Abstract
Initial enteropathogen growth in the microbiota-colonized gut is poorly understood. Salmonella Typhimurium is metabolically adaptable and can harvest energy by anaerobic respiration using microbiota-derived hydrogen (H2) as an electron donor and fumarate as an electron acceptor. As fumarate is scarce in the gut, the source of this electron acceptor is unclear. Here, transposon sequencing analysis along the colonization-trajectory of S.Typhimurium implicates the C4-dicarboxylate antiporter DcuABC in early murine gut colonization. In competitive colonization assays, DcuABC and enzymes that convert the C4-dicarboxylates aspartate and malate into fumarate (AspA, FumABC), are required for fumarate/H2-dependent initial growth. Thus, S.Typhimurium obtains fumarate by DcuABC-mediated import and conversion of L-malate and L-aspartate. Fumarate reduction yields succinate, which is exported by DcuABC in exchange for L-aspartate and L-malate. This cycle allows S.Typhimurium to harvest energy by H2/fumarate respiration in the microbiota-colonized gut. This strategy may also be relevant for commensal E. coli diminishing the S.Typhimurium infection.
Graphical Abstract
eTOC Blurb
Bidong Nguyen and colleagues show how Salmonella Typhimurium grows after arrival in the gut. Salmonella Typhimurium imports aspartate and malate using the DcuABC transporters and converts these substrates into fumarate to fuel anaerobic H2/fumarate respiration. Thereby, Salmonella can grow in face of an intact gut microbiota to establish the infection.
Introduction:
The mammalian gut limits microbial growth via chemical defenses, physical barriers and by depleting nutrients. Most microbial growth is observed in the large intestine, which harbors the bulk of the microbiota. This dense microbial consortium utilizes nutrients that are not depleted by the host, e.g. by fermenting complex carbohydrates. Importantly, the microbiota can limit pathogen growth in the large intestinal lumen (Stecher and Hardt, 2011, Buffie and Pamer, 2013). This colonization resistance (CR) is attributable to diverse mechanisms like microbiota-mediated nutrient depletion, release of pathogen-inhibitors (bacteriocins, lantibiotics, type VI secretion systems), blocking of binding sites, depletion of terminal electron acceptors, or vitamin B6 production and microbiota-mediated modulation of enterocyte metabolism (Fabich et al., 2008, Freter et al., 1983, Sonnenburg et al., 2005, Stecher, 2015, Stecher et al., 2013, Miki et al., 2017, Buffie and Pamer, 2013, Hecht et al., 2016, Byndloss et al., 2017). CR reduces the incidence of enteropathogen colonization in humans and mouse models, alike. Nevertheless, in some cases, enteropathogens succeed in colonizing the gut lumen in face of the microbiota. The underlying mechanisms of how enteropathogens can grow before the onset of overt enteric disease are not completely understood.
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a common enteropathogenic bacterium, which can infect a broad range of hosts, including livestock, humans and mice. While CR suppresses S. Typhimurium growth in the intestine of most exposed hosts, in some cases the pathogen can initiate gut luminal growth (Simonsen et al., 2009) (Stecher and Hardt, 2011, Buffie and Pamer, 2013). Studies from mouse infections and human outbreaks suggest that there is an initial phase of pathogen growth (termed “initial growth”) before the mucosa responds with overt enteropathy, characterized by pronounced phagocyte infiltration, re-modeled mucosal architecture and an altered gut luminal milieu (Wotzka et al., 2017) (Organization and Nations, 2002) (Maier et al., 2013). To fuel initial growth, S. Typhimurium can harvest energy by anaerobic hydrogen (H2)/fumarate respiration (Maier et al., 2013). H2 is a product of anaerobic microbiota metabolism and serves as an electron donor for the hyb hydrogenase (Maier et al., 2013) (Maier et al., 2014a) (Dubini et al., 2002) (Maier, 2005). The resulting protons feed the proton gradient to support ATP production (Unden et al., 2016) and the electrons are transferred to fumarate reductase (Frd) which reduces fumarate to succinate (Maier et al., 2013). However, it remained unclear how S. Typhimurium disposes of succinate and how it obtains fumarate. The free fumarate concentration in the gut lumen might be too low to support this mechanism of energy conservation (Spiga et al., 2017, Cummings et al., 1987).
Here, we employ an unbiased mutagenesis approach, gene deletion epistasis experiments and metabolite analyses to identify DcuABC-dependent uptake of malate and aspartate and their conversion into fumarate as the principle mechanism fueling anaerobic hydrogen (H2)/fumarate respiration during initial growth.
Results:
RB-TnSeq screen implicates C4-dicarboxylate transport in initial growth.
