Supplemental digital content is available in the text.
Key Words: GENE, EXERCISE, HERITABILITY, HIDDEN HERITABILITY
ABSTRACT
Purpose
Polygenic risk scores (PRS) summarize genome-wide genotype data into a single variable that produces an individual-level risk score for genetic liability. PRS has been used for prediction of chronic diseases and some risk factors. As PRS has been studied less for physical activity (PA), we constructed PRS for PA and studied how much variation in PA can be explained by this PRS in independent population samples.
Methods
We calculated PRS for self-reported and objectively measured PA using UK Biobank genome-wide association study summary statistics, and analyzed how much of the variation in self-reported (MET-hours per day) and measured (steps and moderate-to-vigorous PA minutes per day) PA could be accounted for by the PRS in the Finnish Twin Cohorts (FTC; N = 759–11,528) and the Northern Finland Birth Cohort 1966 (NFBC1966; N = 3263–4061). Objective measurement of PA was done with wrist-worn accelerometer in UK Biobank and NFBC1966 studies, and with hip-worn accelerometer in the FTC.
Results
The PRS accounted from 0.07% to 1.44% of the variation (R2) in the self-reported and objectively measured PA volumes (P value range = 0.023 to <0.0001) in the FTC and NFBC1966. For both self-reported and objectively measured PA, individuals in the highest PRS deciles had significantly (11%–28%) higher PA volumes compared with the lowest PRS deciles (P value range = 0.017 to <0.0001).
Conclusions
PA is a multifactorial phenotype, and the PRS constructed based on UK Biobank results accounted for statistically significant but overall small proportion of the variation in PA in the Finnish cohorts. Using identical methods to assess PA and including less common and rare variants in the construction of PRS may increase the proportion of PA explained by the PRS.
Based on family and twin studies, genetic factors underlie an individual’s propensity to participate in physical activity (PA) (1–4). Genome-wide association studies (GWAS) have found a few loci that have genome-wide statistically significant association with PA, but the effect sizes found are small (5–7).
Polygenic risk score (PRS) or also called polygenic score is a score based on variation in multiple genetic loci and their associated weights. It serves as the best prediction for the trait that can be made when accounting for variation in multiple genetic variants (8,9). Genome-wide association analyses comparing disease cases with controls have identified thousands of genetic loci associated with complex disease risk, and genomic information has become a potential candidate for improving disease risk assessment (10). Although PRS has been calculated for many chronic diseases and applied for its predictive value, there is limited amount of research for predicting PA levels using PRS for PA (5,6). In particular, we lack information on how the constructed PRS for PA predict PA levels in independent cohorts with differing ancestry and with differing methods to assess self-reported and objectively measured PA levels. There has been much discussion on the genetic determinants underlying physical inactivity, and it would be helpful to identify those individuals for whom PA participation is difficult. In exercise interventions, these individuals may need tailored exercise programs with more support and supervision to gain the benefits of exercise therapy.
PA is a multifactorial behavior with many environmental and genetic factors influencing the volume of overall PA. Questionnaires and accelerometers are among the most used methods in assessing PA levels, although different methods have their strengths and challenges (11,12). Self-reported leisure-time PA and measured overall PA levels may have same but also different determinants. Age, sex, obesity (13), and chronic diseases (14) are typical examples of other characteristics and traits associated with PA levels, which may modify also the size of the genetic effects on PA in a context-dependent manner.
The main aim of this study was to calculate PRS for PA using UK Biobank GWAS summary statistics and then evaluate their out-of-sample predictive values in the Finnish Twin Cohorts (FTC) and the Northern Finland Birth Cohort 1966 (NFBC1966) using different PA phenotypes. We hypothesized that the PRS will account for a statistically significant proportion of the variation in PA. To deepen our understanding, we also compared the results between single-nucleotide polymorphism (SNP)–based and pedigree-based PA heritability analyses. In addition, to evaluate the usability of PRS for PA in clinical work, we compared how much of the variation in objectively measured PA is explained by simple questionnaire items compared with PRS.
METHODS
Study samples
FTC and NFBC1966 data, in addition to the open UK Biobank summary level data, were used in this study.
