Abstract
A wealth of specialized neuroendocrine command systems intercalated within the hypothalamus control the most fundamental physiological needs1,2. Nevertheless, a developmental blueprint integrating molecular determinants of neuronal and glial diversity along temporal and spatial scales of hypothalamus development remains unresolved3. Here, we combine single-cell RNA-seq on 51,199 cells of ectodermal origin, gene regulatory network (GRN) screens in conjunction with GWAS-based disease phenotyping and genetic lineage reconstruction to show that 9 glial and 33 neuronal subtypes are generated by mid-gestation under the control of distinct GRNs. Combinatorial molecular codes arising from neurotransmitters, neuropeptides and transcription factors are minimally required to decode the taxonomical hierarchy of hypothalamic neurons. Differentiation of GABA and dopamine but not glutamate neurons relies on quasi-stable intermediate states with a pool of GABA progenitors giving rise to dopamine cells4. An unexpected abundance of chemotropic proliferation and guidance cues commonly implicated in dorsal (cortical) patterning5 was found in the hypothalamus. Particularly, Slit/Robo loss-of-function impacted both the production and positioning of periventricular dopamine neurons. Overall, we uncover molecular principles shaping the developmental architecture of the hypothalamus and show how neuronal heterogeneity is transformed into a multimodal neural unit to endow a virtually infinite adaptive potential throughout life.
Concentration of a kaleidoscope of neuroendocrine cell modalities into a minimal brain volume within the hypothalamus is achieved by sometimes only as few as 1,000s of neurons coding essential hormonal output. Therefore, diversification of neuronal subtypes, rather than the numerical expansion of single progenies6,7, might underpin the success of vertebrate evolution to refine metabolic and adaptive capacity. Functional versatility at the level of individual neuroendocrine output neurons is coded by the coincident presence of neurotransmitters and neuropeptides1. Therefore, interrogation of the molecular and positional diversity of hypothalamic neurons by morphological, circuit and endocrine analyses continues to mount a significant challenge. The recent introduction of single-cell RNA-sequencing (scRNA-seq)6,8,9 produced precise molecular insights into the existence of glutamate, GABA, dopamine and even ‘mixed’ neuronal phenotypes4. However, a question of paramount importance that remains systematically unexplored (but see Refs.3,10,11) is how cellular subtypes emerge, migrate, and differentiate during hypothalamus development for neuroendocrine readiness to ensue by birth.
In contrast to a handful of transcription factors (TFs) being sufficient to mark anatomical footprints in cortical structures with a layered organization6,8, the intercalated nature of nuclei poses a formidable challenge to establish an anatomical template within the hypothalamus. Even more so, the breadth of endocrine command neurons and their ability to rapidly undergo cell-state switches (that is, to up-regulate specific hormones or neuropeptides in an on-demand fashion) suggest that what is considered terminally differentiated in the adult brain is in fact a neuronal anagram primarily dictated by the neuronal circuit orchestrating a specific endocrine modality. Therefore, we sought to resolve molecular determinants of ectodermal progenies advancing towards terminal neuroendocrine differentiation. By using a time series of scRNA-seq across critical periods of intrauterine and postnatal hypothalamus development in mouse we read out combinatorial codes for GABA, GABA-derived dopamine and glutamate neurons, catalogued GRNs (regulons) and their dynamic transitions during neurogenesis, directional migration and morphogenesis, and elucidated local chemotropic cues that define anatomical constraints of the hypothalamus.
Results
Emergence of ectoderm-derived cell pools
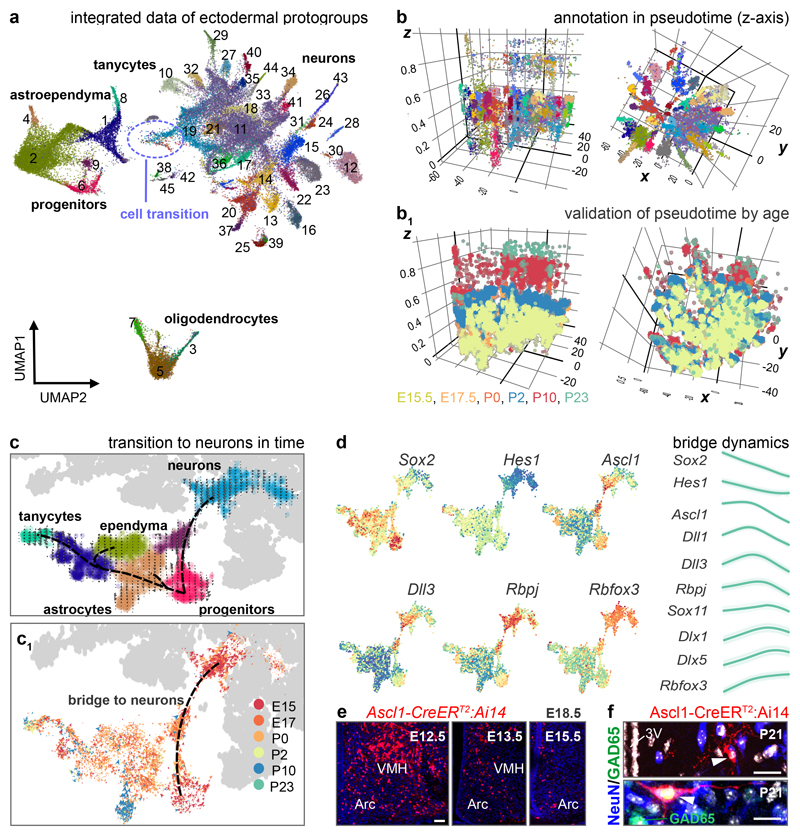
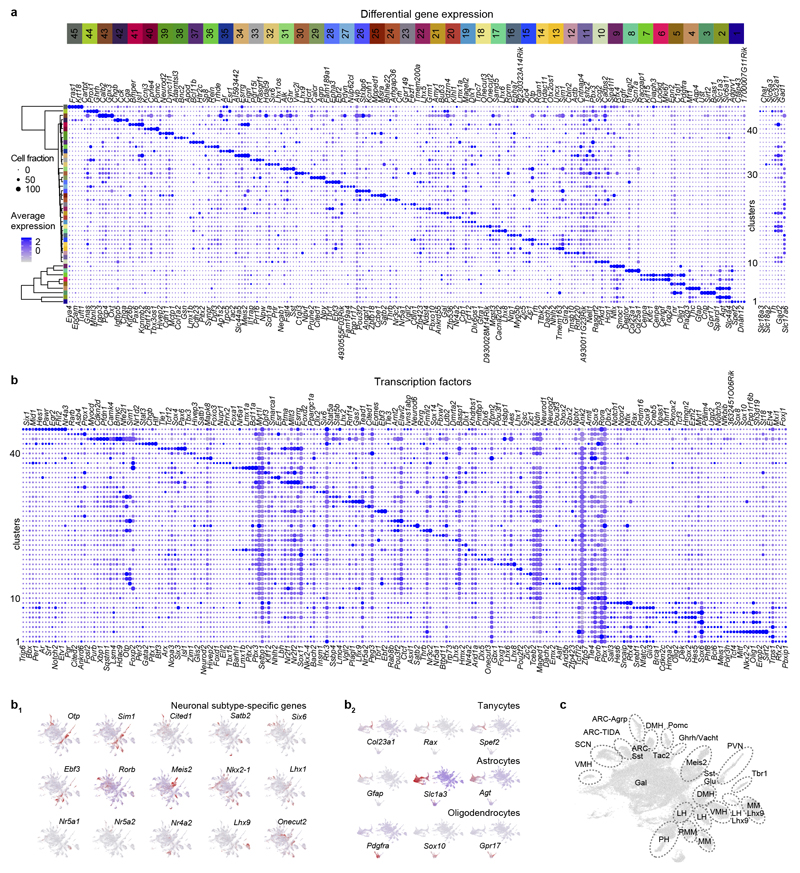
We addressed the differentiation programs for hypothalamic cell pools by parallel scRNA-seq on 51,199 dissociated cells at embryonic days (E)15.5 (8,290), E17.5 (11,213), at birth (7,492), and postnatal day (P)2 (12,824), P10 (8,965) and P23 (2,415; Online Methods and Supplementary Note). Overall, proto-groups of progenitors (2), tanycytes (2), astroependymal cells (2), immature oligodendrocytes (3), cells of the pars tuberalis (3) and neurons (33; Figure 1a), reflecting diversity in adult hypothalamus4,8,9, were specified by differentially-expressed TFs (Figure ED1) during development (Figure 1b,b1).
Figure 1. Developmental diversification of hypothalamic cell lineages.
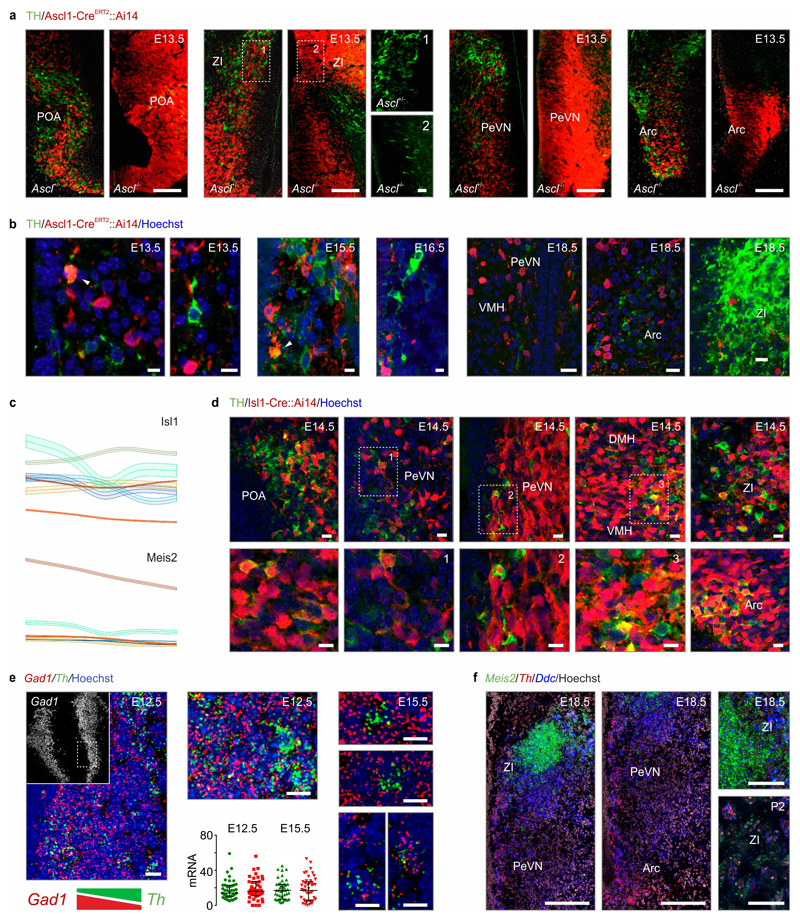
(a) UMAP plot of 51,199 cells of ectodermal origin and integrated by canonical correlation analysis (CCA) to achieve a hypothetical continuum reflecting the progressive attainment of cell identities. Walktrap in iGRAPH distinguished non-mature cells (#11, #19) and neurons (31 proto-groups) at the end of each developmental trajectory. (b,b1) Schemes illustrating the conformity of alignment and clustering in pseudotime (z axis, calculated independently) with biological age (b1). (c,c1) UMAP and PAGA representation of progenitors, glia and immature neurons (‘bridge cells’). RNA velocity12,13 transformed multi-dimensional PAGA data to developmental trajectories. The color of junctions accord with groups (c) and age (c1). Note that a ‘cell bridge’ linking progenitors and immature neurons encompasses cells of early developmental stages even though all time points are minimally represented therein. (d) Imputed expression for the cell groups shown in (c) and pseudotime trajectories of differentiation into neurons starting from a Sox2+ state. (e) Genetic tracing of Ascl1+ progenitors (induction at successive time points) in VMH and Arc. (f) Ascl+ progenitor-derived neurons (arrovheads) generated postnatally. Scale bars = 65 μm (e), 20 μm (f).
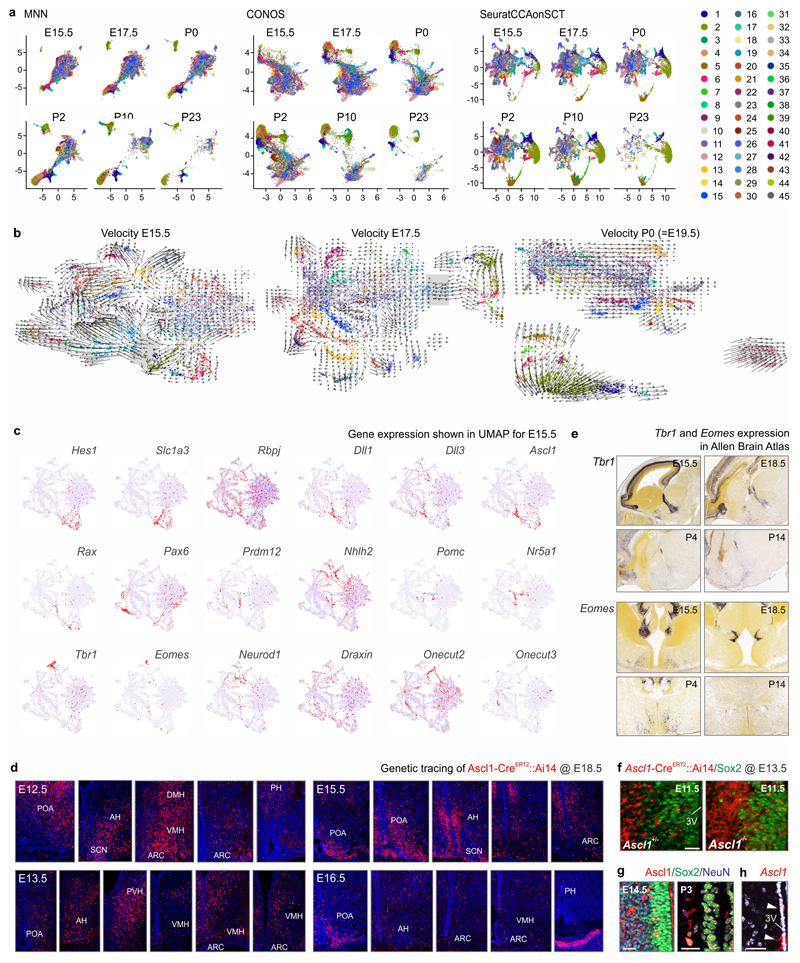
We then asked when and by which progenitors the various cell types are generated. The dynamics of gene expression in hypothalamic progenitors (Figure 1c,c1) to produce astrocytes, ependyma, tanycytes and neurons fit a pseudotime scale on a multidimensional integrated dataset12 (Figure 1c-d), including a bifurcation in cell transition toward glial subtypes or neuronal fates (Figure 1a,c, ED2a,b) that peaked between E15.5-E17.5 (Figure 1c1). RNA-velocity13 (as well as PAGA12; Supplementary Note) demonstrated that the number of neuroblasts (‘bridge cells’) tails off as a factor of age with an appreciable rupture of this cell continuum by birth (Figure ED2b). Semi-supervised analysis of 327 genes for enrichment (for all raw data see DOI:10.6084/m9.figshare.11867889) highlighted that the progression of ‘bridge cells’ relied on the dominance of genes related to the regulation of pluripotency (Sox2), neural stem cell differentiation (Hes1, Ascl1, Rbpj, Dll1/Dll3 for Notch signalling)14,15, Erk signaling (Sox11), neuronal migration (Dlx1/2/5/6) and morphogenesis (Rbfox3; Figure 1d)16,17. Additionally, scRNA-seq suggested an alternative and embryonically restricted (<E15.5) pathway of hypothalamic neurogenesis that centered on Tbr1+/Eomesodermin (Eomes)+ progenitors that reside at the thalamus-hypothalamus dorsal boundary (an Ascl1- territory) and contribute multiple diencephalic neuronal subtypes (Figure ED2d,e). The proposed waves of neurogenesis by self-renewing progenitors and early neuroblasts that ubiquitously express Ascl1 along the 3rd ventricle (Figure 1d) were shown in Ascl1-CreERT2::Ai14 mice at E18.5 with recombination induced during the E12.5-E16.5 period (Figure 1e) and validated in Ascl1-/- mice presenting a restricted cohort of Sox2+ immature precursors (Figure ED2f). tdTomato+ progenies in the many hypothalamic subregions confirmed neurogenesis during mid-gestation with gradual declined after E16.5 (Figure 1e, ED2d). In support of postnatal neurogenesis we show that Sox2+ precursors persist in the wall of the 3rd ventricle and generate progenies that progress through Ascl1+ and Rbfox3+ (NeuN) stages (Figure 1f, ED2g,h).
