Abstract
Antigenic variation and viral evolution have thwarted traditional influenza vaccination strategies. The broad protection afforded by a “universal” influenza vaccine may come from immunogens that elicit humoral immune responses targeting conserved epitopes on the viral hemagglutinin (HA), such as the receptor-binding site (RBS). Here, we engineered candidate immunogens that use non-circulating, avian influenza HAs as molecular scaffolds to present the broadly neutralizing RBS epitope from historical, circulating H1 influenzas. These “resurfaced” HAs (rsHAs) remove epitopes potentially targeted by strain-specific responses in immune-experienced individuals. Through structure-guided optimization we improved two antigenically different scaffolds to bind a diverse panel of pan-H1 and H1/H3 cross-reactive bnAbs with high affinity. Subsequent serological and single germinal center B cell analyses from murine prime-boost immunizations show that the rsHAs are both immunogenic and can augment the quality of elicited RBS-directed antibodies. Our structure-guided, RBS grafting approach provides candidate immunogens for selectively presenting a conserved viral epitope.
Keywords: influenza hemagglutinin, immunogen design, protein engineering, broadly neutralizing antibodies
Graphical Abstract
Influenza evolves primarily at the human population level and within its animal reservoirs (swine and avian)1. Influenza A viruses include 2 groups containing a total of 18 subtypes that are defined by genetic and serologic characteristics of the viral glycoproteins, hemagglutinin (HA) and neuraminidase (NA). Two influenza A subtypes, H1N1 and H3N2, and two influenza B lineages, Victoria and Yamagata, currently cocirculate in the human population1. Host humoral pressure, which predominantly targets the viral HA, selects for influenza mutations that render previous immune responses suboptimal. The humoral response then evolves, through immune memory and further B cell affinity maturation2–5. The net effect of this on-going selection across the entire population exposed to the virus is a virus-immunity “arms race”. The repeated exposure to influenza in the human population results in preexisting immunity which influences subsequent immune responses6–11. This immunological memory12, 13 presents a significant hurdle towards the development of a “universal” influenza vaccine. Strategies that both overcome the recall of refined, strain-specific responses and elicit broadly neutralizing antibodies (bnAbs) are necessary.
bnAbs against influenza HA target two relatively invariant epitopes - the receptor binding site (RBS) on the HA “head” and a surface along the HA “stem”14. While stem-directed immunogens are in clinical development, efforts focusing on the RBS have lagged behind14. A significant challenge for RBS-directed immunogens is presentation of the complex RBS structure that includes multiple segments, separated in linear sequence, but adjacent in conformational space15. While computational design of novel protein scaffolds has been done for HIV and RSV, the grafted epitopes were often less-complex (e.g., a single alpha-helix)16, 17.
To overcome the significant hurdle of de novo protein design we hypothesized that the RBS epitope from one HA subtype could be transplantable onto another antigenically distinct HA. We used non-circulating, avian influenza HAs as molecular scaffolds to present the RBS from circulating H1 influenzas. These resurfaced HA (rsHA) scaffolds present the H1 conserved RBS recognized by bnAbs and alter other epitopes targeted by strain-specific responses in immune-experienced individuals. The crystal structure of one scaffold in complex with a bnAb allowed for further structure-guided optimization of two antigenically distinct scaffolds to bind a diverse panel of pan-H1 and H1/H3 cross-reactive bnAbs. Immunization with a recombinant H1 HA followed by a single, heterologous boost with our rsHA immunogen showed comparable levels of RBS-directed antibody response to the H1 homologous prime-boost regimen. These data suggest that these rsHA immunogens with further optimization of the vaccine regimen may provide a pathway to a universal influenza vaccine, by exploiting the immunogenicity of the conserved RBS.
RESULTS
Grafting the H1 SI-06 RBS onto acceptor HA scaffolds.
As a proof of principle, we chose the circulating H1 RBS epitope as the basis of a donor graft to scaffold onto HA subtypes not currently circulating in the human population (Fig. 1a). H1 influenzas can be grouped into roughly three antigenic clusters with prototypical members represented by H1 Massachusetts/1/1990 (H1 MA-90), H1 Solomon Islands/03/06 (H1 SI-06) and H1 California/04/2009 (H1 CA-09) (Fig. 1b and Fig. S1)6. Importantly, bnAb have been identified that can span these antigenic clusters (e.g., CH6718, 641 I-919, Ab664920 and 5J821). We used the H1 SI-06 as the initial donor and defined four segments, S1–S4 (Fig. 1b–e), comprising the RBS epitope for grafting (Fig. 1b). These segments include 7 of the 13 critical residues that contact the receptor, sialic acid (Fig. 1e); these 13 residues define the RBS “core” (Fig. S1). Many of the remaining residues not included in the graft (e.g., Y95 and W153) are in the base of the RBS and they are nearly invariant across influenza subtypes. For the initial molecular scaffolds, we selected two non-circulating group 2 influenzas, H4N6 A/America black duck/New Brunswick/00464/2010 (H4 NB-10) and H14N6 A/mallard/Wisconsin/10OS3941/2010 (H14 WI-10). We selected them because they have little sequence similarity to circulating H1s (Fig. S2). The acceptor HA S1-S4 boundaries were defined by aligning the H1 SI-06 sequence (Fig. 1c). The rsHAs have the following nomenclature: “rsH4NBvX”; the resurfaced (rs) HA scaffold subtype (H4), with an abbreviated strain name (NB) and different versions (vX). We could successfully overexpress the inter-group transfer of the H1 SI-06 RBS graft onto the H4 and H14 scaffolds resulting in our first generation rsH4NBv1 and rsH14WIv1 scaffolds (Fig. S3).
Figure 1: RBS grafts and sequence alignments.
A, Phylogeny of influenza subtypes. Group 1 and 2 influenzas are annotated. Highlighted in pink are the seasonal subtypes currently circulating in the human population. B, Representative H1 antigenic clusters: H1 Massachusetts/1/1990 (MA-90), H1 Solomon Islands/03/2006 (SI-06) and H1 California/07/2009 (CA-09) are listed. Sequence alignment is in reference to SI-06 and conserved residues are marked as (.); segments defining the H1 RBS graft, S1–4, are colored. C, Residues comprising S1–4 of the acceptor scaffolds (H3 numbering) from non-circulating influenzas H4 New Brunswick/00464/2010 (H4 NB-10), H6 Wisconsin/617/1983 (H6 WI-83), H14 Wisconsin/10OS3941/2006 (H14 WI-06) and H16 Delaware Bay/296/1998 (H16 DB-98). D, Influenza HA trimer (PDB 5UGY) in surface representation. HA1 is in silver, HA2 in dark gray and S1–4 are colored. e, LSTc (sialylneolacto-N-tetraose c, stick representation) modeled in complex with HA. S1–4 is colored and HA is in silver.
Binding affinities of bnAbs to rsHAs.
