Abstract
Aurora kinases are a family of serine/threonine kinases vital for cell division. Because of the overexpression of Aurora kinases in a broad range of cancers and their important roles in mitosis, inhibitors targeting Aurora kinases have attracted attention in cancer therapy. VX-680 is an effective pan-Aurora kinase inhibitor; however, its clinical efficacy was not satisfying. In this study, we performed CRISPR/Cas9 screens to identify genes whose depletion shows synthetic lethality with VX-680. The top hit from these screens was GSG2 (also known as Haspin), a serine/threonine kinase that phosphorylates histone H3 at Thr-3 during mitosis. Moreover, both Haspin knockout and Haspin inhibitor-treated HCT116 cells were hypersensitive to VX-680. Furthermore, we showed that the synthetic lethal interaction between Haspin depletion and VX-680 was mediated by the inhibition of Haspin with Aurora kinase B (AURKB), but not with Aurora kinase A (AURKA). Strikingly, combined inhibition of Haspin and AURKB had a better efficacy than single-agent treatment in both head and neck squamous cell carcinoma and non-small cell lung cancer. Taken together, our findings have uncovered a synthetic lethal interaction between AURKB and Haspin, which provides a strong rationale for this combination therapy for cancer patients.
Keywords: CRISPR screen, Aurora kinase, AURKB, Haspin, Combination therapy
INTRODUCTION
Aurora kinases are a family of serine/threonine kinases comprising Aurora kinase A (AURKA), Aurora kinase B (AURKB), and Aurora kinase C (AURKC). AURKA, AURKB, and AURKC share a highly conserved kinase domain but have quite different subcellular localizations and functions during mitosis (1–3). AURKA mainly localizes at centrosomes and spindle poles and plays a vital role in centrosome maturation and separation as well as bipolar spindle assembly. AURKA deficiency results in abnormal chromosome segregation, mitotic spindles, and eventually aneuploid cells. AURKB is a component of the chromosome passenger complex (CPC), which localizes to chromosomes arms and is highly enriched at the inner centromere from prophase till metaphase. AURKB is essential for the condensation, attachment to kinetochores, and alignment of chromosomes during mitosis. In mammals, the AURKC gene has a high similarity with AURKB, but AURKC expression is mainly detected in meiotically active germ cells. The function of AURKC is less clear than those of AURKA and AURKB (4).
AURKA and AURKB are overexpressed in a number of human cancer types, and their important roles in cell division have led to the development of small molecule Aurora kinase inhibitors for clinical cancer treatment (3,5–8). AURKA is highly overexpressed in primary colorectal carcinoma, glioma, and ovarian, pancreatic, and breast cancers (9–13). Meanwhile, AURKB is overexpressed in glioma, seminoma, colon cancer, thyroid carcinoma, head and neck squamous cell carcinoma (HNSCC), and non-small cell lung cancer (NSCLC) (14–18). Many inhibitors targeting the Aurora kinase family have been assessed in clinical trials, and some exhibited favorable effects on hematologic malignancies. Alisertib and barasertib are potent and specific inhibitors of AURKA and AURKB, respectively that are currently undergoing clinical testing (3,19–22). VX-680 (tozasertib) inhibits all the Aurora kinases, but it is 100 times more selective for AURKA than AURKB or AURKC. Its antitumor efficacy has been confirmed in a human HL-60 xenograft model (7,23). VX-680 has also been tested in phase 2 clinical trials, but its efficacy in patients was not satisfactory (23–25).
VX-680 has potential as a treatment for many types of cancers. However, its clinical trials in colorectal cancer, NSCLC, and advanced solid tumors have been terminated (7). Revealing the genes and/or pathways that exhibit synthetic lethal interaction with VX-680 could potentially improve its clinical efficacy.
With the development of genome-wide clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 screening and analytic tools for their assessment, pooled genome-wide CRISPR/Cas9 screening has been recognized as a powerful and hypersensitive approach to uncover genetic interactions in human cells (26,27). CRISPR/Cas9 has been applied to decipher synthetic lethal relationships with anticancer drugs (28–30), with the hope of uncovering genetic vulnerabilities and/or new combination therapies that can direct future clinical trials. Therefore, in this study we performed single guide RNA (sgRNA) screening of VX-680 in 293A cells and found synthetic lethality relationship between Haspin depletion and VX-680.
Haspin (haploid germ cell-specific nuclear protein kinase, also known as GSG2), an atypical serine/threonine kinase, is the only known kinase that directly phosphorylates threonine 3 of histone H3 (H3T3ph) (31). H3T3ph functions as a docking site for survivin (32–34), a subunit of the CPC complex; facilitates the recruitment of CPC to chromosomes; and subsequently leads to the activation of AURKB and mitosis transition (32). Overexpression or deficiency of Haspin leads to abnormal mitosis (31). However, Haspin knockout (KO) mice appear to be normal and fertile, which implies the existence of compensatory mechanisms that operate in the absence of Haspin (35). Because of Haspin’s roles in H3T3 phosphorylation and mitosis, several Haspin inhibitors have been developed as potential antitumor drugs (36,37). However, it remains to be determined whether these inhibitors are effective in cancer patients.
In this study, we found a previously unknown synthetic lethal interaction between Haspin depletion and VX-680 via unbiased whole genome CRSPR/Cas9 screening. We further confirmed the screen results using both Haspin KO cells and Haspin inhibitors. In addition, we revealed that CHR-6494 specifically potentiated the inhibitory effect of the AURKB inhibitor barasertib but not AURKA inhibitor alisertib, which underlay the synthetic lethality between Haspin depletion and VX-680. Our study provides a rational and promising approach, i.e., combining CHR-6494 (or 5-iodotubercindin) and barasertib, for cancer therapy.
