Abstract
Previously, we described a series of salicylhydrazide compounds with potent anti-cancer activities against a panel of human cancer cell lines derived from different origins. Preclinical evaluation showing efficacy both in vitro and in vivo in human cancer models indicated that these agents may represent a promising class of anticancer drugs. In the present study, we performed an in-depth investigation on the underlying molecular mechanisms of the most potent compounds, SC21 and SC23, using a proteomic method and bioinformatics tools. We demonstrated that SC23 induced apoptosis through multiple signaling pathways. In particular, SC23 regulated the expression of Bcl-2, p21, acetylated histone H3 and β-tubulin and the combined modulation of these proteins may result in the induction of apoptosis. We also examined the effect of SC21 and SC23 on cell cycle progression and found that both compounds arrested cells in S-phase in most cell lines tested. To better understand the signaling networks involved, we analyzed the SC21- and SC23-treated cell lysates by the Kinexus™ 628 antibody microarray. The results were interpreted with the aid of Ingenuity Pathway Analysis (IPA) software. It was found that SC21 interfered with JAK/STAT signaling and elicited apoptosis through Fas and caspases pathways. Unlike SC21, SC23 induced RAR activation and caused cell cycle arrest. The signaling networks identified by this work may provide the basis for future mechanistic studies. The validation of the proposed pathways and the elucidation of the signaling cross-talk are currently under way.
INTRODUCTION
Cancer is the second leading cause of death world-wide. Despite great progress in the development of anticancer drugs, there is still a desperate need for more effective and well-tolerated agents [1, 2]. The discovery of anticancer chemotherapeutics is highly challenging due to the nonselective, multi-target mechanisms employed by most agents [3, 4]. Therefore, a major objective in drug discovery programs is the identification of suitable lead compounds with desired biological activities and selective molecular targets. Insufficient target validation frequently causes failures in the drug development [5, 6]. Previously, we reported a salicylhydrazide class of compounds as potent HIV-1 integrase (IN) inhibitors. The development of this class of compounds as anti-retroviral agents was halted due to their cytotoxicities resulting from a lack of selectivity for integrase [7-10]. The remarkable cytotoxicity of this class of compounds led us to explore their potentials as anticancer agents. In searching our in-house multiconformational database of 5 million compounds for anticancer drug leads, we identified more than 2,200 compounds having common structures and pharmacophore fragments. Compounds satisfying absorption, distribution, metabolism, excretion, and toxicity (ADMET) criteria were obtained from commercial sources and subjected to 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay for initial cytotoxicity screening. Eighteen compounds showed notable activities in the cell-based assays. Among these, SC21 and SC23 were found to be highly potent against a panel of cancer cell lines [11]. Further detailed studies revealed that SC21 exerted its in vitro biological functions by triggering cell cycle arrest and apoptosis in several cancer cell lines derived from different origins. In addition to its in vitro activities, SC21 also exhibited potent in vivo efficacy at dosages as low as 0.3 mg/Kg in a prostate cancer mouse xenograft model [11]. Our preclinical evaluation of SC21 and SC23 suggests that salicylhydrazides may represent a promising class of anticancer drugs. Accordingly, there was a need to better understand signaling networks affected in order to optimize the structures of these compounds and improve their anticancer therapeutic potential.
Bioinformatics technologies allow simplification of complex biological systems to better understand signaling and metabolic pathways. The application of bioinformatics continues to impact drug discovery and it is being widely used in all phases of drug discovery processes [12]. In our current study, we explored the molecular mechanisms of SC21 and SC23 cytotoxicity using proteomics and it bioinformatics analysis. We found several proteins, including β-tubulin, myc promoter-binding protein (MPB-1), and vimentin to be significantly changed by SC23. Validation of β-tubulin up-regulation by Western blotting, confocal microscopy and flow cytometry was consistent with the proteomic observations. In order to understand the underlying mechanisms involved in the activity of SC21 and SC23, we searched for potentially targeted signaling molecules using the Kinexus™ 628 antibody microarray analysis. The expressions of numerous proteins were identified to be significantly altered in response to treatment. We next analyzed the signaling pathways mediated by SC21 and SC23 with the aid of the Ingenuity Pathway Analysis (IPA) platform. Interestingly, it was found that these two compounds have distinct mechanisms of action as proposed by IPA even though they are structurally very similar. In brief, SC21 was suggested to play a role in post-translational modifications and small molecule biochemistry through JAK/STAT signaling and apoptosis pathways, whereas, SC23 was found to act on cell cycle regulation and cancer biology by controlling the cell cycle checkpoint and interfering with retinoic acid receptor (RAR) activation pathways. Validation of the signaling pathways using molecular biological tools is currently under way.
RESULTS AND DISCUSSION
SC21 and SC23 Exhibit Remarkable Potency against a Panel of Cancer Cell Lines
The cytotoxicities of SC21 and SC23 (Fig. 1) were examined in a large panel of human cancer cell lines, including the National Cancer Institute’s NCI60 cell lines and non-small cell lung cancer (NSCLC) cell lines, using MTT assay (Table 1). In all the cell lines tested, both SC21 and SC23 showed potent cytotoxicity in the nanomolar to submicromolar range, an activity comparable to that of camptothecin (CPT) (data not shown). In particular, both compounds exhibited excellent activities against doxorubicin-resistant NCI/ADR-RES cells with IC50 values less than 0.1 μM. Their toxicity in NCI/ADR-RES cells is noteworthy because this cell line overexpresses multi-drug resistant transporters, the most common mechanism for acquired resistance by cancer cells [13]. Considering their broad-spectrum cytotoxicity profiles, we further examined the effects of SC21 and SC23 on tumor cell viability and proliferation using colony formation assays in selected cell lines, including HCT116 p53+/+ and HCT116 p53−/− colon cancer cell lines. Unlike MTT assay, colony formation assay reflects multiple forms of cell death, including necrosis and autophagy, in addition to apoptosis, and, therefore, it is more indicative of the cellular response to chemotherapeutics [14-6]. As expected, the exposure of both cell lines to SC21 (data not shown) and SC23 (Fig. 2A) resulted in significant inhibition of colony formation, further confirming their potent cytotoxic properties. Interestingly, SC23 exhibited a stronger suppression on the colony formation in HCT116 p53−/−, which suggests that not only apoptotic cell death, but other types of cell death may be induced in this cell line [17].
Fig. 1.
Chemical structures of SC21 and SC23.
Table 1.