Data on participants from three FTC (Old twin cohort, Finntwin16, and Finntwin12) (15,16) were included in this study (N = 11,528 for both genetic and self-reported PA data; mean age, 44 yr (range, 18–93 yr); 46% male participants). From a subgroup of the Old twin cohort, 765 individuals had both genetic and objectively measured PA data (MOBILETWIN study; mean age, 73 yr (range, 71–75 yr); 46% male participants) (17). The twin studies were approved by the ethics committees of the University of Helsinki (113/E3/01 and 346/E0/05), Helsinki University Central Hospital (136/E3/01, 01/2011, 270/13/03/01/2008, and 154/13/03/00/2011), and Ethics Committee of the Southwest Finland (MOBILETWIN).
NFBC1966 comprises children born for mothers from Oulu and Lapland (Finland) and who had their expected date of birth between January 1 and Dec 31, 1966 (18). Data on cohort members’ self-reported PA and objectively measured PA were collected at the age of 46 yr (19), and genome-wide data were obtained at the 31-yr follow-up (20). Both genetic data and self-reported PA data were available for 4061 individuals, and genetic data plus objectively measured data from 3263–3437 individuals (48% male participants). The Ethics Committee of the Northern Ostrobothnia Hospital District, Oulu, Finland, approved the study (94/2011).
Open UK Biobank GWAS summary-level data were used in the generation of PRS for PA. The UK Biobank comprises extensive phenotypic data on some 500,000 individuals of the general UK population age between 40 and 69 yr (21). Self-reported PA data for this study were available from 321,309 individuals, and data on objective measurement for PA from 91,105 individuals (44% male participants). The North West Multi-Centre Research Ethics Committee approved the UK Biobank study.
In all study samples, the individuals having data on age, sex, height, weight, genetic data, and data on objectively measured or self-reported PA were included in the analyses of this study. All samples were collected, and studies were conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants.
PA variables
In FTC, the assessment of the leisure-time MET score was based on a series of structured questions on leisure-time PA (monthly frequency, mean duration, and mean intensity of sessions) and PA during commuting (22). The index was calculated by assigning a MET value to each activity and by calculating the product of that activity: intensity–duration–frequency. The MET score was expressed as the sum score of leisure-time PA MET-hours per day (22). In the MOBILETWIN study, PA was measured with a hip-worn triaxial accelerometer (Hookie AM20; Traxmeet Ltd, Espoo, Finland) for 7 d, and the mean duration of daily moderate-to-vigorous PA (MVPA) and mean number of daily steps were calculated; for details, see Waller et al. (17).
In NFBC1966, the leisure-time PA was self-reported with questions on the frequency and duration of light and brisk physical activities during leisure time. Brisk PA was described as causing at least some sweating and breathlessness, whereas light PA was defined as causing no sweating or breathlessness. PA frequency had six response options: 1) once a month or less often, 2) two to three times a month, 3) once a week, 4) two to three times a week, 5) four to six times a week, and 6) daily. PA duration had the following response options: 1) not at all, 2) less than 20 min, 3) 20–39 min, 4) 40–59 min 5) 1–1.5 h, and 6) more than 1.5 h. Daily averages of MET-hour scores of light and brisk PA were calculated by multiplying the PA volume (duration–frequency) by its intensity (light PA, 3 METs; brisk PA, 5 METs) (23). PA was objectively measured with wrist-worn Polar Active monitors (Polar Electro Oy, Kempele, Finland) for 14 d. Polar Active is a waterproof accelerometer providing MET values every 30 s based on daily PA (24). The participants were asked to wear the Polar Active monitor 24 h·d−1 for at least 14 d on the nondominant hand. Measured PA with intensity ≥3.5 METs was classified as MVPA and calculated as daily averages (minutes per day) (19). In addition, mean number of daily steps was calculated.
When constructing PRS, UK Biobank GWAS results of the self-report question on the “number of days/week of moderate PA 10+ min” (PRSreported) and objectively measured overall activity measured with Axivity AX3 wrist accelerometer for 7 d (PRSmeasured) (6) were used. A basic linear regression with sex and the first 10 principal components as covariates was performed on the GWAS used for the construction of the PRSreported and linear mixed-model regression on the GWAS used for PRSmeasured (6). The mean “number of days/week of moderate PA” was 3.7 (SD, 2.3), and the distribution of the variable, that is, UK Biobank Data-Field 884, can be seen from the online showcase of UK Biobank resources (25). The objectively measured overall activity phenotype is a continuous phenotype based on 7-d wrist accelerometry and is the average vector magnitude for each 30-s epoch, and is described in more detail by Doherty et al. (26). The SNP-based genetic correlation (rg) of these UK Biobank PA variables has been reported to be 0.35 (6).
