Abstract
Background
The extent of glioma resection influences the overall survival (OS) and progression-free survival (PFS). Ferroptosis is a newly recognized type of cell death, which may be associated with low-grade glioma border detection and OS. This study is assessed an optimized ferroptosis gene panel for glioma treatment.
Material/Methods
We obtained 45 reports on ferroptosis-related proteins in PubMed and conducted a statistical test of the patients’ overall survival (OS) in the TCGA GBMLGG and CGGA databases. The statistically significant genes were screened for an optimal panel, followed by GO and KEGG analysis and evaluated its correlation with known prognostic factors of glioma, including IDH1 mutation, methylated MGMT, tumor purity, 1p/19q LOH, and methionine cycle.
Results
Eight genes panel (ALOX5, CISD1, FTL, CD44, FANCD2, NFE2L2, SLC1A5, and GOT1) were highly related to OS (P<0.001) and PFS (P<0.001) of low-grade glioma (LGG) patients, out of which 6 genes (ALOX5, CISD1, CD44, FTL, FANCD2, and SLC1A5) were correlated with IDH1_p.R132H (P<0.001) and 5 genes (ALOX5, CD44, FTL, NFE2L2, SLC1A5) showed a correlation with tumor purity (P<0.001). Five genes (ALOX5, CD44, CISD1, FTL, and SLC1A5) were associated with methylated MGMT (P<0.001), out of which 6 genes (ALOX5, CD44, FANCD2, NFE2L2, SLC1A5, and GOT1) had significantly different expression in healthy brain tissue vs. glioma (P<0.001).
Conclusions
Our panel of 8 ferroptosis genes showed a significant correlation with the diagnostic and prognostic factors of low-grade glioma and can be applied in neuroradiology and surgery.
MeSH Keywords: Astrocytoma, Cell Death, Lipid Peroxidation, Prognosis
Background
Cell death is a physiological mechanism of the body to maintain its normal function by necrosis and programmed cell death (autophagy and apoptosis) [1]. With the discovery of necroptosis [2], ferroptosis [3], pyroptosis [4], and other types of cell death, the relationship between cell death and disease has attracted great attention. Ferroptosis is a new type of programmed non-apoptosis cell death explored by Brent Stockwell of Columbia University in the United States [3]. Ferroptosis manifests cell membrane rupture and blebbing, mitochondrial and morphological changes (e.g., mitochondrial atrophy, reduced mitochondrial cristae, and increased density of mitochondrial membranes in the ultra-microstructure); however, the cell nucleus remains intact and is devoid of chromatin clotting, unlike in apoptosis [3,5]. Although it has been reported that the key to ferroptosis is cell membrane destruction mediated by iron-dependent lipid peroxidation [6], the subcellular location and the role of iron and ferroptosis-related proteins are unclear.
Numerous theories about ferroptosis have been explored, including the synthesis of glutathione in cells or the inhibition of GPX4 (glutathione peroxidase 4) [6], p53-mediated ferroptosis [7], and cysteine glutamate transport receptor (System Xc-) inhibition [8]. Erastin is reported to selectively kill tumor cells containing RASV12 protein, which leads to cell death [9]. Studies on ferroptosis in the nervous system were initially focused on epilepsy [10] and stroke [11]. However, a growing number of correlations between neurological diseases and ferroptosis have been found, and the patterns of ferroptosis in such diseases appear to vary widely. In Alzheimer’s disease, the model of ferroptosis is mainly attributed to the dysfunction of APP (amyloid protein precursor) in regulating ferritin levels (Ferroportin) and abnormalities of glutamate transport receptor (System Xc-) pathways[12]. In Parkinson’s disease, ferroptosis is often affected by the abnormal expression of transferrin [12]. Dixon et al. reported that the cysteine glutamate transport receptor (System Xc-) pathway is involved in glutamate-induced deaths in the rat organotypic hippocampal slice culture model (both astrocyte and microglia were included) and ferroptosis [3]. A subsequent study found that the ferroptosis of oligodendrocyte also occurs in periventricular leukomalacia (PVL) [13]. Recently, Ishraq Alim et al. reported that a single dose of selenium could prevent GPX4-dependent ferroptosis in neurons [14]. These results show the presence of cell specificity and diversity in ferroptosis in the brain.