Cytotoxicity of SC21 and SC23 in the NCI 60 Human Cancer Cell Lines
| Cell Line | SC21 | SC23 | ||
|---|---|---|---|---|
| Experiment 1 GI50 |
Experiment 2 GI50 |
Experiment 1 GI50 |
Experiment 2 GI50 |
|
| Leukemia | ||||
| CCRF-CEM | 1.00E-08 | 1.00E-08 | 1.03E-07 | 5.10E-07 |
| HL-60(TB) | 1.00E-08 | 4.95E-08 | 1.79E-07 | |
| K-562 | 1.00E-08 | 1.00E-08 | 1.00E-08 | 3.32E-07 |
| MOLT-4 | 1.00E-08 | 1.00E-08 | 1.35E-07 | 6.17E-07 |
| RPMI-8226 | 1.00E-08 | 1.00E-08 | 1.28E-07 | 6.17E-07 |
| SR | 4.01E-08 | 4.55E-08 | ||
| Non-Small Cell Lung Cancer | ||||
| A549/ATCC | 1.00E-08 | 1.00E-08 | 1.00E-08 | 1.85E-05 |
| EKVX | 1.00E-08 | 2.25E-06 | 1.48E-06 | 8.36E-07 |
| HOP-62 | 1.00E-08 | 1.00E-08 | ||
| HOP-92 | 1.00E-08 | 1.00E-08 | 3.45E-08 | |
| NCI-H226 | 2.10E-05 | 1.00E-08 | 3.15E-08 | |
| NCI-H23 | 1.00E-08 | 1.00E-08 | 2.33E-07 | |
| NCI-H322M | 1.73E-07 | 4.52E-08 | 1.00E-04 | 2.60E-06 |
| NCI-H460 | 1.00E-08 | 1.00E-08 | 1.83E-07 | |
| NCI-H522 | 1.00E-08 | 2.75E-07 | 3.52E-07 | |
| Colon Cancer | ||||
| COLO205 | 3.03E-06 | 4.21E-06 | 1.00E-04 | 1.00E-04 |
| HCC-2998 | 2.14E-06 | 1.00E-08 | 1.00E-04 | 2.41E-05 |
| HCT-116 | 1.00E-08 | 1.00E-08 | 2.12E-07 | 2.97E-07 |
| HCT-15 | 1.00E-08 | 1.00E-08 | 1.49E-07 | 6.04E-07 |
| HT29 | 1.73E-06 | 3.32E-06 | 1.00E-08 | 1.11E-07 |
| KM12 | 1.38E-05 | 7.03E-07 | 8.37E-07 | |
| SW-620 | 1.00E-08 | 1.52E-08 | 2.28E-07 | 2.13E-06 |
| CNS Cancer | ||||
| SF-268 | 1.00E-08 | 1.00E-08 | 2.32E-08 | |
| SF-295 | 1.00E-08 | 1.00E-08 | 1.00E-08 | 6.97E-08 |
| SF-539 | 1.00E-08 | 1.00E-08 | 2.21E-07 | |
| SNB-19 | 1.00E-08 | 5.23E-08 | ||
| SNB-75 | 1.00E-08 | 3.45E-08 | ||
| U251 | 1.00E-08 | 1.00E-08 | 1.00E-08 | 2.73E-06 |
| Melanoma | ||||
| LOX IMVI | 1.00E-08 | 1.00E-08 | 8.31E-08 | |
| MALME-3M | 4.02E-08 | 1.00E-08 | 1.29E-07 | |
| M14 | 2.70E-08 | 1.00E-08 | 1.00E-08 | 7.39E-08 |
| SK-MEL-2 | 1.00E-08 | 1.41E-07 | ||
| SK-MEL-28 | 5.94E-06 | 2.47E-06 | 4.14E-06 | 1.00E-04 |
| SK-MEL-5 | 1.00E-08 | 1.00E-08 | 1.00E-08 | 4.99E-08 |
| UACC-257 | 6.65E-08 | 4.30E-07 | ||
| UACC-62 | 1.00E-08 | 1.00E-08 | 1.00E-08 | 4.17E-08 |
| Ovarian Cancer | ||||
| IGROV1 | 1.00E-08 | 1.00E-08 | 1.00E-08 | 5.03E-08 |
| OVCAR-3 | 1.22E-07 | 1.50E-08 | 3.25E-07 | |
| OVCAR-4 | 1.00E-08 | 1.40E-06 | 3.85E-07 | 1.22E-06 |
| OVCAR-5 | 2.06E-06 | 1.00E-04 | 1.00E-04 | 1.00E-04 |
| OVCAR-8 | 1.00E-08 | 1.71E-08 | 2.33E-07 | |
| SK-OV-3 | 1.00E-08 | 1.00E-08 | ||
| Renal Cancer | ||||
| 786-0 | 1.00E-08 | 1.00E-08 | 1.98E-07 | |
| A498 | 1.00E-08 | |||
| ACHN | 1.00E-08 | 1.00E-08 | 1.00E-08 | 2.61E-08 |
| CAKI-1 | 1.00E-08 | 1.00E-08 | 1.00E-08 | 5.01E-08 |
| RXF 393 | 1.00E-08 | 1.00E-08 | 1.00E-04 | |
| SN12C | 1.00E-08 | 1.00E-08 | 1.00E-04 | |
| TK-10 | 1.00E-08 | 1.00E-08 | 2.18E-05 | 2.97E-07 |
| UO-31 | 1.00E-08 | 1.00E-08 | 1.00E-08 | 1.28E-08 |
| Prostate Cancer | ||||
| PC-3 | 6.25E-08 | 3.47E-06 | 2.14E-05 | |
| DU-145 | 1.00E-08 | 1.00E-08 | 1.00E-04 | 1.00E-04 |
| Breast Cancer | ||||
| MCF7 | 1.32E-08 | 1.00E-08 | 2.37E-07 | 4.94E-07 |
| NCI/ADR-RES | 1.00E-08 | 1.00E-08 | 7.42E-08 | |
| MDA-MB-231/ATCC | 1.00E-08 | 1.00E-08 | ||
| MDA-MB-435 | 1.00E-08 | 1.00E-08 | 1.00E-08 | 3.24E-08 |
| MDA-N | 1.00E-08 | 1.00E-08 | ||
| T-47D | 1.00E-08 | 1.00E-08 | 1.00E-04 | 1.00E-04 |
| BT-549 | 1.00E-08 | 1.00E-04 | ||
| HS 578T | 1.00E-08 | |||
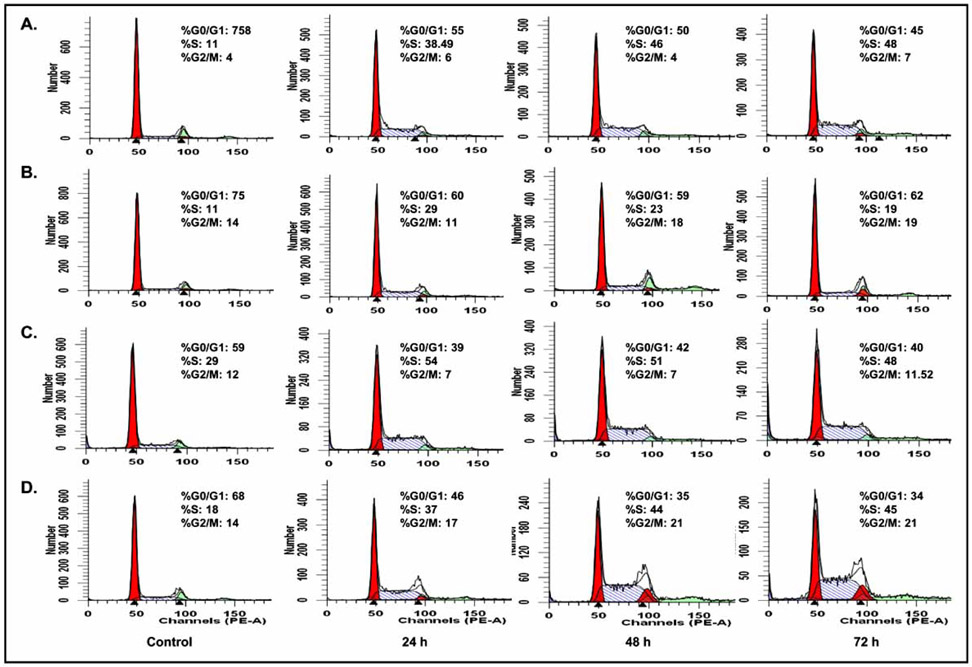
Fig. 2. Flow cytometric analysis of the cell cycle profiles of A549 and H1299 cells treated with SC21 and SC23.
Cells were treated with SC21 (15 μM, A549 and H1299 cells, panels A and C, respectively) and SC23 (10 μM, A549 and H1299 cells, panels B and D, respectively) for 0, 24, 48 and 72 h. The cells were then harvested, stained with propidium iodide and analyzed for cell cycle perturbation.
SC21 and SC23 Treatments Disrupt Cell Cycle Progression in NSCLC
Cell cycle perturbations induced by SC21 and SC23 were examined in several cell lines, including A549 and H1299 non-small cell lung cancer (NSCLC) cells. Cells were treated with SC21 and SC23 for 24, 48, and 72 h. The analysis of DNA profiles by flow cytometry demonstrated that both compounds halted cell cycle progression at S-phase in both cell lines (Fig. 2) In particular, at 48 h, there was an increase of 35% and 22% of cells retained in S-phase in SC21-treated A549 and H1299 cells, respectively, and 12% and 26% of S-phase arrested cells in SC23-treated A549 and H1299 cells, respectively, as compared to their corresponding untreated control cells. Interestingly, SC21 caused G0/G1 phase arrest in DU145 and MDA-MB-435 cells as reported in our previous study [11], while SC23 accumulated cells in S-phase in all the cell lines tested so far, similar to the S-phase arrest properties of CPT [18] (data not shown). The differential cell cycle arrest pattern of SC21 and SC23 indicated that they function through distinct mechanisms. The properties of SC21 and SC23 to disrupt cell cycle progression potentially make them attractive agents for combination treatment with drugs such as paclitaxel that act at different stages of the cell cycle.
SC23 Induces Apoptosis in Human Colon Cancer Cell Lines
Intracellular reactive oxygen species (ROS) trigger apoptosis by promoting the release of cytochrome c, regulating the kinase activity of DNA-PK and stabilizing Topo II-DNA cleavable complexes [19, 20]. To investigate whether ROS contribute to the cytotoxicity observed in SC21- and SC23-treated cells, we measured the changes of mitochondrial ROS production in SKOV-3 cells exposed to these two compounds. Mitochondrial ROS were detected with MitoSOX Red Mitochondrial Superoxide Live Cell Indicator and analyzed by flow cytometry [21]. As shown in Fig. (3B) neither SC21 nor SC23 stimulated ROS production after 24 h treatment. Similar results were also observed in EKVX cells (data not shown). These findings suggested that the two compounds trigger cell death through mechanisms other than ROS induction.
