Abstract
Background and purpose
Machine learning (ML) has attracted much attention with the hope that it could make use of large, routinely collected datasets and deliver accurate personalised prognosis. The aim of this systematic review is to identify and critically appraise the reporting and developing of ML models for predicting outcomes after stroke.
Methods
We searched PubMed and Web of Science from 1990 to March 2019, using previously published search filters for stroke, ML, and prediction models. We focused on structured clinical data, excluding image and text analysis. This review was registered with PROSPERO (CRD42019127154).
Results
Eighteen studies were eligible for inclusion. Most studies reported less than half of the terms in the reporting quality checklist. The most frequently predicted stroke outcomes were mortality (7 studies) and functional outcome (5 studies). The most commonly used ML methods were random forests (9 studies), support vector machines (8 studies), decision trees (6 studies), and neural networks (6 studies). The median sample size was 475 (range 70–3184), with a median of 22 predictors (range 4–152) considered. All studies evaluated discrimination with thirteen using area under the ROC curve whilst calibration was assessed in three. Two studies performed external validation. None described the final model sufficiently well to reproduce it.
Conclusions
The use of ML for predicting stroke outcomes is increasing. However, few met basic reporting standards for clinical prediction tools and none made their models available in a way which could be used or evaluated. Major improvements in ML study conduct and reporting are needed before it can meaningfully be considered for practice.
Introduction
Stroke is the second leading cause of mortality and disability adjusted life years in the world [1,2]. Both the outcomes and presentation of stroke can be extremely varied and timely assessment is essential for optimal management. The complexity of a condition such as stroke potentially lends itself well to the use of ML methods which are able to incorporate a large variety of variables and observations into one predictive framework without the need for preprogrammed rules. There has been increasing interest in the use of ML to predict stroke outcomes, with the hope that such methods could make use of large, routinely collected datasets and deliver accurate personalised prognoses.
While papers applying ML methods to stroke are published regularly, the main focus of these has been on stroke imaging application [3–5]. As far as we are aware, there have been no reviews of studies which have developed ML models to predict stroke outcomes from structured data specifically. The goal of the review was to identify gaps in the literature, critically appraise the reporting and methods of the algorithms and provide the foundation for a wider research program focused on developing novel machine learning based predictive algorithms in stroke care.
Methods
This is a systematic review which was registered with the international prospective register of systematic reviews (PROSPERO) (CRD42019127154): a database of systematic review protocols, maintained by the Centre for Reviews and Dissemination at the University of York. The PRISMA [6] statement was followed as a reporting guideline. Risk of bias and quality of the studies were not assessed because the objective of this paper was to be descriptive, not to draw conclusions about the validity of estimates of predictive accuracy from the included studies. The reporting quality was assessed according to TRIPOD [7] with a few terms adjusted to fit ML methods (See Table 1 for explanations of ML terms).
Table 1. Notations of special machine learning terms.
| Term | Explanation |
|---|---|
| Supervised learning | A subgroup of ML models that requires both predictors and outcomes (labels) |
| Unsupervised learning | A subgroup of ML models meant to find previously unknown patterns in data without pre-existing labels |
| Feature | Predictor or variable in a ML model |
| Feature selection | Variable selection or attribute selection |
| Generalisation ability | The ability of a model to generalise the learned pattern to new data |
| Over-fitting | A model corresponds too closely or exactly to a particular set of data, and may fail to fit new data |
| Missing data mechanism | Three missing-data mechanisms: missing completely at random (MCAR), missing at random (MAR), and missing not at random (MNAR) |
| Imputation | The process of replacing missing data with substituted values |
| Training | The learning process of the data pattern by a model |
| Testing | A validation set used for testing the model |
| LASSO | Least Absolute Shrinkage and Selection Operator: a regression technique that performs both variable selection and regularization |
| Support Vector Machine (SVM) | A supervised classifier that seeks to find the best hyperplane to separate the data |
| Naïve Bayes (NB) | A family of simple "probabilistic classifiers" based on applying Bayes' theorem with strong (naïve) independence assumptions between the features |
| Bayesian Network (BN) | A type of probabilistic graphical model that uses Bayesian inference for probability computations |
| k-nearest neighbours (kNN) | A type of instance-based learning, where the predictionis only approximated locally with the k nearest neighbours |
| Artificial Neural Network (ANN) | A computational model based on a collection of connected units or nodes called artificial neurons, which loosely model the neurons in a biological brain |
| Decision Tree | A tree with a set of hierarchical decisions which eventually gives a final decision |
| Random Forest (RF) | An ensemble learning method that uses a multitude of decision trees |
| Super learner | A stacking algorithm using cross-validated predictions of other models and assigning weights to these predictions to optimise the final prediction |
| Adaptive network based fuzzy inference system (ANFIS) | A fuzzy Sugeno model put in the framework of adaptive systems to facilitate learning and adaptation |
| Xgboost | A decision-tree-based ensemble ML algorithm that uses a gradient boosting framework |
| Adaptive Boosting, (Adaboost) | An algorithm used in combination with others to convert a set of weak classifiers into a strong one |
| Parameters | Coefficients of a model that need to be learned from the data |
| Hyperparameters | Configurations of a model which are often selected and set before training the model |
| Validation | The process of a trained model evaluated with a testing dataset |
| Discrimination | The ability of a model to separate individual observations in multiple classes |
| Calibration | Adjusting the predicted probability from the model to more closely match the observed probability in the test set |
| Cross-validation (CV) | A model validation technique for assessing how the results of a statistical analysis (model) will generalize to an independent data set |
| Leave One Out CV | A performance measurement approach that uses one observation as the validation set and the remaining observations as the training set |
| Leave One Centre Out CV | A performance measurement approach that uses observations from one centre as the validation set and the remaining observations as the training set |
| Bootstrapping | Resampling multiple new datasets with replacement from the original data set |
Search strategy
We searched PubMed and Web of Science for studies on prediction models for stroke outcomes using ML, published in English between 1990 and March 2019. We combined published PubMed search filters for stroke [8], ML [9], and prediction models [10]. To ensure consistency in the searches in both databases, these PubMed filters were translated to Web of Science together with the support of a librarian. We verified the search strategy (S1 Text) with a validation set of seven publications identified manually by the researchers across PubMed and Web of Science and the results of our database queries included all the seven papers in this validation set.