To gain insights into the initial growth physiology of S. Typhimurium in the gut lumen, we have used strain SL1344, a well-established model for Salmonella gut colonization(Barthel et al., 2003) (Maier et al., 2013) (Wotzka et al., 2019) (Sana et al., 2016) and employed a random-barcoded transposon sequencing approach (RB-TnSeq; (Price et al., 2018, Wetmore et al., 2015)). Via transposome transformation (Langridge et al., 2009) (Canals et al., 2012), we generated a library of ≈ 2000 randomly barcoded transposon insertion mutants of S. Typhimurium SL1344 (RB-TnSeq library; Fig. 1A, Fig. S1A). This number of mutants is close to the upper limit for parallel analysis in the same mouse (Maier et al., 2014b) (Maier et al., 2013). The RB-TnSeq mutations were mapped by sequencing of their unique barcodes in conjunction with their transposon’s insertion sites (Fig. S1B). To facilitate quality control, the inoculum pool was spiked with several controls (Fig. 1A; Key Resources Table). Wild type isogenic tagged strains (WITS, Key Resources Table; (Grant et al., 2008)) were used to flag cases where gut inflammation might confound mutant fitness analysis by inflicting bottlenecks in the gut-luminal S. Typhimurium population (Maier et al., 2014b) (Fig. 1A, Fig. S1C). An ampicillin resistant hyb mutant was included to verify H2-dependent S. Typhimurium growth in the gut lumen. Titration series of RB-TnSeq-barcoded wild type S. Typhimurium, and isogenic mutants (hyb, ttss-1/ttss-2, cheY) were also spiked in (Fig. 1A, Key Resources Table). These control strains featured unique RB-TnSeq-barcode sequences, which served as internal standards during the transposon barcode amplification and quantification procedure by tracking fitness of the RB-TnSeq mutants along the colonization trajectory.
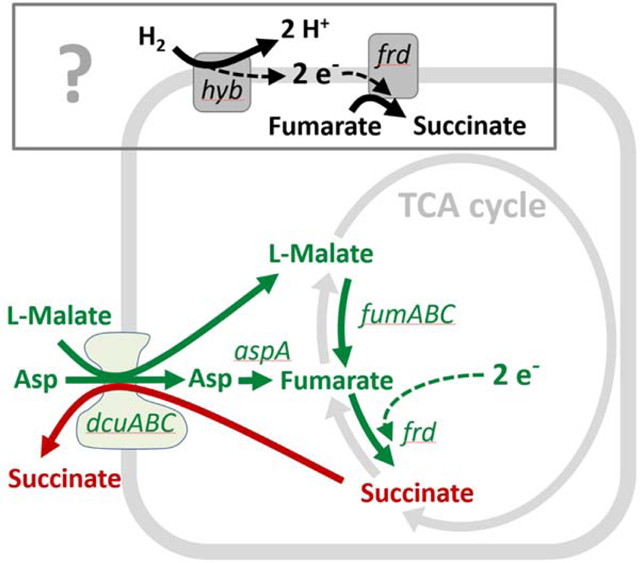
Figure 1. RB-TnSeq screen to identify Salmonella genes involved in growth and colonization of the murine gut.
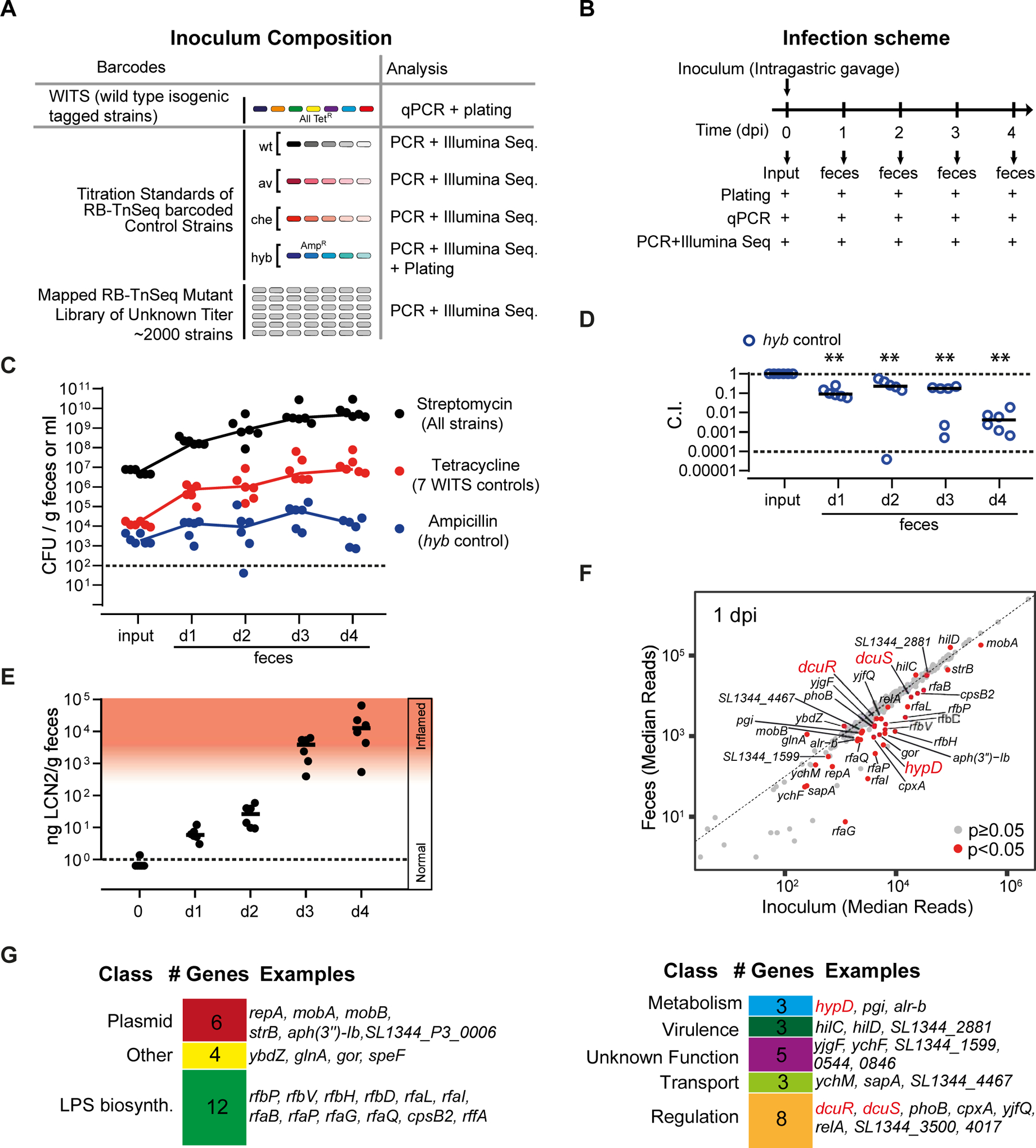
A. Composition of the inoculum pool. WITS-barcoded wild type controls were spiked in at densities of 1/2000 and titration standards of RB-TnSeq barcoded wild type and mutant controls as detailed in Star Methods and the Key Resource Table. B. Mouse infection protocol and sampling procedures. C. S. Typhimurium loads in the inoculum and the feces at days 1–4 pi as determined by plating. Black: Total S. Typhimurium loads. Red: total loads of WITS-barcoded wild type controls. Blue: Loads of the hyb mutant. Dotted line: detection limit. D. Attenuation of the hyb control strain (C.I.). E. Gut inflammation as measured by Lipocalin-2 ELISA. F. Fitness defects of RB-TnSeq mutants. We compared the relative barcode read abundances from the inoculum and the feces (day 1 pi; full data shown in Tab. S3). Red dots: mutants with significantly reduced or enhanced fitness compared to the wild type control strains (p < 0.05). G. Gene classes represented among the 43 RB-TnSeq hits implicated in initial growth.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and Virus Strains | ||
| See Supplemental Table S5 | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Isotopes: [13C4]-fumaric acid (99%), [13C4, 15N]-L-aspartic acid (99%), [1, 4–13C2]-succinic acid (99%) and [13C4]-L-malic acid (98%) | Cambridge Isotope Laboratories | CLM-1529, CNLM-544-H-0.25, CLM-1084-PK CLM-8065-PK, respectively |
