Abstract
Pseudomonas microorganisms are used for bioremediation of soils contaminated with petroleum hydrocarbons. The overall remediation efficiency is largely dependent on the presence of macro- and micronutrients. Widely varying concentrations of available nitrogen and iron (Fe) in soils were shown to affect residual hydrocarbons in the course of biodegradation. The regulatory mechanisms of expression of hydrocarbon catabolic genes in low nitrogen/low iron conditions remain unclear. The catabolism of naphthalene, a two-ring polycyclic aromatic hydrocarbon, has been well studied in pseudomonads in terms of the involvement of specific transcriptional activators, thus making it useful in revealing additional regulatory control of the adaptation of hydrocarbon destructors to a low level of the essential nutrients. The Pseudomonas putida strain BS3701 is a component of the “MicroBak” preparation for soil remediation. Previously, this strain was shown to contain genes encoding the key enzymes for naphthalene catabolism: naphthalene 1,2-dioxygenase, salicylate hydroxylase, catechol 2,3-dioxygenase, and catechol 1,2-dioxygenase. Our study aimed to clarify whether the naphthalene catabolic gene expression is dependent on the amount of nitrogen and iron in the growth culture medium, and if so, at exactly which stages the expression is regulated. We cultivated the strain in low nitrogen/low iron conditions with the concurrent evaluation of the activity of the key enzymes and the mRNA level of genes encoding these enzymes. We are the first to report that naphthalene catabolic genes are subject not only to transcriptional but also post-transcriptional regulation.
Keywords: aromatic compound, oxygenase, nitrogen, iron, nucleoid-associated proteins, RT-qPCR
Introduction
Microorganisms of the genus Pseudomonas are used for bioremediation of soils contaminated with petroleum hydrocarbons. The overall remediation efficiency is largely dependent on the presence of macro- and micronutrients. Widely varying concentrations of available nitrogen and iron (Fe) in soils (Jin et al., 2014; Wang et al., 2018) were shown to affect residual hydrocarbons in the course of biodegradation (Simarro et al., 2011). The regulatory mechanisms of expression of hydrocarbon catabolic genes in low nitrogen/low iron conditions remain unclear. The catabolism of naphthalene, a two-ring polycyclic aromatic hydrocarbon, has been well studied in pseudomonads in terms of the involvement of specific transcriptional activators, thus making it useful in revealing additional regulatory control of the adaptation of hydrocarbon destructors to a low level of the essential nutrients.
The naphthalene transformation to Krebs cycle intermediates was first studied using the Pseudomonas putida PpG1 (Yen and Gunsalus, 1982), also known as the P. putida strain G7 (pNAH7), where the naphthalene conversion proceeded via salicylate (nah-operon, nahABCDEF) and catechol further oxidized by the meta-pathway (sal-operon, nahGHIJKLM). Activation of the nah- and sal-operons occurred with the assistance of the transcriptional regulator NahR (LysR-type transcriptional regulator, LTTR) in the presence of the salicylate ion (Schell and Poser, 1989). Further studies revealed other variants of the genetic organization of naphthalene catabolic genes (Boronin and Kosheleva, 2010), while genes of the catechol oxidation (by both meta- and ortho- pathways) were described in detail as members of other operons (Ladino-Orjuela et al., 2016). The effect of NahR is frequently believed to apply to both the salicylate transformation genes and genes involved in the naphthalene catabolism [e.g., the “classical” scheme of regulation of naphthalene catabolic genes (Schell and Poser, 1989)], although another study has shown that the mRNA level of naphthalene 1,2-dioxygenase remains unchanged in the presence of salicylate (Boronin and Kosheleva, 2010). The protein CatR is known to be involved in the regulation of genes encoding catechol 1,2-dioxygenase (Tover et al., 2000), with the transcription activation induced by the intermediate cis,cis-muconate (Parsek et al., 1992). When considered in the context of naphthalene catabolism, catechol 2,3-dioxygenase is frequently described as non-inducible. However, it is known that the protein XylS that regulates the activity of the toluene/xylene catabolic gene cluster xylDLEGF (the xylE gene encodes catechol 2,3-dioxygenase) acts as an activator in the presence of toluene and meta-benzyl alcohol (Inouye et al., 1987). The level of xylS mRNA depends on XylR which in turn is associated with sigma 54-dependent promoters (Kessler et al., 1994). The nucleoid-associated proteins (NAPs) classified as pleiotropic regulators are also reported to be involved. The integration host factor (IHF) binding site was detected near XylR (sigma-54-dependent Fis family transcriptional regulator that controls toluene/xylene catabolism); the IHF structure appears to be important for transcription activation (Ramos et al., 1997).
The P. putida strain BS3701 is a component of the “MicroBak” preparation for soil remediation (Filonov et al., 2006). Previously, this strain was shown to contain genes encoding the key enzymes for naphthalene catabolism: naphthalene 1,2-dioxygenase, salicylate hydroxylase, catechol 2,3-dioxygenase, and catechol 1,2-dioxygenase (Izmalkova et al., 2006). Our study aimed to clarify whether the naphthalene catabolic gene expression is dependent on the amount of nitrogen and iron in the growth culture medium, and if so, at exactly which stages the expression is regulated. We cultivated the strain in low nitrogen/low iron conditions with the concurrent evaluation of the activity of the key enzymes and the mRNA level of genes encoding these enzymes. The RT-qPCR technique was validated for the chosen growth conditions. Using BestKeeper, the oprI gene was chosen as the reference gene. We are the first to report that naphthalene catabolic genes are subject not only to transcriptional but also post-transcriptional regulation.
Materials and Methods
Growth Media and Conditions
The P. putida strain BS3701 used in this study was grown at 26°C on modified Evans mineral medium supplemented either with glucose (10 g/l) or sodium salicylate (1 g/l) as the sole source of carbon and energy. The modified Evans mineral medium contained (per liter) 50 mM K2HPO4, 5 mM NH4Cl, 0.1 mM Na2SO4, 0.0625 mM MgCl2, 0.018 mM FeCl3, 0.01 mM MnCl2, 0.005 mM ZnO, 0.002 mM CoCl2, 0.001 mM CaCl2, 0.97 μM CuCl2, 0.97 μM H3BO3, and 0.005 μM (NH4)6Mo7O24 (Petrikov et al., 2013). The medium pH was adjusted to 7.5 using concentrated HCl. The low nitrogen growth medium contained 1 mM versus 5 mM of NH4Cl and was supplemented with sodium salicylate (1 g/l) as the sole source of carbon. The low iron growth medium contained 100 μM 2,2′-bipyridyl (Sigma-Aldrich, United States) and was supplemented with sodium salicylate (1 g/l) as the sole source of carbon. BS3701 grown on modified Evans with glucose (10 g/l) for 1 day was further used for inoculation (1 ml per 100 ml of medium). The harvested cells were centrifuged at 10,000 rpm for 10 min at 4°C and re-suspended in appropriate modified Evans at the final concentration of 3 × 108 CFU/ml. The 750-ml flasks used in the experiments contained the appropriate mineral medium and substrate (100 ml each). The growth period lasted until the end of the exponential growth phase. The time of cultivation on all used media was 31 h, the end of exponential growth phase was 21 h.