Intermediate states for GABA neurons
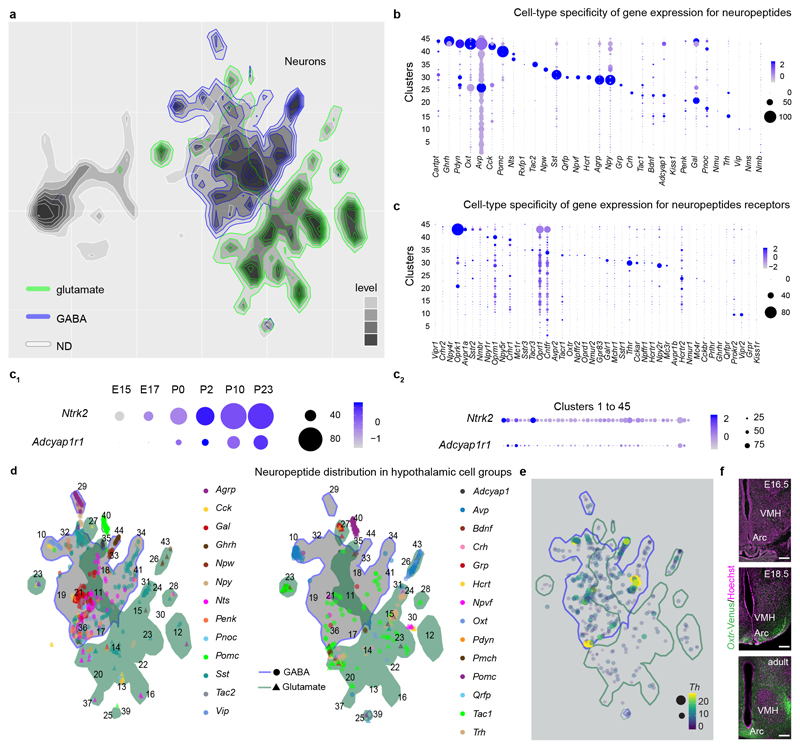
Within our integrated dataset, ~47% of all cells committed to the neuronal lineage were in immature states (#11, #19; Figure 1a and 2a) prior to progressing towards final differentiation, as suggested by the expression of homeobox genes thought to determine GABA identities (#11: Foxg1 and Nkx2-3; #19: Sox2/11, Gsx1/2, E2f1, Arx and Pbx3; Figure ED1). Specifically, cluster #19 contains a continuum of neuroblasts (‘bridge cells’), with a normalized contribution of 36.5% (E15.5), 34.6% (E17.5), 13.2% (birth), 9.2% (P2), 4.8% (P10) and 1.6% (P23), and remains separated from other clusters despite our re-partitioning efforts (Supplementary Note). Cluster #11 is composed of immature neurons that are largely homogeneous with low-level differential gene expression, yet express rate-limiting enzymes and transporters for GABA neurotransmission (Figure ED1a, 3a). When re-partitioning these data, immature GABA neurons were re-assigned to phenotypically-stable groups, emphasizing the intermediate nature of cluster #11. These findings contrast glutamate neurons, which immediately appear as differentiated and spatially segregated groups (Figure 1a) without an intermediate cell pool being detected. Thus, GABA and glutamate neurons seem to adopt principally different developmental programs with immature GABA cells, rather than pre-formed GABA lineages18, serving as precursors for terminal differentiation.
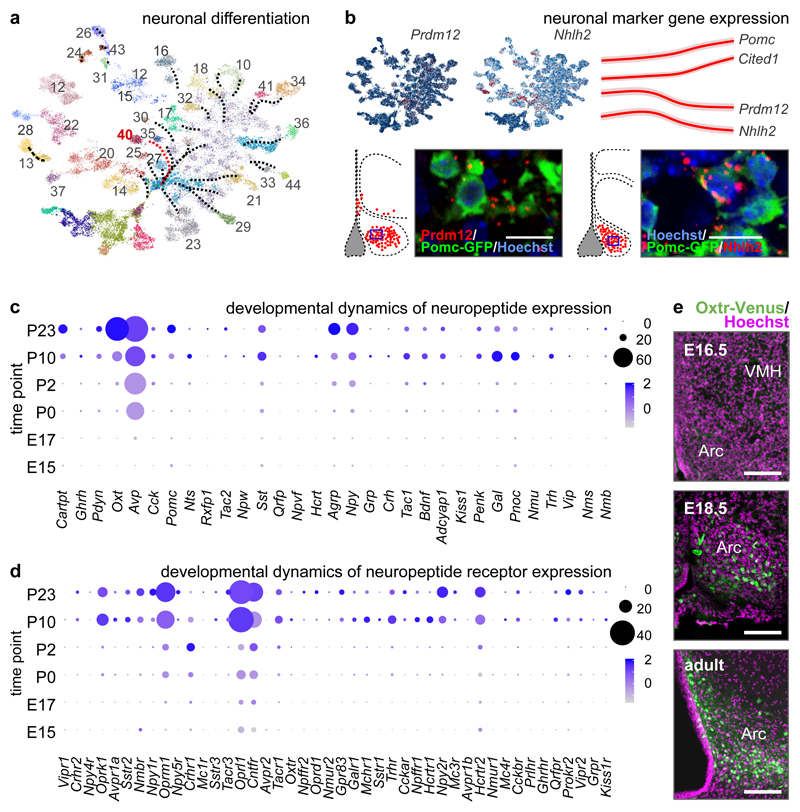
Figure 2. Neuronal differentiation in the hypothalamus.
(a) Cellular clusters from Figure 1a (without #38, #42 and #45 oligodendrocytes) represented as a graph-like map upon transforming UMAP embedding with the PAGA method12 to assess cell differentiation trajectories. Red arrow specifies the trajectory for proopiomelanocortin (Pomc; #40) neurons. (b) Prdm12 and Nhlh2 expression (top left) and their developmental dynamics (pseudotime) relative to Pomc and Cited1, a transcriptional co-activator specifying neurons of the arcuate nucleus (top right). Data in pseudotime were scaled6. Prdm12 and Nhlh2 expression in Pomc+ neurons was validated by in situ hybridization in Pomc-GFP mice (bottom). Blue rectangles in topographical maps show the location of images at single-cell resolution. Dynamics of gene expression for neuropeptides (c) and their receptors (d) during hypothalamus development. Data were shown as dot plots. (e) Developmental mapping of hypothalamic Oxtr expression in OxtrVenus/+ mice. Scale bars = 200 μm (e), 12 μm (b).
TF and neuropeptide codes of diversity
TF-mediated cell-autonomous differentiation programs are key to neuronal specification6,18. Hence, we screened TFs that distinguished hypothalamic cell clusters. We applied a supervised approach sampling stationary states (that is, genes that are spatially-restricted in both pre- and postnatal brains; Figure 1d, ED1) and integrated stages of fate-transition and branching-off of differentiated neurons. Arginine-vasopressin (Avp)+ (#26) and oxytocin (Oxt)+ (#43) magnocellular as well as parvocellular neuroendocrine clusters destined to the paraventricular nucleus (PVN; including Trh+ and Crh+ cells (#24)) exhibited spatial convergence (Figure 2a), and were separated by differentially expressed genes from their non-PVN counterparts: e.g., Mbnl3, Pgf, Irs4, Gpr101, Nr3c2 and Agtr1 demarcated Trh+ neurons in the PVN, whereas Trh+ neurons prospectively populating the dorsomedial hypothalamus (#15) labelled for Onecut2/Onecut3 and Cartpt, and mapped distantly.
Subsequently, we selected pro-opiomelanocortin (Pomc)+ neurons to test if scRNA-seq-based temporal profiling of gene expression allows for the reconstruction of neuronal differentiation. Besides cataloguing Pomc-specific TFs (Figure 2a), we show that, e.g., Prdm12 and Nhlh2, both placed by in situ hybridization into Pomc-GFP+ neurons (Figure 2b), are transiently expressed at early developmental stages followed by gradual expressional decay (pseudotime; Figure 2b). In contrast, Cited1 expression was restricted late gestation when neuronal morphogenesis commences19. Cumulatively, our scRNA-seq data reliably resolved neuronal fate progression along both pseudotime and real-time scales.
Beside fast neurotransmission by GABA and glutamate, dopamine and neuropeptides are chief signalling units in the adult hypothalamus20. Here, we assigned 27 neuropeptides slecifically to GABA, glutamate and dopamine neuronal subtypes (Figure 2c,d and ED3b-e). Our data demonstrate a transient increase in e.g., Sst, Tac1, Bdnf, Adcyap1, Pnoc, Nmu, Trh in juvenile mice (P10)21 (Figure 2c) along with the rapid induction of their cognate receptors during the early postnatal window (Figure 2d). Finally, we used OxtrVenus/+ mice to demonstrate that the onset of Venus expression (used as a surrogate for Oxtr) in e.g., Pomc+ neurons in Arc22, DMH and VMH is ~E18 with a gradual increase postnatally (Figure 2e, ED3f). These findings substantiate the precision of scRNA-seq in resolving hormone receptor expression in even the smallest neuronal contingents.
Regulons typify cell type specificity
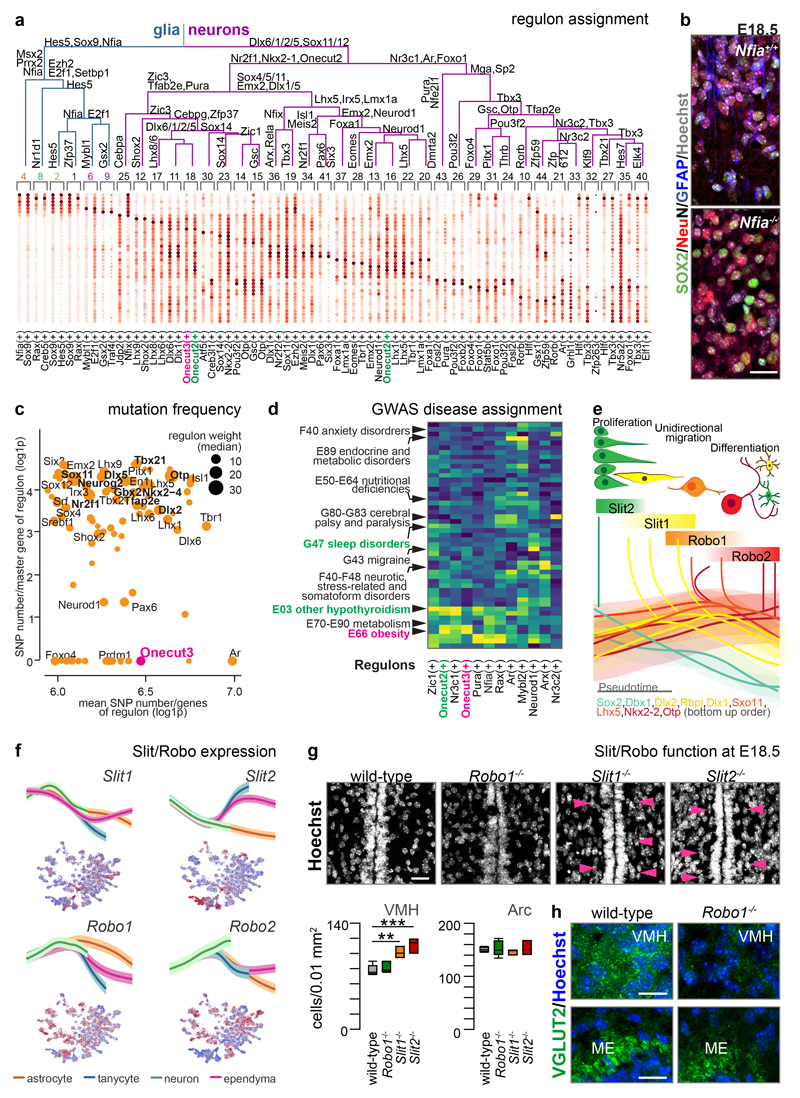
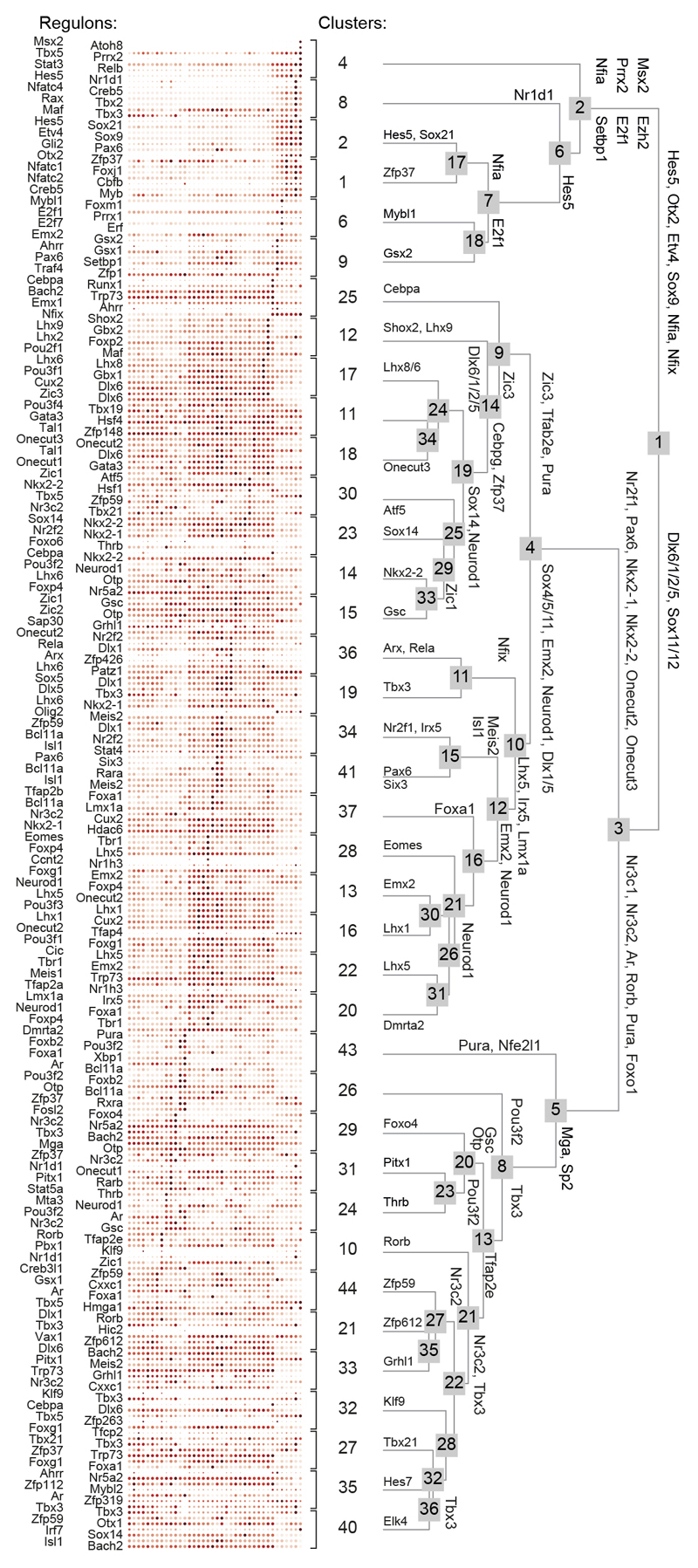
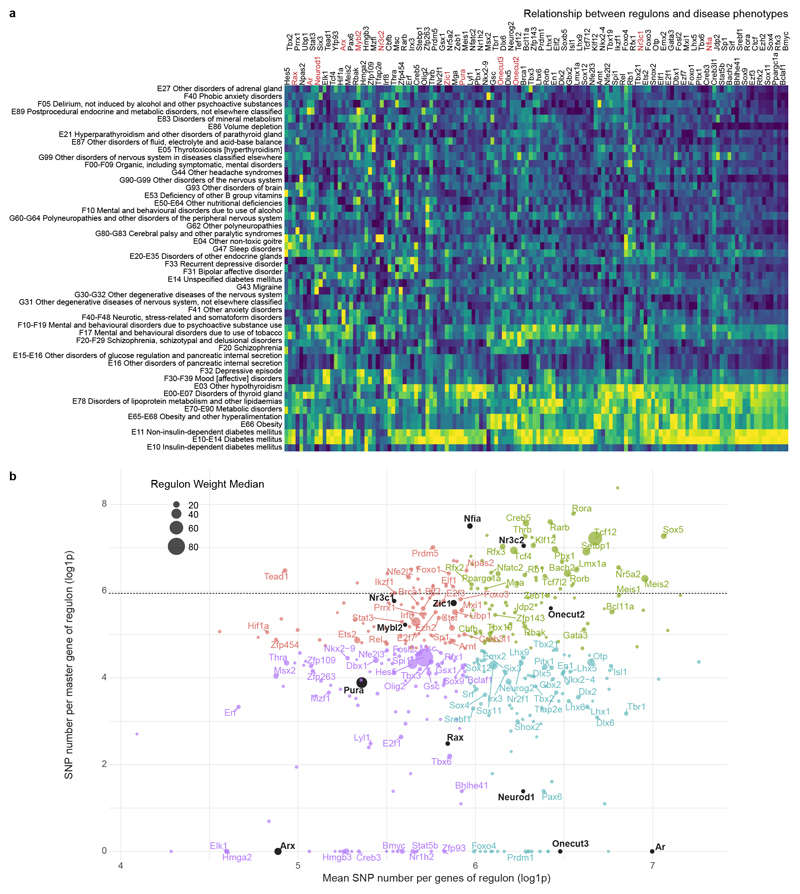
Cellular identities are shaped by developmentally-timed GRNs (‘regulons’) centred on a ‘master’ TF activating its targets through DNA-binding and transcriptional induction23. Therefore, we assigned regulons to each ectodermal cluster by combining 1,962 TF-ChIP-seq for gene interactions and our scRNA-seq data23 (Figures 3a, ED4), yielding positive interactions within 395 active regulons and estimating their prevalence per cell. Pleiotropic regulons with the highest representation define to which major cell lineage a progenitor was destined to: e.g., Hes5, Sox9 and Nfia for prospective astroglia vs. Dlx(1/2/5/6) and Sox11/12 for neurons (Figure 3a). Accordingly, Nfia-/- mice showed impaired formation of hypothalamic tanycytes and astrocytes but not neurons at E18.5 (Figure 3b). Subordinate cell group-specific regulons define select cell clusters (Figure 3a). Co-existent regulons at all levels (e.g. the Pura and Fosl2 regulons in Oxt+ neurons) can produce combinatorial codes for cellular fate decisions (Figure 3a shows at least three, from general to particular developmental processes).