Gliomas, especially glioblastoma, are the most common malignant tumors in the brain. The incidence rate of glioma is about 6/100 000 and is steadily increasing. Cell death is crucial in the development of glioma, and can influence pathological classification and sensitivity to radio-chemotherapy [15,16]. Furthermore, there is an apparent metabolic reprogramming present in gliomas, especially in glioblastoma, which is theoretically associated with ferroptosis in 2 ways [17]. First, the alterations of cellular respiration, especially the efficiency of the tricarboxylic acid cycle, leads to the specific glutamine metabolism in tumor cells after the occurrence of the Warburg effect. Therefore, there could be a unique balance between glutamine and glutamate in the glioma, which is reported to be associated with ferroptosis [18]. Second, lipid metabolism reprogramming is affected by acetyl-coenzyme A [19], which is partly produced from glutamine metabolism, as well as by the efficiency of isocitric acid dehydrogenase (IDH) in the tricarboxylic acid cycle [20]. IDH mutation is an essential event in glioma, especially in low-grade glioma (LGG). The OS of patients with IDH wild-type is much shorter than in those with IDH mutation. IDH mutation results in abnormal production of 2-hydroxyglutarate (2-HG), which significantly inhibits the function of α-KG, which is a natural product of wild-type IDH and affects lipid metabolism [21] and can thus impart unique characteristics of ferroptosis in glioma. With the development of molecular pathology methods, several specific molecular alterations (e.g., 1p/19q LOH, MGMT, and TERT) were found to influence the OS in glioma patients. Some of them have recently been used in clinical practice, such as tumor purity and the proportion of cancer cells in the tumor [22], which identified the tumor border and microenvironment. More importantly, low purity in glioma can exacerbate the poor prognosis and treatment resistance [23]. However, the relationship between ferroptosis and these prognostic/diagnostic factors has not been previously explored, and there have been few studies on the molecular relationship between ferroptosis and glioma. In addition, there has been little research on the association of ferroptosis-related proteins in glioma cells with prognosis or other clinical transformation. Hence, the present study explored the correlation between ferroptosis and glioma prognosis through bioinformatics analysis, which revealed the potential mechanism underlying glioma ferroptosis reprogramming and shows the role of ferroptosis in the process of glioma diagnosis.
Material and Methods
Functional and pathway enrichment analysis
Gene ontology (GO) annotation was integrated into the total DEGs PPI sub-networks for enriched “molecular function” (MF), “biological pathways” (BP), and “cellular component” (CC) of proteins. KEGG is an online database that provides information on high-level functions and utilities of biological systems. We performed GO and KEGG pathway analyses with the affinity propagation algorithm by Webgestalt (http://www.webgestalt.org/), which provides annotated information on systematic and comprehensive biological functions with high-throughput gene expression.
Survival analysis and expression analyses
Two glioma datasets – TCGA and CGGA – were used in this study. The clinical information is shown in Table 1. Single-gene OS and PFS of TCGA LGG, TCGA GBM, and TCGA GBMLGG analyses were made using Linkedomics (http://www.linkedomics.org/login.php) [24]. The optimized gene panel OS and PFS analyses were made via GEPIA2 (http://gepia2.cancer-pku.cn/#index). Single-gene OS of CGGA analyses were performed via CGGA (http://cgga.org.cn/index.jsp). The expression analyses were performed via GEPIA2 (http://gepia2.cancer-pku.cn/#index). All these statistical analyses were set at a significant level of P<0.001.
Table 1.
Clinical information from TCGA GBMLGG and CGGA datasets. All the cases had complete data on OS and age.
| TCGA | CGGA | |||
|---|---|---|---|---|
| GBM | LGG | GBM | LGG | |
| Male | 363 | 284 | 138 | 110 |
| Female | 230 | 230 | 99 | 70 |
| Average age (yr) | 58 | 43 | 49 | 39 |
| Median OS (d) | 369 | 678 | 378 | 1326 |
Statistical analysis of prognostic and therapeutic targets
The correlations among all the genes in the optimized panel, methylated MGMT, tumor purity, and methionine cycle were made using Linkedomics (http://www.linkedomics.org/login.php). The correlation analyses between 1p/19q and the mRNA expression of OS-associated ferroptosis-related proteins were made via TCGAportal (http://tumorsurvival.org/index.html). The statistical analyses were based on a non-parametric test and P<0.001 was considered significantly different.
Results
Nearly 40% of ferroptosis-related proteins were associated with the prognosis of glioma
We obtained 45 reports on ferroptosis-related proteins in PubMed and conducted a statistical test of the patients’ overall survival (OS) in the TCGA GBMLGG and CGGA databases. Among them, there were significant statistical OS differences for AKR1C1, ALOX12B, ALOXE3, CD44, DPP4, FANCD2, GOT1, HSBP1, CRYAB, PTGS2, SAT1, SLC1A5, FDFT1, and TFRC in TCGA glioma (all WHO grades) and CGGA (all WHO grades). There were significant OS differences of ALOX15B in TCGA LGG and CGGA Grade II. ALOX5, CISD1, and FTL showed significant statistical OS differences in TCGA gliomas (all WHO grade), CGGA (all WHO grade), TCGA LGG, and CGGA Grade II at the same time. None of the proteins associated with ferroptosis showed OS differences in TCGA GBM and CGGA GBM (Table 2). These proteins were mainly associated with lipid metabolism (ALOX5, ALOX12B, ALOX15B, ALOXE3, FDFT1, and PTGS2), amino acid metabolism (GOT1, SLC1A5, and SAT1), inflammatory response and DNA damage repair (HSBP1, CRYAB, FANCD2, and CD44), and iron transportation (TFRC, FTL, and CISD1). These results were supported by GO and KEGG analyses with the affinity propagation algorithm, which showed that ferroptosis was affected by lipid metabolism, and it is reported to be related to carboxylic acid metabolism, which suggests that ferroptosis in the central nervous system is unique (Table 2, Figure 1).
Table 2.
