Abstract
Oxidation of cardiolipin (CL) by cytochrome c (cytc) has been proposed to initiate the intrinsic pathway of apoptosis. Domain-swapped dimer (DSD) conformations of cytc have been reported both by our laboratory and others. The DSD is an alternate conformer of cytc that could oxygenate CL early in apoptosis. We demonstrate here that the cytc DSD has a set of properties that would provide tighter regulation of the intrinsic pathway. We show that the human DSD is kinetically more stable than horse and yeast DSDs. Circular dichroism data indicate that the DSD has a less asymmetric heme environment, similar to that seen when the monomeric protein binds to CL vesicles at high lipid-to-protein ratios. The dimer undergoes the alkaline conformational transition near pH 7.0, 2.5 pH units lower than that of the monomer. Data from fluorescence correlation spectroscopy (FCS) and fluorescence anisotropy suggest that the alkaline transition of the DSD may act as a switch from high affinity for CL nanodiscs at pH 7.4 to much lower affinity at pH 8.0. Additionally, the peroxidase activity of the human DSD increases seven-fold compared to the monomer at pH 7 and 8, but by 14-fold at pH 6 when mixed Met80/H2O ligation replaces the lysine ligation of the alkaline state. We also present data that indicate that cytc binding shows a cooperative effect as the concentration of cytc is increased. The DSD appears to have evolved into a pH-inducible switch that provides a means to control activation of apoptosis near pH 7.0.
Graphical Abstract
INTRODUCTION
Cytochrome c (cytc) is a soluble heme protein known for its role in the electron transport chain, shuttling electrons between CoQH2-cytochrome c reductase (Complex 3) and cytochrome c oxidase (Complex 4).1–4 However, cytc has been classified as an extreme multifunctional or “moonlighting” protein because it is involved in other cellular processes besides electron transport.5 Prominent among these moonlighting functions is its role as an initiator of the intrinsic apoptotic pathway.6–9 In this role, cytc migrates from the intermembrane space (IMS) of the mitochondria to the cytosol interacting with apoptotic protease activating factor 1 to form the apoptosome and initiate apoptosis.10 When cytc, which has a +8 net charge at neutral pH, binds the anionic phospholipid cardiolipin (CL), it gains peroxidase activity and oxidizes CL.11–13 The oxidation of CL by cytc has been implicated in permeabilizing the outer mitochondrial membrane (OMM) and in facilitating the release of cytc into the cytosol because of its weaker binding to oxidized CL.11, 14, 15
CL, a potent activator of cytc’s peroxidase activity,16 comprises ~25% of the lipids of the inner mitochondrial membrane.17 Under normal conditions, CL is not present in the OMM.18 Cytc’s function in the electron transport chain is not affected by cytc-CL interaction because there is lower CL availability in the IMS and the local ionic strength is relatively high (150 mM), which inhibits binding.16, 19 Preceding apoptosis, there is migration of CL to the OMM.12, 20–23 Proapoptotic factors, including cytc, are then released from the mitochondria as CL redistribution triggers lipid peroxidation and permeabilization of the OMM.24, 25
An increasing number of domain-swapped protein dimer structures continue to be identified and are believed to have biological roles.26 It has been shown that yeast27 and equine28 cytc can form a domain-swapped dimer (DSD). In the yeast and horse structures, the hinge region between the two subunits is derived from the highly conserved Ω-loop D (residues 70–85).29 This loop acts as the gate-keeper to the heme, providing Met80 to the sixth coordination site of the heme.30 The dynamics of this loop also are critical for providing access to conformers that are competent for peroxidase activity.5 In these crystal structures, the heme is solvent exposed and available for peroxidase activity since the repositioned Ω-loop D leads to a loss of Met80 coordination. We have published DSD structures with lipid surrogates inhabiting a hydrophobic tube connecting the aqueous environment to the heme.27 The binding site is located near Asn52, which was proposed to hydrogen bond to protonated CL31 as part of the C-site for CL binding to cytc. Although recent studies show that CL does not have a protonation equilibrium in the pH range corresponding to the C-site,32, 33 these DSD X-ray structures with lipid bound in a well-defined cavity support the extended lipid anchorage model for cytc/CL binding that was an essential component of the C-site.31 Through docking studies based on the detergent bound DSD structures, we have shown that linoleic acid, the predominate fatty acyl chain found in CL,34 readily fits into this binding site for peroxidase activity.27
There is increasing evidence that DSDs of cytc can form in vivo and thus could be physiologically relevant. The Hirota lab has shown that Hydrogenobacter thermophilus cytochrome c552 can form DSDs and higher order oligomers in vivo when expressed in Escherichia coli.35 The cytoplasmic membrane of E. coli is composed of 20% lipids with negatively-charged headgroups (CL and phosphatidylglycerol).36 In a more recent study, the Hirota lab showed that positively-charged cytochromes c formed DSDs when expressed in E. coli, whereas negatively-charged cytochromes c did not, strongly suggesting that electrostatic interaction with the cellular membrane can catalyze (and is required for) formation of DSDs in vivo.37 Studies on the formation of DSDs during folding of negatively- and positively-charged cytochromes c, in the presence of negatively-charged liposomes provided additional support for this conclusion.37 In vitro formation of DSDs normally requires treatment with ethanol or detergent.28, 38–40 However, crystalized DSD variants have formed directly from monomeric cytc during crystallization,27 and it has been shown that dimeric cytc will form spontaneously, albeit slowly, from a monomeric starting material in solution.27 DSD of cytc can also form during folding in vitro.41 Thus, it is possible that they could also form during folding in vivo.
Ω-loop D is one of the least stable substructures of cytc,42 and thus is a likely mediator of conformational rearrangements of the protein. Interestingly, rearrangement of Ω-loop D of each subunit of the DSD causes the C-terminal helix to splay out from the rest of the polypeptide chain (Figure S1),27, 28 similar to the structure proposed for the extended conformer of cytc at high lipid-to-protein ratios by the Pletneva lab.43 Thus, the extended conformer observed by the Pletneva lab could facilitate formation of the DSD on CL-containing membrane surfaces. This conformational rearrangement also opens up the heme site allowing for increased peroxidase activity.43 The equine cytc-DSD has been shown to have a five-fold increase in peroxidase activity when compared with the monomer.44 The rearrangement of Ω-loop D results in a larger heme cavity and faster formation of Compound I.44 Compound I, the oxoferryl porphyrin π-cation radical, is the initial intermediate of the peroxidase reaction45, 46 and its formation is the rate determining step in the peroxidase reaction of the DSD of cytc.44
Herein, we present human cytc DSD data that support a possible functional role as a switch evolved for controlling the initiation of the intrinsic pathway of apoptosis. Dimer dissociation data show that the human DSD is more kinetically stable than the yeast and horse DSDs. Data for the alkaline conformational transition show that the midpoint pH, pH1/2, is near 7, dramatically lower than that of the monomer. Near pH1/2, the DSD’s binding affinity for CL nanodiscs (NDs), as measured by fluorescence correlation spectroscopy (FCS) and fluorescence anisotropy using a zinc-substituted cytc variant (Zncytc), also increases strongly. Comparing fluorescence binding data and CD binding data, we show that there appears to be a cooperative effect to the binding when the concentration of cytc is increased. Lastly, we show that the dimer has increased peroxidase activity compared to the monomer, and that the relative increase is larger in the presence of CL NDs and at pH values below pH1/2.
MATERIALS AND METHODS
Expression of Human Cytochrome c.
Wild-type human cytc was expressed from the pBTR(HumanCc)47 plasmid after transformation in Ultra BL21 (DE3) E. coli competent cells (EdgeBio, Gaithersburg, MD) using the manufacturer’s protocol. Transformed cells were grown in Terrific Broth medium with the addition of 100 μL of Antifoam C Emulsion (Sigma A8011) per liter of culture.
Cytc extraction and purification were performed as previously described.27, 48–52 However, sonication was used in place of a French press to lyse the cells. Briefly, sonicated cell lysates were treated with 50% ammonium sulfate to precipitate contaminating proteins. CM-sepharose cation-exchange chromatography was used to further purify the protein. Samples were stored at −80 °C until use. Prior to experimentation, samples were thawed for cation-exchange purification with an ÄKTAprime plus (GE Healthcare Life Sciences) and a HiTrap SP HP cation-exchange column (GE Healthcare 17115101). Protein samples were concentrated by ultrafiltration and oxidized with potassium ferricyanide (K3[Fe(CN)6]), followed by separation of oxidized cytc from the oxidizing agent using a Sephadex G25 column.
Human cytc dimer was generated in a similar manner to previously described methods.27, 44 Briefly, after oxidation, cytc was precipitated by the addition of 80% (vol/vol) ethanol. Samples were then incubated at room temperature for 15 minutes, centrifuged for 15 minutes at 4 °C, flash frozen and lyophilized overnight. Lyophilized samples were stored at –80 °C until use. Samples were then resuspended in 50 mM potassium phosphate buffer (pH 7.0) to a concentration of 10 mg/mL and incubated at 37 °C for 1.5 hours with shaking. The dimer species was separated from monomer and higher order oligomers using a BioRad Enrich SEC70 High-Resolution Size-Exclusion column (BioRad 780-1070).
Zinc-substituted Cytochrome c.
The zinc substitution of cytc was adapted from a previously described procedure.53 It should be noted that this is a hazardous procedure performed in a vented fume hood with the operator wearing a respirator, an acid apron and doubled nitrile gloves to minimize the possibility of skin exposure. Calcium gluconate gel should be available in case of skin exposure. Briefly, lyophilized cytc was mixed with HF-pyridine (70% HF) in a Teflon beaker (3 mg of cytc per mL HF-pyridine). All samples were protected from light exposure because of the photosensitivity of the unmetallated cytc and Zncytc. The solution was stirred for 10 minutes and then quenched with 5 mL of 50 mM ammonium acetate (pH 5.0) per 10 mL of HF-pyridine. The solution was kept under a stream of nitrogen gas for 2 hours followed by a 4-hour dialysis against 10 mM sodium acetate (pH 5.0) at room temperature. Metal-free porphyrin cytc was then separated from remaining ferric cytc using a CM Sepharose fast-flow ion-exchange column (GE Healthcare). Demetallated porphyrin cytc was then incubated with a 100-fold excess of zinc acetate for 10 minutes at 70 °C with shaking. Zinc incorporation was monitored spectroscopically: the Soret band at 423 nm sharpens dramatically, and the Q-bands at 549 and 584 nm appear (Figure S2).54 Excess zinc acetate was removed using a G-25 Sephadex size-exclusion column.
Preparation of Cardiolipin Nanodiscs.
A nanodisc (ND) is a discoidal lipid-protein complex whose shape and stability is controlled by the amphipathic α-helical membrane scaffold protein (MSP), which separates the hydrophobic tails of the bilayer from bulk water by forming a belt around the perimeter of the lipid bilayer.55–58 The lipid-to-belt molar ratio is critical to ND formation: 80:1 was used for 1,2-dimyristoyl-sn-glycero-3-phosphocholine) (DMPC) and 25:1 was used for CL. Monounsaturated tetraoleoyl-cardiolipin (TOCL) (1’,3’-bis[1,2-dioleoyl-sn-glycero-3-phospho]-glycerol) (Avanti 710335C) and DMPC (Avanti 850345C) were purchased from Avanti Polar Lipids (Alabaster, AL) and dissolved in HPLC grade chloroform. In this study, we use tetraoleoyl-cardiolipin (TOCL) because CLs with longer unsaturated fatty acyl chains have been shown to be more efficient at binding cytc and inducing peroxidase activity than CLs with shorter saturated fatty acyl chains.59 Similar to ND preparations previously studied,60 appropriate amounts of lipid were dried for 1–2 hours under a steady stream of nitrogen gas. Lipids were then rehydrated in MSP Buffer (50 mM Tris at pH 7.4, 150 mM NaCl, 0.1% NaN3) with 48 mM sodium cholate. The NaCl concentration was increased to 300 mM to facilitate CL ND formation. Following a fifteen-minute cycle of sonication and vortexing, rehydrated lipids were incubated with MSP1D1 Belt protein (Sigma M6574) for 30 minutes. Samples were then incubated with SM-2 Bio-Beads Resin (Bio-Rad 1523920) overnight at 4 °C followed by incubation for 1 hour at room temperature with fresh SM-2 Bio-Beads Resin. Samples were then spun at 15K rpm for 15 minutes to pellet remaining liposomes (Beckman Coulter Microfuge 18 Centrifuge). CL NDs were then purified at 4 °C using a GE Superdex 200 10/300 GL (GE17–5175-01) size-exclusion column and an ÄKTA-FPLC (GE Healthcare). The running buffer was that used for titrations. Purified CL NDs were stored at 4 °C until used. The final concentration of CL NDs was determined using the ε280 of the belt protein (21,430 with HIS tag and 18,450 without tag55) and dividing by two because each ND is composed of two belt proteins.