Determination of the Oxidative Activity of Microorganisms in Relation to Sodium Salicylate
The oxidative activity (OA) of microorganisms was determined from sodium salicylate degradation vs. control (microorganism-free medium):
where Xk is sodium salicylate concentration (mg/l) in the microorganism-free medium and Xi is that in the inoculated flask (mg/l). Residual concentration of sodium salicylate (20 μl sample) was analyzed by liquid chromatography using a silica gel-packed Agilent (1260 infinity) column (length 150 mm, diameter 4 mm, grain size 5 μm) grafted with hexadecyl groups. The used wavelength was 310 nm. The mobile phase was (A) water (0.1% CH3COOH); (B) acetonitrile (0.1% CH3COOH). The gradient mode was as follows: up to 3 min – 20% of eluent B, by 12 min – 50% of eluent B, by 14 min – 100% of eluent B. The eluent flow rate was 1 ml/min. System stabilization took 5 min; the analysis time was 19 min (Liu and Smith, 1996). The column thermostat temperature was 40°C. For sample preparation, 2 ml of the culture was taken from the flask and centrifuged for 10 min at 10,000 rpm. For the measurements, 0.5 ml samples of supernatant were used. If necessary, the supernatant was diluted with distilled water and analyzed as described above. Standards were prepared in distilled water by serial dilution.
Measuring Enzymatic Activity
The biomass was washed twice with chilled 0.05 M phosphate buffer (pH 7.0) and re-suspended in 0.02 M phosphate buffer (pH 7.5). Cell suspension (5 ml sample) was subjected to ultrasonic disintegration (using an MSE150 disintegrator) for 1.5 min (3 × 30 s) at 4°C. Cell debris was removed by centrifugation (a Rotanta 460R centrifuge, Hettich Zentrifugen, Germany) for 20 min at 32,000 × g and 4°C. The supernatant was used as a cell-free extract in measuring enzymatic activity.
Activities of the enzymes were determined as previously described for naphthalene 1,2-dioxygenase in Dua and Meera (1981), for salicylate hydroxylase in Yamamoto et al. (1965), for catechol 1,2-dioxygenase in Hegeman (1966), and catechol 2,3-dioxygenase in Feist and Hegeman (1969). The protein concentration was determined according to Bradford (1976).
RT-qPCR
Specific primers were constructed using the Primer-BLAST tool1 (Table 1). A kit containing SYBR Green (cat. no. R-402, Sintol, Russia) was used for qPCR. For amplification, the following temperature program was used: (1) 95°C for 3 min, (2) 95°C for 20 s, (3) 60°C for 20 s, (4) 72°C for 5 s; 40 cycles included steps (2–4). For each pair of primers, amplification efficiency was determined from the slope of the log-linear portion of the calibration curve. The reaction specificity was confirmed by analysis of the melting curve and by agarose gel electrophoresis Total RNA was isolated using the TRI reagent according to the manufacturer’s instructions (Sigma-Aldrich, United States). A RevertAid RT Reverse Transcription Kit (Thermo, United States) was used according to the manufacturer’s protocol for the reverse transcription reaction including 100 ng of total RNA and specific primers (20 pmol of each reverse primer). To validate this method, a reference gene was selected among 16S rRNA, rpoD, rpoB, rpoS, gyrB, tuf, ppiD, oprI, rpsL, dnaK, fliS, and proC. According to BestKeeper (Pfaffl et al., 2004), the most stable was the oprI gene (SD 0.69, CV 9.8%). The relative amount of transcripts was quantified using the delta-delta Cp method, with amplification efficiency taken into account:
TABLE 1.
Primers for quantitative reverse transcription PCR.
| Gene | Sequence, 5′-3′ | Amplicon length, bp | PCR efficiency | References |
| 16S rRNA | 16S1 | 207 | 2.26 | Li S. et al., 2011 |
| CGGGAATCTGACACAGGTGCT | ||||
| 16S2 | ||||
| GATCCGGACTACGATCGTTTTGT | ||||
| rpoD | rpoD(BS3701)RT788f | 161 | 2.10 | This study |
| AGCTGGTACCGAAGCAGTTC | ||||
| rpoD(BS3701)RT948r | ||||
| CTGGTCGGTTTCGTTGCTTG | ||||
| rpoB | rpoB(BS3701)RT2273f | 220 | 2.03 | This study |
| TGACCAAGTACACCCGTTCG | ||||
| rpoB(BS3701)RT2492r | ||||
| TCTTCCTGAACCACACGCTC | ||||
| rpoS | rpoS(BS3701)RT861f | 107 | 1.95 | This study |
| AGGGCATGAAAGCAGTACCC | ||||
| rpoS(BS3701)RT967r | ||||
| GGATCTCACGCAAACGCTTC | ||||
| gyrB | gyrB(BS3701)RT1585f | 214 | 2.05 | This study |
| ATCTATATTGCCCAGCCGCC | ||||
| gyrB(BS3701)RT1798r | ||||
| ACAGGCGCTTGAGAGTCTTC | ||||
| tuf | tuf(BS3701)RT512f | 221 | 1.98 | This study |
| TCATCGGTTCGGCTCGTATG | ||||
| tuf(BS3701)RT732r | ||||
| ATCCTGAACGCGGACGATAC | ||||
| ppiD | ppiD(BS3701)RT890f | 220 | 1.95 | This study |
| GTGAGGACTTTGCCGCTTTG | ||||
| ppiD(BS3701)RT1109r | ||||
| AAGCTCGGTACTTCTGGTGC | ||||
| oprI | oprI(BS3701)RT97f | 71 | 1.92 | This study |
| GAATCAGCGGAGAACAACGC | ||||