Figure 3. GRNs (regulons), including chemotropic guidance cues, in ectoderm-derived hypothalamic cells.
(a) A dendrogram of regulons for each cell cluster estimated in Figure 1b. TFs at each branching point of the dendrogram are representative for subjacent groups of regulons. Onecut TFs were color-coded. (b) Gfap, Sox2 and Rbfox3 in the anterior arcuate nucleus of wild-type and Nfia-/- mice at E18.5. (c) Ratio between mutability for master genes and their downstream targets in regulons. A quadrant highlighting the Onecut3 regulon is shown (see also Figure ED5). (d) Heat-map of associations between selected regulons and clinical phenotypes (see also Figure ED5). (e) Illustration how regulons that chiefly control Slit/Robo signalling contribute to neuronal differentiation in the hypothalamus. (f) Slit1/2 and Robo1/2 expression in pseudotime (top; means ± s.d.) and on an integrated dataset (bottom). Blue-to-red scale codes for low-to-high RNA expression. (g) Slit1-/- and Slit2-/- mice present increased cell density at the level of the ventromedial hypothalamus (VMH; arrowheads) but not arcuate nucleus (Arc) relative to Robo1-/- and wild-type littermates. (h) In turn, glutamatergic (Slc17a6/Vglut2) synaptogenesis is reduced in the VMH of Robo1-/- mice by E18.5. The median eminence (ME), devoid of Slit2 expression, lacked any phenotype. Scale bars = 20 μm (b,h,i).
We then evaluated the robustness and penetrance of hypothalamic regulons by testing if their mutations (at all gene levels) manifest as clinical perturbations by focusing on metabolic and psychiatric diseases9,24 in the GWAS of the UK biobank (738 phenotypes; Figure ED5a). We adapted existing methods in adult9 by replacing stationary cell identities with regulons. By selecting multiple genes that co-define particular regulons as input we significantly reduced selection bias due to strongly deleterious mutations (‘survivorship bias’25). Regulons driven by pro-neurogenic genes were characterized by the lowest rate of mutated master genes (Figure ED5b) with the Foxo4 (#29) and Onecut3 (#18) clusters completely depleted in mutations (Figure 3a,c). Meanwhile, the Onecut2/3 regulons correlated positively with the incidence of obesity (Figure 3d).
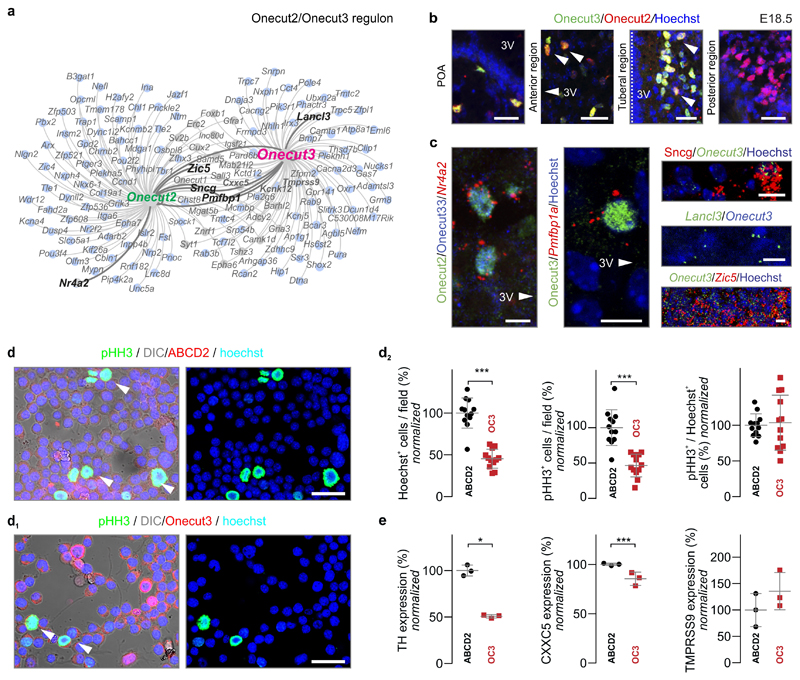
Next, we confirmed that Nr4a2, Ptfmb1a, Sncg, Lancl3 and Zic5, genes in the mutual Onecut2/Onecut3 regulon, co-existed with Onecut3 in differentiated neurons yet restricted to PeVN cell groups (Figure ED6a-c). Additionally, Onecut3 overexpression in Neuro2A cells in vitro caused the cessation of cell proliferation (Figure ED6d-e), substantiating its role in neuronal fate progression. Cumulatively, these data assign regulon screens as a prime strategy to functionally annotate hypothalamic neurons and predict their linkages to metabolic (and psychiatric) disorders. The identification of a spatially-restricted Onecut3+ regulon to the PeVN suggests that neurons specified by the Onecut3 regulon could be sensitive to developmental signalling cues that shape midline structures.
Regulons instruct chemotropic signalling
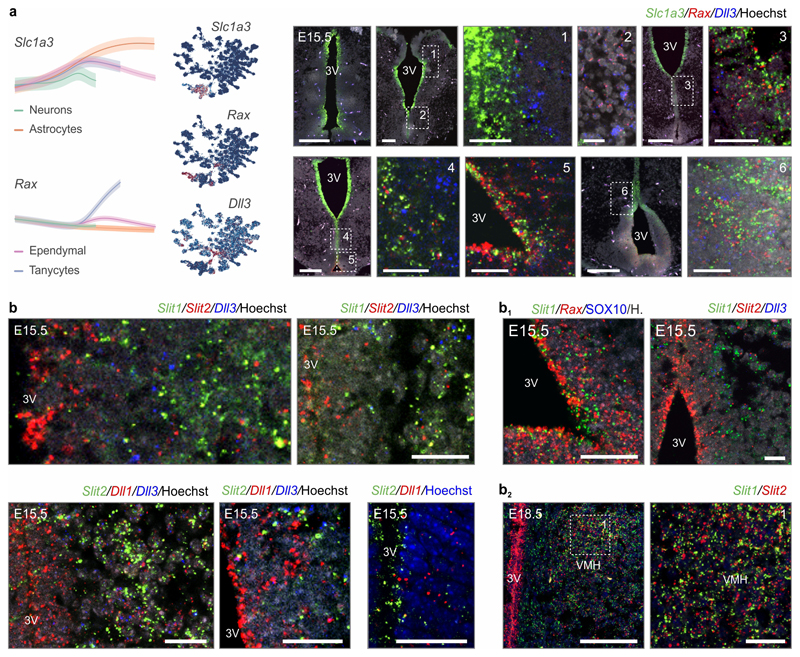
Within laminated structures, progenitor-to-committed progeny transitions occur in a sequential unidirectional order7,26. We asked if similar gene sets14,16,17, intercellular interactions and spatial arrangements could apply to the non-laminar hypothalamus. Early-expressed glial genes (Hes1, Fabp7, Slc1a3, Vim) marked progenitors (e.g. Sox2, Dll1/3) in the innermost (‘ventricular’) zone of the 3rd ventricle at E14.5-E15.5 (Figure 1c and ED7a,b). Committed progeny then unidirectionally distanced themselves laterally (Figure ED2d-g) and expressed protogenes for neuronal migration (Dlx1/2/5/6, Rbfox3; Figure 1d). Plotting regulons along developmental age assigned Sox2 to progenitors (#6,#9), Sox11 to ‘bridge neurons’ (#19) and Dlx1/2 to both ‘bridge’ and immature neurons (#11,19)); Figure 3e), confirming function determination16. These data suggest the temporal and spatial segregation of regulons in an onion skin-like layered configuration (Figure 3e).
Next, we interrogated the complexity of chemotropic signalling systems27 that facilitate neuronal positioning and differentiation, with evidence for Eprinh-ErB, Cbln1/Cbln2, semaphorin-plexin-neuropilin, neurotrophin (Bdnf, Gdnf, Cntf), draxin, netrin (Ntng1/Ntng2) and endocannabinoids. Unexpectedly, we noted the widespread expression of the Slit(1,2)/Robo(1,2) signalling cassette (Figure 3e,f), which dictates direct (vs. indirect) neurogenesis in antagonism with Dll126, and long-range axonal patterning in dorsal (cortical) structures5. Reconstruction of mRNA expression placed Slit2 into neural progenitors at early developmental stages (Figure 3f). Conversely, pseudotime analysis suggested Slit1 expression to dominate early in postmitotic neuroblasts (Figure 3f). Coincidently, Robo2 expression defined a developmental trajectory specific for neurons (Figure 3f). To genetically tie temporal variations in Slit/Robo signalling to neuronal differentiation, we show that major regulons include Slit2 ligand synthesis for cell proliferation and gliogenesis (in the Sox2/Dbx1/Rfx2/3/Myc regulons specific to glia and progenitors) and Slit1 for neuronal migration and morphogenesis (Dlx1/2/Rbpj regulon). Meanwhile, the Sox11 regulon is a chief determinant of both Robo1 and Robo2 expression as early as in ‘bridge neurons’ (#11, #19). At the level of terminal differentiation, Lhx5/Emx2/Lhx1 and Nkx2-1/Otp/Isl1 controlled Robo1 and Robo2 expression, respectively (Figure 3e). In situ hybridization confirmed the reciprocal distribution of Slit2 and Slit1, with the former being restricted to ventricular progenitors (Figure ED7b). Moreover, Slit2 (and to a lesser extent Slit1) gene expression concentrated in the VMH by E17.5 or later (Figure ED7b1). Indeed, an increased cellular density in the subventricular zone at the level of the VMH was seen in both Slit1-/- and Slit2-/- mice, whereas Robo1 inactivation was ineffective (Robo1-/-; Figure 3g). Instead, Robo1-/- mice, in which Slit ligands no longer act as repulsive axon guidance cues5, showed a reduced density of Slc17a6/Vglut2+ synapses in the VMH relative to wild-type controls in contrast to unchanged levels at the median eminence, an area devoid of Slits (Figure 3h). In sum, our data suggest the involvement of Slit/Robo signaling in hypothalamic neurodevelopment, pointing to conserved Slit/Robo functions in ventral brain areas.
Molecular identity of dopamine neurons
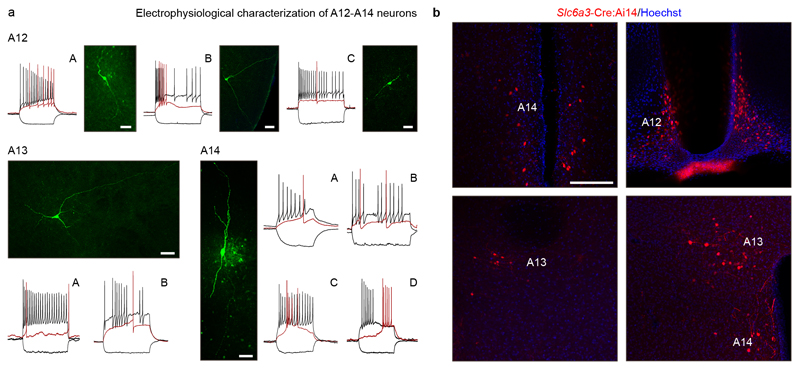
Finally, we asked how molecularly distinct subtypes of phenotypically uniform neurons arise during hypothalamus development. We took advantage of the at least 9 morphologically and electrophysiologically distinct subtypes of parvocellular dopaminergic neurons in A12 (Arc; 3 subtypes), A13 (zona incerta; 2 subtypes) and A14 territories (PeVN, 4 subtypes; Figure ED8) of Thgfp and Slc6a3-Ires-Cre::Ai14 mice; also noting their segregation from midbrain dopamine neurons chiefly regulated by Lmx1a/b and Nr4a2 (Figure 3a)28.
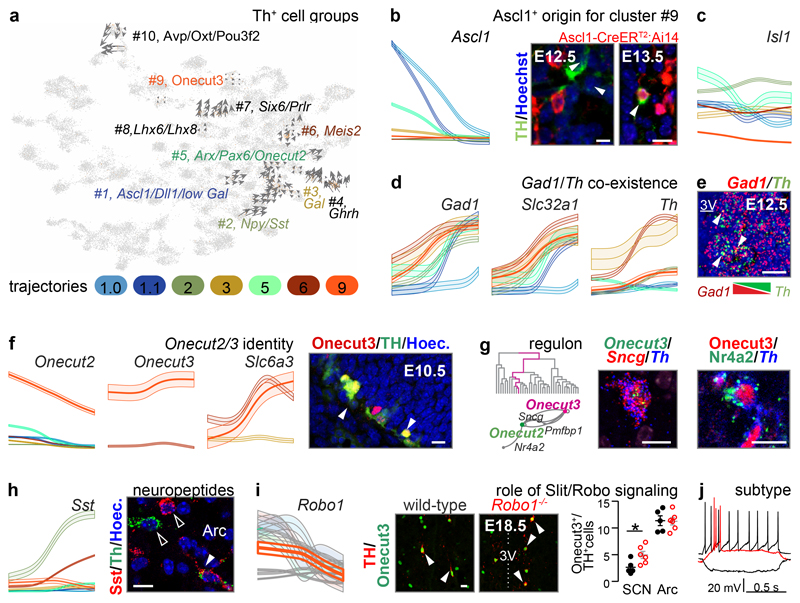
Firstly, we explored if hypothalamic dopamine neurons co-expressing tyrosine hydroxylase (Th), Ddc/dopa decarboxylase and vesicular monoamine transporter 2/Slc18a2 share a developmental trajectory. RNA-velocity vector embedding for all Th+ cells unequivocally identified 10 molecularly far-placed neuronal clusters of which groups #4, #7 and #8 differentiating before E15.5 (Figure 4a and Supplementary Note).
Figure 4. Molecular configuration of hypothalamic dopamine systems.