Dimer Dissociation Kinetics.
Human cytc DSD dissociation kinetics were studied using previous published methods.27 Briefly, following isolation of pure dimer using the BioRad Enrich SEC70 column at 4 °C in 50 mM potassium phosphate (pH 7), solutions of dimer were incubated at 40, 45, 50, 55 and 60 °C in a Julabo F12-ED refrigerated/heating circulator. Dissociation from dimer to monomer was measured at each time point by size-exclusion chromatography of 100–150 μL aliquots using the SEC70 column. The fractions of dimer (fDimer) and monomer (fMonomer) were evaluated from their relative peak heights at 280 nm in the FPLC chromatogram. Plots of fDimer and fMonomer versus time were fit to a single exponential to determine the rate constant for conversion of dimer to monomer (kDM). Eyring plots of kDM were then fit to eq 1 using a reference temperature (T0) of 310.15 K.
| (1) |
In eq. 1 kB and h are the Boltzmann and Planck constants, respectively. and are the difference in enthalpy and entropy between the ground state and the transition state. is the difference in heat capacity between the ground state and the transition state (TS) at constant pressure.
Alkaline Conformation Transition.
The alkaline conformation transition can be followed by monitoring the weak absorbance at 695 nm (A695), which reports on the Met80-heme ligation of the native state of oxidized cytc.2 Absorbance data, collected with a Beckman DU800 spectrophotometer, was corrected for baseline drift using the absorbance at 750 nm (A750) yielding A695corr = (A695 – A750). The pH of samples containing 100 μM heme and 100 mM NaCl was adjusted using NaOH and HCl, using a procedure that keeps protein concentration and NaCl concentration constant during the titration.61 Titration measurements were collected at room temperature (22 ± 3 °C).
The midpoint pH, pH1/2, was determined by fitting plots of A695corr versus pH to a modified form of the Henderson-Hasselbalch equation (eq 2). In eq 2, n represents the number of protons
| (2) |
linked to the alkaline transition. AN represents A695corr in the native state with Met80 bound to the heme and Aalk represents A695corr in the alkaline state with a lysine from Ω-loop D bound to the heme.
Peroxidase Activity Measurements.
Peroxidase activity of cytc was measured at 25 ± 0.1 °C using the previously described guaiacol colorimetric assay.30, 44 Briefly, the formation of tetraguaiacol from guaiacol and hydrogen peroxide in the presence of cytc was monitored at 470 nm, A470, using an Applied Photophysics SX20 stopped-flow spectrometer. Working stocks of H2O2 (ε240 = 41.5 M−1 cm−1),62, 63 guaiacol (ε274 = 2150 M−1 cm−1)64 and cytc were mixed in the stopped-flow apparatus. The final solution concentrations after mixing were 1 μM heme (i.e., 1 μM monomeric cytc or 0.5 μM DSD), 50 mM H2O2, and appropriate amounts of guaiacol in 50 mM buffer. The final CL ND concentration, where appropriate, was 1 μM. Three independent experiments were performed at each pH. Five kinetic traces were collected at each guaiacol concentration during each experiment. MatLab was used to calculate the initial velocity (greatest slope) of the A470 versus time data for each guaiacol concentration. The initial rate of guaiacol consumption (v) was determined by dividing the slope () by the extinction coefficient of tetraguaiacol (ε470 = 26.6 mM−1 cm−1)65 and multiplying by 4 (4 guaiacol consumed per tetraguaiacol produced). This initial rate of guaiacol consumption, v, was then divided by the concentration of cytc and fit to eq 3 in order to obtain Km and kcat values:
| (3) |
Circular Dichroism Spectroscopy.
Stock cytc samples were prepared at twice the experimental concentration and mixed 1:1 with a CL ND solution at twice the desired final CL ND concentration to produce each data point of the titration. The experimental concentration of cytc was 10 μM based on the heme absorbance (i.e., 10 μM monomer or 5 μM DSD). Samples were gently mixed by pipetting up and down then incubated for 30 minutes prior to measurement. Experiments were performed in triplicate.
Soret circular dichroism (CD) spectra (350 to 450 nm) were acquired using an Applied Photophysics Chirascan CD spectrophotometer. Spectra were acquired using a 3 s/nm acquisition time, 1.8-nm bandwidth and 1-nm steps at 25 °C. Each sample was measured five times then averaged. Spectra were smoothed using a sixth-order Savitsky–Golay filter smoothing technique.
Fluorescence Experiments with Human Cytochrome c.
Fluorescence correlation spectroscopy (FCS) and fluorescence anisotropy measurements were obtained by time-correlated single-photon counting (TCSPC) using a PicoQuant MicroTime 200 time-resolved confocal fluorescence microscope (Berlin, Germany) in the BioSpectroscopy Core Research Laboratory at the University of Montana. An Omega DP24-T thermocouple meter was connected to the microscope objective using a glass-braided insulated thermocouple for determination of sample temperature. Samples were excited with a vertically polarized 421-nm pulsed-diode laser (PicoQuant LDH-P-C-420) operating at 3μW average power (measured at the objective), and the vertical and horizontal components of the emission were collected on Hybrid-PMT detectors (PicoQuant). A 40-MHz repetition rate was used for the FCS measurements. Anisotropy measurements were collected at a 20-MHz repetition rate; V and H emission curves were collected for equal lengths of time, until 4×104 counts were obtained at the maximum of the V curve. High density polyethyleneglycol coated coverslips (MicroSurfaces Inc.) were used for sample collection to avoid interaction of cytc with the glass coverslip. A 30-μm pin hole and 488-nm long-pass (LP) filter (AHF/Semrock BLP488-R) were used in the emission paths. A polarizing beam splitter (Ealing), placed behind the 488 LP filter, was used to separate the V- and H-polarized emission components for anisotropy measurements. The confocal volume was determined with Atto 425 dye in ultra-pure water using a diffusion coefficient of 4.07×10−6 cm2 s−1 at 25 °C.66 SymPhoTime64 (PicoQuant version 2.3, build 4724) was used to collect data and analyze the FCS data. FluoFit (PicoQuant version 4.6.6.0, build 20.05.2014) was used to analyze the anisotropy data. Measurements were taken with monomeric and DSD human cytc samples at ~100 nM of Zn-substituted heme (i.e., 100 nM monomer and 50 nM DSD). The titration procedure was the same as used for CD titrations (1:1 mixing of Zncytc and CL ND solutions at twice the desired final concentration). The anisotropy instrument response function (IRF) was obtained as described using Atto 425 quenched with a saturated aqueous KI solution.67
FCS data were fit using SymPhoTime64 version 2.3 (PicoQuant, Berlin). Eq 4, which describes diffusion of several species in the presence of a single triplet state, was used as the fitting model. Figure S3 shows a typical FCS curve of Zncytc binding to CL ND. A more complete treatment of the FCS equations used can be found in the Supporting Information.
| (4) |
In eq. 4, the first part of the equation in brackets describes the triplet state, where T represents the fraction in the triplet (dark) state, τT is the time to relax from triplet back to the singlet state and t is time. The second part of the equation describes the diffusion of molecules in the experiment. The contribution of the ith diffusing species is represented by ρi, τD,i is the diffusion time of the ith species and κ is a geometrical factor describing the length-to-diameter ratio of the confocal volume.
Fluorescence anisotropy data were fit using the analysis software package FluoFit Pro V4.6.6 (PicoQuant, Berlin) according to the relationships described in eqs 5 and 6. A more complete treatment of the anisotropy equations used can be found in the Supporting Information.
| (5) |
| (6) |
In eqs 5 and 6, Gfac is a weighting term that describes the transmission difference between the horizontal and vertical detection paths. The instrument response function (IRF) is used to deconvolute out the excitation pulse and detection electronics. τi is the lifetime and αi is the amplitude of the ith component in the intensity decay. RINF is the anisotropy at infinite time or is due to a homogenous, time-independent background.68 The pre-exponential factors, βj, represent the differences between the excitation and emission transition dipole moments of the probe and the symmetry axes of the protein.69 The sum of βj is the limiting anisotropy at time zero, (ro). The rotation correlation times, ϕj, are directly related to the size and shape of the protein as well as the temperature and viscosity of the aqueous solution.
All data were fit to one ϕ and an RINF, where ϕ is that of the freely diffusing Zncytc. Since the rotational correlation time of a Zncytc bound to an ND is much longer than the fluorescence lifetime of Zncytc, it is fit as RINF; freely diffusing Zncytc in the absence of NDs has RINF = 0. Figure S4 shows examples of fluorescence anisotropy decay curves.
Fitting model for the fluorescence data.
The FCS and anisotropy measurements are from experiments where Zncytc and CL ND are interacting at equilibrium (eq 7). The fractions bound
| (7) |
(fbound) or free (ffree) are directly calculated based on the relative amplitudes of either their FCS diffusion coefficients or anisotropy rotational correlation times. In standard FCS measurements, it is generally accepted that diffusion coefficients must differ by a factor of ~1.6–2 in order to be resolved.70, 71 A rough calculation using an estimate of the protein radius and the Stokes-Einstein relationship indicates that the monomer (~128 μm2/s) or dimer (~101 μm2/s) diffusion coefficients are more than a factor of two larger than when bound to ND (~45 μm2/s, estimated for 1:1 cytc:CL ND complexes), indicating that the free and bound species are resolvable. In anisotropy measurements, the interaction of Zncytc with the membrane surface of the ND will increase the rotational correlation time (ϕ), making it much longer than the fluorescence lifetime, resulting in a resolvable increase in the RINF.72 Once the fbound has been determined, it can be directly plotted against the concentration of CL ND to obtain the association constant (Ka). The binding model is described by eq 8.
| (8) |
The native conformation of monomeric cytc is expected to require 9.1 nm2 of membrane surface to bind.73, 74 A structural rearrangement upon binding could result in an increase of the requirement to ~13 nm2.73–75 The MSP1D1 NDs used in these experiments are expected to have a membrane surface area of ~72 nm2 per side of the ND.58 Thus, there is space for upwards of 5 to 8 cytc to bind per side of the ND assuming no repulsive interactions and ignoring entropy effects for binding on a continuous surface.76 Under the conditions of our FCS experiments, the ND concentration is at least 100-fold higher than the cytc concentration near the midpoint of cytc-ND binding curves such that it is highly unlikely that more than one cytc will be bound per ND Therefore, use of the non-interacting single-site binding equation given by eq. 8 is a reasonable assumption to fit the FCS-monitored Cytc-ND binding data.
RESULTS AND DISCUSSION
Dissociation of Human Cytc DSD to Monomer is Slow Compared to Equine and Yeast DSDs.
We have previously shown that the equine DSD is substantially more stable than the yeast WT* (K72A, C102S) iso-1-cytc dimer.27 We hypothesized that if the DSD of cytc is involved in apoptosis, evolution of increased stability of the DSD could be an adaption favoring more effective induction of the peroxidase activity of cytc as related to apoptosis in mammals. Our previous work27 showed that the thermodynamic stability of monomeric cytc measured by guanidine hydrochloride (GdnHCl) unfolding52 correlated well with the kinetic stability of the cytc DSD.
