Abstract
Background
Interaction of C. albicans with oral bacteria is crucial for its persistence, but also plays a potential role in the infection process. In the oral cavity, it grows as part of dental plaque biofilms. Even though growth and interaction of C. albicans with certain bacterial species has been studied, little is known about its biofilm growth in vitro in the simultaneous presence of Gram-negative and Gram-positive bacteria. The aim was to evaluate the growth of C. albicans in polymicrobial biofilms comprising oral Gram-negative and Gram-positive bacteria. Further, we also aimed to assess the potential of C. albicans in the Candida-bacteria polymicrobial biofilm to elicit cytokine gene expression and cytokine production from human blood cells.
Results
C. albicans cell counts increased significantly up to 48 h in polymicrobial biofilms (p < 0.05), while the bacterial counts in the same biofilms increased only marginally as revealed by qPCR absolute quantification. However, the presence of bacteria in the biofilm did not seem to affect the growth of C. albicans. Expression of IL-8 gene was significantly (p < 0.05) higher upon stimulation from biofilm-supernatants than from biofilms in polymicrobial setting. On the contrary, TNF-α expression was significantly higher in biofilms than in supernatants but was very low (1–4 folds) in the monospecies biofilm of C. albicans. ELISA cytokine quantification data was in agreement with mRNA expression results.
Conclusion
Persistence and enhanced growth of C. albicans in polymicrobial biofilms may imply that previously reported antagonistic effect of A. actinomycetemcomitans was negated. Increased cytokine gene expression and cytokine production induced by Candida-bacteria polymicrobial biofilms and biofilm supernatants suggest that together they possibly exert an enhanced stimulatory effect on IL-8 and TNF-α production from the host.
Keywords: Candida albicans, Polymicrobial biofilms, qPCR, Gram-positive bacteria, Gram-negative bacteria, Cytokines
Background
Candida albicans is a commensal fungus that colonizes the oral cavity and various other sites in human body. Its extensive interaction with oral bacteria might be crucial for its persistence but also potentially contributes to infection process [1]. Despite that complex microbial interactions in dental plaque biofilm have been extensively studied during the past 10–15 years, inter-kingdom interactions have received little attention [2].
C. albicans coexists with a multitude of bacterial species [3] and its interactions with streptococci are often mutually beneficial for their survival in diverse oral niches [2]. On the other hand, not much is known of the interaction between C. albicans and the Gram-negatives such as Aggregatibacter actinomycetemcomitans. Intriguingly, autoinducer 2 (AI-2) of A. actinomycetemcomitans inhibits C. albicans biofilm formation [4], while the same factor produced by streptococci has the opposite effect [5], suggesting differences in roles of the same quorum sensing factors released by each bacterial species.
A critical aspect of microbial infections is the provocation of the host cells to produce inflammatory cytokines. The ability of various Candida species and their biofilms to induce cytokine production from host cells has been reported in a number of studies [6–8] and a prominent role of key cytokines such as TNF-α and IL-8 in candidiasis has been known [7, 9]. Concurrently, diverse oral bacteria and their biofilms orchestrate host cytokine response as documented by a large number of in vitro and in vivo studies [10–14]. In the oral cavity, while encased in complex plaque biofilms, microorganisms can release their cellular components, which can breach host barriers impermeable to whole microbial cells, reach distant sites in the host body and cause tissue destruction. We and others have previously investigated the potential of bacterial biofilm supernatants containing secreted soluble components, in addition to biofilms, to cause cytokine production from host cells [14–17]. Proteomic analyses of biofilm-supernatants has revealed the presence of virulence-related proteins among other secreted proteins [18, 19]. However, there is a dearth of knowledge on cytokine-inducing potential of Candida-bacteria polymicrobial biofilms and biofilm-supernatants containing soluble secreted components from biofilms. While recent research has thrown more light on interactions operating between bacteria and Candida [20–22], little is known how Candida grows in the simultaneous presence of Gram-positive and Gram-negative bacterial partners in polymicrobial biofilms. Further, we also aimed to investigate cytokine gene expression and cytokine production from human blood cells upon challenge with Candida-bacteria polymicrobial biofilms.
Results
Biofilm formation by Candida and the bacterial species
C. albicans and all the test bacteria formed biofilms as evident from confocal laser scanning microscopy images (Fig. 1). The 3-dimensional images in Fig. 1 showed the formation of mat-like biofilms by C. albicans and the three test bacteria individually as well as when grown together.
Fig. 1.
Confocal laser scanning microscopy 3-dimensional (3D) images of monospecies and polymicrobial biofilms of bacteria and C. albicans. A. actinomycetemcomitans (Aa), S. mutans (Sm), S. gordonii (Sg) and C. albicans (Ca) were cultured as monospecies and polymicrobial (mix) in brucella broth in the wells of Millicell® EZ slides (Millipore) in aerobiosis in 5% CO2 at 37 °C for 24 and 48 h and stained with Syto9®. Images were acquired on Carl-Zeiss LSM 700 at × 630 magnification and 3D view reconstructed using the software ZEN 2012
qPCR quantification of Candida and bacteria in monospecies and polymicrobial biofilms
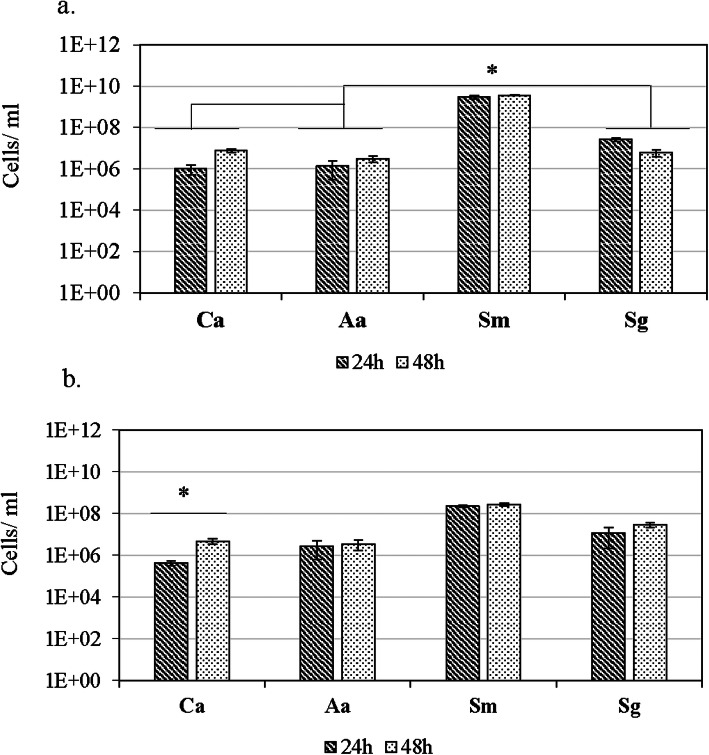
qPCR showed that C. albicans and all the test bacterial strains grew as monospecies biofilms till 48 h of incubation period. In monospecies biofilm, C. albicans median cells per ml increased 7-fold from 9.8 × 105 at 24 h to 6.4 × 106 after 48 h. Among the bacterial species, S. mutans quantities were highest (P < 0.05): 3 × 109 cells per ml at 24 h and remained nearly the same (3.7 × 109) after 48 h. S. gordonii biofilm showed median cells per ml 2.6 × 107 at 24 h and slightly decreased to 6 × 106 after 48 h. Median counts of A. actinomycetemcomitans were 106 cells per ml at 24 h and slightly increased to 3 × 106 cells per ml after 48 h (Fig. 2a).