Eighteen ferroptosis-related proteins associated with the overall survival in one or several grades of glioma, out of which 3 genes (ALOX5, CISD1, FTL) were tightly associated with overall survival in low-grade glioma (LGG). The statistical analysis was based on a non-parametric test.
| Gene symbol | Protein name | Subcellular | TCGA GBMLGG | CGGA ALL Grade | TCGA GBM | CGGA GBM | TCGA LGG | CGGA Grade II |
|---|---|---|---|---|---|---|---|---|
| ACSF2 | Medium-chain acyl-CoA ligase ACSF2 | Mitochondrion | 1 | 0 | 0 | 0 | 1 | 0 |
| ACSL4 | Long-chain-fatty-acid-CoA ligase 4 | Plasma membrane, Endoplasmic reticulum, Peroxisome, Mitochondrion | 1 | 0 | 0 | 0 | 0 | 0 |
| AKR1C1 | Aldo-keto reductase family 1 member C1 | Cytoplasm | 1 | 1 | 0 | 0 | 1 | 0 |
| ALOX12 | Arachidonate 12-lipoxygenase, 12S-type | Cytosol | 0 | 0 | 0 | 0 | 0 | 0 |
| ALOX12B | Arachidonate 12-lipoxygenase, 12R-type | Cytoplasm | 1 | 1 | 0 | 0 | 0 | 0 |
| ALOX15 | Arachidonate 15-lipoxygenase | Plasma membrane, Cytosol | 0 | 0 | 0 | 0 | 0 | 0 |
| ALOX15B | Arachidonate 15-lipoxygenase B | Nucleus, Plasma membrane, Cytoskeleton, Cytosol | 1 | 0 | 0 | 0 | 1 | 1 |
| ALOX5 | Arachidonate 5-lipoxygenase | Nucleus | 1 | 1 | 0 | 0 | 1 | 1 |
| ALOXE3 | Hydroperoxide isomerase ALOXE3 | Cytoplasm | 1 | 1 | 0 | 0 | 0 | 0 |
| ATP5G3 | ATP synthase F(0) complex subunit C3 | Mitochondrion | 1 | 0 | 0 | 0 | 0 | 0 |
| CARS | Cysteine--tRNA ligase | Cytoplasm | 0 | 0 | 0 | 0 | 0 | 0 |
| CBS | Cystathionine beta-synthase | Nucleus, Cytoplasm | 0 | 1 | 0 | 0 | 0 | 0 |
| CD44 | CD44 antigen | Cell membrane | 1 | 1 | 0 | 0 | 1 | 0 |
| CHAC1 | Glutathione-specific gamma-glutamylcyclotransferase 1 | Golgi apparatus, Cytosol | 1 | 0 | 0 | 0 | 0 | 0 |
| CISD1 | CDGSH iron-sulfur domain-containing protein 1 | Mitochondrion | 1 | 1 | 0 | 0 | 1 | 1 |
| CS | Citrate synthase | Mitochondrion | 0 | NA | 0 | 0 | 0 | NA |
| DPP4 | Dipeptidyl peptidase 4 | Extracellular region or secreted, Plasma membrane | 1 | 1 | 0 | 0 | 1 | 0 |
| EMC2 | ER membrane protein complex subunit 2 | Nucleus | 1 | 0 | 0 | 0 | 0 | 0 |
| FANCD2 | Fanconi anemia group D2 protein | Nucleus | 1 | 1 | 0 | 0 | 1 | 0 |
| FPN1/SLC40A1 | Solute carrier family 40 member 1 | Plasma membrane | 1 | 0 | 0 | 0 | 0 | 0 |
| FTH1 | Ferritin heavy chain | Cytosol, Extracellular region or secreted, Lysosome, Nucleus | 1 | 0 | 0 | 0 | 1 | 0 |
| FTL | Ferritin light chain | Cytosol, Extracellular region or secreted, Lysosome, Nucleus | 1 | 1 | 0 | 0 | 1 | 1 |
| GCLC | Glutamate--cysteine ligase catalytic subunit | Cytosol | 1 | 0 | 0 | 0 | 1 | 0 |
| GLS2 | Glutaminase liver isoform | Mitochondrion | 0 | 1 | 0 | 0 | 0 | 0 |
| GOT1 | Aspartate aminotransferase | Cytoplasm | 1 | 1 | 0 | 0 | 0 | 1 |
| GPX4 | Phospholipid hydroperoxide glutathione peroxidase | Mitochondrion | 1 | 0 | 0 | 0 | 0 | 0 |
| GSS | Glutathione synthetase | Cytosol, Extracellular region or secreted | 1 | 0 | 0 | 0 | 1 | 0 |
| HMGCR | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | Endoplasmic reticulum | 0 | 0 | 0 | 0 | 0 | 0 |
| HSPB1 | Heat shock protein beta-1 | Nucleus, Cytoskeleton | 1 | 1 | 0 | 0 | 1 | 0 |
| HSPB5/CRYAB | Alpha-crystallin B chain | Nucleus | 1 | 1 | 0 | 0 | 1 | 0 |
