Abstract
The vast majority of the proteins in nature are under thermodynamic control, consistent with the universally accepted notion that proteins exist in their thermodynamically most stable state. Yet, recently a number of examples of proteins whose fold is under kinetic control have come to light. Their functions and environments vary. The first among these are some proteases, discovered in the early 1990s. There, an N-terminal proregion is self-cleaved after the protein folded, leaving the remainder of the chain in a kinetically trapped state. A related scenario was observed for microcin J25, an antibacterial peptide. This peptide presents a trapped covalently knotted conformation. The third and the most recently discovered case is the multidrug-resistant transporter protein, P-glycoprotein. There, a synonymous ‘silent’ mutation leads to ribosome stalling with a consequent altered kinetically trapped state. Here we argue that in all three examples, the N-terminal plays the role of an intra-molecular chaperone, that is, the N-terminal conformation selects among all competing local conformations of a downstream segment. By providing a pattern, the N-terminal chaperone segment assists the protein folding process. If the N-terminal is subsequently cleaved, the protein can be under kinetic control, since it is trapped in a thermodynamically less-stable state.
1. Introduction
In vivo, via the evolution of natural selection, a newly synthesized polypeptide chain always folds spontaneously into a native functional conformation with or without help from molecular chaperones. In vitro, a small single domain protein with only one hydrophobic folding unit is always able to fold and unfold reversibly [1, 2]. These two observations indicate that the three-dimensional fold of a protein is determined by its one-dimensional amino acid sequence. Yet, folding is not a smooth downhill ride: the traps observed in the protein folding funnels explain experimental and theoretical observations such as two state folding scenario versus folding with intermediates and the effects of mutations which may raise or lower the barrier heights [3]. Nonetheless, some questions are still left unanswered. For example, we still do not completely understand how proteins are able to overcome barriers between local traps with similar thermodynamic stabilities [4, 5]. The hierarchical folding model partially resolves this question: evidence from folding intermediates and transition states suggests that folding begins locally. Local folded building blocks (or foldons) [5-10] with population times higher than alternate conformations hierarchically associate to form hydrophobic folding units [11-16]. Hydrophobic folding units with buried hydrophobic cores are capable of independent, thermodynamically stable existence. The hydrophobic folding units associate into domains, which in turn assemble to form a multi-domain protein fold or multi-subunit quaternary structure. However, the hierarchical model still leaves open the question of how the choice between competing local conformations is made. One way adopted by evolution is the use of an N-terminal ‘chaperone’ segment in the protein chain [17]. By providing a pattern and selecting between local competing conformations [12, 18], an N-terminal chaperone segment can have a key role in assisting the protein folding process. This may be particularly the case under ribosomal pausing regimes. On the other hand, if the N-terminal assists in the folding and is subsequently cleaved [19], the protein may be under kinetic control. Within this general framework, proteins with proregions provide examples where evolution made use of this fragment for specific functions in certain environments [20-22].
2. Mechanisms for increasing the chances of achieving a correctly folded protein
Several strategies have been adopted by evolution to increase the chance of achieving the ‘correct’ native folded local conformations. One of these is to enforce a sequential folding scenario. If building blocks adjacent in the primary sequence of the protein are also adjacent in the three-dimensional structure, then the protein is considered to fold in a sequential manner. In sequential rapidly folding proteins, contiguous fragments may be expected to form 3D contacts. Otherwise, it is a non-sequential folder. An in vivo sequential folding mechanism significantly reduces the possibility of misfolding [23, 24]. On the other hand, in the more ‘complex’ protein folds there are substantial contacts between non-sequential pieces of the chain. It is generally agreed that sequentially folded proteins are more frequent in eukaryotes versus prokaryotes with faster protein synthesis rates. In particular, sequential folding scenarios are also enforced by introducing ribosomal pause sites, allowing the upstream part of the protein chain to fold prior to the synthesis of the downstream part. If the stabilities of the competing conformations are not too different, pause events are expected to decrease the chance of misfolding.
A second strategy adopted by evolution is the use of molecular chaperones or chaperonins. Most studies have focused on two families: the chaperonins which include GroEL/GroES and the heat shock 70 proteins [25-27]. The chaperonins are large, multi-subunit allosteric proteins, with central cavities into which the misfolded proteins enter with subsequent ejection. On the other hand, molecular chaperones of the second family appear to act via binding to the misfolded protein surface. This family consists of small proteins, hence without a cavity large enough to hold misfolded proteins. In vitro, ~40% of the soluble proteins can interact with GroEL. In vivo, it has been estimated that the amount of GroEL in cells is sufficient to assist in the folding of a remarkably small percentage of proteins, only 2–5% under normal conditions [28]. The chaperones lead to macroscopic changes in the energy landscape, by lowering the barrier heights for unfolding, thus preventing misfolding.
