Abstract
Background
The subgenus Megatrypanum Hoare, 1964 of Trypanosoma Gruby, 1843 comprises trypanosomes of cervids and bovids from around the world. Here, the white-tailed deer Odocoileus virginianus (Zimmermann) and its ectoparasite, the deer ked Lipoptena mazamae Rondani, 1878 (hippoboscid fly), were surveyed for trypanosomes in Venezuela.
Results
Haemoculturing unveiled 20% infected WTD, while 47% (7/15) of blood samples and 38% (11/29) of ked guts tested positive for the Megatrypanum-specific TthCATL-PCR. CATL and SSU rRNA sequences uncovered a single species of trypanosome. Phylogeny based on SSU rRNA and gGAPDH sequences tightly cluster WTD trypanosomes from Venezuela and the USA, which were strongly supported as geographical variants of the herein described Trypanosoma (Megatrypanum) trinaperronei n. sp. In our analyses, the new species was closest to Trypanosoma sp. D30 from fallow deer (Germany), both nested into TthII alongside other trypanosomes from cervids (North American elk and European fallow, red and sika deer), and bovids (cattle, antelopes and sheep). Insights into the life-cycle of T. trinaperronei n. sp. were obtained from early haemocultures of deer blood and co-culture with mammalian and insect cells showing flagellates resembling Megatrypanum trypanosomes previously reported in deer blood, and deer ked guts. For the first time, a trypanosome from a cervid was cultured and phylogenetically and morphologically (light and electron microscopy) characterised.
Conclusions
In the analyses based on SSU rRNA, gGAPDH, CATL and ITS rDNA sequences, neither cervids nor bovids trypanosomes were monophyletic but intertwined within TthI and TthII major phylogenetic lineages. One host species can harbour more than one species/genotype of trypanosome, but each trypanosome species/genotype was found in a single host species or in phylogenetically closely related hosts. Molecular evidence that L. mazamae may transmit T. trinaperronei n. sp. suggests important evolutionary constraints making tight the tripartite T. trinaperronei-WTD-deer ked association. In a plausible evolutionary scenario, T. trinaperronei n. sp. entered South America with North American white-tailed deer at the Pliocene-Pleistocene boundary following the closure of the Panama Isthmus.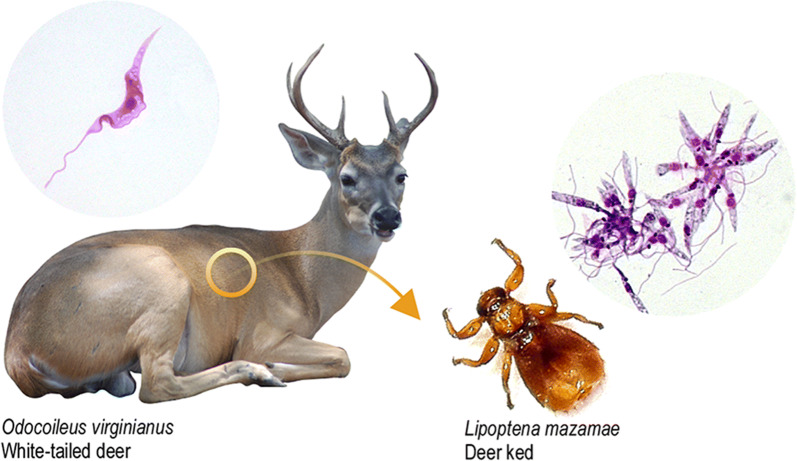
Keywords: Trypanosoma, New species, Cervidae, Deer ked, Phylogeny, Taxonomy, Great American Interchange, Host-parasite restriction
Background
Trypanosomes of the subgenus Megatrypanum Hoare, 1964 of Trypanosoma Gruby, 1843 are distributed worldwide in bovids and cervids. Cervidae comprises 55 species widespread in Eurasia and America of two subfamilies: Cervinae, which comprises mostly deer from Eurasia of the genera Axis, Dama, Elaphurus, Rucervus, Rusa and Cervus (the only genus also present in North America); and Capreolinae, which comprises deer from Eurasia (Capreolus), Eurasia and North America (Alces, Rangifer), and deer endemic to the Americas (Odocoileus) or restricted to South America (Blastocerus, Hippocamelus, Mazama, Ozotoceros, Pudu) [1].
In North American cervids, Megatrypanum trypanosomes have been reported in caribou (Rangifer tarandus caribou (Gmelin)) [2, 3], red deer (Cervus elaphus Linnaeus) [4], roe deer (Capreolus capreolus (Linnaeus)) [5–7], reindeer (Rangifer tarandus Linnaeus) [8], mule deer (Odocoileus hemionus (Rafinesque)) [3, 9], moose (Alces alces (Linnaeus)) [10], white-tailed deer (WTD) (Odocoileus virginianus (Zimmermann)) and elk (Cervus elaphus canadensis (Erxleben)) [3, 11, 12]. In Europe, Megatrypanum trypanosomes have been described in fallow deer (Cervus dama Linnaeus), red deer and roe deer in Germany [6], reindeer and moose in Sweden [13], roe deer in Poland [14], and red deer in Croatia [15]. In Asia, a Megatrypanum trypanosome was reported in the Japanese sika deer Cervus nippon Temminck [16]. Megatrypanum trypanosomes are generally non-pathogenic to domestic and wild ruminants including deer [8].
Contrasting with molecularly characterised Megatrypanum trypanosomes of deer from across the North America and Europe (Sweden, Germany, Croatia and Poland) [12–15], reports of these trypanosomes in South American cervids are limited to the morphology of blood trypomastigotes in WTD, black-tailed deer (Odocoileus hemionus columbianus (Richardson)) and brown brocket deer (Mazama gouazoubira (Fischer)) in Colombia [17, 18], and brocket deer in Argentina and Brazil [19, 20].
Traditionally, the large and broad shape of blood trypomastigotes is the main taxonomic criterion of the subgenus Megatrypanum (type-species Trypanosoma (M.) theileri Laveran, 1902 reported in domestic cattle), whereas species identification relies on data about host restriction provided by field, and cross-experimental infections [13, 21, 22]. This subgenus was revised by Hoare [21] to accommodate trypanosomes of artiodactyls, bats, rodents, non-human primates and marsupials, excluding trypanosomes of non-mammalian hosts formerly included in this subgenus. However, molecular phylogeny revalidated this subgenus as a monophyletic assemblage comprising the type-species T. theileri of cattle and virtually morphologically indistinguishable trypanosomes from bovids and cervids. Presently, this subgenus excludes all non-ruminant trypanosomes, even the poorly investigated closest relatives forming the T. cyclops sister clade [15, 23–25]. Although T. theileri was never observed in blood of hosts other than ruminants, PCR-surveys detected DNA of this trypanosome in bats [26], and in one chimpanzee [27].
Currently, there are three named species of Megatrypanum trypanosomes parasitizing deer, all based just on morphology and host species of origin: Trypanosoma (M.) mazamarum Mazza, Romana & Fiora, 1932 in brocket deer from Argentina [19]; Trypanosoma (M.) cervi Kingston & Morton, 1975 in elk from the USA [28]; and Trypanosoma (M.) stefanskii Kingston, Bobek, Perzanowski, Wita & Maki, 1992 in roe deer from Poland [14]. Molecular studies have uncovered different species/genotypes of Megatrypanum trypanosomes in WTD and elk in the USA [12], red deer in Croatia [15], sika deer in Japan [16], and the invader sika deer plus wisent (European bison), red deer and fallow deer in Poland [29].
Megatrypanum trypanosomes are thought to be cyclically transmitted by tabanid and hippoboscid flies. Trypanosomes similar to T. cervi have been reported in tabanids (deer flies) in the USA, Germany, Russia, and Poland [22, 29–31]. Tabanids harbouring trypanosomes of bovids have been reported in North America [32], South America [24, 33], and Africa [34–36]. The sheep ked Melophagus ovinus (Linnaeus, 1758) transmit T. (M.) melophagium Flu, 1908 exclusive of sheep [15, 21, 37] while T. (M.) theodori Hoare, 1931 of goats is transmitted by Lipoptena capreoli Rondani, 1878 [21]. The finding of trypanosomes in guts of Lipoptena cervi (Linnaeus, 1758) taken from red deer suggests that hippoboscid flies may transmit deer trypanosomes of the subgenus Megatrypanum [38]. However, molecular comparisons of Megatrypanum trypanosomes from deer blood and keds are still lacking.
Comprehensive molecular data on Megatrypanum trypanosomes from wild and domestic ruminants and the proper identification of vectors are essential to understand their evolution, species richness, phylogenetic relationships, range of vertebrate hosts, vectors, and geographical distribution. In the present study, we describe a new species of Megatrypanum in WTD and its deer ked and hypothesise its probable life-cycle and evolutionary history by integrating morphological, behaviour in cultures, biological and phylogeographical data.
Methods
Study area, deer blood and ked collection, and DNA preparation
A total of 75 WTDs were captured at the Anzoátegui state, municipality of Simón Bolívar, an important livestock breeding area at north-eastern Venezuela (10°07′08.95″N, 64°38′23.80″W) (Fig. 1), where the mean annual temperature is 25 °C (21.8–2.2 °C) and rainfall is below 1000 mm, with a dry season from December to April. The captured WTDs (Fig. 2) were kept in quarantine prior to their introduction into a protected reserve. Chemical immobilization and anaesthesia were performed with a combination of 10 mg/kg ketamine and 0.6 mg/kg xylazine intramuscularly and reverted with 0.2 mg/kg of yohimbine intravenously. Anesthetized deer were examined to record heart and breathing frequency rates, and temperature. Institutional and national guidelines for the care and use of wild animals were followed. Deer handling was performed in accordance with the approved protocols, and under the supervision of the MINEC (the Venezuelan Ministerio del Poder Popular para el Ecosocialismo).
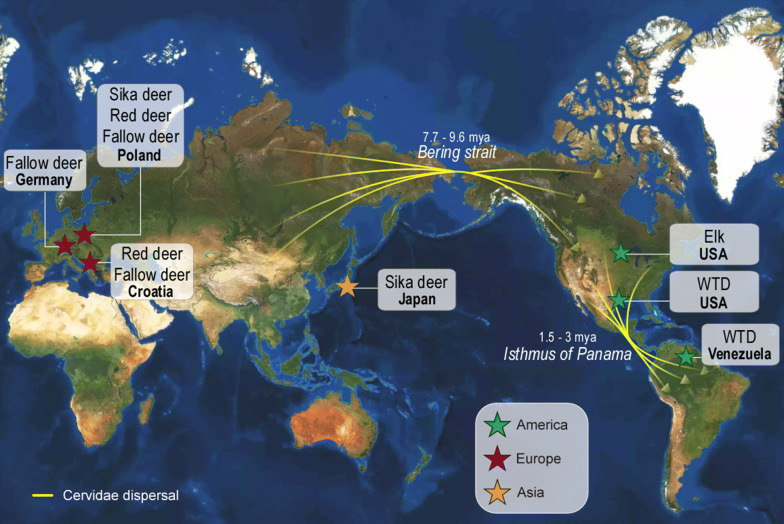
Fig. 1.
