Abstract
Emerging evidence supports the ion channel mechanism for Alzheimer’s disease pathophysiology wherein small β-amyloid (Aβ) oligomers insert into the cell membrane, forming toxic ion channels and destabilizing the cellular ionic homeostasis. Solid-state NMR-based data of amyloid oligomers in solution indicate that they consist of a double-layered β-sheets where each monomer folds into β-strand–turn–β-strand and the monomers are stacked atop each other. In the membrane, Aβ peptides are proposed to be β-type structures. Experimental structural data available from atomic force microscopy (AFM) imaging of Aβ oligomers in membranes reveal heterogeneous channel morphologies. Previously, we modeled the channels in a non-tilted organization, parallel with the cross-membrane normal. Here, we modeled a β-barrel-like organization. β-Barrels are common in transmembrane toxin pores, typically consisting of a monomeric chain forming a pore, organized in a single-layered β-sheet with antiparallel β-strands and a right-handed twist. Our explicit solvent molecular dynamics simulations of a range of channel sizes and polymorphic turns and comparisons of these with AFM image dimensions support a β-barrel channel organization. Different from the transmembrane β-barrels where the monomers are folded into a circular β-sheet with antiparallel β-strands stabilized by the connecting loops, these Aβ barrels consist of multimeric chains forming double β-sheets with parallel β-strands, where the strands of each monomer are connected by a turn. Although the Aβ barrels adopt the right-handed β-sheet twist, the barrels still break into heterogeneous, loosely attached subunits, in good agreement with AFM images and previous modeling. The subunits appear mobile, allowing unregulated, hence toxic, ion flux.
Keywords: toxic amyloid ion channels, β-barrel, U-shaped motif, molecular dynamics simulations, atomic force microscopy
Introduction
β-Barrels are found frequently in the outer membrane of Gram-negative bacteria, mitochondria, and chloroplasts.1 The bacterial outer membrane protein porin has a transmembrane β-barrel pore typically consisting of 8–22 antiparallel β-strands connected by turns.2 Porins act as selective or non-selective channels.3,4 In the prototypical porin β-barrel, hydrophilic side chains orient toward the solvated pore, whereas hydrophobic side chains interact with lipid tails. The β-barrel structure is a large β-sheet composed of an even number of antiparallel β-strands in which the β-sheet adopts a right-handed twist and coils to form an annular structure. Two integer parameters can characterize β-barrels: the number of β-strands in the barrel, N, and the “shear number,” S, which is a measure of the stagger of the β-strands in the β-sheet.5,6 The parameter set (N,S) determines the inclination angle of the β-strands relative to the barrel pore axis.7 The β-barrel structure is rare in eukaryotes but is found in the mitochondria (which is believed to be a bacterial relic) outer membrane and is known as a voltage-dependent anionic channel. It contains ~14–16 β-strands.8–10 The anionic channel has a two-state voltage-dependent gating mechanism opening and closing the pore, inducing cytotoxicity and apoptosis.11,12
It is increasingly accepted that oligomer amyloid seeds are the toxic species13–15 and that the mechanism of toxicity involves small oligomers penetrating into the membrane and forming unregulated ion channels,16–34 which lead to ion leakage and, ultimately, cell death. However, given their dynamic nature, there are no available experimental structures for these channels. Early models ruled out the possibility that the channels consist of β-sheet-rich C-termini and that the disordered β-amyloid (Aβ) N-termini are extramembranous and focused on transmembrane α-helices as a major component.35,36 However, later experimental data37,38 implicated β-structures and confirmed that the N-terminus is not embedded in the bilayer. Moreover, although functional biological channels overwhelmingly consist of α-helices, the only secondary structure type observed in the amyloid oligomers is a β-structure (two strands connected by a turn).39–42 In principle, it is possible that the oligomer conformations in the membrane differ from those determined in solution for the monomeric state. Two recent studies point toward a β-structure in the membrane: in the first, despite the use of a solvent mixture in which Aβ1–40 and Aβ1–42 are largely helical, the Aβ peptides in the hydrophobic core of the bilayers displayed a β-structure with a typical single channel-like Aβ conductance;43 in the second, circular dichroism44 showed that, when incorporated into the lipid bilayer, Aβ1–42 presented more β-sheet structures than observed in the soluble form. This study suggested that the previously observed increased α-helical content could reflect the trifluoroethanol used for incorporation of the Aβ into the bilayer. Even prior to these data, Kagan45 proposed that the propensity of amyloid peptides to form ion channels is a direct result of their physicochemical properties and that β-sheets are suitable for forming pore structures, citing the anthrax toxin protective antigen crystal structure example.46
Non-β-barrel Aβ channels were modeled previously for the N-terminal truncated Aβ peptides Aβ17–42 (known as p3) and Aβ9–42 (called N9) based on β-sheet organization with conventional β-strand insertions parallel with the membrane normal.23,47–50 p3 is produced in vivo by the α-secretase cleavage, whereas Aβ11–42 is formed by β′-secretase. Both are cut at the C-terminus by γ-secretase. Both peptides are found in Alzheimer’s disease amyloid plaques,51–54 and p3 is also the major constituent of preamyloid and neuritic plaques in the brains of Down syndrome patients55; in addition, both are capable of inducing neuronal toxicity.56–58 Significantly, these peptides were previously believed to be non-toxic species in Alzheimer’s disease pathology. With the use of solid-state NMR (ssNMR)-based coordinates of the U-shaped peptides,39,40 explicit solvent molecular dynamics simulations have tested ion channels of different sizes, 12-mer to 36-mer, in the lipid bilayer. The simulations showed that the truncated Aβ channels are ion-permeable and their morphologies and shapes are consistent with atomic force microscopy (AFM) images of full-length Aβ1–40/42 channels.26,29 Subsequent experiments using AFM, planar lipid bilayer recordings, cell calcium imaging, neuritic degeneration, and cell death assays established that these are toxic species as well.49 The channels obtained a preferred size range of 16–24 β-strands lining the pores.23,47–50 This range was also found to hold for other toxic β-sheet channels, the 24-mer K3 channels (β2-microglobulin fragments with a demonstrated U-shaped motif41), also in agreement with AFM.59 In addition, the structurally related antimicrobial peptide protegrin-1 (PG-1) forms channels with 8–10 β-hairpin monomers that laterally arrange to form a pore with 16–20 β-strands.60,61 For the transmembrane β-barrels, although 8–22 β-strands are possible, the channels typically have 12–22 strands depending on the shear.2,62 This range is very similar to the 16–24 that we obtained for the Aβ channels.
Nonetheless, the amyloid landscape is highly polymorphic,63,64 with rugged oligomeric and fibril energy landscapes; thus, a heterogeneous channel population is expected. The β-barrel is a naturally occurring functional motif in prokaryotes and in some organelles in eukaryotes mediating cell toxicity. One can then hypothesize that toxic amyloid channels that consist of β-sheets could adopt a β-barrel motif, which is an intrinsic toxic topology. Here, we modeled such (12-, 16-, and 20-mer) channels (called “Aβ barrels”) for the truncated Aβ peptides p3 and N9 in a lipid bilayer containing 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and compared them with our AFM images of these fragments. Overall, we observed that the Aβ porin-like barrels present a similar subunit organization and dimensions as observed earlier for the β-sheet channels assembled by non-tilted Aβ peptides with a broad channel size range,23,47–50 emphasizing that toxic channels are polymorphic and that amyloids can assemble into the common β-barrel motif in the bilayer.
Results
Design of double-layered β-barrels in the lipid bilayer
Our conceptual designs of Aβ barrels employed the Aβ oligomer coordinate sets.39,40 We extracted the p3 peptide (26 residues; ~2.6 kDa/monomer) from the oligomer coordinates (Protein Data Bank code 2BEG)39 based on two-dimensional (2D) NMR in combination with ssNMR, mutational data, and electron microscopy. The N9 peptide (34 residues; ~3.6 kDa/monomer; adding Ile41 and Ala42 to Aβ9–40 coordinates) coordinates were derived from the ssNMR-based Aβ9–40 oligomer.40 In both coordinate sets, the Aβ peptides retained the U-shaped β-strand–turn–β-strand motif but with disordered N-termini. The U-shaped motif is a general motif in amyloid organization,65–67 first predicted by modeling68 and subsequently found in experiments39,40 in Aβ and in other amyloids [e.g., K3 (a β2-microglobulin fragment)41 and CA150 WW domain42] that could also form channels in the membrane.69–71
Experimentally, the ssNMR-based p3 pentamer39 and Aβ9–40 protofibril40 are currently the only Aβ oligomers with available structures. These data were obtained in solution. To date, determining atomic-level resolution of a lipid-bound Aβ oligomer is very difficult. The structures consist of two parallel, in-register β-sheets connected by a turn. Site-resolved ssNMR suggested that β-sheet-rich oligomeric intermediates are highly populated prior to fibril formation.37 Infrared spectroscopy verified the presence of β-sheets in the oligomers, indicating that they are similar to those of porins and that both parallel and antiparallel β-sheet arrangements of Aβ1–42 exist.72 It was further observed that a β-structure can penetrate the membrane,73 whereas the helical Aβ structure is predominantly on the membrane surface.74 Aβ oligomers were also found to more easily insert into the membrane than a single peptide,75 and more β-sheet structure was observed when embedded in the membrane.44 As pores, the ssNMR U-shaped peptides satisfy the lipid environment: the hydrophobic C-terminal strands interact with the lipid tails, whereas the N-terminal strands containing polar and charged residues face and form the solvated pore.
The shorter p3 peptide has a turn at Ser26–Ile31, whereas the longer N9 turn is at Asp23–Gly29. Although these are the only ones with experiment-based coordinates, other conformational variants are likely. The turn conformation is highly polymorphic.63 Assuming that these sequences are turn-prone, one way to obtain additional polymorphic turn variants is by exchanging turns (Fig. 1a). The first peptide was generated by removing residues 9–16 of the N9 peptide, called a p3 hybrid (p3h) peptide, containing the upstream turn. The second was created by adding residues 9–16 to the p3 peptide, called an N9 hybrid (N9h) peptide. p3h shares sequence with p3 and turn conformation with N9. Similarly, N9h shares sequence and turn conformation with N9 and p3, respectively. Although it is expected that such variants would be less populated, the heterogeneous channel population in the bilayer may include such species, particularly the N9h, because the N-terminus is known to be extramembraneous.
Fig. 1.
