Abstract
Introduction
Chemotherapy resistance is the main cause of poor prognosis in patients with hepatocellular carcinoma (HCC). Therefore, it is important to understand the molecular mechanism of adriamycin (ADM) resistance in HCC. Increasing evidence indicates that circular RNAs (circRNAs) play a crucial regulatory role in different pathological processes. In the current study, we aimed to investigate the roles and the underlying molecular mechanism of circFoxo3 in ADM-resistant HCC.
Materials and Methods
Twenty-five pairs of clinical tumors samples and matched normal tissues were collected from patients with HCC. Gain- and loss-function experiments were performed to investigate the role of circFoxo3 in ADM-resistant cells.
Results
CircFoxo3 expression was increased in ADM-resistant HCC tissues and HCC cell lines and in metastatic tissues compared with non-metastatic tissues. CircFoxo3 knockdown reduces and circFoxo3 overexpression enhances HCC cell invasion and tumor growth. In addition, circFoxo3 interacted with miR-199a-5p and regulated miR-199a-5p expression. Furthermore, ATP Binding Cassette Subfamily C Member 1 (ABCC1) was identified as a new target of miR-199a-5p. CircFoxo3 interacted with miR-199a-5p to positively regulate ABCC1 expression, contributing to epithelial–mesenchymal transition progression.
Conclusion
CircFoxo3 knockdown reduces and circFoxo3 overexpression enhances HCC cell invasion and tumor growth through regulation of miR-199a-5p/ABCC1 axis. Our findings reveal that circFoxo3 may be novel biomarkers and therapeutic target for HCC treatment.
Keywords: hepatocellular cancer, circFoxo3, adriamycin, EMT, biomarker
Introduction
Hepatocellular carcinoma (HCC) has become the leading cause of cancer-related mortality worldwide. There were approximately 854,000 new liver cancer cases in 2015, compared to estimated 810,000 liver cancer-related deaths per year. It is estimated that 72% of cases occur in Asia (more than 50% in China), 10% in Europe.1,2 The therapeutic strategy of HCC is still based on surgery, radiotherapy and chemotherapy, but the prognosis of patients is still very poor, especially in patients with chemotherapy resistance.3 Adriamycin (ADM) is a routine drug chemotherapy treatment for HCC, but multidrug resistance (MDR) of tumor cells seriously affects the effect of chemotherapy for HCC.4 Therefore, a better understanding ADM chemo-resistance mechanism for HCC is needed. The circular RNAs (circRNAs) are stable, abundant, and conserved in eukaryotic cells.5,6 Recent evidence demonstrates that dysregulation of circRNAs can enhance cell proliferation, migration, and metastasis of liver cancer.7,8 Consequently, circRNAs may lead to drug resistance in malignant tumors.
A previous study showed that circFoxo3 was decreased in breast cancer tumor samples and cancer cells. However, circFoxo3 was significantly increased when the cancer cells were induced to apoptosis. They also found that circFoxo3 could decrease p53 levels.9–11 In this study, we revealed that circFoxo3 was over-expressed in HCC tissues and cancer cell lines, especially in ADM-resistant HCC tissues and HCC cell lines. Moreover, we found that circ-Foxo3 promoted HCC cell invasion by regulating ABCC1 expression by sponging miR-199a-5p. Our research revealed that circFoxo3 may offer a novel biomarker and therapeutic target for HCC treatment.
Materials and Methods
Human Clinical Samples
Twenty-five pairs of clinical tumors samples and matched normal tissues were collected from patients with HCC from March 2016 to June 2018, The Third Xiangya Hospital, Central South University. The average age of patients with HCC samples is 50.2 years. Normal tissues were taken from more than 5 cm away from the tumor. All the patients with HCC were treated with Adriamycin before the operation. ADM resistance was identified as more than 30% new lesions or tumor growth after 2 months of ADM chemotherapy, while ADM sensitive was considered to be increased tumor growth less than 20% after ADM chemotherapy. All tissues samples were collected immediately after surgery and stored in liquid nitrogen for further use. All experiments about human samples were approved by the Ethical Committee of The Third Xiangya Hospital, Central South University. Written informed consent was obtained from each subject.
Cell Culture and Cell Transfection
Human normal hepatocellular cells (HL-7702) and two HCC cells: SK-HEP-1, HepG2, SK-HEP-1/ADM and HepG2/ADM were obtained from the Cell Collection Committee of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (Gibco, USA) at 37°C in a 5% CO2 humidified incubator.
To construct circFoxo3-overexpressing plasmids, human circFoxo3 cDNA was synthesized and cloned into pCD25-ciR vector (Geneseed, China). CircFoxo3 siRNA plasmids were constructed and purchased from (GenomediTech, Shanghai, China). Plasmids were transfected into cell with Lipofectamine 3000 (Invitrogen, USA) according to the manufacture’s protocol. The cells transfected with an empty pCD25-ciR vector were used as a control for the circFoxo3-overexpressing group and the cells transfected with scrambled sequence were used as a control for circFoxo3-knockdown group. miR-199a-5p mimic, miR-199a-5p inhibitor and a corresponding NC-miRNA were obtained from RiboBio (Guangzhou, China).