| oprI(BS3701)RT167r | ||||
| ACCAGATCGATGCGGTAACA | ||||
| rpsL | rpsL(BS3701)RT148f | 171 | 2.07 | This study |
| CGTAAAGTATGCCGTGTGCG | ||||
| rpsL(BS3701)RT318f GCCCGAAGTATCCAGAGAGC | ||||
| dnaK | dnaK(BS3701)RT256f | 212 | 1.91 | This study |
| ATCAAGCTGGTGCCGTACAA | ||||
| dnaK(BS3701)RT467r | ||||
| TCTTTGGTAGCCTGACGCTG | ||||
| fliS | fliS(BS3701)RT288f | 80 | 1.95 | This study |
| CGAAGCCAATCTCAAAGGCG | ||||
| fliS(BS3701)RT367r | ||||
| CGTCCCAGCCTTCCTTTACA | ||||
| proC | proC(BS3701)RT267f | 148 | 1.81 | This study |
| TGGCCAGCTGATCGTTTCAA | ||||
| proC(BS3701)RT414r | ||||
| CACTTCAGCGGTAGCGTACA | ||||
| nahAa | nahAa(BS3701)RT202f | 145 | 2.21 | This study |
| CAGTATGTGCTCGCCTGTCA | ||||
| nahAa(BS3701)RT346r | ||||
| AGCGACGGATATCGTGAGTG | ||||
| nahU | nahU(BS3701)RT279f | 112 | 2.40 | This study |
| CAAATACCTCGGTTGCAGCG | ||||
| nahU(BS3701)RT390r | ||||
| CCCGAACTGGGCAATACCTT | ||||
| nahH | nahH(BS3701)RT694f | 129 | 2.03 | This study |
| ATCAGCATGACCGACACCTC | ||||
| nahH(BS3701)RT822r | ||||
| ATAGTTGTAGTTCCCGCCGC | ||||
| catA(0) | catA(0)(BS3701)RT344f | 174 | 1.88 | This study |
| CGATTGCCCAAGGTGAAGTG | ||||
| catA(0)(BS3701)RT517r | ||||
| ACTGGCTTGGGTCGAAGAAC | ||||
| catA(2) | catA(2)(BS3701)RT548f | 85 | 1.85 | This study |
| CCGATGATCAAGGGCGCTAT | ||||
| catA(2)(BS3701)RT632r | ||||
| AGG CAT TCC TGA GTA GGG C | ||||
| catA(8) | catA(8)(BS3701)RT387f | 155 | 2.28 | This study |
| AGGCGTGGTGATGTTCCTTC | ||||
| catA(8)(BS3701)RT541r | ||||
| TAATGATACGCCGACGCAGG | ||||
| ihfA | ihfA(BS3701)RT84f | 159 | 2.20 | This study |
| GCACGCACTTGAAGAGAACG | ||||
| ihfA(BS3701)RT242r | ||||
| TTCAACTTCTGCCCTGGACG | ||||
| ihfB | ihfB(BS3701)RT138f | 135 | 1.90 | This study |
| CGGTTTTGGCAGCTTCTCGC | ||||
| ihfB(BS3701)RT272r | ||||
| TTGACCCGATCACGCAGCTC | ||||
| fis | fis(BS3701)RT122f | 85 | 2.15 | This study |
| TGACGGACGTGTACAACCTG | ||||
| fis(BS3701)RT206r | ||||
| GTCTGGTTGCCCTTCACGTA | ||||
| lrp | lrp(BS3701)RT253f | 158 | 2.32 | This study |
| TCAGGCGATACCTTCGAGGA | ||||
| lrp(BS3701)RT410r | ||||
| AGTTTCAGCAGGATGTCCCC | ||||
| iciA | iciA(BS3701)RT36f | 79 | 1.93 | This study |
| AGTGATCGAACAAGGCGGTT | ||||
| iciA(BS3701)RT114r | ||||
| CTTGATGCGCTGGGAAATGG | ||||
| mvaT10 | mvaT10(BS3701)RT280f | 83 | 2.00 | This study |
| GTGATCGAAACCAAAGGCGG | ||||
| mvaT10(BS3701)RT362r | ||||
| GCCCAGCTTTCAACCACATC | ||||
| mvaT22 | mvaT22(BS3701)RT51f | 209 | 1.93 | this study |
| ACTTCAACAGCTGGATGCGA | ||||
| mvaT22(BS3701)RT259r | ||||
| TCTTGTAGCGCTTGACCTCG | ||||
| mvaT34 | mvaT34(BS3701)RT275f | 86 | 1.97 | This study |
| TCATCGAGACCAAAGGTGGC | ||||
| mvaT34(BS3701)RT360r | ||||
| GGAAACCCAAGACTCGACCA | ||||
| mvaT39 | mvaT39(BS3701)RT231f | 126 | 1.93 | This study |
| GCGGGTGGTCAAGGTCTATC | ||||
| mvaT39(BS3701)RT356r | ||||
| CGTACCCAGCTTTCTACCGT | ||||
| mvaT41 | mvaT41(BS3701)RT130f | 185 | 1.93 | This study |
| CTGGCCAAGTACGGCTACAG | ||||
| mvaT41(BS3701)RT314r | ||||
| TCCTTCAATCGCGAGTGGTT |
where «E» is the amplification efficiency, «Cp» is the crossing point. We used unpaired Student’s t−test for data comparison, p-value < 0.05. All results are derived from three independent replicates and presented as a mean score ± confidence interval.
References to NCBI Databases
The nucleotide sequences of the naphthalene catabolic genes have been deposited in GenBank under accession nos. MN442422.1, MN413629.1, MN442423.1, MN442424.1, MN442425.1, and MN442426.1.
Results
Naphthalene-to-Salicylate Transformation
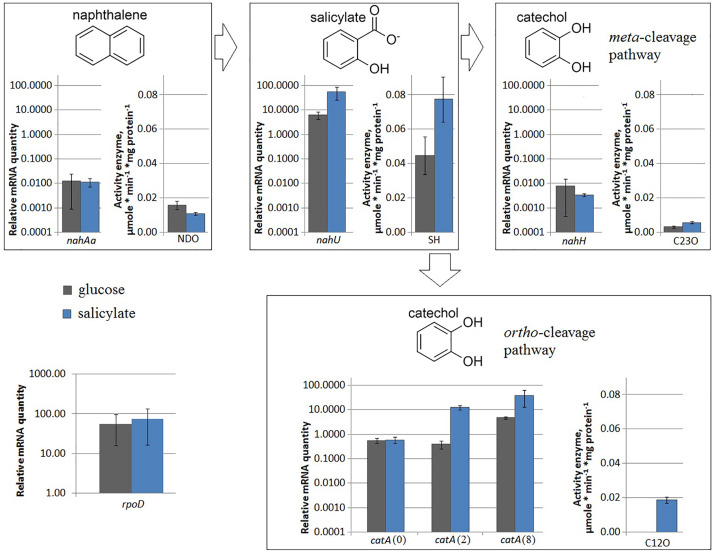
In the P. putida strain BS3701, the transformation of naphthalene to salicylate is assisted by the plasmid-localized gene cluster nahAaAbAcAdBFCQED. BS3701 cultivation in salicylate-supplemented (vs. glucose) Evans medium (Figure 1) did not result in an altered nahAa mRNA level [in contrast to the case of P. putida G7 (pNAH7)] but showed an ∼1.5-fold decrease in naphthalene 1,2-dioxygenase activity (NDO) (from 0.016 ± 0.003 to 0.0107 ± 0.0008 μmol/min∗mg of protein).
FIGURE 1.
The relative level of mRNA and enzymatic activity in Pseudomonas putida BS3701 grown on Evans supplemented either with glucose (dark-gray bars) or salicylate (blue bars). rpoD, sigma-70-dependent RNAP subunit; nahAa, reductase component of naphthalene dioxygenase; nahU, salicylate hydroxylase; nahH, catechol 2,3-dioxygenase; catA(0), catA(2) and catA(8), catechol 1,2-dioxygenase; NDO, activity of naphthalene 1,2-dioxygenase; SH, activity of salicylate hydroxylase; C23O, activity of catechol 2,3-dioxygenase; C12O, activity of catechol 1,2-dioxygenase. The experiments were performed in three biological replicates, the data are presented as a mean score ± confidence interval (p-value < 0.05).