Fig. 2.
Quantities of bacterial species and C. albicans grown as monospecies and polymicrobial biofilms. Standardized numbers of the four species were added to 24-well cell culture plates for monospecies (a) and polymicrobial biofilm (b). The cultures were incubated for 24 and 48 h. At the end of each time point, bacterial viability was checked by plating a small aliquot on brucella blood agar. Bacterial quantities (cells/ml) were determined by using qPCR SYBR Green assay. The results shown are median values from three independent experiments. Abbreviations: Ca = C. albicans, Aa = A. actinomycetemcomitans, Sm = S. mutans, Sg = S. gordonii
In polymicrobial biofilms, qPCR revealed the presence of all the four test species till 48 h of incubation (Fig. 2b). There was a significant (P < 0.05) increase in C. albicans median cells per ml: 13-fold from 3.7 × 105 at 24 h to 4.7 × 106 at 48 h. While S. mutans cell numbers did not seem to increase after 24 h, median cells per ml were about 1 log lower compared to the cell numbers in monospecies biofilm. In polymicrobial biofilms, from 24 h to 48 h, S. gordonii cells increased 3-fold whereas the increase in cell numbers was only one fold each in S. mutans and A. actinomycetemcomitans.
Cytokine gene expression in biofilm- and biofilm-supernatant stimulated human blood cells
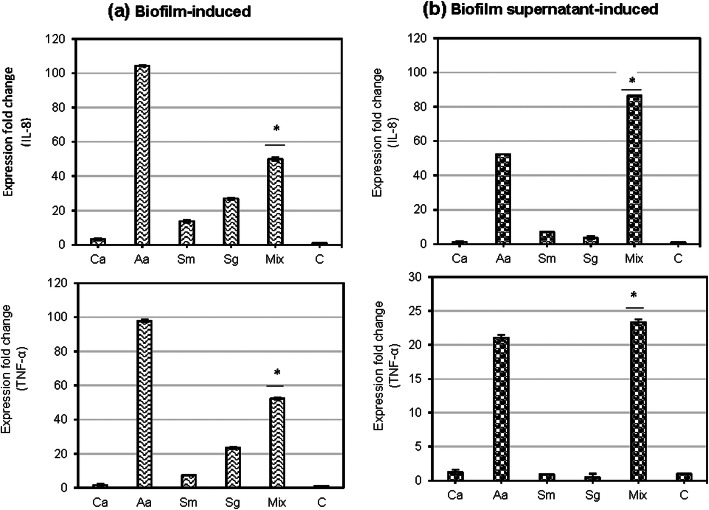
Upon stimulation of blood cells with supernatants from Candida-bacteria polymicrobial biofilm, significantly (P < 0.05) higher fold change in expression of IL-8 genes was observed (86-folds, respectively) than biofilms (50-folds). On the contrary, TNF- α was significantly (P < 0.05) higher in biofilms (52-fold) than in supernatants (23-fold) (Fig. 3).
Fig. 3.
mRNA expression of cytokine genes by biofilms (a) and biofilm supernatants (b). Human blood cells were stimulated by monospecies and polymicrobial biofilm (a) and biofilm supernatants (b) of bacteria and C. albicans. Abbreviations: Ca = C. albicans, Aa = A. actinomycetemcomitans, Sm = S. mutans, Sg = S. gordonii, Mix = Mixture, C=Control, *p < 0.05
Among bacterial biofilms, A. actinomycetemcomitans biofilm induced highest levels of IL-8 and TNF- α, as evident from ~ 100-fold (IL-8, TNF- α) increase in mRNA expression upon stimulation (Fig. 3a). S. gordonii biofilm, but not the supernatant, consistently induced higher folds of all cytokine mRNA expression than S. mutans (Fig. 3a). Cytokine gene expression fold change was very low (in the range of 1–4 folds) in blood cells stimulated with C. albicans monospecies biofilm.
Quantification of select cytokines by ELISA
Mean (SD) levels of IL-8 induced by C. albicans biofilm were 865 (699) pg/ml while C. albicans biofilm-supernatant induced a very low amount of 20 pg/ml. A. actinomycetemcomitans biofilm induced highest levels of IL-8, 26,781 (3307) pg/ml, while the biofilm-supernatant from the same species induced 9354 (2600) pg/ml. S. gordonii and S. mutans did show induction, 9859 (987) and 3315 (2751) pg/ml, respectively but, the levels were significantly (P < 0.05) lower than those induced by the polymicrobial biofilm and the respective supernatant. Biofilm supernatants from S. gordonii and S. mutans failed to induce IL-8. Highest IL-8 levels were induced by biofilms [27,786 (3248) pg/ml] and supernatants [22,312 (11939) pg/ml] of Candida-bacteria polymicrobial biofilm in comparison to monospecies cultures (Fig. 4a).
Fig. 4.