We have extended our studies of dimer dissociation kinetics to include the human cytc DSD, and we can now show that the human cytc DSD is even more stable than the equine cytc DSD, consistent with the great thermodynamic stability observed with GdnHCl unfolding of human cytc.52 The Erying plot of the human DSD is compared to previously reported data for equine and yeast cytc in Figure 1. The human DSD was found to dissociate to monomer at measurable rates in the same temperature range as the equine DSD (Table S1). It was kinetically more stable than the horse DSD of cytc with a factor of two longer lifetime (about two weeks at 40 °C). All data in Figure 1 were fit to eq 1, which allows for curvature in the Eyring plot due to a change in heat capacity required to attain the transition state () for DSD dissociation. Curvature in the plot indicates that and are both temperature dependent.77 We used human physiological temperature (37 °C, 310.15 K) as the reference temperature, To, in the fits of the data in Figure 1 to eq 1 providing the activation parameters, and at 37 °C (Table 1). A large enthalpic barrier for spontaneous dissociation of the DSD to monomer is observed for cytc of all three species.27 However, favorable entropy compensates substantially for this large enthalpy, yielding ∆G‡ near 27 kcal/mol for the mammalian DSD compared to ~19 kcal/mol for the yeast DSD at 37 °C. Plots of ∆G‡ versus T, using the parameters in Table 1, show that the increased kinetic stability of the human DSD compared to the equine DSD arises from a shift in the temperature of maximum ∆G‡ from 16 to 23 °C.
Figure 1.

Eyring plots for dissociation of human WT (green), equine WT (orange) and yeast WT* (K72A, C102S mutations) (red) DSD dimers. The solid curve is a fit to a form of the Eyring equation that accounts for (Eq. 1). The reference temperature (To) was set to 310.15 K (37 °C) for all fits. The equine and yeast data are from previously published results.27
Table 1.
| Species |
(kcal/mol) |
(kcal/mol∙K) |
(kcal/mol∙K) |
∆G‡ (kcal/mol)b |
|---|---|---|---|---|
| Human | 59 ± 4 | 0.10 ± 0.01 | 2.3 ± 0.3 | 27 ± 5 |
| Equine | 66 ± 12 | 0.13 ± 0.04 | 1.8 ± 0.9 | 27 ± 16 |
| Yeast | 107 ± 15 | 0.28 ± 0.05 | 3.1 ± 0.8 | 19 ± 21 |
Similar to the equine DSD,27 the human DSD had less curvature in its Erying plot than the yeast DSD, yielding a smaller than that for the yeast DSD. All three values of indicate significate unfolding of both subunits during dissociation to monomer because the ΔCp for thermal unfolding of the monomeric cytochromes c range from 1.1 to 1.4 kcal/mol∙K.47 The primary evolutionary adaptation that increases the kinetic stability of the DSD of mammalian versus yeast cytc is a decrease in at 37 °C. The larger for yeast DSD dissociation indicates that the TS is more disordered than the TS for dissociation of the mammalian DSDs. The larger for the yeast versus the mammalian cytc also is consistent with a larger degree of unfolding in the TS for the dimer-to-monomer dissociation of the yeast DSD.
For the fluorescence correlation spectroscopy (FCS) experiments described below, we use Zn-substituted monomeric and DSD human cytc. The stability of Zn-substituted monomeric cytc as measured by GdnHCl unfolding is similar to that of the native Fe protein.78 Given the correlation between the kinetic stability of DSD cytc and the stability of monomeric cytc measured by GdnHCl unfolding discussed above, we expect the stability of the Zn-substituted human cytc DSD to be similar to that measured for the Fe form of the human cytc DSD.
The Alkaline Transition of the Human DSD Occurs 2.5 pH Units Lower Than That of the Monomer.
It has been shown that a decrease in the midpoint pH (pH1/2) of the alkaline transition can be correlated with better access to cytc conformational states that promote peroxidase activity.79–81 Below pH1/2, Met80 is the predominate ligand bound to the heme and above pH1/2 Lys72, 73 or 79 from Ω-loop D are predominantly the bound ligands.82 Lys binds more strongly to the Fe(III) of the heme than Met and is therefore a less labile ligand than Met80.65 Thus, the lysine ligation of the alkaline state would be expected to decrease peroxidase activity. However, a decrease in the pH1/2 of the alkaline transition correlates well with a decrease in the stability of Ω-loop D,83, 84 which protects the Met80 side of the heme.
Absorbance changes at 695 nm, an absorbance band that is present when Met80 is bound to the heme, was used to monitor the alkaline transition. A plot of A695corr versus pH in Figure 2 shows that the alkaline transition occurs at much lower pH for the DSD than the monomer. The pH1/2 we observe for the monomer (9.62 ± 0.03) is similar to the previously reported value.52 By contrast, the pH1/2 of the DSD (7.02 ± 0.06) is 2.5 pH units lower. The number of protons linked to the transition for both monomer (1.15 ± 0.09) and DSD (0.90 ± 0.11) was approximately equal to 1, which is consistent with a one proton process, as normally observed for the alkaline transition of cytc.2, 85
Figure 2.
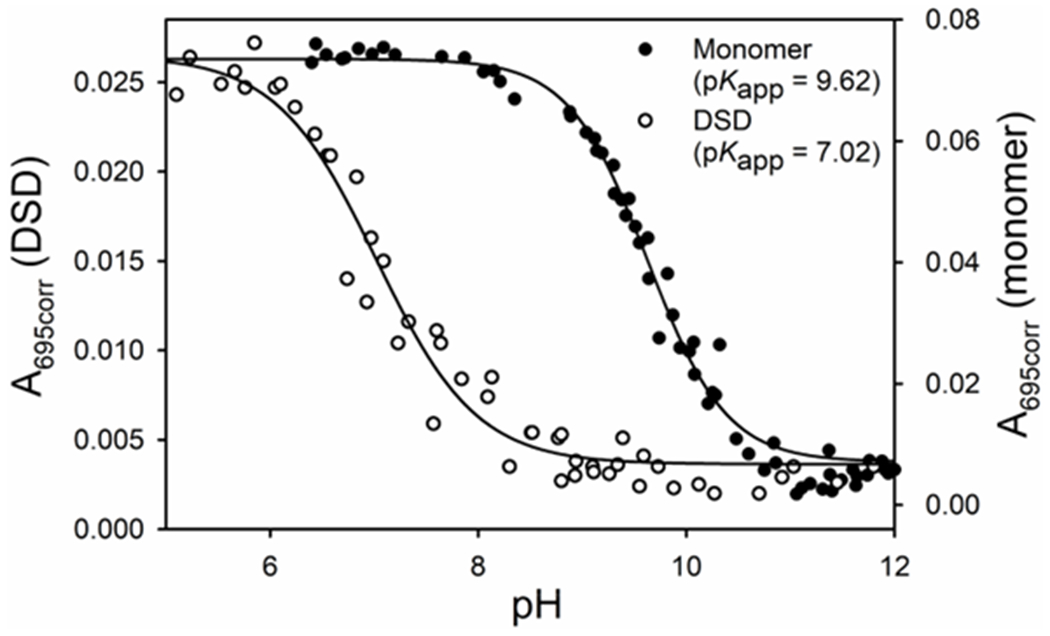
Plot of A695corr versus pH for the alkaline transition for monomeric (closed circles) and DSD (open circles) cytc. Data were collected at room temperature (22 ± 3 °C) in 100 mM NaCl solution with a protein concentration of 100 μM heme (100 μM monomer or 50 μM dimer). Solids lines are fits to eq 2 in materials and methods.
Absorbance changes at 625 nm also give information about dissociation of the Met80 ligand from the heme. An increase in A625 is observed below the pH1/2 for the alkaline transition. This absorbance band has been attributed to formation of a high-spin heme state, resulting from a change in heme coordination to either a pentacoordinate state or a hexacoordinate state with a water molecule bound86 in place of Met80. Whereas spectra of human DSD cytc, acquired near pH 6, show a small peak near 625 nm (Figure S5), the monomeric human cytc does not. Plots of A625corr versus pH show sigmoidal plots for both monomeric human cytc and the DSD (Figure S6). For the DSD, the pH1/2 is slightly lower than that of data monitored at 695 nm, indicating that Met80-heme and H2O-heme ligation states are populating simultaneously as pH is decreased below 7.
Taken together, these data indicate that the two hemes of the DSD are more accessible than the heme of the monomer. Met80, the primary ligand below pH1/2, is located in the hinge-loop region of the dimer structure.27, 28 Population of an available coordination site, whether it is a pentacoordinate heme or a hexacoordinate heme with water bound, is greater for the DSD than for the monomer. The 2.5 pH unit decrease in the pH1/2 of the DSD relative to the monomer places this change in ligation near physiological pH where it could directly regulate peroxidase activity in vivo.
Fluorescence Titrations Show that ND Binding by Human DSD Cytc is Weaker than Monomeric Cytc.
Many studies of binding of cytc to CL-containing liposomes involve titration of cytc, at concentrations that are near the Kd, with increasing concentrations of CL typically in the form of liposomes.75, 87–90 Thus, accurate evaluation of Kd requires correction for bound CL. Estimates of the number of CL bound to each cytc cover a large range.88, 91 Direct experimental determination of the CL:cytc stoichiometry has shown that this stoichiometry depends on ionic strength and requires prior experimental determination of the effective charge on the protein, Z, and an intrinsic binding constant, Ko, for the interaction from binding data as a function of salt concentration under low binding conditions.76 Because of the difficulty of accurately correcting for bound CL, it is often not done. To avoid this complication, we have used CL NDs as our model membrane system and FCS and fluorescence anisotropy as our detection methods. Using a fluorescent form of cytc, Zncytc, the concentration of monomeric and DSD cytc can be kept between 50 and 100 nM, low enough that the total CL ND concentration does not need to be corrected for cytc-bound CL ND. FCS reports on translational diffusion of fluorescent species, while fluorescence anisotropy reports on rotational diffusion. These techniques are orthogonal methods for determining the hydrodynamic radius and therefore are complementary. The experimental setup used allows each individual sample to be measured by both methods; switching measurement modes only requires minor changes (e.g., changing the laser repetition rate and polarization of the fluorescence emission). This dual experimental approach provides an increased level of robustness to the results obtained.
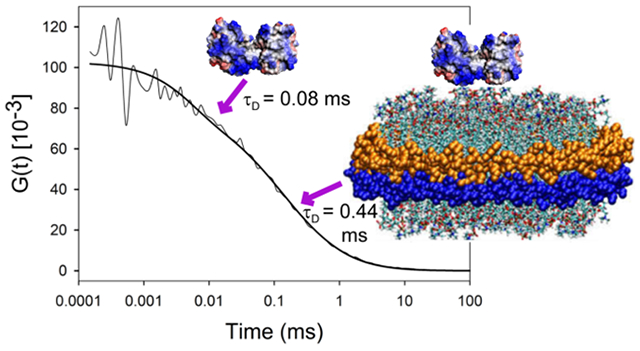
The FCS curves for Zncytc binding (Figure S3) were fit to two diffusion times (τD), a faster one representing a free species (free monomer or free DSD) and a slower one representing the ND-bound species, and a triplet state process (τT). The τD and translational diffusion coefficient (D) are related (τD = ω02/4D) and can be connected to the hydrodynamic radius through the Stokes–Einstein equation; D is temperature and viscosity dependent and τD depends on the size of the Veff, which has been carefully calibrated. The results are presented in Table 2. The free monomer and DSD diffusion coefficients are similar to the calculated values discussed in the Materials and Methods. The experimental diffusion coefficients for cytc/ND complexes, however, are smaller than our calculated estimate in Materials and Methods (45 μm2/s). Thus, the assumption of spherical symmetry may be inadequate for the CL ND complexes. The distinct difference in D for the monomer/ND complex versus the DSD/ND complex indicates that D is sensitive to the difference in size of the DSD versus monomeric forms of cytc bound to CL NDs.
Table 2.
Fluorescence Parameters for Zncytc monomer and DSD.
| FCS |
Fluorescence Anisotropy |
||||||
|---|---|---|---|---|---|---|---|
| τD (ms) | τT (μs) | D (μm2/s) | RH(nm) a | Lifetime (ns) b | ϕ (ns) | RH (nm) a | |
| Monomer | 0.06 ± 0.01 | 12 ± 9 | 125 ± 21 | 1.80 ± 0.25 | 3.27 ± 0.31 | 6.37 ± 0.16 | 1.83 ± 0.05 |
| Monomer Bound | 0.48 ± 0.03 | 3 ± 2 | 33 ± 2.6 | 6.83 ± 0.51 | - | - | - |
| DSD | 0.08 ± 0.01 | 25 ± 5.2 | 92 ± 8.2 | 2.39 ± 0.19 | 4.55 ± 0.04 | 12.7 ± 0.17 | 2.31 ± 0.04 |
| DSD Bound | 0.44 ± 0.02 | 2.7 ± 2.1 | 22 ± 2.4 | 10.1 ± 0.90 | - | - | - |
RH corrected to 25 °C from the experimental temperature.