| IREB2 | Iron-responsive element-binding protein 2 | Cytoplasm | 0 | 0 | 0 | 0 | 0 | 0 |
| LPCAT3 | Lysophospholipid acyltransferase 5 | Endoplasmic reticulum | 1 | 0 | 0 | 0 | 1 | 0 |
| MT1G | Metallothionein-1G | Nucleus | 0 | 0 | 0 | 0 | 0 | 0 |
| NCOA4 | Nuclear receptor coactivator 4 | Lysosome | 1 | 0 | 0 | 0 | 1 | 0 |
| NFE2L2 | Nuclear factor erythroid 2-related factor 2 | Nucleus, Cytosol | 1 | 0 | 0 | 0 | 0 | 0 |
| PTGS2 | Prostaglandin G/H synthase 2 | Endoplasmic reticulum | 1 | 1 | 0 | 0 | 1 | 0 |
| RPL8 | 60S ribosomal protein L8 | Cytoplasm | 0 | 0 | 0 | 0 | 0 | 0 |
| SAT1 | Diamine acetyltransferase 1 | Cytoplasm | 1 | 1 | 0 | 0 | 1 | 0 |
| SLC11A2 | Natural resistance-associated macrophage protein 2 | Plasma membrane, Endosome, Mitochondrion, Plasma membrane | 1 | 0 | 0 | 0 | 0 | 0 |
| SLC1A5 | Neutral amino acid transporter B(0) | Cell membrane | 1 | 1 | 0 | 0 | 1 | 0 |
| SLC3A2 | 4F2 cell-surface antigen heavy chain | Lysosome, Plasma membrane | 1 | 0 | 0 | 0 | 1 | 0 |
| SLC7A11 | Cystine/glutamate transporter | Membrane | 1 | 0 | 0 | 0 | 0 | 0 |
| SQS/FDFT1 | Squalene synthase | Endoplasmic reticulum | 1 | 1 | 0 | 0 | 0 | 0 |
| TFRC | Transferrin receptor protein 1 | Plasma membrane | 1 | 1 | 0 | 0 | 1 | 0 |
1 represents P<0.001 while 0 represents P>0.001.
Figure 1.
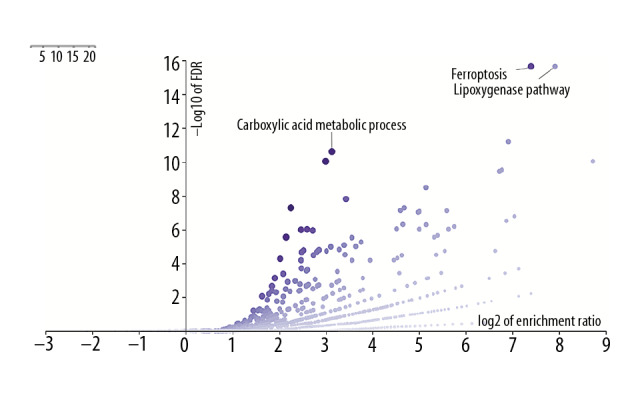
The affinity propagation algorithm of GO and KEGG indicated that the lipoxygenase pathway and carboxylic acid metabolic process (Exemplar) were the critical pathways of 45 proteins.
OS-associated ferroptosis-related proteins could be a potential prognostic panel in low-grade glioma
We made several statistical tests among the OS-associated ferroptosis-related proteins and some known prognostic factors, including IDH1 p.R132H mutation, 1p/19q co-deletion, tumor purity, and histopathology subtype. We found that 16 candidate proteins were highly associated with the IDH1 mutation (p<0.001). Furthermore, the statistical model indicated that the trend of protein expression levels in the long OS group was the same as in the IDH1 mutant group. Although it was almost impossible to discover the casualty based on the data we had, it was still important to show that the ferroptosis can affect IDH1 mutation in glioma, especially low-grade glioma, and it probably plays a role in the tumor immune microenvironment. The 15 candidate proteins were significantly correlated with tumor purity (Table 3).
Table 3.
Five (ALOX5, CD44, FANCD2, NFE2L2, and SLC1A5) of 18 proteins showed a positive tendency with the pathological grade (p<0.001), and GOT1 showed a negative tendency with the pathological grade (p<0.001). Nine out of 18 proteins were highly expressed in astrocytoma (p<0.001), 3 out of 18 proteins were highly expressed in oligodendroglioma (p<0.001), and 6 out of 18 proteins were not correlated to the pathological types. Thirteen out of 18 proteins were associated with tumor purity (p<0.001). Expression conditions of the more extended OS group in 16 out of 18 proteins were the same as IDH1 and pR132H mutation group (p<0.001). Ten out of 18 proteins were significantly associated with the 1p/19q co-deletion (p<0.001).