We determined the binding affinities of a panel of RBS-directed Fabs to these initial scaffolds by using biolayer interferometry (BLI). This panel included four pan-H1 (CH67, 641 I-9, H2526 and H2227) and two H1/H3 cross-reactive bnAbs (K03.12 and C05) that engage the RBS19, 21–25. Each antibody has a footprint that overlaps with the RBS core but has different angles of approach and peripheral contacts (Fig. S4). As seen in Table 1, neither scaffold bound all the RBS-directed Fabs. rsH4NBv1 bound only K03.12 with an equilibrium dissociation constant (KD) ~5.2x greater than wildtype H1 SI-06. rsH14WIv1 bound CH67, K03.12 and C05 with KDs ~17x, ~2.7x and ~1.3x greater than wildtype H1 SI-06. None of the antibodies bound wildtype H4 NB-10 or H14 WI-10. These data suggest that there are peripheral residues in the first-generation scaffolds that impede bnAb binding and/or that the RBS graft is being presented in an altered conformation.
Table 1. Affinity measurements of rsHAs to a panel of RBS-directed Abs.
KDs (in μM) were obtained by applying a 1:1 binding isotherm using vendor-supplied software with at least three independent concentrations. The VH gene usages are listed for each antibody. KDs beyond the limit of detection are reported as >100 μM. Values are for monomeric HA heads and Fabs.
| CH67 VH1~2 |
641 I-9 VH4~59 |
H2526 VH1~69 |
H2227 VH4~4 |
K03.12 VH1~2 |
C05 VH3~23 |
|
|---|---|---|---|---|---|---|
| H1 SI-06 | 0.57 | 0.67 | 0.56 | 0.39 | 0.83 | 1.1 |
| H4 NB-10 | >100 | >100 | >100 | >100 | >100 | >100 |
| H14 WI-10 | >100 | >100 | >100 | >100 | >100 | >100 |
| rsH4NBv1 | >100 | >100 | >100 | >100 | 4.3 | >100 |
| rsH4NBv3 | 1.6 | 17.7 | 0.93 | 0.67 | 0.65 | 0.34 |
| rsH14WIv1 | 9.5 | >100 | >100 | >100 | 2.2 | 1.4 |
| rsH14WIv2 | 0.31 | 2.7 | 0.20 | 1.0 | 1.1 | 0.065 |
Structure of rsH4NBv1 in complex with bnAb, K03.12.
To identify further modifications on the first-generation scaffold that can be engineered to improve affinity to the bnAb panel, we determined the crystal structure of the rsH4NBv1 head in complex with the cross-reactive H1/H3 K03.12 antibody (Fig. 2a, Table S1). Like the previously characterized C05 antibody, K03.12 engages the RBS with almost exclusively CDR H3–dependent contacts22. The antibody contacts 14 of the 26 residues in the S1-S4 RBS graft. Additional contacts are made with conserved residues critical for sialic acid interactions in the base of the RBS including Y95, W153, T155 and H183 (Fig. 2b, H3 numbering). Comparison of the K03.12-rsH4NBv1 structure and the K03.12-H3 Texas/50/2012 complex shows a nearly identical approach with a slight twist and rocking of the VH-VL towards HA about the principle axis (Figs. S5a, b)23. The contacting residues within the antigen-combining sites between the two structures are nearly identical (Figs. S5c, d). A comparison of the rsH4NBv1 HA to a wildtype H4 A/duck/Czechoslovakia/1956 (PDB 5XL3) and wildtype H1 SI-06 (PDB 5UGY) shows a displacement about S2 (150-loop) with a shift of ~3Å, however the overall conformation in the other grafted RBS segments is similar to that of the wildtype H1 SI-06 RBS (Fig. 2c).
Figure 2: Structure of K03.12 in complex with rsH4NBv1 and scaffold improvement.
A, Antibody K03.12 Fab (heavy and light chains are colored blue and green, respectively) in complex with rsH4NBv1 HA1 “head” (silver). The CDR H3 (magenta) is marked. B, Close-up of the antigen-binding site. The CDR H3 (magenta) is shown in sticks with key interacting HA residues (silver). Hydrogen bonds are denoted in yellow, dashed-lines. C, Comparison of the RBS of H1 SI-06, yellow, (PDB 4YJZ), H4 A/duck/Czechoslovakia/1956 (H4 CZ-56), black (PDB 5XL3) and rsH4NBv1, silver. The segments of the grafts are labeled and colored. D, rsH4NBv2 and e, rsH4NBv3 with residues changed for each construct represented by a colored sphere. The original, unchanged segments from rsH4NBv1 are labeled and colored.
Structure-guided improvement of the rsHA scaffolds.
To improve scaffold binding to the RBS-directed bnAb panel, we docked the previously determined Fab structures onto the determined rsH4NBv1 structure to identify residues that may be modified to either alleviate steric clashes and/or reinforce interactions. We engineered two additional (scaffold) versions: 1) rsH4NBv2 had three mutations K131T, T192R and N193A (Fig. 2d) and 2) rsH4NBv3, had four additional mutations, N145S, K196H, N198E and S219K (Fig. 2e); no changes were made in the original grafted segments. We first assayed for scaffold improvement using an enzyme-linked immunosorbent assay (ELISA) (Fig. 3). None of the antibodies bound to wildtype H4 NB-10 (Fig. 3a) consistent with our BLI data using Fabs (Table 1). The second-generation scaffold increased affinity to four of the five Fabs (Fig. 3b, c) while the third generation, rsH4NBv3, resulted in high-affinity binding to all five RBS-directed antibodies (Fig. 3c). Based on this optimized rsH4NBv3 construct, we asked whether the same seven mutations could be made in context of the rsH14WIv1 scaffold to increase its affinity for the entire panel of RBS-directed antibodies. Indeed, when these mutations were engineered into rsH14WIv1 this optimized construct bound the entire panel of RBS-directed antibodies with high affinity (Fig. 3g–i). For reference, H1 SI-06 reactivity to the RBS-directed Ab panel is shown (Fig. 3i). A summary of the KDs are shown in Figs. 3e, 3j.
Figure 3: Reactivity of RBS-directed IgGs for rsHAs.
CH67 (blue), H2526 (red), H2227 (green), 641 I-9 (violet) and K03.12 (orange) IgGs were titrated against A, wildtype H4 NB-10, B, rsH4NBv1, C, rsH4NBv2, D, rsH4NBv3, E, KDs obtained from curves in A-D for H4 constructs displayed as a “heatmap”: warm colors are high affinity and cool colors low affinity. The legend is in M. IgG titrations for F, wildtype H14 WI-10, G, rsH14WIv1, H, rsH14WIv2 and I, control H1 SI-06 HA constructs. J, KDs obtained from curves in F-I for H14 constructs. ELISA measurements were done in duplicate over the concentration range except for A, and F, where only 1μM final concentration of IgG was tested. The wildtype H1 SI-06 values in E and J are both derived from I. Curve fitting was done with a nonlinear regression model and KDs were determined using GraphPad Prism version 8.0.