(a) RNA-velocity vector embedding of tyrosine hydroxylase (Th)+ neurons. Phenotypic convergence even for molecularly-distant neurons is by uniform expression of Th and other enzymes of dopamine synthesis. (b) Th+ neurons invariably rely on Ascl1 as revealed in pseudotime (left). Genetic tracing in Ascl1-CreERT2::Ai14 mice showed the production of Th+/Ascl1+ progeny during the E12-5-E16.5 period. (c) Isl1 is expressed by all Th+ dopamine subgroups (pseudotime; Figure ED9d). (d) Pseudotime trajectories show Gad1, Slc32a1 and Th co-expressed from the early fetal period. (e) Antiparallel expression of Gad1 and Th as a factor of medial-to-lateral positioning (Figure ED9e). (f) Pseudotime trajectories for Onecut2, Onecut3 and Slc6a3 (left) and earliest positions of Th+/Onecut3+ neurons (arrowheads) of prospective PeVN dopamine neurons (#9; right). (g) Validation of target genes for the Onecut3 regulon in PeVN dopamine neurons: scheme to the left identifies hierarchical relationships for Onecut2, Nr4a2, Pmfbp1a and Sncg. (h) Somatostatin (Sst) was enriched in dopamine neurons in the arcuate (Arc) area. (i) Pseudotime trajectory for Robo1 in dopamine neurons (left). Middle: Th+/Onecut3+ neurons in wild-type vs. Robo1-/- mice. Right: quantification of cell numbers at the suprachiasmatic (SCN) and Arc levels (*p < 0.05; Student’s t-test for independent groups). (j) Patch-clamp electrophysiology classifies A14 Onecut3+ dopamine neurons as uniform ‘type C’ cells (see also Figure ED8). Scale bars = 10 μm.
Considering that both pleiotropic and specific genetic programs contribute to molecular diversity among hypothalamic dopamine neurons we addressed the earliest and uniform genetic codes in putative progenitors. Cascading Ascl1 and Isl1 expression was present in all dopamine neurons (Figure 4b,c, ED9a-d), assigning these TFs to defining the entire dopamine class. Indeed, both Ascl1-CreERT2::Ai14 and Isl1-Cre:Ai14 mice produced tdTomato+ dopamine cells, particularly in the PeVN (Figure 4b, ED9a,b,d), when induced at E12-E15. The lack of Th+ neurons in the hypothalamus but not midbrain of Ascl1-/- mice confirmed hypothalamic dependence on an Ascl1-driven transcriptional pathway (Figure 4b,c and ED9a,b).
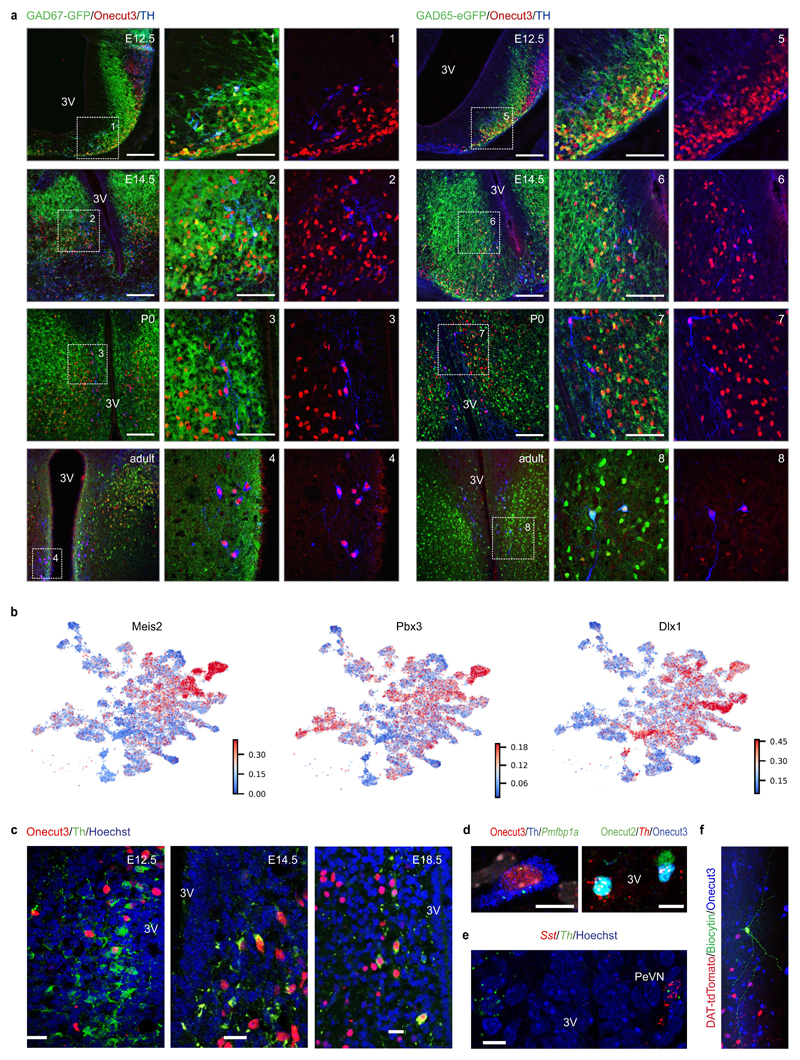
Secondly, we asked if the dopamine phenotype evolves from the GABA lineage (Figure 2d,4a,d), a hypothesis consistent with data in adult4,29. Th+/Ddc+/Slc18a2+ dopamine neurons arise from 7 spatially-segregated GABA groups (#1, #3, #4, #6, #7, #8 and #9; Figure 4a). This was corroborated by the ~90% co-expression of Th and Gad1 in immature neurons (Figure 4d,e and ED9e), including in (BAC)GAD65eGFP and GAD67gfp/+ mice (Figure ED10a). To identify genes that promote GABA-to-dopamine phenotypic transitions, we screened hypothalamic regulons for Th as target and show that the expression patterns of master genes for the Meis2, Nfe2l1, Dlx1 and Pbx3 regulons cover the broad initiation of Th expression at embryonic time points (Figure ED10b).
Thirdly, we searched for TFs that segregate dopamine subclasses. We focused on Onecut3, which specifies dopamine neurons in the PeVN4. Developmentally, Onecut3 serves as the master gene of the regulon that typifies Th/Slc6a3 neurons (#9), and is detectable in the preoptic progenitor area by E10.5 (Figure 4f,g). Histochemistry specifically tied the co-existence of Onecut2/Onecut3 and Sncg, Pmfbp1a, Nr4a2 to PeVN dopamine neurons (Figure 4g and ED10c,d). To further resolve the segregation of A14 neurons, we identified substantial Sst expression prenatally (with a gradual decay after birth) in Onecut3+ dopamine neurons (Figure 4h and ED10e). Based on Robo1 expression in the pseudotime scale, we integrated chemotropic cues for the final positioning of Onecut3+/Th+ neurons by demonstrating a significantly larger contingent of these cells in Robo1-/- mice relative to wild-type littermates (Figure 4i). Finally, Onecut3 expression distinguishes PeVN Th+ neurons that produce a uniform electrophysiological signature amongst the 9 dopamine subtypes tested (Figure 4j and ED8,10f), thus completing a differentiation trajectory segregating PeVN dopamine neurons from all other dopamine subtypes.
Discussion
Our study provides an overview of ectodermal cell identities in the developing hypothalamus during pre- and postnatal periods. We show that a constellation of and temporal dependence on regulon activity, neurotransmitters and neuropeptides shapes ectodermal clusters. Large-scale GWAS-based disease assignment linked GRN activity to the life-long determination of neuronal functionality and consequently to predisposition to metabolic illnesses. Additionally, transient waves of neuropeptide expression were synchronous with critical junctions of neuronal fate progression, thus generating long-lasting imprints on neuronal circuit complexity.
We found a periventricular cellular reserve to persist throughout life to generate hypothalamic neurons with a contingent of GABA progenitors acting as a source for dopamine subtypes. We suggest that the existence of quasi-stable immature intermediates for GABA neurons, their provisional positioning for protracted periods, and sequential depletion until after birth are poised to assure flexibility in expanding functionally distinct neurocircuits by the insertion of neurochemically-specialized cellular subtypes. Thus, fundamental rules of neuronal specification in for the hypothalamus could substantially differ from laminated structures6,7,26. Nevertheless, we find chemogenetic cues that were classically viewed to dominate in cortical areas, particularly Slit/Robo signalling, to also dictate neurogenesis, cell migration5 and synaptogenesis even, if at the microscale, during hypothalamus development. Overall, combining differential gene expression analysis, screens for spatially-restricted genes and GRN profiling into a discovery pipeline showcases the level of precision achievable to disentangle developmental processes that shape neuroendocrine centres and provide a template to study the origins of both hypothalamic circuit operations and molecular underpinnings of congenital and acquired metabolic disorders.
Online-only Methods
Mouse strains
All mice were housed in groups in clear plastic cages on a 12h/12h light/dark cycle (lights on at 08:00 h) and in a temperature (22 ± 2 °C) and humidity (50 ± 10%)-controlled environment. Food and water were available ad libitum. Embryos and tissues were obtained from timed matings with the day of vaginal plug considered as embryonic day (E) 0.5. The day of birth was always registered as postnatal day (P)0. Postnatal animals were weaned on P21. Commercial mouse lines were: C57Bl/6J “wild-type” (RRID:IMSR_JAX:000664), Ai14 (RRID:IMSR_JAX:007914), Ascl1-CreERT2 (RRID:IMSR_JAX:012882), Th-GFP (RRID:IMSR_RBRC03162), (BAC)GAD65-GFP (RRID:MMRRC_011849-UCD), GAD67gfp/+ (RRID:IMSR_RBRC03674), Pomc-GFP (RRID:IMSR_JAX:009593), Slc6a3-Ires-Cre (RRID:IMSR_JAX:006660), Nfia-/- (RRID:MMRRC_010318-UNC), Robo1-/- (RRID:IMSR_APB:5320), Slit1-/- (RRID:MMRRC_030404-MU), Slit2-/- (RRID:MMRRC_030405-MU), Isl1-Cre (RRID:IMSR_JAX:024242) and OxtrVenus/+ (MGI:3838764)30-41. Ascl1-CreERT2 knock-in mice were used as heterozygotes when performing lineage tracing and as homozygotes to study developmental consequences of the lack of Ascl1 since both copies of the gene were replaced by the Cre coding region (referred to as ‘Ascl1 ko’). Nfia-/- mice were provided by J. Bunt and L.J. Richards as a mechanism to re-use tissue (QB/356/17). Nfia-/- mice were bred for work conducted under National Health and Medical Research Council project grant GNT1100443 and Principal Research Fellowship GNT1120615. Tracing experiments for all other Cre lines were performed by using heterozygotes.
Tissue collection and fixation
Whole heads of embryos (E13.5) or dissected brains (E16.5 and older) were collected and fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS, 0.05M, pH 7.4) at 4°C for 4 h for E13.5 and 16-24 h for E16.5 or older. For postnatal stages and adult brain samples, animals were transcardially perfused with 4% PFA and dissected brains post-fixed overnight. Samples were then washed in PBS and cryoprotected by incubating in 30% sucrose in PBS at 4°C overnight.
Ethical approval of animal studies
Experiments on live animals conformed to the 2010/63/EU European Communities Council Directive and were approved by the Austrian Ministry of Science and Research (66.009/0145-WF/II/3b/2014, and 66.009/0277-WF/V/3b/2017). Particular effort was directed towards minimising the number of animals used and their suffering during experiments.
Tamoxifen injection and tissue processing
Ascl1-CreERT2::Ai14 dams were injected with tamoxifen (150 mg/kg) on one of the days of E11.5 to E16.5 to induce Cre-mediated recombination. Brains of the embryos were collected and immersion fixed in 4% PFA in 0.1M phosphate buffer (PB; pH 7.4) for 12-24 h before being immersed into 30% sucrose for cryoprotection (48 h). Embryonic brain tissues were cut at 16 μm thickness and mounted on fluorescence free glasses. Postnatal animals were perfusion-fixed with 50-100 ml of 4% PFA in PB, followed by cryoprotection as above. Brains were then cut on a cryostat as 50 μm-thick serial free-floating coronal sections.
Cell capture, lysis and RNA-seq
C57BL6/N mice (E15.5-P23) of both sexes were used for cell collection. Embryos were removed by Caesarean sections and immersed in ice-cold pre-oxygenated (95% O2/5% CO2) cutting solution containing (in mM): 90 NaCl, 26 NaHCO3, 2.5 KCl, 1.2 NaH2PO4, 10 HEPES-NaOH, 5 Na-ascorbate, 5 Na-pyruvate, 0.5 CaCl2, 8 MgSO4 and 20 glucose. Postnatal animals were deeply anaesthetised (5% isoflurane) and transcardially perfused with 40 ml of the same solution. Entire hypothalami were isolated manually under microscopy guidance from serial 300-μm thick coronal slices and then dissociated using the Papain Dissociation System (Worthington) according to the manufacturer’s recommendations with additional mechanical dissociation using Pasteur pipettes with 600, 300 and 150 μm open tips. After re-suspending the cells in sterile cutting solution supplemented with 0.1% BSA, they were fixed in ice-cold methanol for 10 minutes and stored at -80 °C until library preparation.
For the preparation of cDNA libraries, cells were re-suspended in PBS (0.01M, pH7.4) and concentrated to a range of 105-700 cells/μl. Thirty-three μl of the cell suspension together with 1 μl cellular spike-ins (lymphocytes) were added to the reverse transcription mix. cDNA synthesis, library preparation and sequencing were performed accordingly to the 10x Genomics Chromium Single Cell Kit (version 2). High-throughput RNA sequencing was on an Illumina HiSeq3000 instrument.
10x Genomics data pre-processing
Data derived at each time point were processed independently (Figure S1, Supplementary Note). Raw files were processed with Cell Ranger42 (version 2.2.0) following default arguments for velocyto.py13. Reads were mapped to the Cell Ranger mm10-1.2.0 genome and counted with complimentary annotation (Figure S2 and S3, Supplementary Note). Deriving unique molecule count (UMI) expression matrices, we additionally compared two advanced computational approaches. Firstly, we read raw matrices from the Cell Ranger pipeline (Figure S3, Supplementary Note) into emptyDrops43 implemented in the DropletUtils R package. We used a false discovery rate (FDR) of 0.01 with 2x105 permutations (Figure S4, Supplementary Note). Secondly, we pre-processed raw fastq-files using the dropEst pipeline44 with the UCSC mm10 mouse genome and default dropEst parameters for 10x (Figure S5, Supplementary Note). Briefly, dropEst utilises Bayesian correction of cell barcodes and UMIs, taking into account Hamming distance distribution for cell barcodes and probability distribution by sequential estimation of errors with maximal likelihood between different barcodes within each gene on multiple metadata sources. These include sequencing quality of nucleotide in position (Phred score) and the number of reads for each barcode (coverage) as the most critical parameters. When collision targets are merged, the pipeline estimates damaged and low-quality cells in two steps. Firstly, it automatically assigns cells based on cell-size (Figure S5, Supplementary Note): cell labels were marked with two estimated thresholds: lower than first for ‘low-quality’ (red), then, 75 per cent of cells upper than second as ‘high-quality’ (green) with the remaining cells considered as ‘unknown’ (grey)). Secondly, initial labels together with sets of biological and technical factors (mitochondrial fraction, mean number of reads per UMI, mean number of UMIs per gene, fraction of drop-out genes, fraction of intergenic reads, fraction of not-aligned reads) were deciphered by the kernel density estimate (KDE) classifier to endow each cell with a quality score (0-to-1 range).
emptyDrops and dropEst algorithms hold substantially more cells than the default Cell Ranger approach without a crucial difference between them. Thus, we used the dropEst pipeline44, throughout which additionally provides quality control metrics for the cells albeit at the cost of high computational load. As a result, we used a corrected matrix with cells, which passed filters of both emptyDrops and Cell Ranger and possessing dropEst’s ‘high-quality’ label (upper quartile) together with all cells above the 90-percentile of quality score (Figure S5, Supplementary Note).
