Abstract
Aims
The molecular and cellular mechanisms underlying the antidepressant effects of fluoxetine in the brain are not fully understood. Emerging evidence has led to the hypothesis that chronic fluoxetine treatment induces dematuration of certain types of mature neurons in rodents. These studies have focused on the properties of typical molecular and/or electrophysiological markers for neuronal maturation. Nevertheless, it remains unknown whether dematuration‐related phenomena are present at the genome‐wide gene expression level.
Methods
Based on the aforementioned hypothesis, we directly compared transcriptome data between fluoxetine‐treated adult mice and those of naive infants in the hippocampus and medial prefrontal cortex (mPFC) to assess similarities and/or differences. We further investigated whether fluoxetine treatment caused dematuration in these brain regions in a hypothesis‐free manner using a weighted gene co‐expression network analysis (WGCNA).
Results
Gene expression patterns in fluoxetine‐treated mice resembled those in infants in the mPFC and, to a large extent, in the hippocampus. The gene expression patterns of fluoxetine‐treated adult mice were more similar to those of approximately 2‐week‐old infants than those of older mice. WGCNA confirmed that fluoxetine treatment was associated with maturation abnormalities, particularly in the hippocampus, and highlighted respective co‐expression modules for maturity and immaturity marker genes in the hippocampus in response to fluoxetine treatment.
Conclusions
Our results strongly support the hypothesis that chronic fluoxetine treatment induces dematuration in the adult mouse brain from a transcriptomic standpoint. Detection of discrete transcriptomic regulatory networks related to fluoxetine treatment may help to further elucidate the mechanisms of antidepressant action.
This study compares the transcriptomic profile of adult mice treated with clinically relevant dose of FLX and that of naïve infants in the hippocampus and medial prefrontal cortex (mPFC). We observed that gene expression profiles in FLX‐treated adult mice resembled those of infants in the mPFC and hippocampus. Our results provide support for the hypothesis that FLX can cause dematuration of the adult mouse brain to a more immature phenotype.
1. INTRODUCTION
Antidepressant use has increased over years, and a recent survey estimates that approximately 12.0% of adults in the United States take antidepressants.1 Fluoxetine (FLX), a selective serotonin reuptake inhibitor, is one of the most commonly used antidepressants for the treatment of mood and anxiety disorders worldwide. Despite its widespread use, its underlying molecular and cellular mechanisms of action are not fully understood. Previous studies using animal models suggest that enhanced adult neurogenesis in the hippocampal dentate gyrus (DG) is a potential mechanism of action for antidepressants, including FLX.2, 3 Conversely, other studies showed that the behavioral effects of FLX are not always accompanied by increased neurogenesis.4, 5 It is thus plausible that modifications of preexisting neurons, in addition to generation of new neurons, are important for antidepressant actions.
Emerging evidence has suggested that chronic FLX treatment reverses the established maturation state of preexisting mature neurons in the adult rodent brain, a phenomenon termed “dematuration”.6, 7, 8 In the hippocampal DG, granule cells (GCs) in FLX‐treated adult mice exhibited similarities with typical immature GCs in terms of molecular marker expression (eg, decreased expression of calbindin and increased expression of calretinin, markers of mature and immature GCs, respectively) and electrophysiological characteristics (eg, reduction of basal synaptic transmission and frequency facilitation of the synapses between DG and CA3, and reinstatement of high membrane excitability).6, 9 These results suggest that chronic FLX treatment induces dematuration of GCs in the adult mouse hippocampus. Recent research suggests that FLX induces dematuration‐like phenomena in several brain regions other than the hippocampus. Chronic FLX treatment may convert mature interneurons, in particular parvalbumin (PV)‐positive fast‐spiking (FS) neurons, to a more immature state in the basolateral amygdala10 and visual cortex,11 resulting in reactivation of juvenile‐like plasticity in adulthood. In the medial prefrontal cortex (mPFC) of adult mice, chronic FLX treatment decreased PV expression, a marker of mature FS neurons, and of perineuronal nets (PNNs), extracellular matrices predominantly expressed by mature FS neurons. These findings suggest that FLX induces dematuration of FS neurons.7 Such dematuration‐related phenomena induced by FLX treatment have been evaluated by examining several molecular and/or electrophysiological properties that reflect maturational states of each neuronal type. However, the dematuration effects of FLX have not been assessed at the genome‐wide gene expression level.
We previously provided transcriptomic evidence for pseudo‐immaturity in the PFC and/or hippocampus of patients with schizophrenia12 and alcoholism13 by conducting bioinformatics analyses of microarray data. More specifically, we directly compared patterns of gene expression changes in brain regions of patients with the diseases and typically developing infants, and evaluated the similarities between them using the Running Fisher's algorithm, a nonparametric ranking analysis of gene expression signatures. The results showed striking similarities in gene expression patterns between patients and infants, suggesting pseudo‐immaturity of the PFC/hippocampus in patients with schizophrenia and alcoholism.12, 13 This bioinformatics analytical approach has been found to be powerful for detecting similarities between two transcriptome datasets, even from different experimental platforms (eg, DNA microarray and RNA‐sequencing) or different species (eg, human, mouse, and rat).12, 13, 14, 15, 16 In this study, using the same bioinformatics technique, we first conducted hypothesis‐driven transcriptome analyses, in which genome‐wide gene expression patterns were directly compared between FLX‐treated adult mice and naive infant mice to statistically evaluate their similarities. To further characterize FLX‐induced gene expression perturbations, co‐expression network analysis17 was performed as an unsupervised, hypothesis‐free method, to evaluate whether there were gene sets that were associated with FLX‐induced dematuration in the mouse hippocampus and mPFC.
2. MATERIALS AND METHODS
2.1. Animals and antidepressant treatment
Male C57BL/6J mice were used unless otherwise specified. FLX was administered to 9‐week‐old mice for 3 weeks using FLX‐releasing pellets, aiming for a dose of 15 mg/kg/d, as previously described.18 The dose of FLX used in this study is a clinically relevant dose; 15 mg/kg/d FLX resulted in a range of serum FLX levels in patients taking 20‐80 mg/d of FLX.19 All animal experiments were approved by the Institutional Animal Care and Use Committee of Fujita Health University, based on the Law for the Humane Treatment and Management of Animals (2005) and the Standards Relating to the Care and Management of Laboratory Animals and Relief of Pain (2006). Every effort was made to minimize the number of animals used.