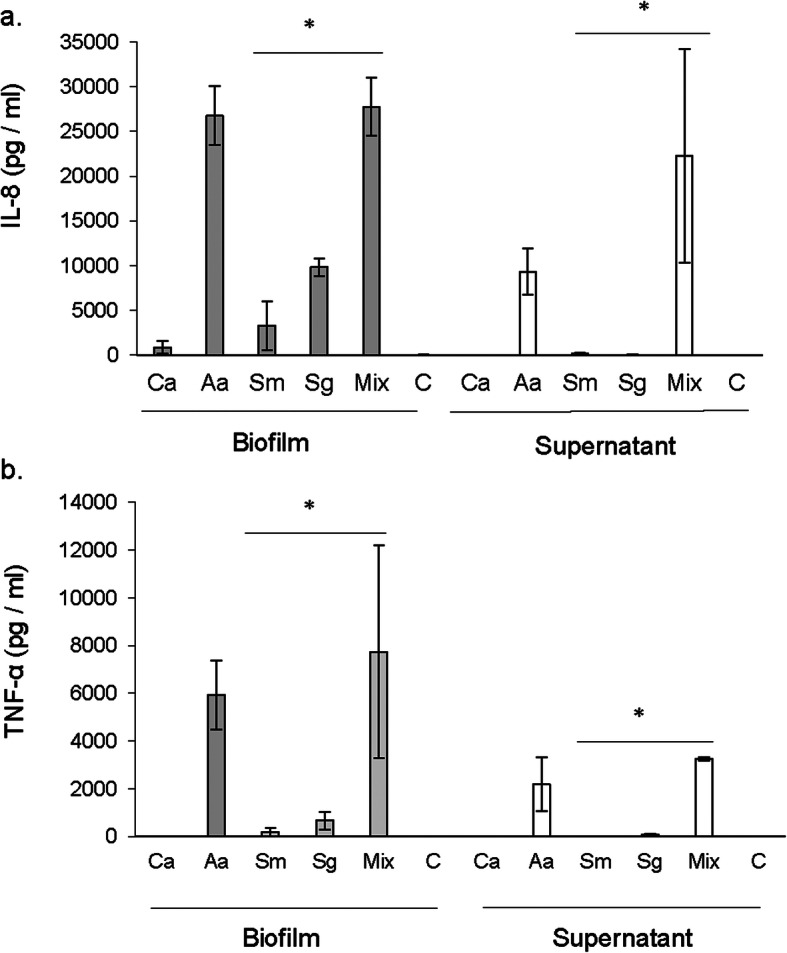
IL-8 and TNF-α induction by biofilms and biofilm supernatants. Human blood cells were stimulated by 24-h old monospecies and polymicrobial biofilms and biofilm supernatants. The stimulated samples were analyzed by ELISA for the quantification of IL-8 (a) and TNF-α (b). Ca = Candida albicans; Aa = Aggregatibater actinomycetemcomitans; Sm = Streptococcus mutans; Sg = Streptococcus gordonii; Mix = Mixture and C=Control, *p < 0.05
Similarly, the levels of TNF- α (Fig. 4b) were significantly higher in polymicrobial biofilm by both, biofilms [7747 (4450) pg/ml] and biofilm-supernatants [3246 (65) pg/ml], in comparison to monospecies biofilms of S. mutans and S. gordonii. TNF- α was under detection levels in both the C. albicans biofilm and biofilm-supernatant stimulated blood cells. Among monospecies biofilms, A. actinomycetemcomitans showed the highest induction levels of TNF- α [5936 (1454) pg/ml for biofilms and 2186 (1129) pg/ml for supernatants]. Biofilm supernatants from S. gordonii and S. mutans failed to induce TNF- α.
Discussion
This study showed that C. albicans did not only persist in the polymicrobial biofilm in the simultaneous presence of Gram-positive and Gram-negative bacteria, but also its growth was significantly enhanced. The biofilms and biofilm-supernatants from 24-h old Candida-bacteria polymicrobial biofilm cultures induced significantly higher levels of gene expression as well as production of the two important cytokines IL-8 and TNF- α. C. albicans monospecies biofilm or the biofilm-supernatant consistently induced only very low levels of these cytokines.
Our qPCR results from the species quantification of Candida-bacteria polymicrobial biofilm cultures showed that each of the Candida and the bacterial species tested were able to grow and persist in the polymicrobial biofilms. Importantly, quantities of C. albicans increased in the polymicrobial biofilm, suggesting that the interactions among these species within biofilms perhaps benefitted Candida growth. Although, different oral bacterial species have been investigated in dual/mixed biofilms with C. albicans in vitro, [2, 23–25] the combination of A. actinomycetemcomitans (Gram-negative) and streptococci (Gram-positives) with C. albicans has not been tested before. We chose the Gram-negative species A. actinomycetemcomitans because of its established implication in periodontitis [26–29]. S. mutans is a major etiologic agent in dental caries while S. gordonii is more associated with health (caries-free) [30–35]. With the known antagonism between A. actinomycetemcomitans and streptococci [36], we were interested in investigating how C. albicans would fare in a polymicrobial biofilm consisting the latter microorganisms. It has been demonstrated that C. albicans presence can enhance S. mutans growth, accumulation, and biofilm formation both in vitro and in vivo [37–43]. S. mutans displayed a strong affinity to C. albicans hyphae, as shown by electron microscopy on both human teeth and hydroxyapatite substrate samples [41]. Existing literature suggests that biofilm interactions of C. albicans with A. actinomycetemcomitans or streptococci are complex. For example, A. actinomycetemcomitans produce autoinducer-2 (AI-2), which inhibits C. albicans biofilm formation [4]. However, AI-2 produced by S. gordonii, increase biofilm formation and filamentation of C. albicans [5]. A small peptide called competence-stimulating peptide (CSP), produced by S. mutans, but not by S. gordonii, inhibits C. albicans hyphal formation in the early stages of biofilm formation [44, 45].
In distinct contrast to earlier studies where C. albicans biofilms involving streptococci or A. actinomycetemcomitans showed either antagonistic or synergistic growth behavior [4, 46], all microbial species tested in our in vitro model grew and persisted in biofilms until 48 h. At this phase we do not know how mutual antagonism was negated in the current Candida-bacteria polymicrobial biofilms. However, one could speculate that C. albicans might regulate antagonistic interactions between A. actinomycetemcomitans and streptococci while evading itself from the inhibitory effect of A. actinomycetemcomitans AI-2, possibly in an indirect way, by taking advantage of S. gordonii AI-2 which enhances C. albicans biofilm [5].
We investigated the inflammatory potential of biofilms and biofilm-secreted components from C. albicans and Candida-bacteria polymicrobial biofilms. This was done at two levels: mRNA and protein. We used human whole blood from a healthy subject because of its easy access and previous use in immunological studies in oral pathogens [6, 47]. The cytokines in this study were chosen because of their significance in oral infections such as periodontitis [48, 49] and caries [50, 51]. IL-8 and TNF- α play a crucial role in innate immune response. While IL-8 is a known neutrophil recruiter, TNF-α triggers robust host immune response by promoting infiltration of inflammatory cells and can directly contribute to osteoclast genesis and bone resorption [52]. In our study, IL-8 and TNF-α were elicited highest in Candida-bacteria polymicrobial biofilm and biofilm-supernatant stimulated blood in comparison to monospecies cultures. Only the bacterial monospecies biofilm cultures, not C. albicans, showed an induction of TNF- α. Contrary to our findings, C. albicans was reported as a better stimulator of TNF- α from blood cells of healthy subjects [6]. The reason that C. albicans stimulated very low levels of cytokines repetitive experiments in our study is possibly variability in responses from individuals. The variations in cytokine production by mononuclear cells in various healthy human volunteers, irrespective of age have been reported earlier [53]. Moreover, host (age, gender etc.) and environmental factors influencing human cytokine responses further supports cytokine response variability in humans [53, 54].