MATERIALS AND METHODS
Cell lines
293A and HCT116 cells were purchased from ATCC. 293A cells were cultured in DMEM (Corning) with 10% fetal calf serum (FCS; HyClone). McCoy’s 5A medium (Gibco) with 10% fetal bovine serum (FBS; Sigma) was used to culture HCT116. SQCCY1 and HN5 cell lines (HPV-negative HNSCC) were gifts from Drs. Jeffrey N. Myers and Faye M. Johnson and were cultured in DMEM/F12 (Corning) supplemented with 10% FBS. A549, H1299, H1975, H358, and H460 cell lines (NSCLC) were gifts from Dr. Jae-Il Park, culturing in RPMI 1640 (Corning) with 10% FBS.
Antibodies and inhibitors
Antibodies against H3T3ph (Cat. No. 13576S) and H3S10ph (Cat. No. 9701S) were purchased from Cell Signaling Technology. Anti-Haspin antibody (Cat. No. A302–241A) was purchased from Bethyl Laboratories. Antibodies against H3 (Cat. No. ab1791) and tubulin (Cat. No. T6199) were purchased from Abcam and Sigma, respectively. These antibodies were used for Western blot analysis.
CHR-6494 (Cat. No. 11478) and VX-680 (Cat. No. 1595–25) were purchased from Cayman Chemical and BioVision, respectively. Alisertib (Cat. No. S1133), barasertib (Cat. No. 1147) and 5-iodotubercidin (Cat. No. S8314) were purchased from Selleck Chemicals.
CRISPR/Cas9-based screening
The TKOv3 library was a gift from Dr. Glen Traver Hart’s laboratory. The library includes 71,090 gRNAs, consisting of 70,948 protein-coding gene-targeted gRNAs (4 gRNAs/gene) and 142 EGFP-, LacZ-, and luciferase-targeted control gRNAs. The library generation and virus preparation were described previously (27).
SgRNA screening was performed as described (29). Briefly, 293A cells were first infected with the TKOv3 library lentivirus. Twenty-four hours later, cells were cultured with fresh medium containing 2 μg/mL puromycin for selection. Then selected cells were treated with dimethyl sulfoxide (DMSO) or VX-680 and incubated for about 20 doubling passages. The IC20 value of VX-680 was used for the screens. Cells from the initial infected cell populations and final cell populations were collected and extracted for genomic DNA (Qiagen kit). Genomic DNA were further amplified and labeled with barcodes via PCR followed by deep sequencing.
Generation of Haspin/GSG2 knockout cells
HCT116 cells were transiently transfected with PX459-GSG2-gRNA. Twenty-four hours after transfection, cells were selected with 2 μg/mL puromycin. Then single cells were seeded into 96-well plates. After 10 days, single clones were examined by Western blot to identify Haspin KO clones. The sequence of GSG2 sgRNA was: TGCACACTTCACCGGATAAG.
Colony formation assay
For clonogenic assay, HCT116 cells were seeded in 6-well plates in triplicate (wild-type: 200 cells/well; Haspin KO: 220 cells/well) and further cultured overnight. Then the cells were treated with CHR-6494, 5-iodotubercindin, VX-680, alisertib, and barasertib, alone or in combination. After 9 days, cells were stained with crystal violet solution (Sigma-Aldrich). Image J software was used to perform unbiased analysis of colony numbers.
Western blot analysis
HCT116 cells were seeded in 24-well plate and treated with inhibitors (VX-680, CHR-6494, alisertib, and barasertib), either single or combined treatment, for 24 hours. Then cells were directly lysed by SDS gel-loading buffer and boiled for further analysis. Samples were separated by SDS-PAGE and analyzed by immunoblotting with indicated antibodies.
Cell viability assay
For short-term survival assays, cells were plated in 96-well plates. The seeded cell numbers ranged from 600 to 1000 cells depending on the cell growth rate. After seeding overnight, cells were cultured with medium plus inhibitors (VX-680, CHR-6494, and barasertib) for 3 days. Cell viability was then evaluated with CellTiter-Glo luminescence assay (Promega).
Statistical analysis
All statistical analyses were performed using GraphPad Prism software, version 8.0.0. The detailed statistical methods are described in the main text and each experiment was repeated twice or more, unless otherwise stated. Differences between 2 groups were analyzed by Student t test analysis. A P value < 0.05 was considered statistically significant.
Live imaging of HCT116 cells
HCT116 cells were incubated with CellLight Histone 2B-GFP BacMam reagent (Life technologies; Cat. No. C10594) overnight and exposed to inhibitors immediately before imaging. Images were acquired with an oil immersion 60× 1.4NA Nikon objective using a Yokogawa CSU-X1 spinning disk that is currently integrated to an inverted Nikon Ti-microscope by 3i (Intelligent Imaging Innovations). Samples were excited with a 3i 488 nm solid state laser line and emission was collected with a 525/30 nm emission filter. Images were collected every 5 minutes for 18 hours, then processed and analyzed with 3i Slide Book software.
RESULTS
CRISPR/Cas9-based genome-wide screens conducted with VX-680 treatment reveal GSG2/Haspin as the top candidate
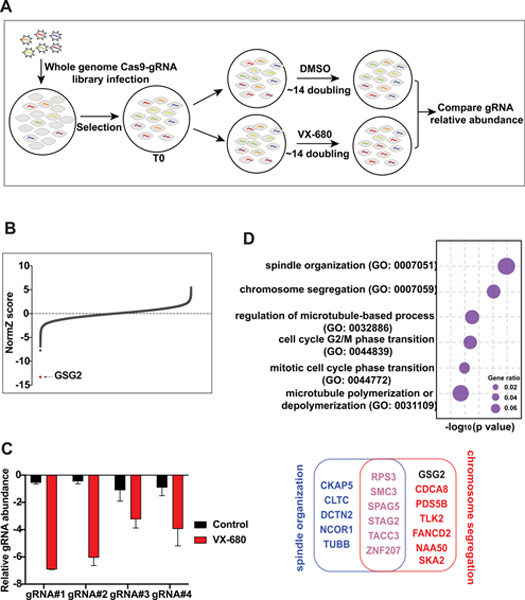
To identify genes whose depletion potentiates sensitivity or resistance to VX-680, we conducted pooled CRISPR/Cas9-based screens in 293A cells, which were treated with DMSO or VX-680. Briefly, cells infected with TKOv3 library virus were selected with puromycin. Then cells were cultured with DMSO or VX-680 and passaged for about 20 doublings. We collected cells at each passage point, and each group had 2 biological replicates. Genomic DNA extracted from the initial infected and final cell populations following DMSO or VX-680 treatment was amplified and labeled with barcodes via PCR. The PCR products were deep-sequenced and analyzed (Fig. 1A). We also performed bioinformatic analysis as described previously (29) to confirm that the screen data were reliable (Supplementary Fig. S1).
