Abstract
Infection of the postpartum uterus with pathogenic bacteria is associated with infertility months later in dairy cattle. However, it is unclear whether these bacterial infections lead to long-term changes in the reproductive tract that might help explain this infertility. Here we tested the hypothesis that infusion of pathogenic bacteria into the uterus leads to changes in the transcriptome of the reproductive tract three months later. We used virgin Holstein heifers to avoid potential confounding effects of periparturient problems, lactation, and negative energy balance. Animals were infused intrauterine with endometrial pathogenic bacteria Escherichia coli and Trueperella pyogenes (n = 4) and compared with control animals (n = 6). Three months after infusion, caruncular and intercaruncular endometrium, isthmus and ampulla of the oviduct, and granulosa cells from ovarian follicles > 8 mm diameter were profiled by RNA-sequencing. Bacterial infusion altered the transcriptome of all the tissues when compared with control. Most differentially expressed genes were tissue-specific, with 109 differentially expressed genes unique to caruncular endometrium, 57 in intercaruncular endometrium, 65 in isthmus, 298 in ampulla, and 83 in granulosa cells. Surprisingly, despite infusing bacteria into the uterus, granulosa cells had more predicted upstream regulators of differentially expressed genes than all the other tissues combined. In conclusion, there were changes in the transcriptome of the endometrium, oviduct and even granulosa cells, three months after intrauterine infusion of pathogenic bacteria. These findings imply that long-term changes throughout the reproductive tract could contribute to infertility after bacterial infections of the uterus.
Keywords: Cow, RNAseq, ovary, bacteria, pathogens
INTRODUCTION
Infection of the uterus with bacteria is ubiquitous after parturition, and approximately 15 per cent of dairy cattle develop clinical endometritis (LeBlanc et al. 2002, Sheldon et al. 2009). Endometritis is important because these animals are less fertile than unaffected animals, even after resolution of disease. When compared with unaffected animals, a history of endometritis on average increases the interval to first insemination by about 11 days, delays conception by 32 days, and nearly doubles involuntary culling (Borsberry and Dobson 1989, LeBlanc et al. 2002). The gap in knowledge is whether infection of the uterus with pathogenic bacteria leads to long-term changes in the reproductive tract that might help explain this reduced fertility.
The pathogenic bacteria that infect the postpartum uterus of dairy cattle include Escherichia coli, Trueperella pyogenes, Fusobacterium necrophorum, and Prevotella and Bacteroides species (Griffin et al. 1974, Huszenicza et al. 1991, Sheldon et al. 2002). Notably, endometrial pathogenic strains of E. coli are adapted to cause uterine disease (Bicalho et al. 2010, Sheldon et al. 2010), and Trueperella pyogenes is the pathogen most associated with the severity of endometritis (Bonnett et al. 1991, Westermann et al. 2010). Moreover, intrauterine infusion of E. coli and/or T. pyogenes into cattle induces clinical endometritis (Rowson et al. 1953, Ayliffe and Noakes 1982, Amos et al. 2014, Piersanti et al. 2019b). Clinical endometritis is characterised by the presence of pus within the uterus and a purulent uterine discharge, which is often detectable in the vagina, 21 days or more postpartum (Sheldon et al. 2019, Sheldon 2020).
When pathogenic bacteria infect the uterus, they cause inflammation and damage in the reproductive tract (Herath et al. 2009, Chapwanya et al. 2012, Bromfield et al. 2015). Endometrial cells, as well as immune cells, mount rapid inflammatory responses to the pathogenic bacteria or their virulence factors, such as lipopolysaccharide (LPS), secreting prostaglandin E2, interleukin (IL)-1, IL-6 and IL-8 (Herath et al. 2006, Cronin et al. 2012, Turner et al. 2014). Furthermore, endometrial cells are susceptible to damage by pore-forming toxins, which are produced by pathogens such as T. pyogenes (Amos et al. 2014, Healy et al. 2014). Epithelial cells from the oviduct also mount inflammatory responses to LPS, including increased expression of TNFA and IL1B (Kowsar et al. 2013, Ibrahim et al. 2015). Similarly, granulosa cells from ovarian follicles secrete IL-1β, IL-6 and IL-8 in response to LPS (Bromfield and Sheldon 2011, Price et al. 2013, Price and Sheldon 2013). As well as these short-term inflammatory responses during bacterial infection, there is in vivo evidence that after infection of the uterus there is long-term perturbation of reproductive tract health (Ribeiro et al. 2016, Piersanti et al. 2019a). However, one of the challenges for determining the long-term effects of bacterial infection on the reproductive tract in cattle is disentangling the effect of the infection from other postpartum effects, including periparturient problems, lactation, and negative energy balance (Chagas et al. 2007, Cerri et al. 2012, LeBlanc 2012, Girard et al. 2015).
The present study aimed to explore whether infection of the uterus with pathogenic bacteria leads to long-term changes in the reproductive tract. Specifically, we tested the hypothesis that infusion of pathogenic bacteria into the uterus leads to changes in the transcriptome of the reproductive tract three months later. To test this hypothesis, we exploited a uterine infection model in virgin Holstein heifers, which allowed us to avoid the potential confounding effects of periparturient problems, lactation, and negative energy balance (Piersanti et al. 2019b). We infused animals intrauterine with pathogenic E. coli and T. pyogenes, and three months later we collected endometrium, oviduct and granulosa cells from these animals and from control animals. We then used RNA-sequencing (RNAseq) and Ingenuity Pathway Analysis to examine alterations in the global patterns of the transcriptome between the bacteria-infused and control animals.
MATERIALS AND METHODS
Ethical statement
The University of Florida Institutional Animal Care and Use Committee approved all animal procedures (protocol number 201508884), which were conducted from June to October 2017 at the University of Florida Dairy Research Unit (Gainesville, FL, USA).
Animal model of bacterial infection of the uterus
The study used ten virgin Holstein heifers, 11 to 13 months old, and the study design, infection of the uterus, and progression of disease was reported in detail previously (Piersanti et al. 2019b). Briefly, animals were clinically healthy prior to the experiment; tested negative for Brucella abortus, Neospora caninum and Leptospirosa; and, had clear vaginal mucus. Oestrous cycles were synchronized using an ovsynch protocol, comprising intramuscular injection with 100 mg gonadotropin-releasing hormone (GnRH; gonadorelin diacetate tetrahydrate; OvaCyst, Bayer HealthCare LLC, Hannover, NJ, USA), followed by 25 mg prostaglandin F2α (dinoprost tromethamine; ProstaMate, Bayer HealthCare LLC) 5 and 6 days after the initial GnRH, and 100 mg GnRH 8 days after the initial GnRH (Lima et al. 2013). Ovarian follicle growth and ovulation was confirmed using ultrasonography during and for two days after the second GnRH injection. Starting the day after the second GnRH injection, animals were injected intramuscularly with 200 mg progesterone in corn oil (Sigma-Aldrich, St Louis, MO, USA) daily for 7 d to ensure elevated progesterone concentrations at the time of intrauterine infusion (Piersanti et al. 2019b). Three days after the second GnRH injection, animals were blocked by age and weight, and randomly assigned to control (n = 6) or bacterial infusion (n = 4). Control animals were infused via a trans-cervical catheter into the body of the uterus with 30 ml sterile Luria-Bertani broth (Sigma-Aldrich); one animal was not fully catheterised due to a tight cervix but was infused intrauterine and included in the experiment. Bacteria-infused animals were infused via a trans-cervical catheter into the body of the uterus with 30 mL Luria-Bertani broth containing 4.64 × 107 CFU/mL E. coli strain MS499 (Goldstone et al. 2014b) and 3.38 × 107 CFU/mL T. pyogenes strain MS249 (Goldstone et al. 2014a), which are pathogens collected from animals with endometritis (Sheldon et al. 2002, Sheldon et al. 2010, Amos et al. 2014). The number of bacteria that were infused was based on preliminary experiments and on previous studies (Ayliffe and Noakes 1982, Semambo et al. 1991, Amos et al. 2014, Lima et al. 2015). Vaginal mucus was collected daily using a Metricheck device (Simcro, Hamilton, New Zealand) from the day before until 7 days after infusion, and endometritis was graded as described previously (Sheldon et al. 2009): grade 0, clear or translucent mucus; grade 1, mucus containing flecks of pus; grade 2, mucus containing ≤ 50% pus; and grade 3, mucus containing > 50% pus.
As reported previously (Piersanti et al. 2019b), endometritis was detected in all bacteria-infused animals 3 days after infusion, with each animal developing grade 3 endometritis by 4 days after infusion. These bacteria-infused animals had pus discharging from the uterus into the vagina, with a median endometritis grade 3; whereas control animals had a median endometritis grade ≤ 1. We also measured an increased abundance of bacterial total 16S RNA in the uterine discharge collected 5 days after infusion from bacteria-infused but not control animals (4.6 vs 0.1 ng/mg discharge). The presence of endometritis was further confirmed using transrectal ultrasonography (Aloka SSD-500, Hitachi Healthcare Americas, Twinsburg, OH, USA). Pus was visible in the uterine lumen of each bacteria-infused animal, whereas control animals had no evidence of fluid or echogenic material in the uterus. The signs of disease detected using the Metricheck device and transrectal ultrasonography mimic the signs seen in field cases of clinical endometritis using the same diagnostic methods about four weeks postpartum in dairy cows (Sheldon 2020).
Collection of reproductive tract tissues
Starting 80 days after infusion, the oestrous cycles of all animals were synchronized using the ovsynch protocol described above, with the second GnRH administered 6 days before slaughter (Lima et al. 2013). Animals were slaughtered by captive bolt and exsanguination 94 days after intrauterine infusion. Reproductive tracts were collected, transported on ice, and processed within 3 h of slaughter. A consistent approach was used to collect samples because there is variation in transcriptome across the reproductive tract in a spatial relationship to the corpus luteum (Locatelli et al. 2019, Sanchez et al. 2019). Caruncular and intercaruncular endometrium was dissected from the underlying myometrium of the uterine horn ipsilateral to the ovary containing the corpus luteum. Ampulla and isthmus were collected by extrusion of the oviduct contralateral to the ovary containing the corpus luteum. Granulosa cells were collected from a single ovarian follicle > 8 mm diameter (range 8 to 24 mm) in the ovary contralateral to the ovary containing the corpus luteum. Granulosa cells were collected by aspiration using a sterile 20-gauge needle and syringe containing collection medium (0.5% BSA, 20 mM HEPES, 2 mM sodium pyruvate, 10 IU/ml heparin, 100 U/ml penicillin, 100 μg/ml streptomycin in Medium 199; Thermo Fisher Scientific, Waltham, MA, USA); and, the follicle aspirates were centrifuged at 500 × g for 10 min to isolate the granulosa cells. Samples of caruncular and intercaruncular endometrium, isthmus, ampulla, and granulosa cells were snap frozen in liquid nitrogen and stored at −80°C. Technical problems prevented collection of the oviduct from one bacteria-infused animal and the intercaruncular endometrium from another bacteria-infused animal.
RNA sequencing
Samples were thawed and suspended in RLT buffer (QIAGEN GmbH, Hilden, Germany). Oviduct and granulosa cell sample RNA was extracted using the RNeasy Micro kit (QIAGEN GmbH), according to the manufacturer’s instructions. Caruncular and intercaruncular endometrial samples were first processed with 2.8 mm ceramic beads in a bead beater tissue homogenizer (Precellys 24; Bertin Technologies SAS, Montigny-le-Bretonneux, France). Samples were processed for 2 cycles of 45 s at 6500 × g, with a 30 s interval in between cycles. After tissue homogenization endometrial RNA was extracted using the RNeasy Mini kit (QIAGEN GmbH), according to the manufacturer’s instructions. Total RNA concentration was determined using a Qubit 2.0 Fluorometer (Thermo Fisher Scientific) and RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Total RNA with a 28S:18S ratio > 1 and RNA integrity number ≥ 7 were used for RNAseq library construction. Unfortunately, granulosa cell RNA for one bacteria-infused animal was below the quality threshold and excluded from further processing.