This small percentage of theoretically estimated proteins whose folding is assisted by chaperonins, and the paradigm that all the information specifying the three-dimensional shape of the protein is encoded in its sequence, emphasizes that the sequence should also encode mechanisms to increase the chance of reaching the native state. Thus, the question arises whether there are elements in the sequence which fulfill the role of an ‘intra-molecular chaperone’ (IMC). The role of an IMC has been recognized for the proregions. We argue that the IMC is a common mechanism; with the proregion being a private case of an IMC.
An IMC is likely to be located at the N-terminal of the molecule. As the first to be translated, it may form a template for the folding of other sequence fragments with which it interacts. Through conformational selection [12, 18, 29], it chooses between competing local downstream conformations. Although all fragments are required for a protein to yield its complete three-dimensional fold, formation and interaction of one or a few of these may be essential for the protein to fold correctly. This may be particularly true for large proteins that fold in a complex manner. To constitute an IMC, a fragment may be expected to fulfill three conditions: it should be in contact with several building blocks in the structure; it is likely to be inserted between sequentially connected building blocks, mediating their tertiary interactions; and in its absence, the remaining building blocks are likely to misassociate. Under such circumstances, the conformations of the assembled building blocks are likely to remain native-like. In the vast majority of the cases, an IMC is not cleaved. Previously, we have identified two IMC candidates: yeast adenylate kinase and dihydrofolate reductase. If the IMC is cleaved following folding the protein may be under kinetic control as in the proregion case.
3. Thermodynamically and kinetically controlled protein conformations
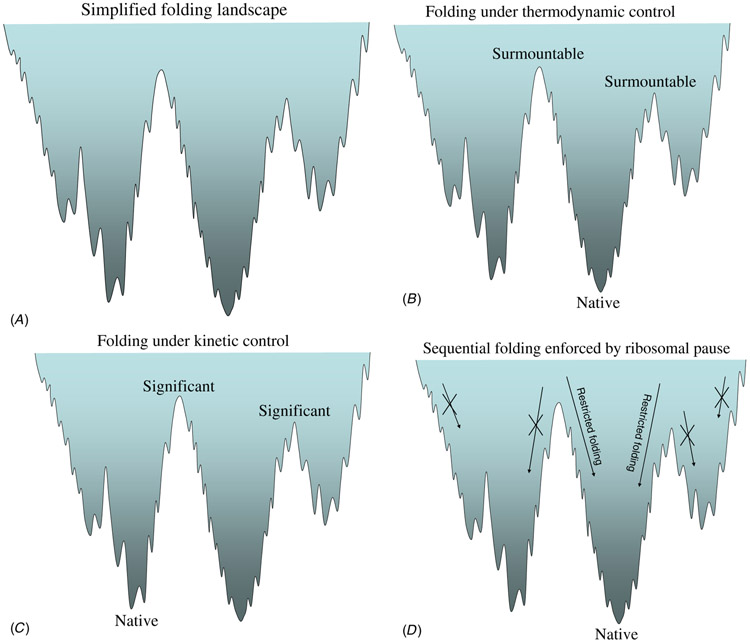
By far, most native protein structures are at their thermodynamically most stable state. In terms of the free energy landscape (figure 1(A)), the native functional state is at the bottom of the funnel, with an energy gap between this conformation and all others (figure 1(B)). These proteins are under thermodynamic control. Considering protein folding as a reaction, under equilibrium conditions the driving force for the formation of the folded protein product reaches the thermodynamically most stable state. If the molecule is trapped in a local minimum, over time it will climb out of the barrier to reach its lowest energy state. Yet, if the barrier is high, the molecule may get stuck over physiological timescales. If the trapped conformation reflects the native state of the molecule, then this molecule is under kinetic control (figure 1(C)). Under such circumstances, the activation energy to reach the transition state is high, thus with low chances to be reached. While thermodynamic control folds pathway independent, kinetic control is path dependent. In general, kinetic control is not a robust mechanism, since barrier heights are sensitive to environmental conditions such as concentration, pH, temperature, ionic strength and mutations. However, if the barrier is either sufficiently high, or aided by covalent linkage, or supported by some surface or the membrane, or caught by certain conditions, and if the native conformation has sufficient stability, it may be the active biological state. A recent study by Agard and his colleagues provided an insight into how kinetic control can be attained [30]. Comparison of the Nocardiopsis alba Protease A (NAPase), an acid-resistant, kinetically stable protease, with a neutrophilic homolog, the alpha-lytic protease (alphaLP), has shown that multiple salt-bridges in the domain interface of alphaLP were relocated to outer regions of NAPase. These suggest a mechanism of acid stability in which acid-sensitive electrostatic interactions are rearranged to similarly affect the energetics of both the native state and the unfolding transition state. In another case [31], a conserved β-hairpin proved crucial to the kinetic stability (figure 2(A)).