Figure 1.
CRSPR/Cas9-based genome-wide screens in VX-680-treated cells reveal GSG2 (also called Haspin) as the top candidate. (A) The flow chart of CRISPR/Cas9 screening in 293A cells treated with DMSO or VX-680. (B) Ranking of the co-essential genes in VX-680-treated samples compared with those treated with DMSO. The Z score from DrugZ analysis of the CRSPR/Cas9 screening results showed the genes with possible synthetic lethal relationships with VX-680. (C) Normalized sgRNA fold changes of GSG2 in DMSO- and VX-680-treated groups from the original screen sequencing data. (D) The top 6 enriched biological processes analyzed through gene ontology (GO; P < 0.01). Genes used for the analysis were high-confidence candidates from (B).
We compared DMSO- and VX-680-treated cells (Fig. 1B). The results showed that the levels of sgRNAs targeting GSG2 were dramatically reduced in VX-680-treated group but not in the DMSO-treated group (Fig. 1C). Moreover, we analyzed gene set enrichment for the genes identified in our screens, and GSG2 was linked with chromosome segregation (Fig. 1D). Of note, many genes identified in this screen were closely related to mitotic functions (Fig. 1D), with GSG2 as the top hit (Fig. 1B). Taken together, our data suggest a synthetic lethal interaction between pan-Aurora inhibitor VX-680 and GSG2 (also called Haspin, which is the common name that will be used below) depletion.
Depletion or inhibition of Haspin sensitizes cells to VX-680 treatment
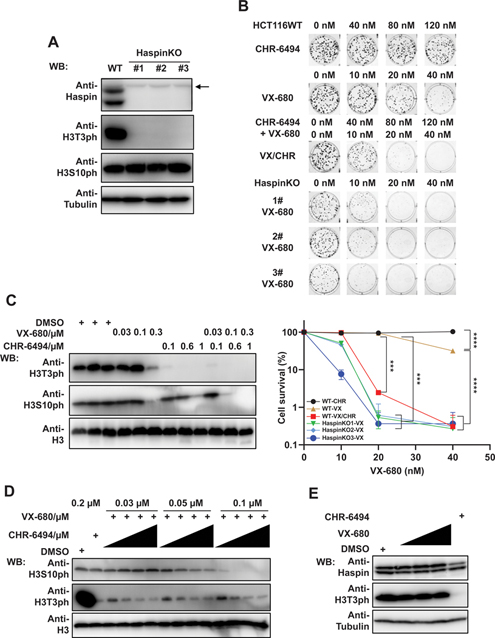
To validate the screening results, we generated HCT116 Haspin KO cells using CRISPR/Cas9 gene editing technology. We chose 3 Haspin KO clones for further analysis (Haspin KO#1, #2, and #3), and Western blot demonstrated the depletion of Haspin in these KO cell lines (Fig. 2A). These clones were also verified by sequencing (Supplementary Fig. S2). As Haspin is known as the serine/threonine kinase that phosphorylates histone H3 at threonine-3 site (31), we examined H3T3ph level in Haspin KO cell lines. Indeed, H3T3ph was significantly reduced or abolished in Haspin KO cells (Fig. 2A). We also examined the level of H3S10ph, the target of Aurora kinase B, to determine whether Haspin KO affects other mitotic phosphorylation events. The data showed that Haspin ablation had no effect on H3S10ph level (Fig. 2A).
Figure 2.
Depletion or inhibition of Haspin sensitizes cells to VX-680 treatment. (A) Confirmation of Haspin depletion in Haspin KO cell lines. HCT116 wild-type and Haspin KO cells were collected and directly lysed by SDS loading buffer. The expression of Haspin, H3T3ph, and H3S10ph were detected with indicated antibodies. Anti-Tubulin and anti-H3 blots, respectively, were included as loading controls for Haspin, H3T3ph and H3S10ph blots. The arrow indicates nonspecific bands. (B) Cells treated with Haspin depletion or inhibition exhibit hypersensitivity to VX-680 treatment. HCT116 wild-type and Haspin KO cells were treated with CHR-6494 and VX-680 for consecutive 9 days, either in combination (VX combined with 0, 40, 80, and 120 nM CHR) or single-agent treatment (CHR; VX). Colonies were fixed, stained, and further analyzed by Image J. Experiments were performed in triplicate with duplicate biological replicates. Representative images and results are shown. Student t tests were performed to estimate differences between 2 groups. Error bar represents SE (n = 3); *** P < 0.001; **** P < 0.0001. (C) HCT116 cells were treated with indicated concentrations of VX-680 and CHR-6494 and further incubated for 24 h. Then cells were lysed by SDS loading buffer and examined by immunoblotting with indicated antibodies. The H3T3ph and H3S10ph levels were determined with indicated antibodies. H3 served as loading control. (D) HCT116 cells were incubated with VX-680 (0.03, 0.05, and 0.1 μM) and CHR-6494 (0.05, 0.1, 0.15, and 0.2 μM) in combination or single agent treatment. After 24 h, cells were lysed and examined by Western blotting with indicated antibodies. (E) Western blots show the effect of single-agent treatment (0.03, 0.05, and 0.1 μM VX; 0.2 μM CHR). Samples were collected and blotted as described in (D).