The expression of the genes encoding IL-8 and TNF-α was investigated by real time PCR. Our results are in line with several studies that demonstrated enhanced expression of host cytokine genes upon stimulation by oral microorganisms including C. albicans, A. actinomycetemcomitans, S. mutans and S.gordonii [6, 7, 13, 14, 55–59]. While IL-8 gene was more induced by the supernatant than the biofilm in the polymicrobial biofilms, it was the opposite in the case of TNF-α gene. Supernatants of Candida-bacteria polymicrobial biofilms were found to induce highest fold change in the expression of cytokine genes than did the supernatants of monospecies biofilms. In a polymicrobial biofilm setting, cell components of one species might affect the cytokine-stimulating potential of other species. For example, S. sanguinis peptidoglycan inhibited the cytokine expression induced by the LPS of periodontopathogens due to the inhibition of LPS binding to LBP and CD14 [55].
An interesting observation was that A. actinomycetemcomitans, which induced highest inflammatory response among monospecies biofilms, appeared to have a lesser immunostimulatory components in its biofilm-supernatants than the biofilms. Even though in silico analysis of the A. actinomycetemcomitans biofilm secretome revealed the presence of a large number of virulence related proteins [18], in our study, the biofilm-secreted components were yet less effective than the biofilm in inducing IL-8 and TNF-α.
In general, the cytokine gene expression results were in agreement with cytokine ELISA quantification. Candida-bacteria polymicrobial biofilms induced higher TNF-α, both at the gene level and at the protein level, than did the respective biofilm-supernatants. The polymicrobial biofilm did induce higher IL-8 than the supernatants at the protein level, but, the expression of IL-8 gene was more in the supernatants compared to biofilms. Further, supernatants from polymicrobial biofilms always triggered more cytokine gene expression and cytokine production than the supernatants from monospecies biofilms, suggesting an enhanced ability to induce host cell inflammatory response, of the microbial secretion products when existing together in the supernatants.
Conclusions
C. albicans grew and persisted as part of polymicrobial biofilms containing the Gram-negative bacterium A. actinomycetemcomitans and the Gram-positive species S. mutans and S. gordonii. These data suggest that hitherto known antagonistic effect of A. actinomycetecomitans on C. albicans growth [4] was negated in the current polymicrobial biofilm setting possibly due to the presence of streptococci. Further, induction of higher levels of certain cytokines and an increased expression of corresponding cytokine genes suggests a possibility that such Candida-bacteria polymicrobial biofilm communities together derive a cumulative potential to exert increased cytokine stimulatory effect on the host.
Methods
Microbial strains and culture conditions
Reference bacterial strains Streptococcus mutans CCUG 11877 T, Streptococcus gordonii CCUG 33482 and a yeast strain Candida albicans ATCC 24433 were purchased from culture collections. Aggregatibacter actinomycetemcomitans strain SA269 was from the strain collection of Sirkka Asikainen (Umeå University, Sweden). Bacterial strains were cultured on brucella blood agar (BBA) for 2 days and C. albicans on sabouraud dextrose agar (SDA) for 24 h in 5% CO2 in air at 37 °C
Biofilm cultures
Established method [60] with some modifications was used for culturing biofilms. First, an inoculum of OD600 = 1 suspension of each test species was prepared. For this the bacterial/yeast colonies were harvested from agar plates with sterile plastic loops and suspended in sterile PBS. The suspensions were washed two times in sterile PBS by centrifuging at 5000×g for 5 min. The cell pellet recovered after centrifugation was suspended in 1-ml brucella broth. Based on the OD measurements of the above suspensions, each test species was adjusted to a standard OD600 = 1. As determined in our laboratory, at OD600 = 1, the CFU/ml were for A. actinomycetemcomitans 1 × 108, S. mutans 1.7 × 108, S. gordonii 1.6 × 108 and C. albicans 6.7 × 106. Further these suspensions (OD600 = 1) were used to culture monospecies and polymicrobial biofilms in 24-well culture plates. Monospecies biofilms were initiated by inoculating 24-well plates containing 900 μl brucella broth with a 100 μl aliquot from the above OD600 = 1 suspension of each species. For polymicrobial biofilm culture, inoculum was prepared in a microfuge tube by combining equal volumes of OD600 = 1 suspensions from each of the four test species. One hundred microliter from this species mixture was transferred into the wells of 24-well plates containing 900 μl of brucella broth. Total three sets of monospecies and polymicrobial biofilms were cultured in parallel. Two sets were cultured in 24-well culture plates for analysis of mRNA expression levels of cytokine genes in human blood cells stimulated with biofilm and biofilm-supernatant, ELISA based quantification of selected cytokines and qPCR quantification of biofilms while third set was cultured on Millicell® EZ slides (EMD Millipore) with detachable wells for confocal laser scanning microscopy of biofilms. For qPCR quantification of biofilms, at the end of the incubation time points, supernatant broth was aspirated and the biofilms were washed once with sterile PBS to remove unattached cells. The biofilms were thoroughly scraped off and the wells were further examined under stereomicroscope to confirm the complete removal of biofilms. Harvested biofilms were resuspended in 100 μl sterile nuclease-free H2O (Ambion, USA) and then immediately preserved at − 20 °C until used for DNA extraction.
Confocal laser scanning microscopy of biofilms
Monospecies and polymicrobial biofilms were cultured on Millicell® EZ slides (EMD Millipore) with detachable wells [15]. 24-and 48 h old biofilms were washed twice with sterile PBS (1 ml) to remove loosely attached and unbound bacteria and/or Candida cells and were then fixed for 30 min at room temperature with 4% freshly prepared paraformaldehyde in PBS (1 ml/cell, pH 7). The biofilms were washed in PBS and stained with Syto® 9 Green Fluorescent Nucleic Acid Stain (Molecular Probes) for 15 min at room temperature in dark according to the manufacturer’s recommendations. The biofilms were washed in PBS again and the polypropylene wells were then detached and the slides were air-dried. The biofilms were covered with a cover glass containing a drop of BacLight® mounting oil. Confocal laser scanning microscope (LSM 700, Carl-Zeiss; software ZEN 2012) was used to analyze and capture the images of stained biofilms. Three dimensional reconstruction of the biofilms was done using Z-stack.