Figure 1.
Simplified free energy landscape to illustrate the folding complexity. Here, the folding landscape is depicted as a funnel shape containing three subdivided funnels with each distinguished by distinct folding pathways (figure 1(A)). If the free energy barriers between sub-funnels are surmountable and the protein native conformation is the corresponding global minimum, there is no misfolding but just various folding rates. Hence, this is the case referred to as protein folding under thermodynamic control (figure 1(B)). On the other hand, if the barriers are significant, misfoldings do happen when a folding path leads into a misfolded funnel. However, if the trapped conformation reflects the native protein, then this kind of protein folding is under kinetic control (figure 1(C)). A sequential folder aided by ribosomal pause to increase the chances of achieving a correctly folded protein is simply to maintain its folding pathways within the correct funnel (figure 1(D)).
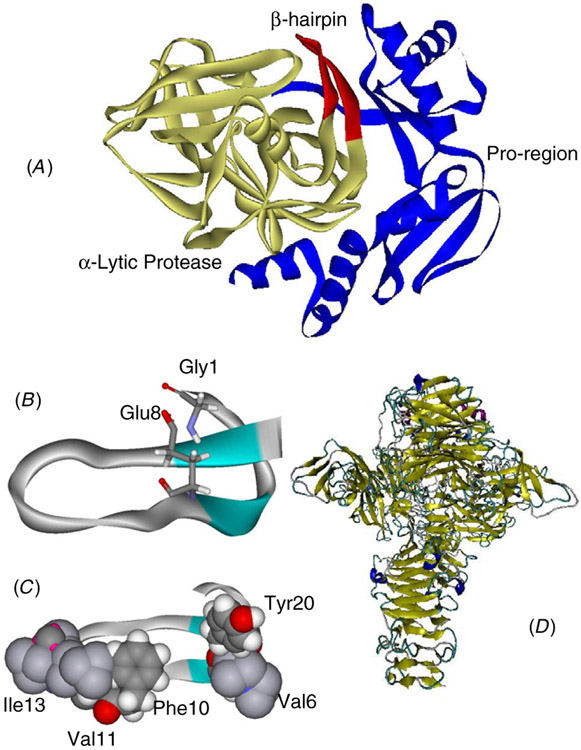
Figure 2.
(A) The complex of α-lytic protease (yellow ribbon) with its proregion (blue ribbon); the β-hairpin (red ribbon) enhances the kinetic stability of α-lytic protease (PDB code: 4pro). (B) Two chemically linked residues in microcin J25 (PDB code: 1pp5). (C) Hydrophobic surface in microcin J25. (D) The dominating β-sheet and β-barrel structure of the endosialidase trimer (PDB code: 1v0e).
An IMC-assisted folding favors certain pathways; thus, it reduces the conformational search and the chances of misfolding. The IMC changes the energy landscape by lowering the barrier heights. In the vast majority of the cases, the final product is a protein at its thermodynamically favored state. On the other hand, if subsequently cleaved and the barrier to unfolding is high, or constrained by the environment, the protein is said to be under kinetic control. Below, we provide three examples of functionally specified kinetic control mediated by an N-terminal IMC: the first is the well-known protease case, the second is a fascinating microbial inhibitor locked by covalent linkage and the third is an uncleaved N-terminal IMC of the human multidrug-resistant transporter, constrained by its membrane environment.
3.1. The serpin family
It has been well known that some proteins are synthesized with an extra fragment (a proregion) at their amino termini [19]. This fragment which is essential for correct folding is degraded after the protein folds. Such cases [32] typically occur in serine proteases, such as subtilisin, α-lytic protease, aqualysin from bacteria and carboxypeptidase Y from yeast. These fragments act as inhibitors, covering the active sites of the enzymes, and hence it is essential that they be cleaved and digested for the enzyme to be functional. However, if the fragment is cleaved in the construct prior to protein synthesis, the newly synthesized chain misfolds; if the cleaved fragment is mixed with the remainder of the chain in a solution, a correctly folded protein is obtained; and if the fragment is added to a solution containing the already misfolded chain, the chain converts to its native fold. A mutation that has been engineered in this 70-residue fragment resulted in an alternately folded subtilisin molecule [33-35]. Hence, it appears that the N-terminal building block segment mediates the interactions of other building blocks. After the active enzyme conformation is obtained, it is self-cleaved, exposing the enzyme active site.