Expression matrix filtering
We performed an exploratory analysis of dropEst output matrices in sequential steps of annotation and filtering. Firstly, we checked known genes, which indicate diverse sources of bias, such as ribosomal, immediate-early stress-responsive and gender-specific genes (Gm42418, Rpl26, Gstp1, Rpl35a, Erh, Slc25a5, Pgk1, Eno1, Tubb2a, Emc4, Scg5, Ehd2, Espl1, Jarid1d, Pnpla4, Rps4y1, Xist, Tsix, Eif2s3y, Ddx3y, Uty, Kdm5d)8,45. Furthermore, we assessed the expression level of HuR (Elavl1) to distinguish damaged neurons46,47. Thus, the above gene profiles were indicative of low-quality (potentially apoptotic) cells as well as identified cells of blood origin. Secondly, after removing biasing genes, we manually explored and annotated cell clusters with pagoda213 as described previously44 by using known marker genes8,9,48–50 (DOI:10.6084/m9.figshare.11867889). Thirdly, we defined differentially expressed genes (DEG) for ‘ribo-rubbish’, ‘excluded’, ‘duplets’, ‘endodermal and mesenchymal related clusters’ by comparison against putative ectodermal cell types using the Model-based Analysis of Single-cell Transcriptomics (MAST) test51,52. We repeated normalization, negative-binomial scaling, PCA dimension reduction, ‘Jackstraw’ pc-selection and kNN-graph construction steps after every cell-molecule matrix subsetting.
Integration of expression profiles against a time factor
We followed the lead design initially implemented for paired canonical correlation analysis (CCA)-based integration of data on embryonic and adult cortical interneurons6. Crucial differences were: 1) interchalated nuclei in hypothalamus vs. laminar cortical architectures; 2) higher adult stage transcriptional heterogeneity of neurons (e.g. mixed GABA/glutamate phenotypes and magnocellular/parvocellular neurons)4,53; 3) distant volume transmission as additional factor54,55; 4) six developmental stages. We aimed to derive a manifold according to known adult cell types and lineages. We integrated datasets from successive time points (E15.5-P23) to discover the succession of developing cell lineages. This allowed us to apply retrospective analysis to distinguish ‘ancestor’ cells. We removed mature and myelin-forming oligodendrocytes from our analysis, which existed only in late postnatal stages. We have additionally filtered cells with < 300 genes or 2.5x104 RNA molecules (taking into account only ectoderm-related genes). Finally, we used a variance-stabilizing transformation of the SCTransform method to find anchor-candidate genes6,51.
Comparison of integration algorithms
Despite a recently published approach6 for the integration of embryonic and adult single-cell RNA-seq data, we additionally tested all presently available algorithms. To match our criteria, the algorithm should 1) provide a mixture of time and batch factor and 2) at the same time keep a defined cell type local structure. Therefore, we benchmarked 11 different solutions of integration using their default parameters (Figure S6, Supplementary Note), default balanced batch k-nearest neighbours (BBKNN)56; BBKNN with a neighbours trimming procedure based on their connectivity scores, which were derived by UMAP algorithm57 (BBKNN_TRM); BBKNN with exact neighbour identification via faiss58 (BBKNN_FAISS); BBKNN based on k-dimensional-tree (cKDTree)59; Scanorama60; LIGER61; Harmony62; mutual nearest neighbour (MNN)63; CONOS64; Seurat 3 CCA-based integration of negative binomial fit scaled matrixes (SeuratCCA)65; Seurat 3 CCA-based integration of scTransform derived matrixes of Pearsons residuals (SeuratCCAonSCT)51. We found that the updated version of the conventional Seurat approach performed similarly to CONOS, Harmony and LIGER, over performed as compared to other algorithms in terms of the MixingMetric and underachieved vs. 1) Harmony (with Harmony over-fitting the batch factor, which we could not optimize against with two factors (time and batch factor) at the same time) and 2) its own more advanced version (Seurat 3.165, unpublished) in terms of the Local structure metric leading us to a more conservative way of integration of adult and embryonic stages.
CCA-based integration with Seurat 3.1
We integrated cells from different time points into a single manifold using the latest version of the Seurat 3.1 CCA-based integration pipeline65. The new version allowed us not to set order of integration explicitly and thus determined the optimal order automatically. This allowed us to obtain the united manifold of 50 CC components from filtered and weighed integration anchors and linearly scaled genes used for integration into the CCA space. Subsequently, we performed cell-cycle difference regression: S-phase score minus G2M-phase score - according to the alternative Seurat workflow to remove differences in cell cycle phases amongst proliferating cells, following the available gene set annotation66. Next, we derived 50 principal component (PC) matrices from the corrected matrix. From this PC matrix, we selected PCs having >25 percentile of s.d. and learned UMAP embedding57,67 (Figure 1a, ED3a,7a,d,e, Supplementary Note). As a result, we obtained a 37-dimensional manifold of all cells and ’its’ two-dimensional embedding.
Annotation of the collective manifold for hypothalamus development
Our next aim was to annotate putative cell lineages pivoting hypothalamus through development to adulthood. Therefore, we used MetaNeighbor68 in supervised mode to evaluate area under the recovery curve (AUC) scores of cross-explanation different cellular annotations4,8,9,69,70 that exist for adult hypothalamic scRNA-seq data (Figure S8, Supplementary Note). Thus, we expected to find putative replicative subtypes, but found an unexpected lack of consensus for cellular annotations, meaning that top calls were assigned to the same studies.
Therefore, we have chosen a recent dataset8 as reference because of its completeness of anatomical sampling (another anatomically complete study of hypothalamus69 was inferior in terms of cell numbers and their variability). Additionally, the label transferring approach of Seurat was used to verify the absence of possible contamination with cells from the thalamus in the filtered dataset. To this end, we integrated the expression profiles with signatures derived from the mouse brain atlas spanning the diencephalon8.
Integration of juvenile data with default parameters for Seurat CCA revealed a comprehensive coverage on the diversity of terminally-differentiated progenies of reference8 (Figure S9a, Supplementary Note). Next, we attempted to incorporate the adult reference to the entire developmental dataset (Figure S9b, Supplementary Note). We observed substantial disparities suggesting the existence of convergence processes during hypothalamus development, which necessitated an unbiased strategy of annotation. For this reason, we utilised clustering factors from the integration manifold by all field prevalent algorithms (graph-based algorithms mostly from the igraph package used in Pagoda2 with default parameters on 37 PCs and separately on the corrected UMI counts matrix, and a small local movement algorithm with different resolutions via Seurat’s FindClusters function) for comparison (Figure S10, Supplementary Note). We observed robustness in the inner structure of the integrated manifold for all used algorithms, except infomap, by estimating information metrics in a cross-annotation manner (due to lacking a correct one) and silhouette scores using R packages aricode71 and clues72. Since most algorithms produced similar results (with only minor differences between them), and our approach proved accurate (except for the infomap), we decided to proceed with walktrap73.
Finally, we decided to first split cells by major lineages: an exploratory analysis was performed in Pagoda213 after transferring our gene-cell integrated matrix and UMAP embedding. We utilized the standard Pagoda2 pipeline with the integrated manifold matrix clustered by walktrap73. We annotated the final 45 clusters by DEG testing with MAST52 (Figure 1a,2a and ED1) on the scTransform-corrected6,51 log-normalized UMI matrix (data slot in a Seurat object).
Dendrogram construction (gene-based)
A dendrogram was constructed for neuronal and glial cells separately with Seurat 3 with different feature sets as input: for the neuronal tree we used a list of genes from Ref.74. The glial tree was based on genes from Ref.75. For both trees, we used gene sets defined in Ref.76, which were filtered by taking only the upper 95-percentile for the corresponding cell types (neurons, astrocytes, oligodendrocytes), the upper 95-percentile for cell types in diencephalon and the upper 50-percentile of MeanExpression. We excluded housekeeping genes77 for both trees. We used dendrograms to order our dot plots (Figure ED1).
Neuropeptide and neurotransmitter assignment
Density plots on UMAP embedding for signalling molecules were assembled by ggalt78 (Figure ED3a,d,e). We manually split neuropeptides to prevalent GABAergic or glutamatergic co-existence. Top-ranked neuropeptides of the corrected UMI matrix were plotted with a threshold of 10 molecules in color and shape-coded manner for two groups separately.
Abstraction graph on repartitioned integrated data
We redistributed ‘bridge’ cells (#19) and immature neuron (#11) populations by applying a combination of two methods: the Leiden network clustering algorithm79 and PAGA12. Moreover, repartitioning immature-to-adult allowed us to negotiate selection bias80. We excluded #3, #5, #7, #38, #39, #42, #45 as oligodendrocytes and pars tuberalis from the integrated manifold matrix and performed clustering with Scanpy81 wrapper with iterations until full optimization. These partitions were used for the construction of an abstraction graph (non-directed) with a threshold of 0.365. Nodes were not established as common clusters. Instead, we derived them to obtain the topological structure of cell ensembles. We analyzed the projection of our preliminary clusters of each developmental stage on partitions of the abstraction graph and transformed it by Leiden algorithm (using time_slices_to_layers() function and then optimise_partition_multiplex() for class la.Optimiser()) to a connection-based annotation of cell lineages with consistent colors. Next, we used abstraction graph nodes as starting points for the UMAP algorithm with maxiter = 1,000, negative_sample_rate = 20, min_dist = 1, spread = 2 parameters to prepare embedding corresponding to cell lineage relations (Figure 1b-d,2a,b,3f,4a and S11, Supplementary Note).
RNA velocity analysis 1
We performed RNA velocity analysis of time points separately; following the original deterministic approach of the velocyto.R/py packages13. Currently, it is not possible to split an UMI-matrix obtained by Bayesian correction of dropestR44. Thus, we exported metadata for filtering of a default matrix and our cellular annotations (walktrap algorithm derived 45 cell groups). They were then sub-grouped to relocate cells, which passed our filters and removed ‘rubbish’ and mesenchymal related genes. Lastly, we applied the original RNA velocity method13 with a few modifications: 1) we filtered out all non-DEGs, which were present in <20 spliced/unspliced molecules, 2) for obtaining a velocyto grid we exported the UMAP embeddings67,73 of our high-quality cells from the previous step for visualization purposes (Figure ED2b,c).
RNA velocity analysis 2
To perform RNA velocity analysis of the integrated dataset, we applied the scvelo python package using a generalised dynamical model82. As input, we used filtered to ectodermal cells and gene loom files, which were merged using loompy (version 2.0.17) and filtered to cells used for PAGA construction (see above). Briefly, spliced and unspliced reads were separately size-normalized to the median of total molecules across cells. Additional gene filtering comprised of those that passed a minimum threshold of 10 expressed counts for spliced and 2 for unspliced mRNA. We quantified a 30-nearest neighbour graph based on Euclidean distances in 30-PC space (PCA performed on logarithmic spliced counts). Therefore, for each cell across its neighbours, we obtained first and second-order moments (means and uncentered variances), then estimated RNA velocity with the explicit fitting of inferred splicing reaction rates. As a result, transition probabilities were estimated forming velocity graph. Thus, we plotted individual cell velocities embedded in UMAP space (Figure 2a,3f and ED7a). Subgraph analysis of both the glial lineage (#1, #10, #18, #24, #34, #51) and ‘bridge’ cells (we subset only the first entering node of the abstraction graph) was performed as described previously12,82. To this end, we subset cells of interest and transformed PAGA to a directed abstraction graph using a default constructed velocity graph from above. Finally, we allocated root cells by using the backward Markov process on the transition probability matrix to define excessive density area, estimated the latent time on learned transcription dynamic model and plotted a velocity grid (Figure 1c) and individual cell velocities embedded in UMAP space (Figure 1c1). We completed all steps using built-in functions with default parameters.
Estimation of developmental regulons
We prepared a subset of putative glial (astroependymal, tanycyte and progenitor) and neuronal clusters (as described above under ‘abstraction graph construction’). A spliced UMI count matrix of the integrated dataset was input in the pySCENIC23,83 pipeline with default settings to infer active transcription factors (TFs) and their target genes. Briefly, the pipeline was implemented in three steps. Firstly, we identified gene co-expression modules of TFs83. Secondly, we pruned each module based on a regulatory motif near a transcription start site (TSS). Cis-regulatory footprints could be obtained with positional sequencing methods (e.g. from ChIP-seq motif calling with an antibody against TF). Binding motifs of TFs across multiple species then built an RCisTarget database. Precisely, modules were retained if the TF-binding motif was enriched among its targets, while target genes without direct TF-binding motifs were removed. Thirdly, we scored the impact of each regulon for each single-cell transcriptome using the AUC score as metric. Each step of this pipeline used rank statistics, and the last classification step ran independently for each cell, avoiding a batch effect.
Moreover, regulons tended to highlight higher-order similarities across cells. Thus, we determined whether the target genes in each regulon were enriched in each cell using the distribution of regulon activity across all cells in the dataset. The input list of TFs was downloaded from the RIKEN database84–86. As a result, we derived the AUC score matrix (AUCell) to validate our clustering and prepared annotation by 395 identified regulons. Inferred regulons and their activity across cells in the integrated dataset are reported in DOI:10.6084/m9.figshare.11867889. Wilcoxon test, Logreg test87 and dot-plot visualization of differentially recruited regulons across 45 identified cell groups (described above) were done in the Scanpy package81 (Figure 3a and ED4). Gene regulatory network (GRN) plots of the Onecut2/3 regulons were done by using the Gephi software package88 (Figure ED6a).
Dendrogram construction (regulon-based)
Next, a dendrogram was constructed for neuronal and glial cells together by Seurat 3 on the AUCell matrix of 395 regulons to observe subtler changes. We deciphered the diverging composite rules of a regulons-based dendrogram by testing each branching node for differential regulon importance. Therefore, we performed the Wilcoxon test and Logreg test87 of every node with min.pct = 0.01, logfc.threshold = 0.01 of Seurat’s function FindAllMarkers to derive the action propagation program of the regulons (DOI:10.6084/m9.figshare.11867889). We used dendrograms to order dot-plots (Figure 3a and ED4).
Regulon assignment to clinical phenotypes
To understand the potential involvement of regulons to human disease phenotypes, we analysed properties of human polymorphic variants (SNPs) located within the regulon genes. Recently, a robust correspondence between human and mouse regulons was reported23. To uncover associations between regulon-specific variants and human phenotypes we used Gene Atlas - a database with summary statistics of genome-wide association studies (GWAS) between millions of variants and hundreds of traits in the UK Biobank cohort (http://geneatlas.roslin.ed.ac.uk/)89. Firstly, we converted a subset of mouse regulon genes with one-to-one orthologs to human Ensembl gene IDs. Here, we used mouse notation for regulons (only the first capital letter and the remaining lowercase letters) to clarify their source. Secondly, we extracted all SNPs belonging to regulon genes and analysed the distribution of their p values. Thirdly, we characterised regulons in terms of the total number of SNPs affecting their master gene, as well as SNPs affecting other regulon-recruited genes. We observed that master genes tend to have a deficit in SNPs as compared to the downstream general representatives of these regulons (median number of SNPs affecting master genes across all regulons is less than the median number of SNPs affecting general representative genes). Some regulons showed zero SNPs (at least in the UK Biobank cohort) in their master genes but significant, close to the median, number of SNPs in regulon-recruited genes. Using the number of SNPs affecting master genes and the number of SNPs affecting other regulon-recruited genes as a metric of evolutionary constraint, we split all regulons into four quadrants reflecting the ratio of constraints between masters and members of regulons (Figure ED5b). Next, we asked where hypothalamus-related regulons appeared in this plot. Thus, to focus on the hypothalamus, we used weights of genes in regulons output by the SCENIC workflow, which were interpreted as an importance of a given gene in a given regulon. To highlight the most important regulons, we chose ones with their median weight higher than the median weights of all individual genes from all regulons.
Henceforth, the logic behind our analysis is similar to pi1 statistics, estimating an excess of low p values90. To make a fast approximation of the excess of low p values for 778 phenotypes and each gene of 395 regulons, we estimated the fraction of p values, which were <0.1. Limited testing demonstrated a strong correlation between our excess of low p values and pi1 estimated in the qvalue R package91. Also, limited testing demonstrated similar trends, observed when we changed the threshold from 0.1 to 0.01. Therefore, we manually selected clinical phenotypes related to the hypothalamus, subset to regulons with importance (weight) higher than the median and aggregate weighted pi1 statistics to the regulon level. For visualisation purposes, we selected three contrast regulons from each quadrant by choosing distant regulons in the two-dimensional PCA space of phenotypes (red labels in Figure ED5a, black spheres in Figure ED5b) and plotted their normalised values via the heatmaply R package (Figure 3d)92.