The RNA library construction and sequencing were performed at the Interdisciplinary Center for Biotechnology Research, University of Florida. To produce RNAseq libraries, 500 ng (endometrium and oviduct) or 200 ng (granulosa) total RNA was used for mRNA isolation using NEBNext Poly(A) mRNA Magnetic Isolation Module, followed by RNA library construction with NEBNext Ultra II Directional Library Preparation (both New England Biolabs, Ipswich, MA, USA), according to the manufacturer’s instructions. Briefly, extracted mRNA was fragmented in NEBNext first-strand synthesis buffer at 94°C, followed by first-strand cDNA synthesis using reverse transcriptase and random primers. Synthesis of double-stranded cDNA was performed using the second strand master mix provided with the kit. The resulting double-stranded cDNA was end-repaired, dA-tailed and ligated with NEBNext adaptors. Finally, libraries were enriched by amplification and purified by Meg-Bind RxnPure Plus beads (Omega Bio-tek, Inc., Norcross, GA, USA). Barcoded libraries were sized on a bioanalyzer, and quantified by Qubit and qPCR (Kapa Biosystems Wilmington, MA, USA). Individual libraries were pooled, and equal molar concentrations were run on an Illumina HiSeq3000 sequencer (Illumina, San Diego, CA, USA). Reads acquired from the sequencing platform were cleaned with the Cutadapt program to trim sequencing adaptors, low-quality bases, and potential errors introduced during sequencing or library preparation (Marcel 2011). Reads with a quality Phred-like score < 20 and read length < 40 bases were excluded from RNAseq analysis, as described previously (Piersanti et al. 2019a).
The transcripts of Bos taurus (76,341 sequences) retrieved from the NCBI genome database (GCF_002263795.1) were used as reference sequences for RNAseq analysis. The cleaned reads of each sample were mapped individually to the reference sequences using the bowtie2 mapper (v2.2.3) with a ‘3 mismatches a read’ allowance (Langmead and Salzberg 2012). The mapping results were processed with the SAMtools (Li et al. 2009), and scripts developed in-house by the Interdisciplinary Centre for Biotechnology Research to remove potential PCR duplicates and select uniquely-mapped reads for gene expression analysis, as described previously (Piersanti et al. 2019a). The RNAseq data were deposited in NCBI’s Gene Expression Omnibus database and is accessible through GEO Series accession number GSE140469 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140469). Gene expression was compared between the bacteria-infused and control animals by counting the number of mapped reads for each transcript (Yao and Yu 2011).
Quantitative PCR
We used qPCR to validate gene expression determined by RNAseq. Primers were designed using the NCBI database to target CD38 (fwd 5’-AATCTCAGCCGTGACCAGTT-3’; rev 5’-CCACCAGCATGTCTCCTGAA-3’), NXPE3 (fwd 5’-GACCTGGATGGAGCAGGAAG-3’; rev 5’-ACAGGGCCTCTGTATCAACC-3’) and CTTN (fwd 5’-ATGGACAAGTCAGCTGTCGG-3’; rev 5’-CAAACTTGCCTCCAAAGCCC-3’) in granulosa cells, ampulla and isthmus, respectively; and the housekeeper gene GAPDH (forward 5’-AGGTCGGAGTGAACGGATTC-3’; reverse 5’-ATGGCGACGATGTCCACTTT-3’). The same samples used for RNAseq were reverse transcribed using the Verso cDNA synthesis kit (Thermo Fisher Scientific) prior to real time qPCR using iTaq Universal SYBR Green Supermix and the CFX Connect Real-Time PCR Detection System (both Bio-Rad, Hercules, CA, USA), with an initial denaturation step at 95°C for 30 sec followed by 40 cycles at 95°C for 5 sec, annealing temperature for 10 sec, and final extension at 60°C for 30 sec. A no-template negative control was used in place of cDNA to determine non-specific amplification for each primer pair. Amplification efficiency for each primer pair was evaluated and met MIQE guidelines of r2 > 0.98 and efficiency of 90 to 110%. Relative expression for genes of interest were calculated using the 2–ΔCt method, relative to the housekeeper gene GAPDH. The expression pattern of CD38 in granulosa cells, NXPE3 in isthmus tissue, and CTTN in ampulla were similar for data obtained by RNAseq and qPCR (Supplemental Figure 1).
Data Analysis
The experimental unit was each animal, and comparisons were made between bacteria-infused and control animals. The RNAseq gene expression data for each tissue was used to generate volcano plots, and for principle component analysis, which was performed using ClustVis (Metsalu and Vilo 2015). Genes that were differentially expressed in bacteria-infused animals compared with control animals were selected using P < 0.05 and are reported as log2 fold-change (log2FC) using a cut-off of > 2 or < –2; adjusted P-values for false discovery rate were not used because the P-values were > 0.10. As described previously (Kong et al. 2017), this approach allowed the generation of informative differentially expressed gene (DEG) lists for each tissue, and the identification of pathways and networks that were affected by bacterial infusion. Heatmaps were generated for the DEGs of individual animals with Heatmapper (Babicki et al. 2016), using Euclidean distance and average linkage (Quackenbush 2001). Venn diagrams were generated using jvenn to compare the DEGs amongst tissues (Bardou et al. 2014). Unless stated otherwise, figures were generated in GraphPad Prism 8.3 (GraphPad Software, La Jolla, CA)
Ingenuity Pathway Analysis (QIAGEN GmbH) was used to identify pathways, gene networks, and upstream regulators of DEGs affected by bacterial infusion (Krämer et al. 2014). Pathways were identified by –log P > 1.3 and log2FC ≤ −2 or ≥ 2, and the software calculated corresponding z-scores to predict activation (z ≥ 2) or inactivation (z ≤ 2). Gene networks were identified by assessing the number of DEGs in each network, and gene network scores were calculated by the software (a network score of ≥ 2 gives 99% confidence the network was not identified by chance). Upstream regulators of DEGs, and predicted diseases and functions, were identified by z-scores ≥ 2 or ≤ −2, and were considered significant predictors of activation or inhibition of DEGs, respectively (Hatzirodos et al. 2014, Piersanti et al. 2019a).
RESULTS
Effects of bacterial infusion on tissue transcriptomes
Caruncular endometrium
produced an average of 31,493 mapped transcripts per sample for analysis, with an average of 31% of the high-quality reads aligned to the Bos taurus reference genome (Supplemental Table 1). There were 87 upregulated and 45 downregulated DEGs in bacteria-infused animals compared with controls, as illustrated in a volcano plot of all the caruncular endometrium genes (Fig. 1A, left panel). Principle component analysis, based on all the genes is presented in Fig. 1A (middle panel), and principal components 1 and 2 explain 20% and 16% of the variance, respectively. A heatmap of the distribution of the DEGs across the samples illustrates the separation between the bacteria-infused and control animals (Fig. 1A, right panel). The five most upregulated DEGs in bacteria-infused animals were SLC45A1, ADARB1, RNF38, DBNDD1 and GOLGA3, and the five most downregulated DEGs were MMP3, SPAG9, NXPE3, ATG10 and VPREB3 (Supplemental Table 2).
Figure 1. Volcano plot, principle component analysis, and heatmap analysis for bacteria-infused and control animals.
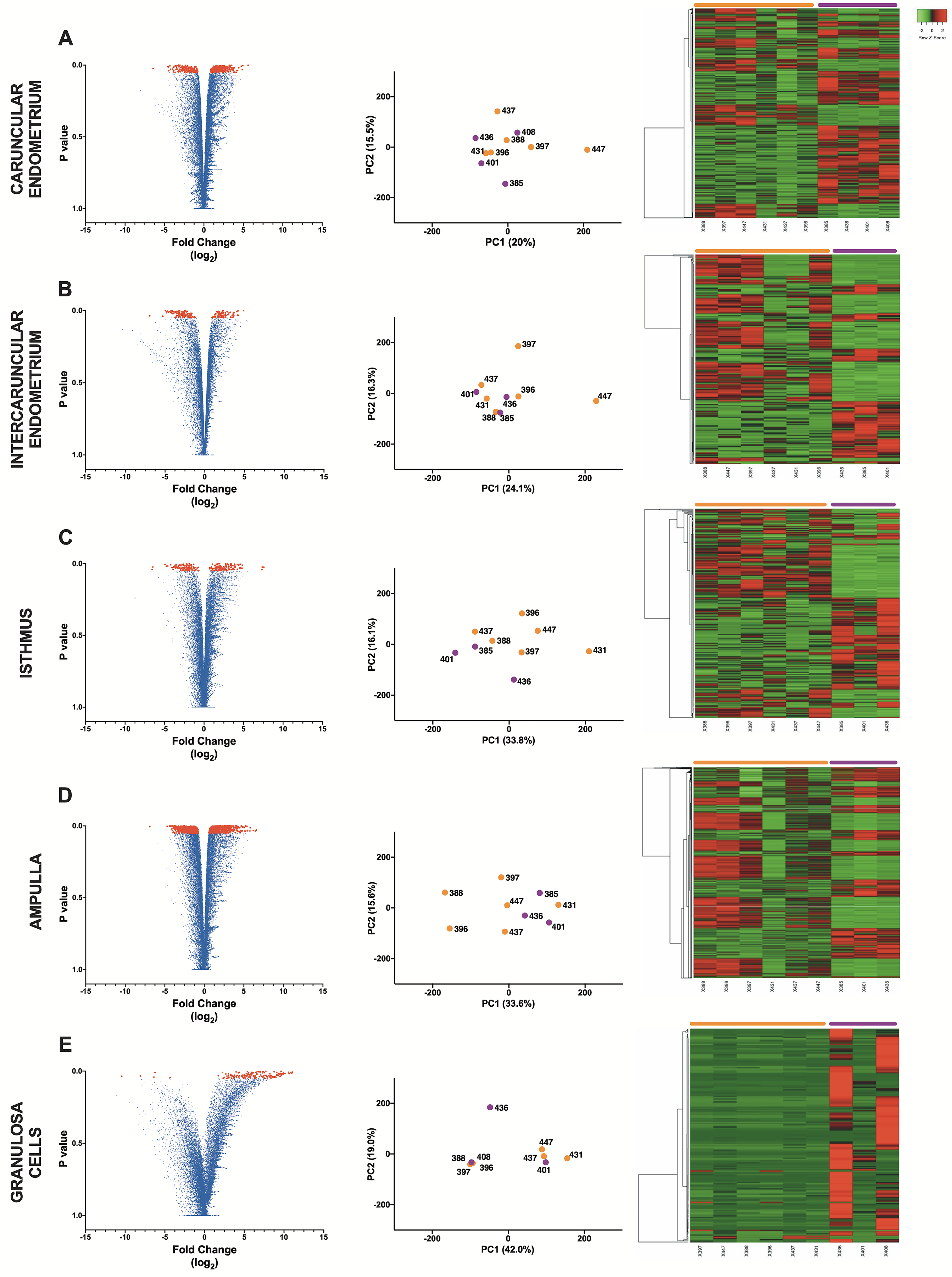
Volcano plots (left panel), principle component analysis plots (middle panel), and heatmap plots (right panel) are presented for (A) caruncular endometrium, (B) intercaruncular endometrium, (C) isthmus, (D) ampulla, and (E) granulosa cells (technical problems led to the exclusion of one bacteria-infused sample from B, C, D and E). The volcano plots present each gene detected by RNAseq, with differentially expressed genes coloured red. Principle component analyses used all the genes detected by RNAseq in each tissue for bacteria-infused (purple) and control animals (orange). Heatmaps present hierarchical clustering of differentially expressed genes, with each column representing an animal and each row a gene; rows were clustered using Euclidian distance and average linkage; gene expression intensities are displayed from green (reduced expression) to red (increased expression) in bacteria-infused (purple) compared with control animals (orange).