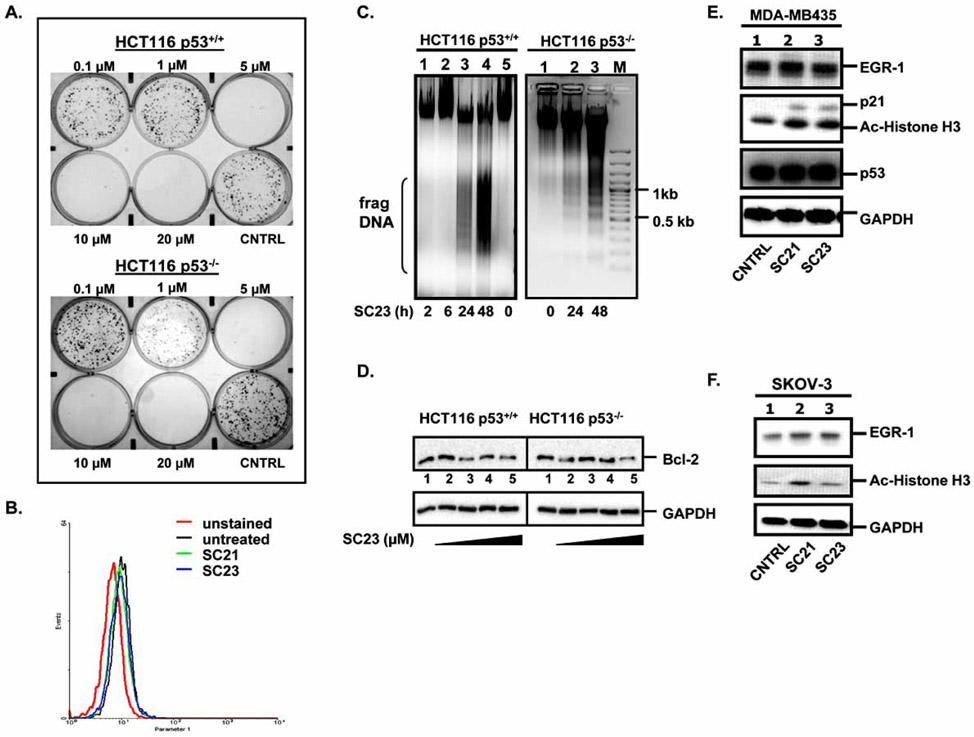
Fig. 3. SC23 triggers apoptosis in HCT116 p53+/+ and HCT116 p53−/− cells.
(A). SC23 exhibits cytotoxicity in the tested cell lines as shown by colony formation assay. (B). Detection of mitochondrial reactive oxygen species (ROS) levels in SKOV-3 cells. SKOV-3 cells were treated with SC21 and SC23 for 24 h. The untreated and treated cells were incubated with MitoSOX, trypsinized, washed with Hank’s Balanced Salt Solution, and subjected to flow cytometric analysis. (C). SC23 causes DNA fragmentation in both HCT116 p53+/+ and HCT116 p53−/− cells. Cells were treated with 5 μM of SC23 for the indicated duration. Cells were then lysed with lysis buffer. Precipitated DNA was resuspended in TE buffer and digested with RNase A and the digested product was separated on 1 % agarose gel. (D). SC23 decreases the anti-apoptotic Bcl-2 protein levels in both cell lines. HCT116 p53+/+ and HCT116 p53−/− cells were treated with increasing concentrations of SC23 for 24 h. Whole cell lysates were extracted and equal amounts of total protein were separated on 10% SDS-PAGE. The proteins were transferred to nitrocellulose membranes and subjected to immunoblotting with anti-Bcl-2 antibodies. (E). SC21 and SC23 up-regulate the expressions of p21 and acetylated histone H3 in MDA-MD-435 cells. Cells were treated with SC21 and SC23 for 24 h. Whole cell lysates were subjected to Western blotting analysis with the appropriate antibodies. (F). SC21 and SC23 up-regulate acetylated histone H3 in SKOV-3 cells. Cells were treated with SC21 and SC23 for 24 h. Whole cell lysates were analyzed with Western blotting with the appropriate antibodies.
Cytotoxic anticancer drugs commonly elicit apoptosis [22]. A hallmark of apoptosis is the breakdown of nuclear DNA into multiple ~200 bp oligonucleosomal DNA fragments. Focusing our studies on SC23, we examined the role of apoptosis in SC23-mediated cell death by nuclear DNA fragmentation assays. HCT116 p53+/+ and HCT116 p53−/− cells were treated with 5 μM of SC23 for various times and the extracted total DNA was separated on a 1% agarose gel. As shown in Fig. (3C), fragmented DNA appeared 24 h after treatment and the amount of DNA fragments increased after 48 h treatment in both cell lines, indicating that SC23 elicits early apoptosis in a time-dependent manner. It is noteworthy that the different p53 status in these two cell lines implies that SC23 utilizes both p53-dependent and –independent apoptotic mechanisms. Previously, we also demonstrated that SC21 activity is mediated by apoptosis in a fashion comparable to that of CPT [11]. Based on these observations, we assumed that apoptosis is one of the mechanisms of SC23- and SC21-induced cytotoxicity and we continued to investigate the roles of several apoptosis-related genes.
A great number of genes involved in apoptosis have been found to be defective in cancer cells. Among them, the anti-apoptotic protein Bcl-2 is known to be overexpressed in most human malignancies [23]. Bcl-2 inhibits the release of cytochrome c through permeability transition pores (PTP) within the mitochondrial membranes [24, 25]. The importance of Bcl-2 in cancer therapy is exemplified by the findings that its overexpression contributes to chemoresistance by blocking the apoptotic pathways involved with the mechanisms of action of many currently available anticancer drugs. Therefore, down-regulation of Bcl-2 might be an attractive strategy for anti-cancer drug development [26]. To understand the mechanisms of SC23-induced apoptosis, we employed Western blotting to examine the protein levels of Bcl-2 in response to SC23 treatment. SC23 decreased Bcl-2 protein levels in both cell lines at concentrations above 0.5 μM (Fig. 3D, lanes 3-5 versus lane 1). The fact that SC23 down-regulated the protein levels of the anti-apoptotic Bcl-2 indicated that it may function as a Bcl-2 inhibitor to induce cell death.
Besides Bcl-2, several other genes, such as EGR-1 and p53, are also closely involved in the process of apoptosis and cell proliferation and their expression has been found to be altered in cancer cells [27-30]. To further delineate the mechanisms of SC23 cytotoxicity, we examined the expression of several signaling molecules in two sensitive cell lines by Western blotting. In MDA-MB-435 cells, SC21 and SC23 increased protein levels of p21 and acetylated histone H3 (Fig. 3E). In SKOV-3 cells, SC21 enhanced protein levels of acetylated histone H3, similar to the result observed in MDA-MB-435 cells, while SC23 failed to do so (Fig. 3F). In both cell lines neither SC21 nor SC23 affected EGR-1 protein levels (Fig. 3E and Fig. 3F). In addition to EGR-1, p53, an important pro-apoptotic protein, was also found to remain unchanged by both compounds in MDA-MB-435 cells (Fig. 3E). We were not able to examine the protein expression of p21 and p53 in SKOV-3 cells due to undetectable levels in this particular cell line [31, 32]. In summary, SC21 and SC23 acted through multiple apoptosis-related signaling pathways. The interplay among these pathways may explain the potent cytotoxicities exhibited by SC21 and SC23.