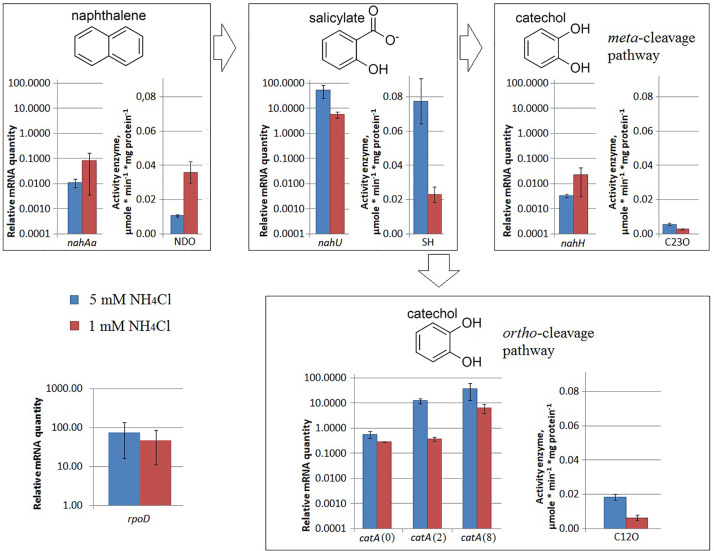
Under nitrogen limitation, the nahAa mRNA level remained unchanged, while the NDO enzyme activity changed (Figure 2). Evans-cultivated BS3701 in low nitrogen conditions (1 mM NH4Cl instead of 5 mM NH4Cl) retained the same nahAa mRNA level, but the NDO activity increased by ∼3.4-fold (from 0.0107 ± 0.0008 to 0.036 ± 0.006 μmol/min∗mg of protein). Previously, the literature offered no reports on the stimulation of NDO synthesis by low nitrogen.
FIGURE 2.
The relative level of mRNA and enzymatic activity in Pseudomonas putida BS3701 grown on Evans with 5 mM NH4Cl (blue bars) or 1 mM NH4Cl (red bars). Each medium was supplemented with salicylate. rpoD, sigma-70-dependent RNAP subunit; nahAa, reductase component of naphthalene dioxygenase; nahU, salicylate hydroxylase; nahH, catechol 2,3-dioxygenase; catA(0), catA(2) and catA(8), catechol 1,2-dioxygenase; NDO, activity of naphthalene 1,2-dioxygenase; SH, activity of salicylate hydroxylase; C23O, activity of catechol 2,3-dioxygenase; C12O, activity of catechol 1,2-dioxygenase. The experiments were performed in three biological replicates, the data are presented as a mean score ± confidence interval (p-value < 0.05).
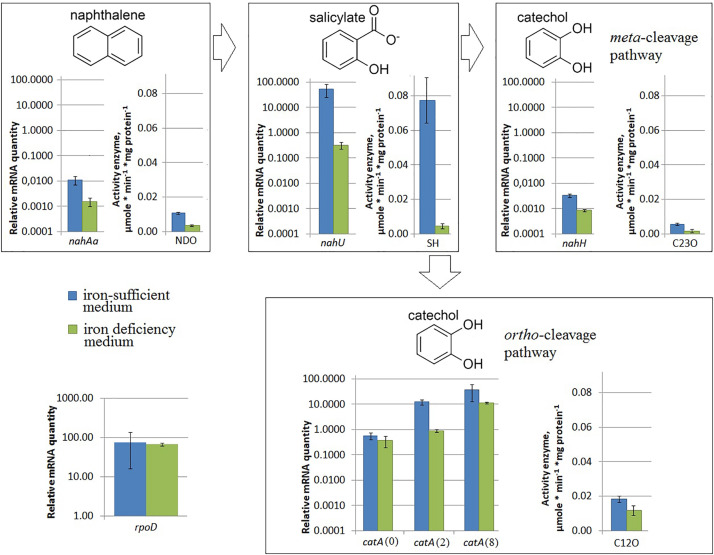
Under iron limitation, both the nahAa mRNA level and the NDO enzyme activity changed (Figure 3). Evans-cultivated BS3701 in low iron conditions (with added 100 μM 2,2′-bipyridyl) showed a decrease of the nahAa mRNA level by ∼7-fold, and a decrease in the NDO activity by ∼3-fold (from 0.0107 ± 0.0008 to 0.0036 ± 0.0006 μmol/min∗mg of protein).
FIGURE 3.
The relative level of mRNA and enzymatic activity in Pseudomonas putida BS3701 grown on Evans with a sufficient iron content (blue bars) or low-iron Evans (green bars). Each medium was supplemented with salicylate. rpoD, sigma-70-dependent RNAP subunit; nahAa, reductase component of naphthalene dioxygenase; nahU, salicylate hydroxylase; nahH, catechol 2,3-dioxygenase; catA(0), catA(2), and catA(8), catechol 1,2-dioxygenase; NDO, activity of naphthalene 1,2-dioxygenase; SH, activity of salicylate hydroxylase; C23O, activity of catechol 2,3-dioxygenase; C12O, activity of catechol 1,2-dioxygenase. The experiments were performed in three biological replicates, the data are presented as a mean score ± confidence interval (p-value < 0.05).
Salicylate-to-Catechol Transformation
The P. putida BS3701 chromosome was found to carry the functional nahU gene encoding salicylate hydroxylase (SH). BS3701 cultivation in Evans medium supplemented with salicylate (vs. glucose) (Figure 1) increased the mRNA level by ∼9-fold and the SH activity by ∼1.7-fold (from 0.045 ± 0.011 to 0.077 ± 0.013 μmol/min∗mg of protein).
Under nitrogen limitation, both the nahU mRNA level and the SH enzyme activity changed (Figure 2). Evans-cultivated BS3701 in low nitrogen conditions showed a decrease in the nahU mRNA level by ∼9.6-fold and in the SH activity by ∼3.3-fold (from 0.077 ± 0.013 to 0.023 ± 0.005 μmol/min∗mg of protein). The extent of salicylate degradation was also reduced (Table 2). The initial concentration of sodium salicylate in the medium was 1000 mg/L. After BS3701 cultivation in Evans supplemented with 5 mM NH4Cl, salicylate was not detected at the end of the exponential growth phase (the residual content was <0.3 mg/l). However, similar to cultivation in 1 mM NH4Cl Evans, the residual content of salicylate was 4.3 ± 0.6 mg/L.
TABLE 2.
The residual salicylate content (for details, see section “Materials and Methods”).
| Medium* | Concentration of sodium salicylate at the end of the exponential growth phase, mg/l |
| Without deficiency | non detected |
| Low nitrogen | 4.3 ± 0.6 |
| Low iron | 69 ± 18 |
*Initial concentration of sodium salicylate was 1000 mg/l.
