 1998; Hardin, Cobelli, and Santambrogio 2015) and they could yield more realistic ECM models compared to biologically inert polymer hydrogels that present synthetic adhesion ligands.(Balakrishnan and Banerjee 2011; Hinderer, Layland, and Schenke-Layland 2016) We designed GAMs with tunable stiffness and cell-adhesion ligand densities and studied their capacity to promote the transfection of SOX9, a pivotal transcription factor in chondrogenesis.(Dy et al. 2012; Bi et al. 1999) IPN stiffness was modulated by changing the amount of the alginate crosslinking agent (i.e. calcium carbonate nanoparticles), while adhesion ligand density was tuned by modifying the weight ratio between alginate (lacking cell adhesion ligands) and collagen (with numerous integrin-binding domains). We characterized the effect of GAM mechanics on SOX9 expression kinetics and nanocomplex internalization and compared the expression levels of chondrogenic markers with those obtained after 2D transfection. These results show how hydrogels can be used to enhance non-viral gene delivery and optimize GAMs for use in regenerative medicine.
2. Materials and methods
2.1. Cell culture
Human adipose derived Mesenchymal Stem Cells (MSCs) and human bone marrow derived MSCs were acquired from ATCC and isolated from fresh unprocessed bone marrow (Lonza), respectively. Briefly, bone marrow samples were diluted in cold PBS pH 7.2 (Gibco) supplemented with 2% v/v heat inactivated fetal bovine serum (Gibco) and filtered (pore size 100 μm). Then, the diluted cell suspension was added over Lymphoprep (Stemcell Technologies) and centrifuged (800xg, 30 min, 20°C) yielding an interphase of mononuclear cells. CD14 microbeads (Miltenyi Biotech) were used for the depletion of CD14+ cells by magnetic separation and MSCs were allowed to adhere to tissue culture flasks. MSCs were cultured in α-MEM (Gibco) supplemented with 10 ng/ml rhFGF basic (Peprotech), 1% penicillin-streptomycin (Gibco) and 10% (adipose MSCs,) or 20% (bone marrow MSCs) heat inactivated fetal bovine serum. Cells were detached from the plates using TrypLE enzyme solution (Gibco) and plated at 5000–7500 cells/cm2. Media was changed every 2–3 days and cells were split at 70–80% confluency. For experiments involving ROCK inhibition, Y-27632 was added to cell culture media at 10 μM.
2.2. Plasmid design and mRNA synthesis
SOX9 plasmid was designed to enable optimal mRNA synthesis by in vitro transcription (IVT) (Fig. S1). To construct the plasmid, the SOX9 CDS, a Kozak consensus sequence and the 3’ UTR of the α globin gene(Kozak 1987; Holcik and Liebhaber 1997; Warren et al. 2010) were synthesized and cloned into a pCMVTnT expression vector (Promega, Madison, WI, USA) by Genewiz (South Plainfield, NJ, USA). The plasmid was sequence-verified (Stab Vida, Caparica, Portugal) and transformed into DH5α E. Coli bacteria for its propagation. Extraction of the plasmid from the bacterial cultures was performed with the NucleoSpin Plasmid Kit (Macherey-Nagel, Dueren, Germany) according to the manufacturer’s instructions. SOX9 plasmid was used as template to synthesize SOX9 mRNA using the mMESSAGE mMACHINE T7 Ultra kit (Ambion, Foster City, CA, USA) following the manufacturer’s protocol. Briefly, SOX9 plasmid was linearized using BamHI restriction enzyme (Promega) and 2–7 enzyme units per μg of pDNA. Endonuclease digestion was confirmed by gel electrophoresis and 1.35 μg of linearized plasmid was used as template for each 20 μl reaction. mRNA was purified via phenol-chloroform extraction using Phase Lock 1.5 ml tubes (5 Prime, Hilden, Germany) followed by ethanol precipitation and quantification by Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA).
2.3. Preparation of 3DFectIN complexes
For 3D transfection experiments, 25 μl of transfection complexes were prepared per 150 μl of total IPN mix. Complexes were assembled in OptiMEM (Gibco) at 3:1 3DFectIN:pDNA/mRNA ratio (μl:μg), following the manufacturer’s instructions. Briefly, a solution containing 1.5 μg of SOX9 pDNA/mRNA was added over a solution containing 4.5 μl of 3DFectIN reagent (OZ Biosciences) in a 1/1 v/v ratio, mixed by pipetting up-down and incubated for 20 min at room temperature. To investigate the distribution of the complexes within the IPNs, SYBRGold-labeled pDNA complexes were prepared. A pDNA aqueous solution was labeled by mixing with a 50X DMSO solution of SYBR®Gold Nucleic Acid Gel Stain (Invitrogen) in a ratio 1:1 (μg:μl) and incubated for 5 min at room temperature before complex formation. Complexes were then encapsulated within the IPNs and visualized under fluorescence microscopy (EVOS FL Cell Imaging system).
2.4. IPNs preparation
Low viscosity, low MW sodium alginate was purchased from Pronova (UP VLVG). Before its use, alginate was dialyzed against deionized water for 2 days (3.5 kDa cutoff) and lyophilized. Dry alginate was then reconstituted at 8.0% w/v in Hank’s Balanced Salt Solution with HEPES (HBSS, 20 mM HEPES) without calcium and magnesium prior to IPN formation. Ice-cold bovine collagen type-I (Advanced Biomatrix) was mixed with 1 M HEPES and 10X HBSS solutions at 1:50 and 1:10 v/v ratios to the amount of collagen needed, respectively, and pH was adjusted to 7.4 with NaOH 1 M. Calcium carbonate nanoparticles (nano-PCC, Multifex-MM, Specialty Minerals) were encapsulated within the IPNs to crosslink the alginate, and Ca2+ release was triggered by a media acidification induced by the addition of glucono delta-lactone (GDL). Before encapsulation, nanoparticles were suspended in RNase free water (Invitrogen) at 1% w/v, sonicated for 15 s at 70% amplitude and 4 °C (Branson Sonifier) and maintained under magnetic stirring to prevent sedimentation. GDL (Merck) was dissolved in HBSS (4% w/v) and incorporated to the IPN mix at a fourfold molar excess with respect to calcium, immediately before IPN molding. For transfection experiments, pDNA and mRNA complexes were prepared as previously described (2.2) and mixed with hMSCs prior to their encapsulation within the IPNs, yielding the GAMs. hMSCs were suspended at 22.5 × 106 cells/ml in OptiMEM and added to the IPN at a density of 1.5 × 106 cells/ml.
All the different components were blended in 2 ml glass vials placed on an ice bath under magnetic stirring. HBSS supplemented with 20 mM HEPES was added first to reach the final volume of IPN mix. This buffer was supplemented with NaOH to yield a 7 mM concentration of NaOH in the final mix. The ice-cold collagen solution was then included, and while it was stirring, transfection complexes were mixed with hMSCs by pipetting up-down and then added to the collagen solution. Once this mix was homogeneous, calcium carbonate nanoparticles were incorporated, followed by the addition of alginate. Finally, the solution of GDL was added and IPNs were casted in 96 or 48 well plates (Thermo Fisher Scientific). IPNs were then allowed to gel for 30 min at room temperature (alginate crosslinking) and 30 min at 37 °C (collagen crosslinking). Due to the drop in pH caused by the hydrolysis of GDL, pH was balanced by the addition of HBSS 20 mM HEPES during the first 4h after IPN crosslinking. Once pH was stabilized, culture media was added on top of the IPNs. Culture media was refreshed every 2–3 days.
2.5. Mechanical Characterization of IPNs
IPN mechanical properties were characterized with an AR-G2 stress controlled rheometer (TA Instruments). IPNs without cells and gene complexes were prepared as described above and placed onto the surface plate of the rheometer immediately before gelation. Right after, a 20 mm 2° aluminum cone was put into contact with the IPN creating a 20 mm gel disk. To prevent sample dehydration, the exposed gel surfaces within the rheometer geometry were surrounded with a wet Kimwipe, and 800 μl of HBSS were carefully added after the first step of IPN gelation. IPNs were allowed to gel at 25 °C for 30 min (alginate crosslinking) followed by 30 min of collagen crosslinking at 37 °C, and mechanical properties were measured over time as previously described(Chaudhuri et al. 2014). The storage modulus at 1% strain and 1 Hz was recorded periodically until it reached its equilibrium value (30–60 min). Then, a strain sweep (0.1–20% strain) was performed to confirm this value was within the linear elastic regime, followed by a frequency sweep (0.1–10 Hz). No pre-stress was applied to the IPNs for these measurements. For stress-relaxation measurements, strain was raised during 1 s to 15% and then held constant, while the load was recorded as a function of time. Both the sweeps and stress-relaxation analysis were performed at 37 °C.
2.6. Cell retrieval for gene expression and flow cytometry analysis
At the desired time points, cells were retrieved from the GAMs before RNA extraction and fluorocytometric analysis. Media was aspirated and GAMs were washed twice with PBS (Gibco). GAMs were then removed from the plates with a spatula and placed in 1.5 ml tubes before a two-step enzymatic digestion. Fresh enzyme solutions were prepared in Ca2+, Mg2+ PBS (Gibco) supplemented with 1% w/v bovine serum albumin (Sigma). In a first step, 350 μl of a solution containing 34 U/ml alginate lyase (Sigma) and 300 U/ml collagenase type I (Gibco) were added per 200 μl of GAMs and incubated for 15 min at 37 °C under horizontal shaking (300 rpm, Heidolph Titramax platform shaker). Next, samples were centrifuged (5 min, 400xg) and the enzyme solution was removed. A second digestion was performed with 350 μl of a solution containing 300 U/ml collagenase and GAMs were further incubated for 15 min and centrifuged to discard the collagenase solution. For RNA extraction, the resulting pellet was mixed with 1 ml of 0.05M ice cold EDTA by pipetting up-down and then centrifuged again. This step was omitted for flow cytometry samples. Finally, the cell pellet was washed with 1 ml ice cold PBS and placed on ice until further analysis.