A Proteomics Approach Reveals that SC23 Regulates the Expression of Multiple Proteins
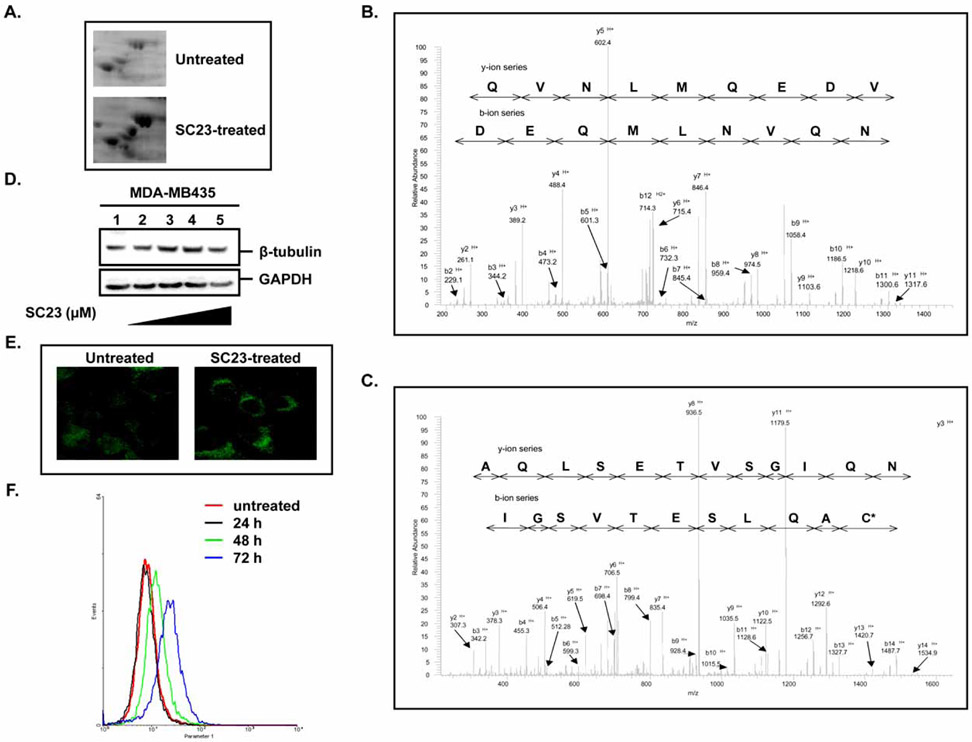
The broad-spectrum cytotoxicity of SC23 and its effects on several apoptosis-related proteins prompted us to further explore its molecular mechanisms of action. Using proteomics- based technology, we compared the protein expression profiling between untreated and SC23-treated cells following 2-D gel electrophoresis. A representative section of 2-D gel is presented in Fig. (3A). The protein spots on 2-D gels were quantified with PDQuest software (Bio-Rad) and approximately 125 spots were identified as being significantly changed (> 2-fold). Proteins regulated by SC23 include β-tubulin, myc promoter-binding protein (MPB-1), retinoblastoma-binding protein 7, vimentin, enolase, phosphopyrubate hydratase β, and mitochondrial ATP synthase β chain. To confirm the proteins identified by PDQuest software, proteins corresponding to β-tubulin and MPB-1 were digested and the digested peptides were analyzed by tandem mass spectrometry. Figs. (3B and 3C) show the spectra of β-tubulin peptide (EVDEQMLNVQNK) and MPB-1 peptide (VNQIGSVTESLQAC*K), respectively. In general, we were able to identify most of the proteins with more than 40% sequence coverage. The spots that did not show good peptide coverage either due to insufficient amount of sample, low protein abundance, or lack of reliable fragments were not explored further. By using this proteomics approach, we identified a set of proteins that were significantly changed by SC23. The characterization of these proteins could provide valuable insights for our future in-depth mechanistic studies.
Among the identified proteins showing significant changes in our proteomic studies, β-tubulin was found to be up-regulated 4-fold by SC23. To validate the results generated by proteomic analysis, we examined β-tubulin protein levels using different approaches. In particular, SC23 increased protein levels of β-tubulin at concentrations above 0.5 μM (Fig. 4D). The Western blotting results were further confirmed by confocal microscopy (Fig. 4E) and flow cytometry, which also demonstrated the time-dependent pattern of β-tubulin induction by SC23 (Fig. 4F). Regulation of β-tubulin protein levels is of biological significance considering the pivotal roles played by microtubules in cell proliferation, motility, cell shape and signaling. The dynamics of microtubules is critical for normal cellular functions and, hence, disruption of the dynamics, such as altering the free tubulin pool, could lead to apoptosis [33]. Based on the above observations, we conclude that modulation of β-tubulin expression and possibly, microtubule dynamics as well, may represent an additional mechanism utilized by SC23 to induce cell death.
Fig. 4. Proteomic analysis of SC23-treated cells.
(A). Selected regions from a 2D gel of untreated and SC23-treated T24 cells. (B). Representative tandem MS spectrum of β-tubulin isolated from 2-D gel electrophoresis analysis of SC23-treated cells. (C). Representative tandem MS spectrum of myc promoter-binding protein (MPB-1) isolated from 2-D gel electrophoresis analysis of SC23-treated cells. (D). SC23 increases β-tubulin expression in MDA-MB-435 cells. MDA-MB-435 cells were treated with the indicated concentrations of SC23 for 24 h. Whole cell lysates were extracted and equal amounts of total protein were subjected to Western blotting analysis. (E). Confocal microscopy indicates that SC23 increases β-tubulin expression in MDA-MB-435 cells. Cells were treated with 5 μM of SC23 for 24 h. After the treatment, cells were fixed with 100% ice-cold methanol. Anti-β-tubulin primary antibody and Cy3-conjugated goat-anti-rabbit secondary antibody were used to visualize the proteins. (F). Flow cytometry analyses show that SC23 increases β-tubulin expression in a time-dependent manner. MDA-MB-435 cells were treated with 5 μM of SC23 for the indicated durations. At the end of the treatment, cells were fixed with 3.7% formaldehyde and stained with the appropriate antibodies. The stained cells were subjected to flow cytometry analyses.
Network Analysis Using Ingenuity Pathway Analysis (IPA) Software
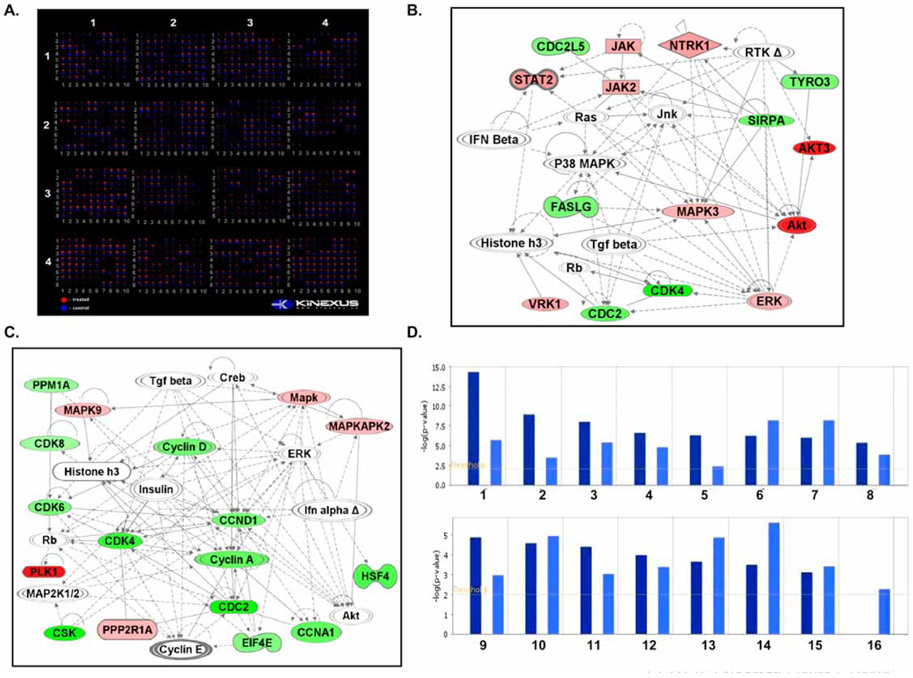
To further understand their mechanisms of action, we subjected SC21- and SC23-treated cell lysates to Kinexus™ 628-antibody microarray analysis to identify potential signaling molecules. 350 pan-specific and 270 phospho-site antibodies were included to track changes in protein expression and phosphorylation status. The cell lysates were run in parallel on the Kinexus™ antibody chip and the differential binding of dye-labeled proteins was detected and quantified (Fig. 5A). Proteins with significantly altered expressions were identified and the results are summarized in Table 2 to Table 5.
Fig. 5. Comparison of SC21- and SC23-mediated signaling pathways using bioinformatics tools.
(A). The Kinex™ antibody microarray of SC21- and SC23-treated MDA-MB-435 cell lysates. MDA-MB-435 cells were treated with 0.1 μM and 1 μM of SC21 and SC23, respectively, for 24 h. The total proteins were harvested and subjected to 628-protein microarray analysis using Kinexus™ Kinex antibody microarray platform. (B). Ingenuity pathway network of SC21 (score 39) obtained on a set of altered proteins. Proteins with their ratio changes and corresponding Swiss-Prot accession numbers were uploaded into IPA software. —: Direct interaction; - - - -: Indirect interaction. (C). Ingenuity pathway network of SC23 (score 41) obtained on a set of altered proteins. Proteins with their ratio changes and corresponding Swiss-Prot accession numbers were uploaded into IPA software. —: Direct interaction; - - - -: Indirect interaction. (D). Biological function comparison of SC21 and SC23. The biological functions of SC21 and SC23 were compared with IPA comparison analysis. 16 biological functions of interest were selected. 1. Cell death, 2. Cell cycle, 3. Cancer, 4. Cellular growth and proliferation, 5. DNA replication, recombination and repair, 6. Post-translational modification, 7. Small molecule biochemistry, 8. Cellular development, 9. Molecular transport, 10. Cell morphology, 11. Gene expression, 12. Cellular movement, 13. Cell signaling, 14. Immunological disease, 15. Cell-cell signaling and interaction, 16. Drug metabolism.