Finally, we tested the optimized scaffolds in BLI to obtain more accurate KDs, free of avidity effects present when using IgGs in ELISA. As seen in Table 1, both optimized scaffolds significantly increased affinities over their first-generation counterparts; rsHAs based on both optimized scaffolds bound all six bnAbs included in the panel. In particular, the rsH14WIv2 scaffold yielded affinities for CH67, H2526 and C05 even greater than those of wildtype H1 SI-06. In some cases (e.g., C05) the optimized resurfaced scaffold bound the Fab >10-fold more tightly than did wildtype H1 SI-06. In general, the rsH14WIv2 had affinities closest to the wildtype H1 SI-06. These data suggest a set of key residues, in addition to the initial RBS-donor grafts, that can be grafted onto other potential scaffolds to present an “optimized” epitope to bind (or elicit) a diverse set of RBS-directed bnAbs.
Serum and germinal center B cell responses to the rsHA immunogen.
To evaluate whether our rsHA immunogens can enrich for RBS-directed responses by allowing the expansion and differentiation of memory B cells, we first generated a knock-in (KI) mouse that has a human JH6 and D2–2 segments (Fig. S7 and Table S2). The human JH6 and D2–2 segments were chosen to mimic two key features of observed human-RBS directed bnAbs that are absent in the wildtype C57BL/6 murine model: 1) a poly-tyrosine motif in the CDR H3 loop and 2) an overall CDR H3 length of 19–20 amino acids necessary for engaging the RBS19. We therefore immunized these generated mice (see Methods) with recombinant H1 SI-06 HA, to prime humoral immunity to an H1, then boosted with our recombinant rsH4NBv3 (Fig. 4a). We compared the resulting serum and germinal center (GC) responses originating from a homologous H1 SI-06/H1 SI-06 or a heterologous H1 SI-06/rsH4NBv3 prime/boost immunization. We found that post-boost, the serum IgG titers, as well as the relative serum reactivity to the initial H1 SI-06 HA were comparable between the two cohorts (Fig. S6a). To deconvolute the contribution of the H4 scaffold-dependent Abs, we performed a serum competition assay with H1 RBS-directed bnAb CH67 (Fig. 4b) and found no demonstrable difference between the two cohorts. At the cellular level, we noted that the GC B cell frequencies were ~3-fold higher in mice boosted with recombinant rsH4NBv3 (Fig. 4c and S8). These data potentially point to disparities in immunogenicity between H1 SI-06 and rsH4NBv3 and highlight the apparent immunodominance of the H4 scaffold. To quantify the serum abundance of RBS-directed antibodies, we screened the sera from the two cohorts against H1 SI-06 and rsH4NBv3 HAs in a Luminex assay (Fig. 4d, e). Sera from the heterologous immunization reacted more strongly with rsH4NBv3 (Fig. 4e and S6b) and as strongly as those from the homologous one with H1 SI-06 (Fig. 4d and S6c). These data highlight the immunogenicity of the resurfaced HA and its potential as a novel antigen.
Figure 4: in vivo assessment of rsHA immunogenicity.
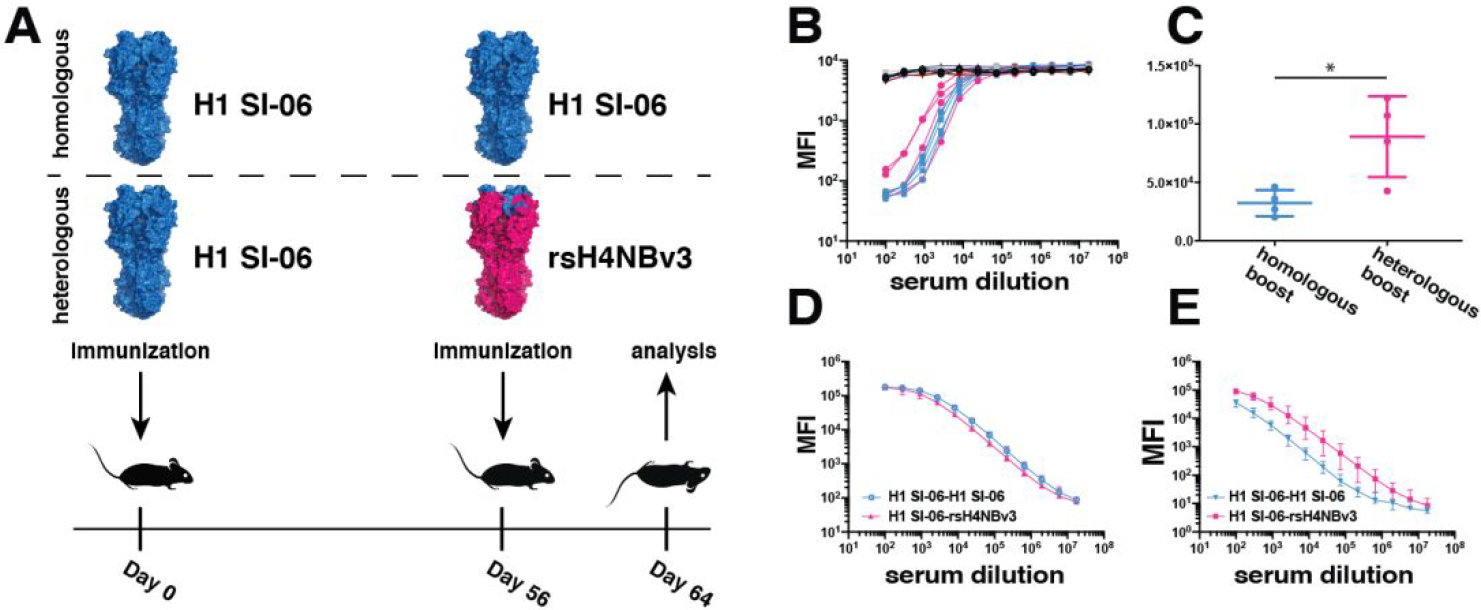
A, schematic representation of the immunization strategy B, CH67 competition to H1 SI-06 FLsE HA against the sera from wild-type non-immunized B6 mice (black), JH6 mice primed and boosted with H1 SI-06 (blue) and mice primed with H1 SI-06, then boosted with rsH4NBv3 (pink), C, total GC B cell counts in the animal cohorts. n=4 mice for B6 and n=4 mice for JH6. *p < 0.05, Welch’s t test. Error bars represent SD, D, serum reactivity against H1 SI-06, e, serum reactivity against rsH4NBv3. The mean values are plotted for d and e with error bars indicating SD.
Comparison of single GC B cell responses to wild-type and resurfaced HAs.