| Critical Commercial Assays | ||
| DuoSet Lipocalin ELISA kit | R&D Systems, Minneapolis, MN, USA | DY1857 |
| AllPrep DNA/RNA Kit | Qiagen | 80204 |
| NEXTflex® 16S V4 Amplicon-Seq Kit 2.0 | Bio Scientific | NOVA-4203–04 |
| Deposited Data | ||
| 16S sequencing data of microbiome in mice | this manuscript | http://www.ebi.ac.uk/ena/data/view/PRJEB35643. |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Mouse: LCM mice (C57BL/6 genetic background) | (Stecher et al., 2010) (Maier et al., 2013) | N/A |
| Mouse: 129SvEv | Jackson Laboratories; bred at EPIC mouse facility of ETH Zurich, Switzerland | N/A |
| Mouse: C57BL/6 | Jackson Laboratories; bred at EPIC mouse facility of ETH Zurich, Switzerland | N/A |
| Oligonucleotides | ||
| See Supplemental Table S6 | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| Graphpad Prism Version 6.0 for Windows | GraphPad Software, La Jolla California USA | N/A |
| eMZed2 | (Kiefer et al., 2013) (Hartl et al., 2017) | http://emzed.ethz.ch/ |
| BLASTp of the NCBI BLAST+ (version 2.8.1) | (Camacho et al., 2009) | N/A |
| Clusters of Orthologous Groups with EggNOG-mapper 2.0.0 | (Galperin et al., 2015) (Huerta-Cepas et al., 2017) |
N/A |
| MassHunter Quantitative Analysis Software | (Agilent, version 7.0) | N/A |
| Other | ||
The screen was performed in gnotobiotic mice associated with a low-complexity microbiota consortium (LCM mice; Star Methods (Maier et al., 2013)). LCM mice provide a well-defined and highly reproducible gut colonization system where the initial H2-dependent growth of S. Typhimurium lasts for 2 days and which permits the fitness analysis of 2000 strains in parallel during this colonization process ((Maier et al., 2013); Fig. S1C). We orally infected 6 LCM mice with our inoculum pool composed of ≈2000 strains (Fig. 1A, 5 × 106 cfu total) and subsequently recovered mutant mixtures from feces collected at days 1–4 (Fig. 1B). We reasoned that the frequencies of mutants impaired in colonization are reduced along the infection trajectory, resulting in decreased frequencies of the corresponding barcodes from feces samples compared to the inoculum pool. Plating on ampicillin agar verified hyb-dependent gut colonization in all mice (competitive index C.I. ≤ 0.1; Fig. 1C,D). Fecal levels of the gut inflammation marker lipocalin-2 verified that initial growth in the absence of overt gut inflammation lasted for 2 days and that pronounced enteropathy commenced by days 3 and 4 post infection (Fig. 1E; Fig. S1D). DNA was prepared from the inoculum pool and from fecal bacteria (see Materials and Methods) to measure fitness phenotypes of mutants and control strains. WITS- and RB-TnSeq controls verified the absence of bottlenecks during days 1 and 2 (Shannon evenness score EH > 0.95, Fig. S1E; linearity of dilution series R > 0.93; Fig. S1F). Using stringent quality controls (Materials and Methods), we could determine the fitness of 1305 mapped RB-TnSeq mutants during initial growth (days 1 and 2 post infection (pi); Tab. S1). This covered 732 genes and 205 intergenic regions. Linearity was reduced in 3/6 mice by day 3 and in 5/6 mice by day 4 pi (R ≥ 0.3; Fig. S1F). Therefore, we limited our mutant phenotype analysis to days 1 and 2 pi.