RNA Isolation and Real-Time PCR
Total RNA isolation was performed with Trizol reagent (Invitrogen, USA). The expression of circFoxo3 was measured by real-time PCR according to the manufacturer’s protocol in CFX96 Real-Time System (Bio-Rad, USA). All primers were synthesized by Sangon Biotech (Shanghai, China) and GAPDH was used as an endogenous control. Divergent primers for circFoxo3: left, CATGGATGCTGATGGGTTGG, right, AGTTCCCTCATTCTGGACCC; primers for Foxo3: forward, CGGGATAACCAACTCTCCTTCT, reverse, GACGAACATTTCCTCGGCTG; ABCC1, forward, ATCACCATCATCCCCCAGGA reverse, TGCAGTCCTCGAACTGTGTC; GAPDH, forward, GAAAGCCTGCCGGTGACTAA, reverse, TTCCCGTTCTCAGCCTTGAC. QPCR was performed at the condition: 95.0°C for 3 min, and 39 circles of 95.0°C for 10 s and 60°C for 30 s. The data were analyzed using the comparative Ct (2−ΔΔCT) method.
RNA Sample Treatment with RNase R and PCR
Total RNA was isolated with TRIzol (Life Technologies) according to the manufacturer’s instructions. RNase R treatment was carried out for 15 min at 37°C using 3 U/mg RNase R (Lucigen, cat. #RNR07250).
RNA Fluorescence in situ Hybridization
CircFOXO3 probes were designed and synthesized by RiboBio. The probe signals were detected with a fluorescence in situ hybridization (FISH) kit (RiboBio) according to the manufacturer’s instructions.
Flow Cytometry Assay
HepG2/ADM and SK-HEP-1/ADM cells transfected with indicated plasmids were stained with propidium iodide using the cycle test plus DNA reagent kit (BD, USA) according to the manufacturer’s protocol. Briefly, the treated cells were digested with trypsin, and collected by centrifuging for 5 min at 1000 g. One mL of ice-cooled PBS was used to resuspend the cells. After centrifugation, the supernatant was carefully aspirated, leaving about 50 μL of PBS. The cells were fixed with ice-cool 70% ethanol at 4°C for 12 h. The cells then were stained with 0.5 mL of propidium iodide at 37°C for 30 min in the dark. The red fluorescence was detected by a flow cytometer (Thermo Fisher, USA) at an excitation wavelength of 488 nm for cell cycle analysis.
Transwell Assay
In vitro invasion of HepG2/ADM and SK-HEP-1/ADM cells were measured by transwell assays. About 1×105 transfected cells were seeded into the upper chambers of each trans-well. The bottom chamber was added 20% FBS for 24 h. Then, the cells on the lower surface of the filter were fixed and stained. Migrating cells were measured by averaging the total numbers of cells from triplicate determinations.
Luciferase Reporter Assay
Wild-type (WT) and mutant (MUT) circFoxo3, and WT and mutant ABCC1 3′-UTR sequences were constructed. Subsequently, we seeded SK-HEP-1/ADM cells into 96-well plates and co-transfected 80 pmol/L miRNAs mimics or negative control with the 100 ng circFoxo3 and ABCC1 wild-type or mutant type plasmids. The miRNA mimics or negative control were acquired from Ribobio (Guangzhou, China). After cultured for 24 h, dual-luciferase reporter assay system (Promega, Madison, WI) was performed to measure the luciferase activity following the manufacturer’s protocol. Renilla activity was used as an internal control.
Pull-Down Assay
The pull-down assay was performed as described in previous study.9 In brief, 107 cells were washed with cold PBS, lysed in 500 μL co-IP buffer, and incubated with 3 μg biotinylated DNA oligo probes against endogenous or ectopically expressed circ-Foxo3, at room temperature for 2 h. A total of 50 μL washed Streptavidin C1 magnetic beads (Invitrogen) were added to each binding reaction for 1 h at room temperature. Then, the RNAs were purified using RNeasy Mini Kit (QIAGEN) for further qPCR analysis.
Western Blot
After indicated treatment, SK-HEP-1/ADM cells were collected for protein extraction using RIPA lysis buffer. BCA Protein Concentration Assay Kit (Thermo Scientific, USA) was used to determine the protein concentration of the sample according to the manufacturer’s protocol. Proteins were separated on 10% SDS gel and transferred into PVDF membranes. Primary antibodies were used as follows: ABCC1 (1:1000 dilution), E-cadherin (1:1000 dilution), N-cadherin (1:1000 dilution), vimentin (1:1000 dilution), GAPDH (1:3000 dilution) (Cell Signaling Technology, USA). After primary antibodies incubation, the membranes were incubated with a secondary anti-rabbit antibody (1:5000, Cell Signaling Technology, USA). The bands were measured using a chemiluminescence system (Bio-Rad, USA) imaging system.