Intercaruncular endometrium
produced an average of 32,818 mapped transcripts per sample for analysis, with an average of 32% of the high-quality reads aligned to the Bos taurus reference genome (Supplemental Table 1; due to technical limitations, sample 408IC was not available for analysis). There were 34 upregulated and 43 downregulated DEGs in bacteria-infused animals, as illustrated in a volcano plot (Fig. 1B, left panel). Principle component analysis is presented in Fig. 1B (middle panel), and principal component 1 and 2 explain 24% and 16% of the variance, respectively. A heatmap of the DEGs illustrates the separation between the bacteria-infused and control animals (Fig. 1B, right panel). The five most upregulated DEGs in bacteria-infused animals were CFAP69, CYP11B2, FBXW7, PDP1 and CREM, and the five most downregulated DEGs were VNN2, FCMR, SMIM12, VPREB3 and POU2AF1 (Supplemental Table 2).
Isthmus
produced an average of 29,470 mapped transcripts per sample for analysis, with an average of 38% of the high-quality reads aligned to the Bos taurus reference genome (Supplemental Table 1; due to technical limitations, sample 408I was not available for analysis). There were 45 upregulated and 34 downregulated DEGs in bacteria-infused animals, as illustrated in a volcano plot (Fig. 1C, left panel). Principle component analysis is presented in Fig. 1C (middle panel), and principal component 1 and 2 explain 34% and 16% of the variance, respectively. A heatmap of the DEGs illustrates the separation between the bacteria-infused and control animals (Fig. 1C, right panel). The five most upregulated DEGs in bacteria-infused animals were CXCR1, CXCL8, ATP13A3, UBTF and PROCR, and the five most downregulated DEGs were GLIPR1L1, TNNI3K, CEP41, FZD5 and NXPE3 (Supplemental Table 2).
Ampulla
produced an average of 29,662 mapped transcripts per sample for analysis, with an average of 39% of the high-quality reads aligned to the Bos taurus reference genome (Supplemental Table 1; due to technical limitations, sample 408A was not available for analysis). There were 119 upregulated and 197 downregulated DEGs in bacteria-infused animals, as illustrated in a volcano plot (Fig. 1D, left panel). Principle component analysis is presented in Fig. 1D (middle panel), and principal component 1 and 2 explain 34% and 16% of the variance, respectively. A heatmap of the DEGs illustrates the separation between the bacteria-infused and control animals (Fig. 1D, right panel). The five most upregulated DEGs in bacteria-infused animals were GSG1L, SLP1, KCNE4, ABCC6 and KERA, and the five most downregulated DEGs were USP46, ZDHHC9, ZBTB24, ZNF319 and KIAA0040 (Supplemental Table 2).
Granulosa cells
produced an average of 16,978 mapped transcripts per sample for analysis, with an average of 30% of the high-quality reads aligned to the Bos taurus reference genome (Supplemental Table 1; due to RNA quality limitations, sample 385GC was not available for analysis). There were 88 upregulated DEGs and 1 downregulated DEG in bacteria-infused animals, as illustrated in a volcano plot (Fig. 1E, left panel). Principle component analysis is presented in Fig. 1E (middle panel), and principal component 1 and 2 explain 42% and 19% of the variance, respectively. A heatmap of the DEGs shows a clear separation between the bacteria-infused and control animals (Fig 1E, right panel). The five most upregulated DEGs in bacteria-infused animals were FAM71A, EOMES, ALKAL2, ADAMTS1 and ARHGAP9, and the only downregulated DEG was CARD9 (Supplemental Table 2).
The shared and unique DEGs amongst the tissues are presented in a Venn diagram (Fig. 2). None of the DEGs were common across all the tissues (Fig. 2, central area). Few DEGs were common between specific tissues (Fig. 2, intersecting areas), and most DEGs were unique to each tissue (Fig. 2, non-overlapping areas). There were 109 unique DEGs in the caruncular endometrium, 57 in the intercaruncular endometrium, 65 in the isthmus, 298 in the ampulla, and 83 in granulosa cells. Taken together, the data in Fig. 1 and Fig. 2 provide evidence that there were tissue-specific alterations in the transcriptome of the reproductive tract three months after bacterial infusion.
Figure 2. Common and unique differentially expressed genes amongst reproductive tract tissues.
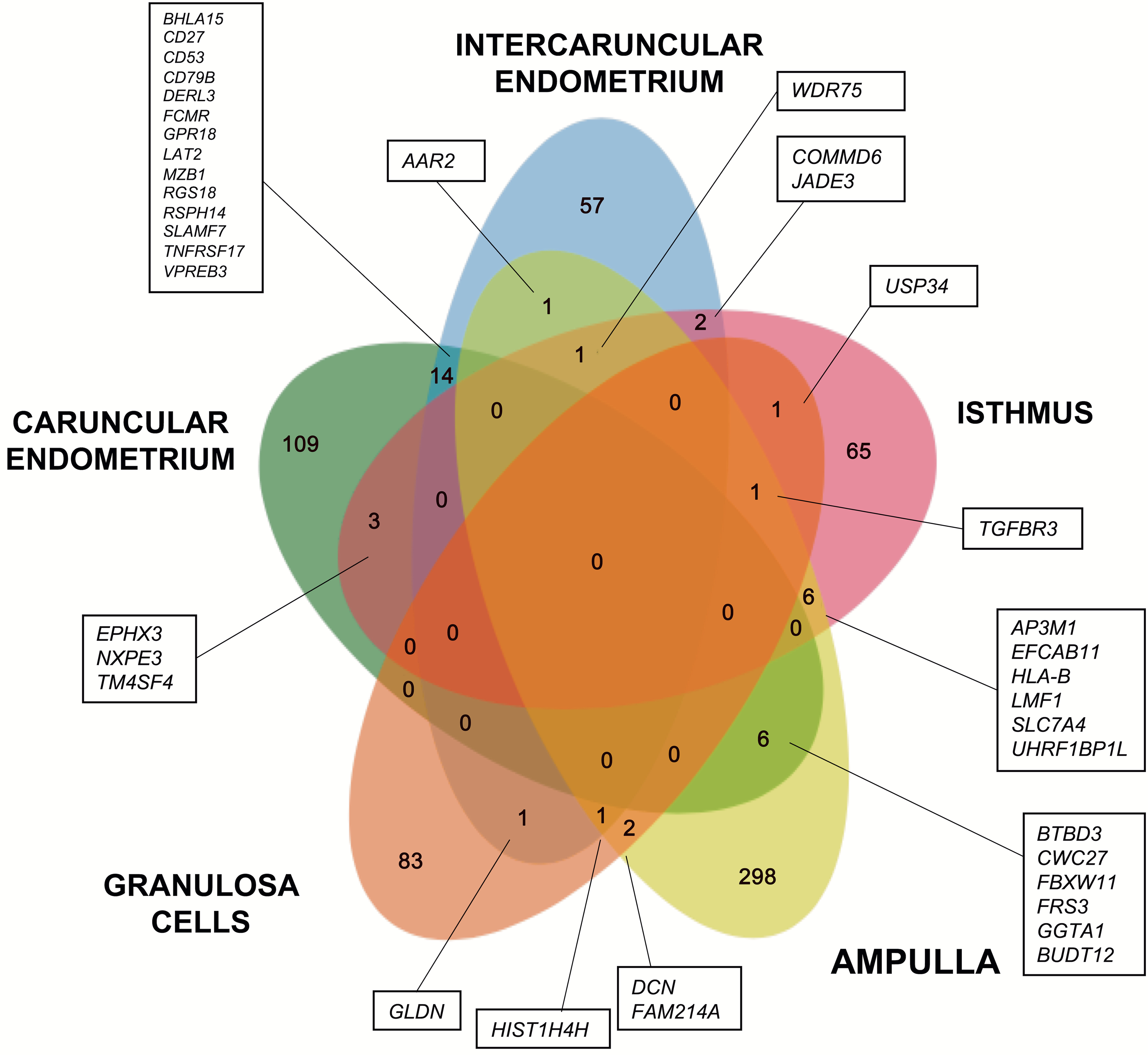
Differentially expressed genes identified in bacteria-infused compared with control animals are shown in a Venn diagram for caruncular endometrium (green), intercaruncular endometrium (blue), isthmus (pink), ampulla (yellow) and granulosa cells (orange). The number of unique differentially expressed genes are reported for each sample and the overlap of common genes are reported at each intersection. The text boxes contain lists of the DEGs for the indicated sectors.
Pathway analysis of differentially expressed genes
Ingenuity Pathway Analysis of DEGs in the bacteria-infused animals identified 18 significantly affected pathways in the caruncular endometrium, 21 in the intercaruncular endometrium, 16 in the isthmus, 24 in the ampulla, and 10 in the granulosa cells (Fig. 3). Amongst these pathways, 5 canonical pathways had z-scores ≤ −2 or ≥ 2, including downregulation of “phospholipase C signalling” in intercaruncular endometrium, upregulation of “3-phosphoinositide biosynthesis” in the caruncular endometrium, and upregulation of the “IL-8 pathway” in the isthmus (Fig. 3).
Figure 3. Ingenuity Pathway Analysis of canonical pathways.