Study selection
We assessed the eligibility of the studies returned by the searches through a two-stage screening process. We first screened the titles and abstracts of all articles. Two authors (WW and MK) independently screened 50% of articles each and a random sample of 10% in duplicate. Any disagreement was solved through discussion, involving a third author (BB) if necessary. For all studies deemed relevant, the full text was reviewed using the same screening procedure as in the first stage.
Studies were eligible if they adhered to the following inclusion criteria:
Focusing on predicting clinical outcomes of stroke, excluding studies predicting the occurrence of stroke
Using structured patient level health data (electronic health records, insurance claims data, registries, cohort studies data, or clinical trials data), excluding studies using text or imaging data
Primary research only, excluding reviews
Complete paper available rather than just an abstract or notes
Reporting quality assessment
Reporting guidelines for ML as prediction models are currently not available. TRIPOD was followed as a reporting standard which was originally developed for regression modelling. As mentioned in TRIPOD’s documentation, most terms apply equally to ML methods developed, validated, or updated as prediction tools. We adopted most terms for reporting of methods and results in TRIPOD with two terms adjusted specifically for ML (S1 Table). Reporting of hyperparameter selection if needed was added to 10b (Specify type of model, all model building procedures) and 15a (Present the full prediction model to allow predictions for individuals) was adjusted for the specification of ML models (links to the final model online, coding of predictors, code, final parameters/coefficients, and with the architecture described in full in the article).
Data extraction
An structured data collection form was developed to aid extraction of items related to: general study characteristics (authors, publication year, type, venue, country under study population, study objective); study population (source of data, single or multi-centre, sample size, features, feature size); data pre-processing methods (handling missing data and unbalanced outcomes, other data pre-processing steps); clinical outcomes; analytical methods (statistical models, ML models, feature selection methods, validation methods, performance measurements); results (feature importance, best performing model) (S2 Table).
Data for all papers were extracted by two authors (WW and MK), with discrepancies resolved by consensus through discussion between and with another author (BB) if necessary.
Results
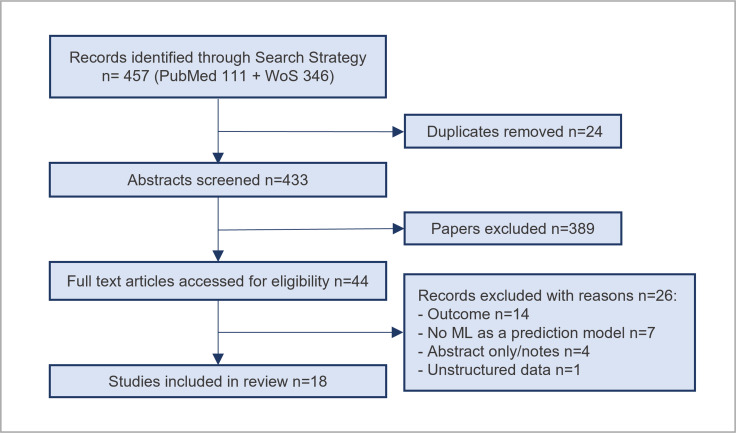
We identified 111 studies from PubMed and 346 studies from Web of Science. After the removal of duplicates, as well as abstract and title screening, 44 studies were considered potentially relevant. After full article screening, 18 studies were identified for information extraction (Fig 1).
Fig 1. PRISMA flowchart.