2.2. DNA microarray analysis
We performed microarray analyses of the following mouse brain samples: the hippocampal DG and mPFC of mice treated with either FLX or vehicle; the mPFC of naive 2‐ and 12‐week‐old mice; and the mPFC of alpha‐calcium/calmodulin‐dependent protein kinase II heterozygous (Camk2a +/−) mice (Table S1) as described previously.14, 20 The microarray data were deposited in the GEO database under accession numbers GSE118667, GSE118668, GSE118669, and GSE118724. Further details are provided in the Appendix S1: Supplementary Methods. Other microarray data used in this study were previously obtained in our laboratory and other laboratories (Table S1).
2.3. Microarray data processing
Using the expression values, we calculated fold changes and t test P‐values between two conditions for each transcript: FLX‐ and vehicle‐treated adult mice, and naive infant and adult mice (Table S1). Fold changes were calculated by dividing the average value of FLX‐treated mice by that of vehicle‐treated mice. The average value of infant mice of each age was divided by that of adult mice. Microarray data on wild‐type mice obtained previously14 were used as data of adult mice to examine developmental gene expression changes in the naive mouse DG (GSE42778 and GSE113727).16 Genes (or transcripts) with absolute fold change >1.2 and P‐value < 0.05 (without correction for multiple testing) were imported to the web‐based bioinformatics tool BaseSpace (formerly named as NextBio; Illumina, San Diego, CA; http://www.nextbio.com)21 according to the manufacturer's instructions. For the microarray data examining time‐course changes in the gene expression patterns of mouse frontal cortex,22 we used the data that had been imported into BaseSpace by its curators.
Regarding the microarray data examining development of different cell types (FS neurons, astrocytes, and oligodendrocytes), we used publicly available data on GSE1780623 and GSE956624 series, as described elsewhere.12 Briefly, fold changes and t test P‐values were calculated between younger and older conditions (Table S1), and genes (transcripts) with absolute fold change >1.5 and a t test P‐value < 0.05 were imported into BaseSpace.12
Microarray analyses of the hippocampal DG and mPFC of Schnurri‐2 knockout (Shn2 KO) mice (GSE42778),14 the hippocampal DG of Camk2a +/− mice,20 and the hippocampus of mutant synaptosomal‐associated protein of 25 kDa knockin (Snap25 KI) mice25 were conducted in the previous studies. Transcripts with absolute fold change >1.2 and P‐value < 0.05 were imported into BaseSpace. We used the publicly available microarray data from epilepsy models (hippocampus of pilocarpine‐treated rats [GSE47752]26 and cortex of kainate‐treated mice [GSE6388]27) that had been imported into BaseSpace by its curators.
Subsequently, the gene expression patterns of the two given gene sets were statistically compared using BaseSpace. Using the bioinformatics tool, similarities were examined using Running Fisher algorithm, a nonparametric rank‐based statistical method, in which information regarding the rank based on absolute value of fold change and the direction of gene expression changes within each gene set was considered.21 The greater the similarity in gene expression patterns between the two conditions, the lower the resulting overlap P‐value. Details of the algorithm have been described previously.14
2.4. Pathway analysis
Pathways/biogroups enriched in the genes of interest were determined through a combination of rank‐based enrichment statistics and biomedical ontologies using BaseSpace.21 Pathways/biogroups from GO and canonical pathways of Broad MSigDB were included in this analysis.
2.5. Weighted gene co‐expression network analysis
Weighted gene co‐expression network analysis was conducted for step‐by‐step block‐wise network construction and module detection using the package implemented in R (ver. 3.1.3) with the code provided by Langfelder and Horvath.17 Microarray data of two brain regions (hippocampal DG [GSE118669] and mPFC [GSE118668]) were separately used for network construction. To remove transcripts that were constitutively expressed, unexpressed, or varied modestly across conditions, a coefficient of variation (CV; CV = μ/σ) filter was applied to the averaged expression values for each transcript across all mice (combined FLX and vehicle‐treated mice) using an R script. The CV cutoff values were set at 0.2 to meet the criteria of maximum block size 5000. The resulting 4686 and 3202 transcripts in the DG and mPFC datasets, respectively, were processed for WGCNA as described in the Appendix S1: Supplementary Methods.
3. RESULTS
3.1. Gene expression patterns in the hippocampal DG and mPFC of adult mice treated with FLX resemble those of naive infant mice
In the microarray analysis, 1051 and 274 of the total 45 037 transcripts were differentially expressed between FLX‐treated and control mice in the DG and mPFC, respectively: Expression of 334 transcripts was upregulated and that of 717 was downregulated in the DG of FLX‐treated mice, and the expression of 138 transcripts was upregulated and that of 136 was downregulated in the mPFC of FLX‐treated mice compared to controls (absolute fold change >1.2, t test P < 0.05; Tables S2 and S3). We used a more liberal threshold (P < 0.05 without any correction for multiple tests) to increase the chance of identifying functionally related genes.
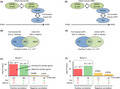
We first assessed whether, or to what extent, the overall gene expression patterns in the DG of adult mice treated with FLX (as compared to vehicle‐treated) were similar to those of naive infant mice (as compared to naive adults; Figure 1A). Similarities in gene expression patterns between the two given datasets were assessed using the Running Fisher algorithm, a nonparametric rank‐based statistical method.21 The bioinformatics analysis revealed a significant similarity in the gene expression patterns between FLX‐treated adult mice and naive infant mice in the DG (denoted as overlap P‐value: P = 1.2 × 10−37, Figure 1B). Of 1051 transcripts whose expression was changed in the FLX‐treated DG, 699 were shared with transcripts altered in infants’ DG (Figure 1B; Table S4). Among the 699 transcripts, 152 were upregulated (P = 9.0 × 10−41) and 382 were downregulated (P = 3.8 × 10−38) in both FLX‐treated and infants’ DG (Figure 1C); these transcripts were denoted as having a positive correlation. Expression of 66 transcripts was upregulated in the FLX‐treated DG and downregulated in the infants’ DG (P = 0.0086), and 99 transcripts were downregulated in the FLX‐treated DG and upregulated in the infants’ DG (P = 0.029; Figure 1C); the transcripts that showed opposite changes between the two conditions were denoted as having a negative correlation.