DNA extraction and purifications
DNA from the harvested biofilms was purified using DNeasy DNA extraction kit (Qiagen). Enzymatic lysis buffer containing Tris EDTA buffer (20 mM Tris, 2 mM EDTA) with 1.2% Triton X-100 and lysozyme was used. Purified DNA was eluted in nuclease free water and concentrations were measured by UV spectrophotometry method using NanoDrop™ 1000.
Quantifying biofilms by qPCR
For qPCR quantification, Species-specific primers for the bacterial 16S rRNA gene and internal transcribed spacer regions 1 of rRNA and universal reverse primer of 26 s rRNA gene of C. albicans were used (Table 1). The reaction mixture containing 12.5 μl SYBR Green master mix (Power SYBR Green® Kit, Applied Biosystems), 1.0 μl each of forward and reverse primer (0.4 μM), 5.0 μl DNA template (equivalent to 50 ng DNA) and 5.5 μl H2O was used in qPCR. A thermal profile with an initial denaturation at 95 °C for 10-min, 40 cycles of 95 °C for 15 s – 30 s, 52–68 °C (depending on the primer pair) for 30 s and 72 °C for 30 s - 1 min was run for carrying out amplifications on ABI Fast RT-PCR machine. The elongation step was considered for the fluorescent signal acquisition. Software SDS v2.3 was used for data analysis. Serially diluted DNA from the above species in the reaction and plotting the Ct values against deduced bacterial cell concentration (cells/ml) for each species were used to construct standard curves using the above software. The standard curves were generated in the above RT-PCR software by computing the bacterial cell numbers. The optimum reaction efficiency of 86–105% (slope − 3.7 to − 3.2) and R2 value (0.97–0.99) for standard curve linearity were considered.
Table 1.
Primers and conditions used in qPCR quantification of microbial cultures
| Species | Primer sequence 5′-3′ | Annealing temperatures (°C) |
References |
|---|---|---|---|
| A. actinomycetemcomitans |
F: TAGCCCTGGTGCCCGAAGC R: CATCGCTGGTTGGTTACCCTCTG |
68 | [61] |
| S. mutans |
F: TCGCGAAAAAGATAAACAAACA R: GCCCCTTCACAGTTGGTTAG |
56 | [62] |
| S. gordonii |
F: GGTGTTGTTTGACCCGTTCAG R: AGTCCATCCCACGAGCACAG |
53 | [63] |
| C. albicans |
F: TCA ACT TGT CAC ACC AGA TTA TT R: TCC TCC GCT TAT TGA TAT GC |
52 | [64] |
Stimulation of human blood cells by monospecies biofilms and Candida-bacteria polymicrobial biofilms
Ethical approval for blood collection from a healthy human volunteer was obtained from Health Science Center Ethical Committee, Kuwait University. One ml of whole blood from a healthy human volunteer was added to the 24-h old biofilms in each well and incubated for 24 h at 37 °C in 5% CO2 in air for stimulation. Well with only blood and no biofilm was used as control. For stimulation with biofilm supernatants, broth supernatants from 24 h old biofilms were filtered through 0.2 μm syringe filters to get rid of the bacterial cells. Two hundred microliters of the filtered supernatant was added into wells containing 1 ml blood. After 24 h of incubation, samples were taken out and centrifuged immediately at 5000×g for 5 min. The supernatant was stored at − 20 °C until use.
Analysis of mRNA expression levels of cytokine genes in human blood cells stimulated with biofilms and biofilm-supernatants
To determine the mRNA expression of IL-8 and TNF-α genes, total cellular RNA was extracted and purified from stimulated blood samples as per manufacturer’s instructions (QIAamp RNA blood Mini kit). Erythrocytes were lysed in buffer EL by incubating on ice for 15 min, vortexing and centrifugation at 400×g at 4 °C for 10 min. Non-nucleated lysed erythrocytes in the supernatant were discarded while leukocytes in the pellet were further disrupted in RLT buffer. The lysate was homogenized by centrifugation at maximum speed in QIAshredder spin column and then precipitated with 70% ethanol and transferred to QIAamp spin column (provided in the kit). The RNA bound to the membrane was further treated with buffers RW1 and RPE and discarded the flow through. Finally bound RNA was eluted in RNase-free water. The concentration of RNA was determined by spectrophotometer NanoDrop™ 1000 and the purity of RNA was assessed by A260/A280 ratio. Purified RNA was converted to cDNA by using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems™) according to manufacturer’s instructions. The concentration of cDNA was determined by spectrophotometer NanoDrop™ 1000 and the purity of DNA was assessed by A260/A280 ratio.
Reverse trascriptase RT-PCR
A reaction mixture of 10 μl SYBR Green master mix (Power SYBR Green® Kit, Applied Biosystems), 1.0 μl each of forward and reverse primer (0.4 μM), 7 μl H2O and 1.0 μl cDNA template (equivalent to 50 ng cDNA) was used to perform Real Time reverse trascriptase PCR reaction on ABI 7500 Fast Real-Time PCR machine. Thermal profile of 10-min initial denaturation at 95 °C, 40 cycles of 95 °C for 15 s – 30 s, 50–60 °C for 30 s (depending on the primer pair) and 72 °C for 30 s – 1 min were run to carry out the amplifications. Primers and conditions used in qPCR for mRNA gene expression analyses are mentioned in Table 2. Expression levels of all the target genes were normalized using endogenous housekeeping gene encoding GAPDH. Fluorescent signal acquisition was set at the elongation step. Data analysis was accomplished using the software SDS v2.3. Analysis of the expression of cytokine controlling genes was performed on cells from untreated control, biofilm and biofilm supernatant treated groups by comparative ∆∆Ct method in ABI 7500 SDS system (Applied Biosystems). The amount of target, normalized to an endogenous reference (GAPDH) and relative to a calibrator (untreated), was given by 2_∆∆Ct and the expression fold change was presented graphically.
Table 2.
Primers and conditions used in RT-PCR for cytokine gene expression analysis
ELISA based quantification of selected cytokines
For cytokine quantifications, ELISA immunoassay kits (Quantikine® ELISA R&D systems) containing a cytokine specific monoclonal antibody pre-coated ELISA plates were used. The assay was based on a quantitative sandwich enzyme immunoassay technique. To determine the absolute quantities of IL-8 and TNF-α in blood upon stimulation with biofilm or biofilm-supernatant, the wells of ELISA plate were pipetted with standards and samples. Any unbound substances in the wells were removed with wash buffer and an enzyme-linked polyclonal antibody specific for cytokine of interest was added to the wells thereafter. Washing steps were followed to remove unbound antibody-enzyme reagent. Finally, a substrate solution was added to the wells and the color development was stopped. The intensity of the color developed in proportion to the amount of bound cytokine of interest was measured in microplate reader (iMark; Bio-rad).