Tumor Xenograft in vivo
Animal experiments were approved by the Ethical Committee for Animal Research of The Third Xiangya Hospital, Central South University. To assess tumor growth, 100 μL of SK-HEP-1/ADM cells (1×106) transfected with circFoxo3 overexpressed or siRNA plasmids was subcutaneously injected into each mouse (BALB/c nude mice, 6-week old, male, five mice per group). The tumor sizes were measured weekly and calculated using the formula: 0.52×L×W2 where L and W are the long and short diameter of the tumor, respectively. Tumor weights were measured 4 weeks later.
Statistical Analysis
Each assay was performed three times. Data were presented as mean ± standard error. All data were analyzed by SPSS 20.0, P < 0.05 was considered as statistically significant. Differences between two groups’ data were analyzed using the Student’s t-test, and difference among three or more groups was compared by One-Way ANOVA with post hoc Bonferroni test. Kaplan–Meier curve and Log-rank test were used for survival analysis, and correlation analysis was performed using Pearson correlation coefficient analysis among circFoxo3, miR-199a-5p, ABCC1.
Results
CircFoxo3 Was Up-Regulated in HCC Tissues and Cell Lines
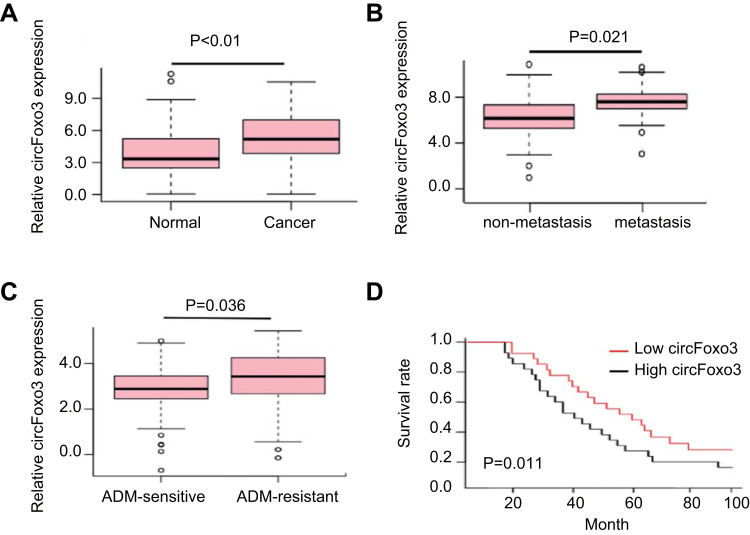
A previous study has showed that circFOXO3 contains one exon that ultimately creates a transcript of 1435 nucleotides by back-splicing in glioblastoma cells.12 In this study, we performed RT-PCR to confirm that the circRNAs were resistant to RNase R treatment, whereas PCR products from linear FOXO3 mRNA were disappeared after RNase R treatment. CircRNAs could not amplify any product from genomic DNA. (suppl. Figure 1). We then detected the expression of circFOXO3 in 25 pairs of HCC tissues and corresponding normal tissues and found that the expression of circFoxo3 in HCC tissues was significantly higher than matched normal tissues (P<0.01, Figure 1A). Moreover, the expression of circFoxo3 was increased in metastatic tissues compared with non-metastatic tissues (P=0.021, Figure 1B). In addition, compared with ADM-sensitive HCC tissues, circFoxo3 expression was significantly up-regulated in ADM-resistant HCC tissues (P=0.036, Figure 1C). Kaplan–Meier curve and Log-rank test evaluated that survival rate in patients with HCC had low expression of circFoxo3 was longer than the patients had high expression of circFoxo3 (P = 0.011, Figure 1D). These findings suggest that circFoxo3 is associated with HCC development.
Figure 1.
CircFoxo3 is up-regulated in hepatocellular carcinoma. (A) qRT-PCR analysis of expression levels of circFoxo3 in HCC tissues compared with matched adjacent normal tissues. (B) The expression levels of circFoxo3 in HCC tissues with lymph node metastasis compared with those without metastasis. (C) The expression levels of circFoxo3 in ADM-sensitive HCC tissues compared with ADM-resistant HCC tissues. (D) The survival rate was evaluated by Kaplan–Meier curve between HCC patients with high or low circFoxo3 expression.