Table 2.
Analysis of Protein Phosphorylation Changes by SC21 in MDA-MB435 Cells Using Kinexus™ Kinex Antibody Microarray Screen
| Target Protein | Phosphorylated Protein | |
|---|---|---|
| Phospho-site | Fold Change | |
| eIF2α | S36 | 0.33 |
| Zap70 | Y315 + Y319 | 0.55 |
| Caveolin | S36 | 0.56 |
| Histone H3 | S28 | 0.56 |
| Rad17 | S645 | 0.62 |
| Hsp27 | S15 | 2.04 |
| PTEN | S370 | 2.06 |
| PKCε | S729 | 2.25 |
Table 5.
Analysis of Total Protein Level Changes by SC23 in MDA-MB435 Cells Using Kinexus™ Kinex Antibody Microarray Screen
| Target Protein | Total Protein |
|---|---|
| Fold Change | |
| Csk | 0.32 |
| CDK1 | 0.33 |
| CDK4 | 0.38 |
| HSF4 | 0.42 |
| Cyclin A | 0.45 |
| PKA Cα/β | 0.47 |
| Cyclin D | 0.49 |
| LAR | 0.49 |
| CDK6 | 0.51 |
| Hsp60 | 0.51 |
| eIF4E | 0.52 |
| PP2Cα/β | 0.60 |
| CDK8 | 0.62 |
| JNK2 | 1.55 |
| MAPKAPK2 | 1.55 |
| PKM2 | 1.55 |
| PP4/’2 | 1.55 |
| STAT5B | 1.57 |
| NME6 | 1.60 |
| PP2A/Aα/β | 1.61 |
| STAT1α/β | 1.68 |
| SOD(Cu/Zn) | 1.88 |
| CASP12 | 1.96 |
| ROKβ (ROCK1) | 2.46 |
| ROR2 | 2.60 |
| Plk1 | 3.36 |
To understand the signaling networks affected by SC21 and SC23, we uploaded the sets of altered pan-specific proteins to the IPA bioinformatics platform. All the proteins uploaded were recognized by the IPA software as being eligible for pathway analysis. Several pathways were proposed as a result of the knowledge base data curation. For SC21, the major canonical pathways proposed by IPA were JAK/STAT signaling and apoptosis pathways, which had the lowest p-values; 9.94 × 10−7 and 4.86 × 10−6, respectively (Fig. 5B). The involvement of JAK/STAT signaling is based on the up-regulation by SC21 of several kinases and transcription factors, including AKT3, signaling transducer and activator of transcription 2 (STAT2), Janus kinase 2 (JAK2), and MAPK3. The JAK/STAT signaling pathway is critical in transmitting external stimuli from the cell membrane to target genes in the nucleus and its key roles in cell growth, cell differentiation and apoptosis are widely recognized [34, 35]. It is noteworthy that every signaling pathway is a component of larger signaling cross-talk networks. Hence, activation of JAK could initiate multiple signals to numerous downstream targets, such as AKT, STAT, GRB2 and MAPK, which serve as principal nodes for other important signaling pathways [36-38]. For example, several kinases whose expression are altered by SC21, including Tyro3, SIRPA, NTRK1 and ERK, elicit their biological functions through the ATK signaling pathway [37, 39-42]. Therefore, the effect of JAK/STAT pathway activation is amplified as a result of cross-talk among signaling pathways and thus it is conceivable that abnormal activation of the JAK/STAT pathway would lead to cellular dysfunction. In addition to JAK/STAT signaling, the apoptosis pathway was also suggested to be involved in SC21-mediated cytotoxicity based on the changes in expression of several apoptosis-related molecules in response to SC21 treatment. In particular, CASP8, CDC2, FasLG and MAPK3 expressions were found to be significantly altered. It is known that activation of Fas triggers apoptosis by initiating the enzymatic activities of caspase8/10 [43], which would further positively regulate the functions of the pro-apoptotic Bid protein [44-46]. Additionally, phosphorylation of the pro-apoptotic Bcl-2 family protein BAD by CDC2 is required for its proper apoptotic functions [47, 48]. Therefore, the effect of these proteins converges on the Bcl-2 family and their combined regulation consequently elicits apoptosis.
For SC23, the major proposed canonical pathways involve RAR activation and cell cycle checkpoint control, with p-values of 2.42 × 10−5 and 2.42 × 10−4, respectively (Fig. 5C). RAR is an important transcription factor that regulates cell growth and survival. In our antibody microarray study, several upstream effectors of nuclear RAR signaling, including STAT5, PKA and MAPKAPK2, were found to be altered by SC23 [49-51]. In addition, CSK, a down-stream target of cytoplasmic RAR, was also shown to be changed by SC23, suggesting that both nuclear and cytoplasmic forms of RAR are involved. The apparent regulation of cell cycle checkpoint by SC23 is based on the expression changes of several cell cycle-related proteins, such as CDC2, cyclin A, CDK4 and CDK6. CDC2 (also known as CDK1) is a member of the Ser/Thr protein kinase family that is essential for G1/S and G2/M phase transitions in the eukaryotic cell cycle, depending on which cyclins it binds to. Specifically, cyclin A/E-CDC2 and cyclin E-CDK4/6 permit the transition from G1 through S [52, 53]. The cell cycle checkpoint pathway proposed by IPA is consistent with our cell cycle analysis, which showed that SC23 arrested cells at S phase (Fig. 2) The different pathway networks mediated by SC21 and SC23 suggested that these two compounds elicit unique signaling effects even though their chemical structures are closely related, and therefore, their ultimate biological effects would presumably be distinct. To understand the biological functions of SC21 and SC23, we applied IPA compare analysis tool and examined their functions in different aspects of biology. As hypothesized, SC21 and SC23 do exert differential cellular functions (Fig. 4D). SC21 affects post-translational modification and small molecule biochemistry, whereas, SC23 elicits cell death, cell cycle arrest and DNA repair. It is apparent from these analyses that SC21 and SC23 have distinct molecular mechanisms and cellular functions and that they may represent different classes of anticancer drugs. The validation of the IPA pathways with biological techniques is currently under way.
EXPERIMENTAL PROCEDURES
Cell Culture
H1299, MDA-MB-435, cells were purchased from the American Type Cell Culture (Manassas, VA). H2228 cells were kindly provided by Dr. Ita Laird-Offringa (USC Norris Cancer Center). HCT116 p53+/+ and HCT116 p53−/− cells were kindly provided by Dr. Bert Vogelstein (Johns Hopkins Medical Institutions, Baltimore, MD). Cells were maintained as monolayer cultures in the appropriate media: RPMI 1640 (NCI60 cell lines, H2228, HCT116 p53+/+, and HCT116 p53−/−) or DMEM (MDA-MB-435) supplemented with 10% fetal bovine serum (FBS) (Gemini-Bioproducts, Woodland, CA) and 2 mmol/L L-Glutamine at 37 °C in a humidified atmosphere of 5% CO2. To remove adherent cells from the flask for subculture and counting, cells were washed with PBS without calcium or magnesium, incubated with a small volume of 0.25% trypsin-EDTA solution (Sigma-Aldrich, St. Louis, MO) for 5-10 min, resuspended with culture medium and centrifuged. All experiments were performed using cells in exponential growth. Cells were routinely checked for Mycoplasma contamination using a PCR-based assay (Stratagene, Cambridge, UK) or PlasmoTest (InvivoGen, San Diego, CA).
Cytotoxicity Assays
Cytotoxicity was assessed by a MTT assay as previously described [54]. Cells were seeded in 96-well microtiter plates and allowed to attach. Cells were subsequently treated with a continuous exposure to the corresponding drugs for 72 h. An MTT solution (at a final concentration of 0.5 mg/mL) was added to each well and cells were incubated for 4 h at 37 °C. After removal of the supernatant, DMSO was added and the absorbance was read at 570 nm. All assays were done in triplicate. An IC50 value was then determined for each drug from a plot of log (drug concentration) versus percentage of cell kill.
Colony Formation Assays
Colony formation assays were also performed to confirm the activity of compounds as previously described [55]. Cells were plated in 6-well plates at a density of 100 cells/well and allowed to attach for 24 h. The following day, serial dilutions of SC23 were added and cells were incubated with treatments for 24 h. After exposure, cells were washed in PBS and cultured in compound-free media until colonies formed (8 to 10 days). Cells were subsequently washed, fixed with a 1% glutaraldehyde solution for 30 min and stained with a solution of crystal violet (2%, 30 min). After staining, cells were thoroughly washed with water and colonies were imaged on a VersaDoc Imaging System (Bio-Rad, Hercules, CA). The data reported are a representative of at least three independent experiments.