The intensity-weighted lifetime is defined as ❬τ❭ =
The anisotropy decay data were fit to a single rotational correlation time ϕ (Table 2) with a limiting anisotropy (RINF) because the ϕ of Zncytc bound to CL ND is much longer than the fluorescence lifetime and therefore difficult to fit with a second exponential. The binding can be followed, however, by the increase in RINF and a decrease in the amplitude of the single ϕ as Zncytc binds to the CL NDs. Our parameters from analysis of the monomer fluorescence decay data (Table 2) agree well with those previously reported by Vanderkooi et al.54 In addition, we observed a 1.3 ns increase in the intensity-weighted lifetime of the dimer compared with that of the monomer. The RH values for the monomeric and DSD cytc extracted from the anisotropy data agree well with those obtained from FCS data (Table 2).
We carried out experiments at pH 8.0 and pH 7.4. At pH 8.0, we have shown that binding is dominated by site A (primarily electrostatic interaction) and appears to be weaker than for studies done at lower pH. At pH 7.4 and lower,92, 93 binding is stronger and has a significant non-electrostatic component. Monomeric human cytc will be completely in the native state at both these pH values. Based on the fit to the data in Figure 2, the human DSD will be 88% in the alkaline state at pH 8 and 69% in the alkaline state at pH 7.4. However, because of the high negative charge at the surface of the 100% CL NDs, the pH near the surface of the ND may be somewhat lower, which might lead to a larger shift toward population of the native conformer at pH 7.4. The data sets collected from FCS and fluorescence anisotropy measurements, respectively, can be fit to a single-site binding model (Eq. 8). The fits to both the FCS and anisotropy data can be seen in Figure 3. Binding constants extracted from these plots are given in Table 3.
Figure 3.
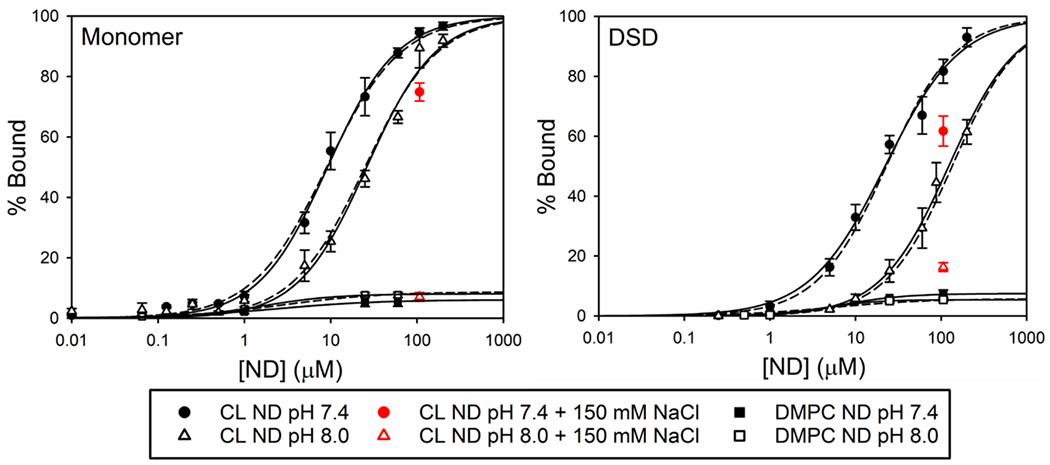
Plots of % bound monomeric (left) and DSD (right) Zncytc versus CL and DMPC nanodisc concentration (logarithmic scale) as determined by FCS and fluorescence anisotropy. Data points shown are from the FCS experiments. The fits of FCS (solid lines) and anisotropy (dashed lines) data sets to eq 8 are shown. Experimental data were corrected to 25 °C to determine binding and performed in 20 mM TES at the indicated pH and salt concentration.
Table 3.
Thermodynamic Parameters for Binding of Zncytc to CL ND from Fluorescence Experiments at 25 °C
| pH 7.4 |
pH 8.0 |
||||
|---|---|---|---|---|---|
| Kd (μM) | ΔG (kcal/mol) | Kd (μM) | ΔG (kcal/mol) | ||
| Monomer | Anisotropy | 9.0 ± 0.2 | 6.88 ± 0.01 | 25 ± 0.6 | 6.28 ± 0.01 |
| FCS | 9.2 ± 0.3 | 6.87 ± 0.02 | 26 ± 1.3 | 6.25 ± 0.03 | |
| Dimer | Anisotropy | 23 ± 2 | 6.32 ± 0.05 | 134 ± 7.1 | 5.28 ± 0.03 |
| FCS | 22 ± 1.2 | 6.35 ± 0.03 | 122 ± 6.0 | 5.34 ± 0.03 | |
Control experiments (Figure 3) show that, regardless of pH, ~5% of the monomers and dimers of Zncytc (100 nM Zn-heme concentration) bind to DMPC ND, which is likely nonspecific binding, perhaps to the belt protein.
We also measured the effect of adding NaCl to a final concentration of 150 mM to solutions containing Zncytc fully bound to the CL NDs. At pH 8.0, when we added NaCl to the 100 μM CL ND titration point, the fraction of bound Zncytc reverted to the level of binding observed with DMPC ND (Figure 3). This observation indicates that at pH 8.0, the cytc−CL ND association is driven by electrostatic interaction at the A site. This result is consistent with our previous study of the binding of human and yeast cytc to 100 nm CL liposomes, which indicated that the predominate binding interaction at pH 8 was through site A.89 At pH 7.4, 150 mM salt causes only partial dissociation of cytc from the CL ND. This behavior has previously been observed by other labs43, 87, 88 and indicates that at pH 7.4 binding sites on cytc that have a significant hydrophobic component also contribute to binding.
The binding data for the Zn-cytc monomer (Figures 3 and S7, Table 3) also show that increasing the pH from 7.4 to 8.0 results in a two-fold increase in the Kd. This result is consistent with reduced influence of binding sites other than the A site at pH 8, as indicated by the lesser effect of 150 mM salt on binding at pH 7.4 versus pH 8.0. These results fit with previously published binding data,89, 90, 94 and they support the interpretation that multiple cytc binding sites are involved in the interaction with CL surfaces. These data also support previous data, which suggest that at lower pH multiple binding locations on cytc work in concert to form a tighter interaction91, 95 that then may be followed by a second unfolding stage.43, 88, 92 These observations could also indicate that the combined effects of these binding sites at lower pH either result from too strong of an electrostatic interaction to be broken at the NaCl levels used in this experiment or rely on hydrophobic interactions usually associated with the C-site.
The dimer binding data (Table 3) show a reduction in the affinity for a CL surface: the Kd of the dimer at pH 7.4 is similar to that of the monomer at pH 8.0, and there is a five-fold decrease in the binding affinity of the dimer when increasing the pH from 7.4 to 8.0. This reduction in affinity at pH 8 indicates that the A site of the dimer makes a weaker contribution to the cytc-CL interaction under these conditions. Thus, formation of the dimer reduces the binding interaction with the CL surface, which could indicate another way in which the dimer has evolved into a dynamic switch to control apoptosis near physiological pH. The decrease in affinity at higher pH may be linked to the alkaline conformational transition of the dimer because the population of this conformer is somewhat higher at pH 8 than at pH 7.4.
The significantly different diffusion coefficients observed for CL ND bound monomeric cytc versus CL ND bound DSD cytc (Table 2) and the considerably different Kd values observed for monomer and DSD cytc binding to CL NDs (Figure 3, Table 3) suggest that human DSD cytc does not dissociate to monomer when bound to CL NDs. These observations are consistent with the high stability of human DSD cytc (Figure 1, Table 1). However, further experiments would be required to demonstrate definitively that human DSD cytc binding to CL NDs does not cause any dissociation of the DSD to monomer.
CD Binding Experiments Show Cooperative Binding with Increased Concentration of Cytc.
It has been shown that ruffling, the perturbation of the heme from perfect D4h symmetry, is influenced by the CXXCH heme attachment sequence and the residues that pack against this sequence.96–98 Breaking of this symmetry leads to a strong circular dichroism (CD) signal in the Soret region.99 The amplitude of the B-band couplet in the Soret CD is dependent on both the splitting of the band and the rotational strength.100 Coupling of the heme π-π* transitions to those of nearby aromatic amino acids is believed to be an important mechanism for induction of rotational strength.101 Mutagenesis studies with yeast iso-1-cytc implicate Phe82, which packs against the heme, as an important contributor to the Soret CD of cytc.102 Thus, local rearrangement of the heme environment can significantly affect the Soret CD. Changes in the CD spectrum have been used successfully to study the rearrangement of the heme environment of cytc that occurs when the protein binds to liposomes.33, 88, 89, 92, 103–106
Human monomer and dimer cytc have markedly different signatures in the Soret CD spectra at pH 8.0 (Figure 4). The degree of B-band splitting reported here for the monomer is similar to that reported in previous studies.89 The lack of a trough in the DSD spectrum is only seen for the monomer at high lipid-cytc ratios, where it indicates a significant perturbation of the heme environment of the monomer.89 The dimer CD spectra are similar to those previously reported for the equine alkaline conformation,107 suggesting that the human dimer is in an alkaline conformation at pH 8. The data in Figure 2 support this conclusion.
Figure 4.

Soret CD spectra of human monomer (solid) and DSD (dashed) cytc at a concentration of 10 μM heme (10 μM monomer or 5 μM DSD). Spectra were acquired in 20 mM TES buffer, 0.1 mM EDTA (pH 8.0) at 25 °C.
The Soret CD spectrum from 350 to 450 nm was monitored as CL NDs were titrated against a fixed concentration of monomeric and DSD human cytc at pH 8, where the electrostatic site A binding site dominates binding.89 Based on the FCS data, the CD experiments were performed at protein concentrations close to the Kd of the monomer, but well below that of the DSD. The CD experiments were performed at 100-fold higher concentration of cytc than the fluorescence experiments. Because of the small surface area of the ND, the higher concentrations of cytc used in the CD experiments could result in crowding that would lead to interactions between individual cytc molecules on the surface of the ND, potentially increasing the apparent affinity of the ND for monomer or DSD forms of cytc.
When titrated against CL ND, the Soret CD signals of the monomer and DSD respond differently at pH 8 (Figure S8). Qualitative binding of the monomer differs with respect to binding to liposomes, too.89 When monomer binds to 100% CL liposomes, there is a strong increase in the amplitude of the 405-nm peak, followed by a red shift of the peak as the trough near 418 nm disappears.89 Unlike the liposome binding data, there is a decrease in the 405-nm peak with the addition of ND. The largest change in monomer Soret CD spectrum is the decrease in the amplitude of the 418-nm trough as CL ND concentration increases. For the DSD, there is only a small increase in the intensity of the Soret CD band centered at 410 nm.
Data at these wavelengths are plotted versus ND concentration in Figure 5. We use a single-site Langmuir-type binding isotherm (eq S14 in the SI) to fit these data, as previously.89 As shown in Table 4, the Kd(app) obtained from the Soret CD data indicates tighter binding than that of the fluorescence data (Table 3). This observation could suggest that interactions between human cytc molecules increase the affinity for CL NDs under crowded conditions (higher cytc concentration coupled to low ND/cytc ratios). The 100-fold higher Cytc concentration leads to about a 10-fold increase in the Kd(app) for monomeric cytc and a >100-fold increase for the DSD. For the DSD, there is likely a significant error in Kd(app) because of the low amplitude of the signal change caused by binding to the CL NDs. At the midpoint of the titration of the CL NDs by the DSD, ∼8 DSD would be bound per ND. Assuming the dimer requires twice the surface area of the monomer, essentially all the available space on both sides of the ND would be occupied (8 monomers can fit on the surface of each side of an ND, see Materials and Methods). While entropy effects might be expected to oppose such high occupancy,76 entropy could be mitigated by interaction between dimers. For monomeric horse cytc binding to phosphatidylglycerol (PG), ligand-ligand interaction was observed to be negligible.76 However, for denatured horse cytc binding to PG, significant ligand-ligand interaction was observed.76 Thus, it is possible that the dimer behaves more like denatured cytc in this regard. Data for the horse cytc dimer binding to PG-containing liposomes, also shows that the dimer binds more tightly than the monomer when cytc concentration is in the μM range.108 The cooperativity parameter, n, is near 1 for both monomer and dimer (Table 4). We are varying ND concentration, so the value of n suggests that there is no cooperativity with respect to the ND (or CL, Figure S9).