CircFoxo3 Regulates Cell Invasion and Cell Cycle in HCC Cells
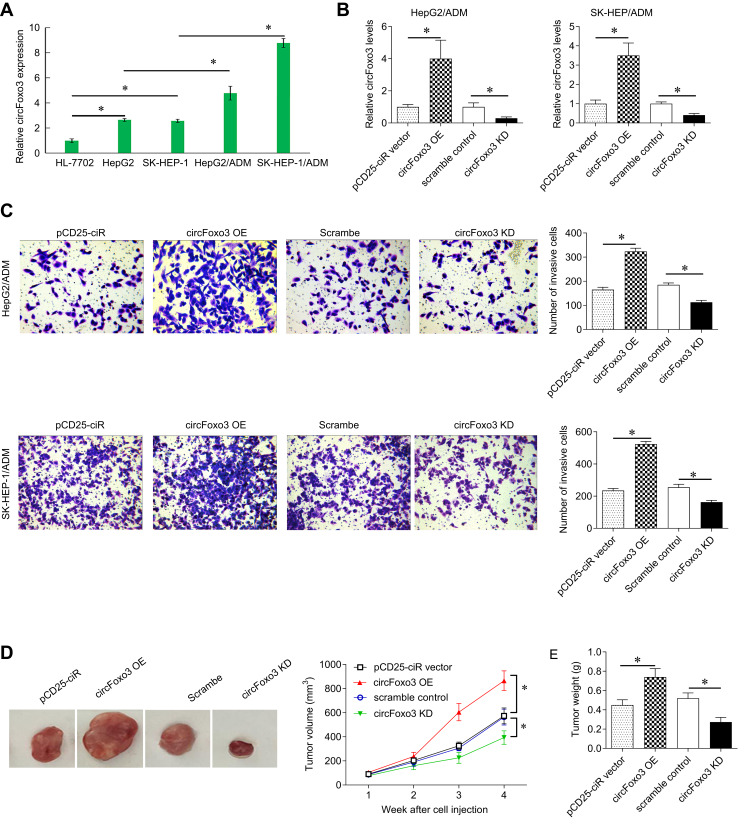
Next, we detected the expression of circFoxo3 in SK-HEP-1, HepG2, SK-HEP-1/ADM and HepG2/ADM and HL-7702. Compared with normal cell lines, the expression of circFoxo3 in SK-HEP-1 and HepG2 cell lines was significantly up-regulated. Moreover, the levels of circFoxo3 in SK-HEP-1/ADM and HepG2/ADM cell lines were significantly raised compared with their parental cells (Figure 2A).
Figure 2.
CircFoxo3 promotes cell invasion, and growth in hepatocellular carcinoma cells. (A) qRT-PCR analysis of expression levels of circFoxo3 in four HCC cell lines (SK-HEP-1,HepG2, SK-HEP-1/ADM and HepG2/ADM) and the normal cell line (HL-7702). *p < 0.05. (B) qRT-PCR analysis for circFoxo3 in HepG2/ADM and SK-HEP-1/ADM cells transfected with circFoxo3 over-expression plasmid or sh-circFoxo3 plasmid. *p < 0.05. (C) Transwell assay was performed in HepG2/ADM and SK-HEP-1/ADM cells transfected with circFoxo3 over-expression plasmid or sh-circFoxo3 plasmid. *p < 0.05. (D) BALB/c nude mice were subcutaneously injected with SK-HEP-1/ADM cells transfected with circFoxo3 over-expression plasmid or sh-circFoxo3 plasmid. The cells transfected with pCD25-ciR or scramble were used as a control for circFoxo3 over-expression plasmid or sh-circFoxo3 plasmid, respectively. Tumor volumes were measured every week. *p < 0.05. (E) The xenografted tumor weights were measured. *p < 0.05.
The circFoxo3 over-expression vector and sh-circFoxo3 were constructed and transfected into SK-HEP-1/ADM and HepG2/ADM cells. CirccFoxo3 was overexpressed and knocked down in SK-HEP-1/ADM and HepG2/ADM cells, respectively (Figure 2B). Transwell assay demonstrated over-expressing circFoxo3 promoted SK-HEP-1/ADM and HepG2/ADM cell migration capacity. However, circFoxo3 knockdown showed the opposite result (Figure 2C). To evaluate the effect of circFoxo3 on the tumor growth in vivo, we subcutaneously injected the transfected SK-HEP-1/ADM cells into nude mice. The tumor volume of the circFoxo3 over-expression group was larger than the control group, whereas circFoxo3 downregulation led to a significant decrease in tumor volume (Figure 2D). In addition, the average tumor weight in the circFoxo3 over-expression group was significantly higher than the control group, and the average tumor weight in the circFoxo3 knockdown group was lighter than control (Figure 2E).