Origin of Trypanosoma (Megatrypanum) trinaperronei n. sp. and other deer trypanosomes. Geographical origin of deer examined in this and in previous studies, and historical dispersion of cervids from Eurasia reaching North America through the Bering Strait, and thereafter through the Panama Isthmus into South America. Abbreviations: WTD, white-tailed deer
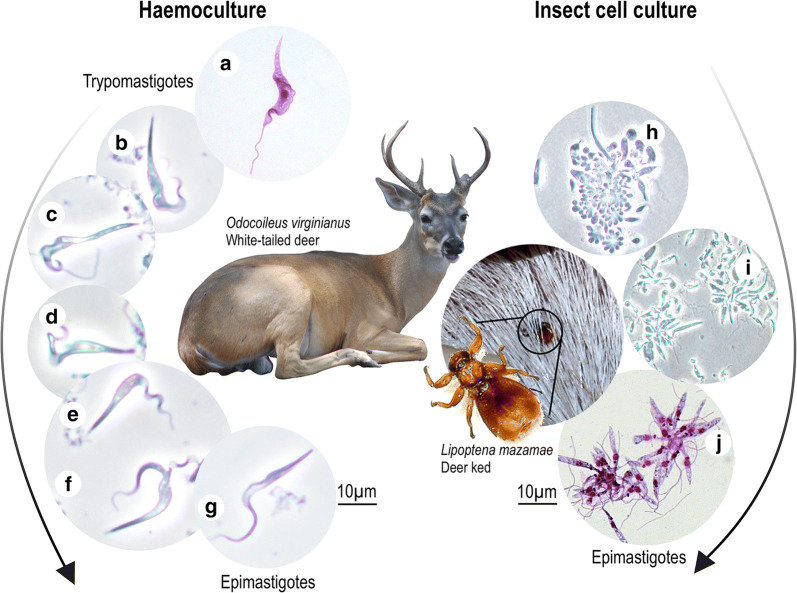
Fig. 2.
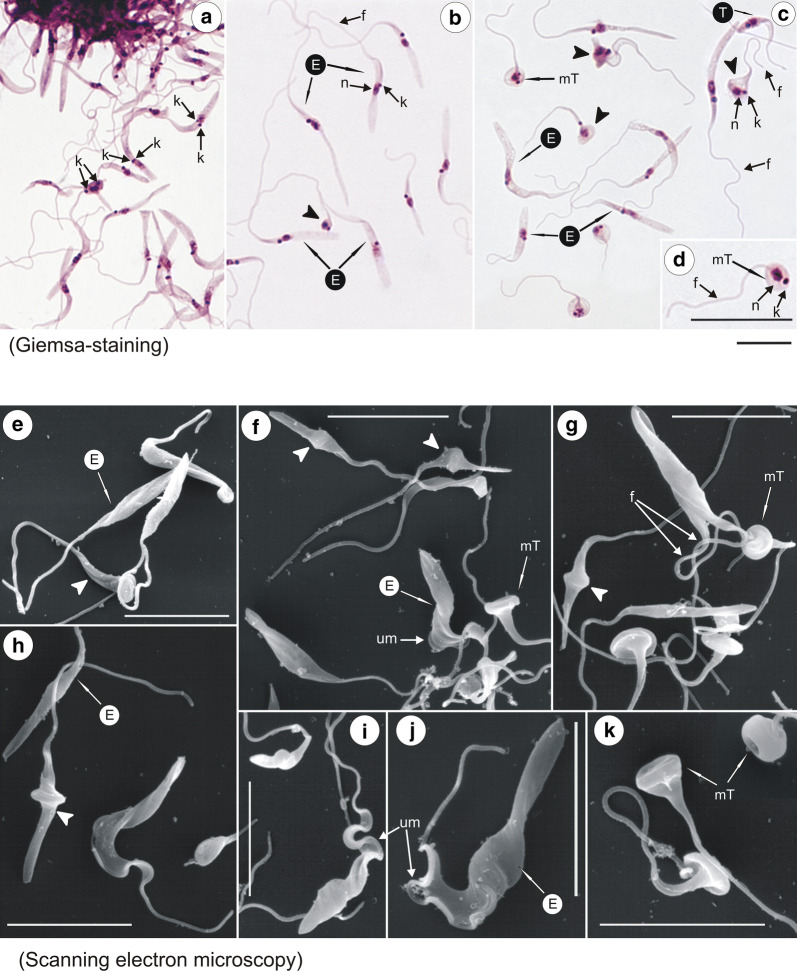
Predicted life-cycle of Trypanosoma (Megatrypanum) trinaperronei n. sp. in its host Odocoileus virginianus (white-tailed deer), and its putative vector the deer ked Lipoptena mazamae inferred from: early haemocultures showing long and slender trypomastigotes (a–c) and epimastigotes (d–g), both forms with noticeable undulant membrane; co-cultures with Hi-5 insect cells exhibiting clumps of small forms adhered to the insect cells (h, i) giving origin to rosettes of epimastigotes (j). Morphology and development of T. trinaperronei n. sp. co-cultivated with Hi-5 (25 °C) and LLCMK2 mammalian (37 °C), from log- to stationary cultures, are detailed in the Figs. 7 and 8. Deer keds become infected by T. trinaperronei n. sp. feeding on deer containing blood trypomastigotes resembling those present in early haemocultures (a-c), which, in their digestive tract, transform and multiply as small forms attached to the cells of the gut wall, as observed in Hi-5 cells (h, i), give origin to rosettes of epimastigotes (j) that multiply and, later, differentiate into metacyclic trypomastigotes. Illustration of T. trinaperronei n. sp. metacyclogenesis in insect cultures are shown in the Fig. 7. Most likely, infective metacyclic trypomastigotes present in the faeces of the vectors are transmitted to WTD by deer keds bite wound or mucosa, thus reaching the bloodstream and transforming into trypomastigotes resembling those detected in early haemocultures (a). Abbreviations: WTD, white-tailed deer
Blood samples were obtained via the jugular vein using tubes with EDTA and aliquots (~ 500 µl) of blood were preserved in 99.5% ethanol (v/v), incubated overnight in 250–500 μl of lysis buffer (20 mM EDTA; 50 mM Tris-HCL; 117 mM NaCl; 1% SDS; 10 mg/ml Proteinase K), precipitated with 400–800 μl ammonium acetate (4M), and centrifuged (10 min at 20,000× g). Then, DNA was precipitated with ethanol, dried at room temperature and resuspended in TE (Tris-EDTA). Deer keds (Fig. 2) were removed from the WTDs, preserved in ethanol, and identified based on morphology [39] and cox1 barcoding using DNA obtained from the gut contents of keds as described previously for tsetse flies [40]. Due to limited field facilities, the guts of deer keds were not examined by microscopy or culturing.
Isolation of deer trypanosomes by hemoculturing
Deer blood samples were examined by microhematocrit, Giemsa-stained blood smears, and hemoculturing [15, 24, 25]. Aliquots (~ 200 µl) of blood were used for hemoculturing in medium consisting of blood agar base containing 15% rabbit blood as a solid phase and an overlay of TC100 medium (Cultilab, São Paulo, Brazil) with 10% FBS, and incubated at 25 °C [23, 24]. Positive haemocultures were transferred to culture flasks containing a monolayer of insect cells (Hi-5 from Trichoplusia ni) in TC100 medium with 10% FBS, and incubated at 25 °C [41]. Cultures were expanded in TC100 for DNA preparation and cryopreservation at the Trypanosomatid Culture Collection (TCC) of the Department of Parasitology, University of São Paulo (USP), São Paulo, Brazil.
Trypanosome diagnosis in deer blood and keds and network of CATL sequences
The Megatrypanum-specific TthCATL-PCR based on Cathepsin-L (CATL) sequences was used for surveys in deer blood and ked guts as described previously [25, 42]. The cdCATL sequences (274 bp) of trypanosomes amplified by TthCATL-PCR were cloned and sequences determined were aligned with those of other Megatrypanum trypanosomes (GenBank, Additional file 1: Table S1), and used for network inferences [25, 42]. Sequences of the whole CATL catalytic domain (477 bp) were determined for the isolate TCC2268 [25, 42].
Phylogenetic analysis of SSU rRNA, gGAPDH and ITS rDNA sequences
DNA from cultured trypanosomes extracted by the phenol-chloroform method was used for PCR amplification of sequences of the variable V7V8 region (~ 728 bp) or entire (2142 bp) SSU rRNA gene, gGAPDH (glycosomal glyceraldehyde 3-phosphate dehydrogenase) gene (~ 847 bp) and ITS1 rDNA sequences (~ 233 bp) [24, 25, 43]. PCR products were purified, cloned and sequences of 5–10 clones from each amplicon were determined, and aligned with sequences obtained from the GenBank using the Clustal X program [44]. We created the following alignments: (i) V7V8 SSU rRNA sequences herein determined and available on GenBank sequences for Megatrypanum trypanosomes; (ii) concatenated V7V8 SSU rRNA and gGAPDH sequences (~ 1575 bp) of Megatrypanum trypanosomes from deer, cattle, water buffalo, antelopes and sheep; trypanosomes of other subgenera were also included, and other trypanosomatids were used as outgroups; (iii) ITS1 rDNA sequences of Megatrypanum trypanosomes. Phylogenies were inferred using parsimony (P), maximum likelihood (ML), and Bayesian inferences (BI). P and bootstrap analyses were carried out using PAUP 4.0b10 [45] with 500 replicates of random addition sequences followed by branch swapping (RAS-TBR). ML was performed using RAxML-VI-HPC v.2.2.3 [46] with tree searches using GTR model with gamma-distributed rate variation across sites and proportion of invariable sites (GTRGAMMA model), and 500 maximum parsimony starting trees; the model parameters were estimated in RAxML for the duration of the tree search. Nodal supports were estimated with 500 bootstrap replicates in RAxML using GTRGAMMA and P starting trees. BI was performed in MrBayes v3.1.2 [47] with GTRGAMMA, and the first 25% of the trees from 1 million generations were discarded as ‛burn-inʼ. Sequences of ITS1 rDNA were employed for network split decomposition using the Neighbor-Net method with Kimura 2-parameter implemented in Splits Tree4 V4.10 [48]. Internode support was estimated with 500 bootstrap replicates using the same parameters optimized for network inferences.
Trypanosomes included in our analyses, and their host species, geographical origins and GenBank accession numbers of DNA sequences are detailed in Additional file 1: Table S1 (CATL sequences), Additional file 2: Table S2 (SSU rRNA gene), Additional file 3: Table S3 (gGAPDH) and Additional file 4: Table S4 (ITS1 rDNA). All newly generated DNA sequences were deposited in the GenBank database under the accessions numbers MN747149-MN747155 (CATL), MN752212 (SSU rRNA gene), MN756794 (gGAPDH) and MN752208-MN752209 (ITS1 rDNA).
Growth behaviour and light and electron microscopy of trypanosomes co-cultivated with insect and mammalian cells
The isolate TCC2268 was co-cultivated with a monolayer of Hi-5 insect cells (TC100 medium) and flagellates from stationary cultures were seeded on monolayers of monkey LLC-MK2 cells, cultivated at 37 °C with 5% CO2, and intracellular parasites were investigated according to Lima et al. [49]. Both cultures were examined daily in an inverted microscopy, and supernatants of 2, 5, 7 and 10 days of culture were smeared on glass slides, fixed with methanol, and stained with Giemsa for light microscopy.
Both scanning electron microscopy (SEM) and transmission electron microscopy (TEM) were performed as detailed in [41]. For SEM, trypanosomes fixed in glutaraldehyde were adhered to poly-L-lysine-coated coverslips and processed for observation on a FEI Quanta 250 (FEI Company, Hillsboro, USA) microscope. For TEM, trypanosomes were fixed in glutaraldehyde, post-fixed with osmium tetroxide, and embedded in Spurr resin. Ultrathin sections were stained with uranyl acetate and lead citrate, and examined with a LEO 906E microscope (Zeiss, Jena, Germany). Images were captured by a CCD camera MegaView III.