Estimation of developmental genes and regulon dynamics
Next, we took the spliced molecule count matrix of the same subset of cells as for the PAGA subgraph analysis of progenitors and their nearest offsprings. Firstly, we size-normalized to the median of total molecules across cells. Secondly, a logarithmic matrix was used to estimate pseudo-time order and probabilities for cells to propagate through the subgraph of glial lineages or ‘bridge’ cells. For this purpose, we used a probabilistic approach, Palantir93, which we implemented as an external module to the Scanpy Python package81 (Figures 1d,4b and ED7a; Figure S12 in Supplementary Note). Similarly, we applied this method to the Pomc-cell group (Figure 2b) and every distinct Th-containing group (Figure 4b-d,f,h,i, ED9c; Figure S13 in Supplementary Note) guided by PAGA topology. In all cases, we selected early cells by taking upper-99-percentile of the Sox2 regulon of AUC scores distribution and used default parameters for estimations with exception for the waypoints parameter: for glia and bridge neurons – 500, for the Pomc+ cells lineage – 1200, and for Th+ trajectories: #1 – 750, #2 – 500, #3 – 1000, #4 – 500, #5 – 350, #6 – 1000, #7 – 200, #8 – 200, #9 – 1000, #10 – 800. Finally, we estimated the impact of regulon dynamics along the identified trajectories using the AUCell matrix as input for Palantir’s function compute_gene_trends, which uses a generalised additive model. Both the trends of genes and regulon actions were clustered for each trajectory with default parameters utilising the Phenograph python package94.
Tissue preparation and immunohistochemistry
After rinsing in 0.1M PB, specimens were exposed to a blocking solution composed of 0.1M PB, 10% normal donkey serum, 5% BSA and 0.3% TX-100 for 3h followed by 48h incubation with select combinations of primary antibodies: rabbit anti-TH (1:500; Millipore AB152, Lot 2593900, 3199177), sheep anti-TH (1:1000, Novus Biologicals, #NB300-110, Lot ajo1217p), sheep anti-ONECUT2 (1:250; R&D Systems, AF6294, Lot CDKS0116081), guinea pig anti-ONECUT3 (1:5,000)95, rabbit anti-VGLUT2 (1:800; a gift from M. Watanabe)96, goat anti-GFP (1:1,000; Abcam, #ab6662, Lot GR311622-15, GR311622-7), chicken anti-GFP (1:500, Aves Labs Inc., #GFP-1020, Lot GFP697986), rabbit anti-SOX2 (1:500, Abcam, #ab97959, Lot GR3244885-1), chicken anti-mCherry (1:1,000; EnCor Biotech, #CPCA-mCHERRY, Lot 7670-4), mouse anti-MASH1 (1:100, BD Pharmingen, 556604, Clone: 24B72D11.1), guinea pig anti-GFAP (1:500, Synaptic Systems, 173004, Lot 2-15, 2-17), rabbit anti-phospho-Histone H3 (1:500; Cell Signaling Technology, 9701, Lot 7), chicken anti-NeuN (1:500, Merck Millipore, ABN91, Lot 3132967), mouse anti-FLAG-tag (1:1,000; Sigma, F1804, Lot SLBR7936V), mouse anti-HA-tag (1:600; Cell Signaling Technology, mAb2367, Lot 1). Secondary antibodies were from Jackson ImmunoResearch, including Alexa Fluor 488-AffiniPure donkey anti-goat (705-545-147, Lot 131669), Alexa Fluor 488 donkey anti-mouse (715-545-151, Lot 127820), Alexa Fluor 488-AffiniPure donkey anti-guinea pig (706-545-148, Lot 138058), Alexa Fluor 647-AffiniPure donkey anti-guinea pig (706-605-148, Lot 135631), Alexa Fluor 647-AffiniPure donkey anti-rabbit (711-605-152, 127614) and carbocyanine (Cy)2-AffiniPure donkey anti-rabbit (711-225-152, Lot 139999), Cy3-AffiniPure donkey anti-chicken (703-165-155, Lot 142225), Cy3-AffiniPure donkey anti-goat (705-165-147, Lot 134527), Cy3-AffiniPure donkey anti-guinea pig (706-165-148, Lot 134844), Cy3-AffiniPure donkey anti-mouse (715-165-150, Lot 116881), Cy3-AffiniPure donkey anti-rabbit (711-165-152, Lot 141941) and applied at a dilution of 1:300 in 0.1M PB supplemented with 2% BSA (20-22 °C, 2h). Nuclei were routinely counterstained with Hoechst 33,342 (1:10,000; Sigma). Tissues were photographed on a Zeiss LSM880 laser-scanning microscope. Images were acquired in the ZEN2010 software package. Multi-panel images were assembled in CorelDraw X7 (Corel Corp.).
RNA-scope® in situ hybridisation
C56Bl6/J mice were used to verify scRNA-seq candidate gene expression as described97. Dissected embryonic mouse heads were fixed in 4% PFA (pH 7.4) overnight. RNAscope® 2.0 was performed according to the manufacturer’s instructions (ISH, RNAscope®, Advanced Cell Diagnostics)98. RNAscope® probes for detection of Slc1a3, Rax, Dll1, Dll3, Neurod1, Slit1, Slit2, Draxin, Prdm12, Nhlh2, Sox10, Ddc, Lancl3, Pmfbp1, Sncg, Sst, Th and Trh were designed commercially by the manufacturer and are available from Advanced Cell Diagnostics. Imaging was performed using an LSM880 Zeiss confocal microscope equipped with a 40x objective.
Fluorescent In Situ hybridization (HCR 3.0)
Stainings were performed on fresh frozen tissues sectioned at 16 μm following the HCR v3.0 protocol for ‘generic sample on the slide’ (Molecular Instruments)99. The pre-treatment of tissue sections included fixation with 4% PFA for 15 min, two washing steps with PBS and dehydration using an ascending EtOH gradient (25%, 50%, 75% and 100%, each step for 5 min with subsequent drying for 15 min). The tissue used for these experiments was obtained from E12.5, E15.5, E16.5, E18.5 embryos or P2, P7 pups. The probes used (Ddc, Gad1, Meis2, Onecut3, Sncg, Th, Trh and Zic5) were designed and purchased from Molecular Instruments.
In vitro overexpression of Onecut3
Neuro2A cells were propagated in DMEM containing 4.5g/L glucose, Glutamax, 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin (all from Gibco). Prior to transfection, cells were plated on glass coverslips (coated with poly-D-lysine (Sigma) at 37 °C overnight) at a density of 75,000 cells/well in a 24-well format. Cells were transfected with 500 ng of either OC3 or ABCD2 (an ATP-binding cassette transporter located on peroxisomes as CMV control) using the jetPRIME transfection system. The medium was replaced after 30 min to growth medium containing 2% FBS (to limit excessive proliferation) and cells were either immersion fixed for immunocytochemistry in 4% PFA in PBS (pH 7.4) for 20 min or lysed for qPCR after 3 days in vitro. Note that no cell death was observed due either to overexpression or the transfection reagent.
For immunocytochemistry, cells were permeabilised and blocked before adding a primary antibody cocktail overnight at 4 °C. Staining was performed with phospho-Histone H3 (rabbit host; 1:500; Cell Signaling Technology), FLAG-tag (mouse host; 1:1,000; Sigma), HA-tag (mouse host; 1:600; Cell Signaling Technology) and counterstained with Hoechst 33,342 (1:10,000; Sigma). After mounting with glycerol gelatine (Sigma), random overview images (20x magnification) were taken on an Zeiss LSM880 confocal microscope. Hoechst+ as well as phospho-Histone H3+ cells were counted with ImageJ1.52a (cell counter plugin) and normalised to Abcd2.
For qPCR quantification, RNA was extracted with the Aurum Total RNA Kit (BioRad). A cDNA library was prepared by transcribing 2μg of RNA with the High Capacity RNA-to-cDNA Kit (Applied Biosystems). For qPCR reactions, 20ng cDNA was amplified using SYBR green (BioRad) on a CFX Real Time Amplifier (BioRad) with [Th forward: TGTTTCAGTGCACACAGTAC]; [Th reverse: CCAATGTCCTGGGAGAACTG], [Cxxc5 forward: AGTGGACAAAAGCAACCCTA]; [Cxxc5 reverse: TTAGCATCTCTGTGGACTGT], [Tmprss9 forward: GCTTGGTGCGACCCATCT]; [Tmprss9 reverse: CATGGAGCCTCCCTCGC] and [Tbp for: CCTTGTACCCTTCACCAATGAC]; [Tbp rev: ACAGCCAAGATTCACGGTAGA] primers as loading control.
Preparation of acute brain slices
All experiments were performed in 300 μm-thick coronal slices prepared on a VT1200S vibratome (Leica) using a protective recovery method for slice preparation100. All constituents were from Sigma-Aldrich. Solutions were aerated with carbogen (5% CO2/95% O2).
Patch-clamp electrophysiology
Whole-cell recordings were carried out using borosilicate glass electrodes (Hilgenberg, Germany) of 3-4 MΩ pulled on a P-1000 instrument (Sutter). Electrodes were filled with an intracellular solution containing (in mM): 130 K-gluconate, 3 KCl, 4 ATP-Na2, 0.35 GTP-Na2, 8 phosphocreatine-Na2, 10 HEPES, 0.5 ethyleneglycol-bis(2-aminoethylether)-N,N,N',N'-tetraacetate (EGTA), (pH 7.2 set with KOH) and 0.5% biocytin (Sigma) for post-hoc cell identification. After recordings, brain slices were immersion-fixed in 4% PFA at 4°C overnight. Recordings were carried out on an EPC-10 triple amplifier (HEKA) controlled by PatchMaster 2.80.
Sample sizes, statistics and reproducibility
Sample sizes for scRNA-seq experiments: n = 8 (E15.5), n = 7 (E17.5), n = 4 (P0), n = 4 (P2), n = 3 (P10), n = 3 (P23). SCTransform corrected UMI-count matrices were statistically tested to obtain DEGs using log-normalized values with pseudocount = 1 for 45 identified cell groups as previously described6,51 using MAST test52. We used the Wilcoxon test and Logreg test87 to define up-regulated regulons. Results of the DGE tests are specified in DOI:10.6084/m9.figshare.11867889.
For Figure 1a, UMAP plot was built for n = 51,199 cells of ectodermal origin integrated by canonical correlation analysis (CCA).
For Figure 1c, n = 5,070 cells were used for UMAP.
For Figure 1d (right), we sampled cell-ordering (n = 5,070 cells) to 500 bins to compute gene expression trends. For each bin, means ± s.e.m. expression was estimated by generalized additive models.
For Figure 1e, n = 2 animals for E12.5, 3 n = animals for E13.5, and n = 3 animals for E15.5 were used for embryonal tracing of Ascl1 at corresponding time points.
For Figure 1f, n = 2 animals were used to trace Ascl1+ cells in (BAC)GAD65-eGFP:Ascl1-CreERT2:Ai14 mice (tamoxifen injection at P14, sample collection at P21).
For Figure 2b (top right), to compute gene trends we sampled the trajectory for POMC neurons (n = 1,643 cells from progenitors to mature POMC cells) to 500 bins. Each single trajectory shows means ± s.e.m. estimated by generalized additive models.
For Figure 2b (bottom), RNA scope in situ hybridization was performed on samples from POMC-GFP mice (n = 4 for Prdm12 and n = 3 for Nhlh2).
For Figure 2e, experiment was performed in triplicate with n = 2 animals/time point.
For Figure 3b, iImages are representative for wild-type (n = 3) and Nfia-/- (n = 3) mice.
For Figure 3f, n = 5,070 cells belong to glia (excluding oligodendrocytes) or bridge neurons. To compute gene expression trends, we sampled cell-ordering to 500 bins. For each bin, means ± s.e.m. was estimated by generalized additive models.
For Figure 3g, n = 5 (wild-type), n = 5 (Robo1-/-), n = 3 (Slit1-/-) and n = 3 (Slit2-/-) mice were used. Data are presented as percentile box-whisker plots (10, 25, 50, 75, 90 percentiles). Data were statistically analysed using one-tailed Student’s t-test of raw data: p = 0.0019 for wild-type vs. Slit1-/-; p = 0.0006 for wild-type vs. Slit2-/-.
For figure 3h, experiment was performed in duplicate with wild-type (n = 3) and Robo1-/- (n = 4) mice.
For Figure 4b,c,d,f,h,i, to compute gene trends we independently sampled 7 differentiation trajectories containing neurons (for group #1: 1,506 cells, for group #2: 997 cells, for group #3: 1,453 cells, for group #5: 948 cells, for group #6: 1,779 cells and for group #9: 1,181 cells) to 500 bins. For each bin, means ± s.e.m. expression was estimated by generalized additive models.
For Figure 4b (right), n = 2 animals for E12.5, n = 3 animals for E13.5. Experiment was performed in duplicate.
For Figure 4e, n = 2 animals were used. Experiment was reproduced 2 times.
For Figure 4f (right), n = 3 animals were used in the experiment.
For Figure 4g (right), n = 2 animals were tested in each independent experiment.
For Figure 4h (right), n = 2 animals were tested in the experiment.
For Figure 4i, n = 5 (wild-type) and n = 6 (Robo1-/-) mice were tested. Data are visualised as error bar plots with individual data point information. Data were statistically evaluated using one-tailed Student’s t-test on raw data: p = 0.015 for wild-type vs. Robo1-/- at the anatomical (rostral-to-caudal) level of the suprachiasmatic nucleus.
For Figure 4k, post-hoc neuroanatomical reconstruction identified that all n = 9 Onecut3+ neurons were of ‘type C’ in A14 (n = 15 cells were characterized as ‘type C’ within 62 cells recorded in total from A14).
For Extended Data Figure 1, we used the dataset of n = 51,199 ectodermal cells for dot-plot representation (a,b) and UMAP visualizations (b1,b2,c).
For Extended Data Figure 2a, we used the same dataset of n = 51,199 ectodermal cells for each alignment algorithm.
For Extended Data Figure 2c, n = 6,314 cells from E 15.5 passed filters of original RNA-velocity analysis and presented at the figure panels for each analysis of gene expression.
For Extended Data Figure 2d, numbers of animals used for embryonal tracing are: n = 2 for E12.5, n = 3 for E13.5, n = 3 for E15.5 and n = 2 for E16.5.
For Extended Data Figure 2d, freely available data from Allen’s Mouse Developmental Brain Atlas were used.
For Extended Data Figure 2f, images are representative for Ascl+/- (n = 3) and Ascl1-/- (n = 3) mice.
For Extended Data Figure 2g, 3 animals were checked for each developmental stage.
For Extended Data Figure 2h, 2 Ascl1+ cells were traced in (BAC)GAD65-eGFP:Ascl1-CreERT2:Ai14 animals for the time-point shown (tamoxifen injection at P14, sample collection at P21).
For Extended Data Figure 3d,e, analysis was done based on n = 33,893 cells of neuronal and glial origin (excl. oligodendrocytes).
For Extended Data Figure 3f, experiment was reproduced 3 times with n = 2 animals/time-point.
For Extended Data Figure 4, n = 33,893 cells in total were tested with two-tailed Wilcoxon rank sum test.
For Extended Data Figure 6b, images are representative for samples from n = 2 animals.
For Extended Data Figure 6c, Images are representative of n = 2 animals/experiment.
For Extended Data Figure 6d,d1, images are representative for the experiment reproduced 2 times with n = 6 (coverslips).
For Extended Data Figure 6d2, the sample size is n = 12 for each group. Data were statistically evaluated using two-tailed Student’s t-test on raw data. p = 0.0000292 for the density of Hoechst positive cells; p = 0,0000000103 for the density of pHH3+ cells.
For Extended Data Figure 6e, there were 3 biological and 3 technical replicates for each probe. Data are visualized as error bar plots (means ± s.e.m.) with individual data point information. Data were statistically evaluated using two-tailed Student’s t-test on raw data. p = 0.0210 for Th expression; p = 0.000153 for Cxxc5 expression.
For Extended Data Figure 7a (UMAP), n = 33,893 cells were used for graph construction.
For Extended Data Figure 7a (bottom), to compute gene expression trends we sampled n = 5,070 cells to 500 bins. For each bin, means ± s.e.m. expression was estimated by generalized additive models
For Extended Data Figure 7a (right), b,b1,b2, all images represent results from n = 3 animals for each experiment.