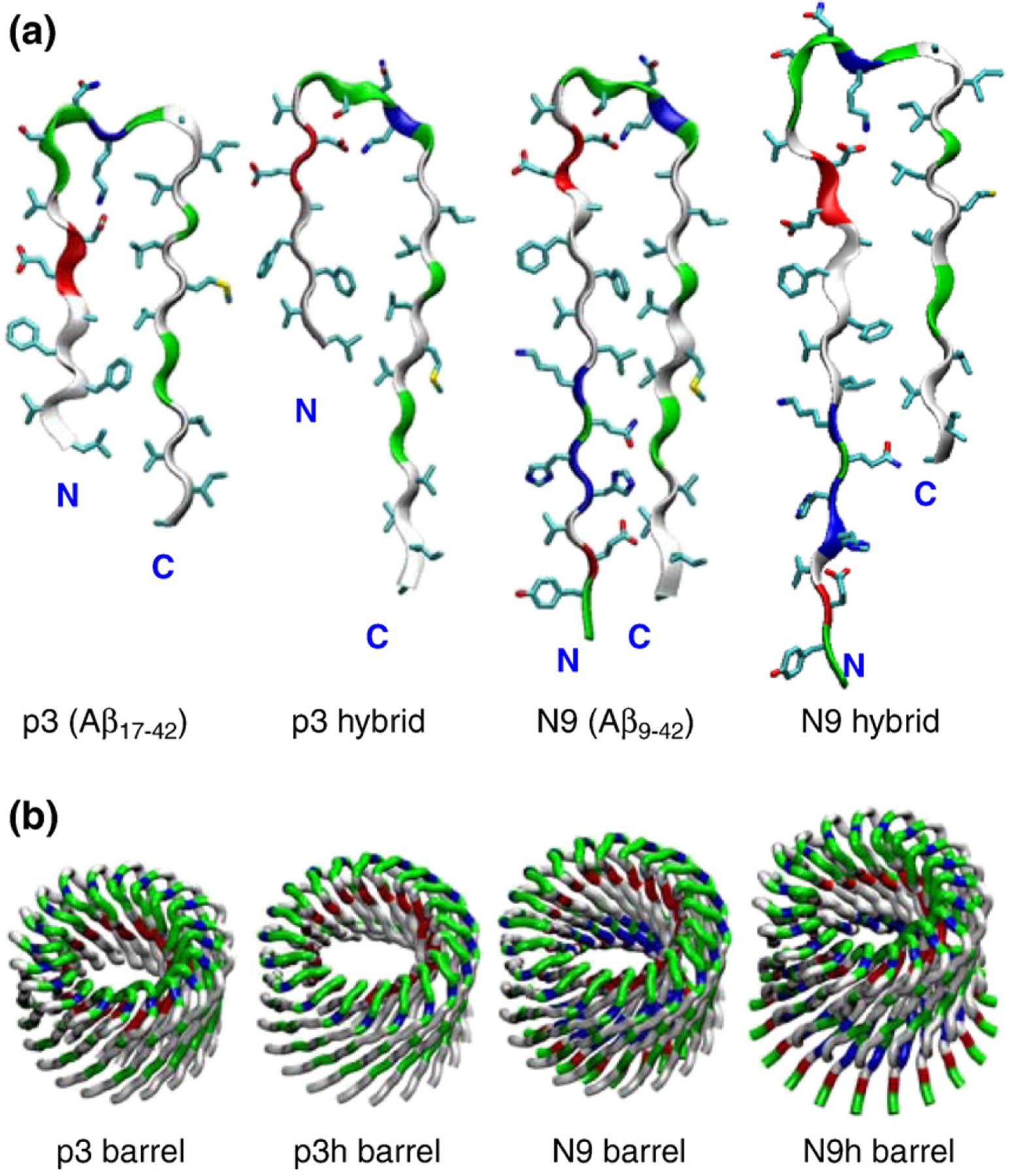
Truncated Aβ peptides with the U-shaped β-strand– turn–β-strand motif. (a) The p3 and N9 monomers extracted from two available experimentally based Aβ oligomer coordinate sets39,40 and their hybrids, p3h (containing the N9 turn conformation) and N9h (containing the p3 turn conformation), are shown by the peptide backbone in a ribbon representation. (b) Conceptual design of annular structure for the 20-mer barrels with the CNpNC topology embedded in the lipid bilayers. In the peptide, hydrophobic residues are shown in white, polar and Gly residues are shown in green, positively charged residues are shown in blue, and negatively charged residues are shown in red. The cartoons are shown for the 20-mer barrels.
Using these peptides, we constructed Aβ barrels with the pore-preserving CNpNC topology (where C and N represent C-terminus and N-terminus, respectively, and p denotes the pore). In the Aβ barrels, the inner barrel enclosing the water pore is composed of polar/charged residues, whereas the outer barrel is composed of mainly hydrophobic residues contacting the lipid tails. In the Aβ barrels, the β-strands are tilted with an inclination angle of ~37° with respect to the membrane normal and coil to create an annular barrel conformation (Fig. 1b). From the equation, , by McLachlan,7 where a is the Cα–Cα distance along the strands (with a=3.3 Å), b is the interstrand distance (with b=4.4 Å), and α is the inclination angle (with α being ~37°), we could estimate the shear number. We have S=N for all Aβ barrels in the simulations. Barrels with either S=N or S=N+4 are most stable with a right-handed β-sheet twist.2,62 Because the peptides are U-shaped, the Aβ barrels have two concentrically layered sheets. The outer sheet may have a shear SNN because the interstrand distance b increases due to the large curvature at the outer periphery. Thus, the double-layered β-sheet architecture in the amyloid channels is reasonable. The dimensions from our AFM images of Aβ channels49 provide outer and inner pore diameters in the ranges of ~6–10 and ~1–2 nm, respectively, indicating a radial channel thickness of ~5–8 nm, whereas for the single-layered β-sheet PG-1 channels,61 the ranges were ~5.13–5.76 and ~1.96– 2.24 nm, respectively, yielding an estimated radial channel thickness of ~3.1–3.5 nm. This suggests that the Aβ channels likely have double-layered β-sheets because they have almost twice the size of the radial channel thickness of the single-layered β-sheet channels. Thus, the double-layered β-barrel topology is employed in the barrel simulations. Although most of the results focus on the 20-mer p3, p3h, N9, and N9h barrels, the results of smaller barrels, 12-mer and 16-mer, are also presented (Fig. S1).
Heterogeneous Aβ barrel conformations
In the lipid bilayer, the Aβ barrels gradually relax during the simulations. We calculated the interaction energy for each peptide with the lipids and then averaged the peptide interaction energy in two ways: over the number of peptides in the channel (Fig. 2a) and over time (Fig. 2b). The time series of the peptide interaction energy with the lipids indicates that relaxed peptides in the lipid environment can be observed after 10 ns. Thus, 50-ns simulations can provide fully relaxed barrel conformations in the lipid bilayer. In the Aβ barrels, the interactions of the individual peptides are non-homogeneous. Even though each peptide is an identical unit in a barrel, the peptides can have different ways of interacting with their surroundings, including other peptides, lipids, water, and ions. The heterogeneity in the overall barrel conformation is consistent with experimental data,26,29,49 suggesting that Aβ barrels are viable channel candidates.
Fig. 2.
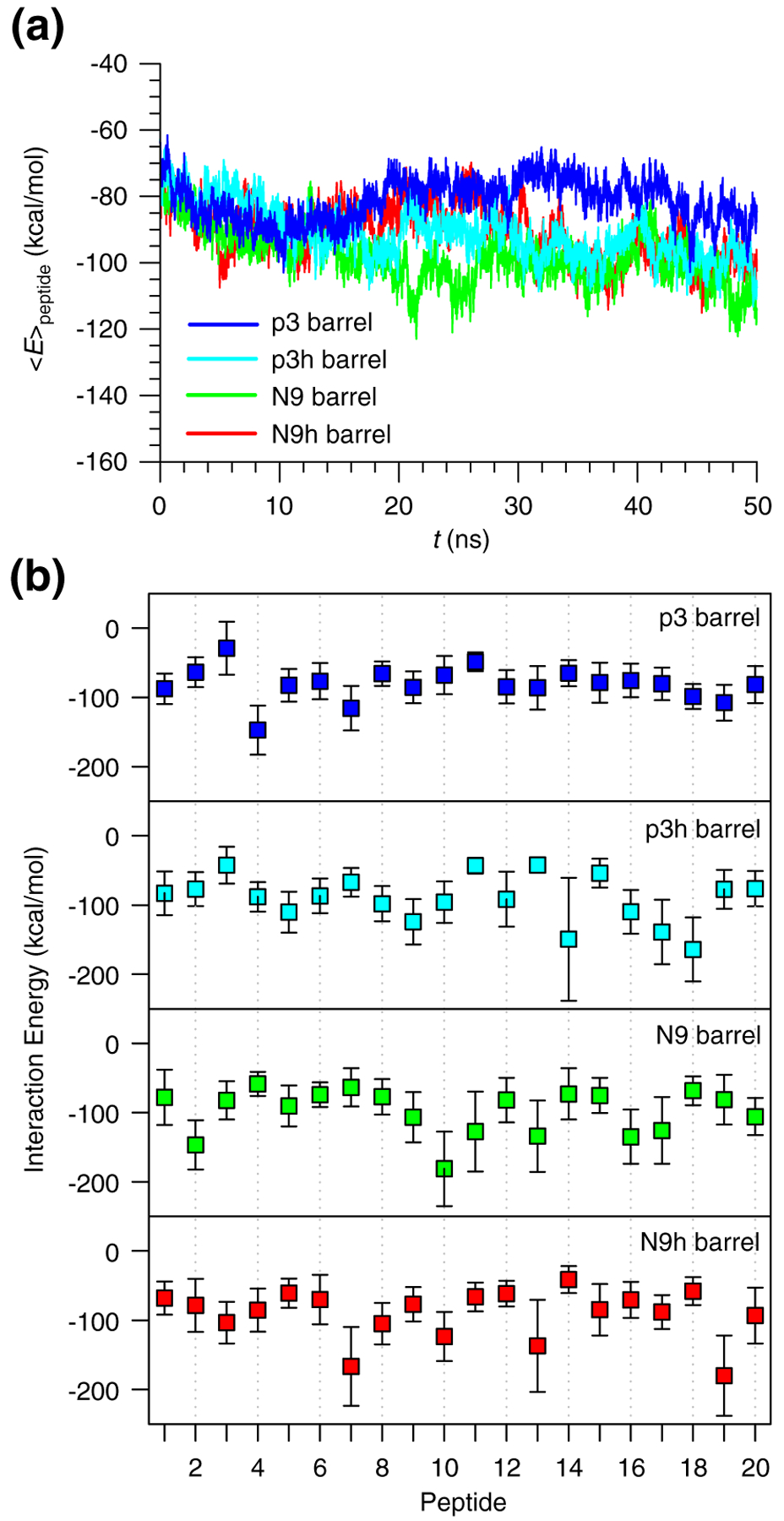
Interaction energies of truncated Aβ peptides with lipids. (a) Time series of averaged peptide interaction energy with DOPC lipids for the 20-mer p3 (blue line), p3h (cyan line), N9 (green line), and N9h (red line) barrels. (b) Interaction energies of each monomer in the 20-mer p3 (blue symbols), p3h (cyan symbols), N9 (green symbols), and N9h (red symbols) barrels averaged over time.
Heterogeneous Aβ barrel structures are presented as cartoons for the 20-mer p3, p3h, N9, and N9h barrels (Fig. 3). The cartoons represent the averaged barrel structures embedding the averaged pore structures as calculated by the HOLE program,76 illustrating the increasing outer and inner pore diameters during the simulations. In the starting conformations, all Aβ barrels have similar dimensions, with the outer diameters ~6.6, ~6.5, ~6.8, and ~6.7 nm and the pore diameters ~1.6, ~1.5, ~1.7, and ~1.7 nm for the p3, p3 h, N9, and N9h barrels, respectively. Following the 50-ns simulation, the averaged outer diameters and pore diameters increased to ~7.9, ~7.3, ~7.7, and ~8.0 nm and to ~2.2, ~1.7, ~1.9, and ~1.9 nm for the p3, p3h, N9, and N9h barrels, respectively. The Aβ barrel dimensions are in good agreement with the non-tilted Aβ channels simulated earlier, where the outer and pore diameters were ~7.4 and ~1.9 nm for the 20-mer p3 channel and ~7.8 and ~ 1.9 n m for the 20 - mer N9 channel , respectively.48,50 In addition, the outer and pore diameters were in the imaged AFM ranges of ~6–10 and ~1–2 nm, respectively.49
Fig. 3.