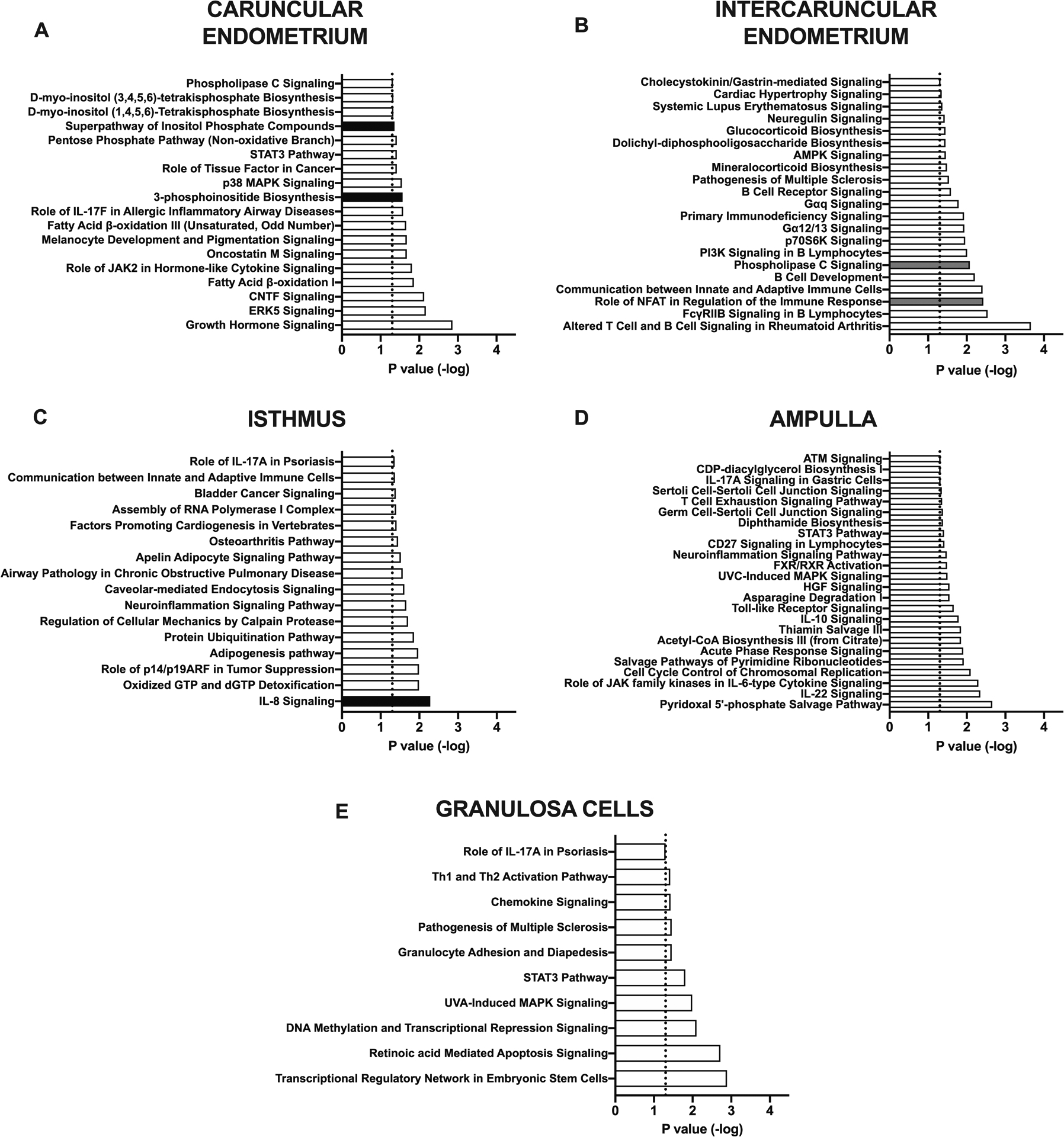
Ingenuity Pathway Analysis of differentially expressed genes identified canonical pathways affected after bacterial infusion in (A) caruncular endometrium, (B) intercaruncular endometrium, (C) isthmus, (D) ampulla, and (E) granulosa cells. Pathways are predicted to be activated (black bars, z-score ≥ 2) or inactivated (grey bars, z-score ≤ −2). White bars represent canonical pathways that were significantly affected (P < 0.05) but did not reach the z-score threshold.
Analysis of gene networks effected by bacterial infusion
Ingenuity Pathway Analysis of DEGs in bacteria-infused animals identified 10 gene networks affected in the caruncular endometrium (Supplemental Table 3) and 8 in the intercaruncular endometrium (Supplemental Table 3). The highest scoring network in both caruncular and intercaruncular endometrium was “cellular development, cellular growth and proliferation and haematological system development and function” (network score = 44 and 36, respectively; Fig. 4A, B). In the isthmus, 8 gene networks were affected (Supplemental Table 3), with “cellular assembly and organization, DNA replication, recombination and repair and cellular development” the highest scoring network (score = 36, Fig. 4C). In the ampulla, 20 gene networks were affected (Supplemental Table 3), with “developmental disorder, post-translational modification, reproductive system development and function” the highest scoring network (score = 48, Fig. 4D). In the granulosa cells, 22 gene networks were affected (Supplemental Table 3), with “endocrine disorders, organ morphology, organismal injury and abnormalities” the highest scoring network (score = 42, Fig. 4E).
Figure 4. Ingenuity Pathway Analysis of gene networks.
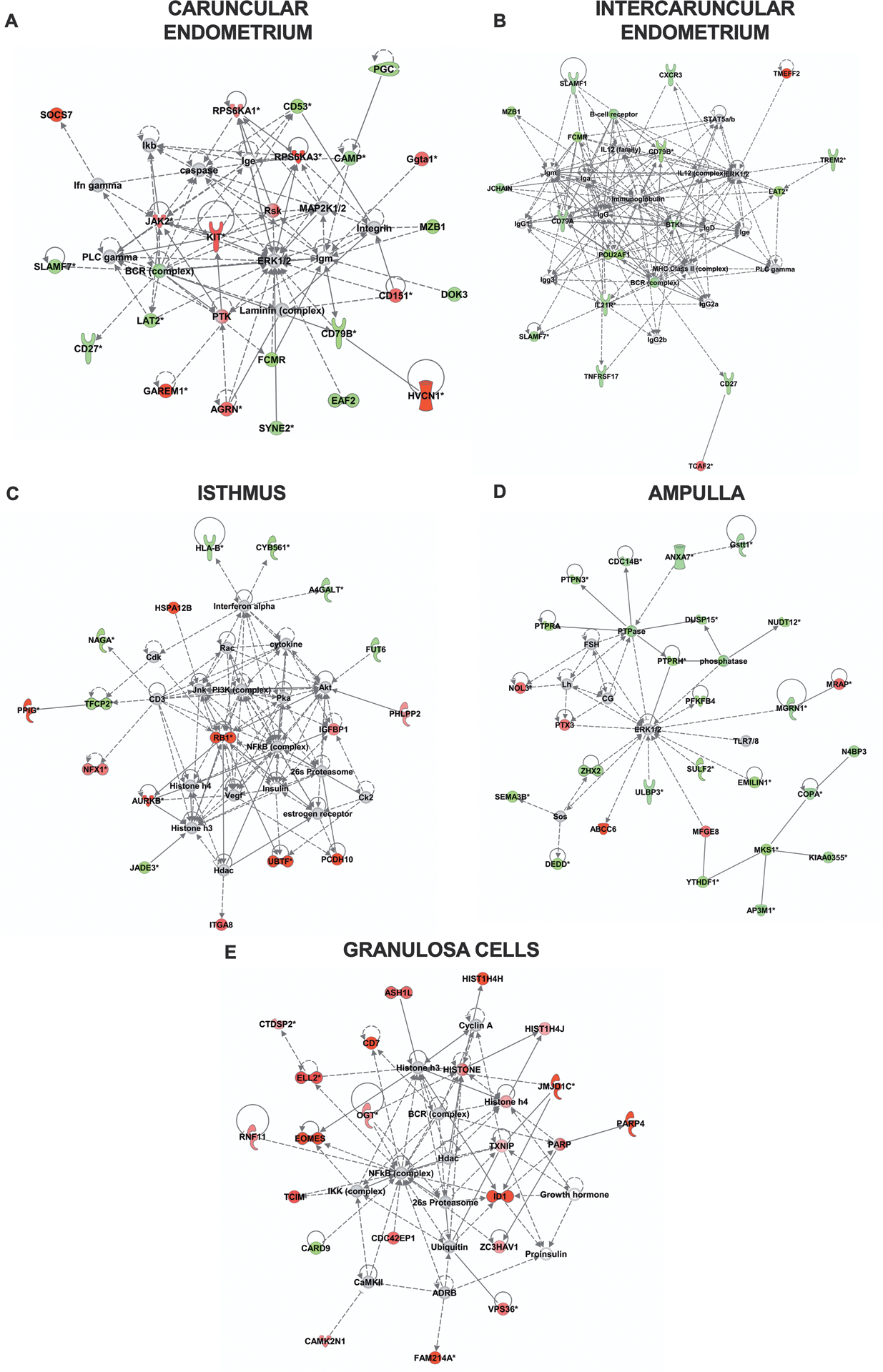
Ingenuity Pathway Analysis of differentially expressed genes identified gene networks affected after bacterial infusion. The highest scoring gene network is presented for (A) caruncular and (B) intercaruncular endometrium: “cellular development, cellular growth and proliferation, haematological system development and function”; (C) isthmus: “cellular assembly and organization, DNA replication, recombination and repair, cellular development”; (D) ampulla: “developmental disorder, post-translational modification, reproductive system development and function”; and, (E) granulosa cells: “cell cycle, DNA replication, recombination and repair”. Gene expression is displayed as green (reduced expression) or red (increased expression) in bacteria-infused animals, compared with control animals; grey indicates genes or molecules that are predicted to be part of the network by Ingenuity Pathway Analysis. Solid lines and dashed lines indicate direct and indirect interactions between nodes, respectively.
Predicted upstream regulators, diseases and functions associated with bacterial infusion
Ingenuity Pathway Analysis of the predicted upstream regulators of DEGs in the bacteria-infused animals are reported in Fig. 5. It was notable that there were 38 upstream regulators in the granulosa cells, including LPS, TLR4, NFκB, IL-6, and IL-1, whereas each of the other tissues only had between 1 and 4 predicted upstream regulators.
Figure 5. Ingenuity Pathway Analysis of predicted upstream regulators.
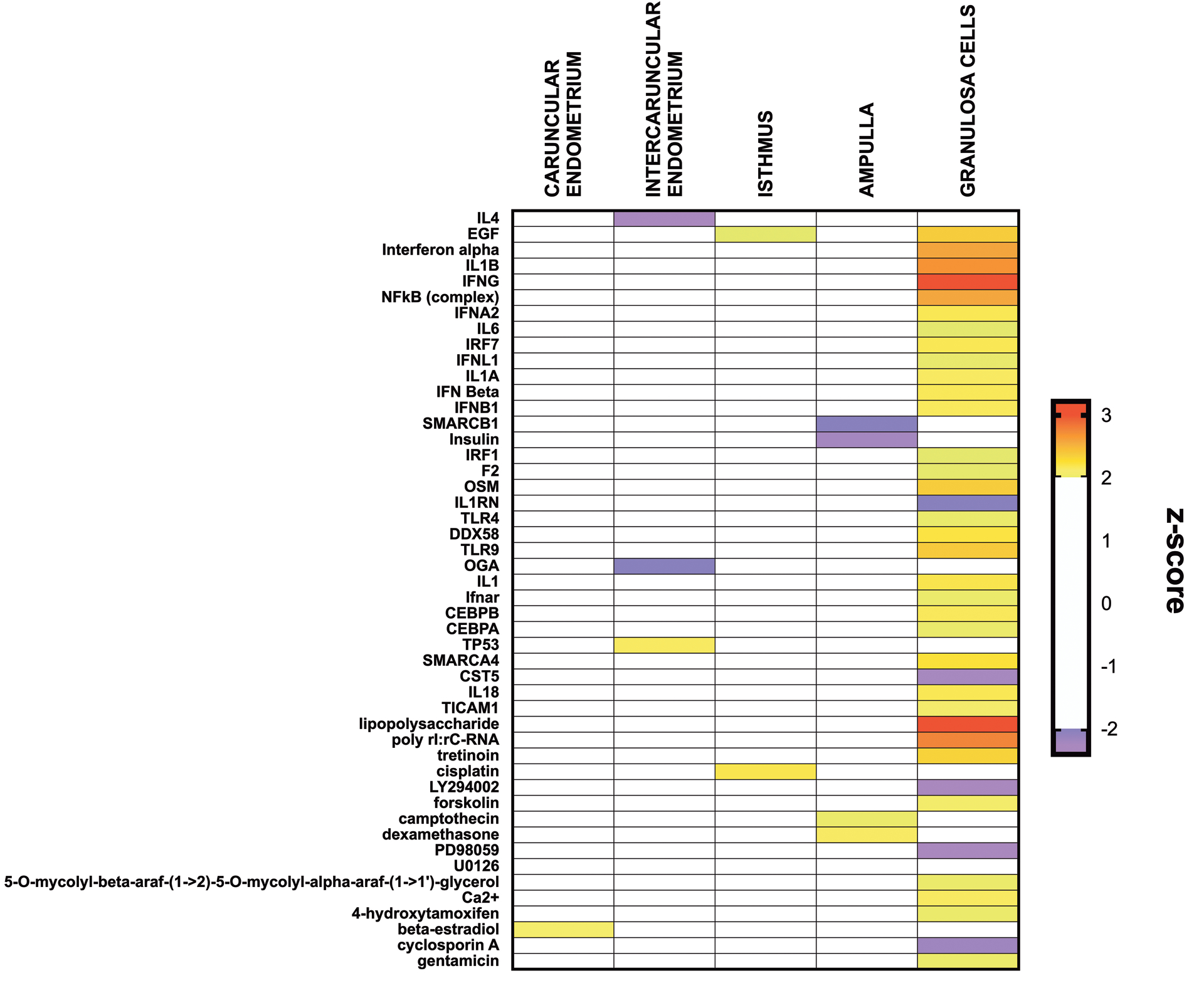
Ingenuity Pathways Analysis identified predicted upstream regulators of differentially expressed genes affected after bacterial infusion in caruncular endometrium, intercaruncular endometrium, isthmus, ampulla, and granulosa cells. The z-score for each regulator is displayed from purple (reduced score) to red (increased score) in bacteria-infused compared with control animals; white blocks represent predicted upstream regulators that did not reach the z-score thresholds.