Figure 5. Oral administration of malate and aspartate promotes initial growth of S. Typhimurium in the gut lumen.
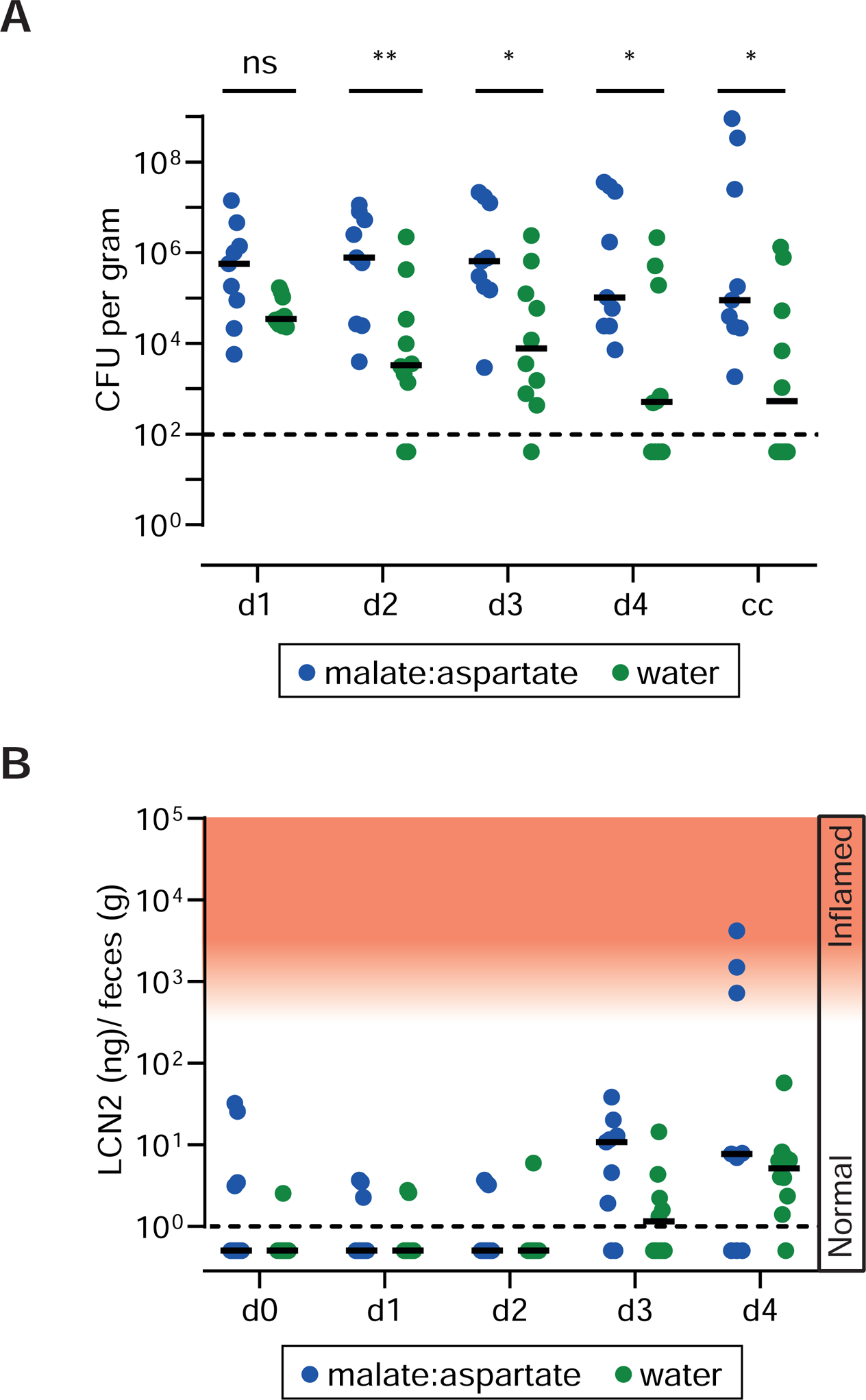
A. CONE mice (N=9) were orally gavaged with 20 mg of an equal mixture (w/w) of L-aspartate and L-malate four hours after infection with a mixed inoculum composed of WITS-barcoded S.Tmwt, S.TmdcuABC, S.TmfumABC aspA, S.Tmhyb, S.TmaspA, S.TmfumABC, S.Tm frd, and untagged wild type S. Typhimurium (5 × 107 cfu; mixed inoculum of 8 strains) and once per day thereafter. Control mice were treated with water (N=10). A. Total S. Typhimurium loads in the feces collected at days 1–4 pi. B. Lipocalin-2 ELISA.
By comparing fitness values across the profiled RB-TnSeq mutants along the infection trajectory, we mapped a set of 44 Tn5 mutants impaired or enhanced in colonization at day 1 and/or 2 pi (p<0.05; Fig. 1F, red dots; Tab. S1). Functional classification revealed that the majority of the identified genes comprise cellular functions such as metabolism, virulence, lipopolysaccharide biosynthesis and gene regulation. The identified gene-set included well-characterized pathogenicity factors, including attenuating mutations in LPS O-sidechain biosynthesis (Ilg et al., 2009) and hypD (Maier et al., 2013) (Fig. 1G; Tab. S1). The latter encodes a maturation factor required for [NiFE] hydrogenases like Hyb, which is used for H2/fumarate respiration (Stripp et al., 2013). We also observed the expected growth acceleration of “cheater mutants” including hilC and hilD defective in the costly expression of the TTSS-1 type III secretion system ((Diard et al., 2013) (Bakkeren et al., 2017); Fig. 1F,G; Tab. S1). Moreover, mutants lacking the two component system DcuR/S, involved in dicarboxylate transport regulation in the closely related bacterium E. coli (Unden et al., 2016), were attenuated (Fig. 1F,G; Tab. S1).