| LGG vs. N | GBM vs. N | Histology | Tumor purity | IDH1_p.R132H (TCGA LGG) | Long OS | 1p/19q LOH | |
|---|---|---|---|---|---|---|---|
| AKR1C1 | NA | − | * | * | ***(wt<mut) | High | * |
| ALOX12B | NA | NA | >***(A<OA<O) | >*** | ***(wt<mut) | High(GBMLGG) | ***(CODEL>NON–CODEL) |
| ALOX15B | NA | NA | * | >*** | ***(wt>mut) | Low | * |
| ALOX5 | + | ++ | >***(A>OA>O) | >*** | ***(wt>mut) | Low | ***(CODEL<NON–CODEL) |
| CD44 | + | ++ | >***(A>OA>O) | >*** | ***(wt>mut) | Low | * |
| CISD1 | NA | NA | >***(A<OA<O) | * | ***(wt<mut) | High | ***(CODEL>NON–CODEL) |
| DPP4 | NA | NA | * | >*** | ***(wt>mut) | Low | * |
| FANCD2 | + | ++ | ***(A>OA=O) | * | ***(wt>mut) | Low | * |
| FTL | NA | ++ | >***(A>OA>O) | >*** | ***(wt>mut) | Low | ***(CODEL<NON–CODEL) |
| GOT1 | − | −− | >***(A<OA<O) | * | * | High(GBMLGG) | ***(CODEL>NON–CODEL) |
| HSPB1 | NA | + | ***(A>OA>O) | >*** | ***(wt>mut) | Low | t* |
| HSPB5/CRYAB | ++ | + | * | * | ***(wt<mut) | High | **(CODEL<NON–CODEL) |
| NFE2L2 | + | ++ | >***(A>OA>O) | >*** | * | Low | **(CODEL<NON–CODEL) |
| PTGS2 | NA | NA | * | >*** | ***(wt>mut) | Low(GBMLGG) | * |
| SAT1 | NA | + | >***(A>OA>O) | >*** | ***(wt>mut) | Low | ***(CODEL<NON–CODEL) |
| SLC1A5 | + | ++ | >***(A>OA>O) | >*** | ***(wt>mut) | Low | ***(CODEL<NON–CODEL) |
| SQS/FDFT1 | NA | NA | >***(A>OA>O) | >*** | ***(wt<mut) | High(GBMLGG) | ***(CODEL>NON–CODEL) |
| TFRC | NA | + | * | >*** | ***(wt>mut) | Low | * |
N – normal brain tissue from GTEx; NA – no alternation; +/++ – increase, −/−− – decrease; A – astrocytoma; OA: oligoastrocytoma; O – oligodendroglioma; wt – wild-type; mut – mutation; wt>/<mut – expression in wt was higher/lower than mut 1p/19q; LOH – loss of heterozygosity of 1p/19q; CODEL – co-deletion; NON-CODEL – non-co-deletion;
p>0.1;
p<0.001.
Basic situation of the optimized panel
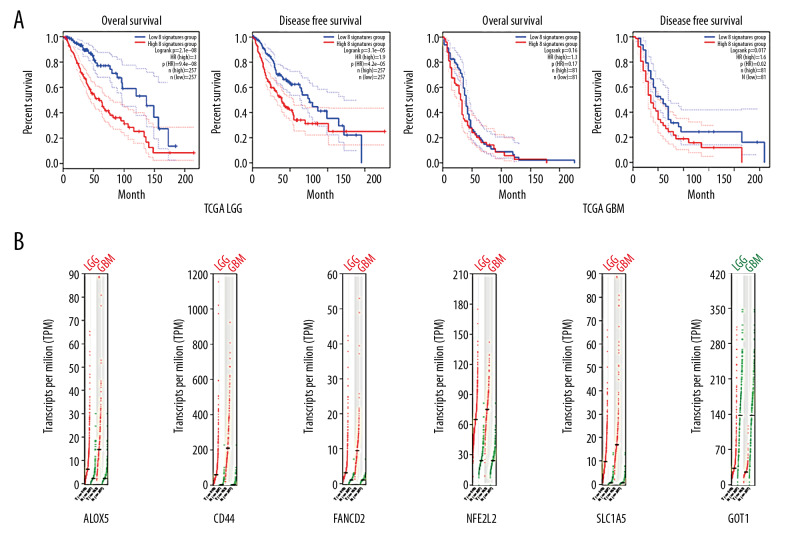
We then optimized the 8-gene panel containing 3 main OS-associated ferroptosis-related proteins (ALOX5, CISD1, and FTL) and 6 main grade-associated ferroptosis-related proteins (ALOX5, CD44, FANCD2, NFE2L2, SLC1A5, and GOT1). This panel was intended to predict the prognosis and detect the border of the tumor at the same time. The survival analysis suggested that this panel was useful in evaluating the more extended OS group and distinguishing the condition of progression-free survival in low-grade glioma. However, this panel was not valid in glioblastoma. As for the tumor border detection, 6 out of 8 genes were found to be significantly progressive and were increased or reduced with malignant states. These 6 genes also showed differences in expression between healthy brain cells and glioma, especially CD44 and SLC1A5. CD44 and SLC1A5 are only located in the cell membrane and are not expressed in healthy brain tissue, but are upregulated in both LGG and GBM. This difference suggests that these 2 proteins play an important role in reprogramming of ferroptosis.