CircFoxo3 Serves as a Sponge for Multiple miRNAs in HCC
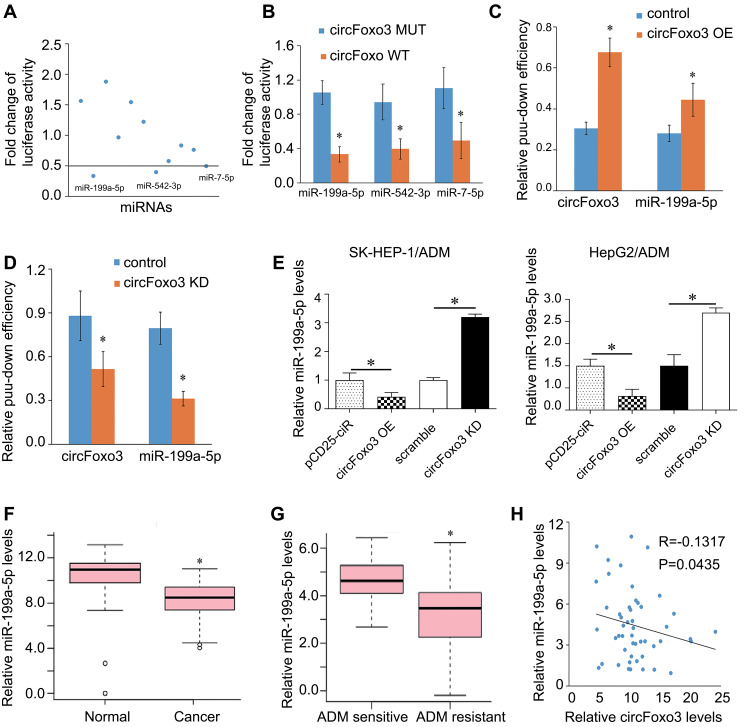
CircRNAs severs as competitive endogenous RNAs (ceRNA) by recruiting miRNA molecules via base-pairing with miRNA-recognition elements (MREs), which subsequently compete with common miRNAs, thus contributing to tumor development, metastasis, and drug resistance.13 We performed FISH analysis to determine the location of circFoxo3 and found that circFOXO3 was abundant and located in the cytoplasm (suppl. Figure 2). Based on this finding, we further explored whether circFOXO3 binds to miRNAs. Online database starBase V2.0 (http://starbase.sysu.edu.cn/) could be used to predict potential miRNA target sites on circRNAs.14 Eleven miRNAs have potential target sites on circFoxo3. We then used luciferase reporter gene assay to screen the miRNAs that bind to circFoxo3 and found that miR-199a-5p, miR-7-5p, and miR-542-3p may target to circFoxo3 (Figure 3A). In addition, co-transfection wild-type circFoxo3, but not mutant circFoxo3, with miR-199a-5p, miR-7-5p, and miR-542-3p led to a significant decrease in luciferase activity (Figure 3B), suggesting a possible interaction between circFoxo3 and these miRNAs. The binding site of miR-199a in circFoxo3 is showed in supplementary Figure 4. However, pull-down assay further confirmed that circFoxo3 could pull down miR-199a-5p (Figure 3C and D), but could not pull down miR-7-5p and miR-542-3p (supplementary Figure 3). Thus, we reveal that circFoxo3 interacts with miR-199a-5p and focus on miR-199a-5p for further functional experiments.
Figure 3.
CircFoxo3 serves as a sponge for miR-199a-5p in HCC cells. (A)11 miRNA mimics were co-transfected with the circFoxo3 vector into SK-HEP-1/ADM cells to screen miRNAs that were able to bind to circFoxo3. The reduction of luciferase reporter activities <0.5 was selected for further analysis. (B) Luciferase reporter assay for the luciferase activity of circFoxo3-MUT or circFoxo3-WT in SK-HEP-1/ADM cells co-transfected with 3 miRNA mimics. *p < 0.05. (C and D) RNA pull-down assay for the amount of circFoxo3 and miR-199a-5p in SK-HEP-1/ADM cells after transfection of circFoxo3 over-expression plasmid or sh-circFoxo3 plasmid. *p < 0.05. (E)Tthe expression levels of miR-199a-5p in SK-HEP-1/ADM and HepG2/ADM cells transfected with circFoxo3 over-expression or sh-circFoxo3. The cells transfected with pCD25-ciR or scramble were used as control for circFoxo3 over-expression plasmid or sh-circFoxo3 plasmid, respectively. *p < 0.05. (F) The expressions of miR-199a-5p in HCC tumor tissues and adjacent normal tissues. *p < 0.05. (G) The expression levels of miR-199a-5p in ADM-sensitive and ADM-resistant HCC tissues. *p < 0.05. (H) CircFoxo3 expression was negatively correlated with miR-199a-5p in HCC patients.
The levels of miR-199a-5p were decreased by circFoxo3 and up-regulated by circFoxo3 knockdown in SK-HEP-1/ADM and HepG2/ADM cells (Figure 3E). In addition, we detected the expressions of miR-199a-5p in 25 pairs of HCC tumor tissues and adjacent normal tissues and found that the expressions of miR-199a-5p decreased in HCC tumor tissues (Figure 3F). Notably, miR-199a-5p was significantly reduced in ADM-resistant HCC tumor tissues compared with ADM-sensitive HCC tissues (Figure 3G). Correlation analysis showed that circFoxo3 expression was negatively correlated with miR-199a-5p (Figure 3H). In summary, circFoxo3 could directly sponge miR-199a-5p.