To validate the importance of the identified colonization factors, we constructed a panel of 29 barcoded in-frame deletion mutants (Key Resources Table). We included 11 genes that had yielded significantly enhanced or reduced C.I.s (C.I. < 0.68 or C.I. > 2.06; p < 0.05, day 1 pi; Tab. S1) in the RB-TnSeq follow-up experiment. They represented the identified gene classes of interest (Tab. 1). In addition, we included 18 controls (Tab. S2). Competitive infections of LCM mice confirmed the reduced C.I.s for 9/9 mutants with reduced and for 2/2 mutants with enhanced C.I.s (C.I. > 2.06); Tab. 1 and Tab. S2) identified by our screen during initial growth. Among the identified colonization factors, the DcuR/S two-component system attracted our particular attention, as two independent hits were identified in our screen, the in-frame deletion verified attenuation (C.I.(dcuR) = 0.34, p = 0.002; day 1 pi) and as DcuR/S is linked to fumarate respiration. If C4-dicarboxylates are available under anaerobic conditions, the DcuR/S signaling system induces the expression of genes involved in the uptake and metabolism of C4-dicarboxylates. Specifically, DcuR/S regulates expression of the C4-dicarboxylate transporter DcuB, of FumB, an enzyme converting L-malate into fumarate, and of fumarate reductase (Kleefeld et al., 2009) (Golby et al., 1999) (Unden et al., 2016). Therefore, the identification of DcuR/S in our RB-TnSeq screen suggested that C4-dicarboxylate transport and the conversion of C4-dicarboxylates into fumarate may contribute to initial growth in the gut lumen.
Table 1.
| day 4 p.i. | ||||||
|---|---|---|---|---|---|---|
| gene | function | gene # | p value | p value | p value | p value |
| rfaP | LPS synth. | 3687 | 0.002 | 1E-03 | 1E-03 | 0.016 |
| gor | other | 3562 | 0.002 | 1E-03 | 1E-03 | 0.016 |
| rfbP | LPS synth. | 2059 | 0.027 | 1E-03 | 2E-03 | 0.016 |
| cpxA | regulation | 4007 | 0.002 | 1E-03 | 1E-03 | 0.016 |
| rfaB | LPS synth. | 3685 | 0.002 | 1E-03 | 1E-03 | 0.016 |
| phoB | regulation | 392 | 0.002 | 1E-03 | 1E-03 | 0.016 |
| yjgF | unknown | 4389 | 0.002 | 1E-03 | 1E-03 | 0.016 |
| yjfQ | regulation | 4314 | 0.002 | 1E-03 | 1E-03 | 0.016 |
| dcuR | regulation | 4240 | 0.002 | 1E-03 | 1E-03 | 0.016 |
| hilC | virulence | 2847 | 0.014 | 1E-02 | 2E-01 | 0.047 |
we selected the hits with C.I.s < 0.68 or >2.06 in RB-TnSeq screen and analyzed 10 (or 9) mice (day 1), 11 mice (day 2, day 3), or 7 mice (day 4) respectively; The full dataset is shown in Tab. S2
The DcuABC C4-dicarboxylate transporters are required for initial growth in the gut lumen.
S. Typhimurium encodes three Dcu transporters (DcuA, DcuB, DcuC). In E. coli, they can functionally complement each other to support anaerobic fumarate-dependent growth on C4-dicarboxylates in vitro (Fig. 2A; (Unden et al., 2016, Zientz et al., 1999, Zientz et al., 1996)). While Dcu transporters have been implicated in S. Typhimurium growth in the inflamed gut (Spiga et al., 2017) (Tchawa Yimga et al., 2006, Mercado-Lubo et al., 2009, Rivera-Chavez et al., 2016) (Wilson et al., 2019), their putative role in the initial gut luminal growth of S. Typhimurium by H2/fumarate respiration has remained unclear. Therefore, we decided to decipher their function. As Dcu transporter genes were not represented in our RB-TnSeq library, we created the respective S. Typhimurium mutants by site-directed mutagenesis (Key Resources Table) and analyzed gut colonization during initial growth at days 1 and 2 of infection, as well as in the overtly inflamed gut (days 3–4 pi, in case of wild type S. Typhimurium infections; Fig. S2).
Figure 2. Dcu transporters are contributing to anaerobic respiration during initial gut luminal growth.