Results and discussion
White-tailed deer and deer keds harbour Megatrypanum trypanosomes uncovered by TthCATL-PCR
Overall, 15 WTDs including 9 males (5 juveniles and 4 adults) and 6 juvenile females captured were clinically healthy, showing normal values of heart frequency (44 to 128, mean of 76, beats/min), breathing frequency (18 to 48, mean of 34, breaths/min), and body temperature (35.4–40.2 °C, mean of 37.4 °C). Inspection of WTDs for ectoparasites revealed an abundance of deer keds in ~ 73% (11/15) of them, mainly in the ventral, inner legs and inguinal regions of the WTDs. No skin or fur damage was observed in ked-infested WTDs. Twenty-nine keds from 11 WTDs preserved in ethanol were identified morphologically as L. mazamae [39], and this identification was confirmed by cox1 DNA barcoding (GenBank: MN756795).
Microscopy of Giemsa-stained blood smears and the microhematocrit technique did not allow detection of trypanosomes in blood samples of the 15 WTDs examined. However, haemoculturing yielded a trypanosome infection rate of 20% (three positive cultures), and one culture (TCC2268) was established, and cryopreserved. These findings are consistent with very low parasitaemia but positive haemocultures as previously reported for other Megatrypanum trypanosomes [23, 25]. The Megatrypanum-specific assay, TthCATL-PCR, was employed aiming at detection of trypanosomes in the blood samples from WTD and in gut contents from the deer keds. PCRs were positive for 7 out of 15 WTDs (~ 47%), and 11 out of 29 deer keds (~ 38%), including keds from the three WTD positive for trypanosomes by haemoculturing.
Characterisation of trypanosomes from blood and keds of white-tailed deer using CATL sequences
CATL DNA sequences (274 bp) obtained by TthCATL-PCR from blood samples and culture (isolate TCC2268) of WTD and gut samples from deer keds share highest similarity with sequences of Megatrypanum trypanosomes on GenBank. The newly generated sequences were aligned with those of other Megatrypanum trypanosomes, and this alignment was used to infer their genetic relatedness. Trypanosome sequences obtained from deer keds were virtually identical to those obtained from WTD blood (TCC2268), indicating that they belong to a single trypanosome species.
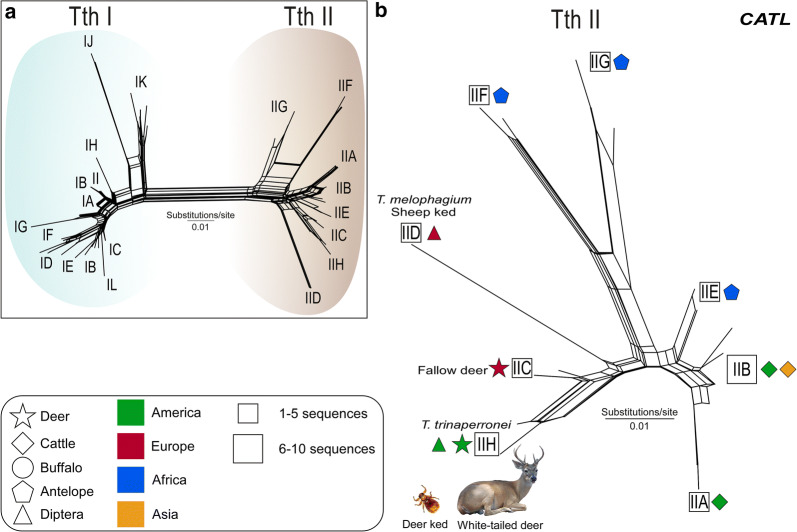
Network inferences of 100 CATL sequences of 274 bp, amplified by TthCATL-PCR (Fig. 3a, b) or 53 larger CATL sequences of 477 bp (data not shown) generated highly similar networks. A divergence of ~ 2.0% in the small fragment of CATL sequences separated TCC2268 from Trypanosoma sp. D30. Although originating from different continents, these two deer isolates were much more related to each other than to trypanosomes from bovids (cattle, buffalo, sheep and antelopes), even those from Venezuelan cattle and buffalo [25]. TCC2268 and Trypanosoma sp. D30 were assigned to different species of close genotypes, TthII H (which includes CATL sequence from deer ked) and TthII C, respectively. TCC2268 diverged by 5.3% from T. melophagium (TthII D) and by 3.3% from T. theileri of cattle nested into TthII (Fig. 3b).
Fig. 3.
Network analysis of cathepsin L (CATL) sequences of Trypanosoma (Megatrypanum) trinaperronei n. sp. from Venezuelan white-tailed deer (WTD), and respective deer keds. a Analysis of Megatrypanum trypanosomes from wild and domestic ruminants evidencing TthI and TthII phylogenetic lineages. b Analysis restricted to the lineage TthII positioning T. trinaperronei n. sp. (TCC2268) tightly clustered with trypanosomes found in guts of deer ked taken from WTD. Analysis was inferred using the Neighbour-Net method with the K2 parameter and nodal support estimated with 500 bootstrap replicates. Abbreviations: TCC, Trypanosomatids Culture Collection; WTD, white-tailed deer
Although useful for assessment of genetic diversity [42, 50–52], the low level of polymorphism detected in short CATL sequences (274 bp) generated by TthCATL-PCR may be insufficient to discriminate reliable lineages and, specially, genotypes. Therefore, the description of novel species and intraspecific genotypes must be supported by additional markers including conserved (SSU rRNA and gGAPDH), and polymorphic (ITS rDNA and spliced leader rRNA) sequences [15, 25, 42, 43, 50, 53].
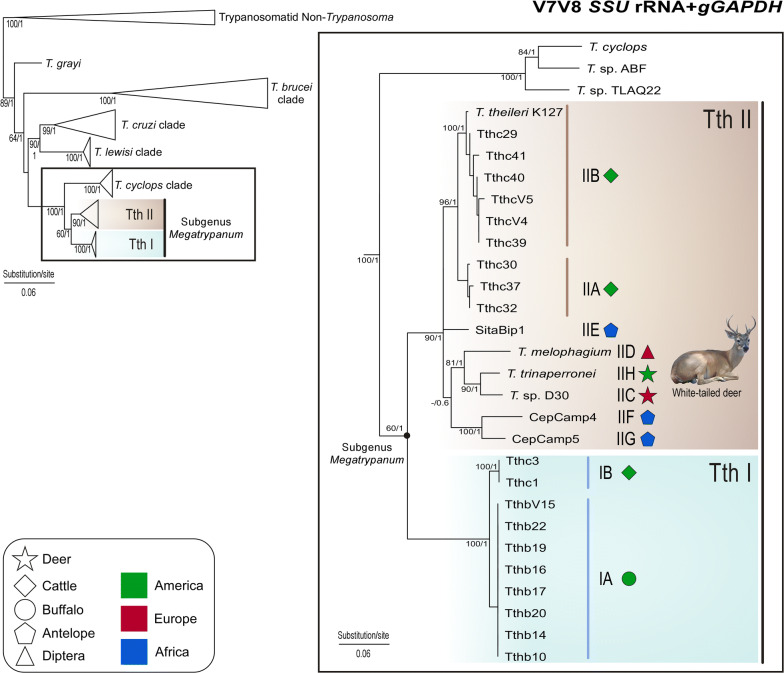
Barcoding of deer trypanosomes through SSU rRNA sequences revealed a new Megatrypanum trypanosome in white-tailed deer
Our comparative analysis of V7V8 SSU rRNA barcodes comprised Megatrypanum trypanosomes from cervids (Venezuela, Germany, Poland, Croatia, Japan and USA), and a large dataset of trypanosome sequences from bovids including cattle (Brazil, Venezuela, Argentina, Colombia, Germany, Poland, Croatia, UK, Japan and USA), water buffaloes (Venezuela, Colombia and Brazil), antelopes (Cameroon and Tanzania) and bison (Poland). In addition, our analyses included sequences of Megatrypanum trypanosomes from the guts of tabanids (Brazil, Africa and Russia), hippoboscids (Croatia and Scotland), tsetse flies (Africa), and sand flies (Italy) (Fig. 4; Additional file 2: Table S2). SSU rRNA barcodes corroborated the distribution of cervid and bovid trypanosomes in both TthI and TthII lineages, whereas the branching patterns of intra-lineages could not be resolved using exclusively these highly conserved sequences (Fig. 4). In addition, two sequences from an elk (elk 317; GenBank: JX178200, JX178201) from the USA diverged by relevant and equidistant genetic distances from TthI (~ 3.0% divergence) and TthII (~ 3.1%), apparently representing a new lineage of Megatrypanum (Fig. 4).
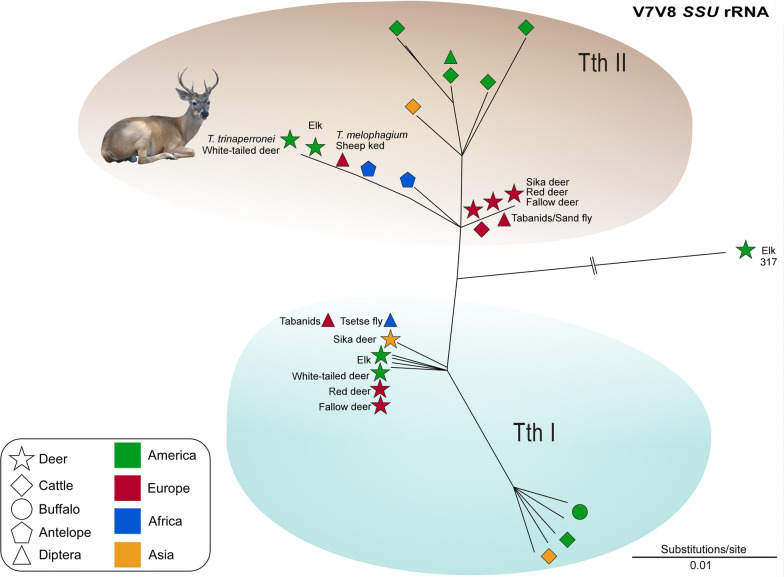
Fig. 4.
Barcoding of Trypanosoma (Megatrypanum) trinaperronei n. sp. Comparison of V7V8 SSU rRNA barcodes from T. trinaperronei n. sp. and trypanosomes of TthI and TthII lineages of the subgenus Megatrypanum from cervids (fallow, red and sika deer, WTD, and elk), and bovids (water buffalo, cattle, bison, and antelopes). Sequences obtained from deer keds, sheep keds, sand flies, tsetse flies and tabanids were also included. Sequences from a distantly related isolate of elk (elk 317) from the USA were placed as a probable new lineage of Megatrypanum. Dendrograms were inferred by Neighbour-Joining using the Kimura 2-parameter algorithm, and nodal supports were estimated with 500 bootstrap replicates. Abbreviations: WTD, white-tailed deer
SSU rRNA sequences of the Venezuelan isolate WTD TCC2268 was highly similar (99.7%) to that of the isolate WTD A3 from the USA, both sharing 99.5% similarity with Trypanosoma sp. D30 of fallow deer from Germany. Besides these three deer trypanosomes, TthII comprised Trypanosoma cf. cervi from North American elk (elk328), and isolates from red deer (Cel34), fallow deer (DdP18) and sika deer (Cn1) from Poland. In addition, TthII included trypanosomes from African antelopes and T. melophagium of sheep, all clustering close to the trypanosomes of deer, whereas trypanosomes of cattle from North and South America, Europe and Asia formed a more separated cluster (Fig. 4). Trypanosomes from these European deer nested into TthII diverged from TCC2268 by 0.3–0.6% in highly conserved SSU rRNA sequences. For comparison, TCC2268 diverged by just 0.6% from the reference sequence for T. theileri TREU124 of cattle, thus reinforcing that these sequences are too conserved to clearly resolve the relationships of the recently diversified Megatrypanum trypanosomes. Regarding possible vectors, trypanosome sequences obtained from the guts of Brazilian, Polish and Russian tabanids, and Central African Republic tsetse flies were all assigned to both TthI and TthII lineages [24, 29, 36].