Total RNA was extracted with the SPEEDTOOLS total RNA extraction kit (Biotools) following the manufacturer’s instructions. RNA concentration was quantified using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific).
2.7. Uptake studies
Uptake of the transfection complexes within the GAMs was evaluated using a Cy5-labeled pDNA. For this, pcDNA3.1(+) (Addgene) was labeled with the Cy®5 Label IT® Tracker™ kit (Mirus) following the manufacturer’s instructions and subsequently loaded in the transfection complexes. Complexes were then encapsulated within the GAMs together with MSCs and cultured for different lengths of time, after which cells were analyzed via flow cytometry and confocal microscopy. For flow cytometry analysis, MSCs were retrieved from the GAMs as previously described (section 2.5) and analyzed on a BD FACSCalibur. Cells were gated to eliminate interferences from remaining traces of polymers (Fig. S2) and the percentage of Cy5+ cells among the gated population was quantified. To evaluate the uptake via confocal microscopy, GAMs were cultured on glass coverslips. At the time of analysis, MSCs were fixed in 10% buffered formalin for 20 min at room temperature followed by permeabilization with Triton X-100 (0.2% v/v in PBS) for 10 min. Cells were then stained with DAPI (Sigma-Aldrich) and Alexa Fluor™ 488 Phalloidin (Thermo Fisher Scientific) and imaged on a Leica TCS SP5 confocal microscope. Cy5 Raw Fluorescence Intensity (RFI) per cell was quantified using ImageJ software after selecting the cell contours in the corresponding confocal images.
2.8. cDNA synthesis and qRT-PCR
Reverse transcription was carried out using 100–500 ng of total RNA per sample in a 30 μl final reaction volume. First, total RNA was mixed with random primers and dNTPs (Invitrogen) and kept at 65 °C for 5 min. Samples were then incubated with a mix of RNase OUT, 5x first strand buffer and DTT (Invitrogen) for 2 min at 37 °C and subsequently placed on ice. Finally, reverse transcriptase (M-MLV, Invitrogen) was included and the cycle was continued as follows: 10 min at 25 °C, 50 min at 37 °C and 15 min at 70 °C. The resulting cDNA (5–10 ng) was used to assemble qRT-PCR reactions in a final volume of 20 μl containing Universal PCR Mastermix and TaqMan assays (Applied Biosystems, Table S1). The thermal cycling was done in a StepOne Real Time PCR System (Applied Biosystems) performing a 10 min hold at 95 °C followed by 40 cycles of 15 sec at 95 °C and 1 min at 60 °C. Gene expression values were normalized to internal controls (Actin β and GAPDH) and presented either as the log2(ΔCt) or the fold change relative to cells plated in 2D before the experiments using the comparative 2−ΔΔCt method.(Pfaffl 2001)
2.9. Chondrogenic differentiation
Chondrogenesis experiments were performed with adipose hMSCs below passage 10. IPNs (200 μl) were casted in 48 well plates and cultured for 21 days in Complete Chondrogenic Medium (CCM) consisting of DMEM high-glucose 1 mM pyruvate (Gibco), 100 nM dexamethasone (Sigma-Aldrich), 50 μg/ml ascorbic acid 2-phosphate (Sigma-Aldrich), 40 μg/ml L-proline (Sigma-Aldrich), 1% ITS Premix supplement (Becton Dickinson), 1% penicillin/streptomycin (Gibco) and 10 ng/ml transforming growth factor-β3 (Peprotech). Media was replaced every 2 days in all experiments. GAMs were activated with 2 μg of SOX9 mRNA per GAM, and their chondrogenic potential was compared to non-activated matrices. At the end of the experiments, GAMs were retrieved for analysis. Gene expression of chondrogenic markers SOX9, aggrecan (ACAN), collagen type-II (COL2A1) and collagen type-X (COLX) was assessed by qRT-PCR. SOX9, aggrecan and collagen type-X were also detected by immunohistochemistry (IHC).
2.10. Immunohistochemistry
GAMs were retrieved from tissue culture plates, washed with PBS twice and fixed with 10% buffered formalin at room temperature for less than 24 h. After fixation, samples were maintained in 70% ethanol until dehydration and paraffin sectioning. Sections of 4 μm thickness were cut and allowed to adhere to poly-L-lysine treated glass slides overnight at 55 °C. Before staining, sections were dewaxed in xylene and hydrated with graded ethanol, and antigens were retrieved in Tris-EDTA buffer during 20 min at 95 °C (PT Link, Agilent). Samples were pretreated with hydrogen peroxide and blocked with serum free protein block (Dako; Agilent) before incubation with primary antibodies against SOX9 (ab5535, Merck), aggrecan (ab3778, Abcam), and collagen type-X (ab49945, Abcam). Primary antibodies were diluted in antibody diluent (Dako) at 1:500 (SOX9), 1:100 (aggrecan) and 1:1000 (collagen type-X) and incubated overnight at 4 °C. Then, samples were incubated with goat secondary antibodies labeled with HRP (Dako) at 1:100 for 1 h and stained with 3,3’-diaminobenzidine (DAB). Negative controls were obtained by omitting the primary antibodies (Fig. S3). All samples were observed using an Olympus BX43 microscope equipped with an Olympus XC50 camera. Mean Intensities were quantified with the IHC macro of the Fiji software(Schindelin et al. 2012) while the percentage of positive cells was calculated by dividing the number of stained cells by the total number of cells.
2.11. Statistical Analysis
The statistical analysis was performed using GraphPad Prism. Data were analyzed with either the D’Agostino & Pearson or the Shapiro-Wilk normality tests. Where applicable, data are reported as the mean ±SD. Data were compared using One-way ANOVA with Tukey’s post-hoc multiple comparison test and unpaired two-tailed t tests and p-values less than 0.05 were considered to be statistically significant.
Results
3.1. Characterization of collagen-I-alginate interpenetrating networks (IPNs)
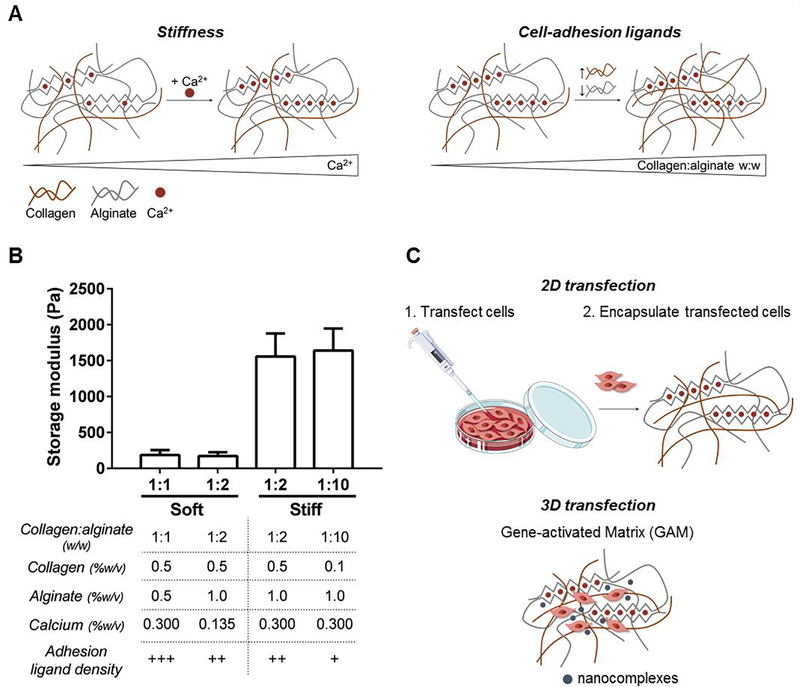
Collagen-I-alginate IPNs with tunable mechanical properties were prepared to investigate the influence of stiffness and adhesion properties on the transfection efficiency of gene-activated matrices (GAMs). To determine the effect of each property independently, we designed a set of IPNs that would allow us to change both variables separately. Analysis with oscillatory rheology was used to tune IPN stiffness based on CaCO3 concentration, and four IPNs were fabricated with storage modulus (G’) from 150 to 1500 Pa at a fixed frequency of 1 Hz and 1% strain after 60 min of gelation (Fig. 1A), S4). A strain sweep was performed to confirm the values were within the linear elastic regime (Fig. S5). Within this range of stiffness, we modulated cell-adhesion ligand density by adjusting the collagen:alginate w:w ratio from 1:1 to 1:10. In contrast to collagen, alginate presents no intrinsic integrin-binding domains for cells and hence, higher collagen:alginate ratios confer higher adhesion ligand density. Accordingly, the resulting IPN set can be divided in four groups: IPNs of low stiffness and very high cell-adhesion ligand density (Soft 1:1), IPNs of low stiffness and high adhesion ligand density (Soft 1:2), IPNs of high stiffness and high adhesion ligand density (Stiff 1:2) and IPNs of high stiffness and low adhesion ligand density (Stiff 1:10). The amount and distribution of collagen fibers across the IPN set was analyzed by confocal reflectance microscopy (Fig. S6). A significant reduction in collagen fibers was observed for the 1:10 condition, which is consistent with the lower percentage of collagen present in these IPNs (0.1 versus 0.5 wt% collagen). The two soft IPNs have the same collagen concentration (0.5 wt%) and showed the same fiber intensity. However, soft 1:1 IPNs have less alginate than soft 1:2 IPN (0.5 versus 1 wt% alginate) and thus a higher collagen-to-alginate ratio. The paired comparison between the different groups enabled us to explore the effect of stiffness (Soft 1:2 vs Stiff 1:2) and cell-adhesion ligand density (Soft 1:1 vs Soft 1:2 and Stiff 1:2 vs Stiff 1:10) on gene transfer efficiency (Fig. 1B).