To complement our serological study and to understand more fully HA RBS targeting, we assessed the reactivity of individual GC B cells isolated post immunization using our single-cell Nojima culture system (Fig. 5). We screened the culture supernatants from each immunization (homologous or heterologous) against both wild-type H1 SI-06 and rsH4 NB-10 in a competition assay with a known RBS-targeting mAb CH67 (Fig. 5a). The competition was performed on Luminex beads and the reduction in fluorescent signal, resulting from antibody competition, was expressed as % inhibition. We defined competitors as antibodies with 75% inhibition of CH67 mAb binding or greater. The data show that the homologous immunization (wild-type H1 SI-06 prime-boost) elicited 7 mAbs with such properties. Those mAbs competed for CH67 binding only on wild-type H1 SI-06 and not on rsH4 NB-10 beads, suggesting that the footprint of these antibodies on HA extends beyond the minimal, conserved RBS present on both H1 SI-06 and rs H4 NB-10 making variable peripheral contacts. While the heterologous immunization elicited a comparable number of antibodies competing with CH67, 5 of 6 antibodies competed on both H1 SI-06 and rsH4 NB-10 beads suggesting that these antibodies engage the minimal grafted RBS epitope, shared between the two HAs. These data point to qualitative differences between antibodies elicited by the two immunization regimens. To further characterize these potential differences, we asked whether there was a difference in breadth of binding to historical H1 HAs. We selected a representative antibody from each immunization group (Ab18: homologous; Ab144: heterologous immunization) to recombinantly express and to test for binding to an HA panel. Figure 5b shows binding data this HA panel using Fabs and monomeric HA heads to avoid any avidity effects obtained from IgGs and trimeric HA FLsE. Ab18 bound only to the immunizing strain H1 SI-06 with modest affinity. However, Ab144 bound to H1 SI-06, rsH4 NB-10 as well as to four additional historical H1 HAs with high affinity. Additionaly, we obtained structural information of antibody Ab133 from the heterologous immunization that is clonally-related to Ab144 (Fig. 5A and S9C) by negative stain EM. The EM 2D class averages show that the antibody binds to the RBS, in a similar way to other HA RBS antibodies26 (Fig. 5C and S9B); a stem-directed mAb FI627 is shown in Fig. S9A for comparison. These data suggest that the heterologous immunization with our resurfaced immunogens offers an advantage over homologous immunization by eliciting antibodies with increased binding breadth; this is most likely a consequence of engaging only the conserved RBS epitope shared between the HAs and minimizing variable, peripheral contacts.
Figure 5: Identification and biochemical characterization of RBS-directed antibodies.
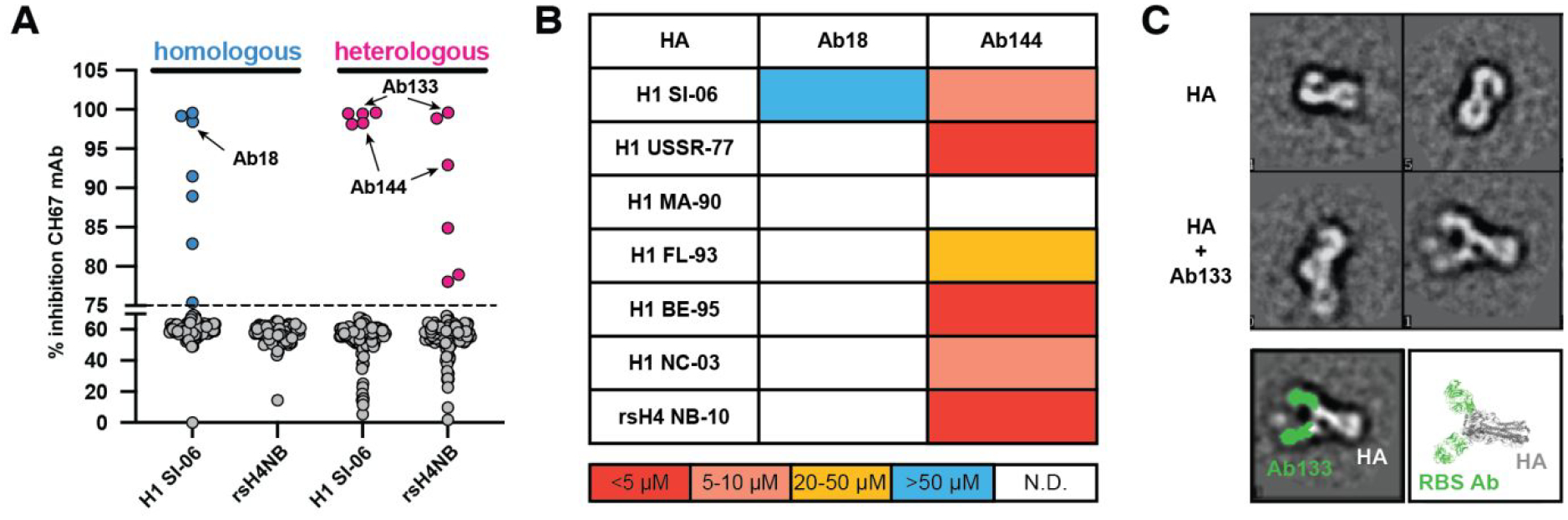
A, CH67 competition experiment against the single B cell culture supernatants from homologous (H1 SI-06; blue) and heterologous (H1 SI-06, rsH4NBv3; pink) immunizations. The competition was performed on Luminex beads coated with either H1 SI-06 or with rsH4 NB-10, antibody competition is expressed as % inhibition (x-axis); antibodies with >75% inhibition are colored. Representative monoclonal antibodies Ab18, Ab133 and Ab144 were selected for further characterization. B, Binding to a panel of historical H1 HAs and the resurfaced rsH4 NB-10 was assayed with recombinant Fabs using bio-layer interferometry. Affinity constants (KDs, in μM) were obtained by applying a 1:1 binding isotherm using vendor-supplied software. The “heatmap” color scheme is a visualization aid: warm colors are high affinity and cool colors low affinity. H1 SI-06 – A/Solomon Islands/03/2006, H1 USSR-77 – A/USSR/90/1977, H1 MA-90 – A/Massachusetts/1/1990, H1 FL-93 – A/Florida/2/1993, H1 BE-95 – A/Beijing/262/1995, H1 NC-03 – A/North Carolina/3/2003, rsH4 NB-10 - resurfaced duck/New Brunswick/00464/2010; N.D. – non-detectable. C, Visualization of the HA-Ab133 complex by negative stain electron microscopy. The top panels are of HA alone with and without crosslinking to Ab133 with glutaraldehyde. Ab133 is colored green in the bottom right panel and compared to an RBS antibody bound to HA (PDB ID: 5UG0). One copy of the RBS antibody is not shown for clarity.
DISCUSSION
Current influenza research has focused on the development of a universal influenza vaccine. Such a vaccine should induce broad immunity a) within seasonal, circulating H1 and H3 subtypes, b) across subtypes (heterosubtypic) and c) pre-pandemic (e.g., H5, H7). The pathway to achieving this broad protection will likely come from eliciting or boosting humoral responses to conserved sites on HA such as the RBS and “stem”. Immunogen design strategies thus far have focused almost exclusively on targeting the conserved stem through either selectively displaying the HA2 stem28–31 or using chimeric HAs that present circulating H1 or H3 stems with a heterologous HA1 “head”14.