The optimized panel is highly associated with several prognostic and therapeutic targets of LGG
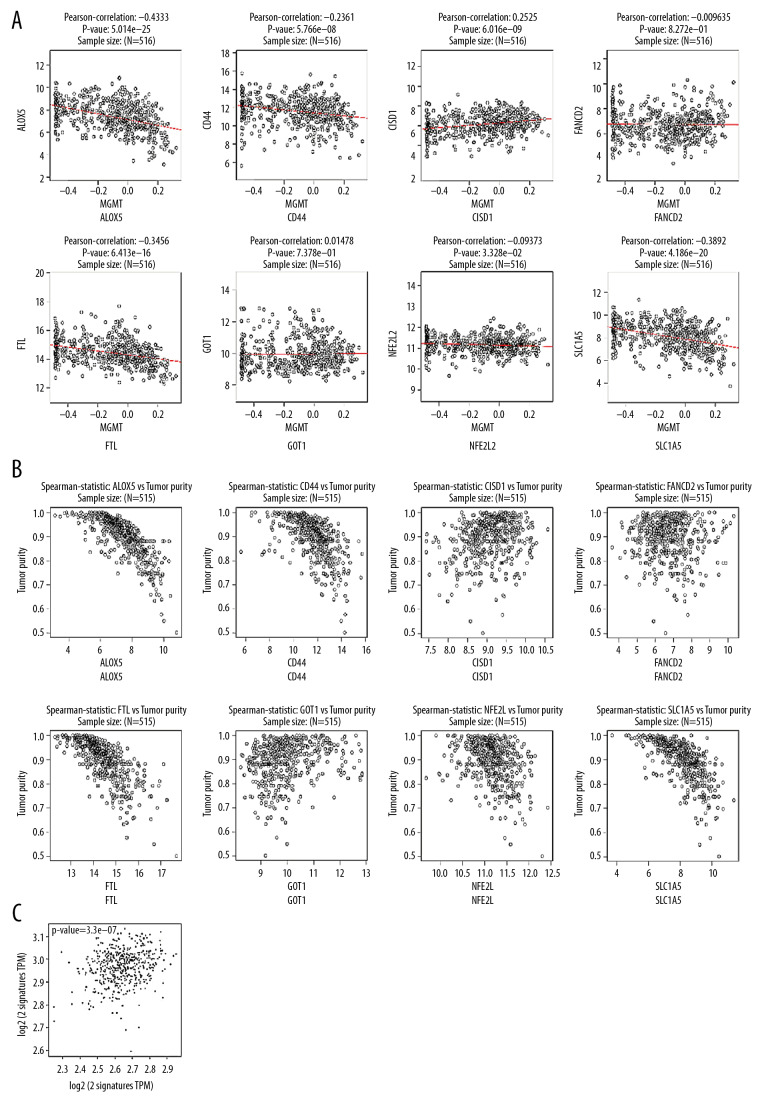
Five out of 8 genes in the optimized panel – ALOX5, CD44, CISD1, FTL, and SLC1A5 – were associated with methylated MGMT. The more extended OS group in each of these 5 genes was associated with a high methylation state of MGMT (Figure 2A). In addition, 5 out of 8 genes (ALOX5, CD44, FTL, NFE2L2, and SLC1A5) showed a correlation with tumor purity (P<0.001). Low tumor purity suggested a high expression of these 5 genes. Moreover, there was a pronounced relationship between the 8-gene panel and 2 key enzymes of the methionine cycle – MAT2A and SAHH (P<0.001).
Figure 2.
(A) 5 of 8 genes in the ptimazed panel including ALOX5, CD44, CISD1, FTL, SLC1A5 were significantly associated to the methylation state of MGMT. CISD1 shows a positive correlation (P<0.001) while the other four genes negative (P<0.001). (B) ALOX5, CD44, FTL, NFE2L2, SLCA5 were negative correlated to the tumor purity (P<0.001). (C) The optimazed panel was statistically associated to the 2 key enzymes of methionine cycle, MAT2A and SAHH (P<0.001).
Optimized panel is a potential target of tumor border localization and detection
We performed statistical analyses of the healthy brain tissues from the GTEx, TCGA LGG, and GBM datasets. ALOX5, CD44, FANCD2, NFE2L2, and SLC1A5 showed an increase of expression from healthy brain tissue to glioblastoma. GOT1 showed the reverse trend with the malignant condition of the tissue. ALOX5, CD44, FANCD2, and SLC1A5 were on a specific low level in the healthy brain tissue. The conditions in low-grade glioma and GBM were at least 3 times higher than usual. SLC1A5 showed a 10-fold increase in glioma. GOT1 showed a nearly 75% decrease in glioma. Combined use of these 6 genes in molecular pathological tests could be useful for identifying the tumor border (Figure 3). Furthermore, 5 genes (all except GOT1) showed a correlation with tumor purity, which is defined as the difference between the healthy samples and cancer samples from the same tissue type (Figure 2B). Such a panel can be utilized to localize the tumor border at both the gene and cell levels.
Figure 3.
(A) The optimized panel was effective in predicting the OS and PFS of low-grade glioma (P<0.001). It was useless in predicting the OS of glioblastoma but might be potentially used for predicting the PFS of glioblastoma (P=0.017). (B): ALOX5, CD44, FANCD2, NFE2L2, and SLC1A5 were increased in glioma, especially in GBM (P<0.001), while GOT1 was decreased in glioma (P<0.001).