3.2. Antibacterial peptide microcin J25
The antibacterial peptide microcin J25 (MccJ25) inhibits bacterial transcription by binding to the RNA polymerase nucleotide uptake channel. MccJ25 is a 21-residue peptide. Interestingly, the backbone of the first residue at the N-terminus of the peptide, glycine is covalently linked to the side chain of glutamic, which is residue 8 in the sequence, forming a covalently sealed circle. The C-terminal is threaded through this circle (figure 2(b)). The covalently locked protoknot is very stable, with a marked resistance to carboxypeptidases and denaturation (8M urea at 95 °C). Phenylalanine at position 19 is below the circle and Tyrosine at position 20 is above it interacting with the circle residues and locking the protoknot structure. The looped-out portion consisting of residues 9–18 contains two short β-strands, β1/β2 which hydrogen bond to each other to form a small antiparallel sheet. As pointed out by Bayro et al [36], this structure implies that the unligated structure must already exist at least transiently in this conformation to allow the covalent linkage to take place. Interestingly, the precursor of MccJ25 is a 58-residue protein. In vivo, the 37 residue fragment at the N-terminal is cleaved and residues 1 and 8 are linked by two enzymes, mcjB and mcjC, though the enzymatic reactions are still not understood. Since when unligated, the population time of a 21-residue peptide in this conformation can be expected to be short, the N-terminal proregion assists in the folding. While no structure exists for the 58-residue precursor, we may expect that the 37-residue N-terminal nests against the exposed hydrophobic surface of the upper side of the threaded peptide (figure 2(c)). When cleaved and ligated, the 21-residue MccJ25 peptide is kinetically trapped. Thus, there are three possible states: the peptide with the proregion, the peptide without the proregion and not ligated, and the peptide with the proregion and ligated. In all cases, the global minimum is the same but the population time is high only in the presence of the proregion or when ligated.
4. What are the functional advantages of kinetic traps?
The α-lytic protease and MccJ25 are both kinetically trapped. In both, the functional conformation was reached with the assistance of an N-terminal proregion that was subsequently cleaved. Nevertheless, there are also differences: in the α-lytic protease, in the absence of a proregion the preferred conformation differs from the functional state; in MccJ25, even in the absence of the proregion the presence of the β-sheet and the hydrogen bonds between Phe19 and Tyr20 and the side chains and backbone of the ring [36, 37] lead to a similar preferred conformational state, however, with a lower population time. For a 21-residue peptide, a fluctuating conformation is expected, since the length is too short to permit a strong hydrophobic core. The two cases also differ from the free energy landscape standpoint. While in the case of the α-lytic protease the cleavage of the proregion does not change significantly the free energy landscape, this is not the case for MccJ25; in MccJ25, the covalent ligation stabilizes the inhibitor, increasing its population time (figure 1).
Kinetic control is sensitive to the environment; thus, it is not a robust mechanism for retaining a native functional state and is rarely used [38]. For the cases discussed here, for the molecules to be under kinetic control, they initially fold into the thermodynamically most stable conformation specified by the entire sequence, and subsequently a segment is cleaved. In the case of the α-lytic protease, the kinetically trapped molecule which in vivo exists under harsh conditions faces high barriers. The function necessitated the cleavage of the N-terminal IMC which acted as an inhibitor, covering the active site. In the case of MccJ25, it is kinetically trapped by the covalent ring closure, which increased the population time of an already preferred state. This leads to a more potent inhibitor yet with a small peptide with an elongated shape sticking out of a lid, an optimal shape for blocking the polymerase channel.
5. The mechanisms of chaperonins and an N-terminal IMC are different
Chaperonins have two roles: to unfold misfolded molecules and to prevent aggregation. If the protein can fold spontaneously, as in the case of the E. coli DHFR (and at low concentration, the eukaryotic DHFR) a chaperonin may help by preventing aggregation; if however it does not fold spontaneously in vitro, the chaperonin may fulfill both roles. Hence, there is a difference between a chaperonin and a proregion. For the α-lytic protease whose proregion has been removed from the nascent chain prior to folding, a chaperonin will be of little use. Had the α-lytic protease been synthesized without the proregion, and a chaperonin was present instead, the native state would not be reached. In terms of the energy landscapes and the folding funnels, an IMC acts by lowering the barrier of a misfolded conformation. It aids the trapped conformation climb out of its minima well. By being chain-linked, this goal is achieved much more efficiently than otherwise. In an IMC, the change in the energy landscape of the protein is both macroscopic and microscopic: IMCs not only lower the barriers allowing the opening of non-native interactions, but assist directly in the folding by providing a template.