The data presented here show an alternative strategy for candidate immunogens to focus the immune response to the broadly neutralizing epitope of the RBS. Grafting of the H1 SI-06 RBS epitope onto two antigenically distinct HA scaffolds exploits the overall architecture of the HA protein circumventing the significant challenge in de novo protein scaffold design. Through structure-guided engineering, the optimized scaffolds bind a diverse panel of pan-H1 and H1/H3 cross-reactive RBS antibodies that represent the type of response one might wish to elicit by a universal influenza vaccine. Collectively, the bnAbs in the panel (Table 1) bind all H1 isolates both pre-pandemic (<2009) and post-pandemic (>2009) as well as circulating H3N2 influenzas. Importantly, this collection of RBS-directed bnAbs tolerate the heterologous peripheries of the scaffolds surrounding the graft. These immunogens, therefore, would stimulate affinity maturation to refine humoral responses to the conserved, RBS core contacts shared between the scaffolds while adapting and accommodating antigenically distinct peripheries.
Our in vivo mouse immunization studies compared the resulting serum and GC responses from mice primed and boosted with either homologous or heterologous HA immunogens. Despite no discernable difference in a competition assay with bulk serum and a standard RBS-directed CH67 mAb, we noted that the heterologous immunization elicited more robust GC B cell responses, owing either to the influx of new, H4-specific clones and/or recall of the H1 RBS-specific memory B cells. Using single-cell isolation, we noted that in the heterologous immunization regimen, very few B cells that were derived from secondary GCs reacted with the grafted RBS; consistent with a recent study on the frequency of memory B cell reentry to GCs32. We did, however, note that the antibodies elicited by the heterologous immunization bound to both the wild-type H1 SI-06 (from which the graft is derived) and the resurfaced HAs, indicating that they engage only the minimal, conserved RBS that minimize, variable peripheral contacts. Consistent with this hypothesis, a representative antibody from the heterologous immunization had broader binding breadth to historical H1 HAs than did a representative antibody from the homologous control group. Although further studies involving fate labeling antigen-experienced cells and single-cell sequencing coupled with serum deconvolution will be necessary to better understand GC reentry and devise more optimal boosting strategies, our data highlight the benefit of boosting with the resurfaced HA to augment the quality of RBS-directed antibodies and mitigate strain-specific responses.
A significant hurdle for the development of a universal influenza vaccine is preexisting immunity present in the human population either through repeated seasonal vaccination or influenza infection. The HA scaffolds derived from avian influenzas that, to date, have not circulated in the human population and thus would likely avoid boosting strain-specific, memory recall responses in immune-experienced individuals. The strategy described here could be used to direct naïve immune responses to the conserved RBS through a prime-boost vaccine approach and/or could boost subdominant RBS-directed bnAbs already present in immune-experienced individuals11,19. More generally, the immunogen design approach of epitope grafting could be used for other rapidly evolving pathogens for which preexisting immunity is present (e.g., RSV and dengue).
METHODS
Expression and purification of HA
rHA1 “head” and rHA full length soluble ectodomains (FLsE) constructs were cloned into pFastBac vector for insect cell expression (Hi5 cells) or pVRC vector for mammalian expression (HEK293F cells). HAs were derived from the following templates: A/America black duck/New Brunswick/00464/2010 (H4N6) (GenBank: AGG81749.1) and A/mallard/Wisconsin/10OS3941/2010 (H14N6) (GenBank: AGE03043) (Table S3). All constructs were confirmed by DNA sequencing at the DNA Sequencing Core Facility at Dana Farber Cancer Institute. For biolayer interferometry (BLI) and crystallography the HA1 head constructs contained an HRV 3C-cleavable C-terminal His6X tag or SBP-His8Xtag. The HA FLsE constructs used in ELISA assays contained a thrombin or HRV 3C-cleavable C-terminal foldon tag with either a His6X or SBP-His8Xtag. All constructs were purified from supernatants by passage over Cobalt-TALON resin (Takara) followed by gel filtration chromatography on Superdex 200 Increase (GE Healthcare) in 10 mM Tris-HCl, 150 mM NaCl at pH 7.5. For BLI and crystallography the tags were removed using HRV 3C protease (ThermoScientific) and the protein repurified using Cobalt-TALON resin to remove the protease, tag and non-cleaved protein.
Fab and IgG expression and purification
For Fab and IgG production the genes for the heavy- and light-chain variable domains were synthesized and codon optimized by Integrated DNA Technologies and subcloned into pVRC protein expression vectors containing human heavy- and light-chain constant domains, as previously described (11, 19). Heavy-chain constructs for Fab production contained a non-cleavable His6X tag; for IgG heavy constructs there was no cleavable purification tag. Constructs were confirmed by sequencing at the DNA Sequencing Core Facility at Dana Farber Cancer Institute. Fabs and IgGs were produced by transient transfection in suspension 293F or adherent HEK 293T cells using Lipofectamine 2000 (Invitrogen) or polyethylenamine (PEI). Supernatants were harvested 4–5 days later and clarified by centrifugation. Fabs were purified using Cobalt-TALON resin (Takara) followed by gel filtration chromatography on Superdex 200 Increase (GE Healthcare) in 10 mM Tris-HCl, 150 mM NaCl at pH 7.5. IgGs were purified using Protein G Plus Agarose (ThermoFisher Scientific). Briefly, IgG supernatants were incubated overnight with agarose slurry, eluted with 0.1M glycine, pH 2.5, normalized with 1M Tris-HCl, pH 8.0 and dialyzed against PBS buffer overnight.
Crystallization and Data Collection.
rsH4NBv1 HA1 head domain and K03.12 Fab (both expressed in HEK293F cells) were incubated at 1:1.5 molar ratio, respectively. The complex was isolated by size exclusion chromatography using a 24 mL Superdex Increase equilibrated in 10 mM Tris-HCl, 150 mM NaCl. Crystallization was achieved by hanging drop vapor diffusion at 18°C. Crystals were grown in 100 mM sodium citrate (pH 4.5), 20% (wt/vol) PEG 4000. Crystals were cryoprotected in mother liquor supplemented with 25% (vol/vol) glycerol and flash-frozen in liquid nitrogen. Data were collected at 0.999 Å with a rotation of 1° per image on the 8.2.2 beamline, Advanced Light Source, at Berkeley National Laboratory.
Structure Determination and Analysis.
X-ray diffraction data were processed with XDS33.The structure was determined by molecular replacement using PHASER34, 35 with the K03.12-A/Texas/50/2012 (H3N2)-head complex (PDB ID 5W08) as a search model23. Density-modified, NCS-averaged electron density maps were generated with DM (CCP4) and were used as a guide for model building. Individual coordinate and group B factor refinement was performed using PHENIX36. Model building was done in COOT37 and assessed with MolProbity38. N-linked glycan stereochemistry was validated with Privateer39. Figures were generated using PyMOL Molecular Graphics System (v1.8.0.0; Schrödinger LLC).