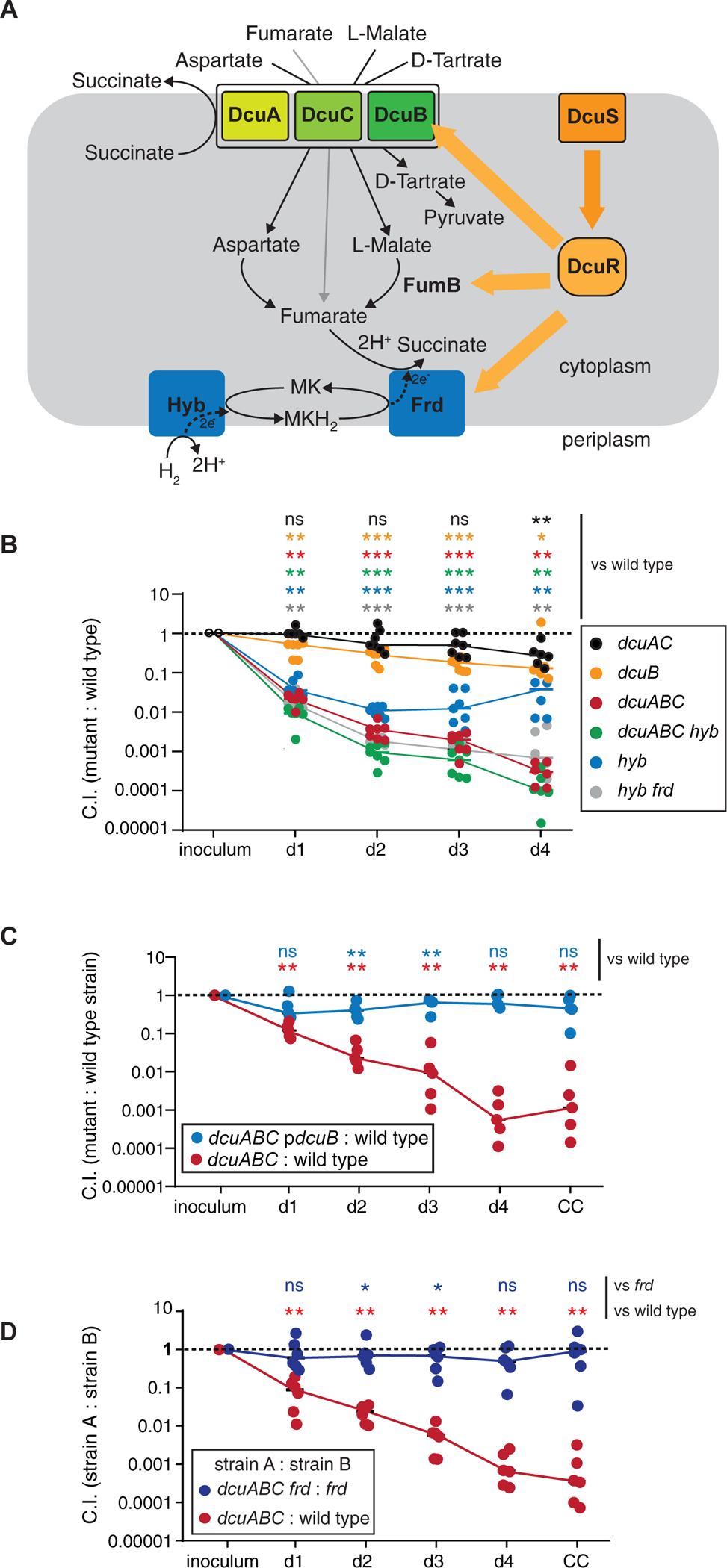
A. Scheme illustrating the regulation by DcuRS and the role of Dcu transporters in C4-dicarboxylate-fueled anaerobic respiration by E. coli. Under anaerobic conditions, and in the presence of suitable substrates, DcuRS activates expression of the indicated proteins. C4-Dicarboxylates, namely aspartate, fumarate, L-malate and L-tartrate, can be imported by Dcu transporters. Under certain conditions, they can be subsequently converted into fumarate to fuel H2/fumarate respiration. The reduction product succinate is secreted in exchange for new aspartate, fumarate, L-malate or L-tartrate. B. Competitive infection with seven different strains. LCM mice were infected with a mixture of untagged wild type S. Typhimurium, WITS-tagged wild type S. Typhimurium and the indicated 6 S. Typhimurium mutants (5×106 cfu in total; by gavage). Strain abundance in the inoculum and the feces was analyzed by qPCR. C. Complementation of S.TmdcuABC. LCM mice were infected with a mixture of wild type S. Typhimurium, S.TmdcuABC and S.TmdcuABC (pdcuB), which carries a plasmid bearing a dcuB gene on a native promotor (5×106 cfu in total; by gavage). Fecal salmonellae were plated on selective media to distinguish between the three strains. D. dcuABC fueled growth requires the fumarate reductase. LCM mice were infected with 1:1 mixtures of wild type S. Typhimurium and S.TmdcuABC (red symbols) or S.TmdcuABC frd and S.Tmfrd (5×106 cfu in total; by gavage). Fecal salmonellae were plated on selective media to distinguish between the three strains. The Mann-Whitney U-test was used for statistical analysis; *: p < 0.05; ** p < 0.01;*** p < 0.001.
DcuB is an antiporter, importing external C4-dicarboxylates (fumarate, malate, aspartate, and D-tartrate) in exchange for cellular succinate and is expressed under anaerobic conditions (Engel et al., 1992) (Engel et al., 1994) (Six et al., 1994) (Bin Kim et al., 2007, Kroger et al., 2013) (Fig. 2A). In competitive infections of LCM mice, a dcuB mutant (S.TmdcuB; Key Resources Table) had a mild gut colonization defect (C.I. = 0.32 at day 2 pi; Fig. 2B, yellow symbols; Fig. S2A–D; Tab. S3). This mild attenuation is similar to that of dcuS and dcuR mutants in our RB-TnSeq screen and the verification experiment (Fig. 1F, Tab. 1), but much smaller than the attenuation of S. Typhimurium mutants lacking the H2-hydrogenase hyb and the fumarate reductase frd (S.Tmfrd, S.Tmhyb; Fig. 2B; blue and grey symbols). Genetic redundancy might mask fitness consequences of individual transporter mutants, as suggested by in vitro studies on E. coli (Zientz et al., 1996, Zientz et al., 1999). Therefore, we assessed the colonization phenotype of double and triple dcu mutants.