Figure 1.
IPNs show tunable stiffness and cell-adhesion ligands and can be loaded with nanocomplexed polynucleotide molecules to generate Gene-Activated Matrices (GAMs). (A) Scheme showing the modulation of IPN mechanical properties through crosslinker concentration and polymer ratio. The IPN is illustrated as brown collagen fibers and grey alginate polymer chains. The calcium ions crosslinking the G-blocks of the alginate network (zig-zag structures) are shown as red dots. Objects are not drawn to scale. (B) Storage modulus at 1 Hz and 1% strain after 1 h of IPN crosslinking and detailed composition and properties of the IPN set. (C) Scheme showing the experimental setups used for the transfection assays.
Frequency dependent storage moduli were characterized over a frequency range of 0.1 to 1 Hz, and indicated that the IPNs exhibited some degree of viscoelasticity, as previously observed(Branco da Cunha et al. 2014) (Fig. S5). IPN viscoelasticity was further characterized by stress relaxation behavior, with half-times (T 1/2) of 10–300 s depending on the collagen:alginate ratio (Fig. S7). A stress relaxation half-time of 100 s is in agreement with previous results for low viscosity, low MW alginate gels, and has been identified as the optimum T 1/2 to allow cell-mediated gel remodeling,(Chaudhuri, Gu, Klumpers, et al. 2015)which is necessary for regeneration of osteochondral tissues like cartilage.(H. P. Lee et al. 2017)
3.2. Increased stiffness and cell-adhesion ligand density enhance IPN transfection efficiency
The transcription factor SOX9 has pivotal roles in chondrogenic lineage specification and has potential for the design of GAMs for cartilage regeneration. To prepare SOX9-GAMs, IPNs were loaded with nanocomplexed SOX9 sequences within the hydrogel matrix, as depicted in Fig. 1C, and their transfection efficiency was assessed. In order to determine the role of cell encapsulation and matrix mechanics on SOX9 expression, we compared the direct transfection of bone marrow MSCs in GAMs with 1) the encapsulation of MSCs in blank IPNs and 2) the transfection of MSCs in 2D and subsequent encapsulation in blank IPNs.
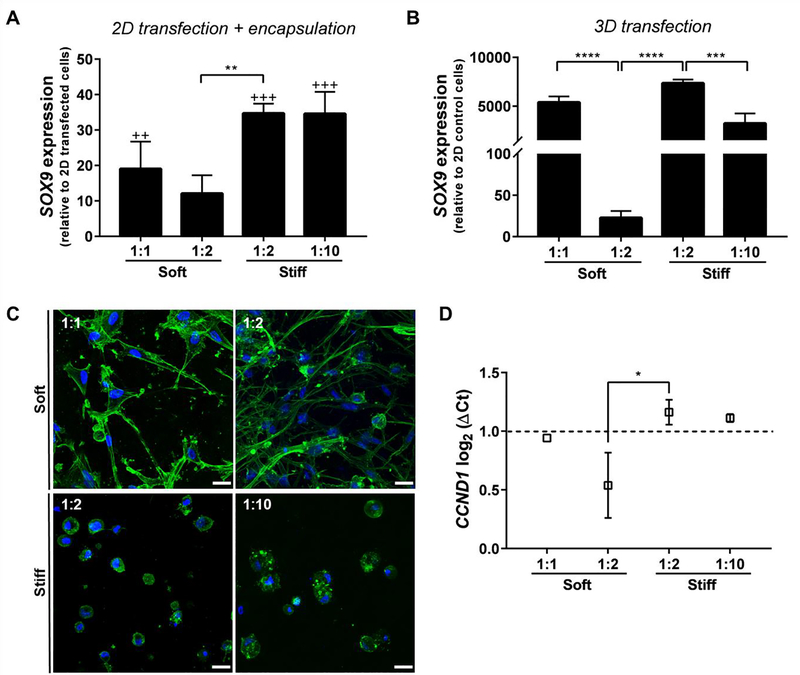
In a first step, bone marrow MSCs were encapsulated in blank IPNs and SOX9 expression was assessed to discard any potential inherent effect of matrix mechanics on SOX9 expression that would impact the transfection results (Fig. S8). In a second step, MSCs were transfected in 2D and then encapsulated within the polymer matrices in order to evaluate whether this encapsulation was able to alter transgene expression levels. To this end, SOX9 pDNA transfections were performed with the commercial reagent Lipofectamine® 2000 and transfected cells were loaded within the IPNs after retrieval from 2D. qRT-PCR analysis revealed that MSCs encapsulated in the IPNs exhibited a higher SOX9 expression relative to the cells after 2D transfection, which indicates some degree of cell transfection occurs within the gels (Fig. 2A), and we hypothesized that remaining pDNA complexes from 2D transfection were encapsulated along with the cells in the IPNs. To examine this hypothesis, Lipofectamine® complexes were loaded with Cy5-labeled pDNA and observed during 2D transfection by fluorescence microscopy, which showed that pDNA complexes remained adhered to the cells even after retrieval by enzymatic separation (Fig. S9). This result confirms that residual complexes were encapsulated within the IPNs together with the cells, and thus 3D transfection was likely occurring. The same result was not observed when 2D transfections were performed with mRNA (Fig. S8), most likely due to the lower stability of mRNA sequences, further confirming that 3D transfection of remaining pDNA complexes was occurring. In these experiments, IPNs of high stiffness promoted the highest fold increase in SOX9 expression, suggesting that IPN mechanics can modulate transfection (Fig. 2A).
Figure 2.
Matrix stiffness and adhesion exert a positive effect on bone marrow MSC transfection. (A) SOX9 overexpression 48 h after encapsulation of pDNA-transfected cells within the IPNs. Gene expression levels were normalized to GAPDH and presented as relative to the levels in 2D transfected cells before encapsulation. One-way ANOVA with a Tukey’s post-hoc multiple comparison test was performed to compare SOX9 expression levels to the levels in transfected cells before encapsulation (++P≤0.01, +++P≤0.001, n=3) and between the IPNs (**P≤0.01, n=3). (B) SOX9 expression 48 h after pDNA transfection within the Gene-activated Matrices (GAMs). Gene expression levels were normalized to GAPDH expression and compared to the levels in MSCs before encapsulation. One-way ANOVA with a Tukey’s post-hoc multiple comparison test was performed to compare gene expression levels between the GAMs (***P≤0.001, ****P≤0.0001, n=3). (C)MSC morphology within the IPN set (F-actin in green, nuclei in blue). Scale bar = 25 μm for all the images. (D) qRT-PCR quantification of Cyclin D1 (CCND1) expression 48h after pDNA 3D transfection. Gene expression levels were normalized to GAPDH and expressed as log2 (ΔCts). The dotted line represents log2 (ΔCt) = 1, and is used to highlight the GAMs with higher CCND1 levels. One-way ANOVA with a Tukey’s post-hoc multiple comparison test was performed to compare gene expression levels between the GAMs (*P≤0.5, n=2). GAMs are described by their stiffness and collagen:alginate w:w ratios (1:1 to 1:10).
To specifically determine the role of mechanics on 3D transfection, SOX9 pDNA sequences were first nanocomplexed with 3DFectIN® and subsequently loaded into the IPNs to prepare SOX9 GAMs. Compared to Lipofectamine® 2000, 3DFectIN® is optimized for transfection within 3D environments and hence the encapsulation of MSCs within these GAMs promoted their 3D transfection by overexpression of SOX9. In agreement with the previous results, qRT-PCR data showed a higher SOX9 expression in matrices of higher stiffness. Additionally, lower collagen:alginate ratios were associated with reduced gene transfer efficiency (Fig. 2B). In the case of stiff GAMs, where the collagen:alginate ratio was modulated from 1:2 to 1:10, this negative impact was found to correlate to an increased destabilization of the nanocomplexes with lower collagen:alginate ratios (Fig. S10). For soft GAMs, changing collagen:alginate ratio from 1:1 to 1:2 did not induce nanocomplex aggregation (Fig. S10), but resulted in a dramatic decrease in SOX9 levels. Soft 1:1 and soft 1:2 IPNs differ in the collagen:alginate ratio but also in the amount of calcium carbonate particles used in their fabrication. Given that inorganic calcium-based particles have been used as complexation reagents for polynucleotides, we performed two control experiments in order to rule out their contribution to the higher transfection efficiency of 1:1 IPNs. We confirmed that the excess of calcium ions did not improve lipoplex transfection efficiency of MSCs plated in 2D (Fig. S10). In this regard, it’s worth mentioning that the calcium carbonate particles are dissolved by glucono delta-lactone during IPN gelation, so one would not expect their effects would persist during transfection. In order to test the effect of adhesion ligands on transfection efficiency, transfection in soft 1:1 IPNs (0.5% collagen, 0.5% alginate, 0.3% calcium carbonate) was compared with a plain 0.5% collagen-only gel. The collagen gel resulted in higher SOX9 expression, which is consistent with higher transfection in the soft 1:1 IPNs that have a higher amount of collagen relative to alginate. We suggest increased collagen ligands of collagen-only or soft 1:1 IPNs promote transfection compared to soft 1:2 IPNs, due to higher availability of adhesive ligands, rather than the presence of calcium carbonate particles (Fig. S11).