An IMC works not via binding to any intermediate conformation of its adjoining building block(s) and thereby inducing it to undergo a conformational change to the native conformation. Rather, just as in inter-molecular binding in general, the binding of the amino-terminus IMC to its adjoining building blocks is via conformational selection [16, 17, 39-42]. Among all available building block conformations, the ones that bind are those whose association is most favorable. In doing so, the equilibrium shifts in the direction of the native building block conformations. Such a proposition is consistent with the effect observed by Shinde et al [33-35]. The mutation they have introduced does not cause a direct, induced, conformational change in the protease. Instead, the point mutation causes a change in the landscape, reflecting the effect of the altered conformation of the amino-terminal building block fragment. The mutant building block conformer preferentially selects different conformers from the populations of conformers of its adjoining building blocks, resulting in an altered subtilisin structure. However, after the proregion containing the mutation is cleaved, with time, we may expect the trapped altered structure to undergo a conformational change, overcoming the barriers, to the thermodynamically more stable native conformation.
Mechanistically, the roles of uncleaved intra-molecular chaperones and proregion segments are similar. However, proteins with uncleaved intra-molecular chaperones are under thermodynamic control; proteins folding with proregion assistance are under kinetic control.
6. Co-translational folding and the case of a transporter protein
The co-translational folding pathway with altered kinetics and intermediate states reduces the chances of misfolding [13, 43]. For the ribosome stalling case [44], the upstream IMC part of the chain folds into its native state prior to the synthesis of the downstream part of the chain. If the downstream part of the chain can fold into two competing conformations, the already folded IMC selects the more favorable conformation even if that conformation is slightly higher in energy. Population shift will propagate the binding reaction. On the other hand, had the ribosome not stalled and the IMC still unfolded, the competing slightly more stable conformation could prevail. In such cases, the barrier between the two protein conformations is not expected to be high and the conformational change between the two is not expected to be large. Nevertheless, it might be sufficiently high over physiological timescales, leading to altered (or diseased) conformations and functions. This might be particularly the case if attached to a surface or embedded in an environment which will support the conformation and increase the barrier. Such a situation was recently observed in a protein transporter, the human P-gp, the multidrug-resistant gene product. There, Kimchi-Tsarfaty et al [45] observed that silent mutations exchanging frequent codons by rare ones, coding for the same amino acid, led to a functional change. Presumably, the rare codon led to ribosome stalling, and hence a scenario as described above. Alternatively, in a more likely scenario the codon substitution led to a change in the mRNA structure, eliciting the ribosome stalling event. Either way, ribosome stalling led to an altered protein conformation [46]. The conformational change in the multidrug-resistant protein was observed through a different pattern of proteolytic cleavage, recognition by conformation-sensitive antibody and some difference in drug binding. On the other hand, some drugs showed a similar pattern, indicating that the conformational change was limited.
7. The C-terminal segment and the contribution of β-sheets to protein kinetic stability
Above, we focused on N-termini intra-molecular chaperones. Yet, it behooves us to note that intra-molecular chaperone functions were also identified for several C-terminal segments. Even though these C-terminal propeptides have not been as thoroughly studied as the N-terminal regions, their function in assisting the folding of other protein segments is documented.
One of the earliest reports of the role of the C-terminal intra-molecular chaperone is the 372-amino acid (aa) precursor of Caldariomyces fumago chloroperoxidase (CPO) which undergoes two proteolytic processing events: removal of a 21-aa N-terminal signal peptide and of a 52-aa C-terminal propeptide [47]. Later, the sucrase domain at the C-terminal end of the sucrase-isomaltase enzyme complex was also found to have a chaperone role. The folding and function of the sucrase domain are independent of the presence of isomaltase. In contrast, isomaltase needs the sucrase domain to mature and be functional. The sucrase domain competes with the nearby non-specific molecular chaperone calnexin at the endoplasmic reticulum [48]. A C-terminal domain chaperone was identified for Aminopeptidase A, a type II integral membrane glycoprotein responsible for the conversion of angiotensin II to angiotensin III in the brain. Deletion of the C-terminal domain causes the N-terminal domain to be retained in the endoplasmic reticulum as an unfolded protein bound to calnexin, and the maturation and enzymatic activity are abolished [49]. The C-terminal domain of endosialidases controls the folding and assembly of endosialidases by increasing the unfolding barrier and trapping the mature trimer in a kinetically stable conformation [50]. Thus, the C-terminal domain of endosialidases not only helps monomeric protein folding, but also mediates the assembly of the endosialidase trimer, similar to the N-terminal chaperone role assisting in oligomerization (for example, the 20 S proteasome [51], caspase-3 [52] and VWF [53]).