Genotyping, quality control, and imputation in FTC
Chip genotyping was done using Illumina Human610-Quad v1.0 B, Human670-QuadCustom v1.0 A, Illumina HumanCoreExome (12 v1.0 A, 12 v1.1 A, 24 v1.0 A, 24 v1.1 A, 24 v1.2 A), and Affymetrix FinnGen Axiom arrays. The algorithms for genotype calling were Illumina’s GenCall for all HumanCoreExome chip genotypes, Illuminus for 610k and 670k chip genotypes, and AxiomGT1 for Affymetrix chip genotypes. Genotype quality control was done in three batches (batch1: 610k + 670k, batch2: HumanCoreExome, and batch3: Affymetrix chip genotypes), with removing variants with call rate less than 97.5% (batch1 and batch3) and 95% (batch2), removing samples with call rate less than 98% (batch1) or 95% (batch2 and batch3), removing variants with its minor allele frequency less than 1%, and Hardy–Weinberg equilibrium P value lower than 1 × 10−6. Also, samples from all batches with heterozygosity test method-of-moments F coefficient estimate value less than −0.03 or higher than 0.05 (batch1 and batch2) or ±4 SD from the mean (batch3) were removed along with the samples that failed sex check or were among the multidimensional scaling principal component analysis outliers. Total amounts of genotyped autosomal variants after quality control were 475,526 (batch1), 239,894 (batch2), and 388,673 (batch3), with following numbers of samples remaining for imputation: 2617 (batch1), 5328 (batch2), and 8218 (batch3).
We then performed prephasing using Eagle v2.3 (27) and imputation with Minimac3 v2.0.1 using the University of Michigan Imputation Server (28). Genotypes of all batches were imputed to Haplotype Reference Consortium release 1.1 reference panel (29).
Genotyping, quality control, and imputation in NFBC1966
Genotyping was performed using Illumina Human CNV370-Duo DNA bead chip as described previously by Sabatti et al. (20). The following quality control steps were applied: SNP with call rate <95% or minor allele frequency <0.05 was excluded from the study along with individuals with genotyping success rate <95%. Imputation to 1000 Genomes phase 3 reference panel (30) was performed using prephasing software SHAPEIT v2 (31) and imputation software IMPUTE2 v2.3.0 (32).
Polygenic scoring
PRS was constructed for self-reported (PRSreported) and objectively measured PA (PRSmeasured); see the “PA variables” section. To obtain PRS, we implemented a Bayesian approach taking account the linkage disequilibrium between each variant (LDpred) (33), and therefore, any pruning or thresholding method to select variants was not used. The infinitesimal model for polygenic scoring was adjusted with LD reference panel, which consists of 27,284 unrelated Finnish samples from the national FINRISK study (34). GWAS summary statistics from the UK Biobank for the risk score calculation were obtained from Neale laboratory repository of summary statistics (35) (questionnaire-based data) or from the data-sharing repository of GWAS of PA measured by accelerometer (6). There were weights from 91,105 to 321,309 samples for the risk score calculation. The LD reference panel, summary statistics, and the target study samples of FTC and NFBC1966 were restricted to HapMap3 (36) variants with European minor allele frequency > 5% and excluding the major histocompatibility complex region from chromosome 6 (GRCh37: 6p22.1–21.3), representing the whole genome capturing the polygenic signal and which tends to be well imputed for samples of European or Finnish ancestry. Total number of variants used for risk score calculation varied from 1,140,182 to 1,142,416 in FTC and from 1,140,159 to 1,142,392 in NFBC1966.
Associations between PRS and self-reported or measured PA
On the basis of previous knowledge on the different types of self-reported and objectively monitored PA variables and their heritability, we focused on analyzing whether PRSreported predicts self-reported PA and whether PRSmeasured predicts either self-reported or objectively measured PA in the Finnish cohorts. The proportion of total variation of PA outcomes explained by the model (R2) was estimated by generalized linear regression models. All PRS values were scaled to obtain standardized normal distribution, with a mean of zero and an SD of 1. Basic models were adjusted for four genetic principal components and sex in FTC and NFBC1966 and also for age in FTC. We also report the change in R2 (∆R2) when PRS was included in the model after the other predictors. Square root transformation of MVPA was used because of the violation of the assumption of normal distribution both in FTC and in NFBC1966 and of daily MET score in NFBC1966. In the linear mixed-model regression of the FTC data, the within-pair dependency was accounted for by using the family identifier as the random effect of the models. Individuals were divided to PRS deciles, and daily MET score, MVPA, and steps were compared between the first and last deciles with independent-samples t-test or Mann–Whitney’s test. The level of significance was set at P < 0.05.