The lineage TthI also harboured trypanosomes (misclassified) referred to as Trypanosoma cf. cervi identified in WTDs (WTD A1, A5, A21, A148 and NL15) and elk (elk142, elk328, elk416 and elk421) from the USA [12], and isolates referred to as T. cervi from Polish red deer (Cel14St), fallow deer (DdP287) and tabanids (Fig. 4). The trypanosome (TSD1) from the Japanese sika deer [16] was the deer trypanosome more genetically distant (~ 1.4% sequence divergence) from the isolate TCC2268 (Fig. 4). In addition, this lineage also comprised T. theileri of cattle from South America, Japan and the USA, and T. theileri-like trypanosomes of South American water buffalo and European bison from Poland [24, 25, 29].
Phylogeny based on SSU rRNA and gGAPDH genes supports a new species of trypanosome from white-tailed deer in the subgenus Megatrypanum
It was previously demonstrated that gGAPDH sequences are generally more variable than SSU rRNA sequences, thus being more suitable for a better differentiation between closely related trypanosomes such as those of the subgenus Megatrypanum [15, 24, 25]. Corroborating results using SSU rRNA sequences, the gGAPDH sequence from the isolate WTD TCC2268 was closest to Trypanosoma sp. D30 (~ 2.2% sequence divergence), and more similar to T. melophagium and trypanosomes of antelopes, all clustering together in the lineage TthII. Cattle isolates formed another group within TthII, separated from WTD TCC2268 by an average of ~ 4.4% sequence divergence.
Here, gGAPDH sequences of TCC2268 were aligned with available sequences from species/genotypes of Megatrypanum trypanosomes, and concatenated with SSU rRNA sequences. The inferred phylogeny (Fig. 5) displayed a highly congruent topology compared with that of independent SSU rRNA (Fig. 4). Together, phylogenetic positioning and genetic distances of WTD TCC2268 from other species of Megatrypanum with sequences available on GenBank allowed for the description of this new isolate as a novel species herein designated Trypanosoma (Megatrypanum) trinaperronei n. sp. Unfortunately, most trypanosomes from cervids have been known merely by partial SSU rRNA sequences such as those reported from elk and WTD from the USA, and deer isolates from Japan and Poland included in the present study in the SSU rRNA analysis (Fig. 4). Results obtained in our analyses are congruent with those from previous studies using SSU rRNA, gGAPDH, CATL and ITS rDNA sequences [15, 24, 25, 42, 50, 53].
Fig. 5.
Phylogenetic positioning of Trypanosoma (Megatrypanum) trinaperronei n. sp. a Phylogenetic tree based on concatenated gGAPDH and SSU rRNA sequences from Megatrypanum trypanosomes and species representative of all other major clades of Trypanosoma, using trypanosomatids of other genera as outgroups. Major phylogenetic clades were collapsed, and the clade Megatrypanum was highlighted. bMegatrypanum clade showing TthI and TthII lineages formed by several genotypes. Trypanosoma trinaperronei n. sp. was assigned to genotype TthII H, sister to TthII C, which comprised the German Trypanosoma sp. D30 of fallow deer and the Croatian TC2 of red deer, altogether forming a clade exclusive of deer trypanosomes from South and North America, and Europe. A concatenated alignment of 1575 characters was employed for maximum likelihood (ML) and Bayesian inference (BI); the numbers at nodes refer to ML/B support values derived from 500 replicates
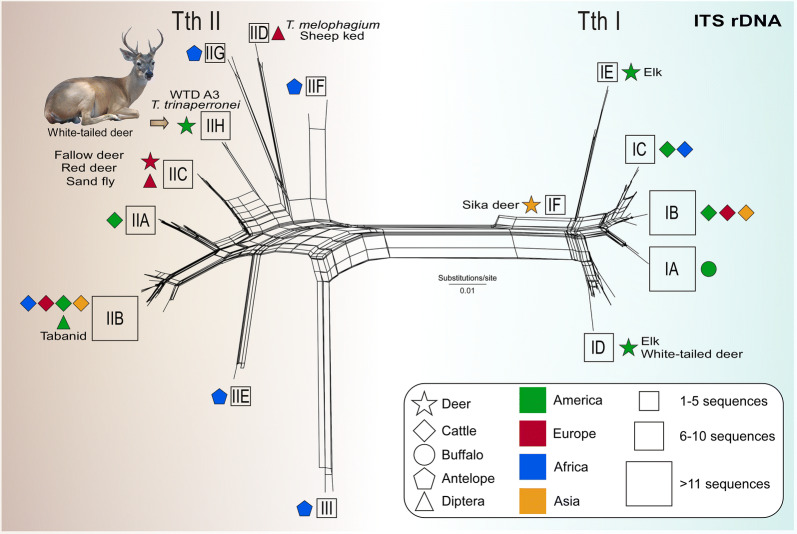
Low levels of polymorphism in ITS1 rDNA sequences of T. trinaperronei n. sp. of white-tailed deer from Venezuela and the USA
To better understand the relatedness of T. trinaperronei n. sp. with its closely related trypanosomes of deer from the USA and Germany, we compared ITS1 rDNA sequences, which are much more polymorphic than SSU rRNA and gGAPDH sequences. A comprehensive analysis of 122 ITS1 rDNA sequences of Megatrypanum trypanosomes (Fig. 6) was carried out including sequences of trypanosomes from a range of deer species and geographical areas: WTD TCC2268 from Venezuela (n = 2 sequences); WTD A3 (n = 2), WTD A1 (n = 2), WTD A21 (n = 3), WTD NL15 (n = 3), elk 416 (n = 1), elk 142 (n = 2), elk 421 (n = 1), and elk 328 (n = 1) from the USA; TSD1 (n = 1) from Japanese sika deer; TC2 (n = 1) from Croatian red deer; and Trypanosoma sp. D30 from German fallow deer (n = 2). These sequences were aligned with those from bovid trypanosomes: T. theileri of cattle (29 sequences of TthI and 37 of TthII lineage); T. theileri-like of water buffalo (n = 13); T. melophagium of sheep ked; and trypanosomes of the antelopes (n = 14) sitatunga, duiker and puku.
Fig. 6.
Lineages and genotypes of trypanosomes of the subgenus Megatrypanum inferred using ITS1 rDNA sequences. Network inferred using polymorphic ITS1 rDNA sequences of trypanosomes representative of genotypes identified in cervids (WTD, follow deer, sika deer and elk), bovids (cattle, water buffalo and antelopes), sheep keds, and sand flies. The analysis was carried out using the Neighbour-Net method with K2P parameter, and nodal support estimated with 500 bootstrap replicates. Trypanosoma trinaperronei n. sp. comprised two highly similar isolates, TCC2268 from Venezuela and WTD A3 from Texas (USA) assigned to the new genotype TthII H, which is closest to TthII C formed by Trypanosoma sp. D30 of fallow deer and TC2 of red deer from German and Croatia, respectively. Abbreviations: TCC, trypanosomatids culture collection; WTD, white-tailed deer
Confirming previous studies, ITS1 rDNA sequences of Megatrypanum trypanosomes were distributed in the TthI and TthII lineages (Fig. 6) regardless of their origin from bovids or cervids, thus corroborating data from the present study (Figs. 3, 4 and 5) and previous studies [12, 15, 24, 25, 53]. Our analysis supported 6 genotypes of TthI (IA-IF), and 9 of TthII (IIA-II I) (Fig. 6). ITS1 rDNA sequences of T. trinaperronei n. sp. from Venezuela and the USA shared highly similar sequences (~ 0.6% divergence), and were tightly clustered together supporting the genotype TthII H. Trypanosomes from European cervids assigned to the lineage TthII C were the German Trypanosoma sp. D30 of fallow deer [12, 25] and the highly similar Croatian trypanosome of red deer [15], and they diverged ~ 17% from T. trinaperronei n. sp. Regarding the relationships between cervid and bovid trypanosomes, ITS1 rDNA sequences of T. melophagium diverged by ~ 18% from T. trinaperronei n. sp. while divergences of above 24% separated T. trinaperronei n. sp. from trypanosomes of cattle (TthII A and TthII B) and antelopes (TthII F, G, E, I) [25]. Interestingly, ITS1 rDNA sequence of a trypanosome obtained from the gut of an Italian sand fly [54] was virtually identical to those of Trypanosoma sp. D30 from Germany [22]. As expected, deer trypanosomes of TthI diverged from T. trinaperronei n. sp. by remarkable divergences in ITS rDNA (Fig. 6): ~ 46% from both TthI D (WTD and elk) and TthI E (elk), which are genotypes identified in deer sympatric with the WTD from which T. trinaperronei n. sp. (isolate WTD A3) was obtained in the USA [12]. As shown with other molecular markers here, the greatest distances of ITS rDNA (~49%) among deer trypanosomes were observed between T. trinaperronei n. sp. and the trypanosome from Japanese sika deer (TthI F).
Host-parasite-vector relationships and evolutionary history of cervid trypanosomes
Although host specificity of Megatrypanum trypanosomes remains to be clearly demonstrated, our findings provide additional support for relevant host-parasite-vector association in the evolution of these trypanosomes. Species diversification was likely shaped by evolutionary constraints exerted by ruminant hosts. In addition, vectors may be also involved in trypanosome host-restriction because deer flies (tabanids) and deer keds (hippoboscids) are strongly associated with their cervid hosts. In agreement with this hypothetical evolutionary scenario, we demonstrated that deer keds taken from WTD exclusively harboured T. trinaperronei n. sp., corroborating a previous suggestion that these flies can transmit, cyclically and/or mechanically, Megatrypanum trypanosomes to cervids [38]. Lipoptena mazamae occurs from south-eastern USA to South America [39, 55] and is tightly linked to WTD, although this ked can eventually jump to phylogenetically close deer species [39]. To date, deer flies, which have been experimentally proven to transmit deer trypanosomes, and deer keds have been implicated as vectors of cervid trypanosomes [21, 29, 31, 36, 38, 56, 57]. Recent studies report on DNA from deer trypanosomes in guts of sand flies and culicids [54, 58], but their roles as vectors remains to be investigated.
Trypanosome cross-infections of cervids and bovids have not been confirmed experimentally or by molecular epidemiology [8, 12, 15, 21, 24, 25, 33, 53, 59]. In Venezuela, we found WTD infected with T. trinaperronei n. sp. while sympatric cattle and water buffalo were found infected with T. theileri and T. theileri-like, respectively [25]. Similarly, Japanese sika deer were found infected exclusively with the Trypanosoma sp. TSD1, whereas sympatric cattle were infected with T. theileri of both TthI and TthII lineages [53]. Deer, cattle and sheep have been reported to harbour host-specific trypanosomes in Croatia [15]. All these findings, coupled with data herein reported, provide strong evidence that Megatrypanum trypanosomes exhibit a narrow host range or even host specificity. Each trypanosome species/genotype was found in a single host species or in closely phylogenetically related hosts, those found in cervids were never detected in bovids, although one host species can harbour trypanosomes of more than one species or genotype [12, 23–25, 53]. To date, reports of elk and WTD sharing trypanosome genotype [12] relied merely on DNA detection, and genuine infections remain to be demonstrated. Reports based on exclusively on morphology of T. cervi, originally in an elk [28], and subsequently in a range of deer including WTD, wapitis [28], mule deer [9], moose [10] and reindeer in the USA [8], and in European fallow, roe and red deer [6] must be all molecularly confirmed.