Ingenuity Pathway Analysis of the predicted diseases and functions associated with the DEGs in the bacteria-infused heifers are reported in Fig. 6. In the caruncular endometrium, functions associated with cancer were predominant with one immune function predicted to be increased, “apoptosis of leukocytes”. In the intercaruncular endometrium, 16 diseases and functions were significant, and 10 were related to inflammation or immune function. However, few significant diseases and functions were identified in the ampulla, and none in the isthmus. In the granulosa cells, 18 disease and functions were significant, and 6 were related to inflammation or immune function.
Figure 6. Ingenuity Pathway Analysis of predicted diseases and functions.
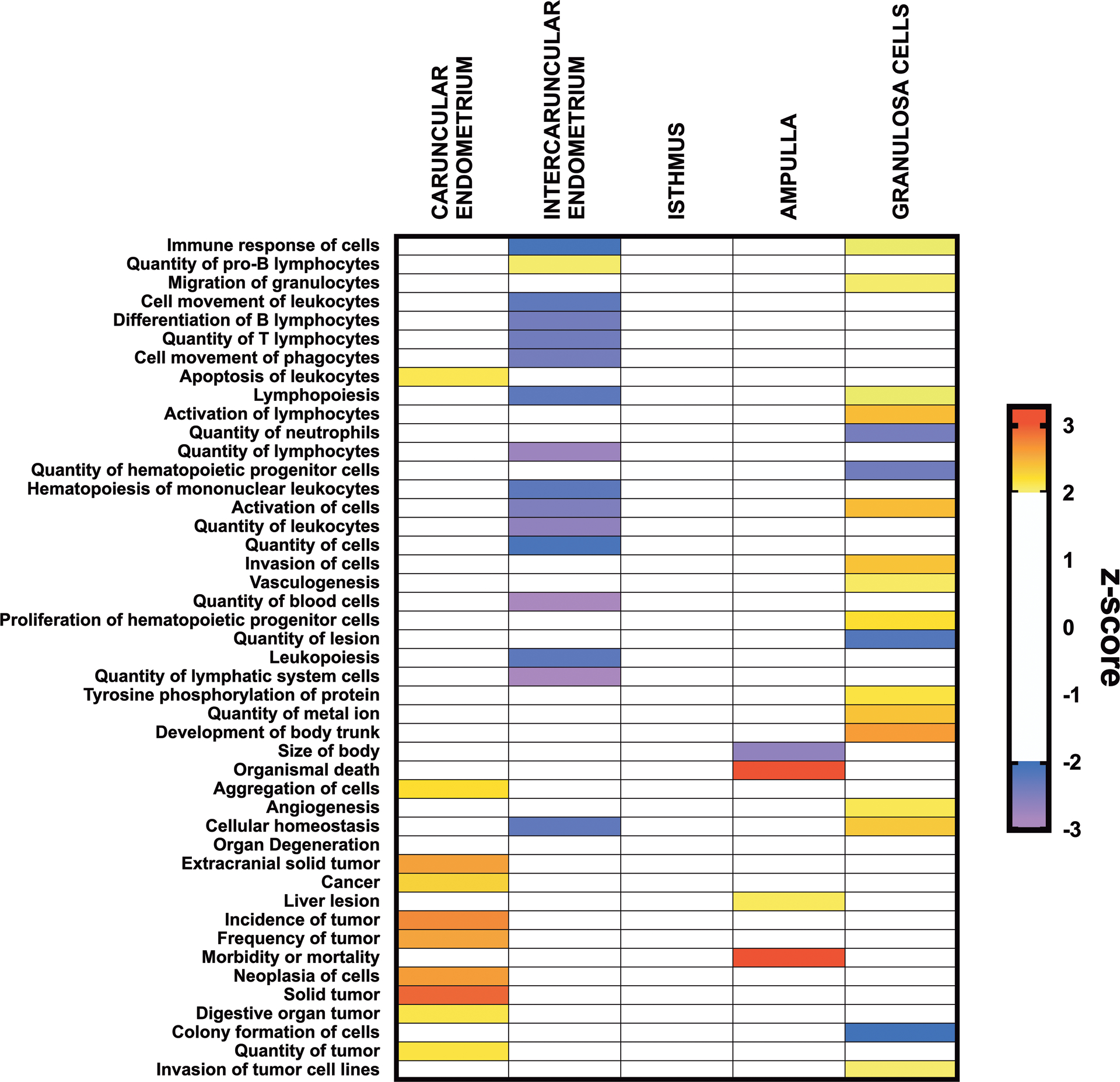
Ingenuity Pathway Analysis of differentially expressed genes identified predicted diseases and functions affected after bacterial infusion in caruncular endometrium, intercaruncular endometrium, isthmus, ampulla, and granulosa cells. The z-score for each disease or function is displayed from purple (reduced score) to red (increased score) in bacteria-infused compared with control animals; white blocks represent predicted upstream regulators that did not meet the z-score thresholds.
Inspection of selected genes involved in inflammation, steroid synthesis and cell viability
Inspection of the RNAseq data for genes associated with inflammation, steroid synthesis, and cell viability, revealed no significant differences between the bacteria-infused and control animals in the caruncular endometrium (Supplementary Table 2) or intercaruncular endometrium (Supplementary Table 2). In the isthmus, as well as the increased expression of CXCL8 (7.36 log2FC, P = 0.02), bacterial infusion was associated with a trend for increased expression of CCL2 (3.47 log2FC, P = 0.1) and IL1A (1.64 log2FC, P = 0.07), compared with control animals (Supplementary Table 2). In the ampulla of bacteria-infused animals, there was increased MHCI expression inreased (2.23 log2FC, P = 0.01) and reduced CASP2 expression (–1.12 log2FC, P = 0.04), and there was a trend for increased expression of CCNB1 (3.25 log2FC, P = 0.09) and CCNB2 (1.12 log2FC, P = 0.07), which encode cyclin B1 and B2, respectively (Supplementary Table 2). In granulosa cells, as well as the reduction in CARD9 expression (–8.09 log2FC, P = 0.03), bacterial infusion was associated with a trend for increased expression of IL1A (4.59 log2FC, P = 0.11) and IL6R (5.61 log2FC, P = 0.08), and reduced expression of HIF1A (–2.17 log2FC, P = 0.10) compared with control (Supplementary Table 2). There was considerable variation in the expression of selected genes amongst the tissues. For example, STAR was highly expressed in granulosa cells but not the other tissues, and IL6R was highly expressed in the endometrium and oviduct but not granulosa cells. It was notable that for both bacteria-infused and control animals there were few transcripts for IL6 or IL10 and there was no expression of TNF across all five tissues.
DISCUSSION
We found that three months after infusion of pathogenic bacteria into the uterus of virgin Holstein heifers there were changes in the transcriptome of the endometrium, oviduct and granulosa cells. The majority of the genes differentially expressed in the bacteria-infused animals were tissue-specific, with few DEGs common amongst the tissues. Many of the gene networks in the endometrium and oviduct that were affected by bacterial infusion are involved in cell growth and repair, rather than infection or inflammation. Despite infusing the bacteria into the uterus, Ingenuity Pathway Analysis revealed more predicted upstream regulators of the DEGs in the granulosa cells of bacteria-infused animals than all the other tissues combined. This is particularly striking because, unlike the other tissues, the granulosa cells were collected from ovarian antral follicles, which would have developed several weeks after the bacterial infusion. Our findings provide evidence that, independent of periparturient problems, lactation and negative energy balance, infecting the uterus with pathogenic bacteria alters the transcriptome of multiple reproductive tract tissues three months later. This includes changes in the transcriptome of granulosa cells, which are distant to the site of infection.
A novelty of our approach was to use virgin Holstein heifers to separate the effects of bacterial infection from other postpartum effects, such as periparturient problems, lactation, and negative energy balance, which can also alter the transcriptome of the reproductive tract (Herath et al. 2009, Wathes et al. 2009, Cerri et al. 2012, Girard et al. 2015). To ensure our bacterial infusion was biologically relevant, we used well-characterised endometrial pathogenic strains of E. coli and T. pyogenes, which had been isolated previously from cows with endometritis (Sheldon et al. 2002, Sheldon et al. 2010, Amos et al. 2014, Goldstone et al. 2014a, Goldstone et al. 2014b). These bacteria induced clinical endometritis in the heifers 3 to 6 days after intrauterine infusion, as determined by transrectal ultrasonographic imaging of pus in the uterus, and using a Metricheck device to detect pus in the vagina, as described in the previous publication (Piersanti et al. 2019b). These clinical signs in the heifers mimic those in dairy cows diagnosed with clinical endometritis about four weeks postpartum using the same techniques of transrectal ultrasonography or a Metricheck device (Sheldon et al. 2006, Sheldon 2020). One consideration is that heifers might have a different immune response to cows. Whether there are differences in immunity between heifers and cows is not fully understood, but age affects cell and antibody-mediated immune responses in dairy heifers (Hine et al. 2011). On the other hand, the innate immune response to E. coli or LPS did not differ significantly between ex vivo endometrium collected from beef heifers and dairy cows (Saut et al. 2014).
The bacterial infusion led to multiple DEGs detectable three months later, in the caruncular and intercaruncular endometrium, isthmus, ampulla, and granulosa cells. Finding DEGs in all the tissues supports the observations that each of these tissues may contribute to infertility after the resolution of uterine disease in cattle (Ribeiro et al. 2016). However, the DEGs in the present study were tissue-specific, with variation amongst the tissues, and no genes common across all the tissues. Furthermore, a striking observation was that there were few RNA transcripts for IL6 or IL10, and no transcripts for TNF, in any of the tissues of bacteria-infused or control animals three months after infusion. This provides evidence that the endometrial infection had resolved by the time of slaughter because it contrasts with the inflammatory response during the first days after exposing the same tissues to bacteria or LPS, when IL6, IL10 and TNF are highly expressed (Herath et al. 2006, Fischer et al. 2010, Gabler et al. 2010, Cronin et al. 2012, Kowsar et al. 2013, Price et al. 2013, Turner et al. 2014). Studies using qPCR also found that whilst there was increased expression of IL6, CXCL8 and TNF transcripts in endometrial samples collected about 2 weeks postpartum, their expression returned to basal levels by 30 to 60 days postpartum (Gabler et al. 2010, Chapwanya et al. 2012).