Western Blotting
Cells were trypsinized with 1X Trypsin/EDTA and collected by centrifugation at 1200 rpm for 5 min. Cells were then lysed in 80 μL of 1X cell lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1% NP-40) and pelleted by centrifugation at 14,000 rpm for 20 min at 4 °C. The protein concentration of the whole cell lysates was measured by a bicinchoninic acid (BCA) protein assay (ThermoFisher, Rockford, IL) and equal amounts of total proteins were resolved on 4-12% gradient Nu-PAGE gel (Invitrogen, Carlsbad, CA). The separated proteins were electroblotted onto nitrocellulose transfer membrane and blocked in 5% BSA/PBS for 1 h at room temperature. The membrane was incubated with anti-β-tubulin (SC-5286, Santa Cruz Biotechnology), anti-Bcl-2 (#2876, Cell Signaling Technology), anti-EGR-1 (SC-110, Santa Cruz Biotechnology), anti-Ac-Histone H3 (#9671, Cell Signaling Technology), anti-p21 (626701, BioLegend), anti-p53 (SC-126, Santa Cruz Biotechnology) or GAPDH (Y3322GAPDH, Biochain Institute, Inc.) antibodies at 4 °C overnight. Horseradish peroxidase conjugated secondary antibodies (Invitrogen, Carlsbad, CA) and SuperSignal Dura (ThermoFisher, Rockford, IL) were used to visualize proteins of interest with a ChemiDoc Imaging system (Bio-Rad, Hercules, CA).
DNA Fragmentation Assay
Cells were treated with 5 μM of SC23 for the indicated durations. After treatment, cells were collected and lysed in 100 μL of lysis buffer (100 mM NaCl, 100 mM Tris-HCl, pH 8.0, 25 mM EDTA, 0.5 % SDS, 100 μL/mL proteinase K) at 50 °C overnight. DNA was precipitated with ethanol and the pellet was dissolved in TE buffer containing 1 μg/mL RNase A and incubated at 30 °C for 1 h. The fragmented DNA was separated on a 1.5% agarose gel.
Detection of Mitochondrial ROS Production
SKOV-3 cells were treated with SC21 and SC23 for 24 h. After drug treatment, a solution of 5 μM MitoSOX Red Mitochondrial Superoxide Indicator (Invitrogen, Carlsbad, CA) was added and cells were incubated for 10 min at 37 °C. Cells were then washed three times with Hank’s Balanced Salt Solution (HBSS) to remove excess MitoSOX. After washing, cells were collected by trypsinization, washed two more times with HBSS and ROS production was measured by a BD LSRII flow cytometer (BD Biosciences, San Jose, CA). Data were analyzed with BD FACSDiva software (BD Biosciences, San Jose, CA) and WinMDI software (Joe Trotter, Scripps Research Institute, San Diego, CA).
Cell Cycle Analysis
Cell cycle perturbations were analyzed by propidium iodide DNA staining. Briefly, exponentially growing cells were treated with indicated concentrations of SC21 and SC23 for various durations. At the end of the treatment, cells were collected and washed with PBS after a gentle centrifugation at 1,200 rpm for 5 min. Cells were thoroughly resuspended in 0.5 mL of PBS and fixed in 70% ethanol for at least 2 h at 4°C. Ethanol-fixed cells were then centrifuged at 1,200 rpm for 5 min and washed twice in PBS to remove residual ethanol. For cell cycle analysis, the pellets were resuspended in 1 mL of PBS containing 0.02 mg/mL of propidium iodide, 0.5 mg/mL of DNase-free RNase A and incubated at 37°C for 30 min. Cell cycle profiles were obtained using BD LSRII flow cytometer (BD Biosciences, San Jose, CA) and data were analyzed with BD FACSDiva software (BD Biosciences, San Jose, CA) and the ModFit LT software package (Verity Software House, Topsham, ME).
2-D Gel Electrophoresis
Two-dimensional gel electrophoresis of SC23-treated T24 cell lysates was performed as below. Cells were treated for 12, 24, 48, and 72 h with SC23 (IC80 dose). The soluble fraction was then extracted and quantified. A total of 50 μg of whole cell lysate was loaded in the first dimension gel at 800 V for 16 h. Gels were then equilibrated, separated on a 12% SDS-PAGE gel for the second dimension, stained with CyproRuby, and imaged by Typhoon 9100 (Amersham Biosciences, Piscataway, NJ). All protein spots were quantified with PDQuest (Bio-Rad, Hercules, CA) and approximately 125 spots were identified as being significantly changed (> 2-fold) compared to the untreated control.
Tandem Mass Spectrometric Identification of Up-Regulated Proteins
The CyproRuby stained gel spots were dissected from the 2-D gel and subjected to in-gel trypsin digestion. At the end of the digestion, the digested peptidic products were extracted and analyzed by a Thermofinnigan LTQ linear ion trap mass spectrometer by loading onto a Michrom Bioresources peptide cap trap at 95% solvent A (2% acetonitrile, 1.0 % formic acid) and 5% solvent B (95% acetonitrile, 1.0% formic acid) and eluting with a linear gradient from 5-90% solvent B. The mass spectrometer was equipped with a nanospray ion source (Thermo Electron) using an uncoated 10 μm–ID SilicaTipTM PicoTipTM nanospray emitter (New Objective, Woburn, MA). The spray voltage of the mass spectrometer was 1.9 kV and the heated capillary temperature was 180 °C. Tandem MS/MS spectra were acquired with Xcalibur 1.4 software. A full MS scan was followed by three consecutive MS/MS scans of the top three ion peaks from the preceding full scan. Dynamic exclusion was enabled after three occurrences of an ion within 1 min, and the ion was placed on the exclusion list for 3 min. Other mass spectrometric data generation parameters were as follows: collision energy 35%, full scan MS mass range 400-1800m/z, minimum MS signal 5×104 counts, minimum MS/MS signal 5×103 counts. At the end of LC/MS/MS analysis, tandem mass spectra were analyzed using Bioworks 3.1, Beta-test site version from ThermoFinnigan, utilizing the SEQUESTTM algorithm to determine cross-correlation scores between acquired spectra and an NCBI mouse protein FASTA database. The following parameters were used for the TurboSEQUEST search analyses: No enzymes were chosen for the protease as not all proteins were digested to completion; molecular weight range: 400-4500; threshold: 1000; monoisotopic; precursor mass: 1.4; group scan: 10; minimum ion count: 20; charge state: auto; peptide: 1.5; fragment ions: 0; and differential amino acid modifications: Cys 57.0520. Results were filtered using SEQUEST cross-correlation scores greater than 1.5 for +1 ions, 2.0 for +2 ions, and 2.5 for +3 ions.
Confocal Microscopy
MDA-MB-435 cells were grown on glass chamber slides (Lab-Tek™, Nunc™, Rochester, NY) overnight at 37 °C and treated with 5 μM SC23 for 24 h. After the treatment, cells were fixed with 3.7% formaldehyde/PBS for 10 min at room temperature and washed with 1× PBS three times for 5 min each. After washing with PBS, cells were permeabilized with 100% ice-cold methanol for 15 min at −20 °C and washed again with 1× PBS for 5 min at room temperature. The permeabilized cells were blocked with 5% goat serum in PBS for 1 h at room temperature and incubated with anti-β-tubulin antibody (Santa Cruz Biotechnology) for 2 h at room temperature, followed by three 1× PBS washes. Cells were then exposed to Cy5-conjugated secondary antibody (Amersham) in the dark for 1 h at room temperature and residual secondary antibody was removed by three PBS washes. The signals from cells were examined using LSM510 image system (Carl Zeiss, Thornwood, NY).
Flow Cytometry
MDA-MB-435 cells were treated with 5 μM of SC23 for various durations. After treatment, cells were collected by centrifugation and fixed in 3.7 % formaldehyde for 10 min at 37 °C and chilled on ice for 1 min. After fixation, the cells were permeabilized by adding ice-cold 100% methanol slowly to a final concentration of 90% methanol and incubated on ice for 30 min. Cells were washed with incubation buffer (0.5% BSA/PBS) two times and then blocked in 100 μL incubation buffer. Cells were then exposed to anti-β-tubulin antibody for 1 h at room temperature, washed with the incubation buffer (three times), exposed to Cy3-conjugated secondary antibody, and washed again in the incubation buffer (three times). The stained cells were resuspended in PBS and analyzed using a BD LSRII flow cytometer (BD Biosciences, San Jose, CA). Data were analyzed with BD FACSDiva (BD Biosciences, San Jose, CA) and WinMDI software packages (Joe Trotter, Scripps Research Institute, San Diego, CA).