It has been demonstrated by isoenzyme and karyotype analyses that the trypanosome found in Swedish reindeer differ from those found in moose, and both differed from cattle isolates, despite all these animals living in sympatry [13]. Similarly, data from zymodemes suggested the existence of different species of Megatrypanum infecting distinct species of deer and cattle in Germany [59]. The isolates of T. trinaperronei n. sp. from Venezuela and the USA are closely related, but not identical. Interestingly, T. trinaperronei n. sp. is more related to deer trypanosomes from Germany, Croatia, Poland and Russia, all nested into TthII lineage [15, 29, 31, 54], than to trypanosomes found in sympatric WTD and elk (USA) nested into TthI, a lineage also harbouring a trypanosome of Japanese sika deer [12, 16].
Our findings agreed with multiple and relatively recent crossings of the Bering Strait by cervids infected with Megatrypanum trypanosomes reaching North America from Eurasia, and from these regions dispersing through the world. Altogether, deer-trypanosome-vector associations and phylogeography support a plausible evolutionary scenario where WTD infected with the ancestor of T. trinaperronei n. sp., likely infested by its tightly linked ectoparasite L. mazamae, were introduced from North America into South America through the Panama Isthmus, reaching this continent at the Pliocene-Pleistocene boundary [1, 60]. Cervidae originated in Asia between 7.7 and 9.6 mya, and according to fossil records, deer did not cross the Bering Land Bridge to North America before 4.2 to 5.7 mya [60]. South American cervids are thought to have originated from at least two invasion events by North American deer: first, by the common ancestor of all deer species endemic to South America during the Great American Interchange at the Early Pliocene (~ 3 mya), and more recently (~ 1.5 mya) by WTD at the Pliocene-Pleistocene boundary [1, 60]. Unfortunately, the only trypanosome reported in a deer species endemic to South America is T. mazamarum, described in the blood of the brocket deer in Argentina [19], and never cultivated or molecularly characterized.
Concordant with our data on Megatrypanum trypanosomes, host-helminth assemblages were also associated with an early dispersion of cervids and bovids from Eurasia into North America, and then into the Neotropics [61]. Also supporting the recent dispersion of cervids and their parasites, Plasmodium sp. from the South American pampas deer (Ozotoceros bezoarticus) is closely related to Plasmodium odocoilei of North American WTD, and these two species are estimated to have diverged just by 0.3–0.9 mya [62].
Development of T. trinaperronei n. sp. in haemocultures and cultures with insect and mammalian cells
In early (7–10 days) haemocultures of WTD blood, live flagellates (phase-contrast microscopy) exhibited a few trypomastigotes (Fig. 2a–c) with large body length, pointed posterior ends and noticeable undulating membrane, alongside large transition forms between trypomastigote and epimastigote forms (Fig. 2d, e, g), and dividing epimastigotes (Fig. 2f). Flagellates of T. trinaperronei n. sp. from early haemocultures seeded on monolayers of insect cells (Hi-5), at 25 °C, initially formed clumps of small and rounded forms attached to insect cell membranes (Fig. 2h, i); these forms increased in length to became epimastigotes that remained adhered by their flagella forming rosettes until released into the supernatant of cultures (Fig. 2j). The developmental forms of T. trinaperronei n. sp. co-cultivated with Hi-5 insect cells very much resembled those reported for T. (Megatrypanum) spp. in the gut of the ked L. cervi taken from red deer [38] and T. melophagium adhered to the cells of gut walls of sheep keds [15, 21].
Epimastigotes of T. trinaperronei n. sp. in log phase Hi-5 cultures (5 days) multiplied intensively attached by their flagella forming large rosettes (Fig. 7a), which initially remained adhered to the insect cells and afterwards were released in the supernatant, where free epimastigotes became progressively abundant (Fig. 7a, b). Giemsa-stained epimastigotes showed the rounded kinetoplast adjacent and lateral to the central nucleus with an almost imperceptible undulating membrane, and a long free flagellum (Fig. 7a, b). In mid-log cultures (7 days), most epimastigotes became longer and thinner with a pointed posterior extremity (Fig. 7b). Stationary phase cultures (10 days) of T. trinaperronei n. sp. exhibited variable forms, all with a long free flagellum, including some wider epimastigotes exhibiting more preeminent undulating membranes (Fig. 7c). Some forms became progressively shortened in their posterior ends giving origin to blunted forms (indicated by arrowheads in Fig. 7c) during the differentiation of epimastigotes to trypomastigotes (Fig. 7b, c) and, finally, to ‛roundedʼ forms with a long flagellum (Fig. 7b, c), which most likely represented metacyclic trypomastigote forms (Fig. 7d). In contrast with the slow movement of the long epimastigotes, these ‛roundedʼ forms were highly mobile, and resembled metacyclic trypomastigotes of T. theileri described previously in the guts of tabanid flies and stationary cultures [57]. Overall, initial co-cultivation of T. trinaperronei n. sp. with insect cells, at 25 °C, showed flagellates resembling those of Trypanosoma (Megatrypanum) spp. present in the guts of L. cervi, the Old-World deer ked, collected from red deer [38].
Fig. 7.
Light and scanning electron microscopy (SEM) of Trypanosoma trinaperronei n. sp. co-cultured with insect cells at 25 °C. Giemsa-stained (a–c) forms of a log-phase (5 days) culture (a) exhibiting flagellates adhered by their flagella forming rosettes, and detached epimastigotes. b Mid-log (7 days) cultures showing long and thin epimastigotes. c Stationary culture (10 days) exhibiting epimastigotes, transition forms between epi- and trypomastigotes (arrow heads), and unique bell-shaped metacyclic trypomastigotes. A typical metacyclic was enlarged in (d). SEM of mid-log cultures (7 days) showing epimastigotes of variable length and width (e–j) including forms with well-developed undulant membranes (f, j), transition forms (f–h), and bell-shaped metacyclic trypomastigotes (f, g, k). Abbreviations: nucleus, n; kinetoplast, k; flagellum, f; undulating membrane, um; epimastigote, E; trypomastigote, T; metacyclic trypomastigotes, mT. Scale-bars: a–k, 10 µm
Scanning electron microscopy (SEM) of the mid-log cultures (7 days) of T. trinaperronei n. sp. in Hi-5 cultures showed flagellates of variable length and shape (Fig. 7e–j): slender epimastigotes without a noticeable undulating membrane (Fig. 7e) became broader epimastigotes exhibiting a conspicuous undulating membrane, easily detectable by SEM (Fig. 7f, i, j). Following the differentiation from epimastigotes to trypomastigotes, a range of transition forms (indicated by arrowheads in Fig. 7f–h) were observed, including flagellates with a pointed posterior end and swollen central region (Fig. 7f–h), which progressively turn into forms with a blunt posterior extremity until whole differentiation into bell-shaped flagellates with long free flagella (Fig. 7f, g, k), which correspond to the apparently ‛roundedʼ metacyclic trypomastigotes observed by light microscopy (Fig. 7c, d).
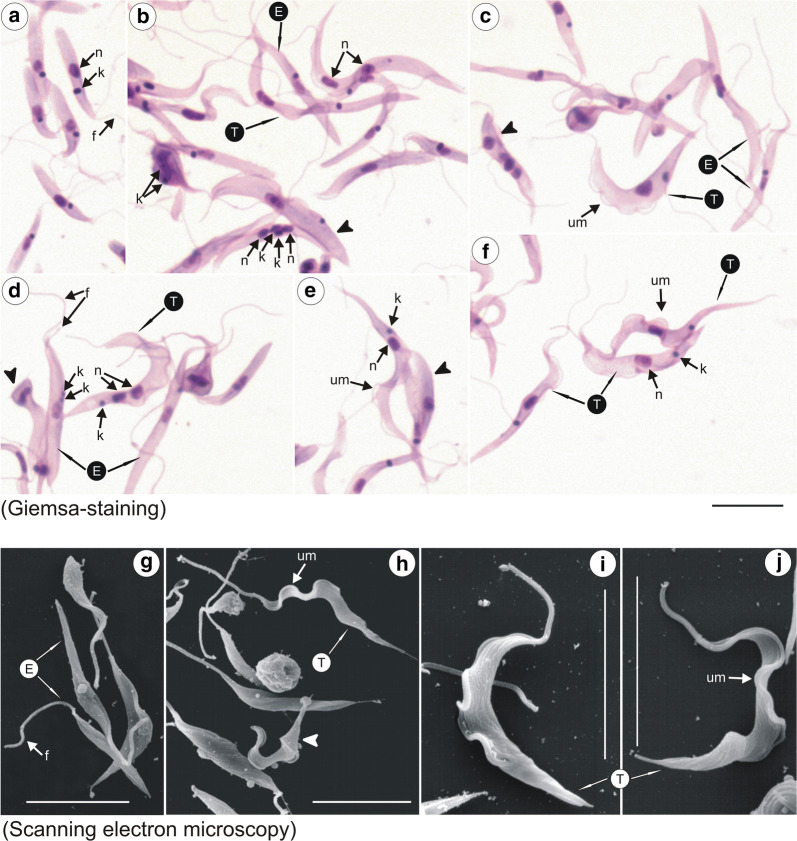
Log-phase epimastigotes from Hi5-cultures were seeded into monolayers of mammalian LLC-MK2 cells, incubated at 37 °C with 5% CO2, and after one to 5 days, cultures of Giemsa-stained flagellates were examined by light microscopy (Fig. 8a–f) and SEM (Fig. 8g–j). In the supernatant of these cultures, slender epimastigotes gradually became wider (Fig. 8a, g; one day culture) and gave origin to large and wide transition forms (indicated by arrowheads in Fig. 8e, h) between epimastigotes and trypomastigotes initially exhibiting wide bodies (Fig. 8b, c, h), and then becoming long and slender showing well-developed undulating membranes and pointed posterior ends (Fig. 8e, f, i, j). Both large epimastigotes and trypomastigotes are multiplicative forms (Fig. 8b, d). Long and slender forms with sharpened posterior ends and prominent undulating membranes (Fig. 8e, f, i, j) resemble those present in early haemocultures (Fig. 2) as well as blood trypomastigotes of T. theileri of cattle and T. theileri -like of water buffalo [6, 21, 23, 53, 63], and T. cervi and T. cervi-like [2, 6, 10, 38, 64]. Intracellular rounded flagellates resembling ‘amastigotes’ [65] could be observed inside mammalian cells (data not shown), but unquestionable demonstration of their intracellular development and differentiation requires further studies.
Fig. 8.
Light (Giemsa-staining) and scanning electron microscopy (SEM) of Trypanosoma trinaperronei n. sp. cultured with monolayers of mammalian (LLCMK2) cells at 37 °C. Giemsa-stained (a–f) and SEM (g–j) showing epimastigotes with one day of culture (a, g), and developmental forms (5 days): large and wide epimastigotes and trypomastigotes (b-d); flagellates with two nuclei and one kinetoplast (b, d); transition forms (arrow heads) between epimastigotes and trypomastigotes (b, d, e); and slender and pointed trypomastigotes with a well-developed undulating membrane (d–f, h–j).Abbreviations: nucleus, n; kinetoplast, k; flagellum, f; undulating membrane, um; epimastigote, E; trypomastigote, T; trypomastigote, T. Scale-bars: a–j,10 µm
Taken together, cultures of T. trinaperronei n. sp. showed large epimastigotes and trypomastigotes typical of Megatrypanum trypanosomes present in both early haemocultures (Fig. 2) and mammalian cell cultures (Fig. 8), similar to previously reported in deer blood [2, 6, 10, 38, 64]. In addition, clumps of small, rounded forms and epimastigotes detected in early co-cultures of T. trinaperronei n. sp. with insect cell (Fig. 7) were quite similar to those reported in guts of deer keds infected with T. (Megatrypanum) spp. [38]. Altogether, these findings allowed for inferences about the morphological differentiation through the life-cycle of T. trinaperronei n. sp. in vertebrate hosts and putative vectors according to the herein predicted life-cycle (Fig. 2). Morphological comparison of T. trinaperronei n. sp. blood trypomastigotes and epimastigotes from vector guts with corresponding forms of previously reported trypanosomes in deer species did not revealed species-specific features, thus corroborating the high morphological resemblance of all Megatrypanum trypanosomes [2, 6, 10, 21, 38, 64].