Figure 1.
Transcriptomic dematuration in the hippocampal dentate gyrus (DG) and medial prefrontal cortex (mPFC) of adult mice chronically treated with fluoxetine (FLX). A and D, The gene expression pattern in the hippocampal DG (A) and mPFC (D) of FLX‐treated adult mice (FLX compared to controls) was compared with that in the corresponding brain regions of naive infant mice (infants compared to adults). B and E, Venn diagram illustrating the overlap in transcriptome‐wide gene expression changes in the DG (B) and mPFC (E) of FLX‐treated adult mice and naive infant mice. C and F, P‐values of overlap between the FLX‐treated adults and naive infants in the DG (C) and mPFC (F) datasets. Bar graphs illustrate the P‐values of overlap of genes upregulated (red arrows) or downregulated (blue arrows) by each condition, between the two conditions. The genes that showed the same directional changes in expression, or positive correlation between two groups, were designated Bioset 1 or Bioset 2 for the DG or mPFC datasets, respectively (surrounded by dotted lines), which were used in the analyses for potential cell‐type contribution (Figure 3)
To confirm these findings, we analyzed different microarray data of FLX‐treated mouse hippocampus obtained in an independent laboratory (GSE6476).28 Similarly, we observed a significant similarity in gene expression patterns between FLX‐treated adult mice and naive infant mice (P = 1.0 × 10−38, Figure S1A‐C, Appendix S1: Supplementary Results). These results suggest that chronic FLX treatment induces dematuration of the hippocampal DG in adulthood at a genome‐wide gene expression level.
We assessed FLX‐induced gene expression changes in the mPFC in the same manner as performed for the hippocampus. The gene expression patterns in the mPFC in FLX‐treated adult mice (as compared to vehicle‐treated adults) and naive infant mice (as compared to naive adults) were significantly similar to each other (overlap P‐value = 2.5 × 10−9, Figure 1D,E). Of 274 transcripts whose expression was altered in the FLX‐treated mPFC, 102 were shared with transcripts altered in the mPFC of naive infant mice (Figure 1E; Table S5). Among the 102 transcripts, 31 were upregulated (P = 2.3 × 10−10) and 39 were downregulated (P = 9.5 × 10−15) in both conditions (Figure 1F), denoting transcripts with a positive correlation. Expression of 13 transcripts was upregulated in the mPFC of FLX‐treated adult mice and downregulated in the mPFC of naive infant mice (P = 0.0001), and 19 transcripts were downregulated in the FLX‐treated mPFC and upregulated in the naive infant mPFC (P = 0.0031; Figure 1F), denoting transcripts with a negative correlation. We further analyzed different microarray data examining development of the mouse PFC obtained in an independent laboratory (GSE4675)22 and found a significant similarity with those of FLX‐treated adult mice (P = 1.2 × 10−6, Figure S1D‐F, Appendix S1: Supplementary Results). The relatively low number of shared transcripts observed in the analyses of PFC than that of DG might be due to the use of relatively young adult mice in the developmental datasets of PFC. Collectively, these results support the hypothesis that chronic FLX treatment induces dematuration of the hippocampal DG and mPFC in adult mice in the context of genome‐wide expression patterns.
The genes showing positive correlations in expression between FLX‐treated adult mice and naive infant mice in the hippocampal DG and mPFC were designated Bioset 1 and 2, respectively (Figure 1C,F; Tables S14 and S5), which represent genes related to FLX‐induced dematuration in each brain region. These Biosets were further processed for potential cell‐type contribution analysis (Figure 3).
3.2. Gene expression patterns in the brains of FLX‐treated adult mice are more similar to those in approximately 2‐week‐old infant mice than those in older mice
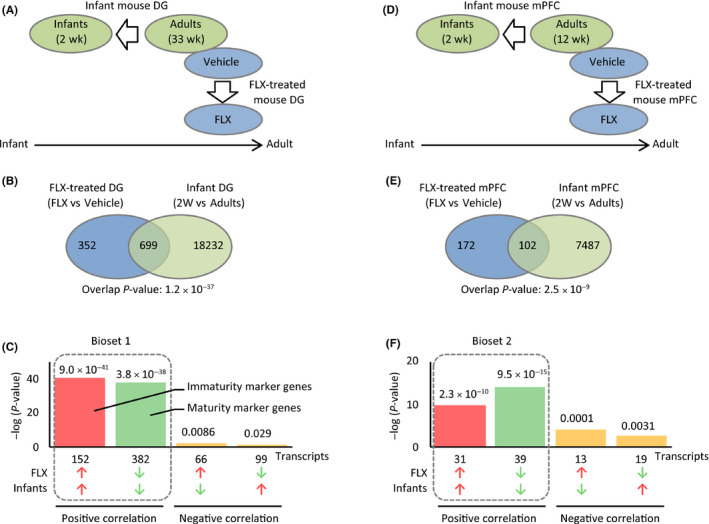
To estimate the age at which FLX treatment may reverse maturational states of adult brain, we compared gene expression patterns in the brains of FLX‐treated adult mice with those of infants at different time points after birth. Gene expression patterns in the DG of FLX‐treated adult mice (as compared to those of vehicle‐treated adults) were compared to those of 1‐, 2‐, 3‐, or 4‐week‐old naive infant mice (as compared to naive adults; Figure 2A). The analyses revealed that gene expression patterns in the DG of FLX‐treated adult mice were more similar to those of 1‐ to 3‐week‐old infant mice than those of the older mice with respect to both overlap P‐value and the number of genes with a positive correlation (Figure 2B,C; Table S6).
Figure 2.