Pedigree and SNP-based heritabilities in FTC
We estimated pedigree- and SNP-based heritabilities simultaneously using the same set of SNP values that were used in the PRS calculations. We implemented a method (GCTA-GREML) according to Zaitlen et al. (37) and Yang et al. (38), where the heritability is calculated using two genetic relatedness matrices where the first takes into account only the heritability caused by family structure and the second takes into account only the genetic part of the heritability, so that in the first matrix, the off-diagonals less than 0.05 were set to 0, letting the second matrix to have values less than 0.5 indicating possible distant genetic relationships between samples. Both of the matrices were used as the fixed effect of the linear mixed model. Using this method, there were no need to drop any related samples from the analysis. The difference between these two heritabilities is called missing (or hidden) heritability (37). SNP-based heritabilities for the UK Biobank PA measures were obtained from association results using LD score regression (39) restricted to the same set of HapMap3 SNP values that were used for PRS calculations.
Questionnaire items versus PRSmeasured in explaining variation in objectively measured PA
In the MOBILETWIN study (17), complete data from 640 individuals were available to analyze how much of the variation in objectively measured PA (MVPA and steps) could be accounted for by questionnaire-based data (age, sex, body mass index, self-reported distance walked or jogged outdoors, self-reported fitness, self-reported mobility-restricting disease, and self-reported PA category) and by PRSmeasured. This methodology and results are described in more detail in Supplemental Digital Content 1, questionnaire items versus PRSmeasured in explaining variation in objectively measured PA, http://links.lww.com/MSS/B929.
RESULTS
PRS
The PRSreported accounted for 0.24% and 0.25% of the variation (R2) in the reported daily MET scores in NFBC1966 and FTC (P values = 0.0017 and <0.0001, respectively; Table 1). The PRSmeasured accounted from 0.07% to 1.44% of the variation in the self-reported and objectively measured PA (P value range = 0.023 to <0.0001; Table 1). For all studied PA volume variables, individuals in the highest PRS deciles compared with the lowest PRS deciles had significantly (11%–28%) higher PA volume both in FTC and in NFBC1966 (P values = 0.017 to <0.0001; Table 2). Figure 1 shows the means of the objectively measured PA variables in MOBILETWIN and NFBC1966 according to PRSmeasured deciles. The associations are quite similar despite differences in age and measurement method between the MOBILETWIN and NFBC1966 studies.
TABLE 1.
Associations between PRS and PA variables in FTC and NFBC1966.
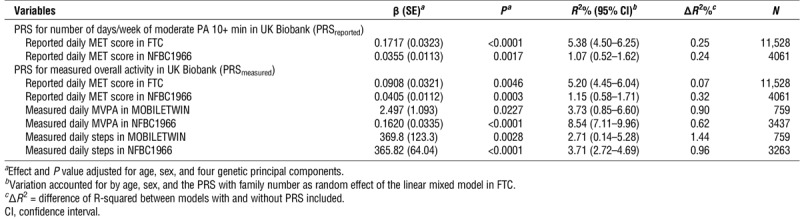
TABLE 2.
Physical activity volumes of Finnish individuals in the lowest and highest PRS deciles.
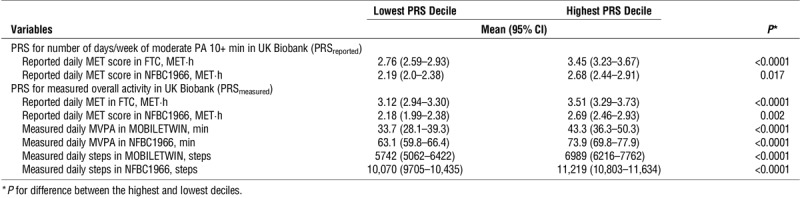
FIGURE 1.