Ultrastructural characterization of Trypanosoma trinaperronei n. sp
Transmission electron microscopy (TEM) of cultured T. trinaperronei n. sp. revealed mitochondrion, Golgi, glycosomes, acidocalcisomes, flagellum, and overall ultrastructural organization typical of trypanosomatids. A set of features can be considered common of Megatrypanum trypanosomes: an abundance of acidocalcisomes (Fig. 9a, b) distributed throughout the cell body; a kinetoplast exhibiting lengthy and weakly compacted DNA fibrils (Fig. 9a–d); a noticeable spongiome comprising a network of tubules and contractile vacuoles near the flagellar pocket (Fig. 9c, e, f); and the absence of cytostome. To our knowledge, this is the first time that a deer trypanosome is characterized by TEM, and the ultra-structural arrangement was similar to that reported previously for T. theileri [23, 65].
Fig. 9.
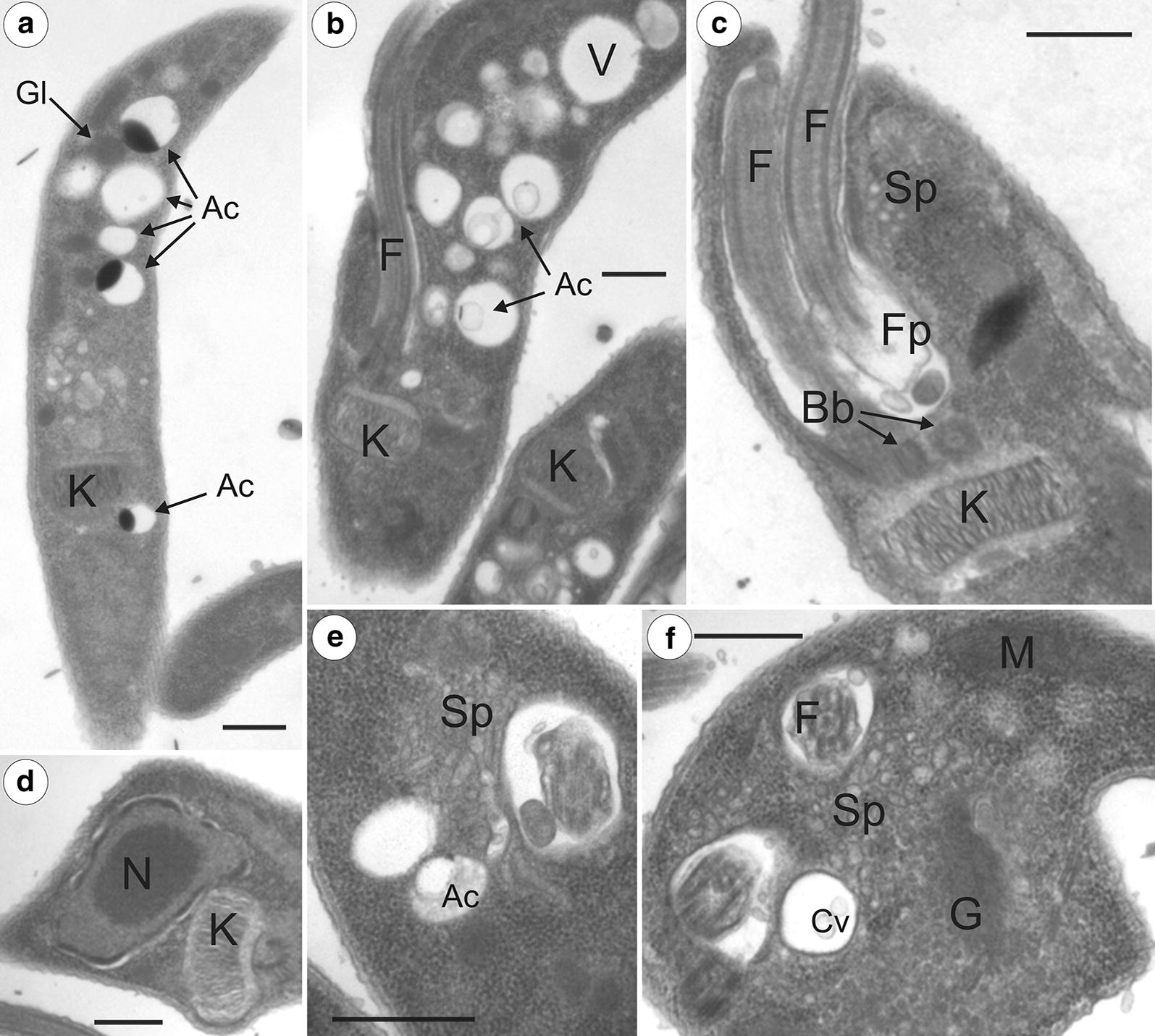
Ultrastructural features of Trypanosoma trinaperronei n. sp. revealed by transmission electron microscopy (TEM). Longitudinal section of epimastigote (a, c) and trypomastigote (b) forms showing many acidocalcisomes (a, b), and kinetoplasts displaying weakly compacted and long DNA fibrils (a–d). Flagellar pocket region and the spongiome, a network of tubules and large contractile vacuole (c, e, f). Longitudinal section of a dividing epimastigote exhibiting two flagella, two basal bodies, and a single kinetoplast (c). Abbreviations: nucleus, N; kinetoplast, K; flagellum, F; Golgi, G; glycosome, GI; contractile vacuole, Cv; basal body, Bb; acidocalcisomes, Ac; Mitochondria, M; spongiome, Sp. Scale-bars: a–f, 0.5 µm
Description of the new species
Family Trypanosomatidae Doflein, 1951
GenusTrypanosomaGruby, 1843
Trypanosoma(Megatrypanum)trinaperroneiTeixeira, Camargo & García n. sp.
Type-host: Odocoileus virginianus Zimmermann (Ruminantia, Cervidae), white-tailed deer.
Type-material: Hapantotype: the culture of the isolate TCC2268. Paratypes: blood samples of WTDs and gut samples of deer keds preserved in ethanol infected with Trypanosoma (M.) trinaperronei n. sp. Culture of T. trinaperronei n. sp. (TCC2268) is cryopreserved at the Trypanosomatid Culture Collection (TCC-USP) of the University of São Paulo, located at the Department of Parasitology, ICB, USP, São Paulo, Brazil, and registered in the World Data Centre for Microorganisms (WDCM611) of the Word Federation for Culture Collection (WFCC, 1981-10-14). TCC/USP also includes Giemsa-stained smears of cultures in glass slides, and blood samples of WTD at BSC (blood sample collection) and gut samples of deer keds at ISC (Insect Sample Collection).
Type-locality: State of Anzoátegui (10°07′08.95″N, 64°38′23.80″W), Venezuela.
Other locality: Texas, USA.
Invertebrate host (putative vector): Lipoptena mazamae Rondani, 1878 (Diptera: Hippoboscidae), deer ked.
Site in vertebrate host: Blood.
Site in invertebrate host: Digestive tract.
Representative DNA sequences: Trypanosoma (M.) trinaperronei n. sp. (TCC2268) DNA sequences deposited in the GenBank database as follows: SSU rRNA (MN752212); V7V8SSU rRNA (MN752143); gGAPDH (MN756794); ITS1 rDNA (MN752208, MN752209); and CATL (MN747149-MN747155). DNA sequences of Trypanosoma sp. PJH-2013a (isolate WTD A3) from the blood of WTD from Texas, USA herein designed as a genotype of T. trinaperronei n. sp.: SSU rRNA and ITS rDNA (JX178172-JX178173).
ZooBank registration: To comply with the regulations set out in Article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN) [66], details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) of the article is urn:lsid:zoobank.org:pub:0F4D44A3-E37D-4687-9DC6-9797E503E3C3.
Etymology: The name “trinaperronei” was given as a tribute to Dr Trina Mercedes Perrone Carmona, a Venezuelan biologist who contributed to the knowledge of animal trypanosomiasis and the progress of the veterinary sciences in Venezuela, and who died unexpectedly in 2008.
Description
Log-phase forms of T. trinaperronei n. sp. co-cultured with Hi-5 insect cells and examined by light microscopy and SEM were epimastigotes (n = 30) with body size averaging 21.61 ± 9.11 µm in length (range 13.42–47.02 µm) and 2.35 ± 1.03 µm in width (range 1.29–5.3 µm), lacking conspicuous undulating membranes and exhibiting a long (mean: 20.15 ± 6.35 µm; range: 11.90–30.3 µm) free flagellum in log-phase cultures (Fig. 7a–d); ‛roundedʼ forms with a posterior kinetoplast and long flagellum were observed in stationary-phase cultures (Fig. 7b–d). Trypanosoma trinaperronei n. sp. co-cultured with mammalian cells showed large and wide epimastigotes and trypomastigotes, and long and slender trypomastigotes with pointed posterior end, all forms exhibiting noticeable undulating membrane. Ultrastructural features (TEM) of T. trinaperronei n. sp. are typical of all Megatrypanum trypanosomes (Fig. 9).
Remarks
Trypanosoma trinaperronei n. sp. was detected in WTD from Venezuela (isolate WTD TCC2268) and USA (isolate WTD A3). SSU rRNA and ITS rDNA sequences of these two isolates diverged by 0.3 and 0.6%, respectively. The new species T. trinaperronei n. sp. and its closest relative Trypanosoma sp. D30 of fallow deer from Germany (both positioned in the TthII lineage of the subgenus Megatrypanum) diverged by 0.5%, 2.2% and 17% on SSU rRNA, gGAPDH and ITS rDNA sequences, respectively. Recent phylogenies detected DNA of a trypanosome of the TthI lineage in WTD and elk from USA [12]. Trypanosoma mazamarum, described in the blood of brocket deer (Mazama sp.) restricted to South America [19] exhibited large blood trypomastigotes typical of all species of the subgenus Megatrypanum, but neither cultures nor DNA sequences of this trypanosome are available for comparison with the molecularly characterized trypanosomes of this subgenus, which are virtually morphologically indistinguishable trypanosomes from bovids and cervids.
Conclusions
In the present study, we combined molecular, morphological, behaviour in cultures, biological and phylogeographical data to describe T. trinaperronei n. sp. detected in WTD blood and deer keds. Phylogeographical histories of cervids and respective trypanosomes from this and previous studies support historical dispersion of cervids and co-migrating Megatrypanum trypanosomes. Altogether, deer-trypanosome-vector associations and phylogeography of both trypanosomes and deer support a plausible evolutionary scenario where WTD infected with the ancestor of T. trinaperronei n. sp., likely infested by its tightly linked ectoparasite L. mazamae, were introduced from North America into South America through the Panama Isthmus. This scenario is strongly reinforced by the discovery of recently diversified geographical variants of the Pan-American T. trinaperronei n. sp. in the USA and Venezuela, compatible with the introduction of T. trinaperronei n. sp. through the recent invasion of South America by WTD from North America at the Pliocene-Pleistocene boundary. Underestimated genetic repertoire and entangled relationships of morphologically indistinguishable trypanosomes from cervids and bovids highlight the need for more comprehensive surveys to assess species richness, and host-parasite-vector associations of Megatrypanum trypanosomes.