Negative Stain EM
Purified SI-06 FLsE trimer was incubated with a 6-fold molar excess of FI6 Fab at 4 °C for 1 h and run over a Superpose 6 Increase (GE Healthcare) in a buffer of 5 mM HEPES, pH 7.5, 150 mM NaCl. Size exclusion chromatography was not run with Ab133 for initial imaging, to prevent Ab133 from dissociating. To keep Ab133 bound, we cross-linked by addition of 0.5% glutaraldehyde (GLA; Electron Microscopy Sciences) at 4 °C for 1 hour, after which the reaction was stopped by addition of 75 mM Tris buffer (pH 7.4) and then run over the Superpose 6 Increase column (GE Healthcare). Fractions containing complex were concentrated to 0.01 mg/mL. A 2.5 μl aliquot of the complex was applied for 30 s onto a carbon-coated 400 Cu mesh grid that had been glow discharged at 5 mA for 2 min, followed by negative staining with 0.7% uranyl formate for 20 s. Samples were imaged using a FEI Tecnai T12 microscope operated at 100 kV and a magnification of ~52,000, yielding a pixel size of 1.66 Å at the specimen plane. Images were acquired with a Gatan 4k × 4k OneView camera using a nominal defocus of 1500 nm at 0°. Particles were picked with EMAN2 and put into a particle stack.
Interferometry binding experiments
Interferometry experiments were performed using a BLItz instrument (fortéBIO, Pall Corporation). Fabs were immobilized on a Ni-NTA biosensor and cleaved rHA heads (made in insect Hi5 cells) were titrated to obtain binding affinities. Initial, single-hit concentrations, were tested at 35μM for binding and then subsequent titrations for at least three different concentrations (chosen depending on the apparent KD from the high concentration); the refined KD was obtained through global fit of the titration curves by applying a 1:1 binding isotherm using vendor-supplied software. All experiments were performed in 10 mM Tris-HCl, 150 mM NaCl at pH 7.5 and at room temperature.
ELISA
5–10ng of rHA FLsE expressed in insect Hi5 cells were adhered to high-capacity binding, 96 well-plates (Corning) overnight in PBS. Plates were blocked with non-fat dried milk in PBS containing Tween-20 (PBS-T) for 1hr at room temperature (RT). Blocking solution was discarded and 10-fold dilutions of RBS-directed IgGs in PBS were added to wells and incubated for 1hr at RT. Plates were then washed three times with PBS-T. Secondary, anti-human IgG-HRP (Abcam), in PBS-T was added to each and incubated for 1hr at RT. Plates were then washed three times with PBS-T. Plates were developed using 1-Step ABTS substrate (ThermoFisher) and immediately read using a plate reader at 410nm. Data were plotted using Prism 8 (GraphPad Software) and affinities determined by applying a nonlinear regression model.
Mice
C57BL/6 mice were obtained from the Jackson Laboratory. JH6 mice were generated as described below. Mice were bred and maintained under specific pathogen-free conditions at the Duke University Animal Care Facility. Mouse immunization experiments were conducted per the approved IRB protocol by the Duke University Institutional Animal Care and Use Committee.
Generation of human JH6 murine model
In the human JH6 mouse model, human JH6 replaces mouse JH1–JH4, and human D2–2 replaces mouse DQ52 segment. The choice of human JH6 and D2–2 was based on the observation that a substantial fraction of human anti-RBS antibodies utilize these gene segments in CDR H3. To generate the hJH6 mouse line, a targeting construct containing the hD2–2 and hJH6 cassette was transfected into an F1 ES cell line that was derived from a cross between the 129/Sv strain and the C57BL/6 strain mice. The cassette was integrated into the mouse DQ52-JH6 locus via homologous integration. The homology arm was derived from genomic DNA from the C57BL/6 mouse strain. Due to sequence polymorphisms between the 129/Sv mouse strain and C57BL/6 strain, homologous recombination preferentially integrated the hD2–2 and hJH6 cassette into the IgHb allele in the ES cell. The ES clone was injected into mouse Rag2 deficient blastocysts to generate chimeric mice. Because Rag2 is essential to V(D)J recombination, all the B and T cells were derived from the injected ES cells, and the chimeric mice can be used directly for analysis. The method is referred to as Rag2 deficient blastocyst complementation40. Fig.S7 shows analysis of the B cells in this type of chimeric mice, as compared with a WT control mouse of mixed 129/Sv and C57BL/6 genetic background. For this analysis, splenic B cells were stained with fluorophore conjugated antibodies, as indicated next to the FACS plots. To analyze the VH, D and JH usage in the Ig heavy chains from IgMb B cells (Table S2), IgMb B cells were sorted by FACS, and 5’RACE was performed to generate a library of μ heavy chains expressed by the sorted B cells. The PCR products were cloned into pGEXT-easy vector, and some of the cloned PCR products were sequenced. The identities of the VH, D and JH segments were determined by Ig Blast.
Immunizations
JH6 mice (female, 8 to 12-week-old) were immunized with 20 μg of H1 SI-06 HAs, expressed in insect Hi5 cells, in Alhydrogel® via footpad. Eight weeks later, cohorts of mice were boosted with 20 μg of either H1 SI-06 HAs or rsH4NBv3 HAs in Alhydrogel® via hock. Mice were sacrificed 8 days after prime or boost immunizations, respectively, and the draining, popliteal lymph nodes and sera were collected to measure GC and antibody responses, respectively.
Flow Cytometry
GC B cells (GL-7+B220hiCD38loIgD−CD93−CD138−) and plasmablasts/-cytes (B220loCD138hi) cells in popliteal LNs were determined as described41, 42. Labeled cells were analyzed/sorted in a FACS Canto (BD Bioscience) or a FACS Vantage with DIVA option (BD Bioscience). Flow cytometric data were analyzed with FlowJo software (Treestar Inc.). Doublets were excluded by FSC-A/FSC-H gating strategy. Cells that take up propidium iodide were excluded from our analyses.
Nojima Culture
Single B cell cultures were performed as described42. Briefly, single GC B cells were sorted for their phenotype and cultured for 10 days in the presence of NB-21.2D9 feeder cells. After culture, the supernatants were harvested for Luminex assays and culture plates were stored at −80°C for V(D)J amplifications.
BCR sequencing
V(D)J rearrangements of cultured B cells were amplified by a nested PCR, cloned, and sequenced as previously described42. The rearranged V, D, and J gene segments were identified using IMGT/V-QUEST (http://www.imgt.org/).
Luminex multiplex assay
Reactivity of clonal IgGs in culture supernatants and of mouse sera was determined by Luminex multiplex assay with modifications41. Briefly, mixtures of antigen-conjugated (H1 SI-06 or rsH4NBv3 HAs, both FLsE, made in insect Hi5 cells) microspheres were incubated with serially diluted serum samples for 2 hours at room temperature or overnight at 4°C (for competition assay) with mild agitation. Samples were diluted in PBS containing 1% cow milk, 1% BSA, 0.05% Tween 20 and 0.05% NaN3. For competition, human monoclonal IgG1 CH67 (2 ng/ml) was added to the plates without washing and plates were incubated for 2 hours at room temperature with mild agitation. After washing, PE goat anti-mouse IgG or PE mouse anti-human IgG (both from Southern Biotech, the latter for competition assay) was added to the plates and incubated for 1 hour at room temperature with mild agitation. After washing, microspheres were resuspended in PBS containing 1% BSA, 0.05% Tween 20 and 0.05% NaN3 and fluorescent signals from each microsphere were measured in a Bio-Plex® 3D machine (Bio-Rad).