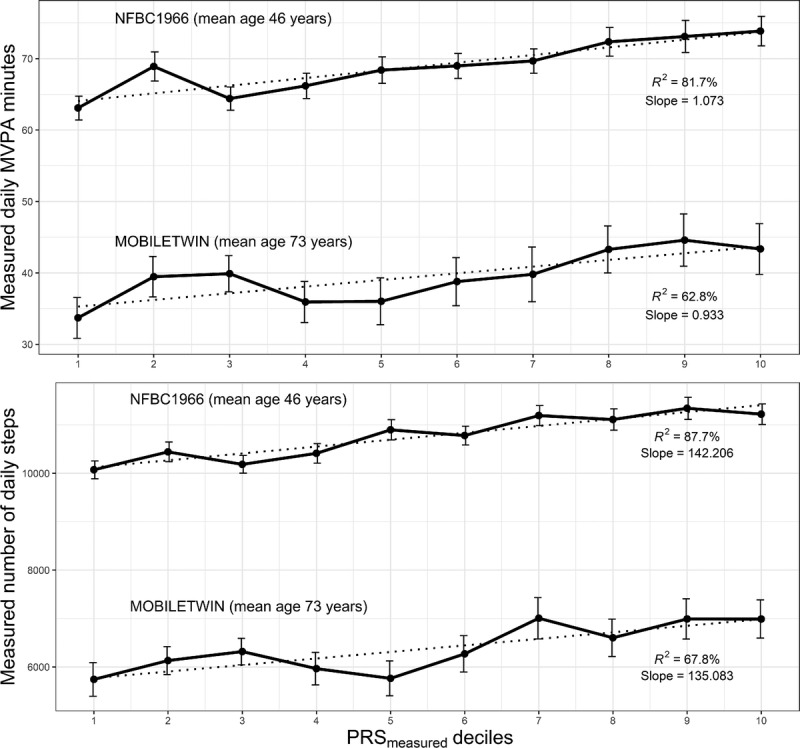
Means and SEMs of the objectively measured daily MVPA minutes (upper panel) and daily steps (lower panel) in the MOBILETWIN and NFBC1966 studies according to PRSmeasured deciles. PRSmeasured constructed based on the objective measurement of overall activity in UK Biobank. Dotted line is the trend line, and R2 is from the bivariate decile mean-decile model to illustrate trend linearity between the PA decile means and the PRS deciles.
Pedigree and SNP heritabilities in FTC
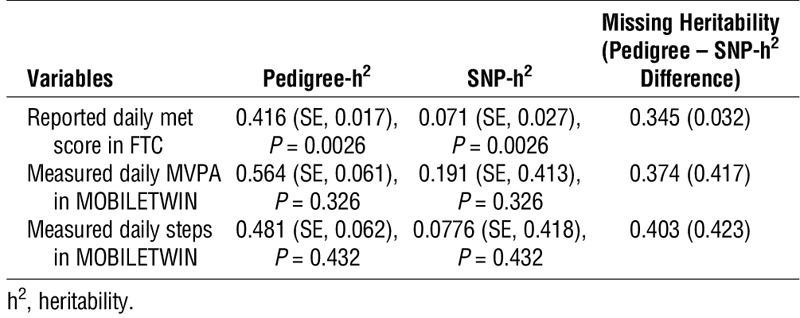
The pedigree heritabilities (37%–56%) and heritability explained by SNP in FTC are shown in Table 3. The results show that the missing (or hidden) heritability estimates (34%–40%) are higher than those of SNP heritabilities (7%–19%). The SNP-based heritabilities (SNP-h2 (LD score regression)) obtained from UK Biobank PA association results were 0.040 (SE, 0.002; P < 0.0001) for reported “number of days/week of moderate PA 10+ min” and 0.143 (SE, 0.008; P < 0.0001) for measured overall PA.
TABLE 3.
Pedigree, SNP, and missing heritabilities in FTC.
Questionnaire items versus PRSmeasured in explaining variation in measured PA in the MOBILETWIN study
For details of the results, see Supplemental Digital Content 1, questionnaire items versus PRSmeasured in explaining variation in objectively measured PA, http://links.lww.com/MSS/B929. Self-reported weekly walking or running distance outdoors accounted for the highest amount of variation in objectively measured MVPA (R2 = 44%) and daily steps (R2 = 36%). Body mass index, self-reported fitness level, mobility restricting disease, and PA category accounted for lower amounts of variation (R2 = 11%–23%). Multivariable model including age, sex, body mass index, and the physical fitness and activity related self-reports accounted for 57% of the variation in MVPA and 47% in daily steps. Adding the PRSmeasured into the models increased the proportion of total variation explained by the model only by 0.03% for MVPA and 0.31% for daily steps.