In the endometrium, the lack of DEGs associated with innate immunity three months after bacterial infusion may reflect resolution of infection, and progression to the repair and maintenance of the tissues. Many significant gene networks and pathways in the endometrium of the animals previously infused with bacteria were typically involved with cellular development, growth and proliferation, although some were associated with inflammation. This may not be surprising as tissue repair, cell growth and inflammation are common transcriptional features of endometrial physiology (Wathes et al. 2011, Chapwanya et al. 2012, Foley et al. 2012, Arai et al. 2013). The 109 unique DEGs in the caruncular endometrium and 57 unique DEGs in the intercaruncular endometrium may also reflect their different physiology. For example, the 3-phosphoinositide biosynthesis pathway was upregulated in caruncular endometrium and the phospholipase C signalling pathway was downregulated in intercaruncular endometrium. Additional evidence for tissue repair was the trend for increased expression in the ampulla of genes encoding cyclin B1 and B2, which help regulate mitosis in proliferating cells (Fung and Poon 2005). However, some of the DEGs also contribute to immunity; VNN2 (Vanin 2) and POU2AF1 (POU Class 2 Homeobox Associating Factor 1) expression was decreased in the intercaruncular endometrium and VPREB3 (Pre-B lymphocyte protein 3) was decreased in both the caruncular and intercaruncular endometrium. Vannin 2 plays a role in leukocyte adhesion and migration to inflammatory sites, VPREB3 is involved in B cell immunoglobulin secretion, and POU2AF1 is essential for B-cell responses to antigens (Suzuki et al. 1999, Rosnet et al. 2004, Corcoran et al. 2005). Interestingly, POU2AF1 was more highly expressed between 7 and 21 days postpartum in Holstein-Friesian cows with clinical endometritis, compared with healthy controls (Foley et al. 2015).
The 65 and 298 DEGs unique to the isthmus and ampulla, respectively, may reflect the different functions of the isthmus, supporting fertilization and early embryonic development, compared with the ampulla, facilitating sperm transport (Maillo et al. 2016). As well as a role in innate immunity and angiogenesis, the predicted activation of IL-8 signalling in the isthmus agrees with observations of increased CXCL8 expression in the human oviduct during the follicular phase of the menstrual cycle (Hess et al. 2013). In the ampulla, there was greater expression of SLPI (secretory leukocyte peptidase inhibitor) in bacteria-infused compared with control animals. As well as a protective role against microbes, SLPI is expressed in the human oviduct, where it protects sperm against the elastase released by neutrophils (Ota et al. 2002). The potassium voltage-gated channel gene (KNCE4) was also upregulated in the ampulla. Potassium channels mediate chloride ion secretion in the oviduct, which is important for the production of oviduct fluid, and dysregulated fluid formation has been linked to reduced fertility (Keating and Quinlan 2012).
The long-term effect of bacterial infusion on the transcriptome was most striking for the granulosa cells derived from antral follicles, because three months earlier these granulosa cells would have been limited to pre-antral follicles (Adams 1999, Britt et al. 2018). Our findings further extend previous observations that 6 weeks after the resolution of postpartum metritis there were changes in the transcriptome of granulosa cells isolated from dominant follicles of lactating dairy cows (Piersanti et al. 2019a). Furthermore, the present study adds knowledge because the effects of infection on the granulosa cell transcriptome were independent of lactation. One potential mechanism for the effect of bacterial infusion on the granulosa cell transcriptome is that around the time of infusion there was an imprinting effect on the cells during primordial to primary follicle development (Adams 1999, Britt et al. 2018). Indeed, there is in vitro evidence that LPS stimulates inappropriate primordial follicle activation (Bromfield and Sheldon 2013). An alternative mechanism is that bacterial virulence factors persisted for several weeks to affect the granulosa cells during antral follicle development. There was evidence for the latter mechanism as the 38 upstream regulators of granulosa cell DEGs included LPS, TLR4, IL-1, and NFκB (nuclear factor kappa-light-chain-enhancer of activated B cells), which are typical of innate immune function (Pahl 1999, Moresco et al. 2011). The latter mechanism is also supported by previous observations that the follicular fluid of antral follicles contains LPS several weeks after uterine infection (Herath et al. 2007, Piersanti et al. 2019a); and, that granulosa cells from antral follicles mount inflammatory responses to LPS (Bromfield and Sheldon 2011, Price et al. 2013, Price and Sheldon 2013).
An unusual feature of the DEGs in the granulosa cells was that only one of the 89 DEGs was downregulated in the bacteria-infused animals compared with control animals. The downregulated gene, CARD9, is a central regulator of innate immunity that is highly expressed in immune cells and is involved in the activation of mitogen-activated protein kinases and NFκB in response to intracellular pathogens (Hsu et al. 2007, Ruland 2008). Upregulated DEGs in the granulosa cells included EOMES (Eomesodermin) and ADAMTS1 (ADAM Metallopeptidase with Thrombospondin Type 1 Motif 1). Eomesodermin is a master regulator of cell-mediated immunity, controlling the production of inflammatory mediators such as interferon-gamma and IL-4 (Shimizu et al. 2019). Eomesodermin is also a component of the pathway that determines the differentiation of human primordial germ cells (Kojima et al. 2017). In granulosa cells, ADAMTS1 expression is induced by luteinizing hormone (Robker et al. 2000), and ADAMTS1–/– mice have impaired follicle development and ovulate fewer oocytes (Mittaz et al. 2004, Shozu et al. 2005).
Our findings of changes in the pattern of the transcriptome across all the tissues imply that infection of the uterus with pathogenic bacteria has widespread and long-term effects on the reproductive tract. These findings agree with observations that the endometrium has a reduced capacity to support an embryo if animals had uterine disease previously (Ribeiro et al. 2016, Gilbert 2019). Further research is needed to complement the findings from the RNAseq analyses and to answer the questions that our findings raise. First, how long do the transcriptome changes persist in the reproductive tract after infection? Second, what are the long-term effects of uterine infection on the reproductive physiology of the animals? Third, which cells and molecules are altered by the long-term effects of uterine infection? These questions are pertinent because there are functional and molecular differences between specific cells, such as the epithelium and stroma, and between different regions of the reproductive tract, such as the glandular and luminal epithelium (Niklaus and Pollard 2006, Spencer et al. 2019). Techniques such as single-cell transcriptomics, immune-localization, and cell sorting will be useful for adding more detail of how infections affect the reproductive tract. Future studies might also extend to the hypothalamus and pituitary, because intravenous LPS infusion suppresses the pulsatile release of GnRH and LH in ruminants, and perturbs ovarian function (Peter et al. 1989, Karsch et al. 2002).
Postpartum uterine infections are a problem for the dairy industry, because they cause infertility, compromise welfare, and reduce profitability (Ribeiro et al. 2016, Gilbert 2019, Sheldon et al. 2019). These infections are likely to continue to be a problem as farmers increase milk yields per cow, to meet the need to feed the expanding world population (Britt et al. 2018). Although uterine infection normally resolves in the first 30 to 60 days following parturition (Sheldon et al. 2009, Gabler et al. 2010, Chapwanya et al. 2012), we found changes in the transcriptome three months after bacterial infusion. Unfortunately, we did not use transrectal ultrasonography or microbiology to monitor the progression of endometritis, but future studies could use these techniques to provide more detail about the relationship between the clinical disease, the pathogens, and the tissue transcriptomes. Whilst postpartum uterine infections are associated with infertility, it is important to recognize that many animals resolve these infections and subsequently conceive. The animal-to-animal variation in DEGs in the present study may reflect this biological variation in the response to infection. Indeed, resilient dairy cows prevent the development of uterine disease using the complementary defensive strategies of tolerating and resisting infection with pathogenic bacteria (Sheldon et al. 2019).
In conclusion, we exploited an infection model in heifers to explore the long-term effects of intrauterine bacterial infusion on the reproductive tract, whilst avoiding the potential confounding effects of periparturient problems, lactation, and negative energy balance. We found that there were changes in the transcriptome of the endometrium, oviduct and granulosa cells three months after bacterial infusion. The majority of the DEGs were tissue-specific, with no genes common across all the tissues. The granulosa cells stood out from the other tissues because they had more predicted upstream regulators of DEGs than all the other tissues combined. The evidence of changes in the transcriptome of the endometrium, oviduct and granulosa cells, imply that long-term changes throughout the reproductive tract could contribute to infertility after bacterial infections of the uterus.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Todd Pritchard, Miguel Torrado and staff at the University of Florida Dairy Research Unit for assistance with animals; Dr Yanping Zhang for assistance with RNAseq; and, Jason Rizo, Paula Molinari and Amanda Teixeira for technical support.