Amyloid structures are dominated by a cross-β-sheet [54]. Remarkably, similar to the kinetically stable amyloid, the β-sheet structure has been identified as a contributor to protein kinetic stability in general [41, 55]. While there are no systematic studies, examination of several intra-molecular chaperones and related proteins illustrate the β-sheet contributions. One example is the microcin J25 mentioned earlier. The proregions and matured proteins of both the α-lytic protease and subtilisin also have large β-sheet contributions. In the case of the well-characterized α-lytic protease, the formation and position of a β-hairpin (residue 118–130, red ribbon, figure 2(A)) in the C-terminal domain contribute to the large energy barrier [31]. This β-hairpin forms a continuous β-sheet with the proregion. The β-propeller and β-barrel contribute to the high kinetic stability of the endosialidases [50] (figure 2(D)).
8. Summary and outlook: depending on the environment, evolution adapts folding scenarios to achieve an optimal function
Current data suggest that evolution does not frequently adopt kinetic control as a regulatory mechanism. Kinetic control rests on barrier heights, which are sensitive to conditions. Over time, it is expected that the protein will flip to its thermodynamically most favored state. Nonetheless, it is interesting to observe where such control scenarios have been adopted by evolution and for what purpose.
The case of the proteases, discovered by David Agard and his colleagues in the early 1990s, is fascinating: there, kinetic control appears to have been selected by evolution for extreme environmental conditions where it holds advantages and special structural features have been implemented to withstand the lack of robustness in this type of control [30]. In this case, the thermodynamically most stable conformation of the mature protease differs from the functional state. Thus, over time, it would flip to the thermodynamically favored state; however, this timescale is irrelevant physiologically. The second case is that of the covalently locked protoknot of the antibacterial peptide microcin J25 (MccJ25). This structure is likely to have been selected by evolution due to its high stability, with a marked resistance to carboxypeptidases and denaturation. Even though here too the N-terminal proregion is cleaved, unlike the protease case, the protoknot is in its thermodynamically favored state; however, without the covalent linkage the stability of this conformation is low. P-gp, the multidrug-resistant protein, provides a yet third scenario (figure 1(D)): first, no cleavage of the N-terminal proregion and, second, ribosome pausing leads to an altered state. However, the conformational change is small. Here, the kinetic control leads to conformations lying nearby on the folding funnel bottom allowing them to bind to a broader range of ligands nicely fitting the P-gp function: the P-gp drug-resistant protein is known to bind to a broad range of drugs.
Proteins are critical for all biological processes. To ensure a proper function, they need to have the ‘correct’ structure in the ‘right’ population, with a ‘favored’ energy landscape. All the necessary information is encoded in the sequence; it is fascinating to observe how nature has engineered the protein folding mechanism and control, tailoring it to specific functional needs. The challenge is to understand folding mechanisms as they relate to a function such that they can be employed in protein and drug design.
Acknowledgments
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Glossary
- Chaperone.
Chaperones are proteins that assist the non-covalent folding/unfolding of other proteins.
- Kinetic control.
Proteins under kinetic control are trapped in a local minimum state, with high barriers separating it from the global thermodynamic minimum.
- Proregion.
A proregion is a sequence at the N-terminal of the sequence that helps in the folding of the protein, and is subsequently cleaved by proteases (proteolytic enzymes).
- Ribosome pausing.
The ribosomes do not translate the mRNA to proteins at uniform rates. It is well known that there are sites on the mRNA where the ribosome pauses. Consequently, depending on the pause timescales, the parts which are already synthesized may fold.
- Hierarchical folding.
The hierarchical protein folding scenario is one of the models for protein folding. In this model, the first step involves folding of local sequence elements; next, the folded elements hierarchically associate until the entire protein is folded.