Gene expression patterns in the dentate gyrus (DG) of fluoxetine (FLX)‐treated adult mice are most similar to those of 1‐wk‐old infants. A, Gene expression patterns in the DG of FLX‐treated adult mice were compared with those in the DG of 1‐, 2‐, 3‐ and 4‐wk‐old infant mice, respectively. D, Gene expression patterns in the medial prefrontal cortex (mPFC) of FLX‐treated adult mice were compared with those in the PFC of 2‐, 3‐, 4‐ and 5‐wk‐old infant mice, respectively. B and E, Bar graphs illustrating the overlap P‐values (−log‐transformed data) between the FLX‐treated adult mice and each group of infants for the DG (B) and mPFC (E) datasets. C and F, Bar graphs illustrating the number of genes with positive correlation between the FLX‐treated adult mice and each group of infants for the DG (C) and mPFC (F) datasets
For the assessment of mPFC, we used microarray data examining developmental expression changes in the mouse PFC (2‐, 3‐, 4‐, 5‐ or 10‐week‐old)22 for comparison with mPFC of FLX‐treated adult mice (Figure 2D). Gene expression patterns in the mPFC of FLX‐treated adult mice were most similar to those of 2‐week‐old infant mice (Figure 2E,F; Table S7). These results suggest that chronic FLX treatment may convert gene expression patterns in the hippocampal DG and mPFC of adult mice to those of approximately 2‐week‐old infants.
3.3. Potential cell‐type contributions to FLX‐induced transcriptomic dematuration in the adult mouse brain
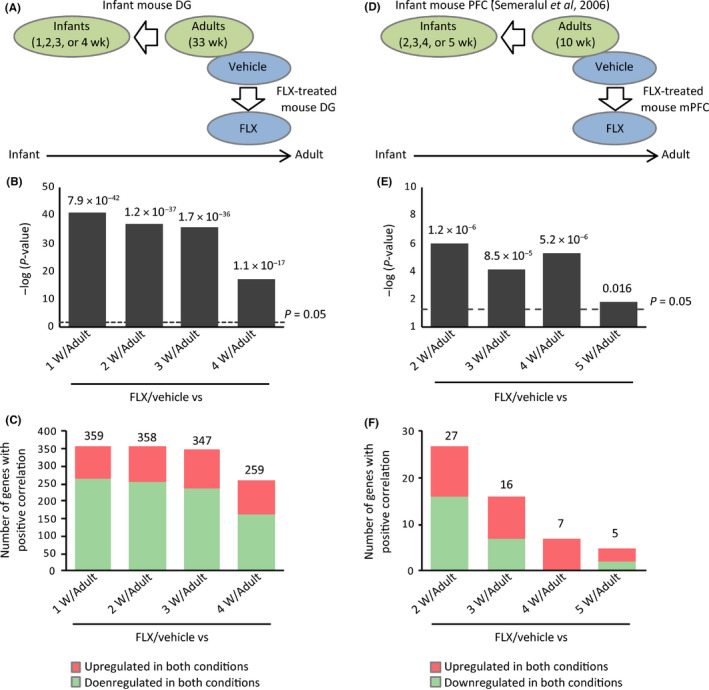
Next, we sought to elucidate which cell types potentially contributed to FLX‐induced transcriptomic dematuration in the hippocampal DG and mPFC. We compared the following two groups of datasets: gene expression patterns of Biosets 1 or 2 (which consisted of genes whose expression was changed in the same direction in FLX‐treated adult and naive infant mice in the hippocampal DG or mPFC, respectively; Figure 1C,F) and those in immature FS neurons, immature astrocytes, and immature oligodendrocytes, respectively (Table S1). Brief descriptions of the microarray data examining development of these cell types are provided in the Appendix S1: Supplementary Methods.
Of 461 genes (encoded by 534 transcripts) in Bioset 1, 143 genes (31.0%) were shared with immature FS neurons (P7 compared to P40; overlap P‐value = 6.0 × 10−8; Figure 3A; Table S8). Furthermore, 95 genes (20.6%) and 151 genes (32.8%) were overlapped between Bioset 1 and datasets of immature astrocytes (overlap P‐value = 1.7 × 10−11; Figure 3B; Table S9) and immature oligodendrocytes (overlap P‐value = 6.6 × 10−5; Figure 3C; Table S10) with a positive correlation, respectively. These results suggest that dematuration may occur in those cell types examined, as well as in GCs,6 which potentially contribute to FLX‐induced transcriptomic dematuration in the hippocampal DG.
Figure 3.
Potential cell‐type contributions to transcriptomic dematuration in the fluoxetine (FLX)‐treated dentate gyrus (DG) and medial prefrontal cortex (mPFC). Genes exhibiting the same directional changes in expression between the normal infant and adult FLX‐treated hippocampal DG (A‐C, Bioset 1 denoted in Figure 1) and mPFC (D‐F, Bioset 2 denoted in Figure 2) were compared to the gene expression changes obtained from cell‐type‐specific developmental experiments (A and D, FS neurons [GSE1780]; B and E, astrocytes [GSE9566]; C and F, oligodendrocytes [GSE9566]). Venn diagrams illustrate the overlap in transcriptome‐wide alterations in gene expression between the two given datasets. Bar graphs illustrate the P‐values of overlap of genes upregulated (red arrows) or downregulated (blue arrows) by each condition. Note the scale of y‐axis is the same in A‐C and D‐F, respectively. Ast, astrocytes; FS neurons, fast‐spiking neurons; OL, oligodendrocytes
We applied the same analysis to Bioset 2 that represents FLX‐induced transcriptomic dematuration in the mPFC. Significant similarities in gene expression patterns were observed between Bioset 2 and immature FS neurons, immature astrocytes, and immature oligodendrocytes, respectively (Figure 3D‐F; Tables S11‐S13).