To test whether depletion of Haspin has a synergistic effect with VX-680, we determined the sensitivity of HCT116 wild-type and Haspin KO#1, #2, and #3 cell lines to VX-680 treatment. As shown in Fig. 2B, Haspin depletion sensitized cells to VX-680 treatment, a finding that was consistent with our screening results. We further compared Haspin expression in normal tissues and cancer samples from Human Protein Atlas (v18.proteinatlas.org) and found no obvious difference of Haspin protein level in cancer cells or tissues (https://www.proteinatlas.org/ENSG00000177602-HASPIN/pathology), suggesting that unlike Aurora kinases, Haspin expression did not vary significantly in cancers. Therefore, we explored whether Haspin inhibition by CHR-6494 would sensitize cells to VX-680. Similar to Haspin KOs, cells treated with both VX-680 and CHR-6494 showed greater sensitivity to treatment than did cells treated with either agent alone (Fig. 2B). In addition, CHR-6494 displayed limited cytotoxicity to cells (Fig. 2B), a finding congruous with apparent normal proliferation of Haspin KO cells. Together, our data suggest that Haspin inhibitor CHR-6494 augments the effect of VX-680 without additional cytotoxicity by itself.
We also performed Western blot analysis to determine the synergistic effect of Haspin inhibition and VX-680 by. We used H3S10ph level, which was dramatically decreased by VX-680 treatment, to assess the inhibitory effect of VX-680. Since high concentrations of inhibitors often result in off-target effect and cytotoxicity, we first tested a broad range of concentrations of VX-680 and CHR-6494, aiming to find proper inhibitor concentrations for follow-up experiments. As shown in Fig. 2C, cells treated with high concentrations of VX-680 (0.3 μM) or CHR-6494 (1 μM) exhibited off-target effects as they started to affect other substrates. Thus, we chose lower concentrations that only target their respective and specific substrates for combinational experiments. Indeed, treatment of cells with VX-680 (0.1 μM) and CHR-6494 (0.6 μM) dramatically inhibited H3S10ph levels compared with cells treated with single agents. We further determined the level of H3S10ph under the combinations of each VX-680 concentration with CHR-6494 concentration gradient increases. Except for the lowest concentration of VX-680 (0.03 μM), cells treated with VX-680 (0.05 and 0.1 μM), when combined with increasing concentrations of CHR-6494 (0.05, 0.1, 0.15, and 0.2 μM), exhibited gradually reduced H3S10ph levels. For example, under the concentration of 0.1 μM VX-680, single-agent treated cells did not display inhibited or reduced H3S10ph levels (Fig. 2C). However, even when combined with lowest CHR-6494 concentration (0.05 μM), treatment with 0.1 μM VX-680 dramatically decreased the H3S10ph level (Fig.2D). The H3T3ph level showed the inhibition effect by CHR-6494 treatment (Fig. 2D). Consistently, even at the highest concentration of single-agent CHR-6494 we used for this assay (0.2 μM), there was no obvious change in H3S10ph level compared with cells treated with DMSO (Fig. 2D). This finding excluded not only the possibility of off-target effect but also the direct impact of CHR-6494 on H3S10ph level.
To assess whether VX-680 affects Haspin expression, we also examined the Haspin protein levels under the aforementioned VX-680 treatment. As shown in Fig. 2E, VX-680 did not affect Haspin protein level, as well as the H3T3ph level. Together, these results confirmed the synergistic effect of VX-680 and Haspin inhibition or depletion.
Haspin inhibitor potentiates the efficacy of AURKB inhibitor but not AURKA inhibitor
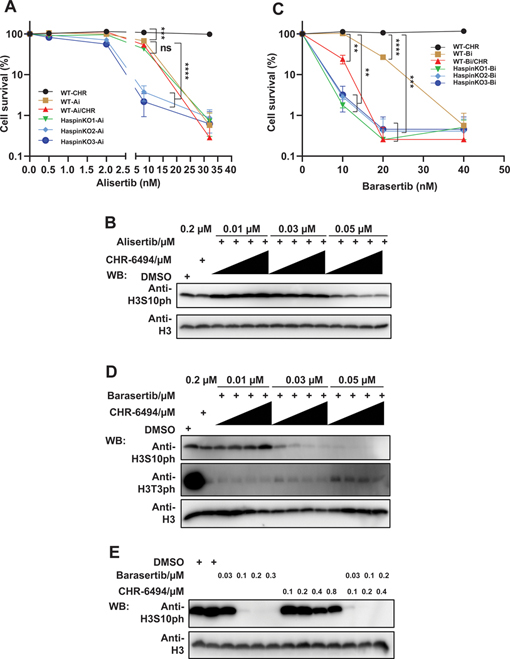
VX-680 inhibits AURKA, AURKB, and AURKC. To gain insights into the specificity of synergistic effect between Haspin inhibition and Aurora kinase inhibition, we performed combinational treatments of CHR-6494 with alisertib or barasertib which respectively inhibit AURKA or AURKB.
We first evaluated the effect of alisertib, alone or combined with CHR-6494, in wild-type or Haspin KO HCT116 cells. The proliferative capacity of cells treated with CHR-6494 and alisertib was compared with cells treated only with alisertib in clonogenic assays. Although Haspin KO cells (Haspin KO#2 and #3) exhibited a little sensitivity to alisertib, there was no obvious difference between combined and single-agent treatments in HCT116 cells (Fig. 3A and Supplementary Fig. S3A). Here we also used the level of H3S10ph as an indirect marker for AURKA inhibition. Consistent with the colony formation assays, Western blot analysis also supported that Haspin inhibition did not enhance the efficacy of alisertib (Fig. 3B).