Statistical analyses
For quantification, all samples were run in duplicate and three independent experiments were performed. Student’s t-tests or Mann-Whitney U tests were used to compare the differences between Candida only, bacteria only and Candida co-cultured with bacteria. A P-value of < 0.05 was considered statistically significant.
Acknowledgements
We thank the Research Core Facility (SRUL02/13) at the Health Sciences Center of Kuwait University for confocal laser scanning microscopy.
Ethical approval and concent to participate
All procedures performed in studies involving human participant were in accordance with the ethical standards of the Health Sciences Center, Kuwait University (VDR/EC/3413) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from the human volunteer in the study.
Abbreviations
- IL-8
Interleukin 8
- TNF-α
Tumor necrosis factor alpha
- ATCC
American Type Culture Collection
- CCUG
Culture Collection University of Gothenburg
- OD
Optical density
- EDTA
Ethylenediamine tetra-acetic acid
- HRP
Horseradish peroxidase
- RT-PCR
Reverse transcription polymerase chain reaction
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
Authors’ contributions
RGB: Design, planning and execution of experiments, data analysis and interpretation, manuscript writing. AE: Data interpretation and manuscript writing. HD: Performed some of the experiments, manuscript writing. MK: Conceived the study, data analysis and interpretation, manuscript writing. All authors have read and approved the manuscript.
Funding
This study was funded by the Research Sector, Kuwait University, Specialized Research Unit Laboratory Grant SRUL 01/14.
Availability of data and materials
We declare that all data is included in this manuscript and there are no supplimentary data files.
Ethics approval and consent to participate
Ethical approval was obtained from the Health Science Center Ethical Committee, Kuwait University (DR/EC/3413).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Krom BP, Kidwai S, Ten Cate JM. Candida and other fungal species: forgotten players of healthy oral microbiota. J Dent Res. 2014;93(5):445–451. doi: 10.1177/0022034514521814. [DOI] [PubMed] [Google Scholar]
- 2.Montelongo-Jauregui D, Srinivasan A, Ramasubramanian AK, Lopez-Ribot JL. An In Vitro model for Oral mixed biofilms of Candida albicans and Streptococcus gordonii in synthetic saliva. Front Microbiol. 2016;7:686. doi: 10.3389/fmicb.2016.00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachtiar EW, Bachtiar BM, Jarosz LM, Amir LR, Sunarto H, Ganin H, Meijler MM, Krom BP. AI-2 of Aggregatibacter actinomycetemcomitans inhibits Candida albicans biofilm formation. Front Cell Infect Microbiol. 2014;4:94. doi: 10.3389/fcimb.2014.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bamford CV, d'Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 2009;77(9):3696–3704. doi: 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nawrot U, Grzybek-Hryncewicz K, Czarny A. The ability of Candida spp. strains to induce production of tumor necrosis factor and interleukin-6 by whole blood cells. Acta Microbiol Pol. 2003;52(1):87–91. [PubMed] [Google Scholar]
- 7.Brieland J, Essig D, Jackson C, Frank D, Loebenberg D, Menzel F, Arnold B, DiDomenico B, Hare R. Comparison of pathogenesis and host immune responses to Candida glabrata and Candida albicans in systemically infected immunocompetent mice. Infect Immun. 2001;69(8):5046–5055. doi: 10.1128/IAI.69.8.5046-5055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Carvalho DK, Barbugli PA, de Patto F, Lordello VB, de Aquino PL, Medeiros AI, Vergani CE. Soluble factors from biofilm of Candida albicans and Staphylococcus aureus promote cell death and inflammatory response. BMC Microbiol. 2017;17(1):146. doi: 10.1186/s12866-017-1031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dongari-Bagtzoglou A, Kashleva H. Candida albicans triggers interleukin-8 secretion by oral epithelial cells. Microb Pathog. 2003;34(4):169–177. doi: 10.1016/s0882-4010(03)00004-4. [DOI] [PubMed] [Google Scholar]
- 10.Ebersole JL, Peyyala R, Gonzalez OA. Biofilm-induced profiles of immune response gene expression by oral epithelial cells. Mol Oral Microbiol. 2019;34(1):14–25. [DOI] [PMC free article] [PubMed]
- 11.Engels-Deutsch M, Pini A, Yamashita Y, Shibata Y, Haikel Y, Scholler-Guinard M, Klein JP. Insertional inactivation of pac and rmlB genes reduces the release of tumor necrosis factor alpha, interleukin-6, and interleukin-8 induced by Streptococcus mutans in monocytic, dental pulp, and periodontal ligament cells. Infect Immun. 2003;71(9):5169–5177. doi: 10.1128/IAI.71.9.5169-5177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takada H, Kawabata Y, Tamura M, Matsushita K, Igarashi H, Ohkuni H, Todome Y, Uchiyama T, Kotani S. Cytokine induction by extracellular products of oral viridans group streptococci. Infect Immun. 1993;61(12):5252–5260. doi: 10.1128/iai.61.12.5252-5260.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levitz SM. North EA: gamma interferon gene expression and release in human lymphocytes directly activated by Cryptococcus neoformans and Candida albicans. Infect Immun. 1996;64(5):1595–1599. doi: 10.1128/iai.64.5.1595-1599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oscarsson J, Karched M, Thay B, Chen C, Asikainen S. Proinflammatory effect in whole blood by free soluble bacterial components released from planktonic and biofilm cells. BMC Microbiol. 2008;8:206. doi: 10.1186/1471-2180-8-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belibasakis GN, Guggenheim B. Induction of prostaglandin E (2) and interleukin-6 in gingival fibroblasts by oral biofilms. FEMS Immunol Med Microbiol. 2011;63(3):381–386. doi: 10.1111/j.1574-695X.2011.00863.x. [DOI] [PubMed] [Google Scholar]