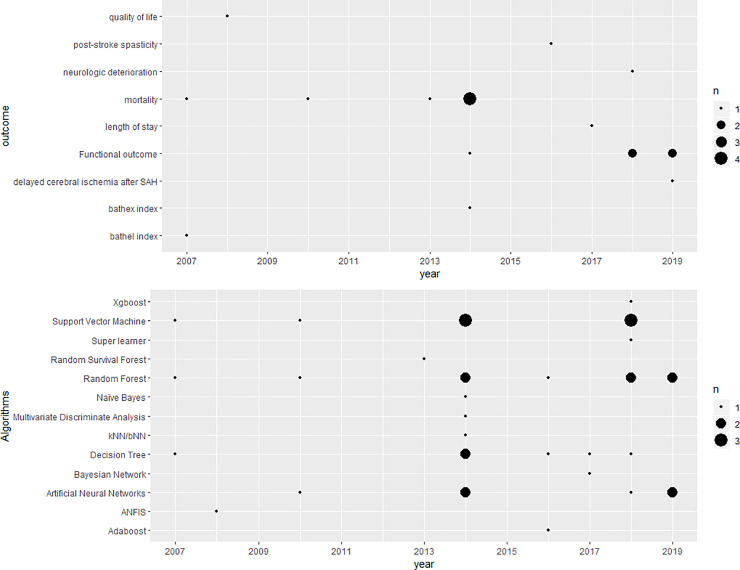
Almost all studies (17) were published as peer reviewed publications in biostatistical or clinical journals. All included studies were published after 2007, with almost half (8) published after 2016, and 3 studies were published in 2018 [11–13] and 2019 [14–16] each (Fig 2). In terms of regions under study, UK (3) [17–19], Germany (2) [20,21], Turkey (2) [22,23] and China (2) [13,14] make up half of the sample. Saudi Arabia [24], Australia [25], Korea [15], USA [26], Denmark [27], Netherlands [11], Portugal [12], Taiwan [28] and Japan [16] had one study each. Single centre studies (10) were slightly more common than multi-centre studies (8). For sources of data, half of the studies (9) used registry data while the rest used EHR (4) [13,22,24,28], cohort (3) [14,15,27], and clinical trial data (2) [11,18]. All the included studies focused on developing new models using ML whilst no study validated existing ML based predictive models on independent data. Most of the studies used only variables collected at admission (Table 2) though three studies [11,12,16] explored model performance with information available at different time points.
Fig 2. Number of papers published according to the algorithms used (top) and outcomes (bottom) predicted at each year.
Table 2. A brief summary of the included studies.
| Reference | Sample (Feature) size | Outcomes | Predictors/variables/features | Missing values handled | Hyperparameter selection | Validation | Calibration | Best Algorithm | Compared algorithms |
|---|---|---|---|---|---|---|---|---|---|
| Al Taleb et al. 2017 | 358 (15) | Length of Stay | At admission | Single imputation | Not reported | 10-fold CV | No | Bayesian Network | DT (C4.5) |
| Asadi et al. 2014 | 107 (8) | 90-day binary and 7 scale mRS | At admission | Not reported | No | Training, test, validation for ANN, Nested CV for SVM | No | SVM | ANN, Linear Regression |
| Liang et al. 2019 | 435 (4) | 90-day binary mRS | Admission, laboratory data | Not reported | Not reported | Training and test split | No | ANN | LR |
| Heo et al. 2019 | 2604 (38) | 90-day binary mRS | Admission | Complete case analysis | No | Training and test split | No | DNN | RF, LR |
| Konig et al. 2007 | 3184 (43) | 100-day Bathel Index | first 72h after admission | Complete case analysis | Yes, Grid search | Temporal and external validation | Yes for LR | - | RF, SVM, LR |
| Celik et al. 2014 | 570 (22) | 10-day mortality | At admission | - | Yes, grid search | 5-fold CV | No | LR | ANN |
| Ho et al. 2014 | 190 (26) | Discharge mortality | Admission and interventions | Complete case analysis | Not reported | 10-fold CV | No | SVM | Naïve Bayes, DT, RF, PCA+SVM, LR |
| Cox et al. 2016 | 2580 (72) | Post stroke spasticity | Not clear | Not reported | Not reported | Training, test and validation split | No | RF | DT (CART), Adaboost |
| Kruppa et al. 2014 | 3184 (43) | 100-day Bathel Index | First 72h after admission data | Complete case analysis | Yes, For KNN, bNN and RF | Temporal and external validation | Yes, Brier score | SVM and LR | K-NN, b-NN, RF |
| Easton et al. 2014 | 933 (-) | Short/very short mortality | Not clear | Not reported | Yes, DT is pruned | Training and test split | No | - | Naïve Bayes, DT, LR |
| Mogensen and Gerds 2013 | 516 (12) | 3-month/1-year/3-year/5-year mortality | Admissiondata | Complete case analysis | No, manually set up | Bootstrap CV | Yes, Brier score | - | Pseudo RF, Cox Regression, and Random survival forest |
| Van Os et al. 2018 | 1383 (83) | Good reperfusion socre, 3-month binary mRS | Admission, laboratory and treatment data | Multiple imputation by chained equations | Yes, nested CV with random grid search | Nested CV | No | - | RF, SVM, ANN, super learner, LR |