Under iron limitation, both the nahU mRNA level and the SH enzyme activity changed (Figure 3). Evans-cultivated BS3701 in low iron conditions showed an approximately 170-fold nahU mRNA decrease with ∼15.4-fold lower activity of SH (from 0.077 ± 0.013 to 0.0050 ± 0.0010 μmol/min∗mg of protein). At the end of the exponential growth phase, the residual salicylate content was 69 ± 18 mg/L (Table 2). The literature describes salicylate-derived siderophores, such as Pseudomonas pyochelin (Crosa and Walsh, 2002) and promysalin (Li W. et al., 2011). As revealed using RAST and Prokka, BS3701 contains homologs of genes involved in the synthesis and transport of pyochelin (fptA, pchR, pchB; unpublished data).
Meta-Cleavage Pathway of Catechol Degradation
Pseudomonas putida BS3701 was found to contain a plasmid-localized gene encoding catechol 2,3-dioxygenase (C23O). During BS3701 cultivation in the presence of salicylate used as a source of carbon (Figure 1), the nahH mRNA level remained unchanged, while the C23O activity increased by ∼1.8-fold (from 0.031 ± 0.008 to 0.057 ± 0.007 μmol/min∗mg of protein).
Under nitrogen limitation, the nahH mRNA level remained unchanged, while the C23O enzyme activity changed (Figure 2). Evans-cultivated BS3701 in low nitrogen conditions showed no change in the nahH mRNA level but the C23O decreased by ∼2-fold (from 0.057 ± 0.007 to 0.028 ± 0.003 μmol/min∗mg of protein). To date, the literature presents no data on the regulatory mechanisms of expression of genes encoding aromatic ring-hydroxylating dioxygenases in low nitrogen conditions. Under iron limitation, both the nahH mRNA level and the C23O enzyme activity changed (Figure 3). Evans-cultivated BS3701 in low iron conditions showed a decrease in the nahH mRNA level by ∼3.7-fold and C23O activity by ∼3.3-fold (from 0.057 ± 0.007 to 0.0175 ± 0.0009 μmol/min∗mg of protein).
Ortho-Cleavage Pathway of Catechol Degradation
The P. putida BS3701 genome contains three copies of the gene encoding catechol 1,2-dioxygenase (C12O) that are localized to the chromosome and denoted as catA(0), catA(2), and catA(8). Although the catA(2) gene is in the immediate vicinity of nahU (the distance between the nucleotide A of the catA(2) start codon ATG and the nucleotide A of the nahU stop-codon TGA is 74 bp), it apparently has its own promoter. In BS3701 grown on Evans supplemented with salicylate (vs. glucose) (Figure 1), the level of catA(0) mRNA remained unchanged, while the content of catA(2)- and catA(8) mRNA increased by ∼32-fold (from 0.39 ± 0.15 to 12.46 ± 3.11) and ∼8-fold (from 4.8 ± 0.6 to 38.2 ± 28.4), respectively. C12O cultivated in the presence of glucose showed no activity which was observed only with salicylate used as a source of carbon (0.0186 ± 0.0018 μmol/min∗mg of protein).
Under nitrogen limitation, the catA mRNA level remained unchanged, while the C12O enzyme activity changed (Figure 2). BS3701 growth on low nitrogen Evans resulted in lower mRNA levels of all three catA copies: for catA(0) it was an approximately 2-fold decrease (from 0.58 ± 0.19 to 0.29 ± 0.01), for catA(2) ∼33-fold (from 12.46 ± 3.11 to 0.38 ± 0.07), and for catA(8) ∼6-fold (from 38.2 ± 28.4 to 6.5 ± 2.8). Though the decrease extents were different, the tendency was the same, reflecting the regulation of catechol 1,2-dioxygenase expression as early as at the level of the varying mRNA content. The activity of C12O was ∼3 times lower (from 0.0186 ± 0.0018 to 0.0064 ± 0.0016 μmol/min∗mg of protein).
Under iron limitation, the catA mRNA level changed for 2 out of 3 copies, and the C23O enzyme activity changed as well (Figure 3). Evans-cultivated BS3701 in low iron conditions retained the same catA(0) mRNA level and showed a decrease in catA(2)- and catA(8) mRNAs by ∼13.5- (from 12.46 ± 3.11 to 0.92 ± 0.15) and ∼33-fold (from 38.2 ± 28.4 to 11.5 ± 0.9), respectively. The activity of C12O decreased by ∼1.5-fold (from 0.0186 ± 0.0018 to 0.012 ± 0.003 μmol/min∗mg of protein).
Bioinformatics Analysis of Regulatory Regions in Genes
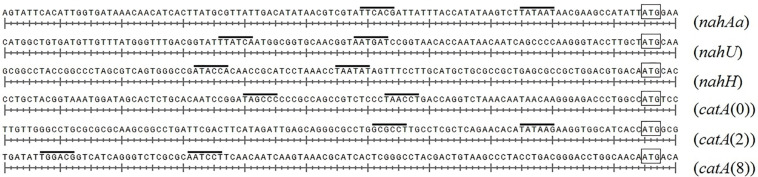
Figure 4 presents DNA fragments with putative promoters of naphthalene catabolic genes. Sigma-70-dependent promoters of the nahAaAbAcAdBFCQED operon, the nahU gene, and nahH-containing gene clusters were identified using BPROM, a SoftBerry prediction program. Localization of the putative promoter of a catA(0)-containing gene cluster was defined using the literature data. Upstream of the benA gene [which probably shares the sole operon with the catA(0) gene] resides a sequence that is 98% identical to the regulatory region adjacent to +1 point of the benABCDKEF operon in P. putida PRS2000 (Cowles et al., 2000). The regions located upstream of the catA(2) gene exhibit homology, though extremely low, to the −10 and −35 boxes of the sigma-70-dependent promoter. Since the catA(2) mRNA content was observed to increase in cells grown in the presence of salicylate, it cannot be ruled out that the transcription activation is mediated by a specific transcription factor that enables RNA polymerase (RNAP) to recognize such a weak promoter. The promoter of the catA(8)-containing catBCA operon is identical to that identified for P. putida catBCA (Chugani et al., 1997). Using the online analyzer Virtual Footprint, the regulatory regions (from ATG to approximately −60 of the putative promoter) were found to have binding patterns of the nucleoid-associated proteins (NAPs) Lrp, Fis, IHF, H-NS, and IciA. Table 3 presents mean scores ± confidence interval typical of operators of these NAPs, as well as scores for operators revealed in the regulatory region of naphthalene catabolic genes.
FIGURE 4.
The structural organization of the regulatory region of gene clusters containing nahAa, nahU, nahH, catA(0), catA(2), and catA(8). The putative –10 and –35 promoter boxes are overlined, the start codon (ATG) closest to the promoter is boxed.
TABLE 3.