FUNDING
Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health (R01HD084316), and a Swansea University Research Scholarship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DISCLOSURE
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
REFERENCES
- Adams GP 1999. Comparative patterns of follicle development and selection in ruminants. Journal of Reproduction and Fertility Supplement 54 17–32. [PubMed] [Google Scholar]
- Amos MR, Healey GD, Goldstone RJ, Mahan SM, Duvel A, Schuberth HJ, Sandra O, Zieger P, Dieuzy-Labaye I, Smith DG et al. 2014. Differential endometrial cell sensitivity to a cholesterol-dependent cytolysin links Trueperella pyogenes to uterine disease in cattle. Biology of Reproduction 90 54 ( 10.1095/biolreprod.113.115972) [DOI] [PubMed] [Google Scholar]
- Arai M, Yoshioka S, Tasaki Y & Okuda K 2013. Remodeling of bovine endometrium throughout the estrous cycle. Animal Reproduction Science 142 1–9. ( 10.1016/j.anireprosci.2013.08.003) [DOI] [PubMed] [Google Scholar]
- Ayliffe TR & Noakes DE 1982. Effects of exogenous oestrogen and experimentally induced endometritis on absorption of sodium benzylpenicillin from the cow’s uterus. Veterinary Record 110 96–98. () [DOI] [PubMed] [Google Scholar]
- Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A & Wishart DS 2016. Heatmapper: web- enabled heat mapping for all. Nucleic Acids Research 44 W147–W153. ( 10.1093/nar/gkw419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardou P, Mariette J, Escudié F, Djemiel C & Klopp C 2014. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15 293 ( 10.1186/1471-2105-15-293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicalho RC, Machado VS, Bicalho ML, Gilbert RO, Teixeira AG, Caixeta LS & Pereira RV 2010. Molecular and epidemiological characterization of bovine intrauterine Escherichia coli. Journal of Dairy Science 93 5818–5830. ( 10.3168/jds.2010-3550) [DOI] [PubMed] [Google Scholar]
- Bonnett BN, Martin SW, Gannon VP, Miller RB & Etherington WG 1991. Endometrial biopsy in Holstein-Friesian dairy cows. III. Bacteriological analysis and correlations with histological findings. Canadian Journal of Veterinary Research 55 168–173. [PMC free article] [PubMed] [Google Scholar]
- Borsberry S & Dobson H 1989. Periparturient diseases and their effect on reproductive performance in five dairy herds. Veterinary Record 124 217–219. ( 10.1136/vr.124.9.217) [DOI] [PubMed] [Google Scholar]
- Britt JH, Cushman RA, Dechow CD, Dobson H, Humblot P, Hutjens MF, Jones GA, Ruegg PS, Sheldon IM & Stevenson JS 2018. Invited review: Learning from the future-A vision for dairy farms and cows in 2067. Journal of Dairy Science 101 3722–3741. ( 10.3168/jds.2017-14025) [DOI] [PubMed] [Google Scholar]
- Bromfield JJ, Santos JE, Block J, Williams RS & Sheldon IM 2015. PHYSIOLOGY AND ENDOCRINOLOGY SYMPOSIUM: Uterine infection: linking infection and innate immunity with infertility in the high-producing dairy cow. Journal of Animal Science 93 2021–2033. ( 10.2527/jas.2014-8496) [DOI] [PubMed] [Google Scholar]
- Bromfield JJ & Sheldon IM 2011. Lipopolysaccharide initiates inflammation in bovine granulosa cells via the TLR4 pathway and perturbs oocyte meiotic progression in vitro. Endocrinology 152 5029–5040. ( 10.1210/en.2011-1124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromfield JJ & Sheldon IM 2013. Lipopolysaccharide reduces the primordial follicle pool in the bovine ovarian cortex ex vivo and in the murine ovary in vivo. Biology of Reproduction 88 98 ( 10.1095/biolreprod.112.106914) [DOI] [PubMed] [Google Scholar]
- Cerri RLA, Thompson IM, Kim IH, Ealy AD, Hansen PJ, Staples CR, Li JL, Santos JEP & Thatcher WW 2012. Effects of lactation and pregnancy on gene expression of endometrium of Holstein cows at day 17 of the estrous cycle or pregnancy. Journal of Dairy Science 95 5657–5675. () [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagas LM, Bass JJ, Blache D, Burke CR, Kay JK, Lindsay DR, Lucy MC, Martin GB, Meier S, Rhodes FM et al. 2007. Invited review: New perspectives on the roles of nutrition and metabolic priorities in the subfertility of high-producing dairy cows. Journal of Dairy Science 90 4022–4032. ( 10.3168/jds.2006-852) [DOI] [PubMed] [Google Scholar]
- Chapwanya A, Meade KG, Foley C, Narciandi F, Evans AC, Doherty ML, Callanan JJ & O’Farrelly C 2012. The postpartum endometrial inflammatory response: a normal physiological event with potential implications for bovine fertility. Reproduction Fertility and Development 24 1028–1039. ( 10.1071/RD11153) [DOI] [PubMed] [Google Scholar]
- Corcoran LM, Hasbold J, Dietrich W, Hawkins E, Kallies A, Nutt SL, Tarlinton DM, Matthias P & Hodgkin PD 2005. Differential requirement for OBF-1 during antibody-secreting cell differentiation. Journal of Experimental Medicine 201 1385–1396. ( 10.1084/jem.20042325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin JG, Turner ML, Goetze L, Bryant CE & Sheldon IM 2012. Toll-like receptor 4 and MYD88-dependent signaling mechanisms of the innate immune system are essential for the response to lipopolysaccharide by epithelial and stromal cells of the bovine endometrium. Biology of Reproduction 86 51 ( 10.1095/biolreprod.111.092718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer C, Drillich M, Odau S, Heuwieser W, Einspanier R & Gabler C 2010. Selected pro-inflammatory factor transcripts in bovine endometrial epithelial cells are regulated during the oestrous cycle and elevated in case of subclinical or clinical endometritis. Reproduction Fertility and Development 22 818–829. ( 10.1071/RD09120) [DOI] [PubMed] [Google Scholar]
- Foley C, Chapwanya A, Callanan JJ, Whiston R, Miranda-CasoLuengo R, Lu J, Meijer WG, Lynn DJ, O’ Farrelly C & Meade KG 2015. Integrated analysis of the local and systemic changes preceding the development of post-partum cytological endometritis. BMC Genomics 16 811 ( 10.1186/s12864-015-1967-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley C, Chapwanya A, Creevey CJ, Narciandi F, Morris D, Kenny EM, Cormican P, Callanan JJ, O’Farrelly C & Meade KG 2012. Global endometrial transcriptomic profiling: transient immune activation precedes tissue proliferation and repair in healthy beef cows. BMC Genomics 13 489 ( 10.1186/1471-2164-13-489) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TK & Poon RYC 2005. A roller coaster ride with the mitotic cyclins. Seminars in Cell & Developmental Biology 16 335–342. () [DOI] [PubMed] [Google Scholar]
- Gabler C, Fischer C, Drillich M, Einspanier R & Heuwieser W 2010. Time-dependent mRNA expression of selected pro-inflammatory factors in the endometrium of primiparous cows postpartum. Reproductive Biology and Endocrinology 8 152 ( 10.1186/1477-7827-8-152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert RO 2019. Symposium review: Mechanisms of disruption of fertility by infectious diseases of the reproductive tract. Journal of Dairy Science 102 3754–3765. ( 10.3168/jds.2018-15602) [DOI] [PubMed] [Google Scholar]
- Girard A, Dufort I & Sirard M-A 2015. The effect of energy balance on the transcriptome of bovine granulosa cells at 60 days postpartum. Theriogenology 84 1350–1361. () [DOI] [PubMed] [Google Scholar]
- Goldstone RJ, Amos M, Talbot R, Schuberth HJ, Sandra O, Sheldon IM & Smith DG 2014a. Draft genome sequence of Trueperella pyogenes, isolated from the infected uterus of a postpartum cow with metritis. Genome Announcements 2 e00194–14. ( 10.1128/genomeA.00194-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone RJ, Talbot R, Schuberth HJ, Sandra O, Sheldon IM & Smith DG 2014b. Draft genome sequence of Escherichia coli MS499, isolated from the infected uterus of a postpartum cow with metritis. Genome Announcements 2 e00217–14. ( 10.1128/genomeA.00217-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JFT, Hartigan PJ & Nunn WR 1974. Non-specific uterine infection and bovine fertility. I. Infection patterns and endometritis during the first seven weeks post-partum. Theriogenology 1 91–106. ( 10.1016/0093-691x(74)90052-1) [DOI] [PubMed] [Google Scholar]
- Hatzirodos N, Irving-Rodgers HF, Hummitzsch K, Harland ML, Morris SE & Rodgers RJ 2014. Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes. BMC Genomics 15 24 ( 10.1186/1471-2164-15-24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy LL, Cronin JG & Sheldon IM 2014. Endometrial cells sense and react to tissue damage during infection of the bovine endometrium via interleukin 1. Scientific Reports 4 7060 ( 10.1038/srep07060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herath S, Fischer DP, Werling D, Williams EJ, Lilly ST, Dobson H, Bryant CE & Sheldon IM 2006. Expression and function of Toll-like receptor 4 in the endometrial cells of the uterus. Endocrinology 147 562–570. ( 10.1210/en.2005-1113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herath S, Lilly ST, Santos NR, Gilbert RO, Goetze L, Bryant CE, White JO, Cronin J & Sheldon IM 2009. Expression of genes associated with immunity in the endometrium of cattle with disparate postpartum uterine disease and fertility. Reproductive Biology and Endocrinology 7 55 ( 10.1186/1477-7827-7-55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herath S, Williams EJ, Lilly ST, Gilbert RO, Dobson H, Bryant CE & Sheldon IM 2007. Ovarian follicular cells have innate immune capabilities that modulate their endocrine function. Reproduction 134 683–693. ( 10.1530/REP-07-0229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess AP, Talbi S, Hamilton AE, Baston-Buest DM, Nyegaard M, Irwin JC, Barragan F, Kruessel JS, Germeyer A & Giudice LC 2013. The human oviduct transcriptome reveals an anti-inflammatory, anti-angiogenic, secretory and matrix-stable environment during embryo transit. Reproductive Biomedicine Online 27 423–435. ( 10.1016/j.rbmo.2013.06.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hine BC, Cartwright SL & Mallard BA 2011. Effect of age and pregnancy status on adaptive immune responses of Canadian Holstein replacement heifers. Journal of Dairy Science 94 981–991. () [DOI] [PubMed] [Google Scholar]
- Hsu YM, Zhang Y, You Y, Wang D, Li H, Duramad O, Qin XF, Dong C & Lin X 2007. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nature Immunology 8 198–205. ( 10.1038/ni1426) [DOI] [PubMed] [Google Scholar]
- Huszenicza G, Fodor M, Gacs M, Kulcsar M, Dohmen MJW, Vamos M, Porkolab L, Legl T, Bartyik J, Lohuis JACM et al. 1991. Uterine bacteriology, resumption of cyclic ovarian activity and fertility in postpartum cows kept in large-scale dairy herds. Reproduction in Domestic Animals 34 237–245. ( 10.1111/j.1439-0531.1999.tb01246.x) [DOI] [Google Scholar]
- Ibrahim S, Salilew-Wondim D, Rings F, Hoelker M, Neuhoff C, Tholen E, Looft C, Schellander K & Tesfaye D 2015. Expression pattern of inflammatory response genes and their regulatory microRNAs in bovine oviductal cells in response to lipopolysaccharide: implication for early embryonic development. PLoS One 10 e0119388 ( 10.1371/journal.pone.0119388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsch FJ, Battaglia DF, Breen KM, Debus N & Harris TG 2002. Mechanisms for ovarian cycle disruption by immune/inflammatory stress. Stress 5 101–112. ( 10.1080/10253890290027868) [DOI] [PubMed] [Google Scholar]
- Keating N & Quinlan LR 2012. Small conductance potassium channels drive ATP-activated chloride secretion in the oviduct. American Journal of Physiology - Cell Physiology 302 C100–109. ( 10.1152/ajpcell.00503.2010) [DOI] [PubMed] [Google Scholar]
- Kojima Y, Sasaki K, Yokobayashi S, Sakai Y, Nakamura T, Yabuta Y, Nakaki F, Nagaoka S, Woltjen K, Hotta A et al. 2017. Evolutionarily distinctive transcriptional and signaling programs drive human germ cell lineage specification from pluripotent stem cells. Cell Stem Cell 21 517–532. () [DOI] [PubMed] [Google Scholar]