| Peng et al. 2010 | 423 (10) | 30-day mortality | Admission, laboratory, radiographic data | No missing values | Yes, empirically | 4-fold CV | No | RF | ANN, SVM, LR |
| Tokmakci et al. 2008 | 70 (6) | Quality of life | Admissiondata | Not reported | Not reported | Training and test split | No | ANFIS | |
| Monteiro et al. 2018 | 425 (152) | 3-month binary mRS | Admission/2 hours/24 hours/7 days data | single imputation | Yes, Grid search | 10-fold CV | No | RF and Xgboost | DT, SVM, RF, LR (LASSO) |
| Tjortjis et al. 2007 | 671 (37) | 2-month mortality | Admission data | Cases discarded with missing outcomes | Yes, pruned | Training and test split | No | DT (T3) | DT (C4.5) |
| Lin et al. 2018 | 382 (5) | Neurologic deterioration | Admission and laboratory data | Not reported | Yes, CV | Training and test split | No | - | SVM |
| Tanioka et al. 2019 | 95 (20) | Delayed cerebral ischemic after SAH | Admission/1-3 days variables | Complete case analysis | Yes, Grid search | Leave one out CV | No | - | RF |
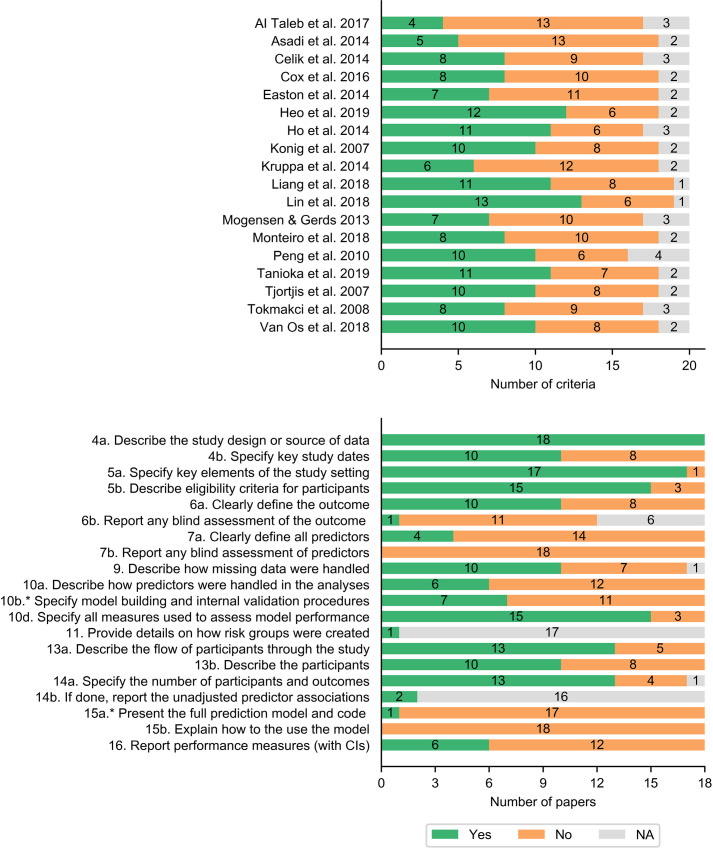
Twenty terms were assessed for each study, including thirteen terms for methods and seven terms for results. Half of the studies reported less than half of the terms in the checklist (excluding NA) and the other half of the studies reported less than around two thirds of the terms (Fig 3). The study design and source of data (4a), study setting (5a), eligibility criteria for participants (5b), measures to assess the model (10d), flow of participants (13a), number of participants and outcomes (14a) were relatively better reported (with more than 13 studies) (Fig 3). Blind assessment of outcome and predictors (6b and 7b), presentation of the full model (15a), and explanation on how to use the model (15b) were not reported in almost any of the studies. Definition of all predictors (7a) and description of how predictors were handled (10a) were reported in four and six studies respectively. Performance measures with confidence intervals (CI) (16) were only reported in 6 studies.
Fig 3.
Number of terms reported in each study (top) and number of studies reported for each assessment term (bottom). * indicates criteria adjusted for ML models.
Mortality (7) was the most frequently predicted clinical outcome. Studies focused on mortality at different time points during follow-up, including short term (10 days [22], 30 days [28], 2 months [19], 3 months [12,27], 100 days [20]) and long term (1/3/5 years) [27]. One study [26] predicted discharge mortality (modified Rankin Score (mRS) = 6). The ML algorithms used for mortality prediction were ANN [22,28], Naïve Bayes [18,26], SVM [26,28], DT [18,19,26], and RF [26–28]. Functional outcome (measure of functional independence which relates to an individual's physical, psychological and social functioning, and the extent to which the depend on assistance from others to fulfill activities of daily living. It is usually measured by mRS) (5) was the second most commonly predicted clinical outcome. Three studies [14,15,25] predicted functional outcome (such as the ability to carry out activities of daily living e.g. washing and dressing) at 90 days, two studies [11,12] predicted it at 3 months. All of those studies used dichotomised mRS (mRS > 2 vs mRS < = 2) whilst Asadi et al. [25] also predicted 7-scale mRS (0–6). The ML algorithms used for predicting functional outcome were ANN [11,14,15,25], SVM [11,12,25], DT [12], RF [11,12,15], Super Learner [11], and Xgboost [12]. Other than mortality and mRS, Barthel Index [20,21] used RF, SVM, and kNN, hospital length of stay [18] used DT and Bayesian Network, post-stroke spasticity [25] used RF, DT, and Adaboost, neurologic deterioration [13] used SVM, quality of life [23] used ANFIS, and delayed cerebral ischemia after aneurysmal subarachnoid haemorrhage (SAH) [16] used RF were also predicted as stroke outcomes.