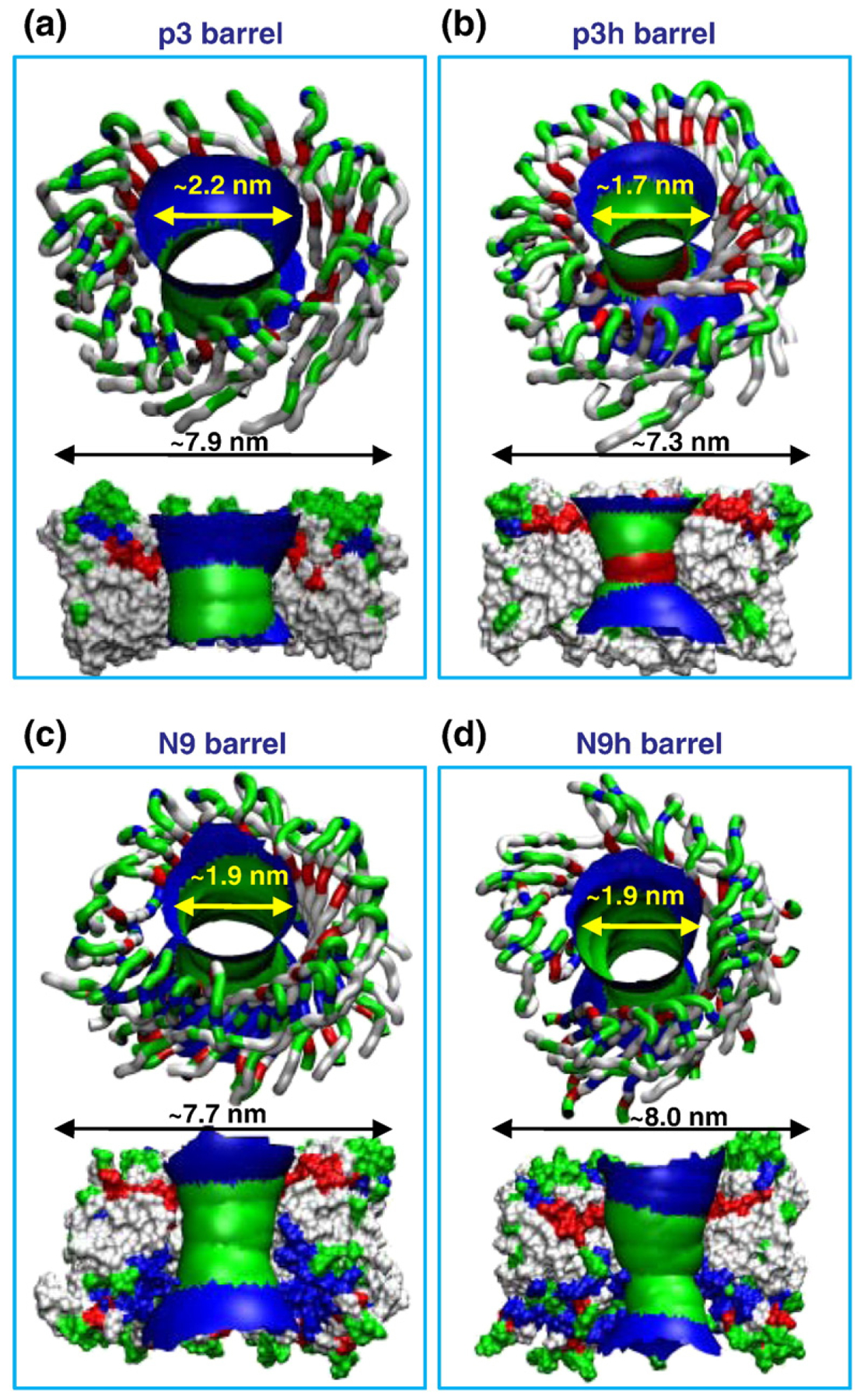
Heterogeneous barrel conformations in the lipid bilayer. Averaged pore structures calculated by the HOLE program76 embedded in the averaged barrel conformations during the simulations for the 20-mer p3 (a), p3h (b), N9 (c), and N9h (d) barrels. In the angle views of the pore structure (upper cartoons in each panel), whole barrel structures are shown with the ribbon representation with the same color representations described in the legend to Fig. 1. In the lateral views of the pore structure (lower cartoons in each panel), cross-sectioned barrels are given in the surface representation with the same color representations used in the legend to Fig. 1. For the pore structures in the surface representation, the degree of the pore diameter is indicated by the color codes in the order of redbgreenbblue, but the scale of these colors is relative to each barrel.
β-Strand reorientations may alter the Aβ barrel shapes
In the initial Aβ barrel design, the β-strands are inserted obliquely into the lipid bilayer, creating a distinct β-barrel conformation. The tilt angle of each β-strand relative to the pore axis is initially set to ~37°. In the non-tilted channel models,23,47–50 the β-strands were inserted parallel with the membrane normal. However, during the simulations, the β-strands do not retain their original orientations in the barrels. We monitored the β-strand tilt angles for the N-terminal and C-terminal β-strands separately and calculated the probability distributions of tilt angles over the simulations (Fig. 4). The N-terminal β-strands decreased the tilt angle, showing a less oblique insertion. A peak in the distribution curve located at a certain tilt angle clearly indicates a high population. In the p3h barrel, the N-terminal β-strand tilt angle has a wide distribution, because the short β-strands may be less constrained by the surroundings (Fig. 1). In contrast, the C-terminal β-strands increase the angle of inclination with a maximum probability at ~50° for all barrels. The β-strand reorientations via decreasing the tilt angle of the N-terminal β-strand tilt angle and increasing that of the C-terminal β-strand tilt angle appear common to all barrels, providing unique characteristics of U-shaped peptide Aβ barrels.
Fig. 4.
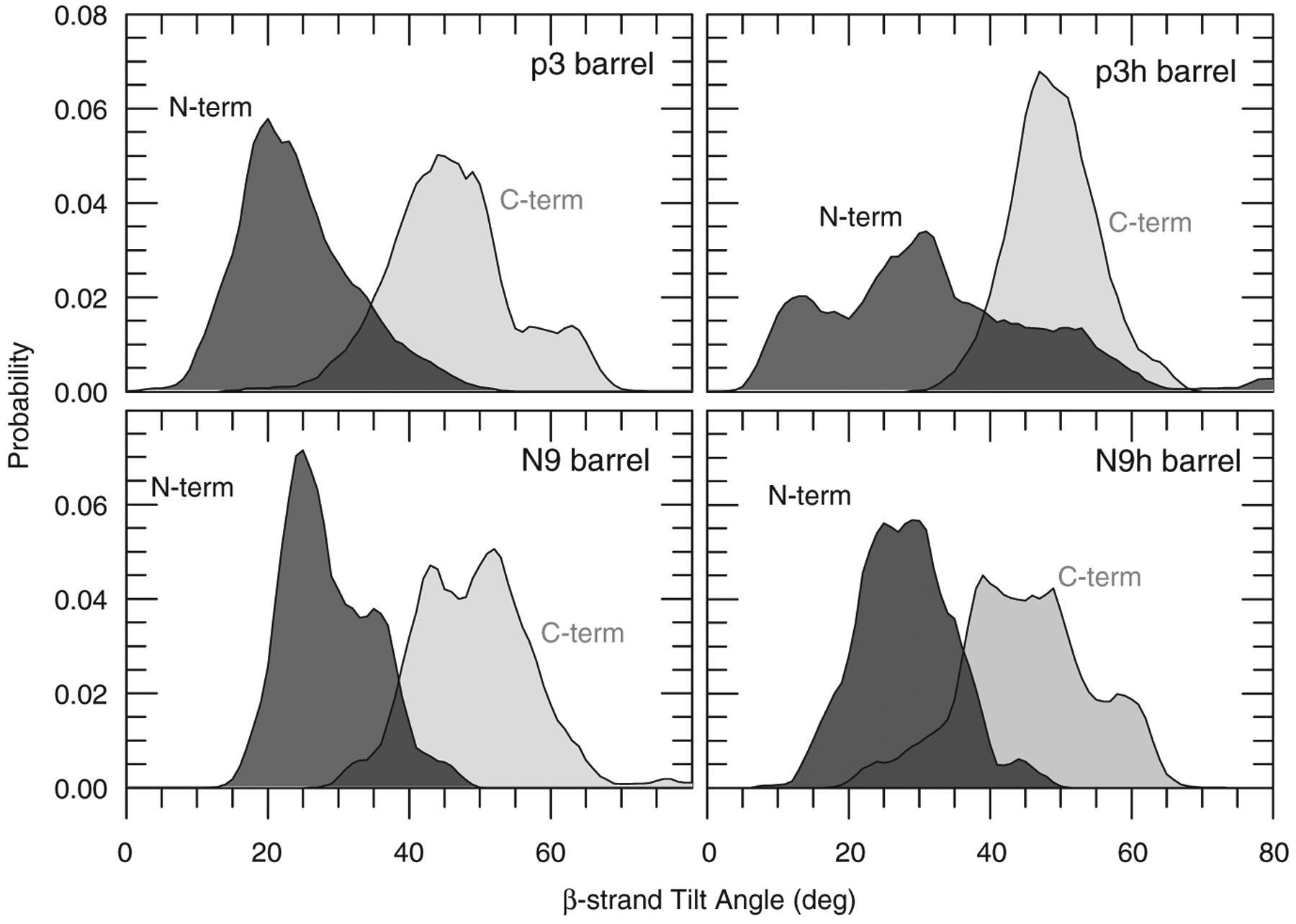
The calculated β-strand inclinations. The probability distributions of the β-strand tilt angle with respect to the bilayer normal are calculated separately for the N-terminal (dark gray area) and C-terminal (light gray area) β-strands in the 20-mer Aβ barrels.
β-Strand optimization plays a key role in the strand tilt and the barrel shape. In the starting Aβ barrel conformations, the inner N-terminal β-strands form a β-sheet with the intermolecular backbone hydrogen bonds (H-bonds) between the β-strands. Unlike the non-tilted Aβ channels,23,47–50 our starting Aβ barrels showed that the outer C-terminal β-strands form a weak β-sheet; however, during the simulations, both inner and outer β-barrels, which are initially circular, break into several small optimized β-sheets. The β-sheet breakages can be observed by monitoring the distance between the β-strands (Fig. 5). The effective β-strand distance, D*, is calculated by where the 〈 〉 brackets denote the time average and DCα is the distance between two intermolecular Cα atoms of the same residues. A larger D* value indicates closer β-strands. The figure shows that the inner N-terminal β-strands (gray circles) are closer than the outer C-terminal β-strands (white triangles). Several β-sheet breakage locations can be observed at low D* points. The discontinuities of the β-sheet network can also be observed in the description of secondary structure by STRIDE77 (Fig. S2).
Fig. 5.
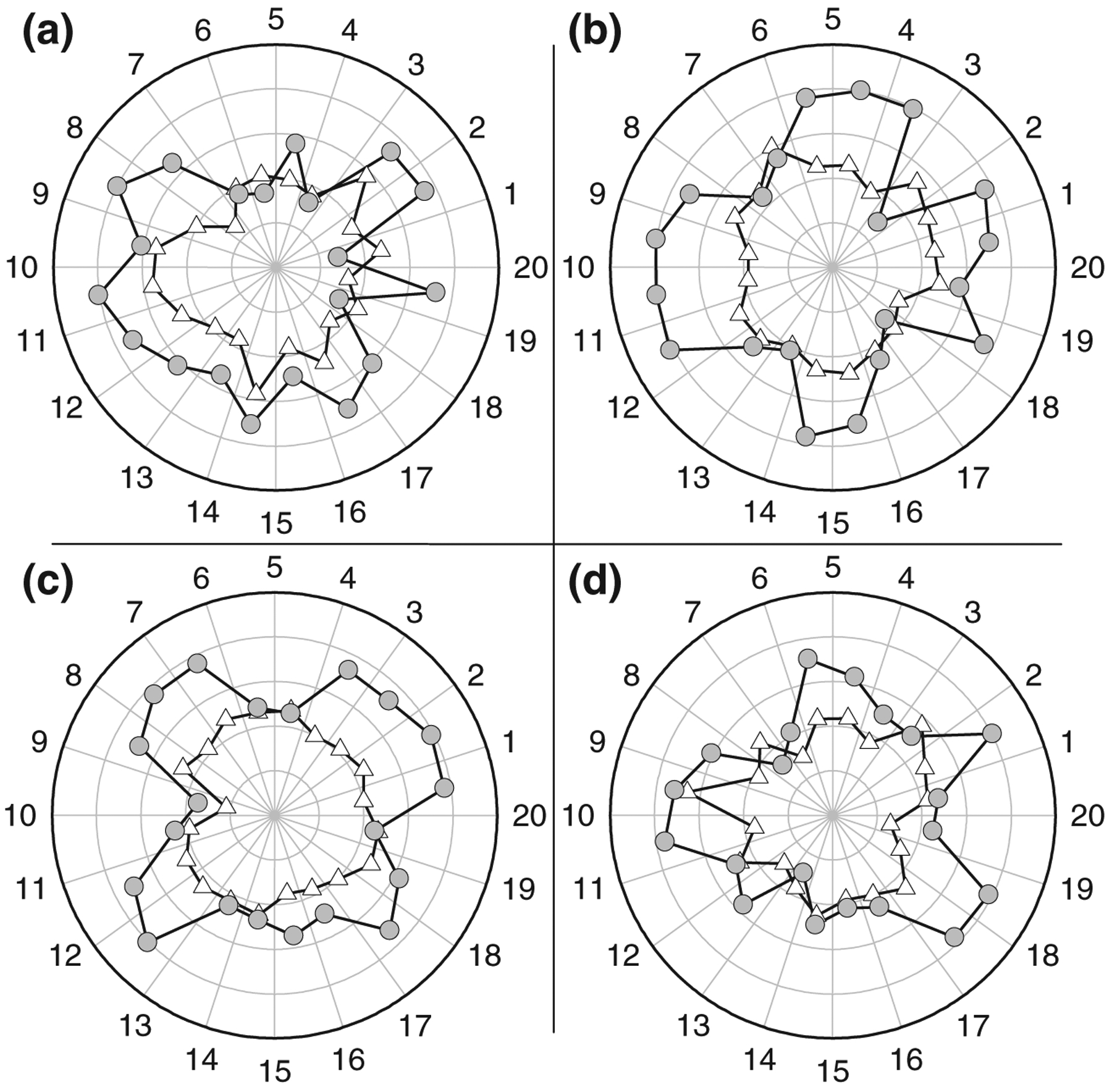
The calculated interstrand distance. The effective β-strand distance, which is an inverse of the distance between two intermolecular Cα atoms of the same residues for the 20-mer p3 (a), p3h (b), N9 (c), and N9h (d) barrels. Gray circles and white triangles denote the effective β-strand distances for the inner N-terminal and outer C-terminal β-strands, respectively. The radial data set of the effective distance is in the scale of 0.25 Å−1. Angular intervals represent each peptide in the Aβ barrels as the number marked on the angular scale.