In accordance with previous studies, we observed that soft matrices promoted spread cell morphologies whereas stiff matrices resulted in rounded cell shapes(Branco da Cunha et al. 2014) (Fig. 2C). We hypothesized that efficient cell attachment might be necessary for cytoskeletal organization, which in turn could modify the intracellular transport of the complexes. Indeed, for both Soft 1:1 and Stiff 1:2 GAMs, transfection was affected by the state of cell cytoskeleton, as demonstrated by a decrease in SOX9 expression after the addition of the ROCK inhibitor Y-27632 (Fig. S11). Together with adhesion ligand density, gene transfer efficacy was also increased with stiffness, as evidenced by the differences between Soft 1:2 and Stiff 1:2 GAMs. Stiffness has been previously shown to promote transfection efficiency in 2D environments by increasing cell proliferation.(Kong et al. 2005) Although an indirect measure of cell proliferation, higher transfection efficiencies were associated with upregulation of cyclin D1 (CCND1) gene expression, which regulates the cell cycle(Shepard et al. 2010; Gojgini, Tokatlian, and Segura 2011) (Fig. 2D).
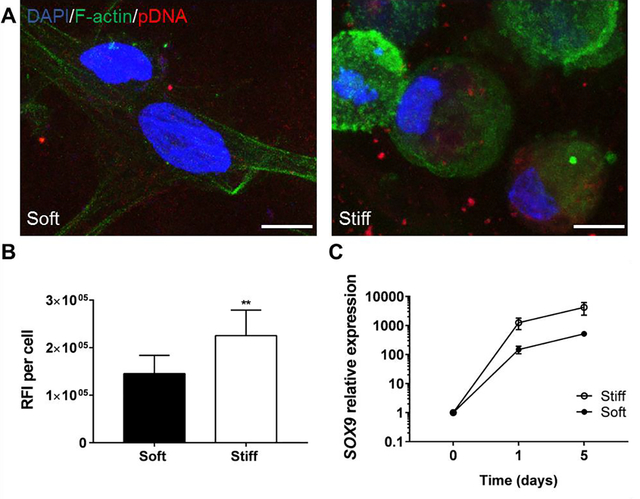
We next hypothesized that changes in transfection efficiency could be related to different cellular internalization rates of the nanocomplexes. Two GAMs with the same collagen:alginate ratio and different stiffness, soft 1:2 and stiff 1:2 GAMs, were selected to investigate this hypothesis. Transfection complexes were prepared with Cy5-labeled pDNA and their localization within the cells was tracked by confocal microscopy, and their amount was quantified by flow cytometry. As observed in Figure 3A and 3B, stiff GAMs induced higher nanocomplex accumulation within the cells compared to soft GAMs. Accordingly, flow cytometry analysis also showed a higher Mean Fluorescence Intensity for bone marrow MSCs transfected within stiff GAMs (Fig. S12).
Figure 3.
IPN stiffness increases the uptake of the pDNA nanocomplexes. (A) Confocal images of bone marrow MSCs within IPNs showing the localization of the pDNA nancomplexes. Scale bars = 10 μm for all the images. (B) Quantification of Raw Fluorescence Intensity (RFI) per cell of A) (Unpaired two-tailed t test, n =3, **P≤0.01). (C) SOX9 expression kinetics after pDNA transfection within stiff and soft GAMs prepared with a 1:2 collagen:alginate w:w ratio. Gene expression levels were measured by qRT-PCR, normalized to GAPDH expression and compared to the levels in MSCs before encapsulation. See also Figure S12.
Further, analysis of the kinetics of transgene expression showed that stiff GAMs maintained over time higher SOX9 expression than soft GAMs (Fig. 3C). This suggests that the effects of GAM mechanics are maintained long-term, and consequently are an important parameter for the design of GAMs.
3.3. Chondrogenic differentiation of aMSCs is enhanced by transfection of SOX9 within stiff GAMs
We next determined if GAM properties could also impact chondrogenic differentiation induced by SOX9 overexpression. Given the influence of matrix mechanics on the modulation of cell phenotype, we first determined whether stiffness affects chondrogenic differentiation. To this end, we encapsulated adipose MSCs within both stiff and soft 1:2 IPNs and subjected them to chondrogenic differentiation in the absence of SOX9 transfection. Additionally, in order to explore the effect of stiffness during SOX9 enforced expression, we transfected the cells in 2D with SOX9 lipoplexes prior to loading within soft and stiff IPNs and compared these Lipofected IPNs with the control (non-transfected) IPNs. mRNA sequences were used for these differentiation experiments instead of pDNA, due to their proven higher potency relative to pDNA.(Sahin, Karikó, and Türeci 2014)−(Warren et al. 2010)
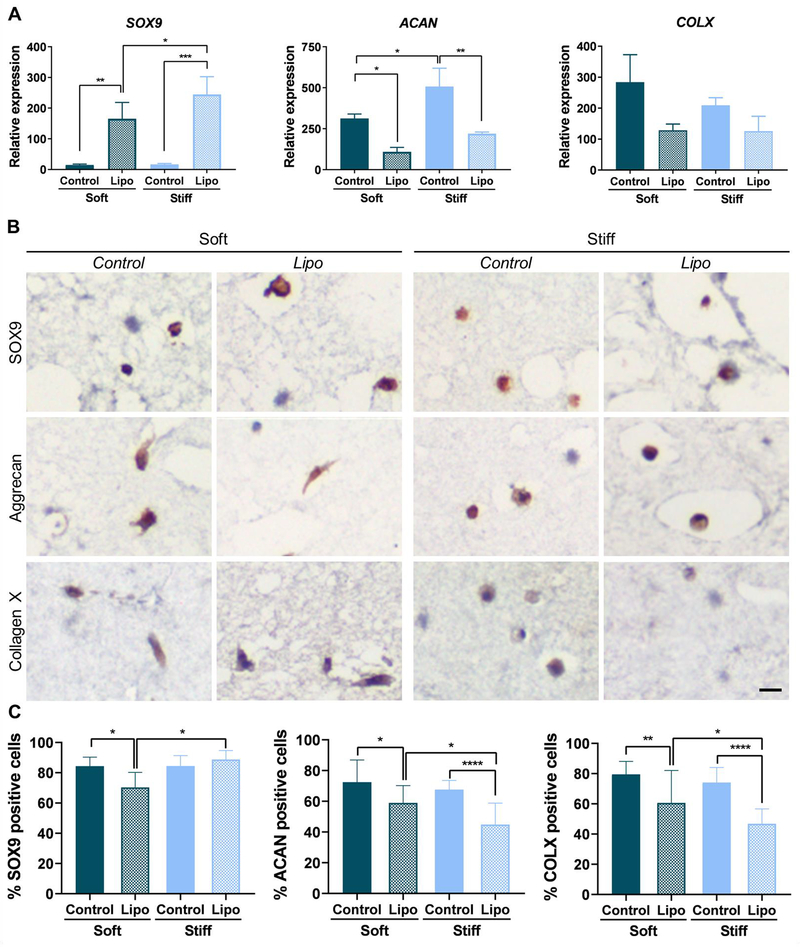
As expected, SOX9-transfected MSCs showed higher levels of SOX9 mRNA expression compared to control (non-transfected) cells (Fig. 4A) after encapsulation within IPNs. However, this higher mRNA expression did not translate into a higher proportion of SOX9 positive cells as assessed by immunohistochemistry (Fig. 4B and C).Contrary to what we expected, aggrecan (ACAN) expression was significantly decreased in SOX9-transfected cells compared to control cells loaded within the IPNs (Fig. 4A and C), suggesting that SOX9 upregulation prior to MSC encapsulation was not suitable for inducing the expression of this ECM protein. Importantly, 2D transfection with SOX9 resulted in the downregulation of the hypertrophic marker collagen type-X at the protein level, suggesting that SOX9 transfection could be a strategy to improve the quality of the regenerated cartilage (Fig. 4B and C). Compared to soft IPNs, MSCs loaded within stiff IPNs preserve a rounded morphology at the end of the chondrogenesis experiments (Fig. S13), which makes it difficult to distinguish nuclear versus cytosolic staining of SOX9, and peri-cellular versus cytosolic staining of aggrecan and collagen type-X. Conversely, soft IPNs show a clear cytosolic staining for both ECM proteins, rather than extracellular. These results are consistent with diffusion limitations of the nano-porous alginate matrix, which are associated with peri-cellular matrix deposition (Loebel, Mauck, and Burdick 2019; H. P. Lee et al. 2017).
Figure 4.
SOX9 2D transfection and subsequent encapsulation of adipose MSCs within the IPNs results in the reduction of collagen type-X expression. (A) Gene expression of chondrogenic markers SOX9, aggrecan (ACAN) and collagen type-X (COLX). Gene expression levels were measured by qRT-PCR, normalized to GAPDH expression and compared to the levels in MSCs before encapsulation (One-way ANOVA, *P≤0.05, **P≤0.01, ***P≤0.001, n=3). (B) Immunostaining for SOX9, aggrecan and collagen type-X in IPN sections.Scale bar = 500 μm for all the images (C) Quantification of positive cells in immunostained sections (One-way ANOVA, ****P≤0.0001, n =3). Soft designates MSCs encapsulated within soft 1:2 IPNs, Control designates MSCs encapsulated within stiff 1:2 IPNs and Lipo designates MSCs transfected with SOX9 in 2D before encapsulation within stiff 1:2 IPNs. Data represent the mean ±SD. Control and Lipo designate the IPNs loaded with non-transfected and transfected cells, respectively. See also figure S13 for lower magnification immunohistochemistry images.