3.4. Pseudo‐immature hippocampus in FLX‐treated mice resembles that in mouse models of neuropsychiatric disorders
Pseudo‐immaturity of particular brain regions, especially of the hippocampal DG, is observed in several strains of mice with good face and concept validities for models of neuropsychiatric disorders including schizophrenia/intellectual disability (Shn2 KO mice14), bipolar disorder (Camk2a +/− mice29, 30), and epilepsy (Snap25 KI mice25; pilocarpine‐treated mice31). We examined similarities in pseudo‐immature phenotypes at a transcriptome level. Gene expression patterns in the DG of FLX‐treated adult mice were significantly similar to each of the model mice examined with positive correlation (overlap P‐value: P = 7.4 × 10−161 for Shn2 KO mice; P = 6.7 × 10−111 for Camk2a +/− mice; P = 6.7 × 10−59 for Snap25 KI mice; P = 1.7 × 10−18 for pilocarpine‐induced epileptic rats; Figure 4A‐D; Tables S14‐S17). We observed that the hippocampal gene expression patterns in these mouse models of neuropsychiatric disorders were similar to each other as well as to those in infants (Figure 4E). These results suggest that divergent causes such as genetic modifications, neuronal hyperexcitation, and antidepressant treatment induce similar maturation‐related abnormalities in the hippocampus, resulting in transcriptomic similarities.
Figure 4.
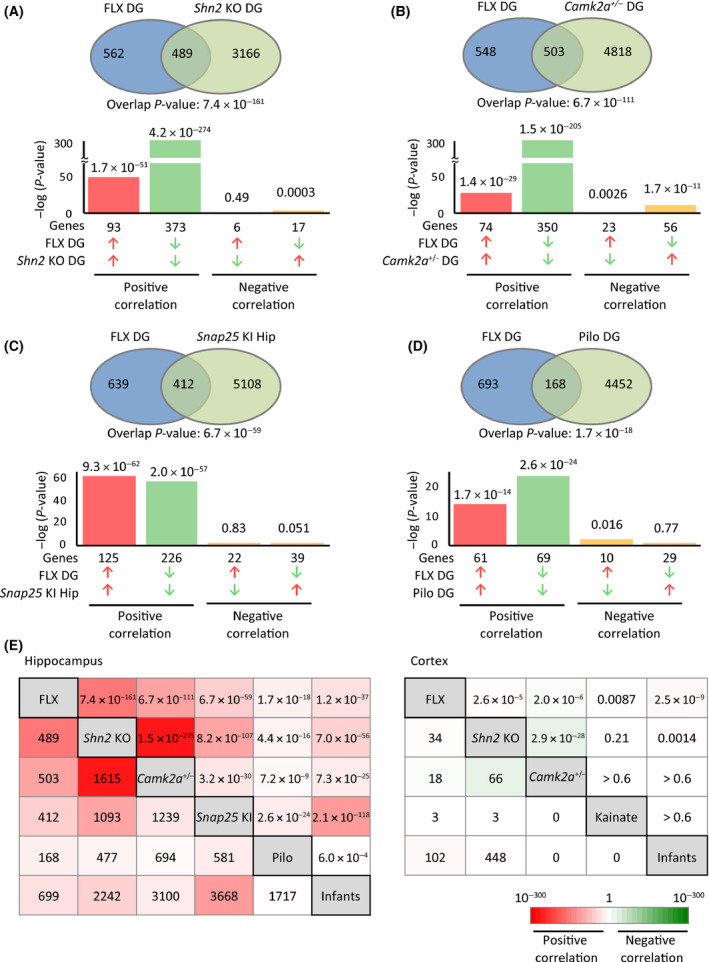
Transcriptomic similarities in the hippocampus between FLX‐treated mice and mice in models of neuropsychiatric disorders showing pseudo‐immaturity phenotypes. A‐D, Gene expression patterns in the hippocampus of FLX‐treated mice were compared with those of Shn2 knockout (KO) mice (dentate gyrus [DG], A), Camk2a +/− mice (DG, B), Snap25 knockin (KI) mice (hippocampus, C), and pilocarpine‐induced epileptic rats (DG, D), respectively. Venn diagrams illustrate the overlap in transcriptome‐wide gene expression changes between the two given datasets. Bar graphs illustrate the P‐values of overlap of genes upregulated (red arrows) or downregulated (blue arrows) by each condition, between the two conditions. E, Overlap P‐values (upper right cells) and the number of common genes/transcripts (lower left cells) responsive to both conditions for each pair of interest. Cells are colored according to the overlap P‐value of the given two datasets. See also Figure S2 for the details of mPFC analyses. Red spectrum colors indicate positive correlations; green spectrum colors indicate negative correlations. DG, dentate gyrus; FLX, fluoxetine; Hip, hippocampus; KA, kainate; Pilo, pilocarpine
In the PFC, low or inverse correlations of gene expression patterns were observed between mice chronically treated with FLX and Shn2 KO mice, Camk2a +/− mice, and kainate‐induced epileptic mice, and among each group (Figure 4E and Figure S2). These results suggest that factors that induce similar transcriptomic alterations in the hippocampus do not necessarily induce similar transcriptomic alterations in the PFC.
3.5. FLX‐related perturbations in maturity marker genes in the hippocampal DG based on co‐expression network analysis
We further investigated whether chronic FLX treatment is associated with maturation abnormalities in the brain regions at a transcriptome level with a hypothesis‐free approach using WGCNA. Hierarchical clustering of our microarray datasets identified ten and five co‐expression modules ranging in size from 65 to 2304 transcripts in the DG and 56 to 2030 in the mPFC, respectively (Figure S3, Tables S18 and S19). Of the 15 modules, three in the DG showed a significant association with FLX treatment (module DG‐brown: r = −0.84, P = 7.0 × 10−5; DG‐red: r = 0.57, P = 0.03; and DG‐green: r = 0.55, P = 0.04; Figure 5A‐D). No modules in the mPFC were associated with FLX treatment at a significance level of P < 0.05 (Figure 5A), suggesting that FLX exerts greater effects on transcriptome alterations in the hippocampal DG compared to the mPFC. Pathway analysis revealed that the module DG‐brown that was most associated with FLX treatment in the DG was enriched for cell differentiation‐ and tissue morphogenesis‐related pathways (Figure 5E, Table S20). Notably, typical marker genes for maturity and immaturity of GCs were included in separate modules: Tdo2, Dsp, and Calb1 (markers of mature GCs6) were included in the module DG‐brown (Figure 5B), and immature marker doublecortin (Dcx)32 was in the module DG‐red (Figure 5C; Figure S4, Table S18). Drd1a, its increase in expression is characteristic of pseudo‐immature DG phenotype8 and is implicated to have critical roles for the actions of chronic FLX treatment,33 was also included in the module DG‐red. Supporting this, gene expression patterns in the module DG‐brown were highly associated with FLX‐induced decrease in expression of maturity marker genes in the DG (Figure 5F). Module DG‐red genes were associated with increased expression of immaturity marker genes (Figure 5F). These results suggest that expression of maturity and immaturity marker genes may be independently regulated in the hippocampal DG under chronic FLX treatment.