Figure 3.
Haspin inhibitor potentiates the efficacy of AURKB inhibitor but not AURKA inhibitor. (A) Depletion or inhibition of Haspin has no effect on cellular sensitivity to alisertib. HCT116 wild-type (WT) or Haspin knockout (KO) cells were treated with CHR-6494 (CHR; 0, 40, 80, 120, or 160 nM) and alisertib (Ai; 0, 0.5, 2, 8, or 32 nM) and further cultured for 9 days followed by fixation and staining. Experiments were performed in triplicate with duplicate biological replicates. Representative results are shown. Student t tests were performed to estimate differences between 2 groups; ns, not significant; *** P < 0.001; **** P < 0.0001. (B) Addition of CHR-6494 has no effect on H3S10ph reduction by alisertib. HCT116 cells were plated in 24-well plate overnight and treated with alisertib, CHR-6494, or the combination of CHR-6494 (0.05, 0.1, 0.15, and 0.2 μM) and alisertib for 24 h. Then cells were lysed and analyzed by Western blotting using indicated antibodies. (C) CHR-6494 potentiates the inhibitory effect of barasertib. Cells treated with barasertib (Bi; 0, 10, 20, or 40 nM) or CHR-6494 (CHR; 0, 40, 80, 120, or 160 nM) were grown for 9 days followed by fixation and staining. Experiments were performed in triplicate with duplicate biological replicates. Representative results were shown. Student t tests were performed to estimate differences between 2 groups; error bar represents SE (n = 3); ** P < 0.01;*** P < 0.001; **** P < 0.0001. (D) Combination of CHR-6494 and barasertib dramatically reduces the level of H3S10ph. Cells were treated with barasertib or CHR-6494 for 24 h, in combination (Bi combined with 0.05, 0.1, 0.15, and 0.2 μM CHR) or alone (CHR; Bi), and followed by lysis with SDS loading buffer. The samples were then examined by immunoblots with indicated antibodies. H3 was included as loading control. (E) Cells were cultured with barasertib or CHR-6494 for 24 h and then analyzed by Western blotting using indicated antibodies.
We then compared cells treated with CHR-6494 and barasertib or barasertib only. Unlike our experiments with alisertib, colony formation assays showed that only half concentration of barasertib (10 nM) combined with CHR-6494 displayed cytotoxicity equal to that of barasertib alone (20 nM). Haspin KO cells also exhibited hypersensitivity to barasertib (Fig. 3C and Supplementary Fig. S3B). In addition, Western blot analysis showed that barasertib treatment drastically reduced the level of H3S10ph when used with increasing concentrations of CHR-6494 (Fig. 3D). Treatment with 0.03 μM barasertib did not affect the level of H3S10ph in cells. However, in cells treated with 0.03 μM barasertib and 0.1 μM CHR-6494, H3S10ph almost disappeared, which was equal to the effect of 0.1 μM barasertib treatment (Fig. 3D and 3E).
To further confirm our results, we chose 5-iodotubercidin (Itu) (38,39), another selective Haspin inhibitor, to test whether Itu would also show a synergistic effect with barasertib. We first compared the inhibitory effect of these two different Haspin inhibitors, CHR-6494 and Itu, on H3T3ph level. The result showed that both inhibitors are very effective in inhibiting H3T3ph level (Fig. S4A). In addition, the broad range of concentrations of Haspin inhibitors (from 0.03 μM to 0.5 μM) used also suggest that they are quite specific, since only H3T3ph, but not H3S10ph, was affected by these treatments (Fig. S4A). Furthermore, we compared the sensitivity of HCT116 cells exposed to combination of Itu and barasertib with cells only exposed to barasertib. Consistent with data obtained using combination with CHR-6494, the concentration of barasertib needed to kill most of the cells could be lowered to 10 nM when it was combined with Itu (Fig. S4B). Together, these results further support our conclusion of the synergistic effect of simultaneously inhibiting Haspin and AURKB.
Since both Haspin and AURKB are mitosis closely related kinases, we determined whether combination of Aurora B and Haspin would affect mitotic progression. HCT116 cells were labeled with GFP-H2B, then treated with either single treatment (0.1 μM CHR-6494 or 0.03 μM barasertib) or combination treatment (0.1 μM CHR-6494 and 0.03 μM barasertib) immediately before live cell imaging acquisition (Videos 1-4). We calculated the percentages of cells exhibiting normal mitotic progression or mitotic defects (Fig. S5A) and the elapsed time from prometaphase to anaphase onset (Fig. S5B). These results showed that combination treatment dramatically affected mitotic progression, including increased percentage of lagging chromosome in anaphase, multipolar spindle configuration as well as prolonged elapsed time from prometaphase to anaphase.
To test whether the inhibitory effect of the combination treatment is permanent or reversible, cells were treated with inhibitors for 9 consecutive days or only 3 days and then allowed to grow for an additional 6 days. Cells treated with inhibitors for 3 days exhibited similar survival rates as cells treated for 9 days, which confirmed that treatment with barasertib alone or with CHR-6494 resulted in irreversible cytotoxicity (Fig. S6).
These results strongly suggest that the synergistic antitumor effect observed in combination treatment with VX680 and CHR-6494 is likely to be mediated by inhibiting AURKB and Haspin. However, since there are no inhibitors targeting AURKC, we could not rule out that AURKC inhibition may also have similar synergistic effect with Haspin inhibition.