Figure 5.
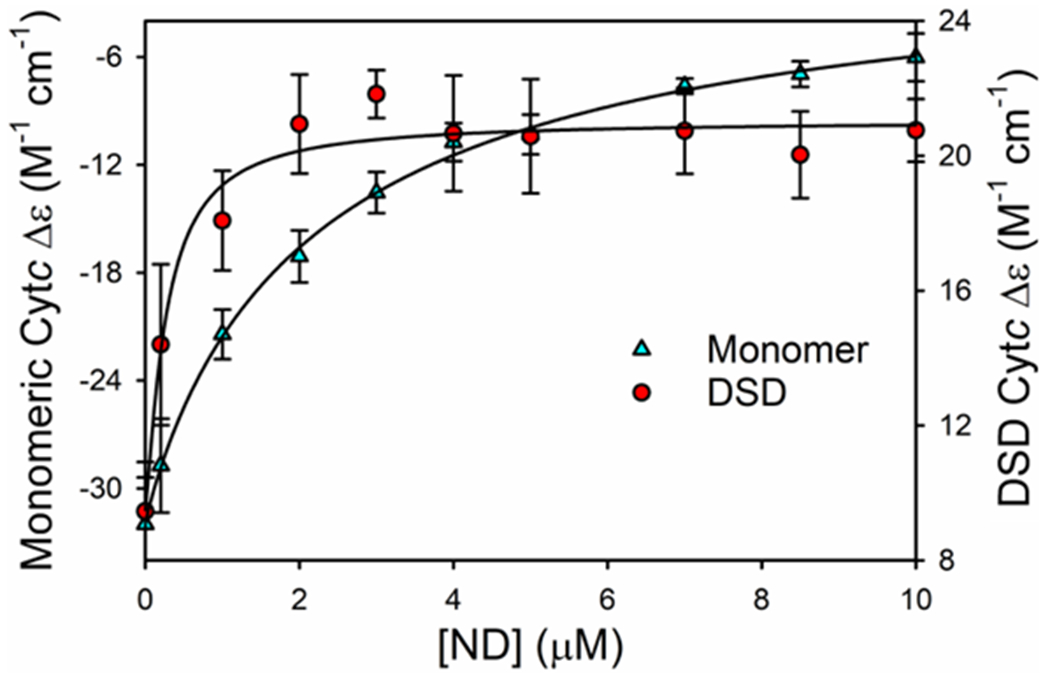
Human cytc Soret CD signal as a function of concentration of cardiolipin ND, [ND]. Data points at each [ND] follow the change in amplitude of the peak of the Soret CD signal. For the monomer the changes at 418-nm and for the dimer the changes at 410-nm are plotted versus [ND]. The experimental concentration of cytc was 10 μM based on the heme absorbance (i.e., 10 μM monomer or 5 μM DSD). Experiments were performed in 20 mM TES buffer and 0.1 mM EDTA (pH 8) at 25 °C. Solid curves are fits to eq S14 in the SI.
Table 4.
Thermodynamic Parameters for Binding of Cytc to CL NDs from CD Data at 25 °C and pH 8.
| Variant | Kd(app) (μM ND)a | n |
|---|---|---|
| Monomer-ND | 2.3 ± 0.1 | 0.9 ±0.3 |
| Dimer-ND | 0.3 ± 0.1 | 1.2 ± 0.4 |
The experimental concentration of cytc was 10 μM based on the heme absorbance (i.e., 10 μM monomer or 5 μM DSD). The error corresponds to the standard error in the fit of the parameter. We use apparent dissociation constant, Kd(app), because we have not corrected ND concentration for ND bound to cytc.
For the monomer, on average ~2 monomers would be bound per ND at the midpoint (one per side). This observation indicates that ligand-ligand interaction is less significant for monomeric cytc compared to the dimer, consistent with previous work.76 When the response of the CD signal is plotted versus lipid-to-protein ratio the binding curves for CL NDs and CL liposomes are identical for the cytc monomer (Figure S9). Therefore, the effect of cytc concentration on Kd(app) cannot be attributed to a difference between NDs and liposomes.
At the Kd measured by single-molecule fluorescence methods at pH 8 (Table 3), the ND/cytc ratio is ~100 for the monomer and >1000 for the DSD, so, it is unlikely that more than one cytc monomer or dimer is bound to each ND. Under conditions of high protein dilution with respect to ND concentration, effects of ligand-ligand interaction would be expected to be minimal.
Human Cytc-DSD Shows Increased Peroxidase Activity in the Absence and Presence of CL NDs.
As previously discussed, cytc’s ability to become a peroxidase is essential for its role as an initiator of the intrinsic apoptotic pathway.10, 109 Increasing the flexibility of Ω-loop D or the removal of the heme-ligand provided by Ω-loop D has been shown to increase peroxidase activity in monomeric cytc.110, 111 In the DSD conformation, Ω-loop D is conformationally distant from the heme and the Met80 ligand is replaced with H2O,27, 28 which should result in an increase in peroxidase activity, as has been shown for the equine DSD of cytc in aqueous solution at pH 7.44 To determine the effect of formation of the alkaline conformer of the DSD on peroxidase activity, we have measured its peroxidase activity from pH 6 to 8. To evaluate the peroxidase activity of monomeric and DSD human cytc bound to NDs though site A, we performed experiments in the presence and absence of CL NDs at pH 8.89
Michaelis-Menten plots of peroxidase activity were generated from data obtained by monitoring the formation of tetraguaiacol from guaiacol at 470 nm in the presence of H2O2. These plots were used to determine the catalytic rate constant (kcat) and the Michaelis-Menten constant (Km) for cytc. The data were normalized based on the amount of heme present (i.e., the concentration of DSD is half that of the monomer). Figure S10 shows Michaelis-Menten plots generated from peroxidase data acquired for the monomer and DSD, with and without CL ND at pH 8.0. While the Km values are relatively similar (also see Table 5), the kcat values increase from monomer to DSD. The kcat values also increase dramatically in the presence of CL NDs.
Table 5.
Peroxidase Activity of Human Cytc Variants as a Function of pH at 25 °C with Guaiacol as Substrate.
| Variant | pH 6.0 |
pH 7.0 |
pH 8.0 |
|||
|---|---|---|---|---|---|---|
| Km (M) | kcat (s−1) | Km (M) | kcat (s−1) | Km (M) | kcat (s−1) | |
| Monomera | 14 ± 2 | 0.11 ± 0.01 | 5.7 ± 0.3 | 0.11 ± 0.002 | 11 ± 2 | 0.06 ± 0.01 |
| Monomer with CL NDb | 6 ± 3 | 0.29 ± 0.03 | ||||
| 11 ± 2 | 0.06 ± 0.01 | |||||
| DSD | 11 ± 1 | 1.52 ± 0.02 | 12 ± 3 | 0.78 ± 0.04 | 18 ± 3 | 0.44 ± 0.02 |
| DSD with CL ND | 24 ± 6 | 1.43 ± 0.12 | ||||
Parameters are from Nold et al.110
Samples with CL ND led to a biphasic dataset
The data in Table 5 show that kcat decreases as pH increases for both monomeric and DSD human cytc. A decrease in kcat as pH increases has been observed for both horse and human monomeric cytc.65, 110, 112 This trend has been attributed to replacement of Met80 by lysine65 because nitrogen donors are much stronger ligands for Fe(III)heme than methionine.113, 114 For the DSD of human cytc, kcat decreases smoothly as the DSD forms an alkaline state (pH1/2 = 7.02 ± 0.06). Thus, the peroxidase activity of the DSD can function as a pH-inducible switch near physiological pH. Also, the peroxidase activity of the human DSD is seven-fold higher than that of the monomer at pH 7 and 8, but 14-fold higher at pH 6 when mixed Met80/H2O ligation replaces the lysine ligation of the alkaline state (see Figures 2, S5 and S6).
The initial velocity (v), was determined by finding the greatest slope in the A470 versus time data after an initial lag. At pH 8, the monomer cytc data in the presence of CL NDs presented a curve with a second obvious slope before approaching saturation, indicating the presence of at least two catalytically distinct species. Both of these velocities were used to determine a Km and kcat. One set of Michaelis-Menten parameters showed a larger kcat (0.29 ± 0.01 s−1) and a smaller Km (6 ± 3 μM) than observed for the monomer in the absence of NDs (see Table 5, Figure 6 and Figure S10). The other set of parameters matched that of free monomeric cytc in solution (see Table 5). The observation of two kinetic phases under the conditions of peroxidase activity measurements (1 μM cytc and 1 μM ND) is consistent with the binding curve in Figure 5, which indicates that monomeric cytc would not be fully bound to the NDs under these conditions. The kcat for the ND bound monomer is based on the total concentration of monomer and thus may be an underestimate.
Figure 6.
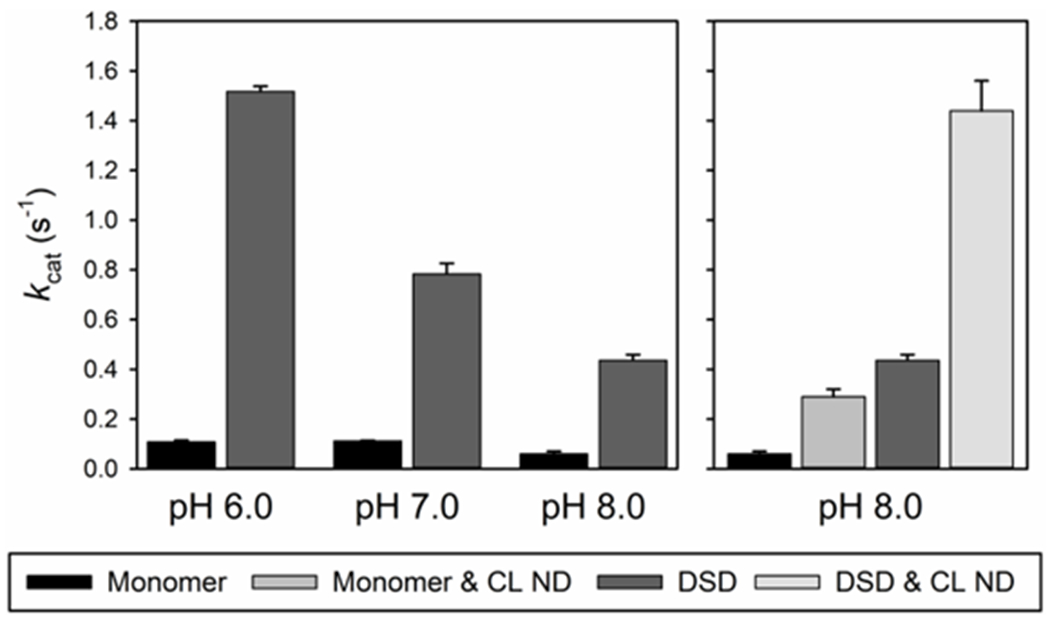
kcat vs pH for monomeric and DSD cytc at 25 °C. Error bars are the standard deviations from three independent experiments. Human data in the absence of CL NDs are from Nold et al.102
The unfolding of cytc by guanidine hydrochloride has been shown to increase the peroxidase activity to rates similar to those observed for isolated heme groups.115 Likewise, the addition of CL membrane mimics has been shown to result in a dramatic increase in the peroxidase activity of monomeric cytc.59 Our data are consistent with these observations, with the DSD showing increased peroxidase activity over the monomer, and the peroxidase activity of both species increasing in the presence of CL NDs at pH 8 when bound through site A.