- Kong B-W, Hudson N, Seo D, Lee S, Khatri B, Lassiter K, Cook D, Piekarski A, Dridi S, Anthony N et al. 2017. RNA sequencing for global gene expression associated with muscle growth in a single male modern broiler line compared to a foundational Barred Plymouth Rock chicken line. BMC Genomics 18 82 ( 10.1186/s12864-016-3471-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowsar R, Hambruch N, Liu J, Shimizu T, Pfarrer C & Miyamoto A 2013. Regulation of innate immune function in bovine oviduct epithelial cells in culture: the homeostatic role of epithelial cells in balancing Th1/Th2 response. Journal of Reproduction and Development 59 470–478. ( 10.1262/jrd.2013-036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Green J, Pollard J & Tugendreich S 2014. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30 523–530. ( 10.1093/bioinformatics/btt703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B & Salzberg SL, 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9 357–359. ( 10.1038/nmeth.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc SJ 2012. Interactions of metabolism, inflammation, and reproductive tract health in the postpartum period in dairy cattle. Reproduction in Domestic Animals 47 Suppl 5 18–30. ( 10.1111/j.1439-0531.2012.02109.x) [DOI] [PubMed] [Google Scholar]
- LeBlanc SJ, Duffield TF, Leslie KE, Bateman KG, Keefe GP, Walton JS & Johnson WH 2002. Defining and diagnosing postpartum clinical endometritis and its impact on reproductive performance in dairy cows. Journal of Dairy Science 85 2223–2236. ( 10.3168/jds.S0022-0302(02)74302-6) [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R & 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 2078–2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima FS, Greco LF, Bisinotto RS, Ribeiro ES, Martinez NM, Thatcher WW, Santos JEP, Reinhard MK & Galvão KN 2015. Effects of intrauterine infusion of Trueperella pyogenes on endometrial mRNA expression of proinflammatory cytokines and luteolytic cascade genes and their association with luteal life span in dairy cows. Theriogenology 84 1263–1272. () [DOI] [PubMed] [Google Scholar]
- Lima FS, Ribeiro ES, Bisinotto RS, Greco LF, Martinez N, Amstalden M, Thatcher WW & Santos JE 2013. Hormonal manipulations in the 5-day timed artificial insemination protocol to optimize estrous cycle synchrony and fertility in dairy heifers. Journal of Dairy Science 96 7054–7065. ( 10.3168/jds.2013-7093) [DOI] [PubMed] [Google Scholar]
- Locatelli Y, Forde N, Blum H, Graf A, Piégu B, Mermillod P, Wolf E, Lonergan P & Saint-Dizier M 2019. Relative effects of location relative to the corpus luteum and lactation on the transcriptome of the bovine oviduct epithelium. BMC Genomics 20 233 ( 10.1186/s12864-019-5616-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillo V, de Frutos C, O’Gaora P, Forde N, Burns GW, Spencer TE, Gutierrez-Adan A, Lonergan P & Rizos D 2016. Spatial differences in gene expression in the bovine oviduct. Reproduction 152 37–46. ( 10.1530/REP-16-0074) [DOI] [PubMed] [Google Scholar]
- Marcel M 2011. Cutadapt removes adapter sequences from high-throughput sequence reads. EMBnet.journal 17 10–12. ( 10.14806/ej.17.1.200) [DOI] [Google Scholar]
- Metsalu T & Vilo J 2015. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Research 43 W566–W570. ( 10.1093/nar/gkv468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittaz L, Russell DL, Wilson T, Brasted M, Tkalcevic J, Salamonsen LA, Hertzog PJ & Pritchard MA 2004. Adamts-1 is essential for the development and function of the urogenital system. Biology of Reproduction 70 1096–1105. ( 10.1095/biolreprod.103.023911) [DOI] [PubMed] [Google Scholar]
- Moresco EM, LaVine D & Beutler B 2011. Toll-like receptors. Current Biology 21 R488–R493. ( 10.1016/j.cub.2011.05.039) [DOI] [PubMed] [Google Scholar]
- Niklaus AL & Pollard JW 2006. Mining the mouse transcriptome of receptive endometrium reveals distinct molecular signatures for the luminal and glandular epithelium. Endocrinology 147 3375–3390. ( 10.1210/en.2005-1665) [DOI] [PubMed] [Google Scholar]
- Ota Y, Shimoya K, Zhang Q, Moriyama A, Chin R, Tenma K, Kimura T, Koyama M, Azuma C & Murata Y 2002. The expression of secretory leukocyte protease inhibitor (SLPI) in the Fallopian tube: SLPI protects the acrosome reaction of sperm from inhibitory effects of elastase. Human Reproduction 17 2517–2522. ( 10.1093/humrep/17.10.2517) [DOI] [PubMed] [Google Scholar]
- Pahl HL 1999. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18 6853–6866. ( 10.1038/sj.onc.1203239) [DOI] [PubMed] [Google Scholar]
- Peter AT, Bosu WT & DeDecker RJ 1989. Suppression of preovulatory luteinizing hormone surges in heifers after intrauterine infusions of Escherichia coli endotoxin. American Journal of Veterinary Research 50 368–373. [PubMed] [Google Scholar]
- Piersanti RL, Horlock AD, Block J, Santos JEP, Sheldon IM & Bromfield JJ 2019a. Persistent effects on bovine granulosa cell transcriptome after resolution of uterine disease. Reproduction 158 35–46. ( 10.1530/REP-19-0037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersanti RL, Zimpel R, Molinari PCC, Dickson MJ, Ma Z, Jeong KC, Santos JEP, Sheldon IM & Bromfield JJ 2019b. A model of clinical endometritis in Holstein heifers using pathogenic Escherichia coli and Trueperella pyogenes. Journal of Dairy Science 102 2686–2697. ( 10.3168/jds.2018-15595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JC, Bromfield JJ & Sheldon IM 2013. Pathogen-associated molecular patterns initiate inflammation and perturb the endocrine function of bovine granulosa cells from ovarian dominant follicles via TLR2 and TLR4 pathways. Endocrinology 154 3377–3386. ( 10.1210/en.2013-1102) [DOI] [PubMed] [Google Scholar]
- Price JC & Sheldon IM 2013. Granulosa cells from emerged antral follicles of the bovine ovary initiate inflammation in response to bacterial pathogen-associated molecular patterns via Toll-like receptor pathways. Biology of Reproduction 89 119 ( 10.1095/biolreprod.113.110965) [DOI] [PubMed] [Google Scholar]
- Quackenbush J 2001. Computational analysis of microarray data. Nature Reviews Genetics 2 418–427. ( 10.1038/35076576) [DOI] [PubMed] [Google Scholar]
- Ribeiro ES, Gomes G, Greco LF, Cerri RLA, Vieira-Neto A, Monteiro PLJ, Lima FS, Bisinotto RS, Thatcher WW & Santos JEP 2016. Carryover effect of postpartum inflammatory diseases on developmental biology and fertility in lactating dairy cows. Journal of Dairy Science 99 2201–2220. ( 10.3168/jds.2015-10337) [DOI] [PubMed] [Google Scholar]
- Robker RL, Russell DL, Espey LL, Lydon JP, O’Malley BW & Richards JS 2000. Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proceedings of the National Academy of Sciences of the United States of America 97 4689–4694. ( 10.1073/pnas.080073497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnet O, Blanco-Betancourt C, Grivel K, Richter K & Schiff C 2004. Binding of free immunoglobulin light chains to VpreB3 inhibits their maturation and secretion in chicken B cells. Journal of Biological Chemistry 279 10228–10236. ( 10.1074/jbc.M312169-A200) [DOI] [PubMed] [Google Scholar]
- Rowson LEA, Lamming GE & Fry RM 1953. The relationship between ovarian hormones and uterine infection. Veterinary Record 65 335–340. [Google Scholar]
- Ruland J 2008. CARD9 signaling in the innate immune response. Annals of the New York Academy of Sciences 1143 35–44. ( 10.1196/annals.1443.024) [DOI] [PubMed] [Google Scholar]
- Sanchez JM, Passaro C, Forde N, Browne JA, Behura SK, Fernandez-Fuertes B, Mathew DJ, Kelly AK, Butler ST, Spencer TE et al. 2019. Do differences in the endometrial transcriptome between uterine horns ipsilateral and contralateral to the corpus luteum influence conceptus growth to day 14 in cattle? Biology of Reproduction 100 86–100. ( 10.1093/biolre/ioy185) [DOI] [PubMed] [Google Scholar]
- Saut JP, Healey GD, Borges AM & Sheldon IM 2014. Ovarian steroids do not affect bovine endometrial cytokine or chemokine responses to Escherichia coli or LPS in vitro. Reproduction 148 593–606. ( 10.1530/rep-14-0230) [DOI] [PubMed] [Google Scholar]
- Semambo DK, Ayliffe TR, Boyd JS & Taylor DJ 1991. Early abortion in cattle induced by experimental intrauterine infection with pure cultures of Actinomyces pyogenes. Veterinary Record 129 12–16. ( 10.1136/vr.129.1.12) [DOI] [PubMed] [Google Scholar]
- Sheldon IM 2020. Diagnosing postpartum endometritis in dairy cattle. Veterinary Record 186 88–90. ( 10.1136/vr.m222) [DOI] [PubMed] [Google Scholar]
- Sheldon IM, Cronin J, Goetze L, Donofrio G & Schuberth HJ 2009. Defining postpartum uterine disease and the mechanisms of infection and immunity in the female reproductive tract in cattle. Biology of Reproduction 81 1025–1032. ( 10.1095/biolreprod.109.077370) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon IM, Cronin JG & Bromfield JJ 2019. Tolerance and innate immunity shape the development of postpartum uterine disease and the impact of endometritis in dairy cattle. Annual Review of Animal Biosciences 7 361–384. ( 10.1146/annurev-animal-020518-115227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon IM, Lewis GS, LeBlanc S & Gilbert RO 2006. Defining postpartum uterine disease in cattle. Theriogenology 65 1516–1530. ( 10.1016/j.theriogenology.2005.08.021) [DOI] [PubMed] [Google Scholar]
- Sheldon IM, Noakes DE, Rycroft AN, Pfeiffer DU & Dobson H 2002. Influence of uterine bacterial contamination after parturition on ovarian dominant follicle selection and follicle growth and function in cattle. Reproduction 123 837–845. ( 10.1530/rep.0.1230837) [DOI] [PubMed] [Google Scholar]
- Sheldon IM, Rycroft AN, Dogan B, Craven M, Bromfield JJ, Chandler A, Roberts MH, Price SB, Gilbert RO & Simpson KW 2010. Specific strains of Escherichia coli are pathogenic for the endometrium of cattle and cause pelvic inflammatory disease in cattle and mice. PLoS One 5 e9192 ( 10.1371/journal.pone.0009192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Sato Y, Kawamura M, Nakazato H, Watanabe T, Ohara O & Fujii SI 2019. Eomes transcription factor is required for the development and differentiation of invariant NKT cells. Communications Biology 2 150 ( 10.1038/s42003-019-0389-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shozu M, Minami N, Yokoyama H, Inoue M, Kurihara H, Matsushima K & Kuno K 2005. ADAMTS-1 is involved in normal follicular development, ovulatory process and organization of the medullary vascular network in the ovary. Journal of Molecular Endocrinology 35 343–355. ( 10.1677/jme.1.01735) [DOI] [PubMed] [Google Scholar]
- Spencer TE, Kelleher AM & Bartol FF 2019. Development and function of uterine glands in domestic animals. Annual Review of Animal Biosciences 7 125–147. ( 10.1146/annurev-animal-020518-115321) [DOI] [PubMed] [Google Scholar]
- Suzuki K, Watanabe T, Sakurai S, Ohtake K, Kinoshita T, Araki A, Fujita T, Takei H, Takeda Y, Sato Y et al. 1999. A novel glycosylphosphatidyl inositol-anchored protein on human leukocytes: a possible role for regulation of neutrophil adherence and migration. Journal of Immunology 162 4277–4284. [PubMed] [Google Scholar]
- Turner ML, Cronin JG, Healey GD & Sheldon IM 2014. Epithelial and stromal cells of bovine endometrium have roles in innate immunity and initiate inflammatory responses to bacterial lipopeptides in vitro via Toll-like receptors TLR2, TLR1, and TLR6. Endocrinology 155 1453–1465. ( 10.1210/en.2013-1822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathes DC, Cheng Z, Chowdhury W, Fenwick MA, Fitzpatrick R, Morris DG, Patton J & Murphy JJ 2009. Negative energy balance alters global gene expression and immune responses in the uterus of postpartum dairy cows. Physiological Genomics 39 1–13. ( 10.1152/physiolgenomics.00064.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathes DC, Cheng Z, Fenwick MA, Fitzpatrick R & Patton J 2011. Influence of energy balance on the somatotrophic axis and matrix metalloproteinase expression in the endometrium of the postpartum dairy cow. Reproduction 141 269–281. ( 10.1530/REP-10-0177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Drillich M, Kaufmann TB, Madoz LV & Heuwieser W 2010. A clinical approach to determine false positive findings of clinical endometritis by vaginoscopy by the use of uterine bacteriology and cytology in dairy cows. Theriogenology 74 1248–1255. ( 10.1016/j.theriogenology.2010.05.028) [DOI] [PubMed] [Google Scholar]
- Yao JQ & Yu F 2011. DEB: A web interface for RNA-seq digital gene expression analysis. Bioinformation 7 44–45. ( 10.6026/97320630007044) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