The locally optimized β-sheets lead to the observed “subunits” in the Aβ barrels. The number of subunits observed in the barrel structure depends on the lipid dynamics during the simulation. In the subunits, the β-strands retain the β-barrel characteristics forming a β-sheet. We obtained five and four subunits for the p3 and p3h barrels, respectively, and five and six subunits for the N9 and N9h barrels, respectively (Fig. 6a–d). However, different numbers of subunit formations may be possible even for the same barrels, because subunit formations characterize an intrinsic feature of the fluidic lipid bilayer dynamics. Compared with the imaged AFM channels,49 high-resolution images of the p3 and N9 channels show pore-like structures with four to five subunits (Fig. 6e–h), consistent with the Aβ barrels.
Fig. 6.
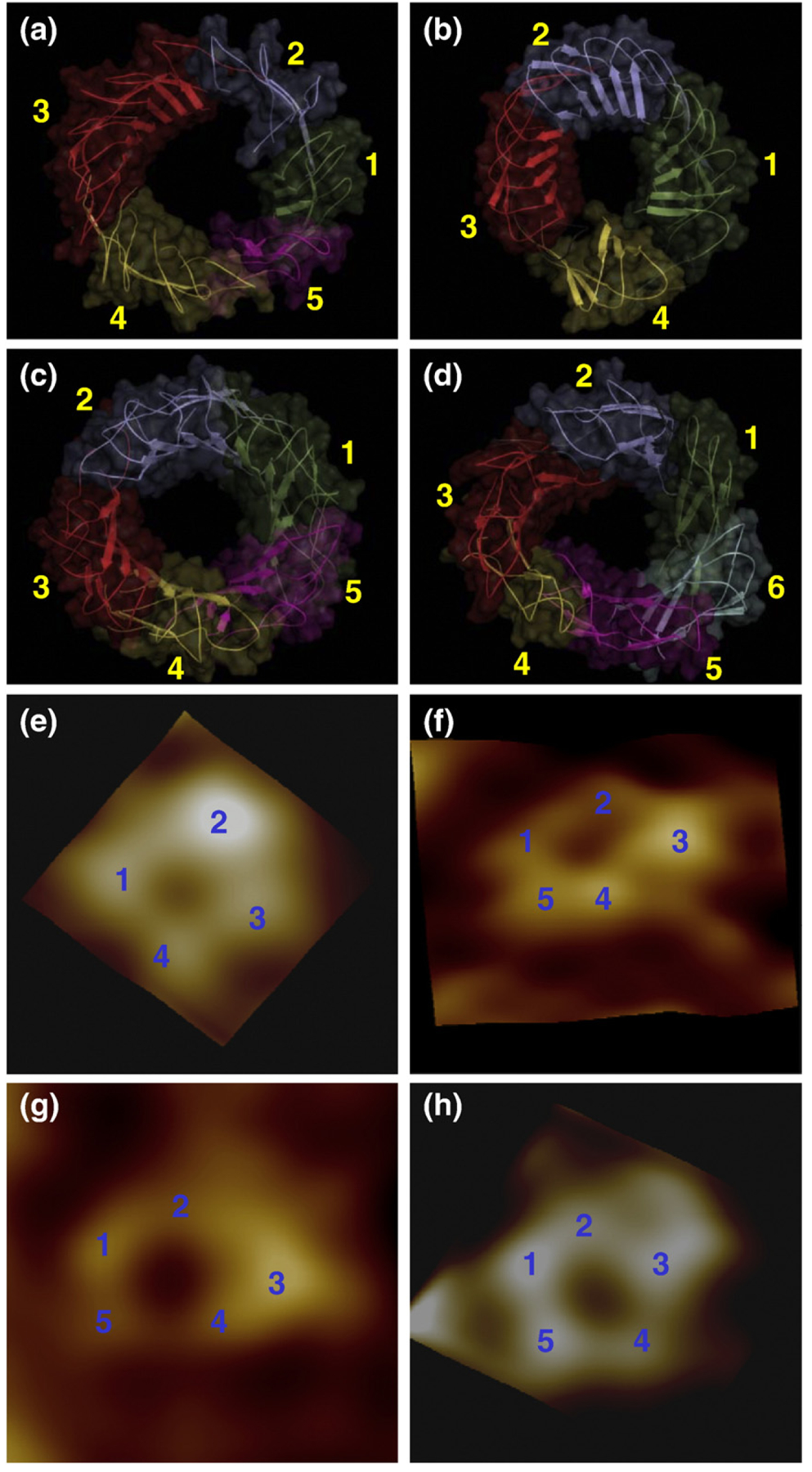
Side-by-side comparison between the computational barrels and the experimental channels. The simulated barrel structures with highlighted subunits for the 20-mer p3 (a), p3h (b), N9 (c), and N9h (d) barrels. The averaged barrels in the surface representation are shown in the view along the membrane normal. AFM images of p3 (e and f) and N9 (g and h) barrels show four or five subunits, consistent with the simulated barrels. Image sizes are 15 × 15 and 23×23 nm2, respectively (Jang et al.49; permission was obtained.)
β-Barrel-like strand arrangement supports the formation of smaller Aβ barrels
To obtain the size effects, in addition to the 20-mer, we also simulated the 12-mer and 16-mer barrels. Both barrels also increased the outer and inner pore diameters during the simulations. The outer and pore diameters for the p3 barrels were ~6.7 and ~1.8 nm for the 12-mer barrel (Fig. 7a) and ~6.8 and ~1.5 nm for the 16-mer barrel (Fig. 7b), respectively. Similarly, for the N9 barrels, the outer and pore diameters were ~6.6 and ~1.3 nm for the 12-mer barrel (Fig. 7c) and ~7.2 and ~1.6 nm for the 16-mer barrel (Fig. 7d), respectively. It is interesting to observe that, unlike the non-tilted β-sheet 12-mer Aβ channels,23,47–50 both 12-mer p3 and N9 barrels preserve the water pore with the size in the range of ~1–2 nm measured for the AFM channels.49 This suggests that the β-strand arrangement may stabilize the β-sheet in the Aβ barrels. The right-handed β-sheet twist in the barrels is known to be an evolutionarily more optimized β-sheet structure. Thus, in this organization, smaller channels can persist.
Fig. 7.
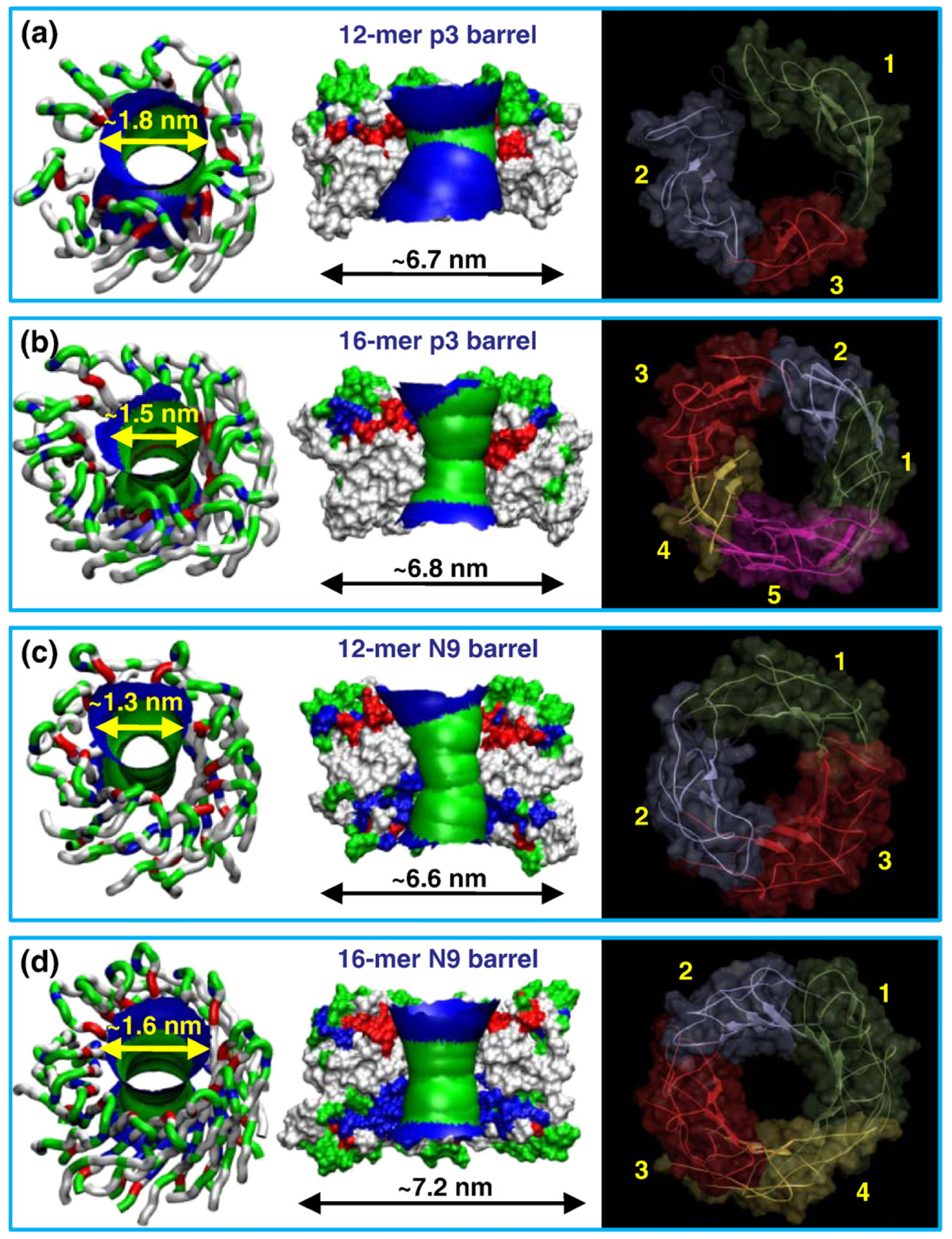
Possible smaller Aβ barrels in the lipid bilayer. Averaged pore structures calculated by the HOLE program76 embedded in the averaged barrel conformations during the simulations for the 12-mer p3 (a), 16-mer p3 (b), 12-mer N9 (c), and 16-mer N9 (d) barrels. In the angle views of the pore structure (left), whole barrel structures are shown with the ribbon representation with the same color representations described in the legend to Fig. 1. In the lateral views of the pore structure (center), cross-sectioned barrels are given in the surface representation with the same color representations used in the legend to Fig. 1. For the pore structures in the surface representation, the degree of the pore diameter is indicated by the color codes in the order of redbgreenbblue, but the scale of these colors is relative to each barrel. The simulated barrel structures (right) with highlighted subunits are shown in the view along the membrane normal.