References
- [1].Anfinsen CB 1973. Principles that govern folding of protein chains Science 181 223–30 [DOI] [PubMed] [Google Scholar]
- [2].Privalov PL 1996. Intermediate states in protein folding J. Mol. Biol 258 707–25 [DOI] [PubMed] [Google Scholar]
- [3].Rothwarf DM and Scheraga HA 1996. Role of non-native aromatic and hydrophobic interactions in the folding of hen egg white lysozyme Biochemistry 35 13797–807 [DOI] [PubMed] [Google Scholar]
- [4].Englander SW, Mayne L and Krishna M M G 2007. Protein folding and misfolding: mechanism and principles Q. Rev. Biophys 40 287–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bedard S et al. 2008. Protein folding: independent unrelated pathways or predetermined pathway with optional errors Proc. Natl Acad. Sci 105 7182–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bedard S et al. 2008. The foldon substructure of staphylococcal nuclease J. Mol. Biol 376 1142–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Inaba K, Kobayashi N and Fersht A R 2000. Conversion of two-state to multi-state folding kinetics on fusion of two protein foldons J. Mol. Biol 302 219–33 [DOI] [PubMed] [Google Scholar]
- [8].Panchenko AR et al. 1997. The foldon universe: a survey of structural similarity and self-recognition of independently folding units J. Mol. Biol 272 95–105 [DOI] [PubMed] [Google Scholar]
- [9].Bhardwaj A et al. 2008. Foldon-guided self assembly of ultra stable protein fibers Protein Sci. 17 1475–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ferreiro DU. et al. The energy landscapes of repeat-containing proteins: topology, cooperativity, and the folding funnels of one-dimensional architectures. PLoS Comput. Biol. 2008;4:e1000070. doi: 10.1371/journal.pcbi.1000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tsai CJ and Nussinov R 1997. Hydrophobic folding units derived from dissimilar monomer structures and their interactions Protein Sci. 6 24–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tsai CJ et al. 1999. Folding funnels, binding funnels, and protein function Protein Sci. 8 1181–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tsai CJ, Maizel JV and Nussinov R 1999. Distinguishing between sequential and nonsequentially folded proteins: implications for folding and misfolding Protein Sci. 8 1591–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tsai CJ and Nussinov R 2001. Transient, highly populated, building blocks folding model Cell Biochem. Biophys 34 209–35 [DOI] [PubMed] [Google Scholar]
- [15].Tsai CJ and Nussinov R 2001. The building block folding model and the kinetics of protein folding Protein Eng. 14 723–33 [DOI] [PubMed] [Google Scholar]
- [16].Tsai CJ et al. 2001. Protein folding: binding of conformationally fluctuating building blocks via population selection Crit. Rev. Biochem. Mol. Biol 36 399–433 [DOI] [PubMed] [Google Scholar]
- [17].Ma BY, Tsai CJ and Nussinov R 2000. Binding and folding: in search of intramolecular chaperone-like building block fragments Protein Eng. 13 617–27 [DOI] [PubMed] [Google Scholar]
- [18].Boehr DD and Wright PE 2008. Biochemistry. How do proteins interact? Science 320 1429–30 [DOI] [PubMed] [Google Scholar]
- [19].Bryan PN 2002. Prodomains and protein folding catalysis Chem. Rev 102 4805–15 [DOI] [PubMed] [Google Scholar]
- [20].Baker D and Agard D A 1994. Kinetics versus thermodynamics in protein-folding Biochemistry 33 7505–9 [DOI] [PubMed] [Google Scholar]
- [21].Baker D, Sohl JL and Agard DA 1992. A protein-folding reaction under kinetic control Nature 356 263–5 [DOI] [PubMed] [Google Scholar]
- [22].Baker D et al. 1993. The role of pro regions in protein folding Curr. Opin. Cell. Biol 5 966–70 [DOI] [PubMed] [Google Scholar]
- [23].Netzer WJ and Hartl FU 1997. Recombination of protein domains facilitated by co-translational folding in eukaryotes Nature 388 343–9 [DOI] [PubMed] [Google Scholar]
- [24].Helenius A 1994. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic-reticulum Mol. Biol. Cell 5 253–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].England J, Lucent D and Pande V 2008. Rattling the cage: computational models of chaperonin-mediated protein folding Curr. Opin. Struct. Biol 18 163–9 [DOI] [PubMed] [Google Scholar]
- [26].Saibil HR 2008. Chaperone machines in action Curr. Opin. Struct. Biol 18 35–42 [DOI] [PubMed] [Google Scholar]
- [27].Sharma S et al. 2008. Monitoring protein conformation along the pathway of chaperonin-assisted folding Cell 133 142–53 [DOI] [PubMed] [Google Scholar]
- [28].Lorimer GH 1996. A quantitative assessment of the role of chaperonin proteins in protein folding in vivo FASEB J. 10 5–9 [DOI] [PubMed] [Google Scholar]
- [29].Lange OF et al. 2008. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution Science 320 1471–5 [DOI] [PubMed] [Google Scholar]