For Extended Data Figure 8a, we recorded n = 20 cells from A12 (including n = 12 of ‘type A’, n = 3 of ‘type B’, n = 5 of ‘type C’), n = 8 cells from A13 (n = 5 cells of ‘type A’ and n = 3 cells of ‘type B’) and n = 62 cells from A14. Specifically for A14, we distinguished 4 electrophysiological profiles: n = 32 cells of ‘type A’, n = 9 cells of ‘type B’, n = 15 cells of ‘type C’ and n = 6 cells of ‘type D’. Post-hoc neuroanatomical reconstruction identified that all n = 9 Onecut3+ neurons were of ‘type C’ in A14.
For Extended Data Figure 8b, images are representative for the experiment reproduced 9 times.
For Extended Data Figure 9a, images are representative for Ascl+/- (n = 3) and Ascl1-/- (n = 3) mice. Immunohistochemical experiment was reproduced 2 times.
For Extended Data Figure 9b, n = 3 animals from E13.5 tamoxifen injection, n = 3 animals from E15.5 tamoxifen injection, and n = 2 mice from E16.5 tamoxifen injection were tested for embryonal tracing of Ascl1-cells at corresponding time points at E18.
For Extended Data Figure 9c, to compute gene trends we independently sampled 7 differentiation trajectories containing neurons (for group #1: 1,506 cells, for group #2: 997 cells, for group #3: 1,453 cells, for group #5: 948 cells, for group #6: 1,779 cells and for group #9: 1,181 cells) to 500 bins. For each bin means ± s.e.m. expression was estimated by generalized additive models.
For Extended Data Figure 9d, n = 3 animals were tested.
For Extended Data Figure 9e (images), n = 2 animals were used for each developmental time-point. Experiment was reproduced twice.
For Extended Data Figure 9e (scatter plot), for both ages n = 2 animals were tested. In total 86 Th-containing (min >2 mRNA molecules) cells were randomly analyzed. Data are visualized as error bar plots (means ± s.e.m.) with individual data point information.
For Extended Data Figure 9f, images are representative for n = 2 animals from each time-point.
For Extended Data Figure 10a, images are representative for the experiment performed in duplicate on n = 4 for each developmental stage of both mouse lines.
For Extended Data Figure 10b, n = 26,316 cells from the neuronal lineage (including progenitors) were used.
For Extended Data Figure 10c, n = 3 animals were tested for each developmental time-point.
For Extended Data Figure 10d, n = 2 animals were tested/independent experiment.
For Extended Data Figure 10e, n = 2 animals were tested for the experiment.
For Extended Data Figure 10f, n = 9 Onecut3+ neurons were reconstructed.
Data availability
Custom code was neither generated nor used. Raw, processed and supplementary datasets were deposited in GEO (accession number: GSE132730). GEO files include: 1) raw fastq files for every sequencing run; 2) filtered matrices for every sample in RDS file format including Seurat 3 objects with all processed cells; 3) original integrated dataset in RDS file format including Seurat 3 objects with all processed cells as well as all used commands; 4) integrated dataset used for dynamics analysis (which passed filtering of RNA Velocity analysis); 5) AUCell matrices from pySCENIC pipeline; 6) full regulon hypothalamic network in GraphML file format; 7) metadata protocol describing all experimental, computational procedures and quality control. An interactive view of the integrated dataset (for processing in Pagoda2) can be accessed through the following URLs: https://dx.doi.org/10.6084/m9.figshare.11867889 (~1.1 GB).
All data presented (e.g., imaging) will be made available by T.Ha. (tibor.harkany@ki.se or tibor.harkany@meduniwien.ac.at) upon reasonable request.
Code availability
Code is available at https://dx.doi.org/10.6084/m9.figshare.11867889
Extended Data
ED Figure 1. Marker genes to define molecular phenotypes.
(a) Differential gene expression by glia (#1-9) and neurons (#10-45). Because of the integration of six stages, early-expressed TFs and spatially-restricted genes amenable to cellular differentiation were identified. For neuronal clusters, fast neurotransmitter specificity is shown to the right. Relative diameter of the solid circles for each cluster is scaled to the fraction of cells that expresses a specific gene. Color coding and numbering at the top correspond to those in Figure 1a. (b) Dot plot representation of differential TF expression in 45 ectoderm-derived cell groups in the hypothalamus. (b1,b2) Subclass-specific TFs recapitulate the UMAP positions of neuronal (b1) and tanycyte (b2) subtypes. (c) Integrated molecular/anatomical annotation of hypothalamic with their specific assignment to hypothalamic areas. Abbreviations: ARC-Agrp, arcuate nucleus-agouti-related peptide+ neurons; ARC-Sst, arcuate nucleus-somatostatin+ neurons; ARC-TIDA, arcuate nucleus-tuberoinfundibular dopamine neurons; DMH, dorsomedial hypothalamus; Gal, galanin; Ghrh/Vacht, growth hormone-releasing hormone/vesicular acetylcholine transporter+ neurons; LH, lateral hypothalamus; LH-Lhx9, lateral hypothalamus-LIM homeobox 9+ cluster; Meis2, meis homeobox 2; MM, ; MM-Lhx9, mammillary nucleus-LIM homeobox 9+ neurons; Pomc, proopiomelanocortin; PH, posterior hypothalamus; PMM, premamillary nucleus; PVN, paraventricular nucleus; SCN, suprachiasmatic nucleus; Tbr1, T-box brain transcription factor 1; VMH, ventromedial hypothalamic nucleus.
ED Figure 2. Molecular analysis of TFs involved in neurogenesis and neuronal differentiation.
(a) Comparative and time-resolved analysis of the ‘cell bridge’ by MNN, CONOS and Seurat alignment. In UMAP space on separate developmental stages, MNN, CONOS and Seurat algorithms were compared for their ability to specifically resolve the transition of progenitors to immature cells (‘bridge’). Color codes correspond to those in Figure 1a). RNA-velocity at E15.5, E17.5 and P0. Color codes are consistent with those in Figure 1a. Note the presence of a ‘bridge’ (grey background) between progenitor/glial and neuronal compartments at early developmental stages with its rupture being evident by birth. (c) Gene expression in UMAP space at E15.5. Note a central role for Notch signalling in neurogenesis. (d) Genetic tracing of Ascl1+ cells produced in the developing hypothalamus during the E12.5-E16.5 period. (e) In situ hybridisation showing the distribution of Tbr1 and Eomes genes as per the open-source Allen brain atlas database (www.brain-map.org). (f) Genetic tracing of Ascl1+ cells in the developing hypothalamus of Ascl+/- and Ascl-/- mice. (g) Sox2, Ascl1 and Rbfox3 localization at successive developmental stages. (h) Genetic tracing of Ascl+ cells postnatally (as in Figure 1f). Scale bars = 200 μm (d), 20 μm (f,g,h).
ED Figure 3. Neurotransmitter and neuropeptide specificity and load in the developing hypothalamus.
(a-c) Coincident profiling of fast neurotransmitters (a), neuropeptides (b) and neuropeptide receptors (c) in 45 cell groups of ectodermal origin. (c1-c2) Given their abundance, Ntrk2 and Adcyap1r1 were plotted separately along the developmental timeline studied with appropriate scaling (c1). Likewise, the distribution of both receptors per cell cluster was mapped and scaled separately (c2). (d) Coincident profiling of neuropeptides in neuronal clusters distinguished as GABA and glutamate phenotypes and graphically identified in blue and grey, respectively. (e) Map of tyrosine hydroxylase (Th) expression in GABA and glutamate neurons. Color coding correspond to that in (d). (f) Developmental mapping of hypothalamic Oxtr expression in OxtrVenus/+ mice. Low-magnification image surveys are shown (see also Figure 2e). Scale bars = 200 μm (f). Data are shown as dot plots and scaled as previously described6,51,65.
ED Figure 4. Hierarchical relationship of GRNs (regulons).
Area under the curve (AUC) separability plot was used to assign regulons that determine cell cluster identities identified in Scenic23. GRNs were reconstructed individually for each cell and then assigned as ‘regulon representation’ (Logreg test) to each cell group. TFs to the left are representative for each regulon. Marked dendrogram branchpoints were estimated by both the Wilcoxon and Logreg tests (see also DOI:10.6084/m9.figshare.11867889).
ED Figure 5. Relationships between regulons and disease phenotypes in humans.
(a) Complete heat map of associations between regulon activity and clinical disease phenotype. Left: classifications of diseases as per phenotypic criteria of the UK biobank registry (www.ukbiobank.ac.uk). Top: master genes for each regulon. Genes presented in Figure 3 are in red and highlighted in (b). Color coding from deep blue to bright yellow show increasing correlation probability. (b) Scatter plot reflecting the ratios of mutability in master genes vs. all downstream target genes per regulon. Mutability and the constrains of TFs were expressed as the total number of mutations. Colors represent four quadrants that were separated on the basis of the total number of mutations per master gene (medians, y axis) vs. target genes (medians, x axis). Horizontal line corresponds to the median of SNPs in all genes. Dot size reflects the median influence of a given regulon on its targets as per Scenic output.
ED Figure 6. Molecular complexity and function of the Onecut3 regulon.
(a) Interlinked Onecut2 and Onecut3 regulons in hypothalamic neurons. Genes that had been biologically validated (see below) are shown in black. (b) Onecut2 and Onecut3 co-expression along the rostrocaudal axis of the hypothalamus. (c) Co-localization of Onecut3 and its target genes (from panel a). (d-d2) Overexpression of Onecut3 and ATP-binding cassette D2 (Abcd2, to control promoter activity) in Neuro2A cells. Representative images by multiple fluorescence labelling-differential interference microscopy. (e) Quantification of Hoechst+ and phosphor-histone H3 (pHH3)+ Neuro2A cells revealed significantly reduced proliferation upon Onecut3 overexpression. No significant cell death was observed by either overexpressed plasmid or the transfection reagent alone. (e) qPCR analysis of genes regulated by Onecut3: CXCC-type zinc finger protein 5 (Cxxc5), transmembrane protease serine 9 (Tmprss9) and tyrosine hydroxylase (Th). All data were normalized to samples transfected with Abcd2, which were taken as technical controls. Scale bars = 50 μm (d,d1) 20 μm (b,f), 10 μm (g).
ED Figure 7. Experimental validation of ventricle-restricted genes identified by scRNA-seq.
(a) Left: Expressional dynamics of ventricle-associated marker genes: Slc1a3 (GLAST), Rax and Dll3 on UMAP embedding (top) and trend lines (bottom). Right: Validation by in situ hybridization. (b,b1) In situ hybridization for the co-existence of Slit2/Rax in ventricular progenitors and consequential medio-to-lateral Slit1/Dll1/Dll3 patterns during neuronal differentiation and migration by E15.5. Left-to-right orientation corresponds to medial-to-lateral hypothalamic positions. (b2) Localization of Slit1 and Slit2 mRNAs in the ventromedial hypothalamus (VMH) at E18.5. Scale bars = 200 μm (a), 20 μm (b,c).
ED Figure 8. Physiological and morphological subtypes of hypothalamic dopamine neurons.
(a) Action potential waveforms of dopamine neurons within the A12-A14 groups. Note the diversification of A14 dopamine cells into subgroups A-D with clearly different action potential signatures. Morphological reconstruction of biocytin-filled neurons is shown to the left of each panel. (b) Distribution of tdTomato+ neurons in the hypothalamus of Slc6a3-Ires-Cre::Ai14 mice. Scale bars = 50 μm (b), 20 μm (a).
ED Figure 9. Transcriptional and physiological features of dopamine neurons in the developing hypothalamus.
(a) Ascl1-CreERT2/+::Ai14 (control) vs. Ascl1-CreERT2/ERT2::Ai14 (a knock-in mouse line with Cre disrupting the Ascl1 gene, referred to as ‘KO’), injected with tamoxifen at E11.5 and analysed at E13.5. Note the accumulation of tdTomato+ cells in the KO relative to controls. (b) Genetic tracing in Ascl1-CreERT2::Ai14 reporter mice identified Ascl1+/Th+ neurons within the preoptic (POA) and periventricular (PeVN) nuclei. Meanwhile, Ascl1-/Th+ neurons populated the arcuate nucleus (Arc) and zona incerta (ZI) by E18.5. (c) Isl1 and Meis2 transcriptional trends of differentiation for trajectories in Th+ groups (#1-9). Amplitudes are shown in log10 scale. Line shading corresponds to means ± s.e.m. (d) Genetic lineage tracing using Isl1-Cre::Ai14 mice. (e) In situ hybridization for Gad1 and Th revealed anti-parallel expressional load for these genes as a factor of medial-to-lateral positioning. Scatter plots show the number of fluorescent puncta per cell (threshold >2). (f) In situ hybridisation for Meis2, Th and Ddc in the hypothalami of E18.5 and P2 mice. Scale bars = 120 μm (a,f/left), 20 μm (k), 12 μm (b,d,e,f/right,i,j).
ED Figure 10. GABA origin of hypothalamic dopamine neurons.
(a) Immunohistochemical analysis of tyrosine hydroxylase (TH) and Onecut3 protein expression in the hypothalamus of (BAC)GAD65-GFP and GAD67gfp/+ mice at the developmental time-points indicated. Note a gradual GABA-to-dopamine transition as a factor of advancing age with OC3 expression preceding that of TH. Dashed rectangles denote the positions of high-resolution insets. (b) Expression patterns of regulon-forming TFs that directly drive Th transcription in the developing hypothalamus. Meis2, Pbx3, Dlx1 were visualised on UMAP embedding for neuronal lineages. (c) Histochemical localization of the migratory route of prospective PeVN dopamine neurons (#9) through the coincident localization of Th and Onecut3 during embryonic development. Dashed lines denote the ventricular surface. (d) Localization of Onecut2 and Pmfbp1a target genes within the Onecut3 regulon to PeVN dopamine neurons by a combination of immunohistochemistry and in situ hybridization. (e) Sst expression in PeVN dopamine neurons. (f) Post-hoc reconstruction of A14 Onecut3+ dopamine neurons after patch-clamp recordings. Abbreviations: 3V, 3rd ventricle; AH, anterior hypothalamus; DMH, dorsomedial hypothalamus; PeVN, periventricular nucleus; VMH, ventromedial hypothalamus. Scale bars = 200 μm (a, overviews), 50 μm (a, insets), 20 μm (f), 12 μm (c,d,e).
Supplementary Material
Supplementary information is available in the online version of the paper.
Acknowledgements
A. Reinthaler is acknowledged for her expert laboratory assistance. We thank the Biomedical Sequencing Facility at the CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences for assistance with next-generation sequencing, G.A. Bazykin for providing access to the 2TB RAM 'Makarich' computational cluster for OMICS data analysis, M. Watanabe for antibodies, and G. Szabó, F. Erdélyi, J. Bunt, L.J. Richards and Y. Yanagawa for transgenic mice. This work was supported by the Swedish Research Council (F.L., I.A., T.Hö., T.Ha.); Novo Nordisk Foundation (T.Hö., T.Ha.); Bertil Hållsten Research Foundation (I.A.); Hjärnfonden (T.Ha.), European Research Council (STEMMING-FROM-NERVE, 2014-CoG-647844; I.A. and SECRET-CELLS, 2015-AdG-695136; T.Ha.), the EMBO Young Investigator Program (I.A.), Åke Wiberg Foundation (I.A.), Wallenberg Academy fellowship (F.L.), a Ming Wai Lau Center investigator grant (F.L.), the Strategic Research program for Brain Sciences (AMED, Japan; K.N.) and intramural funds of the Medical University of Vienna (T.Ha.). M.F. is supported by a special research program of the Austrian Science Fund (FWF-F61). R.A.R. is an EMBO advanced research fellow (ALTF 493-2017). E.O.T. is supported by a scholarship from the Austrian Science Fund (FWF, DOC 33-B27).
Footnotes
Author contributions: T.Ha. and R.A.R. conceived the project, K.N., I.A., T.Hö. and T.Ha. procured funding; R.A.R., I.A., M.F., C.B., T.Hö., E.K. and T.Ha. designed experiments; R.A.R., E.T., M.E.K., M.Z., M.H., S.K., K.P., M.B., P.R. and M.F. performed experiments and analyzed data, F.L., K.N., F.C., W.D.A. and J.G.P. provided unique reagents and mouse models. R.A.R. and T.Ha. wrote the manuscript with input from all co-authors.
Conflict of interest: The authors declare that they have no conflict of interest.