Discussion
Since ferroptosis was discovered 7 years ago, this type of cell death has attracted much attention, especially in the relationship with cancer. Most of the studies focused on malignant tumors like pancreatic ductal adenocarcinoma [25] and breast cancer [26]. Because most of these malignant tumor cells were apoptosis-resistant, radiotherapy-derived ferroptosis could be one of the potential pathways to treat these tumors. Erastin [8], sorafenib [27], siramesine, lapatinib [26], and many small molecules are reported to induce ferroptosis. Temozolomide, the most widely used chemical drug in glioma therapy, is reported to kill tumor cells and glioblastoma stem cells via several pathways, including ferroptosis [28,29]. Based on the evidence above, it is reasonable to assume that Temozolomide can prolong PFS or even OS with use of a ferroptosis-related drug in glioma treatment. Surgery with chemoradiation is the most useful protocol in low-grade glioma. Accurate tumor border localization and detection is the key to achieve “maximal safe resection.” In recent years, there has been much debate about how to identify the tumor border. Surgeons prefer to set the border mainly by T1W1 or a multimodal imaging pattern. Radiotherapists set the gross tumor volume (GTV) by T2WI/FLAIR. Regardless of the method used, some of the gliomas reoccurred, which meant that some of the tumor cells could not be identified in these ways. We suggest that the ferroptosis panel could be a new way to identify the glioma border. Our results suggest that the ferroptosis mechanism might be different between the normal glia cells and the glioma cells. When we assessed the optimized panel, 2 genes (ALOX5, NFE2L2) were associated with the lipid oxidation; 2 genes (FTL, CISD1) were associated with iron combination, and 1 gene (SLC1A5) was associated with glutamine transportation. This evidence indicated that the tumor cells were more sensitive to ferroptosis than the healthy tissues. Therefore, lipid peroxidation can be visualized via the STIR of MRI, and iron stimulation can be visualized by QSM (quantitative susceptibility mapping) of MRI [30]. By combining STIR and QSM, we could localize and detect the glioma border more accurately than before. Moreover, the short OS group had high expression in the optimized panel; therefore, the results of STIR and QSM could potentially show the presence of a malignant tumor. Furthermore, we observed that there was a significant association between ferroptosis and the methionine cycle. Thus, ferroptosis is visualized via 11C-MET PET/CT with a specific algorithm. Moreover, our results indicated that the OS group consisting of low-grade glioma could easily be recognized in 11C-MET PET/CT because of the positive correlation between the optimized panel and the methionine cycle. Most of the genes in the optimized panel showed such a tendency, which indicated that the highly expressed groups are associated with the low methylated MGMT state, IDH wild-type, and the low tumor purity [23], which predicts a poor prognosis. Instead, there could be ferroptosis reprograming or other mechanisms that lead to high expression of ferroptosis-related genes with a poor prognosis. According to traditional thinking, the ferroptosis-sensitive tumors were potentially cured by chemical drugs with ferroptosis inducer. However, our results suggest that ferroptosis is a complicated prognostic factor and excessive ferroptosis treatment could have varied results. Nevertheless, there could be some differences in the mechanisms of ferroptosis among different molecular subtypes of glioma, such as p53 mutation [31]. Therefore, we should not judge the result of ferroptosis treatment based solely on the results of chemical treatment.
Conclusions
Ferroptosis in glioma is an elaborate pathophysiological process. Our results indicated that ferroptosis is unique among the gliomas, healthy tumor cells, and some reported tumors. These characteristics can be applied to tumor border localization and detection. Knowledge of the relationship between ferroptosis and overall survival of glioma can aid further research in glioma, as protocols for chemical therapy with a ferroptosis inducer are largely undefined.