Data Availability
Coordinates and structure factors have been deposited in the Protein Data Bank under accession code PDB 6UR5 for the rsH4NBv1-K03.12 complex.
Supplementary Material
Acknowledgements
We thank Stephen Harrison and members of his laboratory at Harvard Medical School for helpful discussions, in particular Kevin McCarthy. We thank Wenli Zhang, Dongmei Liao, and Xiaoe Liang at Duke University for technical assistance and mouse colony maintenance. BLI affinity measurements were carried out in the Center for Macromolecular Interactions at Harvard Medical School, directed by Kelly Arnett. We thank the beamline staff at the Advanced Light Source (Lawrence Berkeley Laboratory), beamline 8.2.2, for assistance with the recording of X-ray diffraction data. We thank Yoana Dimitrova for help with x-ray data processing. Sequencing reactions were carried out with an ABI3730xl DNA analyzer at the DNA Resource Core of Dana-Farber/Harvard Cancer Center (funded in part by NCI Cancer Center support grant 2P30CA006516-48). This research was supported by NIH grants P01 AI089618 (to S.C. Harrison), U19 AI117892 (to G. Kelsoe) and R01 AI146779 (to A.G. Schmidt). The project described was supported by award Number T32GM007753 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Competing financial interest A.G.S., G.B. and M.J.M. have filed a patent application regarding the work published in this manuscript.
Supporting information
Sequence alignment of historical H1 RBS and critical residues comprising sialic acid (SA) contacts; conservation mapping onto HA structure; Biochemical characterization of the optimized rsHA; Intermolecular contacts (footprints) of RBS-directed antibodies; Structural comparison of mAb K03.12 bound with rsH4NBv1 and H3 TX-12; Total IgG titer measurements; illustration showing the integration of human D2–2*01 (hD2–2*01) and human JH6*03 (hJH6*03) segments into the mouse IgH locus; flow cytometry gating; 2D class averages from negative stain EM are shown for HA in complex with the stem antibody, FI6 as well as a portion of a raw negative stain EM micrograph; crystallographic data collection and model refinement statistics; VH, D, JH usage of IgMb B cells; protein sequences of the different HA constructs. This information is available free of charge on the ACS Publications website.
References
- 1.Taubenberger JK; Morens DM, Influenza: the once and future pandemic. Public Health Rep 2010, 125 Suppl 3, 16–26. [PMC free article] [PubMed] [Google Scholar]
- 2.Dal Porto JM; Haberman AM; Shlomchik MJ; Kelsoe G, Antigen drives very low affinity B cells to become plasmacytes and enter germinal centers. J Immunol 1998, 161 (10), 5373–81. [PubMed] [Google Scholar]
- 3.De Silva NS; Klein U, Dynamics of B cells in germinal centres. Nature reviews. Immunology 2015, 15 (3), 137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foote J; Milstein C, Kinetic maturation of an immune response. Nature 1991, 352 (6335), 530–2. [DOI] [PubMed] [Google Scholar]
- 5.Victora GD; Nussenzweig MC, Germinal centers. Annual review of immunology 2012, 30, 429–57. [DOI] [PubMed] [Google Scholar]
- 6.Bedford T; Suchard MA; Lemey P; Dudas G; Gregory V; Hay AJ; McCauley JW; Russell CA; Smith DJ; Rambaut A, Integrating influenza antigenic dynamics with molecular evolution. eLife 2014, 3, e01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fonville JM; Wilks SH; James SL; Fox A; Ventresca M; Aban M; Xue L; Jones TC; Le NM; Pham QT; Tran ND; Wong Y; Mosterin A; Katzelnick LC; Labonte D; Le TT; van der Net G; Skepner E; Russell CA; Kaplan TD; Rimmelzwaan GF; Masurel N; de Jong JC; Palache A; Beyer WE; Le QM; Nguyen TH; Wertheim HF; Hurt AC; Osterhaus AD; Barr IG; Fouchier RA; Horby PW; Smith DJ, Antibody landscapes after influenza virus infection or vaccination. Science 2014, 346 (6212), 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hensley SE, Challenges of selecting seasonal influenza vaccine strains for humans with diverse pre-exposure histories. Current opinion in virology 2014, 8, 85–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y; Myers JL; Bostick DL; Sullivan CB; Madara J; Linderman SL; Liu Q; Carter DM; Wrammert J; Esposito S; Principi N; Plotkin JB; Ross TM; Ahmed R; Wilson PC; Hensley SE, Immune history shapes specificity of pandemic H1N1 influenza antibody responses. The Journal of experimental medicine 2013, 210 (8), 1493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moody MA; Zhang R; Walter EB; Woods CW; Ginsburg GS; McClain MT; Denny TN; Chen X; Munshaw S; Marshall DJ; Whitesides JF; Drinker MS; Amos JD; Gurley TC; Eudailey JA; Foulger A; DeRosa KR; Parks R; Meyerhoff RR; Yu JS; Kozink DM; Barefoot BE; Ramsburg EA; Khurana S; Golding H; Vandergrift NA; Alam SM; Tomaras GD; Kepler TB; Kelsoe G; Liao HX; Haynes BF, H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PloS one 2011, 6 (10), e25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt AG; Do KT; McCarthy KR; Kepler TB; Liao HX; Moody MA; Haynes BF; Harrison SC, Immunogenic Stimulus for Germline Precursors of Antibodies that Engage the Influenza Hemagglutinin Receptor-Binding Site. Cell reports 2015, 13 (12), 2842–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davenport FM; Hennessy AV; Francis T Jr., Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. The Journal of experimental medicine 1953, 98 (6), 641–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen KE; Davenport FM; Hennessy AV; Francis T Jr., Characterization of influenza antibodies by serum absorption. The Journal of experimental medicine 1956, 104 (2), 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krammer F; Palese P, Advances in the development of influenza virus vaccines. Nature reviews. Drug discovery 2015, 14 (3), 167–82. [DOI] [PubMed] [Google Scholar]
- 15.Skehel JJ; Wiley DC, Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annual review of biochemistry 2000, 69, 531–69. [DOI] [PubMed] [Google Scholar]