CircFoxo3 Positively Regulates ABCC1 Protein Expression via Targeting miR-199a-5p
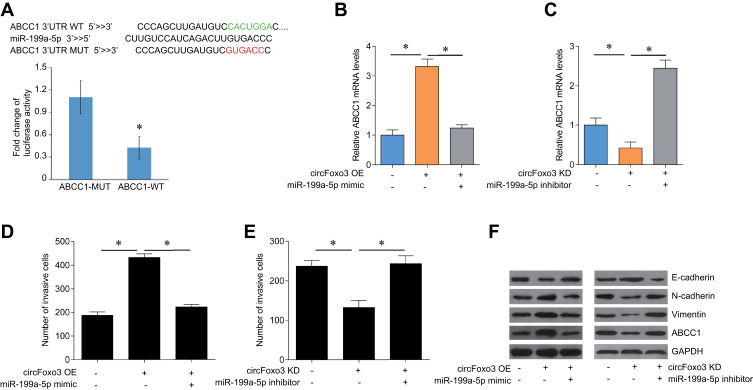
We used online databases (TargetScan, DIANA, and miRanda) to find potential target genes for miR-199a-5p. ABCC1 is a potential target of miR199a-5p and is closely related to drug resistance in several cancers.15 We established plasmids of ABCC1 WT and ABCC1 MUT and transfected into SK-HEP-1/ADM cells. MiR-199a-5p mimic transfection significantly decreased the luciferase activity in co-transfected with ABCC1-WT, but not ABCC1-MUT, in SK-HEP-1/ADM cells (Figure 4A). We detected the expression of ABCC1 mRNA in cirFoxo3-overpressed or -knocked-down SK-HEP-1/ADM cells that co-transfected with miR-199a-5p mimics or inhibitors by qRT-PCR. The results showed that circFoxo3 induced the expression of ABCC1 mRNA, which was inhibited by miR-199a-5p mimics (Figure 4B). However, circFoxo3 knockdown reduced the expression of ABCC1, which up-regulated by miR-199a-5p inhibitor (Figure 4C).
Figure 4.
CircFoxo3 regulates the expression of ABCC1 via targeting miR-199a-5p in HCC cells. (A) Luciferase reporter assay for the luciferase activity of ABCC1-MUT or ABCC1-WT in SK-HEP-1/ADM cells transfected with miR-199a-5p mimics. *p < 0.05. (B) The mRNA expression levels of ABCC1 in SK-HEP-1/ADM cells transfected with circFoxo3 or circFoxo3 plus miR-199a-5p mimics. *p < 0.05. (C) The mRNA expression levels of ABCC1 in SK-HEP-1/ADM cells transfected with sh-circFoxo3 or sh-circFoxo3 plus miR-199a-5p inhibitor. *p < 0.05. (D) Transwell assay was performed to determine the invasion ability of the SK-HEP-1/ADM cells after the transfection of circFoxo3 or circFoxo3 plus miR-199a-5p mimics. *p < 0.05. (E) Transwell assay was performed to determine the invasion ability of the SK-HEP-1/ADM cells after the transfection of sh-circFoxo3 or sh-circFoxo3 plus miR-199a-5p inhibitor. *p < 0.05. (F) Western blot analysis the expression levels of ABCC1, E-cadherin, vimentin and N-cadherin in SK-HEP-1/ADM cells after indicated transfection.
Based on the above results, we suspect that circFoxo3 may regulate the expression of ABCC1 through direct binding miR-199a-5p to promote cell invasion. We found that miR-199a-5p mimic could reverse the effects of circFoxo3 overexpression on SK-HEP-1/ADM cell invasion (Figure 4D). In addition, miR-199a-5p inhibitor could normalize the effects of circFoxo3 knockdown on SK-HEP-1/ADM cell invasion (Figure 4E). EMT is closely related to cancer cell metastasis and drug resistance. In order to explore the possible mechanism of circFoxo3 regulates HCC metastasis, we investigated the markers of EMT and found that circFoxo3 overexpression significantly decreased the expression of E-cadherin but decreased the expression of N-cadherin and Vimentin, indicating a promotion of EMT process, which was suppressed by miR-199a-3p mimic treatment. In contrast, miR-199a-3p inhibitor treatment enhanced EMT process that was impaired by circFoxo3 downregulation (Figure 4F). Our results further confirmed that circFoxo3 overexpression significantly up-regulated ABCC1 expression, which was inhibited by miR-199a-5p mimic. In contrast, circFoxo3 downregulation decreased ABCC1 expression, which was increased by miR-199a-5p inhibitor (Figure 4F).
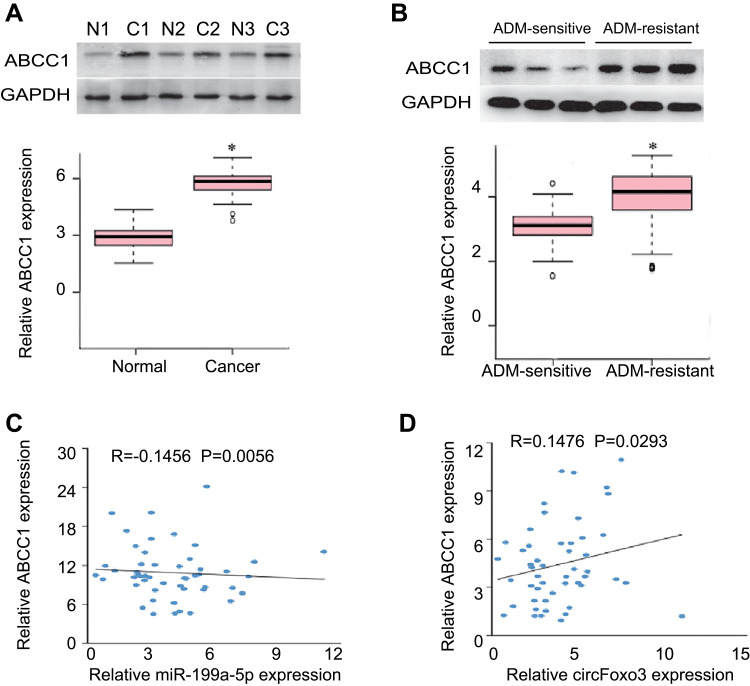
In addition, the expression of ABCC1 was detected in 25 pairs of HCC tumor tissues and adjacent normal tissues. The expression of ABCC1 increased in HCC tumor tissues compared with adjected tissues (Figure 5A). Notably, ABCC1 was significantly up-regulated in ADM-resistant HCC tumor tissues compared with ADM-sensitive HCC tissues (Figure 5B). Correlation analysis showed that miR-199a-5p expression was negatively correlated with ABCC1 expression (Figure 5C) and circFoxo3 expression was positively correlated with ABCC1 expression in HCC patients (Figure 5D). In conclusion, these findings demonstrate that circFoxo3 contributes to ADM resistance by acting as a ceRNA to negatively modulates miR-199a-5p expression to induce EMT in HCC cells.