Combined inhibition of AURKB and Haspin exhibits better anti-tumor efficacy for HNSCC and NSCLC
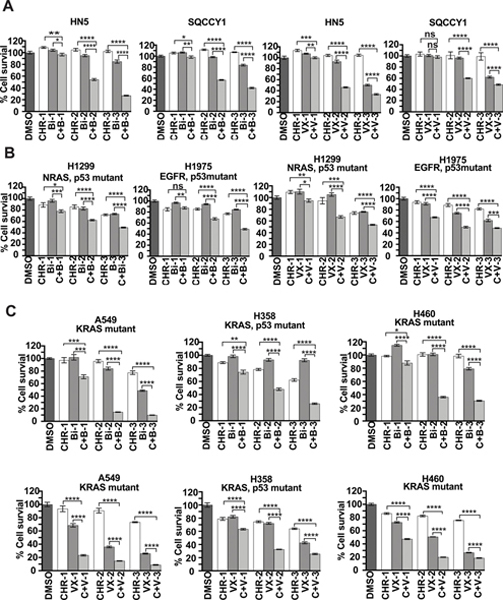
The above data indicate that barasertib with CHR-6494 has the potential to be an effective combination therapy for cancer patients. HNSCC and NSCLC are both common cancers worldwide that lack effective treatment strategies. Moreover, the HPV-negative HNSCC patients show worse prognosis than HPV-positive patients (40). NSCLC, which accounts for about 85% of lung cancer, remains to have low overall survival rate (41). Therefore, developing new therapies for these patients are needed to improve patient outcomes. Therefore, we chose several HNSCC and NSCLC cell lines and evaluated the efficacy of this combination therapy in these cell lines.
We first assessed whether combined treatment of barasertib with CHR-6494 had a better effect than single-agent barasertib on SQCCY1 and HN5 human HPV-negative HNSCC cell lines. We found that combination treatment suppressed cell viability 1.7 to 3 times more efficiently compared with either inhibitor alone in both cell lines (Fig. 4A). We also tested the efficacy of VX-680 with CHR-6494, which had an effect similar to that of barasertib with CHR-6494 (Fig. 4A) in the HNSCC cell lines.
Figure 4.
Combined inhibition of Haspin and AURKB exhibits better antitumor efficacy for HNSCC and NSCLC compared with single-agent treatment. (A) Combination treatment with CHR-6494 with either barasertib or VX-680 shows better antitumor efficacy for HNSCC. SQCCY1 or HN5 cell lines were plated in 96-well plate and treated with DMSO, CHR-6494 (CHR; 50, 100, or 200 nM), VX-680 (VX; 30, 50, or 100 nM) and barasertib (Bi; 10, 30, or 50 nM) for 3 days, in combination (C+V; C+B) or alone. Cells were examined with CellTiter-Glo luminescence assays. Results are representative of duplicate biological replicates. Student t tests were performed to estimate differences between 2 groups; error bar represents SE (n = 6); ns, not significant; *** P < 0.001; **** P < 0.0001. (B and C) Combination treatment with CHR-6494 and barasertib dramatically inhibits cell viability in KRAS-mutated NSCLC cell lines. NSCLC cell lines, including KRAS-mutated cell lines A549, H358, and H460 in (C) and non-KRAS-mutated cell lines (H1299 and H1975 in (B), were treated with indicated inhibitors for 3 days and further analyzed with CellTiter-Glo luminescence assays. Results are representative of duplicate biological replicates. Student t tests were performed to estimate differences between 2 groups; error bar represents SE (n = 6); ns, not significant; * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001.
We then estimated the inhibitory effect these combination treatments in 5 NSCLC cell lines. Intriguingly, NSCLC cell lines harboring KRAS mutation (A549, H358, and H460) were more sensitive to combination treatment than were cell lines with other mutations (H1299 and H1975; Fig. 4B and 4C). This is consistent with the previous report that Aurora kinases are downstream effectors of KRAS and that both AURKA and AURKB were promising targets in KRAS-induced lung cancer (42). Consistent with previous data, combined treatment with CHR-6494 and VX-680 displayed better antitumor effect than single-agent treatment (Fig. 4B and 4C), which confirmed the potential application of this combination therapy.
DISCUSSION
In this study, genome-wide CRSPR/Cas9 screening revealed synthetic lethality between pan-Aurora inhibitor VX-680 and Haspin depletion. Particularly, combined inhibition of Haspin and Aurora kinase B underlays the synthetic lethal relationship. Although clinical trials with barasertib were completed and there was no further clinical progress (7,8), our findings raise a possibility that combined treatment of barasertib and CHR-6494 could be an effective therapeutic option for cancer patients, including HNSCC and NSCLC patients who lack effective treatment strategies.
Here, we demonstrated by colony formation assays that not only Haspin KO cells but also Haspin inhibitor-treated cells are more sensitive to VX-680 than wild-type cells. Western blot analysis confirmed that Haspin inhibitor CHR-6494 potentiates the effect of VX-680 by decreasing H3S10ph levels. Moreover, we showed that CHR-6494 as well as Itu specifically augments the efficacy of AURKB inhibitor barasertib but not AURKA inhibitor alisertib. Notably, CHR-6494 and Itu alone showed very mild cytotoxicity in cells. Based on these data, we conclude that combined inhibition of Haspin and AURKB is a potential new combination for cancer therapy.
Previous studies also indicated the existence of a positive feedback loop involving Haspin and AURKB. Namely, AURKB phosphorylates Haspin directly, which further promotes H3T3 phosphorylation during mitosis (43). Moreover, H3T3ph accurately positions CPC complex and modulates the activation of AURKB (32–34,44–46). Another study showed the concentration of Itu needed to drive mitotic exit could be 10-fold lower when combined with Aurora B inhibitor, again hints the synergistic interaction between Haspin and AURKB (47). On the other hand, AURKA was demonstrated to regulate the association of AURKB and Haspin in early mitosis and to prime the Haspin-AURKB feedback loop in late G2 phase (48). It is possible that AURKA acts upstream of Haspin-AURKB loop (49,50), which may explain why we observed an enhanced inhibition effect with barasertib, but not alisertib, by CHR-6494.