Overall, our results did not show any clear effect of stiffness in the upregulation of chondrogenic differentiation markers (Fig. 4). Accordingly, and considering the better suitability of their mechanical properties relative to loads born by articular cartilage,(Nathaniel, Mow, and Foster 1998; Hardin, Cobelli, and Santambrogio 2015) we selected the stiff 1:2 IPNs to elaborate the GAMs for the chondrogenic differentiation experiments.
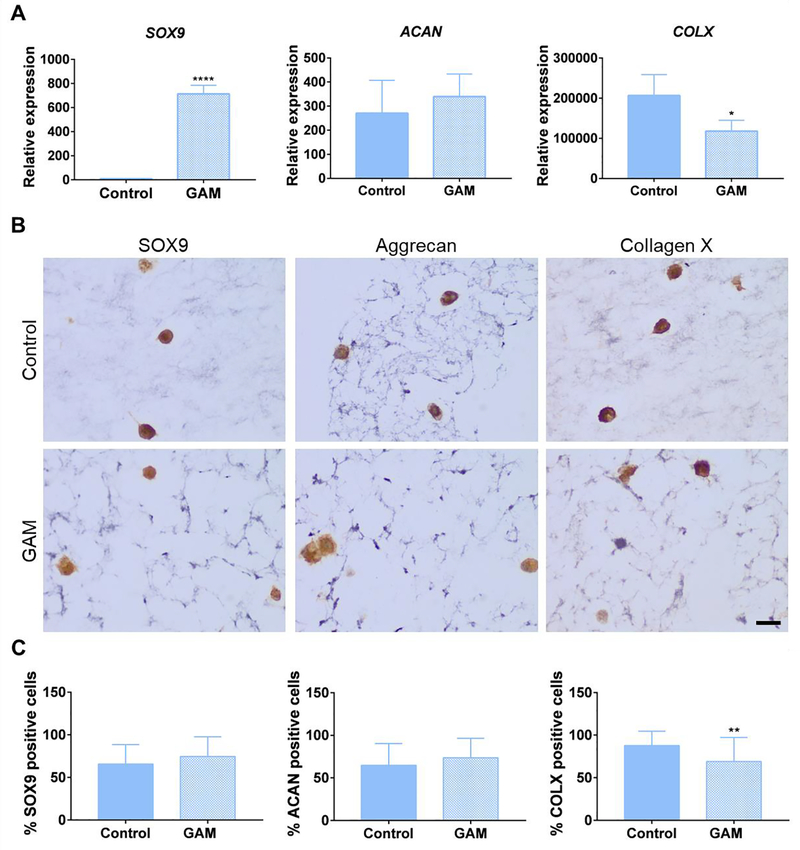
We next evaluated the effect of GAM-mediated SOX9 transfection on the chondrogenic differentiation of encapsulated MSCs. First, we confirmed that SOX9-mRNA-GAMs induced a similar SOX9 expression profile as observed for SOX9-pDNA-GAMs (Fig. S14). Next, adipose MSCs were loaded within stiff 1:2 GAMs (containing SOX9 mRNA nanocomplexes) and stiff 1:2 control IPNs (without nanocomplexes) and subjected to 21 days of chondrogenic differentiation, after which cells were retrieved for assessment of chondrogenic markers. As expected, SOX9-GAMs promoted a higher SOX9 gene expression than control IPNs (Fig. 5A). Although gene expression of chondrogenic marker aggrecan was not statistically higher compared to control IPNs, there was a significant upregulation of collagen type-II (COL2A1) mRNA expression in MSCs encapsulated within SOX9 GAMs (Fig. S15). Conversely, COL2A1 was not detected in control IPNs. In agreement with the results obtained for the 2D transfection procedure, the levels of the hypertrophic marker collagen type-X (COLX) were lower in MSCs within SOX9-GAMs, both at the mRNA and protein level, which illustrates the role of SOX9 overexpression in controlling MSC hypertrophy (Fig. 5A and C). Importantly, no reduction in aggrecan expression was observed in MSCs loaded within SOX9 GAMs, (Fig. 5A), suggesting that the transfection of SOX9 within the matrices, might be more suitable to induce the expression of this chondrogenic marker as compared to the 2D transfection procedure.
Figure 5.
SOX9 3D transfection within the GAMs induces adipose MSC chondrogenic specification and reduces premature hypertrophy. (A) Gene expression of chondrogenic markers SOX9, aggrecan (ACAN) and collagen type-X (COLX). Gene expression levels were measured by qRT-PCR, normalized to GAPDH expression and compared to the levels in MSCs before encapsulation (Unpaired two-tailed t test, *P≤0.05, ****P≤0.001, n=3). (B) Immunostaining for SOX9, aggrecan and collagen type-X in GAM sections at 40x magnification. Scale bar = 20 μm for all the images. (C) Quantification of positive cells in immunostained sections (Unpaired two-tailed t test, n =3, **P≤0.01). Control designates MSCs encapsulated within IPNs and GAM designates MSCs encapsulated within IPNs loaded with SOX9 mRNA nanocomplexes (GAMs). Data represent the mean ±SD. See also Figure S15.
4. Discussion
Our data indicate that both high hydrogel stiffness and adhesion ligand densities (provided by higher collagen:alginate ratios) exert a positive effect on transgene expression. This positive effect is consistent with a similar trend observed for transfections performed in 2D and has been related to an increased cell proliferation rate(Kong et al. 2005) and a more efficient vesicular transport mediated by the microtubular network.(Suh, Wirtz, and Hanes 2003; Dhaliwal, Oshita, and Segura 2013) In our experiments, we observed a higher gene expression of cyclin D1, a cell cycle regulator, within GAMs of higher stiffness and adhesion ligand density. In addition, our data suggest that the increased transfection efficiency of stiff GAMs is related to a higher cell uptake of the nanocomplexes. Although more studies are required to elucidate the underlying mechanism, we anticipate that the endocytic pathway and subsequent intracellular trafficking followed by the nanocomplexes are responsible for this positive effect of stiffness, based on cytoskeletal dynamics.(Dhaliwal, Oshita, and Segura 2013) These transfection results are distinct from previous reports of hyaluronic acid (HA)-based hydrogels, where soft gels induced higher cell proliferation and transgene expression levels than stiff gels.(Shiva, Talar, and Segura 2011) However, those studies tuned hydrogel stiffness with polymer concentration and utilized covalent crosslinking by Michael addition, which yields more elastic materials due to permanent covalent bonds, compared to the reversible ionic crosslinking in our system.(H. P. Lee et al. 2017)
GAM-mediated SOX9 upregulation enhanced the expression of chondrogenic markers aggrecan and collagen type-II compared to control matrices, and reduced premature production of the hypertrophic marker collagen type-X.(Dy et al. 2012) Given that the stability of mRNA inside the cell is known to be limited, particularly for non-modified sequences (Sharova et al. 2009; Tew et al. 2009; Tew and Clegg 2011), SOX9 expression at the end of the chondrogenesis experiments is mostly plausibly driven by endogenous upregulation. Indeed, this positive auto-regulation mechanism has been described in the literature (Mead et al. 2013). Additionally, it has also been described that TGF-β3 has a positive effect on the stabilization of SOX9 protein that may also contribute to this feedback loop (Coricor and Serra 2016; Chavez et al. 2017).
Contrary to what we expected, the higher SOX9 gene expression levels observed in the GAMs did not translate to a higher proportion of SOX9 positive cells compared to control matrices, as detected by immunohistochemistry. Indeed, we observed a high fraction of SOX9-positive cells in both matrices, with nearly all cell nuclei positively stained. Considering that both conditions utilized chondrogenic induction media, it is reasonable to think that this media itself,(Dahlin et al. 2014) together with the cues provided by the hydrogel,(Jiang et al. 2018) is driving a sufficient SOX9 mRNA upregulation to result in high levels of SOX9 protein expression that are difficult to discriminate by semiquantitative immunohistochemistry analysis. This might be also related to the positive auto-regulation mechanism of SOX9 mentioned previously, which may help balance the endpoint SOX9 expression in both conditions after 21 days of chondrogenic differentiation. Although endpoint SOX9 expression was similar between GAMs and control matrices, we observed a significant downregulation of the hypertrophic marker collagen type-X in GAMs compared to controls, which is consistent with previous findings (Dy et al. 2012) and points to an earlier SOX9 upregulation in the GAMs. Given the limitations of current cartilage engineering devices, the observed reduction in collagen type-X is noteworthy, because tissue engineered cartilage usually presents an inadequate balance of collagen type-II and collagen type-X, leading to a premature hypertrophic cartilage of poor mechanical properties.(Huey, Hu, and Athanasiou 2012)
Conversely, we did not observe a significant increase in aggrecan expression in the GAMs compared to the control matrices after 21 days of chondrogenesis, which may be a consequence of the similar amount of SOX9 positive cells at this time point. In this regard, it is worth mentioning that aggrecan expression was downregulated after transfection of SOX9 in 2D, as opposed to the transfection occurring within the GAM. This suggests that the direct transfection of MSCs in the 3D matrix confers an advantage over 2D cell transfection procedures, which is particularly interesting in tissue engineering, since it allows for direct cell modification at the place of injury. Compared to previous mRNA GAMs encoding growth factors, the use of transcription factors may provide a higher potency and specificity for cell reprogramming and directed differentiation (Makiko Iwafuchi-Doi and Kenneth S. Zaret 2014; Takahashi and Yamanaka 2016; Liang et al. 2008; Graf and Enver 2009; Warren et al. 2010). Additionally, the use of mRNA has been proven more efficacious than pDNA to encode transcription factors for cell differentiation. (Warren et al. 2010; Mandal and Rossi 2013; Guo et al. 2014; Aini et al. 2016). For these reasons, and considering their biocompatibility (K. Y. Lee and Mooney 2012; Lin et al. 2019) and injectability(L. Wang et al. 2012), we believe that the IPNs described in this work are promising devices that could potentially simplify the application of cell reprogramming strategies in the clinical setting. Due to the inconsistencies in autologous stem cell populations because of donor-to-donor variability, we propose that these strategies would be better moved forward with biobanked iPSCs(Trounson and DeWitt 2016; Lui et al. 2013; Tani 2015; Kyttälä et al. 2016).