DISCUSSION
Our study adds to the understanding on the out-of-sample predictive value of PRS for PA using different self-report and objectively measured PA methods. PRS constructed on the basis of UK Biobank results explained statistically significantly PA in the Finnish cohorts, but the variation accounted for was small compared with some questionnaire-reported parameters such as physical fitness or health status.
Leisure-time or total PA is influenced by sex, body mass index, education, occupation, family commitments, physical fitness, occurrence of chronic diseases, environmental factors, and many other factors, whose influence also varies with age and with study area (14,40). These variables may mediate or modify the relationship of genetic liability to PA with the observed phenotype.
In addition to the aforementioned determinants of PA, there may be other reasons why our observed explanation rates of PRS constructed based on UK Biobank results to account for the variation in the Finnish cohorts were low. First, UK Biobank and the Finnish cohorts used differing methods both to assess self-reported and to assess objectively measured PA. Second, there may be genetic differences between the studied populations as we are predicting from a general European population to a genetic isolate (41). Third, as the SNP heritabilities were statistically significant but low compared with pedigree heritabilities, there may be hidden genetic factors not covered by the variants used in the calculation of PRS. Among the proposed explanations for this missing (hidden) heritability is the existence of many unidentified common variants with very small effect sizes, rare variants not captured by current genotyping platforms, structural variants, epistatic interactions, gene–environment interactions, or parent-of-origin effects (37,42,43). Interestingly, new research (43) showed that pedigree heritability for height and body mass index seemed to be fully recovered from whole-genome sequence data in the analysis including also rare variants. These hidden genetic factors may also contribute to the differences between the pedigree and SNP heritabilities for PA in our study, as well as the low explanation rates of the constructed PRS to explain variation in PA. SNP heritabilities obtained from UK Biobank association results using only the effect sizes were statistically significant but low compared with pedigree heritabilities obtained from the FTC subjects using the whole genome and phenotypic information together. The SNP heritabilities in FTC were at a similar or slightly higher level compared with those calculated for the UK Biobank PA variables using the same set of SNP (5).
Expectedly, there were simple questions that accounted for more of the variation in measured PA in the MOBILETWIN individuals than PA PRS calculated based on the findings from another cohort. The questionnaire items also contribute to the understanding why PRS accounts for only a small proportion of the variation of the PA measures.
PA measurements differed between the studied populations, which can be considered either as a limitation or as a strength in our study. From UK Biobank results, we selected clinically relevant self-report and objectively monitored PA variables based on which calculation of PA PRS was possible. Then we used clinically relevant variables (self-reported PA volume and measured daily MVPA minutes and steps) from the Finnish cohorts to study the associations of PRS and PA. Although all these indicators of PA describe PA volumes, they are not identical. We did not have access to the original UK Biobank data to construct new PA variables. However, to evaluate the additional value of combining information from the different self-reports, we performed a joint analysis of the UK Biobank GWAS results for reported moderate and reported vigorous activity using multitrait analysis of GWAS (MTAG). However, this new PRSreported MTAG predicted reported daily MET score in FTC only slightly better than the PRSreported with ΔR2 values of 0.32% versus 0.25% (for details, see Supplemental Digital Content 2, polygenic scoring on MTAG-estimated effects, http://links.lww.com/MSS/B930). The PRS constructed on the basis of UK Biobank self-reported PA questions did not predict statistically significantly objective measured PA variables in FTC (results not shown). The used objective PA measurements do not differentiate between leisure-time and work-related or household activities and thus indicate total PA volume in a different way than self-reports. Differing genetic factors may predict work-related and leisure-time PA. An additional difference between self-reported and objectively measured PA volumes is that self-reports take the subjective intensity into account, whereas accelerometer data usually are transformed to PA without considering the individual fitness level (44). Individuals with chronic diseases, low fitness, or advanced biological aging process usually move less and more slowly than healthy and high-fit individuals, although their PA intensity relative to their fitness level may be the same (44), which again may cause challenges in the analysis of genetic factors predicting PA. Despite the aforementioned factors, PRS constructed based on UK Biobank data predicted PA in the Finnish cohorts, which is good news for scientists who are applying Mendelian randomization methods and do not have the exactly same PA phenotypes in their cohorts as the UK Biobank study has.