Aβ barrels as polymorphic variants of Aβ channels
Lipids play an important role in supporting the barrel conformation during the simulations. The individual peptide interaction with the lipids varies, depending on the coupling between the peptide and the environment. In the Aβ barrels, the peptides are mainly organized in subunits, suggesting relatively weak coupling with lipids. In contrast, disordered peptides have relatively strong coupling with lipids when diffusing into the hydrophobic core or weak coupling when located at the solvated pore. The relative free energy changes of the peptide–lipid interaction energy as a function of the root-mean-square deviation (RMSD) from the starting point for the individual peptides are shown in Fig. 8. The free energy profile can be calculated by monitoring the occupancy probability for visiting each grid point on the plane of the interaction energy/RMSD for each peptide.78,79 In the p3 barrel, a highly populated peptide–lipid interaction energy of ~75 kcal/mol for the peptides with an RMSD of ~2.5 Å is enclosed by many contour lines on the plane, suggesting a low free energy profile for the peptides in the barrel (Fig. 8a). Other local minima on the plane with high RMSDs represent the disordered peptides in the barrel. In the p3h barrel, the location of the peptides on the plane presenting low free energy profiles is observed at a grid point of ~70 kcal/mol and ~2.0 Å for the peptide–lipid interaction energy and RMSD, respectively (Fig. 8b). In the N9 barrel, the peptide–lipid interaction energy of ~90 kcal/mol indicates a low free energy profile for peptides with an RMSD of ~3.4 Å (Fig. 8c), whereas in the N9h barrel, it is found at a grid point of ~75 kcal/mol and ~3.2 Å for the peptide–lipid interaction energy and RMSD, respectively (Fig. 8d). Although there are subtle differences in the peptide–lipid interaction between the Aβ barrels, the overall pictures of the free energy surface for the interaction energy/ RMSD are similar. Similar pictures were found for the non-tilted p3 and N9 channel simulations48 (Fig. S3), suggesting that the lipid bilayer can support various sequences, conformations, and arrangements of the peptides in the Aβ channels, providing further support for Aβ channel polymorphism.
Fig. 8.
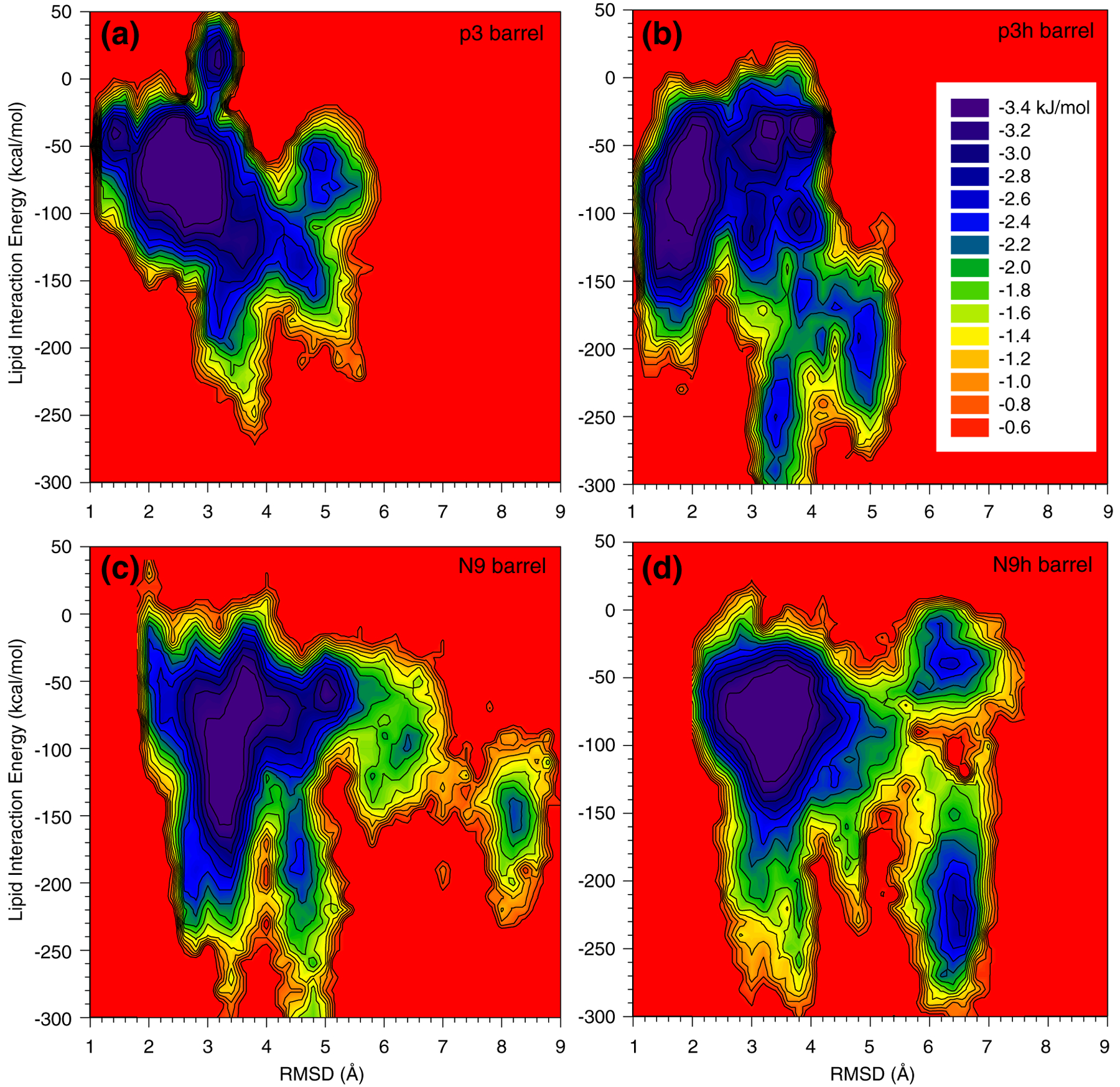
The intensity of peptide interaction with lipid. The relative free energy surface for the peptide–lipid interaction as a function of the RMSD from the starting point for the 20-mer p3 (a), p3h (b), N9 (c), and N9h (d) barrels.
Solvated pores attract ions implicating ion-permeable Aβ barrels
Aβ barrels have charged N-terminal β-strands regardless of the sequence and turn morphology. In the pore of the p3 and its hybrid barrel, the Glu22 side chains circularly cluster, forming a negatively charged ring, serving as cationic binding sites. In addition to the Glu22 cationic binding sites, the pores of the N9 and its hybrid barrel have positively charged His14 and Lys16, creating anionic binding sites. In the simulations of the barrel systems, we observed that the cations Mg2+, K+, Ca2+, and Zn2+ can bind to the cationic binding sites at Glu22 with a strong electrostatic attraction. These cations can also interact with a phosphate group in the lipid head and the C-terminus of peptide. In contrast, the Cl− anion can interact at His14 and Lys16 in the N-terminus of the peptide in the N9 and N9h barrels. To locate the highly populated ion interaction sites, we calculated three-dimensional (3D) density maps around the Aβ barrels (Fig. 9). The projection of a 3D density map onto a 2D plane representing the probability distribution for ions across the bilayer is shown in Fig. 9 (right column). In the p3 and N9 barrel pores, Mg2+ (green mesh), Ca2+ (blue), and Zn2+ (cyan) are highly populated, whereas the probability for K+ (red) in the pore is relatively low. The highly populated ion binding sites are reflected in peaks in the 2D probability distribution curves. At the bottom bilayer leaflet, Cl− (gray mesh) is highly populated, as indicated by peaks at z of ~−1.0 nm in the 2D curves (black lines), but the N9 pore has a much higher probability for Cl− than the p3 pore due to the anionic binding sites.
Fig. 9.
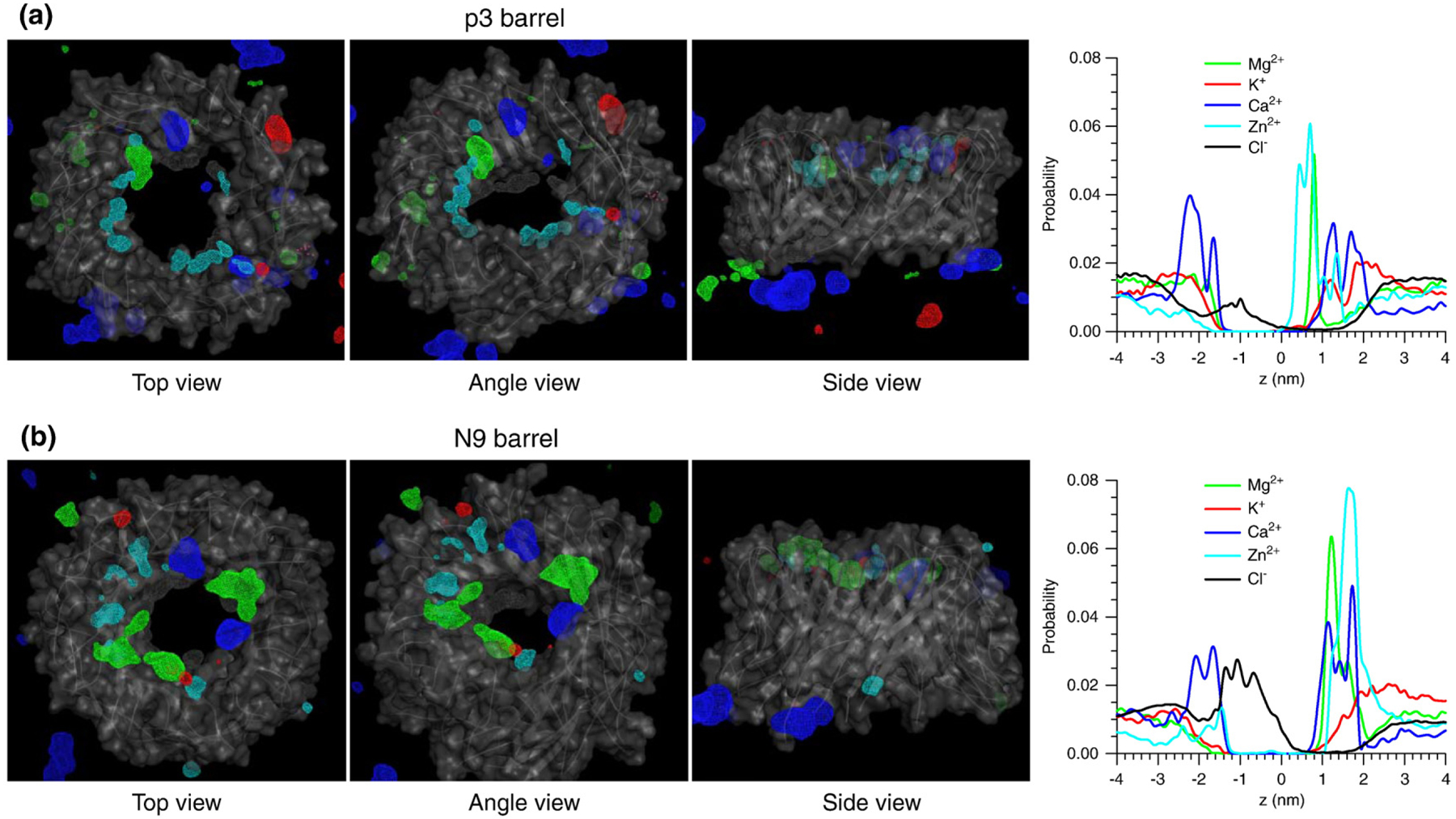
The ionic binding sites in the barrels. Three-dimensional density maps of Mg2+ (green mesh), K+ (red mesh), Ca2+ (blue mesh), Zn2+ (cyan mesh), and Cl− (gray mesh) for the 20-mer p3 (a) and N9 (b) barrels with three views. In the maps, the averaged barrel structures are shown as cartoons in gray. Probability distribution functions for Mg2+ (green line), K+ (red line), Ca2+ (blue line), Zn2+ (cyan line), and Cl− (black line) as a function of the pore axis are shown on the right corresponding to the cartoons on the left.