Nucleoid-associated proteins with binding patterns located in the studied regulatory regions.
| Mean score ± standard deviation | nahAa | nahU | nahH | benA [catA(0)] | catA(2) | catB [catA(8)] | |
| Lrp | 5.79 ± 0.41 | 3.55 | 4.48 | 4.59 | 6.77 | 5.01 | 4.49 |
| 3.52 | 4.43 | 4.41 | 5.70 | 4.74 | 4.17 | ||
| 3.51 | 4.39 | 4.40 | 5.57 | 4.15 | |||
| Fis | 3.56 ± 0.31 | 2.55 | 3.15 | 3.31 | 3.1 | 2.95 | 3.37 |
| 2.47 | 3.11 | 3.22 | 3.0 | 2.91 | 3.20 | ||
| 3.01 | 3.17 | 2.9 | 2.84 | 3.12 | |||
| IHF | 7.67 ± 0.36 | – | 5.79 | – | – | 6.24 | 6.79 |
| 5.86 | 6.05 | ||||||
| H-NS | 6.73 ± 0.07 | 5.27 | 5.72 | – | – | 5.27 | 5.39 |
| 5.58 | 5.22 | ||||||
| 5.25 | 5.04 | ||||||
| IciA | 5.73 ± 0.09 | 5.01 | 4.94 | – | 5.66 | 5.16 | 5.04 |
| 4.64 | 4.89 | ||||||
| 4.77 |
the nahAa region is upstream of the gene encoding reductase component of naphthalene dioxygenase; the nahU region is upstream of the gene encoding salicylate hydroxylase; the nahH region is upstream of the nahH gene; the benA [catA(0)] region is upstream of the benA gene of the cluster containing the catechol 1,2-dioxygenase catA(0) gene; the catA(2) region is upstream of the catechol 1,2-dioxygenase catA(2) gene; the catB [catA(8)] region is upstream of the catB gene of the cluster containing the catechol 1,2-dioxygenase catA(8) gene. The tabulated values are mean scores ± confidence interval typical for each regulator according to Virtual Footprint (the column « mean score ± standard deviation»). For each regulatory region, Table presents scores of all detected binding patterns of each NAP.
The P. putida BS3701 genome contains one copy of genes coding for Fis (factor for inversion stimulation), Lrp (leucine responsive protein), and IciA (inhibitor of chromosome initiation), five copies of the gene coding for H-NS (histone-like nucleoid-structuring protein, MvaT for Pseudomonas, 50–88% homology of amino acid sequences), as well as one copy of genes coding for alpha- and beta-subunits of IHF (integration host factor) (unpublished data).
Changes in the mRNA Level of rpoD and NAPs
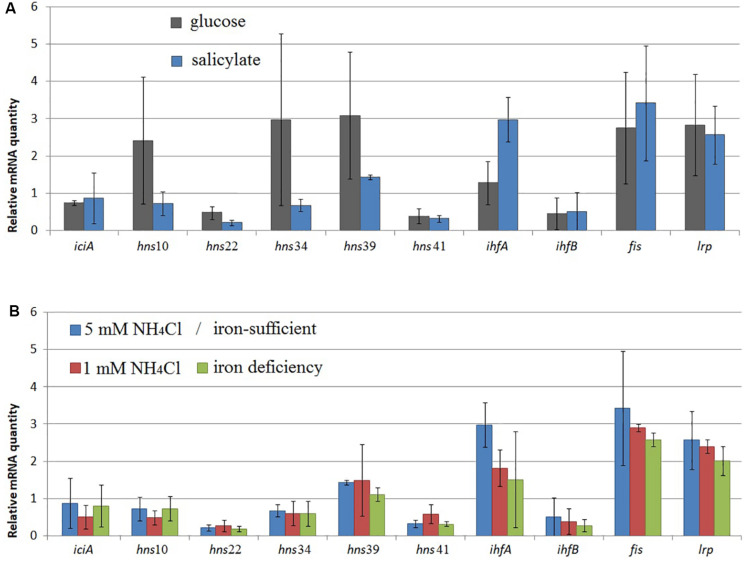
A change was observed in the mRNA level of rpoD encoding RNAP σ70-subunit (Figures 1–3) and in that of iciA, mvaT, ihfA, ihfB, fis, and lrp (Figure 5). Cultivation of BS3701 in Evans supplemented with salicylate (vs. glucose) (Figure 5A) resulted in a statistically significant increase of mRNA of the gene encoding the IHF alpha subunit (by ∼2.3-fold, from 1.28 ± 0.57 to 3.0 ± 0.6). IHF can stimulate the interaction between RNAP and specific translation regulators (Hoover et al., 1990). The literature presents no information concerning the change in IHF amount in response to salicylate. The IHF involvement in the regulation of naphthalene catabolic gene expression certainly requires further experimental confirmation.
FIGURE 5.
The relative mRNA level of genes encoding NAPs in Pseudomonas putida BS3701 grown: (A), on glucose-supplemented Evans (dark-gray bars) or salicylate-supplemented Evans (blue bars); (B), on Evans (Evans, blue bars), low-nitrogen Evans (red bars), and low-iron Evans (green bars). iciA, inhibitor of chromosome initiation; mvaT, histone-like nucleoid-structuring protein (mvaT10, 22, 34, 39, and 41 are homologs); ihfA and ihfB, alpha- and beta-subunits of integration host factor, respectively; fis, factor for inversion stimulation; lrp, leucine responsive protein. The experiments were performed in three biological replicates, the data are presented as a mean score ± confidence interval (p-value < 0.05).
BS3701 grown on Evans in low nitrogen conditions demonstrated (Figure 5B) decreasing mRNA of the gene encoding IHFa (by ∼1.6-fold, from 3.0 ± 0.6 to 1.8 ± 0.5), whereas the literature describes IHF as a co-activator of the transcription of genes involved in nitrogen fixation (Hoover et al., 1990).
Evans-cultivation of BS3701 in low iron conditions produced no effect on the mRNA level of the studied genes coding for NAPs (Figure 5B). We have found that these conditions reduced the mRNA yield of nahAa, nahU, nahH, catA(2), and catA(8), leaving unchanged rpoD mRNA (Figure 3). These results evidence for the existence of other agents whose role in the regulation of naphthalene catabolic gene expression remains unknown.
Discussion
The regulation of expression of genes involved in the naphthalene-salicylate-catechol-Krebs cycle pathway is studied either in terms of the activation in the presence of inductors (in more detail) or in terms of catabolite repression (in part). Here, the activity of key enzymes of the naphthalene catabolic pathway was studied along with the mRNA level of genes encoding these enzymes in P. putida strain BS3701 cultivated in low nitrogen/low iron conditions. In the natural environment, microorganisms have to adapt to nutrient deficiency. In Pseudomonas, NtrC positively regulates the transcription of σ54-dependent target genes under low nitrogen conditions, including the expression of glnA (Hervás et al., 2009) and rhlA (Wenner et al., 2014; Pita et al., 2018), nitrate assimilation operon (Romeo et al., 2012), and nitrogenase genes (Zhan et al., 2019). We observed that mRNAs of nahU (salicylate hydroxylase) and catA (catechol 1,2-dioxygenase) were down-regulated at decreasing NH4Cl concentration from 5 to 1 mM, the rpoD mRNA level remained unchanged, but the ihfA mRNA level altered. Although IHF has been reported to stimulate the RNAP interaction with specific transcriptional regulators (Hoover et al., 1990), our experiments showed a decreased mRNA level. We assume that nahU and catA mRNA levels undergo posttranscriptional regulation through the changing mRNA degradation rate. The mRNAs of nahAa (naphthalene 1,2-dioxygenase) and nahH (catechol 2,3-dioxygenase) remained unchanged, while their enzyme activity changed. Naphthalene 1,2-dioxygenase and catechol 2,3-dioxygenase possibly undergo the posttranscriptional regulation by inhibition/stimulation of translation. The activity of catechol 2,3-dioxygenase decreased. We are the first to report this observation. In contrast, low nitrogen conditions resulted in a higher enzyme activity of naphthalene 1,2-dioxygenase. This effect has not been mentioned in the literature either. Only one fact was found in support of the interconnection between naphthalene 1,2-dioxygenase and nitrogen metabolism. NDO can use indole (a heterocyclic nitrogen-containing compound) as a substrate, having oxidized it to 2,3 dihydroxyindole and further to the salicylate (Ma et al., 2018. It cannot be ruled out that before switching to glutamate catabolism occurring in nitrogen deficiency conditions, P. putida BS3701 uses indole.