We expanded our combination treatment to NSCLC, which accounts for a major fraction of lung cancers and carries poor survival outcomes (41,51). Our data showed that combinatorial inhibition of Haspin and AURKB was more efficacious in NSCLC cell lines harboring KRAS mutation than in cell lines harboring other mutations. Consistently, AURKA and AURKB have been reported as the targets of KRAS in lung cancer (42). Moreover, a previous study reported that H358 and A549 cell lines treated with either knockdown of AURKA or AURKB, or AURKA or AURKB inhibitors, showed decreased cell growth and viability (42). Based on these published data and observations made in our study, we hypothesize that combined treatment with barasertib and CHR-6494 may be an effective approach to improve the outcome of patients with KRAS-mutant lung cancer.
Our study has several limitations. First, although cells treated with CHR-6494 alone showed very mild inhibition of cell survival, we cannot rule out potential cytotoxicity caused by the addition of CHR-6494 when this agent is used in vivo. Second, we only examined H3T3ph and H3S10ph levels after cells were treated with these inhibitors. We cannot exclude the possibility that these inhibitors may also inhibit other kinases and have off-target effects. Third, our conclusions were based on studies of cancer cell lines. It remains to be determined whether similar results will be observed in clinical trials. Nevertheless, our study will provide a rationale to further test combination therapies with barasertib in clinical trials.
Supplementary Material
ACKNOWLEDGEMENT
We thank all the members of the Chen laboratory for their help and constructive discussions and Dr. Faye M. Johnson for her suggestions regarding the experimental design. We thank Drs. Jeffrey N. Myers, Faye M. Johnson, Jae-Il Park, and Glen Traver Hart for providing reagents. We also thank Bryan Tutt in Scientific Publications, Research Medical Library, the University of Texas MD Anderson Cancer Center for help with the scientific editing of the manuscript.
Funding
This work was supported in part by the Pamela and Wayne Garrison Distinguished Chair in Cancer Research to JC. We thank The University of Texas MD Anderson Cancer Center Science Park Next-Generation Sequencing (NGS) core Facility with CPRIT Core Facility Support Award (CPRIT RP170002). JC also received support from NIH (CA193124, CA210929).
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest.
REFERENCES
- 1.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol 2003;4:842–54 [DOI] [PubMed] [Google Scholar]
- 2.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res 2007;5:1–10 [DOI] [PubMed] [Google Scholar]
- 3.Bavetsias V, Linardopoulos S. Aurora Kinase Inhibitors: Current Status and Outlook. Front Oncol 2015;5:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willems E, Dedobbeleer M, Digregorio M, Lombard A, Lumapat PN, Rogister B. The functional diversity of Aurora kinases: a comprehensive review. Cell Div 2018;13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boss DS, Beijnen JH, Schellens JH. Clinical experience with aurora kinase inhibitors: a review. Oncologist 2009;14:780–93 [DOI] [PubMed] [Google Scholar]
- 6.Katayama H, Sen S. Aurora kinase inhibitors as anticancer molecules. Biochim Biophys Acta 2010;1799:829–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borisa AC, Bhatt HG. A comprehensive review on Aurora kinase: Small molecule inhibitors and clinical trial studies. Eur J Med Chem 2017;140:1–19 [DOI] [PubMed] [Google Scholar]
- 8.Tang A, Gao K, Chu L, Zhang R, Yang J, Zheng J. Aurora kinases: novel therapy targets in cancers. Oncotarget 2017;8:23937–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J 1998;17:3052–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet 1998;20:189–93 [DOI] [PubMed] [Google Scholar]
- 11.Tanaka T, Kimura M, Matsunaga K, Fukada D, Mori H, Okano Y. Centrosomal kinase AIK1 is overexpressed in invasive ductal carcinoma of the breast. Cancer Res 1999;59:2041–4 [PubMed] [Google Scholar]
- 12.Gritsko TM, Coppola D, Paciga JE, Yang L, Sun M, Shelley SA, et al. Activation and overexpression of centrosome kinase BTAK/Aurora-A in human ovarian cancer. Clin Cancer Res 2003;9:1420–6 [PubMed] [Google Scholar]
- 13.Reichardt W, Jung V, Brunner C, Klein A, Wemmert S, Romeike BF, et al. The putative serine/threonine kinase gene STK15 on chromosome 20q13.2 is amplified in human gliomas. Oncol Rep 2003;10:1275–9 [PubMed] [Google Scholar]
- 14.Araki K, Nozaki K, Ueba T, Tatsuka M, Hashimoto N. High expression of Aurora-B/Aurora and Ipll-like midbody-associated protein (AIM-1) in astrocytomas. J Neurooncol 2004;67:53–64 [DOI] [PubMed] [Google Scholar]
- 15.Chieffi P, Troncone G, Caleo A, Libertini S, Linardopoulos S, Tramontano D, et al. Aurora B expression in normal testis and seminomas. J Endocrinol 2004;181:263–70 [DOI] [PubMed] [Google Scholar]
- 16.Sorrentino R, Libertini S, Pallante PL, Troncone G, Palombini L, Bavetsias V, et al. Aurora B overexpression associates with the thyroid carcinoma undifferentiated phenotype and is required for thyroid carcinoma cell proliferation. J Clin Endocrinol Metab 2005;90:928–35 [DOI] [PubMed] [Google Scholar]
- 17.Mehra R, Serebriiskii IG, Burtness B, Astsaturov I, Golemis EA. Aurora kinases in head and neck cancer. Lancet Oncol 2013;14:e425–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeshita M, Koga T, Takayama K, Ijichi K, Yano T, Maehara Y, et al. Aurora-B overexpression is correlated with aneuploidy and poor prognosis in non-small cell lung cancer. Lung Cancer 2013;80:85–90 [DOI] [PubMed] [Google Scholar]