Among the included studies, ten studies reported having missing values, one study [28] reported no missing values, and seven studies did not mention missing values. In terms of imputation methods, complete case analysis (6) was the most commonly used among the ten studies that included information on how missing data were handled. Other imputation methods included single imputation (2) [12,24] and multiple imputation (1) [11]. For dealing with imbalanced data distributions, three studies [16,22,26] reported addressing it, of which two studies [16,26] used Synthetic Minority Over-sampling Technique (SMOTE) [16] and one study [26] did not report the method used. Four studies [12,15,16,27] did not report performing feature selection and fourteen studies reported that the features were selected before applying their algorithms.
The most commonly used ML methods were RF (9), SVM (8), DT (6), and ANN (6). The following algorithms were each used in one study: kNN [20], NB [18], BN [24], boosting [12,17], Super learner [11], and ANFIS [23]. Details of ML models is shown in S2 Text. There were fourteen models used across the studies as comparators, including logistic regression (10), Cox regression (1) [27], linear regression (1) [25], random survival forest (1) [27], and multivariate discriminant analysis (MDA) (1) [22].
For hyperparameter selection (Table 2), five studies [14,17,23,24,26] did not mention the method or rationale for hyperparameter choice, three studies [15,25,27] subjectively set a value for the hyperparameters, and ten studies performed hyperparameter tuning using the development data. Among these ten studies, grid search (5) [11,12,16,21,22] was the most widely used tuning method. Four studies [18–20,28] reported tuning hyperparameters empirically without a specific method. One study [13] used CV on the training set.
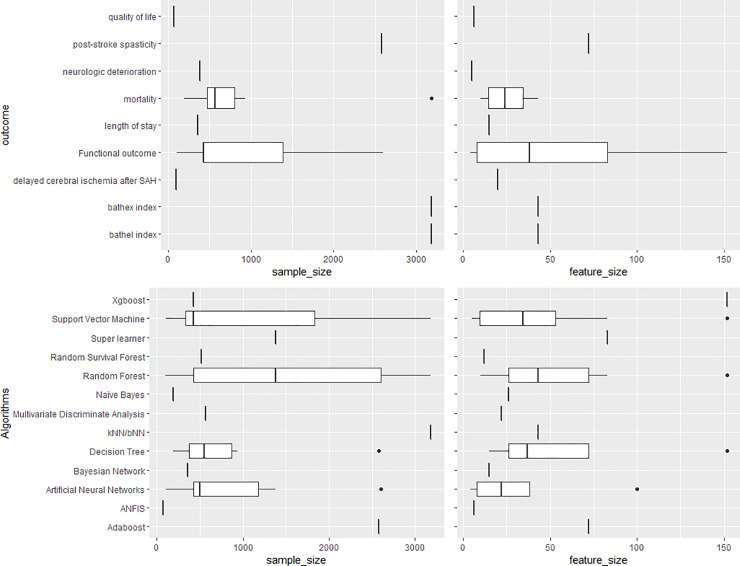
There was no apparent relationship between the algorithms used and the sample size or number of features (Fig 4). Only one dataset had a sample size bigger than 3000 patients and was used by two studies [20,21]. The median sample size was 475 and the smallest was 70. The median number of features was 22 [range: 4–152].
Fig 4. Boxplots showing the distribution of sample size and feature size according to algorithms used (top) and outcomes predicted (bottom).
Twelve studies compared the performance of regression models with ML algorithms (Table 2), of which six studies [12,14,15,25,26,28] reported that ML models outperformed the compared regression models and five studies [11,18,20,21,27] concluded that there was no significant difference between the ML and statistical models. One study [22] reported that LR outperformed ANN. In total, SVM outperformed the comparison algorithms in three studies [20,25,26], ANN outperformed the comparison algorithms in two studies [14,19], RF outperformed the comparisons in two studies [12,28], and LR outperformed competing algorithms in two studies [20,22].
With regards to validation methods (Table 2), CV was the most commonly used method (10) for internal validation. Eight studies split the data into training and test (and/or validation) sets. Only two studies [20,21] used external validation.
For discrimination measures, AUC (13) was the most commonly used among the classification models. Nine studies (9/13) used AUC accompanied by other discrimination measures. Four studies used only AUC. Other commonly used discrimination measures were accuracy (9), sensitivity (8), and specificity (7). Calibration was assessed in three studies [20,21,27]. One study [21] assessed calibration by plotting the observed outcome frequencies against the predicted probabilities. Two studies [20,27] used the Brier score.
Discussion
This is the first systematic review on the application of ML methods using structured data to predict outcomes of stroke. Our results show that the interest in using ML to predict stroke outcomes using structured data has markedly increased in recent years: almost all studies in this review were published since 2014. The data sizes used in many included studies are relatively small to fully explore the potential of ML methods. Only one dataset had a sample size of over 3000 patients with a feature size of 43.
For handling missing values, almost all of the studies used only relatively simple methods such as complete case analysis and single imputation. Only one study [11] used multiple imputation. Previous studies have shown that more complicated imputation methods such as multiple imputation [29,30] are better at restoring the natural variability of the missing values than single imputation and retain more useful information than complete case analysis [31]. Future studies in the application of ML methods to stroke outcome prediction would benefit from using more sophisticated imputation methods to handle missing values.