Fur (Ferric uptake regulator) acts as a repressor under iron-rich conditions (Ochsner et al., 1995), but with deficient iron, in Pseudomonas, Fur-dependent ncRNA of PrrF, a RyhB homolog, reduces mRNA stability (Massé et al., 2003; Wilderman et al., 2004). Our study has shown a decrease in the mRNA level of nahAa, nahU, nahH, catA(2), and catA(8), though that of rpoD and NAPs remained unchanged. We assume that the mRNA level undergoes post-transcriptional regulation by changing the mRNA degradation rate. Low iron conditions can produce a critical effect on the functioning of dioxygenases with iron-sulfur clusters (naphthalene 1,2-dioxygenase, catechol 2,3-dioxygenase, and catechol 1,2-dioxygenase). As shown previously (Dinkla et al., 2001), these conditions result in a lower activity of catechol 2,3-dioxygenase. Iron-storage proteins are known to undergo post-transcriptional regulation through an ncRNA-induced increase in the mRNA degradation rate (Massé et al., 2003). The expression of salicylate hydroxylase free of iron-sulfur clusters also varied in response to the decreasing concentration of available iron, which can be a result of interconnection between salicylate and salicylate-derived siderophores [e.g., Pseudomonas pyochelin (Crosa and Walsh, 2002) and promysalin (Li W. et al., 2011)]. Annotation using RAST and Prokka revealed that BS3701 contains homologs of genes involved in the synthesis and transport of pyochelin (fptA, pchR, pchB; unpublished data).
In Pseudomonas, naphthalene catabolism has been best studied in terms of the effects of specific transcription activators. We propose that under low nitrogen and low iron conditions the naphthalene catabolic genes are subject not only to transcriptional but also post-transcriptional regulation. These results form a basis for studying the mechanisms of additional regulatory control of genes involved in aromatic hydrocarbon catabolism.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.ncbi.nlm.nih.gov/genbank/, MN442422.1; https://www.ncbi.nlm.nih. gov/genbank/, MN413629.1; https://www.ncbi.nlm.nih.gov/gen bank/, MN442423.1; https://www.ncbi.nlm.nih.gov/genbank/, MN44P2424.1; https://www.ncbi.nlm.nih.gov/genbank/, MN44 2425.1; https://www.ncbi.nlm.nih.gov/genbank/, MN442426.1.
Author Contributions
IP-F, KP, AV, and MZ contributed to the experiment design. IP-F, KP, AV, AF, and RS conducted all experimental work and analyses. IP wrote the manuscript with critical review and inputs from KP, AV, and MZ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Ksenia Skulkina for assistance in primer designing.
Funding. The reported study was funded by the Russian Foundation for Basic Research (#18-29-05071).
References
- Boronin A., Kosheleva I. (2010). “Diversity of naphthalene biodegradation systems in soil bacteria,” in Handbook of Hydrocarbon and Lipid Microbiology, ed. Timmis K. N. (Berlin: Springer; ), 1155–1163. 10.1007/978-3-540-77587-4_80 [DOI] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Chugani S. A., Parsek M. R., Hershberger C. D., Murakami K., Ishihama A., Chakrabarty A. M. (1997). Activation of the catBCA promoter: probing the interaction of CatR and RNA polymerase through in vitro transcription. J. Bacteriol. 179 2221–2227. 10.1128/jb.179.7.2221-2227.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles C. E., Nichols N. N., Harwood C. S. (2000). BenR, a XylS homologue, regulates three different pathways of aromatic acid degradation in Pseudomonas putida. J. Bacteriol. 182 6339–6346. 10.1128/jb.182.22.6339-6346.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa J. H., Walsh C. T. (2002). Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66 223–249. 10.1128/mmbr.66.2.223-249.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkla I. J., Gabor E. M., Janssen D. B. (2001). Effects of iron limitation on the degradation of toluene by Pseudomonas strains carrying the tol (pWWO) plasmid. Appl. Environ. Microbiol. 67 3406–3412. 10.1128/AEM.67.8.3406-3412.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua R. D., Meera S. (1981). Purifiation and characterisation of naphthalene oxygenase from Corynebacterium renale. Eur. J. Biochem. 120 461–465. 10.1111/j.1432-1033.1981.tb05724.x [DOI] [PubMed] [Google Scholar]
- Feist C. F., Hegeman G. D. (1969). Phenol and benzoate metabolism by Pseudomonas putida: regulation of tangential pathways. J. Bacteriol. 100 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov A. E., Kosheleva I. A., Samojlenko V. A., Shkidchenko A. N., Nechaeva I. A., Puntus I. F., et al. (2006). Biological Preparation for Cleaning of Soils From Contaminations With Oil and Oil Products, Method of Its Production and Application. Technical Report No. RU 2 378 060 C2. Moscow: Federal service for intellectual property, patents and trademarks. [Google Scholar]
- Hegeman G. D. (1966). Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida I. Synthesis of enzymes by the wild type. J. Bacteriol. 91 1140–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervás A. B., Canosa I., Little R., Dixon R., Santero E. (2009). NtrC-dependent regulatory network for nitrogen assimilation in Pseudomonas putida. J. Bacteriol. 191 6123–6135. 10.1128/JB.00744-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover T. R., Santero E., Porter S., Kustu S. (1990). The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell 63 11–22. 10.1016/0092-8674(90)90284-L [DOI] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. (1987). Overproduction of the xylS gene product and activation of the xylDLEGF operon on the TOL plasmid. J. Bacteriol. 169 3587–3592. 10.1128/jb.169.8.3587-3592.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izmalkova T. Y., Mavrodi D. V., Sokolov S. L., Kosheleva I. A., Smalla K., Thomas C. M., et al. (2006). Molecular classification of IncP-9 naphthalene degradation plasmids. Plasmid 56 1–10. 10.1016/J.PLASMID.2005.12.004 [DOI] [PubMed] [Google Scholar]