Indeed, while S.TmdcuAC and S.TmdcuB were only mildly attenuated, S.TmdcuABC exhibited ≈100 to 1000-fold attenuated initial growth compared to wild type S. Typhimurium (C.I. = 0.01 – 0.001; Fig. 2B, red symbols; Tab. S3). This colonization defect could be complemented (Fig. 2C, blue symbols). Deleting hyb did not further attenuate S.TmdcuABC at days 1–4 pi (compare to S.TmdcuABC (red symbols) and S.TmdcuABC hyb mutants (green symbols) in Fig. 2B), indicating that dcu genes are epistatic to hyb, at least during initial growth.
The data presented above indicated that DcuABC transporters are required for the H2/fumarate-dependent initial growth in the gut lumen of LCM mice. Accordingly, competitive infections with a 1:1 mixture of S.Tmfrd and S.TmdcuABC frd verified that both strains have similar gut luminal growth defects (C.I. ≈1; Fig. 2D, blue symbols; Fig. S2E–H). Compared to mice infected with strain mixtures that include wild type S. Typhimurium (e.g. S.Tm and S.TmdcuAB, Fig. 2D and Fig. S2E–H, red symbols) mice infected with a mixture of S.Tmfrd and S.TmdcuABC frd showed a delayed rise of gut luminal pathogen densities and delayed gut inflammation kinetics (Fig. S2E,G,H, blue symbols). This prolonged the phase of initial growth and delayed progression towards the next stage of the gut infection process. Thus, DcuABC promote gut colonization and accelerate mucosal inflammation in LCM mice.
The LCM gut microbiota composition shows little change during initial S. Typhimurium growth
While overt gut inflammation is known for its dramatic effects on the microbiota, it remained to be established how the microbiota composition is affected during initial growth. Therefore, we infected LCM mice with wild type S. Typhimurium. To assess the effects of C4-dicarboxylate-fuelled pathogen growth, we infected a second group with S.TmdcuABC. In line with the data presented above, wild type S. Typhimurium featured faster initial growth and elicited gut inflammation about 1 day earlier than S.TmdcuABC, i.e. at day 3 vs. day 4 pi (Fig. 3A,B,C).
Figure 3. Dcu transporters promote gut colonization and accelerate the kinetics of mucosal inflammation and of microbiota perturbation.
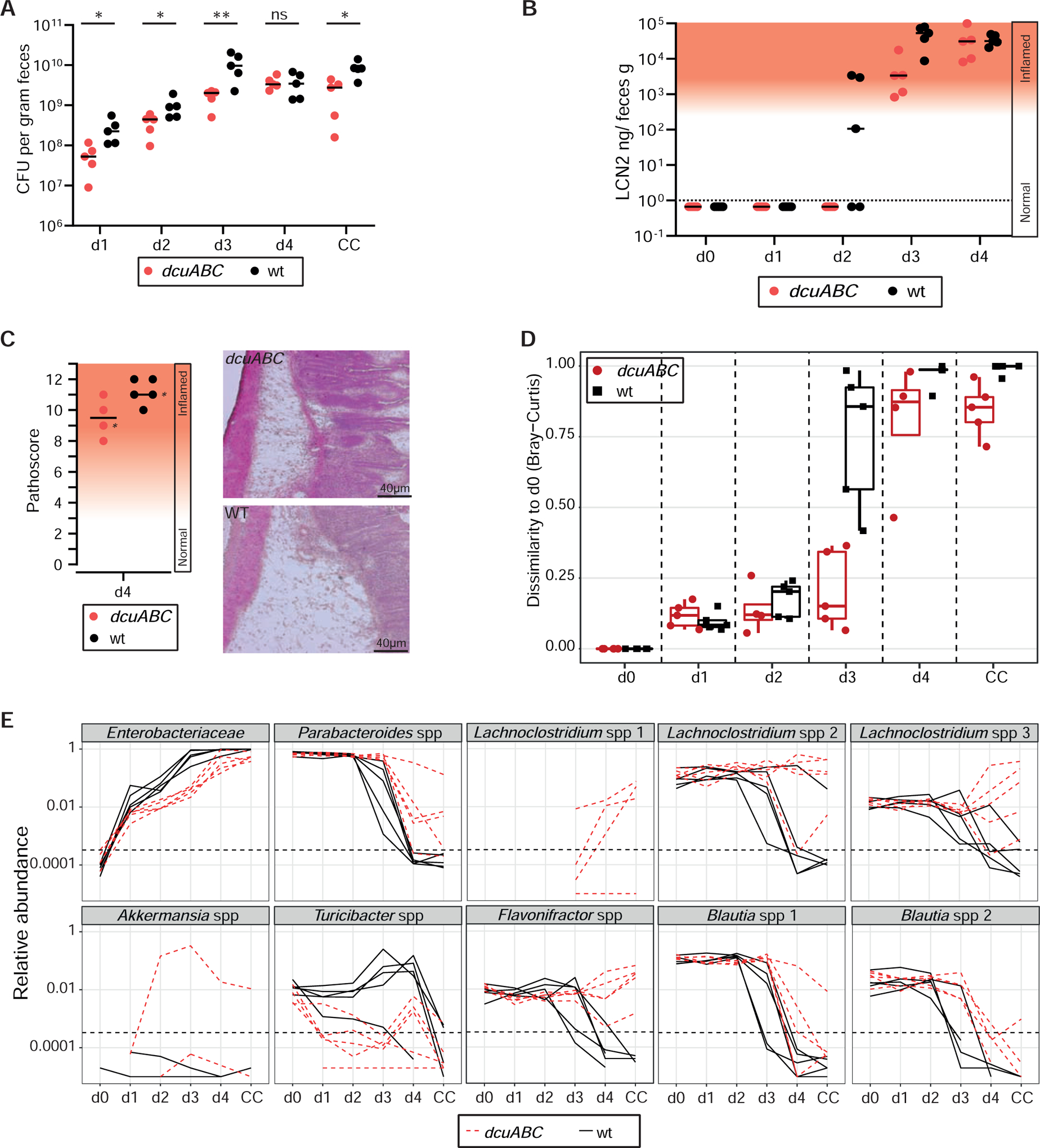
A. Fecal pathogen loads in LCM mice infected with wt S. Typhimurium (black) or S.TmdcuABC (red; n=5 per group; 5×106 cfu; by gavage) as determined by plating. B. Gut inflammation was measured by Lipocalin-2 ELISA. C. Histopathology of the infected cecum tissue at day 4 pi. D,E. 16S rDNA analysis of the fecal microbiota composition before the infection and at days 1,2,3,4 pi. D. Bray-Curtis dissimilarity scores (black = wild type S. Typhimurium infection; red: S.TmdcuABC infection). E. Relative abundance of the identified phylotypes (rarefied to 100.000 16S sequences). Solid lines = wild type S.Tm infection; dashed lines = S.TmdcuABC infection.