- 16.Bostanci N, Meier A, Guggenheim B, Belibasakis GN. Regulation of NLRP3 and AIM2 inflammasome gene expression levels in gingival fibroblasts by oral biofilms. Cell Immunol. 2011;270(1):88–93. doi: 10.1016/j.cellimm.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Bhardwaj RG, Al-Khabbaz A, Karched M. Cytokine induction of peripheral blood mononuclear cells by biofilms and biofilm supernatants of Granulicatella and Abiotrophia spp. Microb Pathog. 2018;114:90–94. doi: 10.1016/j.micpath.2017.11.037. [DOI] [PubMed] [Google Scholar]
- 18.Zijnge V, Kieselbach T, Oscarsson J. Proteomics of protein secretion by Aggregatibacter actinomycetemcomitans. PLoS One. 2012;7(7):e41662. doi: 10.1371/journal.pone.0041662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao K, Bostanci N, Thurnheer T, Belibasakis GN. Proteomic shifts in multi-species oral biofilms caused by Anaeroglobus geminatus. Sci Rep. 2017;7(1):4409. doi: 10.1038/s41598-017-04594-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bor B, Cen L, Agnello M, Shi W, He X. Morphological and physiological changes induced by contact-dependent interaction between Candida albicans and Fusobacterium nucleatum. Sci Rep. 2016;6:27956. doi: 10.1038/srep27956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu T, Cen L, Kaplan C, Zhou X, Lux R, Shi W, He X. Cellular components mediating Coadherence of Candida albicans and Fusobacterium nucleatum. J Dent Res. 2015;94(10):1432–1438. doi: 10.1177/0022034515593706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koo H, Andes DR, Krysan DJ. Candida-streptococcal interactions in biofilm-associated oral diseases. PLoS Pathog. 2018;14(12):e1007342. doi: 10.1371/journal.ppat.1007342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sztajer H, Szafranski SP, Tomasch J, Reck M, Nimtz M, Rohde M, Wagner-Dobler I. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 2014;8(11):2256–2271. doi: 10.1038/ismej.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thein ZM, Samaranayake YH, Samaranayake LP. Effect of oral bacteria on growth and survival of Candida albicans biofilms. Arch Oral Biol. 2006;51(8):672–680. doi: 10.1016/j.archoralbio.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Xu H, Jenkinson HF, Dongari-Bagtzoglou A. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 2014;29(3):99–116. doi: 10.1111/omi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saarela MH, Dogan B, Alaluusua S, Asikainen S. Persistence of oral colonization by the same Actinobacillus actinomycetemcomitans strain(s) J Periodontol. 1999;70(5):504–509. doi: 10.1902/jop.1999.70.5.504. [DOI] [PubMed] [Google Scholar]
- 27.Arenas Rodrigues VA, de Avila ED, Nakano V, Avila-Campos MJ. Qualitative, quantitative and genotypic evaluation of Aggregatibacter actinomycetemcomitans and Fusobacterium nucleatum isolated from individuals with different periodontal clinical conditions. Anaerobe. 2018;52:50–58. doi: 10.1016/j.anaerobe.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 28.Suprith SS, Setty S, Bhat K, Thakur S. Serotypes of Aggregatibacter actinomycetemcomitans in relation to periodontal status and assessment of leukotoxin in periodontal disease: a clinico-microbiological study. J Indian Soc Periodontol. 2018;22(3):201–208. doi: 10.4103/jisp.jisp_36_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 30.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellen RP, Banting DW, Fillery ED. Streptococcus mutans and Lactobacillus detection in the assessment of dental root surface caries risk. J Dent Res. 1985;64(10):1245–1249. doi: 10.1177/00220345850640101301. [DOI] [PubMed] [Google Scholar]
- 32.Faustova MO, Ananieva MM, Basarab YO, Dobrobolska OV, Vovk IM, Loban GA. Bacterial factors of cariogenicity (literature review) Wiad Lek. 2018;71(2 pt 2):378–382. [PubMed] [Google Scholar]
- 33.Li Y, Saraithong P, Chen Z, Leung E, Pattanaporn K, Dasanayake A. Comparison of real-time quantitative PCR with a chairside test for Streptococcus mutans assessment. Chin J Dent Res. 2017;20(4):199–210. doi: 10.3290/j.cjdr.a39219. [DOI] [PubMed] [Google Scholar]
- 34.Srilatha A, Doshi D, Kulkarni S, Reddy MP, Reddy BS, Satyanarayana D. Determination and comparison of Dermatoglyphic patterns and salivary Streptococcus mutans counts and its correlation with dental caries among 3- to 6-year-old children. Oral Health Prev Dent. 2018;16(3):291–297. doi: 10.3290/j.ohpd.a40720. [DOI] [PubMed] [Google Scholar]
- 35.Tanner A, Kent R, Maiden MF, Taubman MA. Clinical, microbiological and immunological profile of healthy, gingivitis and putative active periodontal subjects. J Periodontal Res. 1996;31(3):195–204. doi: 10.1111/j.1600-0765.1996.tb00484.x. [DOI] [PubMed] [Google Scholar]
- 36.Kozlovsky A, Wolff A, Saminsky M, Mazor Y, Venezia E, Bar-Ness Greenstein R. Effect of Aggregatibacter actinomycetemcomitans from aggressive periodontitis patients on Streptococcus mutans. Oral Dis. 2015;21(8):955–961. doi: 10.1111/odi.12362. [DOI] [PubMed] [Google Scholar]
- 37.de Carvalho FG, Silva DS, Hebling J, Spolidorio LC, Spolidorio DM. Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch Oral Biol. 2006;51(11):1024–1028. doi: 10.1016/j.archoralbio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Ellepola K, Liu Y, Cao T, Koo H, Seneviratne CJ. Bacterial GtfB augments Candida albicans accumulation in cross-kingdom biofilms. J Dent Res. 2017;96(10):1129–1135. doi: 10.1177/0022034517714414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai CH, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 2014;82(5):1968–1981. doi: 10.1128/IAI.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klinke T, Guggenheim B, Klimm W, Thurnheer T. Dental caries in rats associated with Candida albicans. Caries Res. 2011;45(2):100–106. doi: 10.1159/000324809. [DOI] [PubMed] [Google Scholar]
- 41.Metwalli KH, Khan SA, Krom BP, Jabra-Rizk MA. Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog. 2013;9(10):e1003616. doi: 10.1371/journal.ppat.1003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira-Cenci T, Deng DM, Kraneveld EA, Manders EM, Del Bel Cury AA, Ten Cate JM, Crielaard W. The effect of Streptococcus mutans and Candida glabrata on Candida albicans biofilms formed on different surfaces. Arch Oral Biol. 2008;53(8):755–764. doi: 10.1016/j.archoralbio.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Raja M, Hannan A, Ali K. Association of oral candidal carriage with dental caries in children. Caries Res. 2010;44(3):272–276. doi: 10.1159/000314675. [DOI] [PubMed] [Google Scholar]