The reporting and conducting of hyperparameter selection in the studies were often neglected though the choice of hyperparameters can greatly impact the model’s performance [32,33]. To the best of our knowledge, there exist no guidelines on reporting the hyperparameter tuning result/procedure for ML as clinical prediction models.
The most commonly used ML methods were RF, SVM, ANN, and DT. In this review, we did not compare the performance of algorithms across studies due to the different characteristics of each study. SVM performed the best in 3 studies. ANN and RF outperformed the comparison algorithms in 2 studies. Even though DTs were commonly used, they did not outperform other algorithms in the reviewed studies. The performance of ML models compared to regression models was found to be mixed, which is consistent with other ML related systematic reviews [34,35].
Performance evaluation can typically be thought to include discrimination and calibration. All studies reported discrimination whilst only three studies discussed calibration. This is concerning because poor calibration can lead to harmful decisions [36] and reporting both is essential for prediction models [37].
Validation is a crucial step for obtaining a model that can be generalised beyond the sample population. A majority of studies used internal validation methods (training and test split and CV), whilst only two studies used external validation [20,21]. External validation is an invaluable part of implementing the model in routine clinical practice–it assesses the transportability of the predictions to new data (and hence the generalisability of the model) and should be undertaken before clinical use [21,38].
None of the studies reported decision-analytic measures to assess the clinical utility of prediction models [37,39,40]. Also, no study discussed real-life implementation of the model in clinical practice even though the ultimate goal is presumably to assist the clinicians making treatment decisions and estimating prognoses. There are also several reasons why implementing ML models could be challenging in clinical settings. ML algorithms are typically not very transparent in terms of how the prediction has been made and how individual predictors have contributed to the overall prediction. This may limit the acceptability and face validity of the predictions generated by the model for clinical decision makers. In addition, we found that the reporting of the models and model building was not clear enough in most studies to enable the models to be replicated in other datasets or externally validated. This means that the models will have limited evidence of accuracy in different settings or may not be implementable at all in real-world settings.
Thus, guidelines and reporting standards for implementing ML algorithms might improve the utility of studies and future studies would benefit from attempting to evaluate potential impact and clinical utility [41]. Reporting guidelines for developing and validating clinical predictions models [7,40] provide a good starting point at this stage. Potential ethical challenges of implementing ML models was also addressed in recent studies [42]. Making algorithms and the developed models fully and publicly available with transparent and full reporting is imperative to allow independent external validation across various settings and facilitate clinical utility [43].
This systematic review has its strength and limitations. It is the first systematic review that has reviewed not only the reporting quality of the ML studies, but also the development of the ML models. Yet, even though we used published search filters for stroke, prediction models and ML, we might not have found all studies in PubMed and Web of Science, or studies that are not included in these databases and not published in English. For conference proceedings, Web of Science does include proceedings of major international conferences on machine learning such as International Conference on Machine Learning (ICML); European Conference on Machine Learning and Principles and Practices of Knowledge Discovery in Databases (ECMLPKDD); Asian Conference on Machine Learning (ACML); and International Conference on Machine Learning and Machine Intelligence (MLMI). However, there could still be smaller conferences that are not included in Web of Science.
Conclusions
As the first systematic review on current applications of ML methods using structured data to predict outcomes of stroke, we see increasing interest in using ML for predicting stroke outcome. However, despite a surge of research articles, few met basic reporting standards for clinical prediction tools, and none of them made their models available in a way which could be used or evaluated. There is significant scope for improvement in how ML prediction algorithms are developed and validated, including using larger, richer, and more diverse data sources, improvements in model design, and fully reporting on the development process as well as the final model. As a result, it cannot be confidently said whether ML is any better than traditional statistical approaches. Major improvements in ML study conduct and reporting are needed before these methods could be meaningfully considered for practice. Guidelines and reporting standards of implementing ML algorithms could improve the utility of studies in this regard and future studies would benefit from attempting to evaluate potential impact and clinical utility.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