Based on the 2D ion probability distributions, the potential of mean force (PMF) representing the relative free energy profile for each ion across the bilayer can be calculated using the equation, ΔGPMF=−kBTln(ρz/ρbulk). Here, kB is the Boltzmann constant, T is the simulation temperature, ρz is the ion density at position z along the pore axis, and ρbulk is the ion density in the bulk region. The peaks in the 2D probability distribution curves provide a low free energy profile for ions at the binding sites (Fig. 10). All Aβ barrels confirm that cations are easily trapped by the negatively charged Glu22 side chains at the top bilayer leaflet, creating a cationic ring.23,47–50 The 3D density maps also show an additional low free energy profile for the interactions of cations with the peptides’ C-termini at the bottom bilayer leaflet. Cl− yields a relatively low free energy profile at the peptides’ N-termini for all Aβ barrels and at the anionic binding sites for the N9 and N9h barrels (Fig. 10c and d). For the smaller Aβ barrels (12-mer and 16-mer), the PMF curves also provide the ion binding sites (Fig. S4), which are similar to those of the 20-mer Aβ barrels. In contrast to the 12-mer Aβ non-tilted channels, which have a collapsed pore, the 12-mer Aβ barrel pores are wide enough for conducting ions and water, suggesting that the Aβ barrels are better optimized for pore solvation.
Fig. 10.
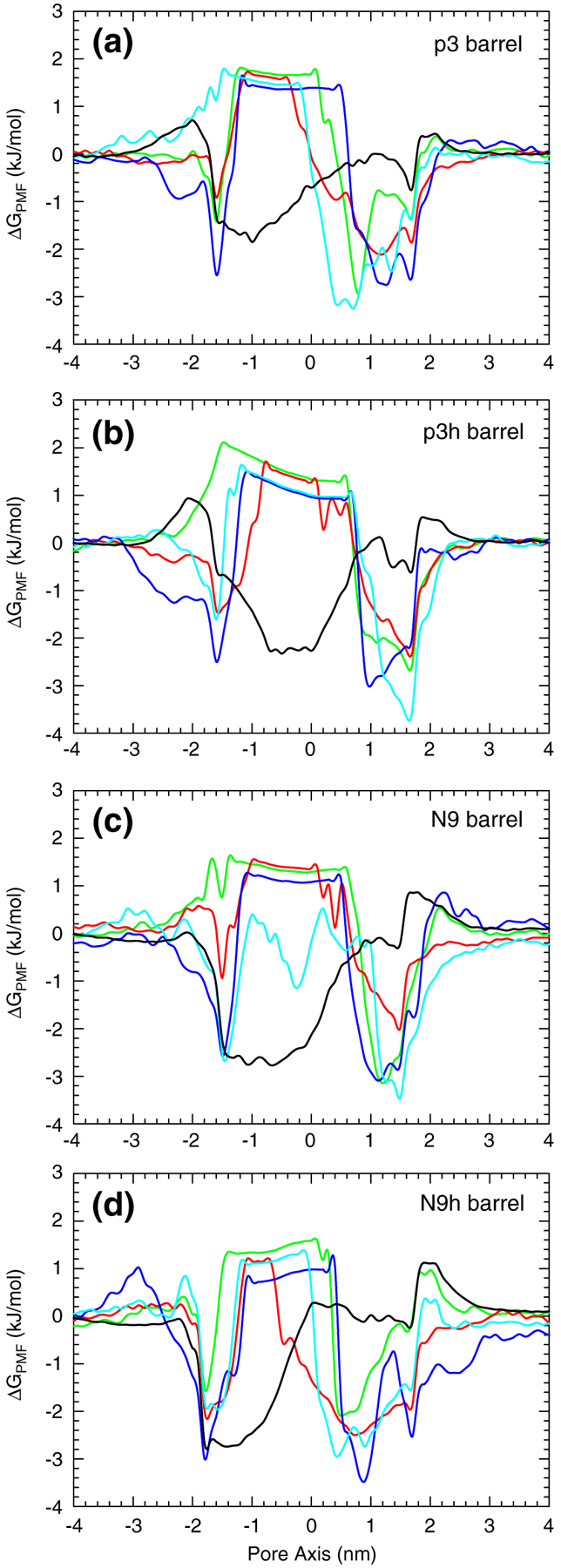
Ion-permeable Aβ barrels. PMF, ΔGPMF, calculated using the equation ΔGPMF=−kBTln(ρz/ρbulk) , where kB is the Boltzmann constant, T is the simulation temperature, ρz is the ion density at position z along the pore axis, and ρbulk is the ion density in the bulk region, representing the relative free energy profile for Mg2+ (green lines), K+ (red lines), Ca2+ (blue lines), Zn2+ (cyan lines), and Cl− (black lines) as a function of the distance along the pore center axis for the 20-mer p3 (a), p3h (b), N9 (c), and N9h (d) barrels.
Discussion and Conclusions
β-Barrel is a well-known motif in transmembrane channels, optimized by evolution. Because toxic amyloid channels also consist of a β-structure, we modeled the truncated Aβ peptides into a range of β-barrel-like structures and compared the results with our AFM images for the same p3 and N9 fragments (Fig. 6). We simulated different barrel sizes (12-mer, 16-mer, and 20-mer) and topologies to obtain the preferred ranges of outer and inner pore dimensions. For the p3 barrels, we obtained outer and pore diameters in the ranges of ~6.7–7.9 and ~1.5–2.2 nm, respectively. Similarly, the N9 barrel simulations provided outer and pore diameters in the ranges of ~6.6–7.7 and ~1.3–1.9 nm, respectively. In the 20-mer simulations, we introduced the hybrid Aβ peptides by exchanging the turn conformations and the sequences of p3 and N9 peptides, generating the p3h and N9h barrels. The hybrid 20-mer simulations also provided channel characteristics and dimensions in good agreement with AFM images.49 The barrel shapes were also in agreement with AFM subunit organizations.
It is interesting to note that the 12-mer Aβ barrels produce a well-defined pore, large enough for ion crossing, which is not the case for the 12-mer Aβ non-tilted channels whose pore collapsed.48,50 This suggests that the β-barrel-like strand arrangement in the Aβ barrels with a right-handed twist provides a more optimized pore organization, similar to the functionally optimized β-barrel porins. The right-handed β-sheet twist is also commonly observed in the cross-β-structure of amyloid fibrils.80 We note, however, that the β-sheet twist in the fibrils may differ from the twist in the Aβ barrels. In fibrils, residues in the twisted β-sheet are in register with adjacent strands, whereas in the Aβ barrels, the β-strands in the twisted β-sheet adopt a parallel β-structure where the strands are shifted with respect to each other. However, although the β-barrel strand arrangement holds well in the monomeric porins, which present an intact β-sheet, the amyloid barrels still break into dynamic subunits, suggesting that the membrane does not support a continuous circular β-sheet network formed by multimeric chains. During the simulations, Aβ barrels abandon the perfect annular organization and the relaxed barrels exhibit heterogeneous shapes: from triangular with three subunits to hexagonal with six, all with loosely attached mobile subunits in agreement with AFM images49 and previous simulations for the β-sheet channels assembled by non-tilted Aβ peptides.23,47–50 The β-barrel-like strand organization in the channels seems to provide more optimized pore structure than the channels with non-tilted Aβ peptides, but the subunit comparison between two channel topologies shows very similar results.
In β-barrels, a positive shear number S greater than or equal to the number of barrel-forming β-strands N (i.e., S≥N) stabilizes the barrel structure; β-barrels with S = N or S = N + 4 are the most stable.2,62 In the Aβ barrels presented here, the choice of the β-strand inclination angle (~37°) allows us to estimate the shear number, which is equal to N. In the case of β-strand insertion without tilt angle, the β-sheet channels have S=0, indicating that Aβ barrels are intrinsically more stable than our previously modeled β-sheet channels.23,47–50,59–61 In the Aβ barrel simulations, the inner β-strands decrease the inclination angle, whereas the outer β-strands increase the angle. Consequently, the shear number decreases at the inner barrels and increases at the outer barrels. With smaller S, the inner barrels are unstable, resulting in their breaking into several β-sheets; at the same time, the outer barrels with larger S try to stabilize by increasing the β-strand tilt angle, producing further β-sheet optimization. These forces on the inner and outer barrels impact the overall barrel structure. This suggests why Aβ barrels still break, reorganizing into multimeric subunit assemblies, which is reflected in the heterogeneous channel morphologies observed under AFM imaging and channel conductance observed in electrical recording.
Our modeled Aβ barrels follow the universal morphological features of amyloid oligomers,41,42,65 suggesting that similar toxic pores could be formed by other amyloids.29,70,71,81–84 The Aβ barrel is similar to the cytotoxic channel formed by the β-cytolytic antimicrobial PG-1 peptides. Recent studies showed that octameric and decameric β-sheet channels of PG-1 divided into four to five subunits,61,85 as observed in many amyloid channels.29,49 Aβ barrel formation is cytotoxic, inducing membrane defects. Our simulations suggest that the Aβ barrels induce a barrel-stave membrane pore. However, this does not apply to all amyloid channels. Islet amyloid polypeptide (also known as amylin) can induce other membrane pore types, such as toroidal membrane pores,82 depending on the outward-facing amino acid sequence interacting with the bilayer. Amylin also forms ion channels, as demonstrated in earlier work.29
In conclusion, here we used molecular dynamics simulations to examine the conformational heterogeneity of toxic β-barrel ion channels in Alzheimer’s Aβ using N-terminal truncated peptides. Our Aβ barrel simulations provide a range of sizes and morphologies. The 12-mer, 16-mer, and 20-mer simulations obtain similar subunit organization and dimensions as observed in the Aβ channels by AFM49 and in conventional β-sheet channels without tilt.23,47–50 In the 20-mer simulations, peptides with hybrid turns also form ion channel-like structures with similar channel morphologies and dimensions. Aβ barrels present a β-barrel-like strand arrangement with right-handed strand twist, which is the optimized β-sheet structure. However, the Aβ barrels are distinct from the transmembrane β-barrels. The β-barrel porins have an antiparallel β-strand arrangement, a single chain, and a single-layered circular β-sheet. In contrast, Aβ barrels have a parallel β-strand arrangement, multimeric chains, and two-layered circular β-sheets. Nevertheless, the presence of β-barrel-like conformation and the formation of ion-permeable barrels with hybrid monomer conformation suggest that Aβ barrel is a populated polymorphic variant of Aβ channels. In nature, β-barrel conformations may present an intrinsic toxic topology when introduced to eukaryotes. Thus, this study leads to two major conclusions: first, it shows that Aβ barrels can serve as candidates for toxic Alzheimer’s ion channels and, second, it highlights the possibility of conformational heterogeneity and polymorphism of amyloid channels in the membranes—an issue that would need careful consideration in drug design targeting amyloid ion channels.
Materials and Methods
Two monomer conformations of Aβ, p3 as defined in the Aβ1–42 pentamer based on hydrogen/deuterium-exchange NMR data, side-chain packing constraints from pairwise mutagenesis, ssNMR, and electron microscopy (Protein Data Bank code 2BEG; residues 1–16 are missing due to disorder)39 and N9 based on the ssNMR model of small Aβ9–40 protofibril coordinates40 adding Ile41 and Ala42, were used to construct β-barrel-like channels with annular shapes. To construct the β-barrel structure, we inclined the U-shaped Aβ peptides ~37° relative to the pore axis and then rotated them 12, 16, and 20 times with respect to the pore axis, creating different sizes of Aβ barrels. In the 20-mer simulations, hybrid peptides, p3h (p3 peptide adapting the N9 turn conformation) and N9h (N9 peptide adapting the p3 turn conformation), were used to construct the barrels using the same method for the peptides with intrinsic turns.