Kinexus™ Antibody Microarray
MDA-MB-435 cells were treated with 0.1 μM and 1 μM of SC21 and SC23, respectively, for 24 h. After treatment, cells were washed in ice-cold PBS to remove residual medium. The cells were then lysed in 200 μL of lysis buffer (20 mM MOPS, pH 7.0, 2 mM EGTA, 5 mM EDTA, 30 mM sodium fluoride, 60 mM β-glycerophosphate, pH 7.2, 20 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonylfluoride, 3 mM benzamidine, 5 μM pepstatin A, 10 μM leupeptin, 1% Triton X-100, 1 mM dithiothreitol) and collected. The cell lysates were sonicated four times (10 s each) with 15 s intervals on ice to rupture the cells and shear DNA. After sonication, the homogenates were centrifuged at 90,000×g for 30 min at 4 °C. The supernatants were transferred to a clean microcentrifuge tube and the protein concentrations were measured with the BCA protein assay (Thermo Fisher Scientific, Rockford, IL). A 250 μL aliquots of the whole cell lysates were submitted to Kinexus™ for 628-antibody microarray analysis.
Ingenuity Pathway Analysis
Potential signaling pathways induced by SC21 and SC23 were analyzed by the IPA software based on the Kinexus™ antibody microarray results. Statistically significant up-regulated or down-regulated pan-specific proteins with their Swiss-Prot accession numbers and the ratio changes were uploaded as an Excel spreadsheet file to the IPA server. SC21- and SC23-mediated signaling pathways were analyzed individually by core analysis. To compare their biological functions, the signaling networks were further analyzed by the IPA compare analysis.
Table 3.
Analysis of Total Protein Level Changes by SC21 in MDA-MB435 Cells Using Kinexus™ Kinex Antibody Microarray Screen
| Target Protein | Total Protein |
|---|---|
| Fold Change | |
| CDK4 | 0.42 |
| BUB1A | 0.52 |
| CDK1 | 0.57 |
| PCK2 | 0.60 |
| SIRPα1 | 0.60 |
| CASK/Lin2 | 0.62 |
| FasL | 0.64 |
| Tyro3 | 0.65 |
| CDC2L5 | 0.66 |
| Erk1/2 | 1.54 |
| JAK2 | 1.67 |
| Vrk1 | 1.72 |
| TrkA | 1.82 |
| STAT2 | 1.88 |
| CASP8 | 2.35 |
| PKBg | 3.2 |
Table 4.
Analysis of Protein Phosphorylation Changes by SC23 in MDA-MB435 Cells Using Kinexus™ Kinex Antibody Microarray Screen
| Target Protein | Phosphorylated Protein | |
|---|---|---|
| Phospho-site | Fold Change | |
| JNK | T183 + Y185 | 0.35 |
| Dab1 | Y198 | 0.38 |
| Rb | T356 | 0.54 |
| PKCh | T655 | 0.58 |
| MLC | S20 | 0.60 |
| PKA Cα/β | T197 | 0.60 |
| PKA Cβ1/2 | T500 | 0.61 |
| IR (INSR) | Y999 | 0.65 |
| MEK1 | S297 | 1.56 |
| FAK | Y577 | 1.60 |
| BLNK | Y84 | 1.75 |
| Shc1 | Y349 + Y350 | 1.83 |
| Mnk1 | T209 + T214 | 2.30 |
| PTEN | S370 | 3.14 |
| B23 (NPM) | T234 + T237 | 3.39 |
ACKNOWLEDGEMENT
This work was supported by funds from the Susan G. Komen Breast Cancer Foundation and the American Lung Association to NN and the work in TRB’s laboratory was supported by the Intramural Research Program of the NIH, Center for Cancer Research, NCI-Frederick.
ABBREVIATIONS
- CPT
camptothecin
- IN
HIV-1 integrase
- IPA
ingenuity pathway analysis
- JAK/STAT
Janus kinase-signal transducer and activator of transcription
- MPB-1
myc promoter-binding protein
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NSCLC
non-small cell lung cancer
- RAR
Retinoic acid receptor
- ROS
Reactive oxygen species
REFERENCES
- [1].Dy GK; Adjei AA Obstacles and opportunities in the clinical development of targeted therapeutics. Prog. Drug Res 2005, 63, 19–41. [DOI] [PubMed] [Google Scholar]
- [2].Murdoch D; Sager J Will targeted therapy hold its promise? An evidence-based review. Curr. Opin. Oncol 2008, 20, 104–111. [DOI] [PubMed] [Google Scholar]
- [3].Kola I; Landis J Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov 2004, 3, 711–715. [DOI] [PubMed] [Google Scholar]
- [4].Zheng C; Han L; Yap CW; Xie B; Chen Y Progress and problems in the exploration of therapeutic targets. Drug Discov. Today 2006, 11,412–420. [DOI] [PubMed] [Google Scholar]
- [5].Smith C Drug target validation: Hitting the target. Nature 2003, 422, 341, 343, 345. passim. [DOI] [PubMed] [Google Scholar]
- [6].Zhu F; Zheng CJ; Han LY; Xie B; Jia J; Liu X; Tammi MT; Yang SY; Wei YQ; Chen YZ Trends in the exploration of anticancer targets and strategies in enhancing the efficacy of drug targeting. Curr. Mol. Pharmacol 2008,1, 213–232. [DOI] [PubMed] [Google Scholar]
- [7].Hong H; Neamati N; Wang S; Nicklaus MC; Mazumder A; Zhao H; Burke TR Jr.; Pommier Y; Milne GW Discovery of HIV-1 integrase inhibitors by pharmacophore searching. J. Med. Chem 1997, 40, 930–936. [DOI] [PubMed] [Google Scholar]
- [8].Zhao H; Neamati N; Sunder S; Hong H; Wang S; Milne GW; Pommier Y; Burke TR Jr. Hydrazide-containing inhibitors of HIV-1 integrase. J. Med. Chem 1997, 40, 937–941. [DOI] [PubMed] [Google Scholar]
- [9].Neamati N; Hong H; Owen JM; Sunder S; Winslow HE; Christensen JL; Zhao H; Burke TR Jr.; Milne GW; Pommier Y Salicylhydrazine-containing inhibitors of HIV-1 integrase: implication for a selective chelation in the integrase active site. J. Med. Chem 1998, 41, 3202–3209. [DOI] [PubMed] [Google Scholar]
- [10].Neamati N; Lin Z; Karki RG; Orr A; Cowansage K; Strumberg D; Pais GC; Voigt JH; Nicklaus MC; Winslow HE; Zhao H; Turpin JA; Yi J; Skalka AM; Burke TR Jr.; Pommier Y Metal-dependent inhibition of HIV-1 integrase. J. Med. Chem 2002, 45, 5661–5670. [DOI] [PubMed] [Google Scholar]
- [11].Plasencia C; Dayam R; Wang Q; Pinski J; Burke TR Jr.; Quinn DI; Neamati N Discovery and preclinical evaluation of a novel class of small-molecule compounds in hormone-dependent and -independent cancer cell lines. Mol. Cancer Ther 2005, 4, 1105–1113. [DOI] [PubMed] [Google Scholar]
- [12].Chen YP; Chen F Identifying targets for drug discovery using bioinformatics. Expert. Opin. Ther. Targets 2008,12, 383–389. [DOI] [PubMed] [Google Scholar]
- [13].Gottesman MM Mechanisms of cancer drug resistance. Annu. Rev. Med 2002, 53, 615–627. [DOI] [PubMed] [Google Scholar]
- [14].Brown JM; Attardi LD The role of apoptosis in cancer development and treatment response. Nat. Rev. Cancer 2005, 5, 231–237. [DOI] [PubMed] [Google Scholar]
- [15].Abend M Reasons to reconsider the significance of apoptosis for cancer therapy. Int. J. Radiat. Biol 2003, 79, 927–941. [DOI] [PubMed] [Google Scholar]
- [16].Okada H; Mak TW Pathways of apoptotic and non-apoptotic death in tumour cells. Nat. Rev. Cancer 2004, 4, 592–603. [DOI] [PubMed] [Google Scholar]
- [17].Brown JM Cell status--dead or alive? Nat. Med 1996, 2, 1055–1056. [DOI] [PubMed] [Google Scholar]
- [18].Ishii T; Teramoto S; Matsuse T GSTP1 affects chemoresistance against camptothecin in human lung adenocarcinoma cells. Cancer Lett. 2004, 216, 89–102. [DOI] [PubMed] [Google Scholar]
- [19].Pierce GB; Parchment RE; Lewellyn AL Hydrogen peroxide as a mediator of programmed cell death in the blastocyst. Differentiation 1991, 46, 181–186. [DOI] [PubMed] [Google Scholar]
- [20].Lu HR; Zhu H; Huang M; Chen Y; Cai YJ; Miao ZH; Zhang JS; Ding J Reactive oxygen species elicit apoptosis by concurrently disrupting topoisomerase II and DNA-dependent protein kinase. Mol. Pharmacol 2005, 68, 983–994. [DOI] [PubMed] [Google Scholar]