Figure 5.
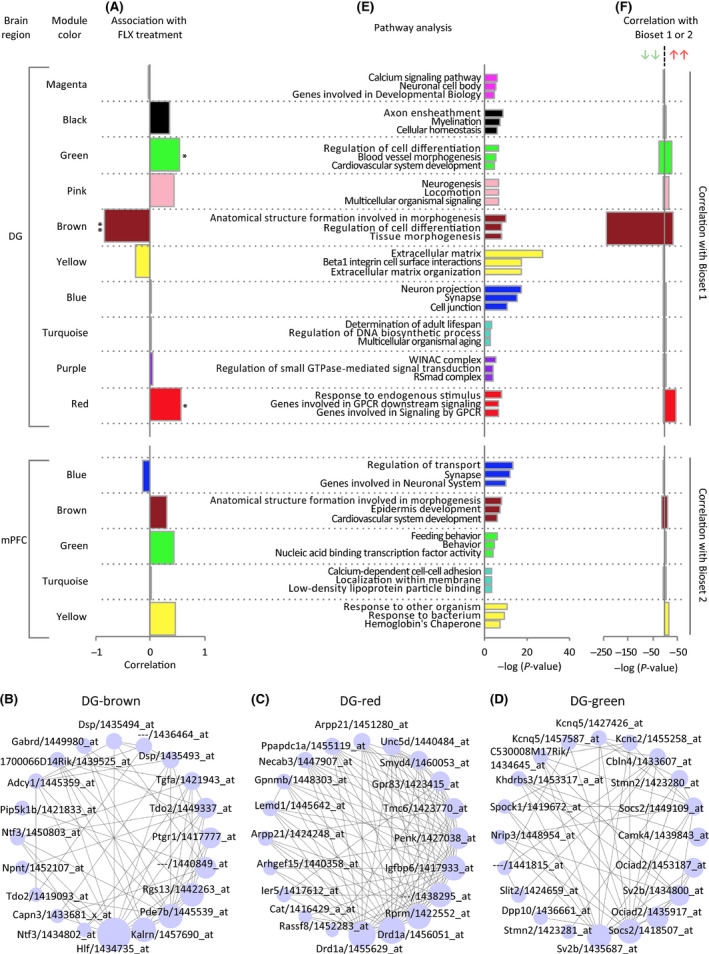
Network analysis identifies modules of co‐expressed genes associated with FLX‐induced dematuration. A, Correlation of each module eigengene with FLX treatment. B‐D, Networks of the connected transcripts in the brown (B), red (C), and green modules (D) for the dataset DG. The networks are visualized with the “Degree sorted circle layout” of Cytoscape. Each node is one transcript, represented by the gene name and Affymetrix ProbeSet ID corresponding to the transcript. Circles are sized by the within module connectivity. Each edge represents the topological overlap or interconnectedness between two nodes. The top 30 hub genes with high connectivity in the module are shown. E, Gene Ontology enrichment analysis (top three pathways are shown for each module). F, Comparisons of expression patterns of the modules with global markers for maturity or immaturity marker genes (Bioset 1 and 2 defined in Figure 1C,F, respectively)
4. DISCUSSION
This study demonstrated that the hippocampal DG and mPFC of FLX‐treated adult mice resembles that of naive infant mice with respect to genome‐wide gene expression patterns. We provide evidence supporting the notion that mature neurons, that is, GCs in the hippocampus and FS neurons in the mPFC, become more immature with FLX treatment.6, 7, 18 In addition to these cell types, the present study highlighted potential contributions of astrocytes and oligodendrocytes to dematuration‐related gene expression changes induced by chronic FLX treatment in the adult hippocampus and mPFC. Furthermore, the hypothesis‐free approach characterizing FLX‐induced gene expression alterations revealed that FLX treatment is associated with maturation abnormalities, especially in the hippocampus.
Based on typical maturation‐related molecular/electrophysiological cellular markers, it has been suggested that chronic FLX may reverse maturation states of preexisting GCs in the hippocampal DG6 and PV‐positive FS neurons in the hippocampus and mPFC7 in adult mice to those found in approximately 10‐day‐old infant mice. Our results are fairly consistent with previous findings, indicating that gene expression patterns of FLX‐treated adult mice were more similar to those of 1‐ to 3‐week‐old hippocampus and 2‐week‐old mPFC than those of older mice. In the mouse hippocampus, 1 and 2 weeks of age corresponds to the peak period for neuronal differentiation and synapse formation.34, 35 Astrocytes in the hippocampus exhibit immature morphological properties during this period.36 In the mouse PFC, 2 weeks of age is considered the middle of maturation of FS neurons and oligodendrocytes, based on the expression of conventional genes for maturation of these cell types that is drastically increased between 1 and 3 weeks of age.37 Considering these findings, our results suggest that chronic FLX treatment may induce dematuration of astrocytes, oligodendrocytes, GCs, and FS neurons in the adult hippocampus and mPFC. This may reopen developmental or juvenile‐like states in these cell types as previously proposed for FS neurons in the visual cortex and lateral amygdala.10, 11, 38
Striking similarities in gene expression patterns were observed in mice chronically treated with FLX, several rodent models of neuropsychiatric disorders, and naive infant mice, especially in the hippocampus. GCs in the hippocampal DG of rodent models of these diseases commonly exhibit molecular, morphological, and/or electrophysiological features that are similar to those of naive infant mice.8 Indeed, the existence of shared endophenotypes related to immaturity of the hippocampus was confirmed at a transcriptome level in this study. In contrast, gene expression patterns in the PFC of these models were not necessarily similar to each other. Behaviorally, these rodent models of disease exhibit a decrease in depression‐like behaviors as measured by forced swim test,14, 25, 29, 39, 40 consistent with the findings found in FLX‐treated mice.41 Such shared and distinct endophenotypes in the brain, and their unique combinations may underlie mechanisms of common and specific behavioral abnormalities in rodent models of these disorders.8
We confirmed transcriptomic dematuration induced by chronic FLX treatment especially in the hippocampal DG with a hypothesis‐free approach using WGCNA. The WGCNA identified gene modules associated with FLX treatment in the DG, with the most relevant module being cell/tissue maturation‐related. Importantly, the module most relevant to FLX reflected a decrease in the maturity of GCs. Previous WGCNA studies using whole hippocampal samples also identified gene modules, one of which was downregulated by chronic FLX treatment (as well as by voluntary exercise, another antidepressant intervention) and was enriched for genes involved in neuronal differentiation and maturation.42 More than half of the top 10 genes significantly correlated with FLX treatment in the module were maturity marker genes of the DG, whose expression was increased in adult mice compared to 2‐week‐old infants (ie, Igfbp2, Mkl2, Hap1, Smarca2, Ntng1, and Rasgrf2)42 (GSE42778, GSE113727). This module may reflect decreased maturity of the hippocampus induced by chronic FLX treatment, consistent with our findings.