- Jin C. W., Ye Y. Q., Zheng S. J. (2014). An underground tale: contribution of microbial activity to plant iron acquisition via ecological processes. Ann. Bot. 113 7–18. 10.1093/aob/mct249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler B., Herrero M., Timmis K. N., de Lorenzo V. (1994). Genetic evidence that the XylS regulator of the Pseudomonas TOL meta operon controls the Pm promoter through weak DNA-protein interactions. J. Bacteriol. 176 3171–3176. 10.1128/jb.176.11.3171-3176.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladino-Orjuela G., Gomes E., da Silva R., Salt C., Parsons J. R. (2016). “Metabolic pathways for degradation of aromatic hydrocarbons by bacteria,” in Reviews of Environmental Contamination and Toxicology, Vol. 237 ed. de Voogt W. P. (Cham: Springer International Publishing; ), 105–121. 10.1007/978-3-319-23573-8_5 [DOI] [PubMed] [Google Scholar]
- Li S., Li X., Zhao H., Cai B. (2011). Physiological role of the novel salicylaldehyde dehydrogenase NahV in mineralization of naphthalene by Pseudomonas putida ND6. Microbiol. Res. 166 643–653. 10.1016/J.MICRES.2011.01.003 [DOI] [PubMed] [Google Scholar]
- Li W., Estrada-de los Santos P., Matthijs S., Xie G.-L., Busson R., Cornelis P., et al. (2011). Promysalin, a salicylate-containing Pseudomonas putida antibiotic, promotes surface colonization and selectively targets other Pseudomonas. Chem. Biol. 18 1320–1330. 10.1016/J.CHEMBIOL.2011.08.006 [DOI] [PubMed] [Google Scholar]
- Liu J.-H., Smith P. C. (1996). Direct analysis of salicylic acid, salicyl acyl glucuronide, salicyluric acid and gentisic acid in human plasma and urine by high-performance liquid chromatography. J. Chromatogr. B Biomed. Sci. Appl. 675 61–70. 10.1016/0378-4347(95)00337-1 [DOI] [PubMed] [Google Scholar]
- Ma Q., Zhang X., Qu Y. (2018). Biodegradation and biotransformation of indole: advances and perspectives. Front. Microbiol. 9:2625. 10.3389/fmicb.2018.02625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E., Escorcia F. E., Gottesman S. (2003). Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17 2374–2383. 10.1101/gad.1127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U. A., Vasil A., I, Vasil M. L. (1995). Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin a expression: purification and activity on iron-regulated promoters. J. Bacteriol. 177 7194–7201. 10.1128/jb.177.24.7194-7201.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek M. R., Shinabarger D. L., Rothmel R. K., Chakrabarty A. M. (1992). Roles of CatR and cis, cis-muconate in activation of the catBC operon, which is involved in benzoate degradation in Pseudomonas putida. J. Bacteriol. 174 7798–7806. 10.1128/jb.174.23.7798-7806.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrikov K., Delegan Y., Surin A., Ponamoreva O., Puntus I., Filonov A., et al. (2013). Glycolipids of Pseudomonas and Rhodococcus oil-degrading bacteria used in bioremediation preparations: formation and structure. Process Biochem. 48 931–935. 10.1016/J.PROCBIO.2013.04.008 [DOI] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper – excel-based tool using pair-wise correlations. Biotechnol. Lett. 26 509–515. 10.1023/B:BILE.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- Pita T., Feliciano R. J., Leitão H. J. (2018). Small noncoding regulatory RNAs from Pseudomonas aeruginosa and burkholderia cepacia complex. Int. J. Mol. Sci. 19:3759. 10.3390/ijms19123759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L., Marqués S., Timmis K. N. (1997). Transcriptional control of the pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu. Rev. Microbiol. 51 341–373. 10.1146/annurev.micro.51.1.341 [DOI] [PubMed] [Google Scholar]
- Romeo A., Sonnleitner E., Sorger-Domenigg T., Nakano M., Eisenhaber B., Bläsi U. (2012). Transcriptional regulation of nitrate assimilation in Pseudomonas aeruginosa occurs via transcriptional antitermination within the nirBD–PA1779–cobA operon. Microbiology 158 1543–1552. 10.1099/mic.0.053850-0 [DOI] [PubMed] [Google Scholar]
- Schell M. A., Poser E. F. (1989). Demonstration, characterization, and mutational analysis of NahR protein binding to nah and sal promoters. J. Bacteriol. 171 837–846. 10.1128/jb.171.2.837-846.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simarro R., González-Benítez N., Bautista L. F., Sanz R., Molina M. C. (2011). Optimisation of key abiotic factors of PAH (Naphthalene, Phenanthrene and Anthracene) biodegradation process by a bacterial consortium. Water, Air, Soil Pollut. 217, 365–374. 10.1007/s11270-010-0593-8 [DOI] [Google Scholar]
- Tover A., Zernant J., Chugani S. A., Chakrabarty A. M., Kivisaar M. (2000). Critical nucleotides in the interaction of CatR with the pheBA promoter: conservation of the CatR-mediated regulation mechanisms between the pheBA and catBCA operons. Microbiology 146 173–183. 10.1099/00221287-146-1-173 [DOI] [PubMed] [Google Scholar]
- Wang Q., Wang C., Yu W., Turak A., Chen D., Huang Y., et al. (2018). Effects of nitrogen and phosphorus inputs on soil bacterial abundance, diversity, and community composition in Chinese Fir plantations. Front. Microbiol. 9:1543. 10.3389/fmicb.2018.01543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenner N., Maes A., Cotado-Sampayo M., Lapouge K. (2014). NrsZ: a novel, processed, nitrogen-dependent, small non-coding RNA that regulates Pseudomonas aeruginosa PAO1 virulence. Environ. Microbiol. 16 1053–1068. 10.1111/1462-2920.12272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman P. J., Sowa N. A., FitzGerald D. J., FitzGerald P. C., Gottesman S., Ochsner U. A., et al. (2004). Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U.S.A. 101 9792–9797. 10.1073/pnas.0403423101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Katagiri M., Maeno H., Hayaishi O. (1965). Salicylate hydroxylase amonooxygenase requiring flavin adenine dinucleotide. I. Purification general properties. J. Biol. Chem. 240, 3408–3413. [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. (1982). Plasmid gene organization: naphthalene/salicylate oxidation. Proc. Natl. Acad. Sci. U S A 79, 874–878. 10.1073/pnas.79.3.874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y., Deng Z., Yan Y., Zhang H., Lu C., Yang Z., et al. (2019). NfiR, a new regulatory noncoding RNA (ncRNA), is required in concert with the NfiS ncRNA for optimal expression of nitrogenase genes in Pseudomonas stutzeri A1501. Appl. Environ. Microbiol. 85:e00762-19. 10.1128/AEM.00762-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.ncbi.nlm.nih.gov/genbank/, MN442422.1; https://www.ncbi.nlm.nih. gov/genbank/, MN413629.1; https://www.ncbi.nlm.nih.gov/gen bank/, MN442423.1; https://www.ncbi.nlm.nih.gov/genbank/, MN44P2424.1; https://www.ncbi.nlm.nih.gov/genbank/, MN44 2425.1; https://www.ncbi.nlm.nih.gov/genbank/, MN442426.1.