In conclusion, PRS constructed based on UK Biobank results accounted for statistically significant but overall small proportion of the variation in PA in the Finnish cohorts. However, there were significant differences in the PA levels between the individuals who were in the highest PRS deciles and those in the lowest PRS deciles. In future studies, using identical questions or objective measurement methods to assess detailed PA behaviors and possibly including rare variants in the construction of PRS may increase the proportion of PA explained by the PRS, which may increase the usability of PRS for identifying individuals at risk for physical inactivity.
Supplementary Material
Acknowledgments
We wish to thank the FTC and NFBC1966 participants, and UK Biobank.
Financial support of this work included the Finnish Ministry of Education and Culture (grant OKM/56/626/2013 to U. M. K.), European Regional Development Fund (grant number 539/2010 A31592), Ministry of Education and Culture in Finland (grant numbers OKM/86/626/2014, OKM/43/626/2015, OKM/17/626/2016, OKM/54/626/2019 to R. K. and T. J.). NFBC1966 data collection at 46 y received financial support from the University of Oulu (grant no. 24000692) and Oulu University Hospital (grant no. 24301140). Phenotype and genotype data collection in the twin cohort has been supported by the Wellcome Trust Sanger Institute, the Broad Institute, ENGAGE – European Network for Genetic and Genomic Epidemiology, FP7-HEALTH-F4-2007, grant agreement number 201413, National Institute of Alcohol Abuse and Alcoholism (grants AA-12502, AA-00145, and AA-09203 to R. J. Rose and AA15416 and K02AA018755 to D. M. Dick), and the Academy of Finland (grants 100499, 205585, 118555, 141054, 264146, 308248, and 312073 to J. K.). J. K. has been supported by the Academy of Finland (grants 265240, 263278, 308248, 312073).
The funding sources had no roles in the study design, collection, analysis, or interpretations of the data or in the publication process.
The authors have no conflicts of interest to report. The results of the present study do not constitute endorsement by the American College of Sports Medicine. All authors declare that the results of the study are presented clearly, honestly, and without fabrication, falsification, or inappropriate data manipulation.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.acsm-msse.org).
REFERENCES
- 1.Kaprio J, Koskenvuo M, Sarna S. Cigarette smoking, use of alcohol, and leisure-time physical activity among same-sexed adult male twins. Prog Clin Biol Res. 1981;69(Pt C):37–46. [PubMed] [Google Scholar]
- 2.Lauerdale DS, Fabsitz R, Meyer JM, Sholinsky P, Ramakrishnan V, Goldberg J. Familial determinants of moderate and intense physical activity: a twin study. Med Sci Sports Exerc. 1997;29(8):1062–8. [DOI] [PubMed] [Google Scholar]
- 3.Stubbe JH, Boomsma DI, Vink JM, et al. Genetic influences on exercise participation in 37,051 twin pairs from seven countries. PLoS One. 2006;1:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.den Hoed M, Brage S, Zhao JH, et al. Heritability of objectively assessed daily physical activity and sedentary behavior. Am J Clin Nutr. 2013;98(5):1317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klimentidis YC, Raichlen DA, Bea J, et al. Genome-wide association study of habitual physical activity in over 277,000 UK Biobank participants identifies multiple variants including CADM2 and APOE. Int J Obes (Lond). 2018;42(6):1161–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doherty A, Smith-Byrne K, Ferreira T, et al. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat Commun. 2018;9(1):5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hara M, Hachiya T, Sutoh Y, et al. Genomewide association study of leisure-time exercise behavior in Japanese adults. Med Sci Sports Exerc. 2018;50(12):2433–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee N, Shi J, Garcia-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17(7):392–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khera AV, Chaffin M, Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19(9):581–90. [DOI] [PubMed] [Google Scholar]
- 11.Ainsworth B, Cahalin L, Buman M, Ross R. The current state of physical activity assessment tools. Prog Cardiovasc Dis. 2015;57(4):387–95. [DOI] [PubMed] [Google Scholar]
- 12.Sievänen H, Kujala UM. Accelerometry—simple, but challenging. Scand J Med Sci Sports. 2017;27(6):574–8. [DOI] [PubMed] [Google Scholar]
- 13.Hukkanen H, Husu P, Sievänen H, et al. Aerobic physical activity assessed with accelerometer, diary, questionnaire, and interview in a Finnish population sample. Scand J Med Sci Sports. 2018;28(10):2196–206. [DOI] [PubMed] [Google Scholar]