In conclusion, the results of bioinformatics analyses of genome‐wide gene expression patterns support the dematuration hypothesis of FLX in the hippocampal DG and the mPFC. Further studies are needed to investigate whether the dematuration effects of FLX in these brain regions mediate its therapeutic actions.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA REPOSITORY
The microarray data obtained in this study were deposited in the GEO database. Details are shown in Table S1.
APPROVAL OF THE RESEARCH PROTOCOL BY AN INSTITUTIONAL REVIEWER BOARD
n/a.
INFORMED CONSENT
n/a.
REGISTRY AND THE REGISTRATION NO. OF THE STUDY/TRIAL
n/a.
ANIMAL STUDIES
All animal experiments were approved by the Institutional Animal Care and Use Committee of Fujita Health University.
Supporting information
ACKNOWLEDGMENTS
We thank the members of our laboratory for their support during the course of these studies, especially to Rika Takeuchi for her technical support in microarray experiments. This work was supported by JSPS Grant‐in‐Aid for Scientific Research on Innovative Areas Grant Number 25116526, 15H01297, and 16H06462, JSPS KAKENHI Grant Number 25242078, and AMED Strategic Research Program for Brain Sciences Grant Number 579.
Hagihara H, Ohira K, Miyakawa T. Transcriptomic evidence for immaturity induced by antidepressant fluoxetine in the hippocampus and prefrontal cortex. Neuropsychopharmacol Rep. 2019;39:78–89. 10.1002/npr2.12048
REFERENCES
- 1. Moore TJ, Mattison DR. Adult utilization of psychiatric drugs and differences by sex, age, and race. JAMA Intern Med. 2017;177(2):274–5. [DOI] [PubMed] [Google Scholar]
- 2. Malberg JE, Eisch AJ, Nestler EJ, Duman RS. Chronic antidepressant treatment increases neurogenesis in adult rat hippocampus. J Neurosci. 2000;20(24):9104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Santarelli L, Saxe M, Gross C, et al. Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science. 2003;301(5634):805–9. [DOI] [PubMed] [Google Scholar]
- 4. Holick KA, Lee DC, Hen R, Dulawa SC. Behavioral effects of chronic fluoxetine in BALB/cJ mice do not require adult hippocampal neurogenesis or the serotonin 1a receptor. Neuropsychopharmacology. 2007;33(2):406–17. [DOI] [PubMed] [Google Scholar]
- 5. Huang G‐J, Bannerman D, Flint J. Chronic fluoxetine treatment alters behavior, but not adult hippocampal neurogenesis, in BALB/cJ mice. Mol Psychiatry. 2008;13(2):119–21. [DOI] [PubMed] [Google Scholar]
- 6. Kobayashi K, Ikeda Y, Sakai A, et al. Reversal of hippocampal neuronal maturation by serotonergic antidepressants. Proc Natl Acad Sci USA. 2010;107(108):8434–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ohira K, Takeuchi R, Iwanaga T, Miyakawa T. Chronic fluoxetine treatment reduces parvalbumin expression and perineuronal netsin gamma‐aminobutyric acidergic interneurons of the frontal cortex in adult mice. Mol Brain. 2013;6:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hagihara H, Takao K, Walton NM, Matsumoto M, Miyakawa T. Immature dentate gyrus: an endophenotype of neuropsychiatric disorders. Neural Plast. 2013;2013:e318596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kobayashi K, Ikeda Y, Suzuki H. Behavioral destabilization induced by the selective serotonin reuptake inhibitor fluoxetine. Mol Brain. 2011;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karpova NN, Pickenhagen A, Lindholm J, et al. Fear erasure in mice requires synergy between antidepressant drugs and extinction training. Science. 2011;334(6063):1731–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vetencourt JFM, Sale A, Viegi A, et al. The antidepressant fluoxetine restores plasticity in the adult visual cortex. Science. 2008;320(5874):385–8. [DOI] [PubMed] [Google Scholar]
- 12. Hagihara H, Ohira K, Takao K, Miyakawa T. Transcriptomic evidence for immaturity of the prefrontal cortex in patients with schizophrenia. Mol Brain. 2014;7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murano T, Koshimizu H, Hagihara H, Miyakawa T. Transcriptomic immaturity of the hippocampus and prefrontal cortex in patients with alcoholism. Sci Rep. 2017;7:44531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takao K, Kobayashi K, Hagihara H, et al. Deficiency of Schnurri‐2, an MHC enhancer binding protein, induces mild chronic inflammation in the brain and confers molecular, neuronal, and behavioral phenotypes related to schizophrenia. Neuropsychopharmacology. 2013;38(8):1409–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci USA. 2015;112(4):1167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murano T, Hagihara H, Tajinda K, Matsumoto M, Miyakawa T. Transcriptomic immaturity inducible by neural hyperexcitation is shared by multiple neuropsychiartic disorders. Commun Biol. 2019;2:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ohira K, Miyakawa T. Chronic treatment with fluoxetine for more than 6 weeks decreases neurogenesis in the subventricular zone of adult mice. Mol Brain. 2011;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dulawa SC, Holick KA, Gundersen B, Hen R. Effects of chronic fluoxetine in animal models of anxiety and depression. Neuropsychopharmacology. 2004;29(7):1321–30. [DOI] [PubMed] [Google Scholar]