Supplementary information
Additional file 1: Table S1. Isolates of trypanosomes of the subgenus Megatrypanum employed for network inferences using cathepsin L sequences (CATL).
Additional file 2: Table S2. Isolates of trypanosomes of the subgenus Megatrypanum employed for phylogenetic inferences using SSU rRNA sequences.
Additional file 3: Table S3. Isolates of trypanosomes of the subgenus Megatrypanum employed for phylogenetic inferences using gGAPDH sequences.
Additional file 4: Table S4. Isolates of trypanosomes of the subgenus Megatrypanum employed for Network inferences using ITS1 rDNA sequences.
Acknowledgements
We are thankful to “Empresas Polar”, Barcelona, state of Anzoátegui in Venezuela for authorization, logistic support and help in deer management. The authors would like to thank CE Jared and MM Antoniazzi for the access to electron microscopic facilities of The Institute Butantan, Brazil.
Abbreviations
- TCC
Trypanosomatid Culture Collection of the Department of Parasitology, ICB, University of São Paulo, SP, Brazil
- SSU rRNA
small subunit of ribosomal RNA
- gGAPDH
glycosomal glyceraldehyde-3-phosphate dehydrogenase
- CATL
cathepsin L
- ITS rDNA
internal transcribed spacer of rDNA
- PCR
polymerase chain reaction
- Hi-5
insect (Trichoplusia sp.) cell
- WTD
white-tailed deer
- mya
million years ago
Authors’ contributions
HAG, MMGT and EPC conceived and supported the study. HAG, CMFR and PAB designed and coordinated field and laboratory work. HAG and PAB performed the fieldwork in Venezuela and participated in parasite isolation and morphological analysis. HAG, CMFR and ACR were responsible for the molecular diagnosis and phylogenetic analyses. MC carried out culturing, behavioural and morphological analyses. CSAT performed electron microscopic analyses. HAG, MMGT and EPC wrote the manuscript. All authors contributed to the revisions of the manuscript. All authors read and approved the final manuscript.
Funding
This study received financial support through Brazilian grants from the PROSUL program (MCTI-CNPq) and FAPESP (Process no. 2016/07487-0). CMFR and HAG are post-doctoral fellows of CNPq (INCT-EpiAmo) and FAPESP (Process no. 2016/03028-1), respectively.
Availability of data and materials
Cultured flagellates cryopreserved and in glass slides, and DNA samples of Trypanosoma trinaperronei n. sp. are deposited at the Trypanosomatid Culture Collection (TCC-USP) of the Department of Parasitology, ICB, USP, São Paulo, Brazil, under the accession number TCC2268. The newly generated DNA sequences were deposited in the GenBank database under the accession numbers MN752212 (SSU rRNA), MN752143 (V7V8 SSU rRNA), MN756794 (gGAPDH), MN752208, MN752209 (ITS1 rDNA) and MN747149-MN747155 (CATL gene).
Ethics approval and consent to participate
All field procedures and deer handling were performed in accordance with the approved protocols, and under the supervision of the MINEC (the Venezuelan Ministerio del Poder Popular para el Ecosocialismo).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Herakles A. Garcia, Email: heraklesantonio@gmail.com
Pilar A. Blanco, Email: albla69@yahoo.com.mx
Adriana C. Rodrigues, Email: fuzatoadriana@gmail.com
Carla M. F. Rodrigues, Email: cmonadeli@hotmail.com
Carmen S. A. Takata, Email: ctakata@usp.br
Marta Campaner, Email: camp@usp.br.
Erney P. Camargo, Email: erney@usp.br
Marta M. G. Teixeira, Email: mmgteix@icb.usp.br
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-020-04169-0.
References
- 1.Heckeberg NS, Erpenbeck D, Wörheide G, Rössner GE. Systematic relationships of five newly sequenced cervid species. PeerJ. 2016;4:e2307. doi: 10.7717/peerj.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefebvre MF, Semalulu SS, Oatway AE, Nolan JW. Trypanosomiasis in woodland caribou of northern Alberta. J Wildl Dis. 1997;33:271–277. doi: 10.7589/0090-3558-33.2.271. [DOI] [PubMed] [Google Scholar]
- 3.Kutz SJ, Ducrocq J, Verocai GG, Hoar BM, Colwell DD, Beckmen KB, et al. Parasites in ungulates of Arctic North America and Greenland: a view of contemporary diversity, ecology, and impact in a world under change. Adv Parasitol. 2012;79:99–252. doi: 10.1016/B978-0-12-398457-9.00002-0. [DOI] [PubMed] [Google Scholar]
- 4.Kingston N, Drozdz J, Rutkowska M. Trypanosoma spp. in red deer (Cervus elaphus) and elk (Alces alces) in Poland. Proc Helminthol Soc Wash. 1985;52:144–145. [Google Scholar]
- 5.Morton JK, Kingston N. Further studies on trypanosomes in game animals in Wyoming. J Wildl Dis. 1976;12:233–236. doi: 10.7589/0090-3558-12.2.233. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann M, Büscher G, Friedhoff KT. Stercorarian trypanosomes from deer (Cervidae) in Germany. J Protozool. 1984;31:581–584. doi: 10.1111/j.1550-7408.1984.tb05509.x. [DOI] [PubMed] [Google Scholar]
- 7.Kingston N, Bobek B. A trypanosome in roe deer, Capreolus capreolus, in southern Poland. Proc Helminthol Soc Wash. 1985;52:143. [Google Scholar]
- 8.Kingston N, Morton JK, Dieterich R. Trypanosoma cervi from Alaskan reindeer, Rangifer tarandus. J Protozool. 1982;29:588–591. doi: 10.1111/j.1550-7408.1982.tb01342.x. [DOI] [PubMed] [Google Scholar]
- 9.Matthews MJ, Kingston N, Morton JK. Trypanosoma cervi Kingston and Morton, 1975 from mule deer, Odocoileus hemionus, in Wyoming. J Wildl Dis. 1977;13:33–39. doi: 10.7589/0090-3558-13.1.33. [DOI] [PubMed] [Google Scholar]
- 10.Kingston N, Franzmann A, Maki L. Redescription of Trypanosoma cervi (Protozoa) in moose, Alces alces, from Alaska and Wyoming. Proc Helminthol Soc Wash. 1985;52:54–59. [Google Scholar]
- 11.Telford SR, Forrester DJ, Wright SD, Roelke ME, Ferenc SA, McCown JW. The identity and prevalence of Trypanosoma in white-tailed deer (Odocoileus virginianus) from South Florida. J Helminthol Soc Wash. 1991;58:19–23. [Google Scholar]
- 12.Fisher AC, Schuster G, Cobb WJ, James AM, Cooper SM, Peréz de León AA, et al. Molecular characterization of Trypanosoma (Megatrypanum) spp. infecting cattle (Bos taurus), white-tailed deer (Odocoileus virginianus), and elk (Cervus elaphus canadensis) in the United States. Vet Parasitol. 2013;197:29–42. doi: 10.1016/j.vetpar.2013.04.037. [DOI] [PubMed] [Google Scholar]
- 13.Dirie MF, Bornstein S, Wallbanks KR, Molyneux DH, Steen M. Comparative studies on Megatrypanum trypanosomes from cervids. Trop Med Parasitol. 1990;41:198–202. [PubMed] [Google Scholar]
- 14.Kingston N, Bobek B, Perzanowski K, Wita I, Maki L. Description of Trypanosoma (Megatrypanum) stefanskii n. sp. from roe deer (Capreolus capreolus) in Poland. J Helminthol Soc Wash. 1992;59:89–95. [Google Scholar]
- 15.Martinković F, Matanović K, Rodrigues AC, Garcia HA, Teixeira MMG. Trypanosoma (Megatrypanum) melophagium in the sheep ked Melophagus ovinus from organic farms in Croatia: phylogenetic inferences support restriction to sheep and sheep keds and close relationship with trypanosomes from other ruminant species. J Eukaryot Microbiol. 2012;59:134–144. doi: 10.1111/j.1550-7408.2011.00599.x. [DOI] [PubMed] [Google Scholar]
- 16.Hatama S, Shibahara T, Suzuki M, Kadota K, Uchida I, Kanno T. Isolation of a Megatrypanum trypanosome from sika deer (Cervus nippon yesoensis) in Japan. Vet Parasitol. 2007;149:56–64. doi: 10.1016/j.vetpar.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 17.D’Alessandro A, Wells EA. Trypanosome infections in the family Cervidae. Trans R Soc Trop Med Hyg. 1971;65:845–846. doi: 10.1016/0035-9203(71)90108-8. [DOI] [PubMed] [Google Scholar]
- 18.Ayala SC, DAlessandro A, Mackenzie R, Angel D. Hemoparasite infections in wild animals from the eastern llanos of Colombia. J Parasitol. 1973;59:52–9. [PubMed]
- 19.Mazza S, Romana C, Fiora A. Algunos hemoparásitos de mamíferos del Norte. VII. Reunion Soc Argent Patol Reg Norte. 1932;2:990–997. [Google Scholar]
- 20.Deane LM. Tripanosomideos de mamíferos da região Amazônica. I. Alguns flagelados encontrados no sangue de mamíferos silvestres do Estado do Pará. Rev Inst Med Trop S Paulo. 1961;3:15–28. [Google Scholar]
- 21.Hoare CA. The trypanosomes of mammals: a zoological monograph. Oxford: Blackwell Scientific Publications; 1972. [Google Scholar]
- 22.Böse R, Friedhoff KT, Olbrich S, Büscher G, Domeyer I. Transmission of Trypanosoma theileri to cattle by Tabanidae. Parasitol Res. 1987;73:421–424. doi: 10.1007/BF00538199. [DOI] [PubMed] [Google Scholar]
- 23.Rodrigues AC, Campaner M, Takata CS, Dell’Porto A, Milder RV, Takeda GF, et al. Brazilian isolates of Trypanosoma (Megatrypanum) theileri: diagnosis and differentiation of isolates from cattle and water buffalo based on biological characteristics and randomly amplified DNA sequences. Vet Parasitol. 2003;116:185–207. doi: 10.1016/s0304-4017(03)00236-x. [DOI] [PubMed] [Google Scholar]
- 24.Rodrigues AC, Paiva F, Campaner M, Stevens JR, Noyes HA, Teixeira MMG. Phylogeny of Trypanosoma (Megatrypanum) theileri and related trypanosomes reveals lineages of isolates associated with artiodactyl hosts diverging on SSU and ITS ribosomal sequences. Parasitology. 2006;132:215–224. doi: 10.1017/S0031182005008929. [DOI] [PubMed] [Google Scholar]
- 25.Garcia HA, Rodrigues AC, Martinković F, Minervino AH, Campaner M, Nunes VL, et al. Multilocus phylogeographical analysis of Trypanosoma (Megatrypanum) genotypes from sympatric cattle and water buffalo populations supports evolutionary host constraint and close phylogenetic relationships with genotypes found in other ruminants. Int J Parasitol. 2011;41:1385–1396. doi: 10.1016/j.ijpara.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Ramírez JD, Tapia-Calle G, Muñoz-Cruz G, Poveda C, Rendón LM, Hincapié E, et al. Trypanosome species in neo-tropical bats: biological, evolutionary and epidemiological implications. Infect Genet Evol. 2014;22:250–256. doi: 10.1016/j.meegid.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jirků M, Votýpka J, Petrželková KJ, Jirků-Pomajbíková K, Kriegová E, Vodička R, et al. Wild chimpanzees are infected by Trypanosoma brucei. Int J Parasitol Parasites Wildl. 2015;4:277–282. doi: 10.1016/j.ijppaw.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kingston N, Morton JK. Trypanosoma cervi n. sp. from elk (Cervus canadensis) in Wyoming. J Parasitol. 1975;61:17–23. [PubMed] [Google Scholar]
- 29.Werszko J, Szewczyk T, Steiner-Bogdaszewska Ż, Wróblewski P, Karbowiak G, Laskowski Z. Molecular detection of Megatrypanum trypanosomes in tabanid flies. Med Vet Entomol. 2020;34:69–73. doi: 10.1111/mve.12409. [DOI] [PubMed] [Google Scholar]