- 19.Hoar K, Chakravarty A, Rabino C, Wysong D, Bowman D, Roy N, et al. MLN8054, a small-molecule inhibitor of Aurora A, causes spindle pole and chromosome congression defects leading to aneuploidy. Mol Cell Biol 2007;27:4513–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manfredi MG, Ecsedy JA, Meetze KA, Balani SK, Burenkova O, Chen W, et al. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. Proc Natl Acad Sci U S A 2007;104:4106–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sells TB, Chau R, Ecsedy JA, Gershman RE, Hoar K, Huck J, et al. MLN8054 and Alisertib (MLN8237): Discovery of Selective Oral Aurora A Inhibitors. ACS Med Chem Lett 2015;6:630–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helfrich BA, Kim J, Gao D, Chan DC, Zhang Z, Tan AC, et al. Barasertib (AZD1152), a Small Molecule Aurora B Inhibitor, Inhibits the Growth of SCLC Cell Lines In Vitro and In Vivo. Mol Cancer Ther 2016;15:2314–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med 2004;10:262–7 [DOI] [PubMed] [Google Scholar]
- 24.Bebbington D, Binch H, Charrier JD, Everitt S, Fraysse D, Golec J, et al. The discovery of the potent aurora inhibitor MK-0457 (VX-680). Bioorg Med Chem Lett 2009;19:3586–92 [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Zhang ZF, Chen J, Huang D, Ding Y, Tan MH, et al. VX680/MK-0457, a potent and selective Aurora kinase inhibitor, targets both tumor and endothelial cells in clear cell renal cell carcinoma. Am J Transl Res 2010;2:296–308 [PMC free article] [PubMed] [Google Scholar]
- 26.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343:84–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 2015;163:1515–26 [DOI] [PubMed] [Google Scholar]
- 28.Burdova K, Yang H, Faedda R, Hume S, Chauhan J, Ebner D, et al. E2F1 proteolysis via SCF-cyclin F underlies synthetic lethality between cyclin F loss and Chk1 inhibition. EMBO J 2019:e101443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Wang G, Feng X, Shepherd P, Zhang J, Tang M, et al. Genome-wide CRISPR screens reveal synthetic lethality of RNASEH2 deficiency and ATR inhibition. Oncogene 2019;38:2451–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potting C, Crochemore C, Moretti F, Nigsch F, Schmidt I, Manneville C, et al. Genome-wide CRISPR screen for PARKIN regulators reveals transcriptional repression as a determinant of mitophagy. Proc Natl Acad Sci U S A 2018;115:E180–E9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai J, Sultan S, Taylor SS, Higgins JMG. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Gene Dev 2005;19:472–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 2010;330:235–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, et al. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 2010;330:231–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science 2010;330:239–43 [DOI] [PubMed] [Google Scholar]
- 35.Shimada M, Goshima T, Matsuo H, Johmura Y, Haruta M, Murata K, et al. Essential role of autoactivation circuitry on Aurora B-mediated H2AX-pS121 in mitosis. Nat Commun 2016;7:12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kestav K, Uri A, Lavogina D. Structure, Roles and Inhibitors of a Mitotic Protein Kinase Haspin. Curr Med Chem 2017;24:2276–93 [DOI] [PubMed] [Google Scholar]
- 37.Amoussou NG, Bigot A, Roussakis C, Robert JH. Haspin: a promising target for the design of inhibitors as potent anticancer drugs. Drug Discov Today 2018;23:409–15 [DOI] [PubMed] [Google Scholar]
- 38.Balzano D, Santaguida S, Musacchio A, Villa F. A general framework for inhibitor resistance in protein kinases. Chem Biol 2011;18:966–75 [DOI] [PubMed] [Google Scholar]
- 39.Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein RL, Murray JW, et al. Structure and functional characterization of the atypical human kinase haspin. Proc Natl Acad Sci U S A 2009;106:20198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marur S, Forastiere AA. Head and Neck Squamous Cell Carcinoma: Update on Epidemiology, Diagnosis, and Treatment. Mayo Clin Proc 2016;91:386–96 [DOI] [PubMed] [Google Scholar]
- 41.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446–54 [DOI] [PubMed] [Google Scholar]
- 42.Dos Santos EO, Carneiro-Lobo TC, Aoki MN, Levantini E, Basseres DS. Aurora kinase targeting in lung cancer reduces KRAS-induced transformation. Mol Cancer 2016;15:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang F, Ulyanova NP, van der Waal MS, Patnaik D, Lens SM, Higgins JM. A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr Biol 2011;21:1061–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeyaprakash AA, Basquin C, Jayachandran U, Conti E. Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex. Structure 2011;19:1625–34 [DOI] [PubMed] [Google Scholar]
- 45.Du J, Kelly AE, Funabiki H, Patel DJ. Structural basis for recognition of H3T3ph and Smac/DIABLO N-terminal peptides by human Survivin. Structure 2012;20:185–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niedzialkowska E, Wang F, Porebski PJ, Minor W, Higgins JM, Stukenberg PT. Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres. Mol Biol Cell 2012;23:1457–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang F, Ulyanova NP, Daum JR, Patnaik D, Kateneva AV, Gorbsky GJ, et al. Haspin inhibitors reveal centromeric functions of Aurora B in chromosome segregation. J Cell Biol 2012;199:251–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu F, Jiang Y, Lu L, Cao M, Qiao Y, Liu X, et al. Aurora-A promotes the establishment of spindle assembly checkpoint by priming the Haspin-Aurora-B feedback loop in late G2 phase. Cell Discov 2017;3:16049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou L, Tian X, Zhu C, Wang F, Higgins JM. Polo-like kinase-1 triggers histone phosphorylation by Haspin in mitosis. EMBO Rep 2014;15:273–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghenoiu C, Wheelock MS, Funabiki H. Autoinhibition and Polo-dependent multisite phosphorylation restrict activity of the histone H3 kinase Haspin to mitosis. Mol Cell 2013;52:734–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res 2016;5:288–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.