5. Conclusions
The results in this study show that matrix stiffness and cell-adhesion ligand density are key factors promoting gene delivery in collagen-I-alginate GAMs. These GAMs can be activated with SOX9 and promote MSC chondrogenesis with low hypertrophy levels, resulting in an improved chondrogenic marker expression compared to the encapsulation of 2D transfected MSCs in control matrices. These results indicate that matrix mechanics can be adjusted in order to design more efficient GAMs for in situ cell reprogramming in tissue engineering.
Supplementary Material
Statement of significance.
Gene-activated matrices (GAMs) are biodegradable polymer networks integrating gene therapies, and they are promising technologies for supporting tissue regeneration. Despite this interest, there is still limited information on how to rationally design these systems. Here, we provide a systematic study of the effect of matrix stiffness and cell adhesion ligands on gene transfer efficiency. We show that high stiffness and the presence of cell-binding sites promote transfection efficiency and that this result is related to more efficient internalization and trafficking of the gene therapies. GAMs with optimized mechanical properties can induce cartilage formation and result in tissues with better characteristics for articular cartilage tissue engineering as compared to previously described standard methods.
Acknowledgements
This work has been funded by Ministerio de Economía y Competitividad (MINECO-RETOS, Grant MAT2017-84361-R, Feder Funds), Fundación BBVA 2014-PO0110, Xunta de Galicia (Grupos de Referencia Competitiva, Feder Funds) and the National Institute of Dental and Craniofacial Research of the National Institutes of Health under Award Number R01DE013033. A. M. L. was supported by a FPU fellowship from the Spanish Ministry of Education (FPU12/05528). K. H. V. was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under Award Number K08DE025292. Research reported in this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank Thomas Ferrante for assistance with confocal reflectance microscopy, Ana Senra for assistance with immunohistochemical analysis and Dr. J. Li, Dr. S. T. Koshy and Dr. A. S. Cheung for scientific discussions.
Footnotes
Conflict of interest
The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aini Hailati, Itaka Keiji, Fujisawa Ayano, Uchida Hirokuni, Uchida Satoshi, Fukushima Shigeto, Kataoka Kazunori, Saito Taku, Chung Ung-il, and Ohba Shinsuke. 2016. “Messenger RNA Delivery of a Cartilage-Anabolic Transcription Factor as a Disease-Modifying Strategy for Osteoarthritis Treatment.” Scientific Reports 6 (November 2015). Nature Publishing Group: 18743. doi: 10.1038/srep18743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson Erin M, Silva Eduardo A, Hao Yibai, Martinick Kathleen D, Lewin Sarah A, Stafford Alexander G, Doherty Elisabeth G, Wang Lin, Doherty Edward J, Grossman Paul M, Mooney Dave J, and D Ph. 2017. “VEGF and IGF Delivered from Alginate Hydrogels Promotes Stable Perfusion Recovery in Ischemic Hindlimbs of Aged Mice and Rabbits” 54 (5): 288–98. doi: 10.1159/000479869.VEGF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan Biji, and Banerjee R 2011. “Biopolymer-Based Hydrogels for Cartilage Tissue Engineering.” Chemical Reviews 111 (8): 4453–74. doi: 10.1021/cr100123h. [DOI] [PubMed] [Google Scholar]
- Balmayor Elizabeth R., Geiger Johannes P., Koch Christian, Aneja Manish K., Martijn van Griensven Carsten Rudolph, and Plank Christian. 2016. “Modified MRNA for BMP-2 in Combination with Biomaterials Serves as a Transcript-Activated Matrix for Effectively Inducing Osteogenic Pathways in Stem Cells.” Stem Cells and Development 26 (1): 25–34. doi: 10.1089/scd.2016.0171. [DOI] [PubMed] [Google Scholar]
- Bi Weimin, Jian Min Deng Zhaoping Zhang, Behringer Richard R, and De Crombrugghe Benoit. 1999. “Sox9 Is Required for Cartilage Formation.” Nature Genetics 22 (may): 85–89. [DOI] [PubMed] [Google Scholar]
- Branco da Cunha Cristiana, Klumpers Darinka D., Li Weiwei A., Koshy Sandeep T., Weaver James C., Chaudhuri Ovijit, Granja Pedro L., and Mooney David J. 2014. “Influence of the Stiffness of Three-Dimensional Alginate/Collagen-I Interpenetrating Networks on Fibroblast Biology.” Biomaterials 35 (32): 8927–36. doi: 10.1016/j.biomaterials.2014.06.047. [DOI] [PubMed] [Google Scholar]
- C Hansen KC and Tyler JK, Das Lucia M S. 2017. “Substrate Stress-Relaxation Regulates Scaffold Remodeling and Bone Formation in Vivo.‖ Advanced Healthcare Materials 176 (3): 139–48. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Tingjie, Xie Jie, Li Hongzhuo, Li Dong, Liu Ping, and Hu Yihe. 2016. “MicroRNA-30a Promotes Extracellular Matrix Degradation in Articular Cartilage via Downregulation of Sox9.” Cell Proliferation 49 (2): 207–18. doi: 10.1111/cpr.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri Ovijit, Gu Luo, Darnell Max, Klumpers Darinka, Bencherif Sidi A., Weaver James C., Huebsch Nathaniel, and Mooney David J. 2015. “Substrate Stress Relaxation Regulates Cell Spreading.” Nature Communications 6 Nature Publishing Group: 6365. doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri Ovijit, Gu Luo, Klumpers Darinka, Darnell Max, Bencherif Sidi A., Weaver James C., Huebsch Nathaniel, Lee Hong-pyo, Lippens Evi, Duda Georg N., and Mooney David J. 2015. “Hydrogels with Tunable Stress Relaxation Regulate Stem Cell Fate and Activity.” Nature Materials 15 (3): 326–34. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri Ovijit, Koshy Sandeep T., Da Cunha Cristiana Branco, Shin Jae Won, Verbeke Catia S., Allison Kimberly H., and Mooney David J. 2014. “Extracellular Matrix Stiffness and Composition Jointly Regulate the Induction of Malignant Phenotypes in Mammary Epithelium.” Nature Materials 13 (10): 970–78. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- Chavez RD, Coricor G, Perez J, Seo HS, and Serra R 2017. “SOX9 Protein Is Stabilized by TGF-β and Regulates PAPSS2 MRNA Expression in Chondrocytes.” Osteoarthritis and Cartilage 25 (2). Elsevier Ltd: 332–40. doi: 10.1016/j.joca.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Cathy, and Kong Hyunjoon. 2012. “Interplay of Cell Adhesion Matrix Stiffness and Cell Type for Non-Viral Gene Delivery.” Acta Biomaterialia 8 (7). Acta Materialia Inc.: 2612–19. doi: 10.1016/j.actbio.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Coricor George, and Serra Rosa. 2016. “TGF-β Regulates Phosphorylation and Stabilization of Sox9 Protein in Chondrocytes through P38 and Smad Dependent Mechanisms.” Scientific Reports 6 (August). Nature Publishing Group: 1–11. doi: 10.1038/srep38616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin Rebecca L., Ni Mengwei, Meretoja Ville V., Kasper F. Kurtis, and Mikos Antonios G. 2014. “TGF-Β3-Induced Chondrogenesis in Co-Cultures of Chondrocytes and Mesenchymal Stem Cells on Biodegradable Scaffolds.” Biomaterials 35 (1). Elsevier Ltd: 123–32. doi: 10.1016/j.biomaterials.2013.09.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVine John, Dettori Joseph, France John, Brodt Erika, and McGuire Robert. 2012. “The Use of RhBMP in Spine Surgery: Is There a Cancer Risk?” Evidence-Based Spine-Care Journal 3 (02): 35–41. doi: 10.1055/s-0031-1298616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaliwal Anandika, Maldonado Maricela, Han Zenas, and Segura Tatiana. 2010. “Differential Uptake of DNA-Poly(Ethylenimine) Polyplexes in Cells Cultured on Collagen and Fibronectin Surfaces.” Acta Biomaterialia 6 (9). Acta Materialia Inc.: 3436–47. doi: 10.1016/j.actbio.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaliwal Anandika, Oshita Victor, and Segura Tatiana. 2013. “Transfection in the Third Dimension.” Integr Biol (Camb.) 5 (10): 1206–16. doi: 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy Peter, Wang Weihuan, Bhattaram Pallavi, Wang Qiuqing, Wang Lai, Ballock R Tra, and Lefebvre Véronique. 2012. “Sox9 Directs Hypertrophic Maturation and Blocks Osteoblast Differentiation of Growth Plate Chondrocytes.” Developmental Cell 22 (3): 597–609. doi: 10.1016/j.devcel.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elangovan Satheesh, Khorsand Behnoush, Anh Vu Do Liu Hong, Dewerth Alexander, Kormann Michael, Ross Ryan D., Sumner D. Rick, Allamargot Chantal, and Salem Aliasger K. 2015. “Chemically Modified RNA Activated Matrices Enhance Bone Regeneration.” Journal of Controlled Release 218 Elsevier B.V.: 22–28. doi: 10.1016/j.jconrel.2015.