- 30.Krinsky WL, Pechuman LL. Trypanosomes in horse flies and deer flies in central New York State. J Parasitol. 1975;61:12–16. [PubMed] [Google Scholar]
- 31.Ganyukova AI, Zolotarev AV, Malysheva MN, Frolov AO. First record of Trypanosoma theileri-like flagellates in horseflies from Northwest Russia. Protistology. 2018;12:223–230. [Google Scholar]
- 32.Clark GG. Trypanosomes from mule deer in New Mexico and Colorado. J Wildl Dis. 1972;8:325–326. doi: 10.7589/0090-3558-8.4.325. [DOI] [PubMed] [Google Scholar]
- 33.Jaimes-Dueñez J, Triana-Chávez O, Mejía-Jaramillo AM. Spatial-temporal and phylogeographic characterization of Trypanosoma spp. in cattle (Bos taurus) and buffaloes (Bubalus bubalis) reveals transmission dynamics of these parasites in Colombia. Vet Parasitol. 2018;249:30–42. doi: 10.1016/j.vetpar.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Taioe MO, Motloang MY, Namangala B, Chota A, Molefe NI, Musinguzi SP, et al. Characterization of tabanid flies (Diptera: Tabanidae) in South Africa and Zambia and detection of protozoan parasites they are harbouring. Parasitology. 2017;144:1162–1178. doi: 10.1017/S0031182017000440. [DOI] [PubMed] [Google Scholar]
- 35.Votýpka J, Rádrová J, Skalický T, Jirků M, Jirsová D, Mihalca AD, et al. A tsetse and tabanid fly survey of African great apes habitats reveals the presence of a novel trypanosome lineage but the absence of Trypanosoma brucei. Int J Parasitol. 2015;45:741–748. doi: 10.1016/j.ijpara.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Votýpka J, Brzoňová J, Ježek J, Modrý D. Horse flies (Diptera: Tabanidae) of three West African countries: a faunistic update, barcoding analysis and trypanosome occurrence. Acta Trop. 2019;197:105069. doi: 10.1016/j.actatropica.2019.105069. [DOI] [PubMed] [Google Scholar]
- 37.Gibson W, Pilkington JG, Pemberton JM. Trypanosoma melophagium from the sheep ked Melophagus ovinus on the island of St Kilda. Parasitology. 2010;137:1799–1804. doi: 10.1017/S0031182010000752. [DOI] [PubMed] [Google Scholar]
- 38.Böse R, Petersen K. Lipoptena cervi (Diptera), a potential vector of Megatrypanum trypanosomes of deer (Cervidae) Parasitol Res. 1991;77:723–725. doi: 10.1007/BF00928691. [DOI] [PubMed] [Google Scholar]
- 39.Skvarla MJ, Machtinger ET. Deer Keds (Diptera: Hippoboscidae: Lipoptena and Neolipoptena) in the United States and Canada: new state and county records, pathogen records, and an illustrated key to Species. J Med Entomol. 2019;56:744–760. doi: 10.1093/jme/tjy238. [DOI] [PubMed] [Google Scholar]
- 40.Garcia HA, Rodrigues CMF, Rodrigues AC, Pereira DL, Pereira CL, Camargo EP, et al. Remarkable richness of trypanosomes in tsetse flies (Glossina morsitans morsitans and Glossina pallidipes) from the Gorongosa National Park and Niassa National Reserve of Mozambique revealed by fluorescent fragment length barcoding (FFLB) Infect Genet Evol. 2018;63:370–379. doi: 10.1016/j.meegid.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 41.Fermino BR, Paiva F, Viola LB, Rodrigues CMF, Garcia HA, Campaner M, et al. Shared species of crocodilian trypanosomes carried by tabanid flies in Africa and South America, including the description of a new species from caimans, Trypanosoma kaiowa n. sp. Parasit Vectors. 2019;12:225. doi: 10.1186/s13071-019-3463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigues AC, Garcia HA, Ortiz PA, Cortez AP, Martinković F, Paiva F, et al. Cysteine proteases of Trypanosoma (Megatrypanum) theileri: cathepsin L-like gene sequences as targets for phylogenetic analysis, genotyping diagnosis. Parasitol Int. 2010;59:318–325. doi: 10.1016/j.parint.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Rodrigues AC, Garcia HA, Batista JS, Minervino AH, Góes-Cavalcante G, Maia da Silva F, et al. Characterization of spliced leader genes of Trypanosoma (Megatrypanum) theileri: phylogeographical analysis of Brazilian isolates from cattle supports spatial clustering of genotypes and parity with ribosomal markers. Parasitology. 2010;137:111–122. doi: 10.1017/S0031182009991053. [DOI] [PubMed] [Google Scholar]
- 44.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;11:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Massachusetts: Sinauer Associates; 2002. [Google Scholar]
- 46.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 47.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 48.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 49.Lima L, Espinosa-Álvarez O, Hamilton PB, Neves L, Takata CS, Campaner M, et al. Trypanosoma livingstonei: a new species from African bats supports the bat seeding hypothesis for the Trypanosoma cruzi clade. Parasit Vectors. 2013;6:e221. doi: 10.1186/1756-3305-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia HA, Kamyingkird K, Rodrigues AC, Jittapalapong S, Teixeira MMG, Desquesnes M. High genetic diversity in field isolates of Trypanosoma theileri assessed by analysis of cathepsin L-like sequences disclosed multiple and new genotypes infecting cattle in Thailand. Vet Parasitol. 2011;180:363–367. doi: 10.1016/j.vetpar.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 51.Ybañez AP, Sivakumar T, Ybañez RH, Vincoy MR, Tingson JA, Perez ZO, et al. Molecular survey of bovine vector-borne pathogens in Cebu, Philippines. Vet Parasitol. 2013;196:13–20. doi: 10.1016/j.vetpar.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Weerasooriya G, Sivakumar T, Lan DT, Long PT, Takemae H, Igarashi I, et al. Epidemiology of bovine hemoprotozoa parasites in cattle and water buffalo in Vietnam. J Vet Med Sci. 2016;78:1361–1367. doi: 10.1292/jvms.16-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suganuma K, Kondoh D, Sivakumar T, Mizushima D, Elata ATM, Thekisoe OMM, et al. Molecular characterization of a new Trypanosoma (Megatrypanum) theileri isolate supports the two main phylogenetic lineages of this species in Japanese cattle. Parasitol Res. 2019;118:1927–1935. doi: 10.1007/s00436-019-06313-x. [DOI] [PubMed] [Google Scholar]
- 54.Calzolari M, Rugna G, Clementi E, Carra E, Pinna M, Bergamini F, et al. Isolation of a trypanosome related to Trypanosoma theileri (Kinetoplastea: Trypanosomatidae) from Phlebotomus perfiliewi (Diptera: Psychodidae) Biomed Res Int. 2018;15:e2597074. doi: 10.1155/2018/2597074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lloyd JE. Louse flies, keds, and related flies (Hippoboscoidea) In: Mullen G, Durden L, editors. Medical and veterinary entomology. San Diego: Academic Press; 2002. pp. 349–362. [Google Scholar]
- 56.Böse R, Friedhoff KT, Olbrich S. Transmission of Megatrypanum trypanosomes to Cervus dama by Tabanidae. J Protozool. 1987;34:110–113. doi: 10.1111/j.1550-7408.1987.tb03143.x. [DOI] [PubMed] [Google Scholar]
- 57.Böse R, Heister NC. Development of Trypanosoma (M.) theileri in tabanids. J Eukaryot Microbiol. 1993;40:788–792. doi: 10.1111/j.1550-7408.1993.tb04475.x. [DOI] [PubMed] [Google Scholar]
- 58.Schoener E, Uebleis SS, Cuk C, Nawratil M, Obwaller AG, Zechmeister T, et al. Trypanosomatid parasites in Austrian mosquitoes. PLoS ONE. 2018;13:e0196052. doi: 10.1371/journal.pone.0196052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Böse R, Petersen K, Pospichal H, Buchanan N, Tait A. Characterization of Megatrypanum trypanosomes from European Cervidae. Parasitology. 1993;107:55–61. doi: 10.1017/s0031182000079403. [DOI] [PubMed] [Google Scholar]
- 60.Gilbert C, Ropiquet A, Hassanin A. Mitochondrial and nuclear phylogenies of Cervidae (Mammalia, Ruminantia): systematics, morphology, and biogeography. Mol Phylogenet Evol. 2006;40:101–117. doi: 10.1016/j.ympev.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 61.Hoberg EP, Galbreath KE, Cook JA, Kutz SJ, Polley L. Northern host-parasite assemblages: history and biogeography on the borderlands of episodic climate and environmental transition. Adv Parasitol. 2012;79:1–97. doi: 10.1016/B978-0-12-398457-9.00001-9. [DOI] [PubMed] [Google Scholar]
- 62.Asada M, Takeda M, Tomas WM, Pellegrin A, de Oliveira CHS, Barbosa JD, et al. Close relationship of Plasmodium sequences detected from South American pampas deer (Ozotoceros bezoarticus) to Plasmodium spp. in North American white-tailed deer. Int J Parasitol Parasites Wildl. 2018;7:44–47. doi: 10.1016/j.ijppaw.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wells EA. Subgenus Megatrypanum. In: Lumsden WHR, Evans DA, editors. Biology of the kinetoplastida. London: Academic Press; 1976. pp. 257–275. [Google Scholar]
- 64.Krinsky WL. Trypanosomes from white-tailed deer (Odocoileus virginianus) in New York. J Parasitol. 1975;61:145–146. [PubMed] [Google Scholar]
- 65.Lee YF, Cheng CC, Chen JS, Lin NN, Hung YW, Wang JM, et al. Evidence of intracellular stages in Trypanosoma (Megatrypanum) theileri in non-phagocytic mammalian cells. Vet Parasitol. 2013;191:228–239. doi: 10.1016/j.vetpar.2012.08.027. [DOI] [PubMed] [Google Scholar]
- 66.ICZN International Commission on Zoological Nomenclature: amendment of articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to expand and refine methods of publication. Bull Zool Nomencl. 2012;69:161–169. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Isolates of trypanosomes of the subgenus Megatrypanum employed for network inferences using cathepsin L sequences (CATL).
Additional file 2: Table S2. Isolates of trypanosomes of the subgenus Megatrypanum employed for phylogenetic inferences using SSU rRNA sequences.
Additional file 3: Table S3. Isolates of trypanosomes of the subgenus Megatrypanum employed for phylogenetic inferences using gGAPDH sequences.
Additional file 4: Table S4. Isolates of trypanosomes of the subgenus Megatrypanum employed for Network inferences using ITS1 rDNA sequences.
Data Availability Statement
Cultured flagellates cryopreserved and in glass slides, and DNA samples of Trypanosoma trinaperronei n. sp. are deposited at the Trypanosomatid Culture Collection (TCC-USP) of the Department of Parasitology, ICB, USP, São Paulo, Brazil, under the accession number TCC2268. The newly generated DNA sequences were deposited in the GenBank database under the accession numbers MN752212 (SSU rRNA), MN752143 (V7V8 SSU rRNA), MN756794 (gGAPDH), MN752208, MN752209 (ITS1 rDNA) and MN747149-MN747155 (CATL gene).