- 20. Hagihara H, Toyama K, Yamasaki N, Miyakawa T. Dissection of hippocampal dentate gyrus from adult mouse. J Vis Exp. 2009;33:1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kupershmidt I, Su QJ, Grewal A, et al. Ontology‐based meta‐analysis of global collections of high‐throughput public data. PLoS One. 2010;5(9):e13066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Semeralul MO, Boutros PC, Likhodi O, Okey AB, Van Tol HHM, Wong AHC. Microarray analysis of the developing cortex. J Neurobiol. 2006;66(14):1646–58. [DOI] [PubMed] [Google Scholar]
- 23. Okaty BW, Miller MN, Sugino K, Hempel CM, Nelson SB. Transcriptional and electrophysiological maturation of neocortical fast‐spiking GABAergic interneurons. J Neurosci. 2009;29(21):7040–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cahoy JD, Emery B, Kaushai A, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohira K, Kobayashi K, Toyama K, et al. Synaptosomal‐associated protein 25 mutation induces immaturity of the dentate granule cells of adult mice. Mol Brain. 2013;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dingledine R, Coulter DA, Fritsch B, et al. Transcriptional profile of hippocampal dentate granule cells in four rat epilepsy models. Sci Data. 2017;4:170061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Akahoshi N, Murashima YL, Himi T, Ishizaki Y, Ishii I. Increased expression of the lysosomal protease cathepsin S in hippocampal microglia following kainate‐induced seizures. Neurosci Lett. 2007;429(2):136–41. [DOI] [PubMed] [Google Scholar]
- 28. Miller BH, Schultz LE, Gulati A, Cameron MD, Pletcher MT. Genetic regulation of behavioral and neuronal responses to fluoxetine. Neuropsychopharmacology. 2007;33(6):1312–22. [DOI] [PubMed] [Google Scholar]
- 29. Yamasaki N, Maekawa M, Kobayashi K, et al. Alpha‐CaMKII deficiency causes immature dentate gyrus, a novel candidate endophenotype of psychiatric disorders. Mol Brain. 2008;1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hagihara H, Horikawa T, Nakamura HK, et al. Circadian gene circuitry predicts hyperactive behavior in a mood disorder mouse model. Cell Rep. 2016;14(12):2784–96. [DOI] [PubMed] [Google Scholar]
- 31. Shin R, Kobayashi K, Hagihara H, et al. The immature dentate gyrus represents a shared phenotype of mouse models of epilepsy and psychiatric disease. Bipolar Disord. 2013;15(4):405–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brown JP, Couillard‐Després S, Cooper‐Kuhn CM, Winkler J, Aigner L, Kuhn HG. Transient expression of doublecortin during adult neurogenesis. J Comp Neurol. 2003;467(1):1–10. [DOI] [PubMed] [Google Scholar]
- 33. Shuto T, Kuroiwa M, Sotogaku N, et al. Obligatory roles of dopamine D1 receptors in the dentate gyrus in antidepressant actions of a selective serotonin reuptake inhibitor, fluoxetine. Mol Psychiatry. 2018; 10.1038/s41380-018-0316-x Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mody M, Cao Y, Cui Z, et al. Genome‐wide gene expression profiles of the developing mouse hippocampus. Proc Natl Acad Sci USA. 2001;98(15):8862–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deguchi Y, Donato F, Galimberti I, Cabuy E, Caroni P. Temporally matched subpopulations of selectively interconnected principal neurons in the hippocampus. Nat Neurosci. 2011;14(4):495–504. [DOI] [PubMed] [Google Scholar]
- 36. Bushong EA, Martone ME, Ellisman MH. Maturation of astrocyte morphology and the establishment of astrocyte domains during postnatal hippocampal development. Int J Dev Neurosci. 2004;22(2):73–86. [DOI] [PubMed] [Google Scholar]
- 37. Ueda S, Niwa M, Hioki H, et al. Sequence of molecular events during the maturation of the developing mouse prefrontal cortex. Mol Neuropsychiatry. 2015;1(2):94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Umemori J, Winkel F, Didio G, Pou ML, Castrén E. iPlasticity: induced juvenile‐like plasticity in the adult brain as a mechanism of antidepressants. Psychiatry Clin Neurosci. 2018;72(9):633–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oliveira CV, Grigoletto J, Funck VR, et al. Evaluation of potential gender‐related differences in behavioral and cognitive alterations following pilocarpine‐induced status epilepticus in C57BL/6 mice. Physiol Behav. 2015;143:142–50. [DOI] [PubMed] [Google Scholar]
- 40. dos Santos JG, Longo BM, Blanco MM, Menezes de Oliveira MG, Mello LE. Behavioral changes resulting from the administration of cycloheximide in the pilocarpine model of epilepsy. Brain Res. 2005;1066(1):37–48. [DOI] [PubMed] [Google Scholar]
- 41. Cryan JF, Page ME, Lucki I. Differential behavioral effects of the antidepressants reboxetine, fluoxetine, and moclobemide in a modified forced swim test following chronic treatment. Psychopharmacology. 2005;182(3):335–44. [DOI] [PubMed] [Google Scholar]
- 42. Huang G‐J, Ben‐David E, Piella AT, Edwards A, Flint J, Shifman S. Neurogenomic evidence for a shared mechanism of the antidepressant effects of exercise and chronic fluoxetine in mice. PLoS One. 2012;7(4):e35901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