- [21].Mukhopadhyay P; Rajesh M; Hasko G; Hawkins BJ; Madesh M; Pacher P Simultaneous detection of apoptosis and mitochondrial superoxide production in live cells by flow cytometry and confocal microscopy. Nat. Protoc. 2007, 2, 2295–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lowe SW; Lin AW Apoptosis in cancer. Carcinogenesis 2000, 21, 485–495. [DOI] [PubMed] [Google Scholar]
- [23].Zhang JY Apoptosis-based anticancer drugs. Nat. Rev. Drug Discov 2002, 1, 101–102. [DOI] [PubMed] [Google Scholar]
- [24].Zou H; Henzel WJ; Liu X; Lutschg A; Wang X Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 1997, 90, 405–413. [DOI] [PubMed] [Google Scholar]
- [25].Green DR; Reed JC Mitochondria and apoptosis. Science 1998, 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- [26].Reed JC Apoptosis-targeted therapies for cancer. Cancer Cell 2003, 3, 17–22. [DOI] [PubMed] [Google Scholar]
- [27].Huang RP; Darland T; Okamura D; Mercola D; Adamson ED Suppression of v-sis-dependent transformation by the transcripttion factor, Egr-1. Oncogene 1994, 9, 1367–1377. [PubMed] [Google Scholar]
- [28].Calogero A; Cuomo L; D'Onofrio M; de Grazia U; Spinsanti P; Mercola D; Faggioni A; Frati L; Adamson ED; Ragona G Expression of Egr-1 correlates with the transformed phenotype and the type of viral latency in EBV genome positive lymphoid cell lines. Oncogene 1996, 13, 2105–2112. [PubMed] [Google Scholar]
- [29].Liu C; Rangnekar VM; Adamson E; Mercola D Suppression of growth and transformation and induction of apoptosis by EGR-1. Cancer Gene Ther. 1998, 5, 3–28. [PubMed] [Google Scholar]
- [30].Traverse S; Gomez N; Paterson H; Marshall C; Cohen P Sustained activation of the mitogen-activated protein (MAP) kinase cascade may be required for differentiation of PC12 cells. Comparison of the effects of nerve growth factor and epidermal growth factor. Biochem. J 1992, 288(Pt. 2), 351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yaginuma Y; Westphal H Abnormal structure and expression of the p53 gene in human ovarian carcinoma cell lines. Cancer Res, 1992, 52, 4196–4199. [PubMed] [Google Scholar]
- [32].Lincet H; Poulain L; Remy JS; Deslandes E; Duigou F; Gauduchon P; Staedel C The p21(cip1/waf1) cyclin-dependent kinase inhibitor enhances the cytotoxic effect of cisplatin in human ovarian carcinoma. cells Cancer Lett. 2000, 161, 17–26. [DOI] [PubMed] [Google Scholar]
- [33].Jordan MA; Wilson L Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 2004, 4, 253–265. [DOI] [PubMed] [Google Scholar]
- [34].Klampfer L Signal transducers and activators of transcription (STATs): Novel targets of chemopreventive and chemotherapeutic drugs. Curr. Cancer Drug Targets 2006, 6, 107–121. [DOI] [PubMed] [Google Scholar]
- [35].Nefedova Y; Gabrilovich DI Targeting of Jak/STAT pathway in antigen presenting cells in cancer. Curr. Cancer Drug Targets 2007, 7, 71–77. [DOI] [PubMed] [Google Scholar]
- [36].van der Geer P; Hunter T; Lindberg RA Receptor protein-tyrosine kinases and their signal transduction pathways. Annu. Rev. Cell Biol 1994, 10, 251–337. [DOI] [PubMed] [Google Scholar]
- [37].Dijkers PF; van Dijk TB; de Groot RP; Raaijmakers JA; Lammers JW; Koenderman L; Coffer PJ Regulation and function of protein kinase B and MAP kinase activation by the IL-5/GM-CSF/IL-3 receptor. Oncogene 1999, 18, 3334–3342. [DOI] [PubMed] [Google Scholar]
- [38].Yamauchi T; Yamauchi N; Ueki K; Sugiyama T; Waki H; Miki H; Tobe K; Matsuda S; Tsushima T; Yamamoto T; Fujita T; Taketani Y; Fukayama M; Kimura S; Yazaki Y; Nagai R; Kadowaki T Constitutive tyrosine phosphorylation of ErbB-2 via Jak2 by autocrine secretion of prolactin in human breast cancer. J. Biol Chem 2000, 275, 33937–33944. [DOI] [PubMed] [Google Scholar]
- [39].Lan Z; Wu H; Li W; Wu S; Lu L; Xu M; Dai W Transforming activity of receptor tyrosine kinase tyro3 is mediated, at least in part, by the PI3 kinase-signaling pathway. Blood 2000, 95, 633–638. [PubMed] [Google Scholar]
- [40].Fukuda R; Kelly B; Semenza GL Vascular endothelial growth factor gene expression in colon cancer cells exposed to prostaglandin E2 is mediated by hypoxia-inducible factor 1. Cancer Res. 2003, 63, 2330–2334. [PubMed] [Google Scholar]
- [41].Gryz EA; Meakin SO Acidic substitution of the activation loop tyrosines in TrkA supports nerve growth factor-dependent, but not nerve growth factor-independent, differentiation and cell cycle arrest in the human neuroblastoma cell line, SY5Y. Oncogene 2003, 22, 8774–8785. [DOI] [PubMed] [Google Scholar]
- [42].Neznanov N; Neznanova L; Kondratov RV; Burdelya L; Kandel ES; O'Rourke DM; Ullrich A; Gudkov AV Dominant negative form of signal-regulatory protein-alpha (SIRPalpha /SHPS-1) inhibits tumor necrosis factor-mediated apoptosis by activation of NF-kappa B. J. Biol. Chem 2003, 278, 3809–3815. [DOI] [PubMed] [Google Scholar]
- [43].Wang J; Chun HJ; Wong W; Spencer DM; Lenardo MJ Caspase-10 is an initiator caspase in death receptor signaling. Proc. Natl. Acad. Sci. USA 2001, 98, 13884–13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Li H; Zhu H; Xu CJ; Yuan J Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 1998, 94, 491–501. [DOI] [PubMed] [Google Scholar]
- [45].Yin XM Bid, a critical mediator for apoptosis induced by the activation of Fas/TNF-R1 death receptors in hepatocytes. J. Mol. Med 2000, 78, 203–211. [DOI] [PubMed] [Google Scholar]
- [46].Milhas D; Cuvillier O; Therville N; Clave P; Thomsen M; Levade T; Benoist H; Segui B Caspase-10 triggers Bid cleavage and caspase cascade activation in FasL-induced apoptosis. J. Biol. Chem 2005, 280, 19836–19842. [DOI] [PubMed] [Google Scholar]
- [47].Konishi Y; Lehtinen M; Donovan N; Bonni A Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Mol. Cell 2002, 9, 1005–1016. [DOI] [PubMed] [Google Scholar]
- [48].Zhang J; Liu J; Yu C; Lin A BAD Ser128 is not phosphorylated by c-Jun NH2-terminal kinase for promoting apoptosis. Cancer Res. 2005, 65, 8372–8378. [DOI] [PubMed] [Google Scholar]
- [49].Huggenvik JI; Collard MW; Kim YW; Sharma RP Modification of the retinoic acid signaling pathway by the catalytic subunit of protein kinase-A. Mol. Endocrinol 1993, 7, 543–550. [DOI] [PubMed] [Google Scholar]
- [50].Rochette-Egly C; Oulad-Abdelghani M; Staub A; Pfister V; Scheuer I; Chambon P; Gaub MP Phosphorylation of the retinoic acid receptor-alpha by protein kinase A. Mol. Endocrinol 1995, 9, 860–871. [DOI] [PubMed] [Google Scholar]
- [51].Zhao Q; Tao J; Zhu Q; Jia PM; Dou AX; Li X; Cheng F; Waxman S; Chen GQ; Chen SJ; Lanotte M; Chen Z; Tong JH Rapid induction of cAMP/PKA pathway during retinoic acid-induced acute promyelocytic leukemia cell differentiation. Leukemia 2004, 18, 285–292. [DOI] [PubMed] [Google Scholar]
- [52].Aleem E; Kiyokawa H; Kaldis P Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol 2005, 7, 831–836. [DOI] [PubMed] [Google Scholar]
- [53].Morgan DO Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol 1997, 13, 261–291. [DOI] [PubMed] [Google Scholar]
- [54].Carmichael J; DeGraff WG; Gazdar AF; Minna JD; Mitchell JB Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987, 47, 936–942. [PubMed] [Google Scholar]
- [55].Munshi A; Hobbs M; Meyn RE Clonogenic cell survival assay Methods Mol. Med. 2005, 110, 21–28. [DOI] [PubMed] [Google Scholar]