CONCLUSIONS
The data presented provide evidence for how the DSD conformer of cytc may be an evolved switch to allow tighter control of the intrinsic apoptotic pathway. The human cytc DSD is substantially more stable to dissociation than that of yeast. Because yeast lacks a complete apoptotic pathway,116 the increased stability of human and equine cytc DSDs could indicate evolution in mammals to regulate the intrinsic apoptotic pathway. The DSD of human cytc undergoes an alkaline conformational transition with a midpoint near neutral pH. The structural reorganization seems to act as a conformational switch that increases the affinity for CL NDs by 5-fold from pH 8 to pH 7.4 and also leads to a progressive increase in peroxidase activity as pH is lowered from 8 to 6. The alkaline conformer of the DSD converts to a mix of Met80-ligated and high-spin (H2O-ligated) heme by pH 6, leading to a larger enhancement in peroxidase activity as pH decrease from 8 to 6 than for that of monomeric human cytc. The DSD of human cytc is well-adapted to act as a pH-sensitive functional switch near the pH of the IMS (pH = 6.88 ± 0.08).117 During apoptosis, the pH of the cytoplasm decreases below 6.118 If the same is true of the IMS, the intrinsic peroxidase activity of the DSD would continue to increase (Figure 6).
Our results also suggest that self-interaction between cytc molecules may enhance the affinity of cytc for CL-containing membranes under crowded conditions. DSD affinity appears to be more strongly enhanced than that of the monomer. This effect of cytc concentration on the strength of the interaction of cytc with CL membranes bears further investigation given the millimolar concentration of cytc in mitochondria.119, 120
Supplementary Material
Funding
This work was supported by grants from the NSF [CHE-1609720 and CHE-1904895 (B.E.B)]. The BioSpectroscopy Core Research Laboratory at the University of Montana was supported by a CoBRE grant from the National Institute of General Medical Sciences [P20GM103546].
ABBREVIATIONS
- IMS
intermembrane space
- CL
cardiolipin
- DSD
domain-swapped dimer
- CD
circular dichroism
- FCS
fluorescence correlation spectroscopy
- cytc
cytochrome c
- Zncytc
zinc-substituted cytc
- ND
nanodisc
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine)
- kDM
rate constant for conversion of DSD to monomer
- GdnHCl
guanidine hydrochloride
Footnotes
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website.
A PDF file is provided containing a more detailed derivation of the equations used to fit single-molecule fluorescence data, figures showing the structure of one subunit of the cytc DSD, the intermediates in the production of zinc-substituted cytc, a typical FCS correlation function plot for cytc/ND binding, typical fluorescence anisotropy data, spectra showing the 695- and 625-nm bands as a function of pH for monomeric and DSD cytc, a plot of absorbance at 625 nm versus pH for monomeric and DSD human WT cytc, binding curves for the interaction of monomeric and DSD cytc with CL NDs, CD spectra as a function of CL ND concentration and Michaelis-Menten plots for monomeric and DSD cytc in the presence and absence of CL NDs at pH 8 and a Table containing the kDM values for the human DSD used in Figure 1.
Accession Codes
UniProt ID: human cytochrome c, P99999.
The authors declare no competing financial interest.
REFERENCES
- 1.Dickerson RE, and Timkovich R (1975) Cytochromes c, In The Enzymes (Boyer PD, Ed.) 3rd ed., pp 397–547, Academic Press, New York. [Google Scholar]
- 2.Moore GR, and Pettigrew GW (1990) Cytochromes c: Evolutionary, Structural and Physicochemical Aspects, Springer-Verlag, New York; 10.1007/978-3-642-74536-2. [DOI] [Google Scholar]
- 3.Winge DR (2012) Sealing the mitochondrial respirasome, Mol. Cell. Biol 32, 2647–2652. 10.1128/MCB.00573-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berghuis AM, and Brayer GD (1992) Oxidation state-dependent conformational changes in cytochrome c, J. Mol. Biol 223, 959–976. 10.1016/0022-2836(92)90255-I. [DOI] [PubMed] [Google Scholar]
- 5.Deacon OM, Karsisiotis AI, Moreno-Chicano T, Hough MA, Macdonald C, Blumenschein TMA, Wilson MT, Moore GR, and Worrall JAR (2017) Heightened dynamics of the oxidized Y48H variant of human cytochrome c increases its peroxidatic activity, Biochemistry 56, 6111–6124. 10.1021/acs.biochem.7b00890. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Kim CN, Yang J, Jemmerson R, and Wang X (1996) Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c, Cell 86, 147–157. 10.1016/S0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 7.Fulda S, and Debatin KM (2006) Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy, Oncogene 25, 4798–4811. 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- 8.van Loo G, Saelens X, van Gurp M, MacFarlane M, Martin SJ, and Vandenabeele P (2002) The role of mitochondrial factors in apoptosis: a Russian roulette with more than one bullet, Cell Death Differ. 9, 1031–1042. 10.1038/sj.cdd.4401088. [DOI] [PubMed] [Google Scholar]
- 9.Huberts DHEW, and van der Klei IJ (2010) Moonlighting proteins: an intriguing mode of multitasking, Biochim. Biophys. Acta - Mol. Cell Res 1803, 520–525. 10.1016/j.bbamcr.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 10.Jiang X, and Wang X (2004) Cytochrome c-mediated apoptosis, Annu. Rev. Biochem 73, 87–106. 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 11.Orrenius S, and Zhivotovsky B (2005) Cardiolipin oxidation sets cytochrome c free, Nat. Chem. Biol 1, 188–189. 10.1038/nchembio0905-188. [DOI] [PubMed] [Google Scholar]
- 12.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, and Borisenko GG (2005) Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors, Nat. Chem. Biol 1, 223–232. 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 13.Kagan VE, Borisenko GG, Tyurina YY, Tyurin VA, Jiang J, Potapovich AI, Kini V, Amoscato AA, and Fujii Y (2004) Oxidative lipidomics of apoptosis: redox catalytic interactions of cytochrome c with cardiolipin and phosphatidylserine, Free Radical Biol. Med 37, 1963–1985. 10.1016/j.freeradbiomed.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Ott M, Robertson JD, Gogvadze V, Zhivotovsky B, and Orrenius S (2002) Cytochrome c release from mitochondria proceeds by a two-step process, Proc. Natl. Acad. Sci. U.S.A 99, 1259 http://www.pnas.org/content/99/3/1259.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shidoji Y, Hayashi K, Komura S, Ohishi N, and Yagi K (1999) Loss of molecular interaction between cytochrome c and cardiolipin due to lipid peroxidation, Biochem. Biophys. Res. Commun 264, 343–347. 10.1006/bbrc.1999.1410. [DOI] [PubMed] [Google Scholar]
- 16.Kagan VE, Tyurina YY, Bayir H, Chu CT, Kapralov AA, Vlasova II, Belikova NA, Tyurin VA, Amoscato A, Epperly M, Greenberger J, DeKosky S, Shvedova AA, and Jiang J (2006) The “pro-apoptotic genies” get out of mitochondria: oxidative lipidomics and redox activity of cytochrome c/cardiolipin complexes, Chem. Biol. Interact 163, 15–28. 10.1016/j.cbi.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 17.McMillin JB, and Dowhan W (2002) Cardiolipin and apoptosis, Biochim. Biophys. Acta, Mol. Cell. Biol. Lipids 1585, 97–107. 10.1016/S1388-1981(02)00329-3. [DOI] [PubMed] [Google Scholar]
- 18.Horvath SE, and Daum G (2013) Lipids of mitochondria, Prog. Lipid Res 52, 590–614. 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Nicholls P (1974) Cytochrome c binding to enzymes and membranes, Biochim. Biophys. Acta, Rev. Bioenerg 346, 261–310. 10.1016/0304-4173(74)90003-2. [DOI] [PubMed] [Google Scholar]
- 20.Scorrano L, Ashiya M, Buttle K, Weiler S, Oakes SA, Mannella CA, and Korsmeyer SJ (2002) A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis, Dev. Cell 2, 55–67. 10.1016/S1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- 21.Montero J, Mari M, Colell A, Morales A, Basañez G, Garcia-Ruiz C, and Fernández-Checa JC (2010) Cholesterol and peroxidized cardiolipin in mitochondrial membrane properties, permeabilization and cell death, Biochim. Biophys. Acta, Bioenerg 1797, 1217–1224. 10.1016/j.bbabio.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckmann J, Eckert SH, Leuner K, Muller WE, and Eckert GP (2013) Mitochondria: mitochondrial membranes in brain ageing and neurodegeneration, Int. J. Biochem. Cell Biol 45, 76–80. 10.1016/j.biocel.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez MG, Troiano L, Moretti L, Nasi M, Pinti M, Salvioli S, Dobrucki J, and Cossarizza A (2002) Early changes in intramitochondrial cardiolipin distribution during apoptosis, Cell Growth Differ. 13, 449–455. http://cgd.aacrjournals.org/cgi/content/abstract/13/9/449. [PubMed] [Google Scholar]
- 24.Gonzalvez F, and Gottlieb E (2007) Cardiolipin: setting the beat of apoptosis, Apoptosis 12, 877–885. 10.1007/s10495-007-0718-8. [DOI] [PubMed] [Google Scholar]
- 25.Epand RF, Martinou JC, Fornallaz-Mulhauser M, Hughes DW, and Epand RM (2002) The apoptotic protein tBid promotes leakage by altering membrane curvature, J. Biol. Chem 277, 32632–32639. 10.1074/jbc.M202396200. [DOI] [PubMed] [Google Scholar]
- 26.Gronenborn AM (2009) Protein acrobatics in pairs - dimerization via domain swapping, Curr. Opin. Struct. Biol 19, 39–49. 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClelland LJ, Steele HBB, Whitby FG, Mou T-C, Holley D, Ross JBA, Sprang SR, and Bowler BE (2016) Cytochrome c can form a well-defined binding pocket for hydrocarbons, J. Am. Chem. Soc 138, 16770–16778. 10.1021/jacs.6b10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirota S, Hattori Y, Nagao S, Taketa M, Komori H, Kamikubo H, Wang Z, Takahashi I, Negi S, Sugiura Y, Kataoka M, and Higuchi Y (2010) Cytochrome c polymerization by successive domain swapping at the C-terminal helix, Proc. Natl. Acad. Sci. U.S.A 107, 12854–12859. 10.1073/pnas.1001839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fetrow JS, Dreher U, Wiland DJ, Schaak DL, and Boose TL (1998) Mutagenesis of histidine 26 demonstrates the importance of loop-loop and loop-protein interactions for the function of iso-1-cytochrome c, Protein Sci. 7, 994–1005. 10.1002/pro.5560070417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClelland LJ, Mou T-C, Jeakins-Cooley ME, Sprang SR, and Bowler BE (2014) Structure of a mitochondrial cytochrome c conformer competent for peroxidase activity, Proc. Natl. Acad. Sci. U.S.A 111, 6648–6653. 10.1073/pnas.1323828111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rytömaa M, and Kinnunen PKJ (1994) Evidence for two distinct acidic phospholipid-binding sites in cytochrome c, J. Biol. Chem 269, 1770–1774. https://www.jbc.org/content/269/3/1770. [PubMed] [Google Scholar]
- 32.Kooijman EE, Swim LA, Graber ZT, Tyurina YY, Bayır H, and Kagan VE (2017) Magic angle spinning 31P NMR spectroscopy reveals two essentially identical ionization states for the cardiolipin phosphates in phospholipid liposomes, Biochim. Biophys. Acta - Biomembr 1859, 61–68. 10.1016/j.bbamem.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malyshka D, Pandiscia LA, and Schweitzer-Stenner R (2014) Cardiolipin containing liposomes are fully ionized at physiological pH. An FT-IR study of phosphate group ionization, Vib. Spectrosc 75, 86–92. 10.1016/j.vibspec.2014.10.003. [DOI] [Google Scholar]