Data availability: ScRNA-seq data were deposited in Gene Expression Omnibus (accession number: GSE132730) without restriction for review or analysis. We refer to the Supplementary Information for downloadable datasets.
References
- 1.Saper CB, Lowell BB. The hypothalamus. Current biology : CB. 2014;24:R1111–1116. doi: 10.1016/j.cub.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 2.Dulac C, O'Connell LA, Wu Z. Neural control of maternal and paternal behaviors. Science. 2014;345:765–770. doi: 10.1126/science.1253291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Y, Dorsky RI. Development of the hypothalamus: conservation, modification and innovation. Development. 2017;144:1588–1599. doi: 10.1242/dev.139055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romanov RA, et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nature Neuroscience. 2017;20:176–188. doi: 10.1038/nn.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews W, et al. The role of Slit-Robo signaling in the generation, migration and morphological differentiation of cortical interneurons. Developmental biology. 2008;313:648–658. doi: 10.1016/j.ydbio.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 6.Mayer C, et al. Developmental diversification of cortical inhibitory interneurons. Nature. 2018;555:457–462. doi: 10.1038/nature25999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nature reviews. Neuroscience. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeisel A, et al. Molecular Architecture of the Mouse Nervous System. Cell. 2018;174:999–1014 e1022. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell JN, et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat Neurosci. 2017;20:484–496. doi: 10.1038/nn.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toda C, Santoro A, Kim JD, Diano S. POMC Neurons: From Birth to Death. Annual review of physiology. 2017;79:209–236. doi: 10.1146/annurev-physiol-022516-034110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burbridge S, Stewart I, Placzek M. Development of the Neuroendocrine Hypothalamus. Comprehensive Physiology. 2016;6:623–643. doi: 10.1002/cphy.c150023. [DOI] [PubMed] [Google Scholar]
- 12.Wolf FA, et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome biology. 2019;20:59. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aujla PK, Naratadam GT, Xu L, Raetzman LT. Notch/Rbpjkappa signaling regulates progenitor maintenance and differentiation of hypothalamic arcuate neurons. Development. 2013;140:3511–3521. doi: 10.1242/dev.098681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNay DE, Pelling M, Claxton S, Guillemot F, Ang S-L. Mash1 is required for generic and subtype differentiation of hypothalamic neuroendocrine cells. Molecular Endocrinology. 2006;20:1623–1632. doi: 10.1210/me.2005-0518. [DOI] [PubMed] [Google Scholar]
- 16.De Marco Garcia NV, Karayannis T, Fishell G. Neuronal activity is required for the development of specific cortical interneuron subtypes. Nature. 2011;472:351–355. doi: 10.1038/nature09865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cobos I, Borello U, Rubenstein JL. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron. 2007;54:873–888. doi: 10.1016/j.neuron.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wamsley B, Fishell G. Genetic and activity-dependent mechanisms underlying interneuron diversity. Nature reviews Neuroscience. 2017;18:299–309. doi: 10.1038/nrn.2017.30. [DOI] [PubMed] [Google Scholar]
- 19.Gerstner JR, Landry CF. Expression of the transcriptional coactivator CITED1 in the adult and developing murine brain. Developmental neuroscience. 2007;29:203–212. doi: 10.1159/000096389. [DOI] [PubMed] [Google Scholar]
- 20.Hokfelt T, Meister B, Melander T, Everitt B. Coexistence of classical transmitters and peptides with special reference to the arcuate nucleus--median eminence complex. Advances in biochemical psychopharmacology. 1987;43:21–34. [PubMed] [Google Scholar]
- 21.Motoike T, et al. Transient expression of neuropeptide W in postnatal mouse hypothalamus--a putative regulator of energy homeostasis. Neuroscience. 2015;301:323–337. doi: 10.1016/j.neuroscience.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Maejima Y, et al. Oxytocinergic circuit from paraventricular and supraoptic nuclei to arcuate POMC neurons in hypothalamus. FEBS Lett. 2014;588:4404–4412. doi: 10.1016/j.febslet.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nature Methods. 2017;14:1083. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyle EA, Li YI, Pritchard JK. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popadin K, et al. Slightly deleterious genomic variants and transcriptome perturbations in Down syndrome embryonic selection. Genome Res. 2018;28:1–10. doi: 10.1101/gr.228411.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardenas A, et al. Evolution of Cortical Neurogenesis in Amniotes Controlled by Robo Signaling Levels. Cell. 2018;174:590–606 e521. doi: 10.1016/j.cell.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chedotal A, Richards LJ. Wiring the brain: the biology of neuronal guidance. Cold Spring Harb Perspect Biol. 2010;2 doi: 10.1101/cshperspect.a001917. a001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arenas E, Denham M, Villaescusa JC. How to make a midbrain dopaminergic neuron. Development. 2015;142:1918–1936. doi: 10.1242/dev.097394. [DOI] [PubMed] [Google Scholar]
- 29.Tritsch NX, Ding JB, Sabatini BL. Dopaminergic neurons inhibit striatal output through non-canonical release of GABA. Nature. 2012;490:262–266. doi: 10.1038/nature11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsushita N, et al. Dynamics of tyrosine hydroxylase promoter activity during midbrain dopaminergic neuron development. J Neurochem. 2002;82:295–304. doi: 10.1046/j.1471-4159.2002.00972.x. [DOI] [PubMed] [Google Scholar]
- 31.Cowley MA, et al. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature. 2001;411:480–484. doi: 10.1038/35078085. [DOI] [PubMed] [Google Scholar]
- 32.Kim EJ, Ables JL, Dickel LK, Eisch AJ, Johnson JE. Ascl1 (Mash1) defines cells with long-term neurogenic potential in subgranular and subventricular zones in adult mouse brain. PLoS ONE. 2011;6:e18472. doi: 10.1371/journal.pone.0018472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature Neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shu T, Butz KG, Plachez C, Gronostajski RM, Richards LJ. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J Neurosci. 2003;23:203–212. doi: 10.1523/JNEUROSCI.23-01-00203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bali B, Erdélyi F, Szabó G, Kovács KJ. Visualization of stress-responsive inhibitory circuits in the GAD65-eGFP transgenic mice. Neurosci Lett. 2005;380:60–65. doi: 10.1016/j.neulet.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Tamamaki N, et al. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. The Journal of Comparative Neurology. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- 37.Andrews W, et al. Robo1 regulates the development of major axon tracts and interneuron migration in the forebrain. Development. 2006;133:2243–2252. doi: 10.1242/dev.02379. [DOI] [PubMed] [Google Scholar]
- 38.Plump AS, et al. Slit1 and Slit2 cooperate to prevent premature midline crossing of retinal axons in the mouse visual system. Neuron. 2002;33:219–232. doi: 10.1016/s0896-6273(01)00586-4. [DOI] [PubMed] [Google Scholar]
- 39.Yang L, et al. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development. 2006;133:1575–1585. doi: 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida M, et al. Evidence that oxytocin exerts anxiolytic effects via oxytocin receptor expressed in serotonergic neurons in mice. J Neurosci. 2009;29:2259–2271. doi: 10.1523/JNEUROSCI.5593-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bäckman CM, et al. Characterization of a mouse strain expressing Cre recombinase from the 3' untranslated region of the dopamine transporter locus. Genesis. 2006;44:383–390. doi: 10.1002/dvg.20228. [DOI] [PubMed] [Google Scholar]
- 42.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lun ATL, et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biology. 2019;20:63. doi: 10.1186/s13059-019-1662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petukhov V, et al. dropEst: pipeline for accurate estimation of molecular counts in droplet-based single-cell RNA-seq experiments. Genome biology. 2018;19:78. doi: 10.1186/s13059-018-1449-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Brink SC, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nature Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- 46.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo S, Blackshaw S. Regulation and function of neurogenesis in the adult mammalian hypothalamus. Progress in neurobiology. 2018;170:53–66. doi: 10.1016/j.pneurobio.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miranda-Angulo AL, Byerly MS, Mesa J, Wang H, Blackshaw S. Rax regulates hypothalamic tanycyte differentiation and barrier function in mice. The Journal of Comparative Neurology. 2014;522:876–899. doi: 10.1002/cne.23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimogori T, et al. A genomic atlas of mouse hypothalamic development. Nature Neuroscience. 2010;13:767–775. doi: 10.1038/nn.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. bioRxiv. 2019 doi: 10.1186/s13059-019-1874-1. 576827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finak G, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biology. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 54.Alpár A, Benevento M, Romanov RA, Hökfelt T, Harkany T. Hypothalamic cell diversity: non-neuronal codes for long-distance volume transmission by neuropeptides. Curr Opin Neurobiol. 2019;56:16–23. doi: 10.1016/j.conb.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 55.Romanov RA, Alpár A, Hökfelt T, Harkany T. Unified classification of molecular, network, and endocrine features of hypothalamic neurons. Annu Rev Neurosci. 2019;42:1–26. doi: 10.1146/annurev-neuro-070918-050414. [DOI] [PubMed] [Google Scholar]
- 56.Polański K, et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 2020;36:964–965. doi: 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McInnes L, Healy J, Saul N, Großberger L. UMAP: uniform manifold approximation and projection. JOSS. 2018;3:861. doi: 10.21105/joss.00861. [DOI] [Google Scholar]
- 58.Johnson J, Douze M, Jegou H. Billion-scale similarity search with GPUs. IEEE Trans Big Data. 2019 doi: 10.1109/TBDATA.2019.2921572. 1–1. [DOI] [Google Scholar]
- 59.Maneewongvatana S, Mount D. In: Data structures, near neighbor searches, and methodology: fifth and sixth DIMACS implementation challenges. DIMACS series in discrete mathematics and theoretical computer science. Goldwasser Michael, Johnson David, McGeoch Catherine., editors. Vol. 59. American Mathematical Society; 2002. pp. 105–123. [Google Scholar]
- 60.Hie B, Bryson B, Berger B. Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat Biotechnol. 2019;37:685–691. doi: 10.1038/s41587-019-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welch JD, et al. Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell. 2019;177:1873–1887 e1817. doi: 10.1016/j.cell.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korsunsky I, Fan J, Slowikowski K, Zhang F, Wei K. Fast, sensitive, and accurate integration of single cell data with harmony. bioRxiv. 2018 doi: 10.1038/s41592-019-0619-0. 461954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barkas N, et al. Joint analysis of heterogeneous single-cell RNA-seq dataset collections. Nat Methods. 2019;16:695–698. doi: 10.1038/s41592-019-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kowalczyk MS, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Becht E, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nature Biotechnology. 2018 doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 68.Crow M, Paul A, Ballouz S, Huang ZJ, Gillis J. Characterizing the replicability of cell types defined by single cell RNA-sequencing data using MetaNeighbor. Nature Communications. 2018;9 doi: 10.1038/s41467-018-03282-0. 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen R, Wu X, Jiang L, Zhang Y. Single-Cell RNA-Seq Reveals Hypothalamic Cell Diversity. Cell reports. 2017;18:3227–3241. doi: 10.1016/j.celrep.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moffitt JR, et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science. 2018;362 doi: 10.1126/science.aau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vinh NX, Epps J, Bailey J. Information Theoretic Measures for Clusterings Comparison: Variants, Properties, Normalization and Correction for Chance. Journal of Machine Learning Research. 2010 [Google Scholar]
- 72.Chang F, Qiu W, Zamar RH, Lazarus R, Wang X. clues : an r package for nonparametric clustering based on local shrinking. J Stat Softw. 2010;33 doi: 10.18637/jss.v033.i04. [DOI] [Google Scholar]
- 73.Pons P, Latapy M. International symposium on computer and information sciences; Springer; pp. 284–293. [Google Scholar]
- 74.Kurrasch DM, et al. The neonatal ventromedial hypothalamus transcriptome reveals novel markers with spatially distinct patterning. J Neurosci. 2007;27:13624–13634. doi: 10.1523/JNEUROSCI.2858-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kelley KW, Nakao-Inoue H, Molofsky AV, Oldham MC. Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nature Neuroscience. 2018;21:1171–1184. doi: 10.1038/s41593-018-0216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin Y, et al. Evaluating stably expressed genes in single cells. Gigascience. 2019;8 doi: 10.1093/gigascience/giz106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Extra Coordinate Systems, 'Geoms', Statistical Transformations, Scales and Fonts for 'ggplot2' [R package ggalt version 0.4.0] v. 0.4.0. 2017 https://CRAN.R-project.org/package=ggalt.
- 79.Traag V, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Scientific reports. 2019;9 doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang JM, Kamath GM, Tse DN. Valid Post-clustering Differential Analysis for Single-Cell RNA-Seq. Cell Syst. 2019 doi: 10.1016/j.cels.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biology. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. BioRxiv. 2019 doi: 10.1101/820936. [DOI] [PubMed] [Google Scholar]
- 83.Moerman T, et al. GRNBoost2 and Arboreto: efficient and scalable inference of gene regulatory networks. Bioinformatics. 2019;35:2159–2161. doi: 10.1093/bioinformatics/bty916. [DOI] [PubMed] [Google Scholar]
- 84.Kanamori M, et al. A genome-wide and nonredundant mouse transcription factor database. Biochem Biophys Res Commun. 2004;322:787–793. doi: 10.1016/j.bbrc.2004.07.179. [DOI] [PubMed] [Google Scholar]
- 85.Ravasi T, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Consortium F, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ntranos V, Yi L, Melsted P, Pachter L. Identification of transcriptional signatures for cell types from single-cell RNA-Seq. BioRxiv. 2018 doi: 10.1101/258566. [DOI] [Google Scholar]
- 88.Bastian M, Heymann S, Jacomy M. Gephi: An Open Source Software for Exploring and Manipulating Networks. 2009 [Google Scholar]
- 89.Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat Genet. 2018;50:1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.qvalue: Q-value estimation for false discovery rate control v. 2.18.0 (R package) 2019.
- 92.Galili T, O'Callaghan A, Sidi J, Sievert C. heatmaply: an R package for creating interactive cluster heatmaps for online publishing. Bioinformatics. 2018;34:1600–1602. doi: 10.1093/bioinformatics/btx657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Setty M, et al. Characterization of cell fate probabilities in single-cell data with Palantir. Nature Biotechnology. 2019;37:451–460. doi: 10.1038/s41587-019-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Levine JH, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Espana A, Clotman F. Onecut transcription factors are required for the second phase of development of the A13 dopaminergic nucleus in the mouse. The Journal of Comparative Neurology. 2012;520:1424–1441. doi: 10.1002/cne.22803. [DOI] [PubMed] [Google Scholar]
- 96.Miyazaki T, Fukaya M, Shimizu H, Watanabe M. Subtype switching of vesicular glutamate transporters at parallel fibre-Purkinje cell synapses in developing mouse cerebellum. Eur J Neurosci. 2003;17:2563–2572. doi: 10.1046/j.1460-9568.2003.02698.x. [DOI] [PubMed] [Google Scholar]
- 97.Bartesaghi L, et al. PRDM12 Is Required for Initiation of the Nociceptive Neuron Lineage during Neurogenesis. Cell reports. 2019;26:3484–3492.e3484. doi: 10.1016/j.celrep.2019.02.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang F, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choi HMT, et al. Mapping a multiplexed zoo of mRNA expression. Development. 2016;143:3632–3637. doi: 10.1242/dev.140137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao S, et al. Cell type–specific channelrhodopsin-2 transgenic mice for optogenetic dissection of neural circuitry function. Nature Methods. 2011;8:745–752. doi: 10.1038/nmeth.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Custom code was neither generated nor used. Raw, processed and supplementary datasets were deposited in GEO (accession number: GSE132730). GEO files include: 1) raw fastq files for every sequencing run; 2) filtered matrices for every sample in RDS file format including Seurat 3 objects with all processed cells; 3) original integrated dataset in RDS file format including Seurat 3 objects with all processed cells as well as all used commands; 4) integrated dataset used for dynamics analysis (which passed filtering of RNA Velocity analysis); 5) AUCell matrices from pySCENIC pipeline; 6) full regulon hypothalamic network in GraphML file format; 7) metadata protocol describing all experimental, computational procedures and quality control. An interactive view of the integrated dataset (for processing in Pagoda2) can be accessed through the following URLs: https://dx.doi.org/10.6084/m9.figshare.11867889 (~1.1 GB).
All data presented (e.g., imaging) will be made available by T.Ha. (tibor.harkany@ki.se or tibor.harkany@meduniwien.ac.at) upon reasonable request.