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Kierszenbaum AL. Histology and cell biology: An introduction to pathology. 2nd ed. Philadelphia, PA: Mosby Elsevier; 2007. [Google Scholar]
- 2.Vanden Berghe T, Linkermann A, Jouan-Lanhouet ST, et al. Regulated necrosis: the expanding network of non-apoptotic cell death pathways. Nat Rev Mol Cell Biol. 2014;15(2):135–47. doi: 10.1038/nrm3737. [DOI] [PubMed] [Google Scholar]
- 3.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–72. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cookson BT, Brennan MA. Pro-inflammatory programmed cell death. Trends Microbiol. 2001;9(3):113–14. doi: 10.1016/s0966-842x(00)01936-3. [DOI] [PubMed] [Google Scholar]
- 5.Stockwell BR, Friedmann Angeli JP, Bayir H, et al. Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–85. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao JY, Dixon SJ. Mechanisms of ferroptosis. Cell Mol Life Sci. 2016;73(11–12):2195–209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang L, Kon N, Li T, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520(7545):57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15(3):234–45. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolma S, Lessnick SL, Hahn WC, Stockwell BR, et al. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3(3):285–96. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 10.Ye Q, Zeng C, Dong L, et al. Inhibition of ferroptosis processes ameliorates cognitive impairment in kainic acid-induced temporal lobe epilepsy in rats. Am J Transl Res. 2019;11(2):875–84. [PMC free article] [PubMed] [Google Scholar]
- 11.Zille M, Karuppagounder SS, Chen Y, et al. Neuronal death after hemorrhagic stroke in vitro and in vivo shares features of ferroptosis and necroptosis. Stroke. 2017;48(4):1033–43. doi: 10.1161/STROKEAHA.116.015609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li LB, Chai R, Zhang S, et al. Iron exposure and the cellular mechanisms linked to neuron degeneration in adult mice. Cell. 2019;8(2) doi: 10.3390/cells8020198. pii: E198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skouta R, Dixon SJ, Wang J, et al. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J Am Chem Soc. 2014;136(12):4551–56. doi: 10.1021/ja411006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alim I, Caulfield JT, Chen Y, et al. Selenium drives a transcriptional adaptive program to block ferroptosis and treat stroke. Cell. 2019;177(5):1262–79.e25. doi: 10.1016/j.cell.2019.03.032. [DOI] [PubMed] [Google Scholar]
- 15.Wechsler-Reya R, Scott MP. The developmental biology of brain tumors. Annu Rev Neurosci. 2001;24:385–428. doi: 10.1146/annurev.neuro.24.1.385. [DOI] [PubMed] [Google Scholar]
- 16.Schiffer D, Annovazzi L, Casalone C, et al. Glioblastoma: Microenvironment and niche concept. Cancers (Basel) 2018;11(1) doi: 10.3390/cancers11010005. pii: E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agnihotri S, Zadeh G. Metabolic reprogramming in glioblastoma: The influence of cancer metabolism on epigenetics and unanswered questions. Neuro Oncol. 2016;18(2):160–72. doi: 10.1093/neuonc/nov125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garnier D, Renoult O, Alves-Guerra MC, et al. Glioblastoma stem-like cells, metabolic strategy to kill a challenging target. Front Oncol. 2019;9:118. doi: 10.3389/fonc.2019.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams NC, O’Neill LAJ. A Role for the Krebs cycle intermediate citrate in metabolic reprogramming in innate immunity and inflammation. Front Immunol. 2018;9:141. doi: 10.3389/fimmu.2018.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L, Wang Z, Hu C, et al. Integrated metabolomics and lipidomics analyses reveal metabolic reprogramming in human glioma with IDH1 mutation. J Proteome Res. 2019;18(3):960–69. doi: 10.1021/acs.jproteome.8b00663. [DOI] [PubMed] [Google Scholar]
- 21.Turkalp Z, Karamchandani J, Das S, et al. IDH mutation in glioma: new insights and promises for the future. JAMA Neurol. 2014;71(10):1319–25. doi: 10.1001/jamaneurol.2014.1205. [DOI] [PubMed] [Google Scholar]
- 22.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C, Cheng W, Ren X, et al. Tumor purity as an underlying key factor in glioma. Clin Cancer Res. 2017;23(20):6279–91. doi: 10.1158/1078-0432.CCR-16-2598. [DOI] [PubMed] [Google Scholar]
- 24.Vasaikar SV, Straub P, Wang J, Zhang B, et al. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–63. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eling N, Reuter L, Hazin J, et al. Identification of artesunate as a specific activator of ferroptosis in pancreatic cancer cells. Oncoscience. 2015;2(5):517–32. doi: 10.18632/oncoscience.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma S, Henson ES, Chen Y, Gibson SB. Ferroptosis is induced following siramesine and lapatinib treatment of breast cancer cells. Cell Death Dis. 2016;7:e2307. doi: 10.1038/cddis.2016.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lachaier E, Louandre C, Godin C, et al. Sorafenib induces ferroptosis in human cancer cell lines originating from different solid tumors. Anticancer Res. 2014;34(11):6417–22. [PubMed] [Google Scholar]
- 28.Sehm T, Rauh M, Wiendieck K, et al. Temozolomide toxicity operates in a xCT/SLC7a11 dependent manner and is fostered by ferroptosis. Oncotarget. 2016;7(46):74630–47. doi: 10.18632/oncotarget.11858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buccarelli M, Marconi M, Pacioni S, et al. Inhibition of autophagy increases susceptibility of glioblastoma stem cells to temozolomide by igniting ferroptosis. Cell Death Dis. 2018;9(8):841. doi: 10.1038/s41419-018-0864-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schweser F, Deistung A, Lehr BW, Reichenbach JR, et al. Quantitative imaging of intrinsic magnetic tissue properties using MRI signal phase: An approach to in vivo brain iron metabolism? Neuroimage. 2011;54(4):2789–807. doi: 10.1016/j.neuroimage.2010.10.070. [DOI] [PubMed] [Google Scholar]
- 31.Lin CH, Lin PP, Lin CY, et al. Decreased mRNA expression for the 2 subunits of system xc(−), SLC3A2 and SLC7A11, in WBC in patients with schizophrenia: Evidence in support of the hypo-glutamatergic hypothesis of schizophrenia. J Psychiatr Res. 2016;72:58–63. doi: 10.1016/j.jpsychires.2015.10.007. [DOI] [PubMed] [Google Scholar]