- 34.Houtkooper RH, and Vaz FM (2008) Cardiolipin, the heart of mitochondrial metabolism, Cell. Mol. Life Sci 65, 2493–2506. 10.1007/s00018-008-8030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayashi Y, Yamanaka M, Nagao S, Komori H, Higuchi Y, and Hirota S (2016) Domain swapping oligomerization of thermostable c-type cytochrome in E. coli cells, Sci. Rep 6, 19334 10.1038/srep19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin T-Y, and Weibel DB (2016) Organization and function of anionic phospholipids in bacteria, Appl. Microbiol. Biotechnol 100, 4255–4267. 10.1007/s00253-016-7468-x. [DOI] [PubMed] [Google Scholar]
- 37.Yang H, Yamanaka M, Nagao S, Yasuhara K, Shibata N, Higuchi Y, and Hirota S (2019) Protein surface charge effect on 3D domain swapping in cells for c-type cytochromes, Biochim. Biophys. Acta: Proteins Proteomics 1867, 140265 10.1016/j.bbapap.2019.140265. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi Y, Nagao S, Osuka H, Komori H, Higuchi Y, and Hirota S (2012) Domain swapping of the heme and N-terminal α-helix in Hydrogenobacter thermophilus cytochrome c552 dimer, Biochemistry 51, 8608–8616. 10.1021/bi3011303. [DOI] [PubMed] [Google Scholar]
- 39.Yamanaka M, Nagao S, Komori H, Higuchi Y, and Hirota S (2015) Change in structure and ligand binding properties of hyperstable cytochrome c555 from Aquifex aeolicus by domain swapping, Protein Sci. 24, 366–375. 10.1002/pro.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagao S, Ueda M, Osuka H, Komori H, Kamikubo H, Kataoka M, Higuchi Y, and Hirota S (2015) Domain-swapped dimer of Pseudomonas aeruginosa cytochrome c551: structural insights into domain swapping of cytochrome c family proteins, PLoS ONE 10, e0123653 10.1371/journal.pone.0123653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parui PP, Deshpande MS, Nagao S, Kamikubo H, Komori H, Higuchi Y, Kataoka M, and Hirota S (2013) Formation of oligomeric cytochrome c during folding by intermolecular hydrophobic interaction between N- and C-terminal α-helices, Biochemistry 52, 8732–8744. 10.1021/bi400986g. [DOI] [PubMed] [Google Scholar]
- 42.Maity H, Maity M, Krishna MM, Mayne L, and Englander SW (2005) Protein folding: the stepwise assembly of foldon units, Proc. Natl. Acad. Sci. U.S.A 102, 4741–4746. 10.1073/pnas.0501043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanske J, Toffey JR, Morenz AM, Bonilla AJ, Schiavoni KH, and Pletneva EV (2012) Conformational properties of cardiolipin-bound cytochrome c, Proc. Natl. Acad. Sci. U.S.A 109, 125–130. 10.1073/pnas.1112312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Matsuo T, Nagao S, and Hirota S (2011) Peroxidase activity enhancement of horse cytochrome c by dimerization, Org. Biomol. Chem 9, 4766–4769. 10.1039/c1ob05552f. [DOI] [PubMed] [Google Scholar]
- 45.Dawson JH (1988) Probing structure-function relations in heme-containing oxygenases and peroxidases, Science 240, 433 https://science.sciencemag.org/content/240/4851/433.abstract. [DOI] [PubMed] [Google Scholar]
- 46.Hersleth H-P, Ryde U, Rydberg P, Görbitz CH, and Andersson KK (2006) Structures of the high-valent metal-ion haem–oxygen intermediates in peroxidases, oxygenases and catalases, J. Inorg. Biochem 100, 460–476. 10.1016/j.jinorgbio.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 47.Olteanu A, Patel CN, Dedmon MM, Kennedy S, Linhoff MW, Minder CM, Potts PR, Deshmukh M, and Pielak GJ (2003) Stability and apoptotic activity of recombinant human cytochrome c, Biochem. Biophys. Res. Commun 312, 733–740. 10.1016/j.bbrc.2003.10.182. [DOI] [PubMed] [Google Scholar]
- 48.Duncan MG, Williams MD, and Bowler BE (2009) Compressing the free energy range of substructure stabilities in iso-1-cytochrome c, Protein Sci. 18, 1155–1164. 10.1002/pro.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Redzic JS, and Bowler BE (2005) Role of hydrogen bond networks and dynamics in positive and negative cooperative stabilization of a protein, Biochemistry 44, 2900–2908. 10.1021/bi048218b. [DOI] [PubMed] [Google Scholar]
- 50.Wandschneider E, Hammack BN, and Bowler BE (2003) Evaluation of cooperative interactions between substructures of iso-1-cytochrome c using double mutant cycles, Biochemistry 42, 10659–10666. 10.1021/bi034958t. [DOI] [PubMed] [Google Scholar]
- 51.Cherney MM, Junior C, and Bowler BE (2013) Mutation of trimethyllysine-72 to alanine enhances His79-heme mediated dynamics of iso-1-cytochrome c, Biochemistry 52, 837–846. 10.1021/bi301599g. [DOI] [PubMed] [Google Scholar]
- 52.Goldes ME, Jeakins-Cooley ME, McClelland LJ, Mou T-C, and Bowler BE (2016) Disruption of a hydrogen bond network in human versus spider monkey cytochrome c affects heme crevice stability, J. Inorg. Biochem 158, 62–69. 10.1016/j.jinorgbio.2015.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ensign AA, Jo I, Yildirim I, Krauss TD, and Bren KL (2008) Zinc porphyrin: a fluorescent acceptor in studies of Zn-cytochrome c unfolding by fluorescence resonance energy transfer, Proc. Natl. Acad. Sci. U.S.A 105, 10779–10784. 10.1073/pnas.0802737105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanderkooi JM, Adar F, and Erecińska M (1976) Metallocytochromes c : characterization of electronic absorption and emission spectra of Sn4+ and Zn2+ cytochromes c, Eur. J. Biochem 64, 381–387. 10.1111/j.1432-1033.1976.tb10312.x. [DOI] [PubMed] [Google Scholar]
- 55.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, and Sligar SG (2009) Reconstitution of membrane proteins in phospholipid bilayer nanodiscs Methods Enzymol. 464, 211–231. 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nath A, Atkins WM, and Sligar SG (2007) Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins, Biochemistry 46, 2059–2069. 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 57.Bayburt TH, Grinkova YV, and Sligar SG (2002) Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins, Nano Lett. 2, 853–856. 10.1021/nl025623k. [DOI] [Google Scholar]
- 58.Denisov IG, Grinkova YV, Lazarides AA, and Sligar SG (2004) Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size, J. Am. Chem. Soc 126, 3477–3487. 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 59.Belikova NA, Vladimirov YA, Osipov AN, Kapralov AA, Tyurin VA, Potapovich MV, Basova LV, Peterson J, Kurnikov IV, and Kagan VE (2006) Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes, Biochemistry 45, 4998–5009. 10.1021/bi0525573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Her C, Filoti Dana I., McLean Mark A., Sligar Stephen G., Alexander Ross, J. B., Steele H, and Laue Thomas M. (2016) The charge properties of phospholipid nanodiscs, Biophys. J 111, 989–998. 10.1016/j.bpj.2016.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baddam S, and Bowler BE (2005) Thermodynamics and kinetics of formation of the alkaline state of a Lys 79->Ala/Lys 73->His variant of iso-1-cytochrome c, Biochemistry 44, 14956–14968. 10.1021/bi0515873. [DOI] [PubMed] [Google Scholar]
- 62.Noble RW, and Gibson QH (1970) The reaction of ferrous horseradish peroxidase with hydrogen peroxide, J. Biol. Chem 245, 2409–2413. https://www.jbc.org/content/245/9/2409. [PubMed] [Google Scholar]
- 63.Nelson DP, and Kiesow LA (1972) Enthalpy of decomposition of hydrogen peroxide by catalase at 25 °C (with molar extinction coefficients of H2O2 solutions in the UV), Anal. Biochem 49, 474–478. 10.1016/0003-2697(72)90451-4. [DOI] [PubMed] [Google Scholar]
- 64.Goldschmid O (1953) The effect of alkali and strong acid on the ultraviolet absorption spectrum of lignin and related compounds, J. Am. Chem. Soc 75, 3780–3783. 10.1021/ja01111a052. [DOI] [Google Scholar]
- 65.Diederix REM, Ubbink M, and Canters GW (2001) The peroxidase activity of cytochrome c-550 from Paracoccus versutus, Eur. J. Biochem 268, 4207–4216. 10.1046/j.1432-1327.2001.02335.x. [DOI] [PubMed] [Google Scholar]
- 66.Buschmann V, Krämer B, Koberling F, Macdonald R, and Rüttinge S (2009) Quantitative FCS: determination of the confocal volume by FCS and bead scanning with the MicroTime 200, In Application Note, pp 1–8, PicoQuant GmbH, Berlin. [Google Scholar]
- 67.Szabelski M, Luchowski R, Gryczynski Z, Kapusta P, Ortmann U, and Gryczynski I (2009) Evaluation of instrument response functions for lifetime imaging detectors using quenched Rose Bengal solutions, Chem. Phys. Lett 471, 153–159. 10.1016/j.cplett.2009.02.001. [DOI] [Google Scholar]
- 68.Dale R, Chen L, and Brand L (1977) Rotational relaxation of the ‘microviscovity’ probe diphenylhexatriene in paraffin oil and egg lecithin vesicles, J. Biol. Chem 252, 7500–7510. https://www.jbc.org/content/252/21/7500.abstract. [PubMed] [Google Scholar]
- 69.Barkley MD, Kowalczyk AA, and Brand L (1981) Fluorescence decay studies of anisotropic rotations of small molecules, J. Chem. Phys 75, 3581–3593. 10.1063/1.442468. [DOI] [Google Scholar]
- 70.Meseth U, Wohland T, Rigler R, and Vogel H (1999) Resolution of fluorescence correlation measurements, Biophys. J 76, 1619–1631. 10.1016/S0006-3495(99)77321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saffarian S, and Elson EL (2003) Statistical analysis of fluorescence correlation spectroscopy: the standard deviation and bias, Biophys. J 84, 2030–2042. 10.1016/S0006-3495(03)75011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lakowicz JR (2006) Principles of Fluorescence Spectroscopy, 3rd ed., Springer, New York. [Google Scholar]
- 73.Bernad S, Oellerich S, Soulimane T, Noinville S, Baron MH, Paternostre M, and Lecomte S (2004) Interaction of horse heart and Thermus thermophilus type c cytochromes with phospholipid vesicles and hydrophobic surfaces, Biophys. J 86, 3863–3872. 10.1529/biophysj.103.025114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hong Y, Muenzner J, Grimm SK, and Pletneva EV (2012) Origin of the conformational heterogeneity of cardiolipin-bound cytochrome c, J. Am. Chem. Soc 134, 18713–18723. 10.1021/ja307426k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elmer-Dixon MM, and Bowler BE (2018) Electrostatic constituents of cardiolipin interaction with site A of cytochrome c, Biochemistry 57, 5683–5695. 10.1021/acs.biochem.8b00704. [DOI] [PubMed] [Google Scholar]
- 76.Heimburg T, and Marsh D (1995) Protein surface-distribution and protein-protein interactions in the binding of peripheral proteins to charged lipid membranes, Biophys. J 68, 536–546. https://dx.doi.org/10.1016%2FS0006-3495(95)80215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hobbs JK, Jiao W, Easter AD, Parker EJ, Schipper LA, and Arcus VL (2013) Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates, ACS Chemical Biology 8, 2388–2393. 10.1021/cb4005029. [DOI] [PubMed] [Google Scholar]
- 78.Lee JC, Chang IJ, Gray HB, and Winkler JR (2002) The cytochrome c folding landscape revealed by electron-transfer kinetics, J. Mol. Biol 320, 159–164. 10.1016/S0022-2836(02)00466-7. [DOI] [PubMed] [Google Scholar]