- 14.Kujala UM, Hautasaari P, Vähä-Ypyä H, et al. Chronic diseases and objectively measured physical activity among aged individuals—a cross-sectional twin cohort study. Ann Med. 2019;51(1):78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaprio J. The Finnish twin cohort study: an update. Twin Res Hum Genet. 2013;16(1):157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaprio J, Bollepalli S, Buchwald J, et al. The older Finnish Twin Cohort—45 years of follow-up. Twin Res Hum Genet. 2019;22(4):240–54. [DOI] [PubMed] [Google Scholar]
- 17.Waller K, Vähä-Ypyä H, Törmäkangas T, et al. Long-term leisure-time physical activity and other health habits as predictors of objectively monitored late-life physical activity—a 40-year twin study. Sci Rep. 2018;8(1):9400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oulu University Web site [Internet]. Oulu (Finland): Oulu University; [cited 2019 Oct 19]. Available from: https://www.oulu.fi/nfbc. [Google Scholar]
- 19.Niemelä M, Kangas M, Ahola RJ, et al. Dose-response relation of self-reported and accelerometer-measured physical activity to perceived health in middle age—the Northern Finland Birth Cohort 1966 Study. BMC Public Health. 2019;19(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabatti C, Service SK, Hartikainen AL, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41(1):35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kujala UM, Kaprio J, Sarna S, Koskenvuo M. Relationship of leisure-time physical activity and mortality: the Finnish twin cohort. JAMA. 1998;279(6):440–4. [DOI] [PubMed] [Google Scholar]
- 23.Suija K, Timonen M, Suviola M, Jokelainen J, Järvelin M, Tammelin T. The association between physical fitness and depressive symptoms among young adults: results of the northern Finland 1966 birth cohort study. BMC Public Health. 2013;13(1):535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hautala A, Martinmaki K, Kiviniemi A, et al. Effects of habitual physical activity on response to endurance training. J Sports Sci. 2012;30(6):563–9. [DOI] [PubMed] [Google Scholar]
- 25.UK Biobank Web site [Internet]. UK: UK Biobank; [cited 2019 Dec 3]. Available from: http://biobank.ctsu.ox.ac.uk/crystal/field.cgi?id=884. [Google Scholar]
- 26.Doherty A, Jackson D, Hammerla N, et al. Large scale population assessment of physical activity using wrist worn accelerometers: the UK Biobank Study. PLoS One. 2017;12(2):e0169649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loh PR, Daqnecek P, Palamara PF, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48(11):1443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das S, Forer L, Schonherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(19):1284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaneau O, Zagury J-F, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. [DOI] [PubMed] [Google Scholar]
- 32.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vilhjalmsson BJ, Yang J, Finucane HK, et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97(4):576–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borodulin K, Vartiainen E, Peltonen M, et al. Forty-year trends in cardiovascular risk factors in Finland. Eur J Public Health. 2015;25(3):539–46. [DOI] [PubMed] [Google Scholar]
- 35.UK Biobank Web site [Internet]. UK: UK Biobank; [cited 2019 Oct 19]. Available from: http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-phenotypes-for-337000-samples-in-the-uk-biobank. [Google Scholar]
- 36.International HapMap3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaitlen N, Kraft P, Patterson N, et al. Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 2013;9(5):e1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD score distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bauman AE, Reis RS, Sallis JF, et al. Correlates of physical activity: why are some people physically active and others not? Lancet. 2012;380(9838):258–71. [DOI] [PubMed] [Google Scholar]
- 41.Kerminen S, Martin AR, Koskela J, et al. Geographic variation and bias in the polygenic scores of complex diseases and traits in Finland. Am J Hum Genet. 2019;104(6):1169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11(6):446–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geddes L. Genetic study homes in on height’s heritability mystery. Nature. 2019;568:444–5. [DOI] [PubMed] [Google Scholar]
- 44.Kujala UM, Pietilä J, Myllymäki T, et al. Physical activity: absolute intensity versus relative-to-fitness-level volumes. Med Sci Sports Exerc. 2017;49(3):474–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