In our simulations, all Aβ barrels were simulated in the zwitterionic DOPC bilayer. Our choice of DOPC closely follows the experimental conditions employed during AFM imaging.25–27,29,49 At room temperature, DOPC is well above its transition temperature (~−20 °C). Its fluidity increases the probability of proteins and peptides to get incorporated into the bilayer during reconstitution, and the absence of phase-separated domains provides flat and uniform lipid supported bilayers on mica surfaces such that the incorporated peptides can be readily distinguished during imaging.27,29 Although some amyloids prefer to interact with anionic bilayers86–89 as compared with the zwitterionic phosphatidylcholine lipids,43,90 anionic lipids interact repulsively with the negatively charged mica surface used as substrate in AFM experiments, thus complicating the formation of a supported bilayer. In our previous simulations, Aβ channels were modeled in both the zwitterionic DOPC bilayer and the anionic bilayer containing POPC and POPG.48,50 However, no major differences in the results of the subunit formation in the channel conformation were found.
The barrels were then minimized with a rigid-body motion for the peptides in order to enhance the formation of backbone hydrogen bonds (H-bonds) within a β-sheet and then embedded in the DOPC bilayer. A unit cell containing two layers of lipids with 95,000–160,000 atoms, depending on the size, was constructed. In the bilayer construction, our method closely follows previous β-sheet channel simulations.23,47–50,59–61 For the lipid bilayer, 200–400 lipids constituted the unit cell with TIP3P water molecules added at both sides. The system contained MgCl2, KCl, CaCl2, and ZnCl2 at the same concentration of 25 mM to satisfy a total cation concentration near 100 mM.
The CHARMM program91 was used to construct the set of starting points and to relax the systems to a production-ready stage. In the pre-equilibrium stages, the initial configurations were gradually relaxed, with the barrels held rigid. A series of dynamic cycles were performed with the harmonically restrained peptides in the channels, and then the harmonic restraints were gradually diminished with the full Ewald electrostatics calculation and constant temperature (Nosé–Hoover) thermostat/barostat at 300 K. The entire pre-equilibration cycle took 5 ns to yield the starting point. Simulations for the initial construction and the pre-equilibration were performed on the NPAT (constant number of atoms, pressure, surface area, and temperature) ensemble. For production runs to 50 ns for all barrels, the NAMD code92 on a Biowulf cluster at the National Institutes of Health† (Bethesda, MD) was used for the starting point. Averages were taken after 10 ns, discarding initial transients. AFM images were obtained as described by Jang et al.49
Supplementary Material
Acknowledgements
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health (NIH), under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the National Cancer Institute Center for Cancer Research, NIH. This research was also supported by the National Institute on Aging, NIH, extramural program (R.L.). All simulations had been performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the NIH.
Abbreviations used:
- 2D
two-dimensional
- 3D
three-dimensional
- Aβ
β-amyloid
- AFM
atomic force microscopy
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- N9
Aβ9–42
- N9h
N9 hybrid
- p3
Aβ17–42
- p3h
p3 hybrid
- PG-1
protegrin-1
- PMF
potential of mean force
- ssNMR
solid-state NMR
Footnotes
Supplementary Data
Supplementary data to this article can be found online at doi:10.1016/j.jmb.2010.10.025
References
- 1.Wimley WC (2003). The versatile β-barrel membrane protein. Curr. Opin. Struct. Biol 13, 404–411. [DOI] [PubMed] [Google Scholar]
- 2.Schulz GE (2002). The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta, 1565, 308–317. [DOI] [PubMed] [Google Scholar]
- 3.Benz R, Schmid A & Hancock RE (1985). Ion selectivity of Gram-negative bacterial porins. J. Bacteriol 162, 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jap BK & Walian PJ (1990). Biophysics of the structure and function of porins. Q. Rev. Biophys 23, 367–403. [DOI] [PubMed] [Google Scholar]
- 5.Murzin AG, Lesk AM & Chothia C (1994). Principles determining the structure of beta-sheet barrels in proteins: I. A theoretical analysis. J. Mol. Biol 236, 1369–1381. [DOI] [PubMed] [Google Scholar]
- 6.Murzin AG, Lesk AM & Chothia C (1994). Principles determining the structure of beta-sheet barrels in proteins: II. The observed structures. J. Mol. Biol 236, 1382–1400. [DOI] [PubMed] [Google Scholar]
- 7.McLachlan AD (1979). Gene duplications in the structural evolution of chymotrypsin. J. Mol. Biol 128, 49–79. [DOI] [PubMed] [Google Scholar]
- 8.Thinnes FP, Gotz H, Kayser H, Benz R, Schmidt WE, Kratzin HD & Hilschmann N (1989). Identification of human porins: I. Purification of a porin from human B-lymphocytes (Porin 31HL) and the topochemical proof of its expression on the plasmalemma of the progenitor cell. Biol. Chem. Hoppe Seyler, 370, 1253–1264. [PubMed] [Google Scholar]
- 9.Bay DC & Court DA (2002). Origami in the outer membrane: the transmembrane arrangement of mitochondrial porins. Biochem. Cell Biol 80, 551–562. [DOI] [PubMed] [Google Scholar]
- 10.Casadio R, Jacoboni I, Messina A & De Pinto V (2002). A 3D model of the voltage-dependent anion channel (VDAC). FEBS Lett. 520, 1–7. [DOI] [PubMed] [Google Scholar]
- 11.Shulga N, Wilson-Smith R & Pastorino JG (2009). Hexokinase II detachment from the mitochondria potentiates cisplatin induced cytotoxicity through a caspase-2 dependent mechanism. Cell Cycle, 8, 3355–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abu-Hamad S, Arbel N, Calo D, Arzoine L, Israelson A, Keinan N et al. (2009). The VDAC1 N-terminus is essential both for apoptosis and the protective effect of anti-apoptotic proteins. J. Cell Sci 122, 1906–1916. [DOI] [PubMed] [Google Scholar]
- 13.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J et al. (2002). Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature, 416, 507–511. [DOI] [PubMed] [Google Scholar]
- 14.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS et al. (2002). Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature, 416, 535–539. [DOI] [PubMed] [Google Scholar]
- 15.Cheng IH, Scearce-Levie K, Legleiter J, Palop JJ, Gerstein H, Bien-Ly N et al. (2007). Accelerating amyloid-β fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J. Biol. Chem 282, 23818–23828. [DOI] [PubMed] [Google Scholar]
- 16.Arispe N, Pollard HB & Rojas E (1993). Giant multilevel cation channels formed by Alzheimer disease amyloid β-protein [AβP-(1–40)] in bilayer membranes. Proc. Natl Acad. Sci. USA, 90, 10573–10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arispe N, Rojas E & Pollard HB (1993). Alzheimer disease amyloid β protein forms calcium channels in bilayer membranes: blockade by tromethamine and aluminum. Proc. Natl Acad. Sci. USA, 90, 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arispe N, Pollard HB & Rojas E (1994). β-Amyloid Ca(2+)-channel hypothesis for neuronal death in Alzheimer disease. Mol. Cell. Biochem 140, 119–125. [DOI] [PubMed] [Google Scholar]
- 19.Arispe N, Pollard HB & Rojas E (1996). Zn2+ interaction with Alzheimer amyloid β protein calcium channels. Proc. Natl Acad. Sci. USA, 93, 1710–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhatia R, Lin H & Lal R (2000). Fresh and globular amyloid β protein (1–42) induces rapid cellular degeneration: evidence for AβP channel-mediated cellular toxicity. FASEB J. 14, 1233–1243. [DOI] [PubMed] [Google Scholar]
- 21.Hirakura Y, Lin MC & Kagan BL (1999). Alzheimer amyloid Aβ1–42 channels: effects of solvent, pH, and Congo red. J. Neurosci. Res 57, 458–466.10440895 [Google Scholar]
- 22.Hirakura Y, Yiu WW, Yamamoto A & Kagan BL (2000). Amyloid peptide channels: blockade by zinc and inhibition by Congo red (amyloid channel block). Amyloid, 7, 194–199. [DOI] [PubMed] [Google Scholar]
- 23.Jang H, Zheng J, Lal R & Nussinov R (2008). New structures help the modeling of toxic amyloidβ ion channels. Trends Biochem. Sci 33, 91–100. [DOI] [PubMed] [Google Scholar]
- 24.Kawahara M, Arispe N, Kuroda Y & Rojas E (1997). Alzheimer’s disease amyloid β-protein forms Zn(2+)-sensitive, cation-selective channels across excised membrane patches from hypothalamic neurons. Biophys. J 73, 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lal R, Lin H & Quist AP (2007). Amyloid beta ion channel: 3D structure and relevance to amyloid channel paradigm. Biochim. Biophys. Acta, 1768, 1966–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin H, Bhatia R & Lal R (2001). Amyloid β protein forms ion channels: implications for Alzheimer’s disease pathophysiology. FASEB J. 15, 2433–2444. [DOI] [PubMed] [Google Scholar]
- 27.Lin H, Zhu YJ & Lal R (1999). Amyloid β protein (1−40) forms calcium-permeable, Zn2+-sensitive channel in reconstituted lipid vesicles. Biochemistry, 38, 11189–11196. [DOI] [PubMed] [Google Scholar]
- 28.Lin MC & Kagan BL (2002). Electrophysiologic properties of channels induced by Aβ25–35 in planar lipid bilayers. Peptides, 23, 1215–1228. [DOI] [PubMed] [Google Scholar]
- 29.Quist A, Doudevski I, Lin H, Azimova R, Ng D, Frangione B et al. (2005). Amyloid ion channels: a common structural link for protein-misfolding disease. Proc. Natl Acad. Sci. USA, 102, 10427–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee SK, Quist AP & Lal R (1998). Amyloid β protein-(1–42) forms calcium-permeable, Zn2+- sensitive channel. J. Biol. Chem 273, 13379–13382. [DOI] [PubMed] [Google Scholar]
- 31.Zhu YJ, Lin H & Lal R (2000). Fresh and nonfibrillar amyloid β protein(1–40) induces rapid cellular degeneration in aged human fibroblasts: evidence for AβP-channel-mediated cellular toxicity. FASEB J. 14, 1244–1254. [DOI] [PubMed] [Google Scholar]
- 32.Diaz JC, Simakova O, Jacobson KA, Arispe N & Pollard HB (2009). Small molecule blockers of the Alzheimer Abeta calcium channel potently protect neurons from Abeta cytotoxicity. Proc. Natl Acad. Sci. USA, 106, 3348–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kayed R, Pensalfini A, Margol L, Sokolov Y, Sarsoza F, Head E et al. (2009). Annular protofibrils are a structurally and functionally distinct type of amyloid oligomer. J. Biol. Chem 284, 4230–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capone R, Garcia Quiroz F, Prangkio P, Sauer AM, Bautista MR, Turner RS et al. (2009). Amyloid-β-induced ion flux in artificial lipid bilayers and neuronal cells: resolving a controversy. Neurotox. Res 16, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mobley DL, Cox DL, Singh RR, Maddox MW & Longo ML (2004). Modeling amyloid β-peptide insertion into lipid bilayers. Biophys. J 86, 3585–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durell SR, Guy HR, Arispe N, Rojas E & Pollard HB (1994). Theoretical models of the ion channel structure of amyloid β-protein. Biophys. J 67, 2137–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chimon S & Ishii Y (2005). Capturing intermediate structures of Alzheimer’s β-amyloid, Aβ(1–40), by solid-state NMR spectroscopy. J. Am. Chem. Soc 127, 13472–13473. [DOI] [PubMed] [Google Scholar]