- 79.Rajagopal BS, Silkstone GG, Nicholls P, Wilson MT, and Worrall JAR (2012) An investigation into a cardiolipin acyl chain insertion site in cytochrome c, Biochim. Biophys. Acta 1817, 780–791. 10.1016/j.bbabio.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 80.Josephs TM, Liptak MD, Hughes G, Lo A, Smith RM, Wilbanks SM, Bren KL, and Ledgerwood EC (2013) Conformational change and human cytochrome c function: mutation of residue 41 modulates caspase activation and destabilizes Met-80 coordination, JBIC, J. Biol. Inorg. Chem 18, 289–297. 10.1007/s00775-012-0973-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rajagopal BS, Edzuma AN, Hough MA, Blundell KLIM, Kagan VE, Kapralov AA, Fraser LA, Butt JN, Silkstone GG, Wilson MT, Svistunenko DA, and Worrall JAR (2013) The hydrogen-peroxide-induced radical behaviour in human cytochrome c–phospholipid complexes: implications for the enhanced pro-apoptotic activity of the G41S mutant, Biochem. J 456, 441–452. 10.1042/BJ20130758. [DOI] [PubMed] [Google Scholar]
- 82.Pollock WB, Rosell FI, Twitchett MB, Dumont ME, and Mauk AG (1998) Bacterial expression of a mitochondrial cytochrome c. Trimethylation of Lys72 in yeast iso-1-cytochrome c and the alkaline conformational transition, Biochemistry 37, 6124–6131. 10.1021/bi972188d. [DOI] [PubMed] [Google Scholar]
- 83.Kristinsson R, and Bowler BE (2005) Communication of stabilizing energy between substructures of a protein, Biochemistry 44, 2349–2359. 10.1021/bi048141r. [DOI] [PubMed] [Google Scholar]
- 84.Maity H, Rumbley JN, and Englander SW (2006) Functional role of a protein foldon - an Ω-loop foldon controls the alkaline transition in ferricytochrome c, Proteins: Struct., Funct., Genet 63, 349–355. 10.1002/prot.20757. [DOI] [PubMed] [Google Scholar]
- 85.Cherney MM, and Bowler BE (2011) Protein dynamics and function: making new strides with an old warhorse, the alkaline conformational transition of cytochrome c, Coord. Chem. Rev 255, 664–677. 10.1016/j.ccr.2010.09.014. [DOI] [Google Scholar]
- 86.Milorey B, Schweitzer-Stenner R, Kurbaj R, and Malyshka D (2019) pH-induced switch between different modes of cytochrome c binding to cardiolipin-containing liposomes, ACS Omega 4, 1386–1400. 10.1021/acsomega.8b02574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pandiscia LA, and Schweitzer-Stenner R (2014) Salt as a catalyst in the mitochondria: returning cytochrome c to its native state after it misfolds on the surface of cardiolipin containing membranes, Chem. Commun 50, 3674–3676. 10.1039/c3cc48709a. [DOI] [PubMed] [Google Scholar]
- 88.Pandiscia LA, and Schweitzer-Stenner R (2015) Coexistence of native-like and non-native cytochrome c on anionic liposomes with different cardiolipin content, J. Phys. Chem. B 119, 12846–12859. 10.1021/acs.jpcb.5b07328. [DOI] [PubMed] [Google Scholar]
- 89.Elmer-Dixon MM, and Bowler BE (2017) Site A-mediated partial unfolding of cytochrome c on cardiolipin vesicles is species-dependent and does not require Lys72, Biochemistry 56, 4830–4839. 10.1021/acs.biochem.7b00694. [DOI] [PubMed] [Google Scholar]
- 90.Kawai C, Prado FM, Nunes GLC, Di Mascio P, Carmona-Ribeiro AM, and Nantes IL (2005) pH-dependent interaction of cytochrome c with mitochondrial mimetic membranes: the role of an array of positively charges amino acids, J. Biol. Chem 280, 34709–34717. 10.1074/jbc.M412532200. [DOI] [PubMed] [Google Scholar]
- 91.Mohammadyani D, Yanamala N, Samhan-Arias AK, Kapralov AA, Stepanov G, Nuar N, Planas-Iglesias J, Sanghera N, Kagan VE, and Klein-Seetharaman J (2018) Structural characterization of cardiolipin-driven activation of cytochrome c into a peroxidase and membrane perturbation, Biochim. Biophys. Acta - Biomembr 1860, 1057–1068. 10.1016/j.bbamem.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 92.Pandiscia LA, and Schweitzer-Stenner R (2015) Coexistence of native-like and non-native partially unfolded ferricytochrome c on the surface of cardiolipin-containing liposomes, J. Phys. Chem. B 119, 1334–1349. 10.1021/jp5104752. [DOI] [PubMed] [Google Scholar]
- 93.Sinibaldi F, Fiorucci L, Patriarca A, Lauceri R, Ferri T, Coletta M, and Santucci R (2008) Insights into the cytochrome c-cardiolipin interaction. Role played by ionic strength, Biochemistry 47, 6928–6935. [DOI] [PubMed] [Google Scholar]
- 94.Mahajan N, Hoover B, Rajendram M, Shi HY, Kawasaki K, Weibel DB, and Zhang M (2019) Maspin binds to cardiolipin in mitochondria and triggers apoptosis, FASEB J. 33, 6354–6364. 10.1096/fj.201802182R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kobayashi H, Nagao S, and Hirota S (2016) Characterization of the cytochrome c membrane-binding site using cardiolipin-containing bicelles with NMR, Angew. Chem. Int. Ed 55, 14019–14022. 10.1002/anie.201607419. [DOI] [PubMed] [Google Scholar]
- 96.Bren KL (2016) Going with the electron flow: heme electronic structure and electron transfer in cytochrome c, Isr. J. Chem 56, 693–704. 10.1002/ijch.201600021. [DOI] [Google Scholar]
- 97.Michel LV, Ye T, Bowman SEJ, Levin BD, Hahn MA, Russell BS, Elliott SJ, and Bren KL (2007) Heme attachment motif mobility tunes cytochrome c redox potential, Biochemistry 46, 11753–11760. 10.1021/bi701177j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kleingardner JG, Levin BD, Zoppellaro G, Andersson KK, Elliott SJ, and Bren KL (2018) Influence of heme c attachment on heme conformation and potential, JBIC, J. Biol. Inorg. Chem 23, 1073–1083. 10.1007/s00775-018-1603-3. [DOI] [PubMed] [Google Scholar]
- 99.Blauer G, Sreerama N, and Woody RW (1993) Optical activity of hemoproteins in the Soret region. Circular dichroism of the heme undecapeptide of cytochrome c in aqueous solution, Biochemistry 32, 6674–6679. 10.1021/bi00077a021. [DOI] [PubMed] [Google Scholar]
- 100.Schweitzer-Stenner R (2008) Internal electric field in cytochrome c explored by visible electronic circular dichroism spectroscopy, J. Phys. Chem. B 112, 10358–10366. 10.1021/jp802495q. [DOI] [PubMed] [Google Scholar]
- 101.Woody RW, and Hsu M-C (1971) The origin of heme cotton effects in myoglobin and hemoglobin, J. Am. Chem. Soc 93, 3515–3525. 10.1021/ja00743a036. [DOI] [PubMed] [Google Scholar]
- 102.Pielak GJ, Oikawa K, Mauk AG, Smith M, and Kay CM (1986) Elimination of the native Soret Cotton effect of cytochrome c by replacement of the invariant phenylalanine using site-directed mutagenesis, J. Am. Chem. Soc 108, 2724–2727. 10.1021/ja00270a035. [DOI] [Google Scholar]
- 103.Sinibaldi F, Droghetti E, Polticelli F, Piro MC, Di Pierro D, Ferri T, Smulevich G, and Santucci R (2011) The effects of ATP and sodium chloride on the cytochrome c–cardiolipin interaction: The contrasting behavior of the horse heart and yeast proteins, J. Inorg. Biochem 105, 1365–1372. 10.1016/j.jinorgbio.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 104.Sinibaldi F, Howes BD, Droghetti E, Polticelli F, Piro MC, Di Pierro D, Fiorucci L, Coletta M, Smulevich G, and Santucci R (2013) Role of lysines in cytochrome c−cardiolipin interaction, Biochemistry 52, 4578–4588. 10.1021/bi400324c. [DOI] [PubMed] [Google Scholar]
- 105.Sinibaldi F, Howes BD, Piro MC, Polticelli F, Bombelli C, Ferri T, Coletta M, Smulevich G, and Santucci R (2010) Extended cardiolipin anchorage to cytochrome c: a model for protein-mitochondrial membrane binding, JBIC, J. Biol. Inorg. Chem 15, 689–700. 10.1007/s00775-010-0636-z. [DOI] [PubMed] [Google Scholar]
- 106.Elmer-Dixon MM (2018) Elucidation of the physical and chemical properties of cytochrome c - cardiolipin interactions, In Biochemistry & Biophysics, University of Montana, Missoula, Montana. [Google Scholar]
- 107.Verbaro D, Hagarman A, Soffer J, and Schweitzer-Stenner R (2009) The pH dependence of the 695 nm charge transfer band reveals the population of an intermediate state of the alkaline transition of ferricytochrome c at low ion concentrations, Biochemistry 48, 2990–2996. 10.1021/bi802208f. [DOI] [PubMed] [Google Scholar]
- 108.Junedi S, Yasuhara K, Nagao S, Kikuchi J-I, and Hirota S (2014) Morphological change of cell membrane by interaction with domain-swapped cytochrome c oligomers, ChemBioChem 15, 517–521. 10.1002/cbic.201300728. [DOI] [PubMed] [Google Scholar]
- 109.Green DR (2005) Apoptotic pathways: ten minutes to dead, Cell 121, 671–674. 10.1016/j.cell.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 110.Nold SM, Lei H, Mou T-C, and Bowler BE (2017) Effect of a K72A mutation on the structure, stability, dynamics and peroxidase activity of human cytochrome c, Biochemistry 56, 3358–3368. 10.1021/acs.biochem.7b00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Muenzner J, Toffey JR, Hong Y, and Pletneva EV (2013) Becoming a peroxidase: cardiolipin-induced unfolding of cytochrome c, J. Phys. Chem. B 117, 12878–12886. 10.1021/jp402104r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Radi R, Thomson L, Rubbo H, and Prodanov E (1991) Cytochrome c-catalyzed oxidation of organic molecules by hydrogen peroxide, Arch. Biochem. Biophys 288, 112–117. 10.1016/0003-9861(91)90171-E. [DOI] [PubMed] [Google Scholar]
- 113.Xu Y, Mayne L, and Englander SW (1998) Evidence for an unfolding and refolding pathway in cytochrome c, Nat. Struct. Biol 5, 774–778. 10.1038/1810. [DOI] [PubMed] [Google Scholar]
- 114.Adams PA, Baldwin DA, and Marques HM (1996) The hemepeptides from cytochrome c: Preparation, physical and chemical properties, and their use as model compounds for the hemoproteins, In Cytochrome c: A Multidisciplinary Approach (Mauk AG, and Scott RA, Eds.), pp 635–692, University Science Books, Sausalito, CA. [Google Scholar]
- 115.Diederix REM, Ubbink M, and Canters GW (2002) Peroxidase activity as a tool for studying the folding of c-type cytochromes, Biochemistry 41, 13067–13077. 10.1021/bi0260841. [DOI] [PubMed] [Google Scholar]
- 116.Laun P, Buettner S, Rinnerthaler M, Burhans WC, and Breitenbach M (2012) Yeast Aging and Apoptosis, In Subcellular Biochemistry: Aging Research in Yeast (Breitenbach M, Jazwinski SM, and Laun P, Eds.), pp 207–232, Springer, Netherlands. [DOI] [PubMed] [Google Scholar]
- 117.Porcelli AM, Ghelli A, Zanna C, Pinton P, Rizzuto R, and Rugolo M (2005) pH difference across the outer mitochondrial membrane measured with a green fluorescent protein mutant, Biochem. Biophys. Res. Commun 326, 799–804. 10.1016/j.bbrc.2004.11.105. [DOI] [PubMed] [Google Scholar]
- 118.Nilsson C, Kågedal K, Johansson U, and Öllinger K (2003) Analysis of cytosolic and lysosomal pH in apoptotic cells by flow cytometry, Methods Cell Sci. 25, 185–194. 10.1007/s11022-004-8228-3. [DOI] [PubMed] [Google Scholar]
- 119.Hackenbrock CR, Chazotte B, and Gupte SS (1986) The random collision model and a critical assessment of diffusion and collision in mitochondrial electron transport, J. Bioenerg. Biomembr 18, 331–368. 10.1007/BF00743010. [DOI] [PubMed] [Google Scholar]
- 120.Morales JG, Holmes-Hampton GP, Miao R, Guo Y, Münck E, and Lindahl PA (2010) Biophysical characterization of iron in mitochondria isolated from respiring and fermenting yeast, Biochemistry 49 5436–5444. 10.1021/bi100558z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