Figure 5.
The expression levels of ABCC1 in HCC. (A) The protein levels of ABCC1 in HCC tumor tissues and adjacent normal tissues. *p < 0.05. N, matched adjacent normal tissues; C, cancer tissues. (B) The protein levels of ABCC1 in ADM-sensitive and ADM-resistant HCC tissues. *p < 0.05. (C) miR-199a-5p expression was negatively correlated with ABCC1 expression in HCC patients. (D) CircFoxo3 expression was positively correlated with ABCC1 expression in HCC patients.
Discussion
At present, significant progress has been made in the treatment of HCC, but it is indispensable to explore the molecular mechanism of chemoresistance and to identify new targets for chemoresistance therapy.16 In this study, we demonstrated that circFoxo3 is elevated in HCC metastatic tissues and in ADM-resistant HCC tumor tissue and cell lines. In addition, we also revealed that circFoxo3 regulated HCC cell invasion and tumor growth through regulation of miR-199a-5p/ABCC1 axis.
As new gene regulators, circRNAs are involved in a variety of biological and pathological processes.17–19 With the advances of genome research technology, it has been recognized that circRNA is widely expressed in human cells. The two most important characteristics of circRNA are highly conserved and stable in mammalian cells.20 There are many reports about the occurrence of circRNA in the development of HCC.21,22 However, the role of circFoxo3 in liver cancer is little known. Thus, our results suggest that circFoxo3 may be an oncogene in HCC. Previous studies have shown that dysregulation of circFoxo3 is associated with the occurrence and development of many diseases.23 Du WW et al demonstrated that circFoxo3 could regulate the cancer cell cycle and promote cardiac senescence.11 Recently, Zhang et al find that circFoxo3 knockdown reduces and circFoxo3 overexpression enhances glioblastoma cell proliferation and invasion.12
In this study, we found that circFoxo3 expression was significantly increased in ADM-resistant HCC tissues and also in SK-HEP-1/ADM and HepG2/ADM cells. However, the mechanism in which circFoxo3 leads to ADM resistance remains unclear in HCC. We revealed that circFoxo3 functionally acts as a sponge for miR-199a-5p. Previous studies have shown that miR-199a-5p decreases drug resistance by regulating ATG7 in HCC cells.24 MiR-199a-5p has been reported to mediate suppression in cell proliferation and invasion in ovarian cancer, cervical carcinoma, clear cell renal cell carcinoma and papillary thyroid carcinoma by directly targeting certain genes, such as PIAS3, DDR1, NF-κB1, MAP4K3 and SNAI1.25–28 This study identified a new target of miRNA-199a-5p, ATP Binding Cassette Subfamily C Member 1 (ABCC1), which also named multidrug resistance-associated protein 1 (MRP1). ABCC1 led to chemotherapy resistance in many cancers.29 More importantly, ABCC1 may contribute to chemotherapy resistance in HCC. Our results showed that circFoxo3 knockdown reduces and circFoxo3 overexpression enhances the expression of ABCC1, promoting epithelial–mesenchymal transition (EMT) progression. CircFoxo3 expression was negatively correlated with miR-199a-5p and positively correlated with ABCC1. These results suggest that circFoxo3 contributes to HCC cell invasion through regulation of miR-199a-5p and ABCC1.
In conclusion, our study revealed that circFoxo3 increased in ADM-resistant HCC tissues and cell lines. CircFoxo3 knockdown reduces and circFoxo3 overexpression enhances HCC cell invasion and tumor growth through regulation of miR-199a-5p/ABCC1 axis. Our findings reveal that circFoxo3 may be novel biomarkers and therapeutic target for HCC treatment.
Ethics Approval and Informed Consent
This study was approved by the Ethics Committee of The Third Xiangya Hospital, Central South University. Informed consent was obtained from each subject in accordance with the Declaration of Helsinki. Animal experiments were approved by the Ethics Committee for Animal Research of the Third Xiangya Hospital of Central South University following the National Research Council’s Guide for the Care and Use of Laboratory Animals (Eight Edition).