- 38.Singer SJ & Dewji NN (2006). Evidence that Perutz’s double-beta-stranded subunit structure for beta-amyloids also applies to their channel-forming structures in membranes. Proc. Natl Acad. Sci. USA, 103, 1546–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Dobeli H et al. (2005). 3D structure of Alzheimer’s amyloid-β(1–42) fibrils. Proc. Natl Acad. Sci. USA, 102, 17342–17347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petkova AT, Yau WM & Tycko R (2006). Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry, 45, 498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwata K, Fujiwara T, Matsuki Y, Akutsu H, Takahashi S, Naiki H & Goto Y (2006). 3D structure of amyloid protofilaments of β2-microglobulin frag ment probed by solid-state NMR. Proc. Natl Acad. Sci. USA, 103, 18119–18124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson N, Becker J, Tidow H, Tremmel S, Sharpe TD, Krause G et al. (2006). General structural motifs of amyloid protofilaments. Proc. Natl Acad. Sci. USA, 103, 16248–16253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Planque MR, Raussens V, Contera SA, Rijkers DT, Liskamp RM, Ruysschaert JM et al. (2007). β-Sheet structured β-amyloid(1–40) perturbs phosphatidylcholine model membranes. J. Mol. Biol 368, 982–997. [DOI] [PubMed] [Google Scholar]
- 44.Lau TL, Ambroggio EE, Tew DJ, Cappai R, Masters CL, Fidelio GD et al. (2006). Amyloid-β peptide disruption of lipid membranes and the effect of metal ions. J. Mol. Biol 356, 759–770. [DOI] [PubMed] [Google Scholar]
- 45.Kagan BL (2005). Amyloidosis and protein folding. Science, 307, 42–43; author reply, 42–43. [DOI] [PubMed] [Google Scholar]
- 46.Petosa C, Collier RJ, Klimpel KR, Leppla SH & Liddington RC (1997). Crystal structure of the anthrax toxin protective antigen. Nature, 385, 833–838. [DOI] [PubMed] [Google Scholar]
- 47.Jang H, Zheng J & Nussinov R (2007). Models of β-amyloid ion-channels in the membrane suggest that channel formation in the bilayer is a dynamic process. Biophys. J 93, 1938–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jang H, Arce FT, Capone R, Ramachandran S, Lal R & Nussinov R (2009). Misfolded amyloid ion channels present mobile β-sheet subunits in contrast to conventional ion channels. Biophys. J 97, 3029–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang H, Arce FT, Ramachandran S, Capone R, Azimova R, Kagan BL et al. (2010). Truncated β-amyloid peptide channels provide an alternative mechanism for Alzheimer’s disease and Down syndrome. Proc. Natl Acad. Sci. USA, 107, 6538–6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jang H, Arce FT, Ramachandran S, Capone R, Lal R & Nussinov R (2010). Structural convergence among diverse toxic β-sheet ion channels. J. Phys. Chem. B, 114, 9445–9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Motte J & Williams RS (1989). Age-related changes in the density and morphology of plaques and neurofibrillary tangles in Down syndrome brain. Acta Neuropathol. 77, 535–546. [DOI] [PubMed] [Google Scholar]
- 52.Gowing E, Roher AE, Woods AS, Cotter RJ, Chaney M, Little SP & Ball MJ (1994). Chemical characterization of Aβ17–42 peptide, a component of diffuse amyloid deposits of Alzheimer disease. J. Biol. Chem 269, 10987–10990. [PubMed] [Google Scholar]
- 53.Higgins LS, Murphy GM, Forno LS, Catalano R & Cordell B (1996). p3 β-amyloid peptide has a unique and potentially pathogenic immunohisto-chemical profile in Alzheimer’s disease brain. Am. J. Pathol 149, 585–596. [PMC free article] [PubMed] [Google Scholar]
- 54.Wisniewski T, Lalowski M, Bobik M, Russell M, Strosznajder J & Frangione B (1996). Amyloid β1–42 deposits do not lead to Alzheimer’s neuritic plaques in aged dogs. Biochem. J 313, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lalowski M, Golabek A, Lemere CA, Selkoe DJ, Wisniewski HM, Beavis RC et al. (1996). The “nonamyloidogenic” p3 fragment (amyloid β17–42) is a major constituent of Down’s syndrome cerebellar preamyloid. J. Biol. Chem 271, 33623–33631. [DOI] [PubMed] [Google Scholar]
- 56.Wei W, Norton DD, Wang X & Kusiak JW (2002). Aβ17–42 in Alzheimer’s disease activates JNK and caspase-8 leading to neuronal apoptosis. Brain, 125, 2036–2043. [DOI] [PubMed] [Google Scholar]
- 57.Szczepanik AM, Rampe D & Ringheim GE (2001). Amyloid-β peptide fragments p3 and p4 induce pro-inflammatory cytokine and chemokine production in vitro and in vivo. J. Neurochem 77, 304–317. [DOI] [PubMed] [Google Scholar]
- 58.Pike CJ, Overman MJ & Cotman CW (1995). Amino-terminal deletions enhance aggregation of β-amyloid peptides in vitro. J. Biol. Chem 270, 23895–23898. [DOI] [PubMed] [Google Scholar]
- 59.Mustata M, Capone R, Jang H, Arce FT, Ramachandran S, Lal R & Nussinov R (2009). K3 fragment of amyloidogenic β2-microglobulin forms ion channels: implication for dialysis related amyloidosis. J. Am. Chem. Soc 131, 14938–14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jang H, Ma B & Nussinov R (2007). Conformational study of the protegrin-1 (PG-1) dimer interaction with lipid bilayers and its effect. BMC Struct. Biol 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Capone R, Mustata M, Jang H, Arce FT, Nussinov R & Lal R (2010). Antimicrobial prote-grin-1 (PG-1) forms ion channels: MD simulation, AFM, and electrical conductance studies. Biophys. J 98, 2644–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sansom MS & Kerr ID (1995). Transbilayer pores formed by beta-barrels: molecular modeling of pore structures and properties. Biophys. J 69, 1334–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller Y, Ma B & Nussinov R (2009). Polymorphism of Alzheimer’s Aβ17–42 (p3) oligomers: the importance of the turn location and its conformation. Biophys. J 97, 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller Y, Ma B & Nussinov R (2010). Polymorphism in Alzheimer Aβ amyloid organization reflects conformational selection in a rugged energy landscape. Chem. Rev 110, 4820–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng J, Ma B & Nussinov R (2006). Consensus features in amyloid fibrils: sheet–sheet recognition via a (polar or nonpolar) zipper structure. Phys. Biol 3, P1–P4. [DOI] [PubMed] [Google Scholar]
- 66.Zheng J, Jang H, Ma B, Tsai CJ & Nussinov R (2007). Modeling the Alzheimer Aβ17–42 fibril architecture: tight intermolecular sheet–sheet association and intramolecular hydrated cavities. Biophys. J 93, 3046–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng J, Jang H & Nussinov R (2008). β2- Microglobulin amyloid fragment organization and morphology and its comparison to Aβ suggests that amyloid aggregation pathways are sequence specific. Biochemistry, 47, 2497–2509. [DOI] [PubMed] [Google Scholar]
- 68.Ma B & Nussinov R (2002). Stabilities and conformations of Alzheimer’s β-amyloid peptide oligomers (Aβ16–22, Aβ16–35, and Aβ10–35): sequence effects. Proc. Natl Acad. Sci. USA, 99, 14126–14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin MC, Mirzabekov T & Kagan BL (1997). Channel formation by a neurotoxic prion protein fragment. J. Biol. Chem 272, 44–47. [DOI] [PubMed] [Google Scholar]
- 70.Hirakura Y, Azimov R, Azimova R & Kagan BL (2000). Polyglutamine-induced ion channels: a possible mechanism for the neurotoxicity of Huntington and other CAG repeat diseases. J. Neurosci. Res 60, 490–494. [DOI] [PubMed] [Google Scholar]
- 71.Hirakura Y & Kagan BL (2001). Pore formation by β2-microglobulin: a mechanism for the pathogenesis of dialysis associated amyloidosis. Amyloid, 8, 94–100. [DOI] [PubMed] [Google Scholar]
- 72.Cerf E, Sarroukh R, Tamamizu-Kato S, Breydo L, Derclaye S, Dufrene YF et al. (2009). Antiparallel beta-sheet: a signature structure of the oligomeric amyloid beta-peptide. Biochem. J 421, 415–423. [DOI] [PubMed] [Google Scholar]
- 73.McLaurin J & Chakrabartty A (1997). Characterization of the interactions of Alzheimer beta-amyloid peptides with phospholipid membranes. Eur. J. Biochem 245, 355–363. [DOI] [PubMed] [Google Scholar]
- 74.Shao H, Jao S, Ma K & Zagorski MG (1999). Solution structures of micelle-bound amyloid β-(1–40) and β-(1–42) peptides of Alzheimer’s disease. J. Mol. Biol 285, 755–773. [DOI] [PubMed] [Google Scholar]
- 75.Ege C & Lee KY (2004). Insertion of Alzheimer’s A beta 40 peptide into lipid monolayers. Biophys. J 87, 1732–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smart OS, Goodfellow JM & Wallace BA (1993). The pore dimensions of gramicidin A. Biophys. J 65, 2455–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frishman D & Argos P (1995). Knowledge-based protein secondary structure assignment. Proteins, 23, 566–579. [DOI] [PubMed] [Google Scholar]
- 78.Smart JL, Marrone TJ & McCammon JA (1997). Conformational sampling with Poisson–Boltzmann forces and a stochastic dynamics/Monte Carlo method: application to alanine dipeptide. J. Comput. Chem 18, 1750–1759. [Google Scholar]
- 79.Jang H & Woolf TB (2006). Multiple pathways in conformational transitions of the alanine dipeptide: an application of dynamic importance sampling. J. Comput. Chem 27, 1136–1141. [DOI] [PubMed] [Google Scholar]
- 80.Sipe JD & Cohen AS (2000). Review: history of the amyloid fibril. J. Struct. Biol 130, 88–98. [DOI] [PubMed] [Google Scholar]
- 81.Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M et al. (1998). A transmembrane form of the prion protein in neuro-degenerative disease. Science, 279, 827–834. [DOI] [PubMed] [Google Scholar]
- 82.Soong R, Brender JR, Macdonald PM & Ramamoorthy A (2009). Association of highly compact type II diabetes related islet amyloid polypeptide intermediate species at physiological temperature revealed by diffusion NMR spectroscopy.J. Am. Chem. Soc 131, 7079–7085. [DOI] [PubMed] [Google Scholar]
- 83.Brender JR, Hartman K, Nanga RP, Popovych N, de la Salud Bea R, Vivekanandan S et al. (2009). Role of zinc in human islet amyloid polypeptide aggregation. J. Am. Chem. Soc 132, 8973–8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brender JR, Lee EL, Cavitt MA, Gafni A, Steel DG & Ramamoorthy A (2008). Amyloid fiber formation and membrane disruption are separate processes localized in two distinct regions of IAPP, the type-2-diabetes-related peptide. J. Am. Chem. Soc 130, 6424–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jang H, Ma B, Lal R & Nussinov R (2008). Models of toxic β-sheet channels of protegrin-1 suggest a common subunit organization motif shared with toxic Alzheimer β-amyloid ion channels. Biophys. J 95, 4631–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wong PT, Schauerte JA, Wisser KC, Ding H, Lee EL, Steel DG & Gafni A (2009). Amyloid-beta membrane binding and permeabilization are distinct processes influenced separately by membrane charge and fluidity. J. Mol. Biol 386, 81–96. [DOI] [PubMed] [Google Scholar]
- 87.Chauhan A, Ray I & Chauhan VP (2000). Interaction of amyloid beta-protein with anionic phospholipids: possible involvement of Lys28 and C- terminus aliphatic amino acids. Neurochem. Res 25, 423–429. [DOI] [PubMed] [Google Scholar]
- 88.Zhao H, Tuominen EK & Kinnunen PK (2004). Formation of amyloid fibers triggered by phosphatidylserine-containing membranes. Biochemistry, 43, 10302–10307. [DOI] [PubMed] [Google Scholar]
- 89.Maltseva E, Kerth A, Blume A, Mohwald H & Brezesinski G (2005). Adsorption of amyloid beta (1–40) peptide at phospholipid monolayers. ChemBioChem, 6, 1817–1824. [DOI] [PubMed] [Google Scholar]
- 90.Yoda M, Miura T & Takeuchi H (2008). Nonelectrostatic binding and self-association of amyloid beta-peptide on the surface of tightly packed phosphatidylcholine membranes. Biochem. Biophys. Res. Commun 376, 56–59. [DOI] [PubMed] [Google Scholar]
- 91.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S & Karplus M (1983). CHARMM — a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem 4, 187–217. [Google Scholar]
- 92.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E et al. (2005). Scalable molecular dynamics with NAMD. J. Comput. Chem 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