CDW, BB, NP, VC, AGR hold the award from the Health Foundation. Award Number is 553013. CDW, AD, VC, and YW acknowledge support from the National Institute for Health Research (NIHR) Biomedical Research Centre (BRC) based at Guy’s and St Thomas’ National Health Service (NHS) Foundation Trust and King’s College London, and the NIHR Collaboration for Leadership in Applied Health Research and Care (ARC) South London at King’s College Hospital NHS Foundation Trust. NP acknowledges support from the NIHR Manchester BRC. IJM is funded by the Medical Research Council (MRC), through its Skills Development Fellowship program, fellowship MR/N015185/1. The views expressed are those of the authors and not necessarily those of the NHS, the BRC or ARC. No sponsors or funders play any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Johnson CO, Nguyen M, Roth GA, Nichols E, Alam T, Abate D, et al. Global, regional, and national burden of stroke, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. The Lancet Neurology. 2019;18: 439–458. 10.1016/S1474-4422(19)30034-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global Burden of Disease Collaborative Network. Global Burden of Disease Study 2017. Institute for Health Metrics and Evaluation (IHME); 2018. Available: http://ghdx.healthdata.org/gbd-results-tool
- 3.Kamal H, Lopez V, Sheth SA. Machine Learning in Acute Ischemic Stroke Neuroimaging. Front Neurol. 2018;9: 945 10.3389/fneur.2018.00945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng R, Badgeley M, Mocco J, Oermann EK. Deep learning guided stroke management: a review of clinical applications. J NeuroIntervent Surg. 2018;10: 358–362. 10.1136/neurintsurg-2017-013355 [DOI] [PubMed] [Google Scholar]
- 5.Lee E-J, Kim Y-H, Kim N, Kang D-W. Deep into the Brain: Artificial Intelligence in Stroke Imaging. J Stroke. 2017;19: 277–285. 10.5853/jos.2017.02054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. International Journal of Surgery. 2010;8: 336–341. 10.1016/j.ijsu.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 7.Moons KGM, Altman DG, Reitsma JB, Ioannidis JPA, Macaskill P, Steyerberg EW, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): Explanation and Elaboration. Ann Intern Med. 2015;162: W1 10.7326/M14-0698 [DOI] [PubMed] [Google Scholar]
- 8.Flinders University. Stroke Search Filters. Available: https://www.flinders.edu.au/flinders-digital-health-research-centre/flinders-filters/stroke-search-filters
- 9.Senders JT, Staples PC, Karhade AV, Zaki MM, Gormley WB, Broekman MLD, et al. Machine Learning and Neurosurgical Outcome Prediction: A Systematic Review. World Neurosurgery. 2018;109: 476–486.e1. 10.1016/j.wneu.2017.09.149 [DOI] [PubMed] [Google Scholar]
- 10.Geersing G-J, Bouwmeester W, Zuithoff P, Spijker R, Leeflang M, Moons K. Search Filters for Finding Prognostic and Diagnostic Prediction Studies in Medline to Enhance Systematic Reviews. Smalheiser NR, editor. PLoS ONE. 2012;7: e32844 10.1371/journal.pone.0032844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Os HJA, Ramos LA, Hilbert A, van Leeuwen M, van Walderveen MAA, Kruyt ND, et al. Predicting Outcome of Endovascular Treatment for Acute Ischemic Stroke: Potential Value of Machine Learning Algorithms. Front Neurol. 2018;9: 784 10.3389/fneur.2018.00784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monteiro M, Fonseca AC, Freitas AT, Pinho E Melo T, Francisco AP, Ferro JM, et al. Using Machine Learning to Improve the Prediction of Functional Outcome in Ischemic Stroke Patients. IEEE/ACM Trans Comput Biol Bioinform. 2018;15: 1953–1959. 10.1109/TCBB.2018.2811471 [DOI] [PubMed] [Google Scholar]
- 13.Lin J, Jiang A, Ling M, Mo Y, Li M, Zhao J. Prediction of neurologic deterioration based on support vector machine algorithms and serum osmolarity equations. Brain Behav. 2018;8: e01023 10.1002/brb3.1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang Y, Li Q, Chen P, Xu L, Li J. Comparative Study of Back Propagation Artificial Neural Networks and Logistic Regression Model in Predicting Poor Prognosis after Acute Ischemic Stroke. Open Med (Wars). 2019;14: 324–330. 10.1515/med-2019-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heo J, Yoon JG, Park H, Kim YD, Nam HS, Heo JH. Machine Learning-Based Model for Prediction of Outcomes in Acute Stroke. Stroke. 2019;50: 1263–1265. 10.1161/STROKEAHA.118.024293 [DOI] [PubMed] [Google Scholar]
- 16.Tanioka S, Ishida F, Nakano F, Kawakita F, Kanamaru H, Nakatsuka Y, et al. Machine Learning Analysis of Matricellular Proteins and Clinical Variables for Early Prediction of Delayed Cerebral Ischemia After Aneurysmal Subarachnoid Hemorrhage. Mol Neurobiol. 2019. 10.1007/s12035-019-1601-7 [DOI] [PubMed] [Google Scholar]
- 17.Cox AP, Raluy-Callado M, Wang M, Bakheit AM, Moore AP, Dinet J. Predictive analysis for identifying potentially undiagnosed post-stroke spasticity patients in United Kingdom. J Biomed Inform. 2016;60: 328–333. 10.1016/j.jbi.2016.02.012 [DOI] [PubMed] [Google Scholar]
- 18.Easton JF, Stephens CR, Angelova M. Risk factors and prediction of very short term versus short/intermediate term post-stroke mortality: A data mining approach. Computers in Biology and Medicine. 2014;54: 199–210. 10.1016/j.compbiomed.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 19.Tjortjis C, Saraee M, Theodoulidis B, Keane JA. Using T3, an improved decision tree classifier, for mining stroke-related medical data. Methods Inf Med. 2007;46: 523–529. 10.1160/me0317 [DOI] [PubMed] [Google Scholar]