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China, 2013. Cancer Lett. 2017;401:63–71. doi: 10.1016/j.canlet.2017.04.024 [DOI] [PubMed] [Google Scholar]
- 3.Innes H, Barclay ST, Hayes PC, et al. The risk of hepatocellular carcinoma in cirrhotic patients with hepatitis C and sustained viral response: role of the treatment regimen. J Hepatol. 2018;68(4):646–654. doi: 10.1016/j.jhep.2017.10.033 [DOI] [PubMed] [Google Scholar]
- 4.Qian JQ, Sun P, Pan ZY, Fang ZZ. Annonaceous acetogenins reverses drug resistance of human hepatocellular carcinoma BEL-7402/5-FU and HepG2/ADM cell lines. Int J Clin Exp Pathol. 2015;8(9):11934–11944. [PMC free article] [PubMed] [Google Scholar]
- 5.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141–157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 7.Kumar A, Cherukumilli M, Mahmoudpour SH, Brand K, Bandapalli OR. ShRNA-mediated knock-down of CXCL8 inhibits tumor growth in colorectal liver metastasis. Biochem Biophys Res Commun. 2018;500(3):731–737. doi: 10.1016/j.bbrc.2018.04.144 [DOI] [PubMed] [Google Scholar]
- 8.Kim DH, Kim KS, Ramakrishna S. NFAT5 promotes in vivo development of murine melanoma metastasis. Biochem Biophys Res Commun. 2018;505(3):748–754. doi: 10.1016/j.bbrc.2018.09.171 [DOI] [PubMed] [Google Scholar]
- 9.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi: 10.1038/cdd.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):1402–1412. doi: 10.1093/eurheartj/ehw001 [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Liao K, Miao Z, et al. CircFOXO3 promotes glioblastoma progression by acting as a competing endogenous RNA for NFAT5. Neuro Oncol. 2019;21(10):1284–1296. doi: 10.1093/neuonc/noz128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–D97. doi: 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel TH, Norman L, Chang S, et al. European mtDNA variants are associated with differential responses to cisplatin, an anticancer drug: implications for drug resistance and side effects. Front Oncol. 2019;9:640. doi: 10.3389/fonc.2019.00640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fekir K, Dubois-Pot-Schneider H, Desert R, et al. Retrodifferentiation of human tumor hepatocytes to stem cells leads to metabolic reprogramming and chemoresistance. Cancer Res. 2019;79(8):1869–1883. doi: 10.1158/0008-5472.CAN-18-2110 [DOI] [PubMed] [Google Scholar]
- 17.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 18.Tang CM, Zhang M, Huang L, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. 2017;7(1):40342. doi: 10.1038/srep40342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5(1):12453. doi: 10.1038/srep12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu S, Zhou L, Ponnusamy M, et al. A comprehensive review of circRNA: from purification and identification to disease marker potential. PeerJ. 2018;6:e5503. doi: 10.7717/peerj.5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Shen M. Circular RNA hsa_circ_103809 suppresses hepatocellular carcinoma proliferation and invasion by sponging miR-620. Eur Rev Med Pharmacol Sci. 2019;23(2):555–566. doi: 10.26355/eurrev_201902_16868 [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Deng T, Ge S, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38(15):2844–2859. doi: 10.1038/s41388-018-0619-z [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847–859. doi: 10.1038/nrc2223 [DOI] [PubMed] [Google Scholar]
- 24.Shen Q, Cicinnati VR, Zhang X, et al. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Mol Cancer. 2010;9(1):227. doi: 10.1186/1476-4598-9-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Yao B, Wu Z. miRNA-199a-5p suppresses proliferation and invasion by directly targeting NF-κB1 in human ovarian cancer cells. Oncol Lett. 2018;16(4):4543–4550. doi: 10.3892/ol.2018.9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma S, Jia W, Ni S. miR-199a-5p inhibits the progression of papillary thyroid carcinoma by targeting SNAI1. Biochem Biophys Res Commun. 2018;497(1):181–186. doi: 10.1016/j.bbrc.2018.02.051 [DOI] [PubMed] [Google Scholar]
- 27.Krazinski BE, Kiewisz J, Sliwinska-Jewsiewicka A, et al. Altered expression of DDR1 in clear cell renal cell carcinoma correlates with miR-199a/b-5p and patients’ outcome. Cancer Genomics Proteomics. 2019;16(3):179–193. doi: 10.21873/cgp.20124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L, Lu L, Zheng A, et al. MiR-199a-5p and let-7c cooperatively inhibit migration and invasion by targeting MAP4K3 in hepatocellular carcinoma. Oncotarget. 2017;8(8):13666–13677. doi: 10.18632/oncotarget.14623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao J, Yao X, Tian T, et al. ABCB5-ZEB1 axis promotes invasion and metastasis in breast cancer cells. Oncol Res. 2017;25(3):305–316. doi: 10.3727/096504016X14734149559061 [DOI] [PMC free article] [PubMed] [Google Scholar]