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler Adam J., Sen Shamik, Sweeney H. Lee, and Discher Dennis E. 2006. “Matrix Elasticity Directs Stem Cell Lineage Specification.” Cell 126 (4): 677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Epstein NancyE. 2013. “Complications Due to the Use of BMP/INFUSE in Spine Surgery: The Evidence Continues to Mount.” Surgical Neurology International 4 (6): 343. doi: 10.4103/2152-7806.114813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojgini Shiva, Tokatlian Talar, and Segura Tatiana. 2011. “Utilizing Cell-Matrix Interactions to Modulate Gene Transfer to Stem Cells inside Hyaluronic Acid Hydrogels.” Molecular Pharmaceutics 8 (5): 1582–91. doi: 10.1021/mp200171d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf Thomas, and Enver Tariq. 2009. “Forcing Cells to Change Lineages.” Nature 462 (7273). Nature Publishing Group: 587–94. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Guilak Farshid, Cohen DM, and Estes BT. 2009. “Control of Stem Cell Fate by Physical Interactions with the Extracellular Matrix.” Cell Stem Cell 5 (1): 17–26. doi: 10.1016/j.stem.2009.06.016.Control. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Xing Rong, Li Wang Xiao, Li Man Chol, Yuan Ya Hong, Chen Yun, Zou Dan Dan, Bian Liu Jiao, and Li Dong Sheng. 2014. “PDX-1 MRNA-Induced Reprogramming of Mouse Pancreas-Derived Mesenchymal Stem Cells into Insulin-Producing Cells in Vitro.” Clinical and Experimental Medicine 15 (4): 501–9. doi: 10.1007/s10238-014-0319-0. [DOI] [PubMed] [Google Scholar]
- Hardin John A., Cobelli Neil, and Santambrogio Laura. 2015. “Consequences of Metabolic and Oxidative Modifications of Cartilage Tissue.” Nature Reviews Rheumatology 11 (9). Nature Publishing Group: 521–29. doi: 10.1038/nrrheum.2015.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderer Svenja, Lee Layland Shannon, and Schenke-Layland Katja. 2016. “ECM and ECM-like Materials - Biomaterials for Applications in Regenerative Medicine and Cancer Therapy.” Advanced Drug Delivery Reviews 97 The Authors: 260–69. doi: 10.1016/j.addr.2015.11.019. [DOI] [PubMed] [Google Scholar]
- Holcik M, and Liebhaber S a. 1997. “Four Highly Stable Eukaryotic MRNAs Assemble 3’ Untranslated Region RNA-Protein Complexes Sharing Cis and Trans Components.” Proceedings of the National Academy of Sciences of the United States of America 94 (6): 2410–14. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch Nathaniel, Arany Praveen R., Mao Angelo S., Shvartsman Dmitry, Ali Omar A., Bencherif Sidi A., Rivera-Feliciano José, and Mooney David J. 2010. “Harnessing Traction-Mediated Manipulation of the Cell/Matrix Interface to Control Stem-Cell Fate.” Nature Materials 9 (6). Nature Publishing Group: 518–26. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey Daniel J., Hu Jerry C., and Athanasiou Kyriacos A. 2012. “Unlike Bone, Cartilage Regeneration Remains Elusive.” Science 338 (6109): 917–21. doi: 10.1126/science.1222454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im Gun I., Kim Hye Joung, and Lee Jin H. 2011. “Chondrogenesis of Adipose Stem Cells in a Porous PLGA Scaffold Impregnated with Plasmid DNA Containing SOX Trio (SOX-5,−6 and −9) Genes.” Biomaterials 32 (19). Elsevier Ltd: 4385–92. doi: 10.1016/j.biomaterials.2011.02.054. [DOI] [PubMed] [Google Scholar]
- Jiang Xianfang, Huang Xianyuan, Jiang Tongmeng, Zheng Li, Zhao Jinmin, and Zhang Xingdong. 2018. “The Role of Sox9 in Collagen Hydrogel-Mediated Chondrogenic Differentiation of Adult Mesenchymal Stem Cells (MSCs).” Biomaterials Science 6 (6). Royal Society of Chemistry: 1556–68. doi: 10.1039/c8bm00317c. [DOI] [PubMed] [Google Scholar]
- Jopling Chris, Boue Stephanie, and Belmonte Juan Carlos Izpisua. 2011. “Dedifferentiation, Transdifferentiation and Reprogramming: Three Routes to Regeneration.” Nature Reviews Molecular Cell Biology 12 (2). Nature Publishing Group: 79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- Kang Daniel G., Hsu Wellington K., and Lehman Ronald A. 2017. “Complications Associated With Bone Morphogenetic Protein in the Lumbar Spine.” Orthopedics 40 (2): e229–37. doi: 10.3928/01477447-20161213-06. [DOI] [PubMed] [Google Scholar]
- Khorsand Behnoush, Elangovan Satheesh, Hong Liu, Dewerth Alexander, Kormann Michael S. D., and Salem Aliasger K. 2017. “A Comparative Study of the Bone Regenerative Effect of Chemically Modified RNA Encoding BMP-2 or BMP-9.” The AAPS Journal 19 (2). The AAPS Journal: 438–46. doi: 10.1208/s12248-016-0034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Hyun Joon, Liu Jodi, Riddle Kathryn, Matsumoto Takuya, Leach Kent, and Mooney David J. 2005. “Non-Viral Gene Delivery Regulated by Stiffness of Cell Adhesion Substrates.” Nature Materials 4 (6): 460–64. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- Kozak Marilyn. 1987. “An Analysis of 5’-Noncoding Sequences from 699 Vertebrate Messenger RNAS.” Nucleic Acids Research 15 (20): 8125–48. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyttälä Aija, Moraghebi Roksana, Valensisi Cristina, Kettunen Johannes, Andrus Colin, Kalyan Kumar Pasumarthy Mahito Nakanishi, Nishimura Ken, Ohtaka Manami, Weltner Jere, Ben Van Handel Olavi Parkkonen, Sinisalo Juha, Jalanko Anu, Hawkins R. David, Woods Niels Bjarne, Otonkoski Timo, and Trokovic Ras. 2016. “Genetic Variability Overrides the Impact of Parental Cell Type and Determines IPSC Differentiation Potential.” Stem Cell Reports 6 (2): 200–212. doi: 10.1016/j.stemcr.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Hong Pyo, Gu Luo, Mooney David J., Levenston Marc E., and Chaudhuri Ovijit. 2017. “Mechanical Confinement Regulates Cartilage Matrix Formation by Chondrocytes.” Nature Materials 16 (12): 1243–51. doi: 10.1038/nmat4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Kuen Yong, and Mooney David J. 2012. “Alginate: Properties and Biomedical Applications.” Progress in Polymer Science (Oxford) 37 (1): 106–26. doi: 10.1016/j.progpolymsci.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Pedro, Padmashali Roshan M., and Andreadis Stelios T. 2009. “Cell-Controlled and Spatially Arrayed Gene Delivery from Fibrin Hydrogels.” Biomaterials 30 (22). Elsevier Ltd: 3790–99. doi: 10.1016/j.biomaterials.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Qing Le, Wang Xiao Xun, Yan Xiao Fei, Yang Li Jun, Tang Dong Qi, and Li Dong Sheng. 2008. “Transcription Factor Directed Differentiation of Human Embryonic Stem Cells into the Pancreatic Endocrine Lineage.” Cell Research 18: S109–S109. doi: 10.1038/cr.2008.199. [DOI] [Google Scholar]
- Lin Kaili, Zhang Dawei, Macedo Maria Helena, Cui Wenguo, Sarmento Bruno, and Shen Guofang. 2019. “Advanced Collagen-Based Biomaterials for Regenerative Biomedicine.” Advanced Functional Materials 29 (3): 1–16. doi: 10.1002/adfm.201804943. [DOI] [Google Scholar]
- Lo Kevin W-H, Ulery Bret D, Ashe Keshia M, and Laurencin Cato T. 2012. “Studies of Bone Morphogenetic Protein-Based Surgical Repair.” Advanced Drug Delivery Reviews 64 (12): 1277–91. doi: 10.1016/j.addr.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel Claudia, Mauck Robert L., and Burdick Jason A. 2019. “Local Nascent Protein Deposition and Remodelling Guide Mesenchymal Stromal Cell Mechanosensing and Fate in Three-Dimensional Hydrogels.” Nature Materials 18 (8). Springer US: 883–91. doi: 10.1038/s41563-019-0307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui Kathy O., Zangi Lior, Silva Eduardo A., Bu Lei, Sahara Makoto, Li Ronald A., Mooney David J., and Chien Kenneth R. 2013. “Driving Vascular Endothelial Cell Fate of Human Multipotent Isl1 + Heart Progenitors with VEGF Modified MRNA.” Cell Research 23 (10). Nature Publishing Group: 1172–86. doi: 10.1038/cr.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi Makiko and Kennet h S. Zaret. 2014. “Pioneer Transcription Factors in Cell Reprogramming.” Genes and Development 28 (May): 2679–92. doi: 10.1210/me.2014-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal Pankaj K, and Rossi Derrick J. 2013. “Reprogramming Human Fibroblasts to Pluripotency Using Modified MRNA.” Nature Protocols 8 (3): 568–82. doi: 10.1038/nprot.2013.019. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanchez Aida, and Murphy Chris L. 2013. “MiR-1247 Functions by Targeting Cartilage Transcription Factor SOX9.” Journal of Biological Chemistry 288 (43): 30802–14. doi: 10.1074/jbc.M113.496729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead Timothy J, Wang Qiuqing, Bhattaram Pallavi, Dy Peter, and Afelik Solomon. 2013. “A Far-Upstream ( −70 Kb ) Enhancer Mediates Sox9 Auto-Regulation in Somatic Tissues during Development and Adult Regeneration.” Nucleic Acids Research 41 (8): 4459–69. doi: 10.1093/nar/gkt140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell Allyson J., and Stanger Ben Z. 2016. “Adult Cell Plasticity in Vivo: De-Differentiation and Transdifferentiation Are Back in Style.” Nature Reviews Molecular Cell Biology 17 (7). Nature Publishing Group: 413–25. doi: 10.1038/nrm.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzamohammadi Fatemeh, Papaioannou Garyfallia, and Kobayashi Tatsuya. 2014. “MicroRNAs in Cartilage Development, Homeostasis, and Disease.” Current Osteoporosis Reports 12 (4): 410–19. doi: 10.1007/s11914-014-0229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry Charles E., and Keller Gordon. 2008. “Differentiation of Embryonic Stem Cells to Clinically Relevant Populations: Lessons from Embryonic Development.” Cell 132 (4): 661–80. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Nakamura Yukio, He Xinjun, Kato Hiroyuki, Wakitani Shigeyuki, Kobayashi Tatsuya, Watanabe Sumiko, Iida Atsumi, Tahara Hideaki, Warman Matthew L., Watanapokasin Ramida, and Postlethwait John H. 2012. “Sox9 Is Upstream of MicroRNA-140 in Cartilage.‖ Applied Biochemistry and Biotechnology 166 (1): 64–71. doi: 10.1007/s12010-011-9404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathaniel P, Mow Van C, and Foster Robert J. 1998. “Composition and Dynamics of Articular Cartilage : Structure, Function, and Maintaining a Healthy State.” Journal of Ortophaedic & Sports Physical Therapy 28 (4): 203–15. [DOI] [PubMed] [Google Scholar]
- Perlstein I, Connolly JM, Cui X, Song C, Li Q, Jones PL, Lu Z, DeFelice S, Klugherz B, Wilensky R, and Levy RJ 2003. “DNA Delivery from an Intravascular Stent with a Denatured Collagen-Polylactic-Polyglycolic Acid-Controlled Release Coating: Mechanisms of Enhanced Transfection.” Gene Therapy 10 (17): 1420–28. doi: 10.1038/sj.gt.3302043. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW 2001. “A New Mathematical Model for Relative Quantification in Real-Time RT-PCR.” Nucleic Acids Research 29 (9): 45e – 45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisin Sophie, Belamie Emmanuel, and Morille Marie. 2016. “Non-Viral Gene Activated Matrices for Mesenchymal Stem Cells Based Tissue Engineering of Bone and Cartilage.” Biomaterials 104 Elsevier Ltd: 223–37. doi: 10.1016/j.biomaterials.2016.07.017. [DOI] [PubMed] [Google Scholar]
- Sahin Ugur, Karikó Katalin, and Türeci Özlem. 2014. “MRNA-Based Therapeutics — Developing a New Class of Drugs.” Nature Reviews Drug Discovery 13 (10). Nature Publishing Group: 759–80. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- Schindelin Johannes, Ignacio Arganda-Carreras Erwin Frise, Kaynig Verena, Longair Mark, Pietzsch Tobias, Preibisch Stephan, Rueden Curtis, Saalfeld Stephan, Schmid Benjamin, Jean Yves Tinevez Daniel James White, Hartenstein Volker, Eliceiri Kevin, Tomancak Pavel, and Cardona Albert. 2012. “Fiji: An Open-Source Platform for Biological-Image Analysis.” Nature Methods 9 (7): 676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova Lioudmila V., Sharov Alexei A., Nedorezov Timur, Piao Yulan, Shaik Nabeebi, and Ko Minoru S H. 2009. “Database for MRNA Half-Life of 19 977 Genes Obtained by DNA Microarray Analysis of Pluripotent and Differentiating Mouse Embryonic Stem Cells.” DNA Research 16 (1): 45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard Jaclyn A., Huang Alyssa, Shikanov Ariella, and Shea Lonnie D. 2010. “Balancing Cell Migration with Matrix Degradation Enhances Gene Delivery to Cells Cultured Three-Dimensionally within Hydrogels.” Journal of Controlled Release. doi: 10.1016/j.jconrel.2010.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiva Gojgini, Talar Tokatlian, and Segura Tatiana. 2011. “Utilizing Cell-Matrix Interactions to Modulate Gene Transfer to Stem Cells Inside Hyaluronic Acid Hydrogels.” Molecular Pharmaceutics 8 (5): 1582–91. doi: 10.1016/j.humov.2008.02.015.Changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J, Wirtz D, and Hanes J 2003. “Efficient Active Transport of Gene Nanocarriers to the Cell Nucleus.” Proceedings of the National Academy of Sciences 100 (7): 3878–82. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Kazutoshi, and Yamanaka Shinya. 2016. “A Decade of Transcription Factor-Mediated Reprogramming to Pluripotency.” Nature Reviews Molecular Cell Biology 17 (3). Nature Publishing Group: 183–93. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- Tani Kenzaburo. 2015. “Towards the Safer Clinical Translation of Human Induced Pluripotent Stem Cell–Derived Cells to Regenerative Medicine.” Molecular Therapy - Methods and Clinical Development 2. Official journal of the American Society of Gene & Cell Therapy: 15032. doi: 10.1038/mtm.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tew Simon R., and Clegg Peter D. 2011. “Analysis of Post Transcriptional Regulation of SOX9 MRNA during in Vitro Chondrogenesis.” Tissue Engineering - Part A 17 (13–14): 1801–7. doi: 10.1089/ten.tea.2010.0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tew Simon R., Peffers Mandy J., McKay Tristan R., Lowe Emma T., Khan Wasim S., Hardingham Timothy E., and Clegg Peter D. 2009. “Hyperosmolarity Regulates SOX9 MRNA Posttranscriptionally in Human Articular Chondrocytes.” American Journal of Physiology - Cell Physiology 297 (4): 898–906. doi: 10.1152/ajpcell.00571.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounson Alan, and DeWitt Natalie D. 2016. “Pluripotent Stem Cells Progressing to the Clinic.” Nature Reviews Molecular Cell Biology 17 (3). Nature Publishing Group: 194–200. doi: 10.1038/nrm.2016.10. [DOI] [PubMed] [Google Scholar]
- Vining Kyle H., and Mooney David J. 2017. “Mechanical Forces Direct Stem Cell Behaviour in Development and Regeneration.” Nature Reviews Molecular Cell Biology 18 (12). Nature Publishing Group: 728–42. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Dan, Tai Phillip W.L., and Gao Guangping. 2019. “Adeno-Associated Virus Vector as a Platform for Gene Therapy Delivery.” Nature Reviews Drug Discovery 18 (5). Springer US: 358–78. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Ying A, Yu Xiong, Silverman Philip M, Harris Robin L, and Edward H. 2007. “Nanoscale Cell Adhesion Ligand Presentation Regulates Non- Viral Gene Delivery and Expression Hyun.” Nano Letters 7 (1): 161–66. doi: 10.1016/j.jmb.2008.10.054.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren Luigi, Manos Philip D., Ahfeldt Tim, Yuin Han Loh Hu Li, Lau Frank, Ebina Wataru, Mandal Pankaj K., Smith Zachary D., Meissner Alexander, Daley George Q., Brack Andrew S., Collins James J., Cowan Chad, Schlaeger Thorsten M., and Rossi Derrick J. 2010. “Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified MRNA.” Cell Stem Cell 7 (5): 618–30. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamizu Kohei, Piao Yulan, Sharov Alexei A., Zsiros Veronika, Yu Hong, Nakazawa Kazu, Schlessinger David, and Ko Minoru S.H. 2013. “Identification of Transcription Factors for Lineage-Specific ESC Differentiation.” Stem Cell Reports 1 (6). Elsevier: 545–59. doi: 10.1016/j.stemcr.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannas IV, Lee E, Orgill DP, Skrabut EM, and Murphy GF. 1989. “Synthesis and Characterization of a Model Extracellular Matrix That Induces Partial Regeneration of Adult Mammalian Skin.” Proceedings of the National Academy of Sciences of the United States of America 86 (3): 933–37. doi: 10.1073/pnas.86.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Haiyuan, Lee Moo-yeal, Hogg Michael G, Dordick Jonathan S, and Sharfstein Susan T. 2010. “Gene Delivery in Three-Dimensional Nanoparticles.” ACS Nano 4 (8): 4733–43. [DOI] [PubMed] [Google Scholar]
- Zhang Haiyuan, Moo Yeal Lee Michael G. Hogg, Dordick Jonathan S., and Sharfstein Susan T. 2012. “High-Throughput Transfection of Interfering RNA into a 3D Cell-Culture Chip.” Small 8 (13): 2091–98. doi: 10.1002/smll.201102205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.