- [30].Kelch BA et al. 2007. Structural and mechanistic exploration of acid resistance: kinetic stability facilitates evolution of extremophilic behavior J. Mol. Biol 368 870–83 [DOI] [PubMed] [Google Scholar]
- [31].Truhlar SME and Agard DA 2005. The folding landscape of an alpha-lytic protease variant reveals the role of a conserved beta-hairpin in the development of kinetic stability Proteins Struct. Funct. Bioinf 61 105–14 [DOI] [PubMed] [Google Scholar]
- [32].Jacob R, Peters K and Naim HY 2002. The prosequence of human lactase-phlorizin hydrolase modulates the folding of the mature enzyme J. Biol. Chem 277 8217–25 [DOI] [PubMed] [Google Scholar]
- [33].Shinde U and Inouye M 1995. Folding pathway mediated by an intramolecular chaperone—characterization of the structural-changes in pro-subtilisin-E coincident with autoprocessing J. Mol. Biol 252 25–30 [DOI] [PubMed] [Google Scholar]
- [34].Shinde U and Inouye M 1995. Folding mediated by an intramolecular chaperone—autoprocessing pathway of the precursor resolved via a substrate assisted catalysis mechanism J. Mol. Biol 247 390–5 [DOI] [PubMed] [Google Scholar]
- [35].Shinde UP, Liu JJ and Inouye M 1997. Protein memory through altered folding mediated by intramolecular chaperones Nature 389 520-2 [DOI] [PubMed] [Google Scholar]
- [36].Bayro MJ et al. 2003. Structure of antibacterial peptide microcin J25: a 21-residue lariat protoknot J. Am. Chem. Soc 125 12382–3 [DOI] [PubMed] [Google Scholar]
- [37].Wilson KA et al. 2003. Structure of microcin J25, a peptide inhibitor of bacterial RNA polymerase, is a lassoed tail J. Am. Chem. Soc 125 12475–83 [DOI] [PubMed] [Google Scholar]
- [38].Kelch BA and Agard DA 2007. Mesophile versus thermophile: insights into the structural mechanisms of kinetic stability J. Mol. Biol 370 784–95 [DOI] [PubMed] [Google Scholar]
- [39].Kumar S et al. 2000. Folding and binding cascades: dynamic landscapes and population shifts Protein Sci. 9 10–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kumar S et al. 2001. Protein folding and function: the N-terminal fragment in adenylate kinase Biophys. J 80 2439–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Xia K et al. 2007. Identifying the subproteome of kinetically stable proteins via diagonal 2D SDS/PAGE Proc. Natl Acad. Sci 104 17329–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ma BY et al. 2002. Multiple diverse ligands binding at a single protein site: a matter of pre-existing populations Protein Sci. 11 184–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hsu STD et al. 2007. Structure and dynamics of a ribosome-bound nascent chain by NMR spectroscopy Proc. Natl Acad. Sci 104 16516–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sivan G and Stein OE 2008. Regulation of mRNA translation during cellular division Cell Cycle 7 741–4 [DOI] [PubMed] [Google Scholar]
- [45].Kimchi-Sarfaty C et al. 2007. A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity Science 315 525–8 [DOI] [PubMed] [Google Scholar]
- [46].Tsai CJ et al. 2008. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima J. Mol. Biol 383 281–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Conesa A et al. 2001. C-terminal propeptide of the Caldariomyces fumago chloroperoxidase: an intramolecular chaperone? FEBS Lett. 503 117–20 [DOI] [PubMed] [Google Scholar]
- [48].Jacob R, Purschel B and Naim HY 2002. Sucrase is an intramolecular chaperone located at the C-terminal end of the sucrase-isomaltase enzyme complex J. Biol. Chem 277 32141–8 [DOI] [PubMed] [Google Scholar]
- [49].Rozenfeld R et al. 2004. The C-terminal domain of aminopeptidase A is an intramolecular chaperone required for the correct folding, cell surface expression, and activity of this monozinc aminopeptidase J. Biol. Chem 279 43285–95 [DOI] [PubMed] [Google Scholar]
- [50].Schwarzer D et al. 2007. Characterization of a novel intramolecular chaperone domain conserved in endosialidases and other bacteriophage tail spike and fiber proteins J. Biol. Chem 282 2821–31 [DOI] [PubMed] [Google Scholar]
- [51].Li X et al. 2007. beta-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint EMBO J. 26 2339–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Feeney B and Clark AC 2005. Reassembly of active caspase-3 is facilitated by the propeptide J. Biol. Chem 280 39772–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rosenberg J B et al. 2002. The role of the D1 domain of the von Willebrand factor propeptide in multimerization of VWF Blood 100 1699–706 [DOI] [PubMed] [Google Scholar]
- [54].Ma B Y and Nussinov R 2002. Molecular dynamics simulations of alanine rich beta-sheet oligomers: insight into amyloid formation Protein Sci. 11 2335–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Manning M and Colon W 2004. Structural basis of protein kinetic stability: resistance to sodium dodecyl sulfate suggests a central role for rigidity and a bias toward beta-sheet structure Biochemistry 43 11248–54 [DOI] [PubMed] [Google Scholar]