- 20.Kruppa J, Liu Y, Diener H-C, Holste T, Weimar C, Konig IR, et al. Probability estimation with machine learning methods for dichotomous and multicategory outcome: applications. Biom J. 2014;56: 564–583. 10.1002/bimj.201300077 [DOI] [PubMed] [Google Scholar]
- 21.Konig IR, Malley JD, Weimar C, Diener H-C, Ziegler A. Practical experiences on the necessity of external validation. Stat Med. 2007;26: 5499–5511. 10.1002/sim.3069 [DOI] [PubMed] [Google Scholar]
- 22.Celik G, Baykan OK, Kara Y, Tireli H. Predicting 10-day mortality in patients with strokes using neural networks and multivariate statistical methods. J Stroke Cerebrovasc Dis. 2014;23: 1506–1512. 10.1016/j.jstrokecerebrovasdis.2013.12.018 [DOI] [PubMed] [Google Scholar]
- 23.Tokmakci M, Unalan D, Soyuer F, Ozturk A. The reevaluate statistical results of quality of life in patients with cerebrovascular disease using adaptive network-based fuzzy inference system. Expert Systems with Applications. 2008;34: 958–963. 10.1016/j.eswa.2006.10.026 [DOI] [Google Scholar]
- 24.Al Taleb AR, Abul Hasanat MH, Khan MB. Application of Data Mining Techniques to Predict Length of Stay of Stroke Patients. 2017. Available: https://ieeexplore.ieee.org/stamp/stamp.jsp?tp=&arnumber=7899004 [Google Scholar]
- 25.Asadi H, Dowling R, Yan B, Mitchell P. Machine learning for outcome prediction of acute ischemic stroke post intra-arterial therapy. PLoS One. 2014;9: e88225 10.1371/journal.pone.0088225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho KC, Speier W, El-Saden S, Liebeskind DS, Saver JL, Bui AAT, et al. Predicting discharge mortality after acute ischemic stroke using balanced data. AMIA Annu Symp Proc. 2014;2014: 1787–1796. [PMC free article] [PubMed] [Google Scholar]
- 27.Mogensen UB, Gerds TA. A random forest approach for competing risks based on pseudo-values. Statistics in Medicine. 2013;32: 3102–3114. 10.1002/sim.5775 [DOI] [PubMed] [Google Scholar]
- 28.Peng S-Y, Chuang Y-C, Kang T-W, Tseng K-H. Random forest can predict 30-day mortality of spontaneous intracerebral hemorrhage with remarkable discrimination. Eur J Neurol. 2010;17: 945–950. 10.1111/j.1468-1331.2010.02955.x [DOI] [PubMed] [Google Scholar]
- 29.Sterne JAC, White IR, Carlin JB, Spratt M, Royston P, Kenward MG, et al. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ. 2009;338: b2393–b2393. 10.1136/bmj.b2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubin DB. Multiple Imputation after 18+ Years. Journal of the American Statistical Association. 1996;91: 473–489. 10.1080/01621459.1996.10476908 [DOI] [Google Scholar]
- 31.Kang H. The prevention and handling of the missing data. Korean J Anesthesiol. 2013;64: 402 10.4097/kjae.2013.64.5.402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li T, Convertino G, Wang W, Most H. HyperTuner: Visual Analytics for Hyperparameter Tuning by Professionals. 2018; 11. [Google Scholar]
- 33.Luo G. A review of automatic selection methods for machine learning algorithms and hyper-parameter values. Netw Model Anal Health Inform Bioinforma. 2016;5: 18 10.1007/s13721-016-0125-6 [DOI] [Google Scholar]
- 34.Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. Journal of Clinical Epidemiology. 2019;110: 12–22. 10.1016/j.jclinepi.2019.02.004 [DOI] [PubMed] [Google Scholar]
- 35.Senanayake S, White N, Graves N, Healy H, Baboolal K, Kularatna S. Machine learning in predicting graft failure following kidney transplantation: A systematic review of published predictive models. International Journal of Medical Informatics. 2019;130: 103957 10.1016/j.ijmedinf.2019.103957 [DOI] [PubMed] [Google Scholar]
- 36.Van Calster B, Vickers AJ. Calibration of Risk Prediction Models: Impact on Decision-Analytic Performance. Med Decis Making. 2015;35: 162–169. 10.1177/0272989X14547233 [DOI] [PubMed] [Google Scholar]
- 37.Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, et al. Assessing the Performance of Prediction Models: A Framework for Traditional and Novel Measures. Epidemiology. 2010;21: 128–138. 10.1097/EDE.0b013e3181c30fb2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins GS, de Groot JA, Dutton S, Omar O, Shanyinde M, Tajar A, et al. External validation of multivariable prediction models: a systematic review of methodological conduct and reporting. BMC Med Res Methodol. 2014;14: 40 10.1186/1471-2288-14-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah ND, Steyerberg EW, Kent DM. Big Data and Predictive Analytics: Recalibrating Expectations. JAMA. 2018;320: 27 10.1001/jama.2018.5602 [DOI] [PubMed] [Google Scholar]
- 40.Steyerberg EW, Vergouwe Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. European Heart Journal. 2014;35: 1925–1931. 10.1093/eurheartj/ehu207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shillan D, Sterne JAC, Champneys A, Gibbison B. Use of machine learning to analyse routinely collected intensive care unit data: a systematic review. Crit Care. 2019;23: 284 10.1186/s13054-019-2564-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Char DS, Shah NH, Magnus D. Implementing Machine Learning in Health Care—Addressing Ethical Challenges. N Engl J Med. 2018;378: 981–983. 10.1056/NEJMp1714229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Calster B, Wynants L, Timmerman D, Steyerberg EW, Collins GS. Predictive analytics in health care: how can we know it works? Journal of the American Medical Informatics Association. 2019;26: 1651–1654. 10.1093/jamia/ocz130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.