- 16.Ofek G; Guenaga FJ; Schief WR; Skinner J; Baker D; Wyatt R; Kwong PD, Elicitation of structure-specific antibodies by epitope scaffolds. Proceedings of the National Academy of Sciences of the United States of America 2010, 107 (42), 17880–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Correia BE; Bates JT; Loomis RJ; Baneyx G; Carrico C; Jardine JG; Rupert P; Correnti C; Kalyuzhniy O; Vittal V; Connell MJ; Stevens E; Schroeter A; Chen M; Macpherson S; Serra AM; Adachi Y; Holmes MA; Li Y; Klevit RE; Graham BS; Wyatt RT; Baker D; Strong RK; Crowe JE Jr.; Johnson PR; Schief WR, Proof of principle for epitope-focused vaccine design. Nature 2014, 507 (7491), 201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt AG; Xu H; Khan AR; O’Donnell T; Khurana S; King LR; Manischewitz J; Golding H; Suphaphiphat P; Carfi A; Settembre EC; Dormitzer PR; Kepler TB; Zhang R; Moody MA; Haynes BF; Liao HX; Shaw DE; Harrison SC, Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proceedings of the National Academy of Sciences of the United States of America 2013, 110 (1), 264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt AG; Therkelsen MD; Stewart S; Kepler TB; Liao HX; Moody MA; Haynes BF; Harrison SC, Viral receptor-binding site antibodies with diverse germline origins. Cell 2015, 161 (5), 1026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raymond DD; Bajic G; Ferdman J; Suphaphiphat P; Settembre EC; Moody MA; Schmidt AG; Harrison SC, Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc Natl Acad Sci U S A 2018, 115 (1), 168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong M; Lee PS; Hoffman RM; Zhu X; Krause JC; Laursen NS; Yoon SI; Song L; Tussey L; Crowe JE Jr.; Ward AB; Wilson IA, Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. Journal of virology 2013, 87 (22), 12471–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekiert DC; Kashyap AK; Steel J; Rubrum A; Bhabha G; Khayat R; Lee JH; Dillon MA; O’Neil RE; Faynboym AM; Horowitz M; Horowitz L; Ward AB; Palese P; Webby R; Lerner RA; Bhatt RR; Wilson IA, Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 2012, 489 (7417), 526–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy KR; Watanabe A; Kuraoka M; Do KT; McGee CE; Sempowski GD; Kepler TB; Schmidt AG; Kelsoe G; Harrison SC, Memory B Cells that Cross-React with Group 1 and Group 2 Influenza A Viruses Are Abundant in Adult Human Repertoires. Immunity 2018, 48 (1), 174–184 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whittle JR; Zhang R; Khurana S; King LR; Manischewitz J; Golding H; Dormitzer PR; Haynes BF; Walter EB; Moody MA; Kepler TB; Liao HX; Harrison SC, Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proceedings of the National Academy of Sciences of the United States of America 2011, 108 (34), 14216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarthy KR; Raymond DD; Do KT; Schmidt AG; Harrison SC, Affinity maturation in a human humoral response to influenza hemagglutinin. Proc Natl Acad Sci U S A 2019, 116 (52), 26745–26751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y; Pan J; Jenni S; Raymond DD; Caradonna T; Do KT; Schmidt AG; Harrison SC; Grigorieff N, CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface. J Mol Biol 2017, 429 (12), 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corti D; Voss J; Gamblin SJ; Codoni G; Macagno A; Jarrossay D; Vachieri SG; Pinna D; Minola A; Vanzetta F; Silacci C; Fernandez-Rodriguez BM; Agatic G; Bianchi S; Giacchetto-Sasselli I; Calder L; Sallusto F; Collins P; Haire LF; Temperton N; Langedijk JP; Skehel JJ; Lanzavecchia A, A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011, 333 (6044), 850–6. [DOI] [PubMed] [Google Scholar]
- 28.Yassine HM; Boyington JC; McTamney PM; Wei CJ; Kanekiyo M; Kong WP; Gallagher JR; Wang L; Zhang Y; Joyce MG; Lingwood D; Moin SM; Andersen H; Okuno Y; Rao SS; Harris AK; Kwong PD; Mascola JR; Nabel GJ; Graham BS, Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nature medicine 2015, 21 (9), 1065–70. [DOI] [PubMed] [Google Scholar]
- 29.Sagawa H; Ohshima A; Kato I; Okuno Y; Isegawa Y, The immunological activity of a deletion mutant of influenza virus haemagglutinin lacking the globular region. J Gen Virol 1996, 77 (Pt 7), 1483–7. [DOI] [PubMed] [Google Scholar]
- 30.Steel J; Lowen AC; Wang TT; Yondola M; Gao Q; Haye K; Garcia-Sastre A; Palese P, Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 2010, 1 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyoglu-Barnum S; Hutchinson GB; Boyington JC; Moin SM; Gillespie RA; Tsybovsky Y; Stephens T; Vaile JR; Lederhofer J; Corbett KS; Fisher BE; Yassine HM; Andrews SF; Crank MC; McDermott AB; Mascola JR; Graham BS; Kanekiyo M, Glycan repositioning of influenza hemagglutinin stem facilitates the elicitation of protective cross-group antibody responses. Nat Commun 2020, 11 (1), 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mesin L; Schiepers A; Ersching J; Barbulescu A; Cavazzoni CB; Angelini A; Okada T; Kurosaki T; Victora GD, Restricted Clonality and Limited Germinal Center Reentry Characterize Memory B Cell Reactivation by Boosting. Cell 2020, 180 (1), 92–106 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W, Xds. Acta Crystallogr D Biol Crystallogr 2010, 66 (Pt 2), 125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy AJ, Solving structures of protein complexes by molecular replacement with Phaser. Acta crystallographica. Section D, Biological crystallography 2007, 63 (Pt 1), 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy AJ; Grosse-Kunstleve RW; Adams PD; Winn MD; Storoni LC; Read RJ, Phaser crystallographic software. Journal of applied crystallography 2007, 40 (Pt 4), 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy KRW,A; Kuraoka M; Do KT; McGee CE; Sempowski GD; Schmidt AG; Kelsoe G; Harrison SC, Human memory B cells that cross-react with group 1 and group 2 influenza A viruses. Submitted 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P; Cowtan K, Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography 2004, 60 (Pt 12 Pt 1), 2126–32. [DOI] [PubMed] [Google Scholar]
- 38.Chen VB; Arendall WB 3rd; Headd JJ; Keedy DA; Immormino RM; Kapral GJ; Murray LW; Richardson JS; Richardson DC, MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography 2010, 66 (Pt 1), 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winn MD; Ballard CC; Cowtan KD; Dodson EJ; Emsley P; Evans PR; Keegan RM; Krissinel EB; Leslie AG; McCoy A; McNicholas SJ; Murshudov GN; Pannu NS; Potterton EA; Powell HR; Read RJ; Vagin A; Wilson KS, Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography 2011, 67 (Pt 4), 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J; Lansford R; Stewart V; Young F; Alt FW, RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proceedings of the National Academy of Sciences of the United States of America 1993, 90 (10), 4528–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bajic G; Maron MJ; Adachi Y; Onodera T; McCarthy KR; McGee CE; Sempowski GD; Takahashi Y; Kelsoe G; Kuraoka M; Schmidt AG, Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe 2019, 25 (6), 827–835 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuraoka M; Schmidt AG; Nojima T; Feng F; Watanabe A; Kitamura D; Harrison SC; Kepler TB; Kelsoe G, Complex Antigens Drive Permissive Clonal Selection in Germinal Centers. Immunity 2016, 44 (3), 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank under accession code PDB 6UR5 for the rsH4NBv1-K03.12 complex.