- 44.Jack AA, Daniels DE, Jepson MA, Vickerman MM, Lamont RJ, Jenkinson HF, Nobbs AH. Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology. 2015;161(Pt 2):411–421. doi: 10.1099/mic.0.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jarosz LM, Deng DM, van der Mei HC, Crielaard W, Krom BP. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell. 2009;8(11):1658–1664. doi: 10.1128/EC.00070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diaz PI, Xie Z, Sobue T, Thompson A, Biyikoglu B, Ricker A, Ikonomou L, Dongari-Bagtzoglou A. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 2012;80(2):620–632. doi: 10.1128/IAI.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murciano C, Villamon E, Yanez A, Murciano J, Mir A, O'Connor JE, Gozalbo D, Gil ML. In vitro response to Candida albicans in cultures of whole human blood from young and aged donors. FEMS Immunol Med Microbiol. 2007;51(2):327–335. doi: 10.1111/j.1574-695X.2007.00309.x. [DOI] [PubMed] [Google Scholar]
- 48.Ertugrul AS, Sahin H, Dikilitas A, Alpaslan N, Bozoglan A. Comparison of CCL28, interleukin-8, interleukin-1beta and tumor necrosis factor-alpha in subjects with gingivitis, chronic periodontitis and generalized aggressive periodontitis. J Periodontal Res. 2013;48(1):44–51. doi: 10.1111/j.1600-0765.2012.01500.x. [DOI] [PubMed] [Google Scholar]
- 49.Jiang ZL, Cui YQ, Gao R, Li Y, Fu ZC, Zhang B, Guan CC. Study of TNF-alpha, IL-1beta and LPS levels in the gingival crevicular fluid of a rat model of diabetes mellitus and periodontitis. Dis Markers. 2013;34(5):295–304. doi: 10.3233/DMA-130974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cogulu D, Onay H, Ozdemir Y, IA G, Ozkinay F, Kutukculer N, Eronat C. Associations of interleukin (IL)-1beta, IL-1 receptor antagonist, and IL-10 with dental caries. J Oral Sci. 2015;57(1):31–36. doi: 10.2334/josnusd.57.31. [DOI] [PubMed] [Google Scholar]
- 51.Gornowicz A, Bielawska A, Bielawski K, Grabowska SZ, Wojcicka A, Zalewska M, Maciorkowska E. Pro-inflammatory cytokines in saliva of adolescents with dental caries disease. Ann Agric Environ Med. 2012;19(4):711–716. [PubMed] [Google Scholar]
- 52.Suttles J, Miller RW, Tao X, Stout RD. T cells which do not express membrane tumor necrosis factor-alpha activate macrophage effector function by cell contact-dependent signaling of macrophage tumor necrosis factor-alpha production. Eur J Immunol. 1994;24(8):1736–1742. doi: 10.1002/eji.1830240803. [DOI] [PubMed] [Google Scholar]
- 53.Mozes T, Barath I, Gornicsar K, Grosz A, Mozes T, Gondocs C, Szephalmi P, Gaal K, Madarasz E. Deviations in circulating TNFalpha levels and TNFalpha production by mononuclear cells in healthy human populations. Mediat Inflamm. 2011;2011:972609. doi: 10.1155/2011/972609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ter Horst R, Jaeger M, Smeekens SP, Oosting M, Swertz MA, Li Y, Kumar V, Diavatopoulos DA, Jansen AFM, Lemmers H, et al. Host and environmental factors influencing individual human cytokine responses. Cell. 2016;167(4):1111–1124. doi: 10.1016/j.cell.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee SH. Antagonistic effect of peptidoglycan of Streptococcus sanguinis on lipopolysaccharide of major periodontal pathogens. J Microbiol. 2015;53(8):553–560. doi: 10.1007/s12275-015-5319-6. [DOI] [PubMed] [Google Scholar]
- 56.Mostefaoui Y, Bart C, Frenette M, Rouabhia M. Candida albicans and Streptococcus salivarius modulate IL-6, IL-8, and TNF-alpha expression and secretion by engineered human oral mucosa cells. Cell Microbiol. 2004;6(11):1085–1096. doi: 10.1111/j.1462-5822.2004.00420.x. [DOI] [PubMed] [Google Scholar]
- 57.Nagata E, de Toledo A, Oho T. Invasion of human aortic endothelial cells by oral viridans group streptococci and induction of inflammatory cytokine production. Mol Oral Microbiol. 2011;26(1):78–88. doi: 10.1111/j.2041-1014.2010.00597.x. [DOI] [PubMed] [Google Scholar]
- 58.Schaller M, Mailhammer R, Korting HC. Cytokine expression induced by Candida albicans in a model of cutaneous candidosis based on reconstituted human epidermis. J Med Microbiol. 2002;51(8):672–676. doi: 10.1099/0022-1317-51-8-672. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka YZL, Ikuta T, Omori J, Omine H, Mega J, Kuboyama N, Abiko Y. TNF-α expression in Oral Candida albicans -infected human gingival epithelial cells. Int J Oral-Med Sci. 2011;10(2):77–82. [Google Scholar]
- 60.Karched M, Bhardwaj RG, Inbamani A, Asikainen S. Quantitation of biofilm and planktonic life forms of coexisting periodontal species. Anaerobe. 2015;35(Pt A):13–20. doi: 10.1016/j.anaerobe.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 61.Kim SG, Kim SH, Kim MK, Kim HS, Kook JK. Identification of Actinobacillus actinomycetemcomitans using species-specific 16S rDNA primers. J Microbiol. 2005;43(2):209–212. [PubMed] [Google Scholar]
- 62.Chen Z, Saxena D, Caufield PW, Ge Y, Wang M, Li Y. Development of species-specific primers for detection of Streptococcus mutans in mixed bacterial samples. FEMS Microbiol Lett. 2007;272(2):154–162. doi: 10.1111/j.1574-6968.2007.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzuki N, Nakano Y, Yoshida A, Yamashita Y, Kiyoura Y. Real-time TaqMan PCR for quantifying oral bacteria during biofilm formation. J Clin Microbiol. 2004;42(8):3827–3830. doi: 10.1128/JCM.42.8.3827-3830.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li YL, Leaw SN, Chen JH, Chang HC, Chang TC. Rapid identification of yeasts commonly found in positive blood cultures by amplification of the internal transcribed spacer regions 1 and 2. Eur J Clin Microbiol Infect Dis. 2003;22(11):693–696. doi: 10.1007/s10096-003-1020-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We declare that all data is included in this manuscript and there are no supplimentary data files.