At days 1 and 2 pi, we observed only little change in the relative abundance of the different microbiota phylotypes (Fig. 3D,E). This pertained to wild type S.Tm and S.TmdcuABC infected mice, as indicated by the relative abundance of the different detectable OTUs and by the low Bray-Curtis dissimilarity scores (Fig. 3D,E). Please note that the Bray-Curtis dissimilarity scores of the feces before the infection (day 0) were defined as the reference (Bray-Curtis dissimilarity score at day 0 = 0). The scores of ≤0.2 at days 1 and 2 pi are likely attributable to the normal variability of the microbiota community composition that is also observed in unperturbed LCM mice (see below in Fig. S6B; approximately 20% variation in water-control group). The newly introduced pathogen (which is not present at day 0) may also contribute to some extent. Similarly, low-grade mucosal responses observed at days 1–2 pi might contribute a little (lipocalin 100-102 ng/g; Fig. 3B, Fig. 1E, Fig. S2C, Fig. S3). These low-grade responses are characterized by ≤10-fold induction of Il6, Il1b, nos2, and a mucosal morphology equivalent to uninfected controls (Fig. S3D,E, days 1 and 2 pi). However, these mucosal responses and inflammation-boosted pathogen growth (e.g. via cheY-driven chemotaxis, NO3−- and O2-respiration) are lower than those observed after the onset of overt gut inflammation (i.e. days 3–4 pi with wild type S.Tm, Fig. S3B). Thus, during the first 1–2 days of infection in LCM mice, the initial growth of S. Typhimurium proceeds while mucosal responses remain sub-acute and the microbiota shows little change.
The end of the initial growth phase is defined functionally by the onset of overt gut inflammation, as indicated by histopathological analysis and fecal lipocalin-2 concentrations above 3×102 ng/g (Fig. 3B,C). In the case of wild type S. Typhimurium, this occurs typically by day 3 pi. At the same time, many strains of the microbiota are suppressed, as indicated by the elevated Bray-Curtis dissimilarity scores (Fig. 3D). Suppression is particularly pronounced in the cases of the Parabacteroides, Lachnoclostridium, Blautia and Flavonifractor spp. (Fig. 3E). In line with the delayed onset of overt gut inflammation, microbiota disruption occurs later (day 4 pi) and is less pronounced in the case of S.TmdcuABC (Fig. 3D,E). Thus, DcuABC accelerates the infection by promoting initial growth in face of the microbiota. However, the nature of the relevant C4-dicarboxylate substrates of DcuABC remained unclear.
Gut luminal availability of malate, aspartate, citrate and succinate
DcuABC transporters can import several C4-dicarboxylates, namely L-malate, L-aspartate, fumarate or D-tartrate and exhibit affinities in the micromolar range (DcuAB, Km(fumarate) ≤ 100 μM; (Unden et al., 2016, Six et al., 1994, Janausch et al., 2002, Engel et al., 1994, Bin Kim et al., 2007)). To gain further insights into the relevant substrates fueling the DcuABC-dependent initial growth, we focused on the cecum lumen. This is the main site of S. Typhimurium growth in the murine gut. We measured the extracellular metabolites by targeted liquid chromatography/mass spectrometry (Materials and Methods). In line with previous work (Spiga et al., 2017, Cummings et al., 1987), we observed low amounts of fumarate (1.27–3.75 nmol/g cecum content; Fig. 4A) in the cecum lumen of LCM mice and mice harboring a complex pathogen-free microbiota (CONE). This residual fumarate might originate from bacterial lysis (see lysis control experiment in Fig. S4B) or malate degradation, as germ free mice featured even lower amounts of free fumarate (<0.8 nmol/g cecum content). Overall, we conclude that extracellular fumarate in the cecum lumen is likely insufficient to support anaerobic H2/fumarate respiration.
Figure 4. Aspartate and malate are important nutrients used to fuel anaerobic respiration.
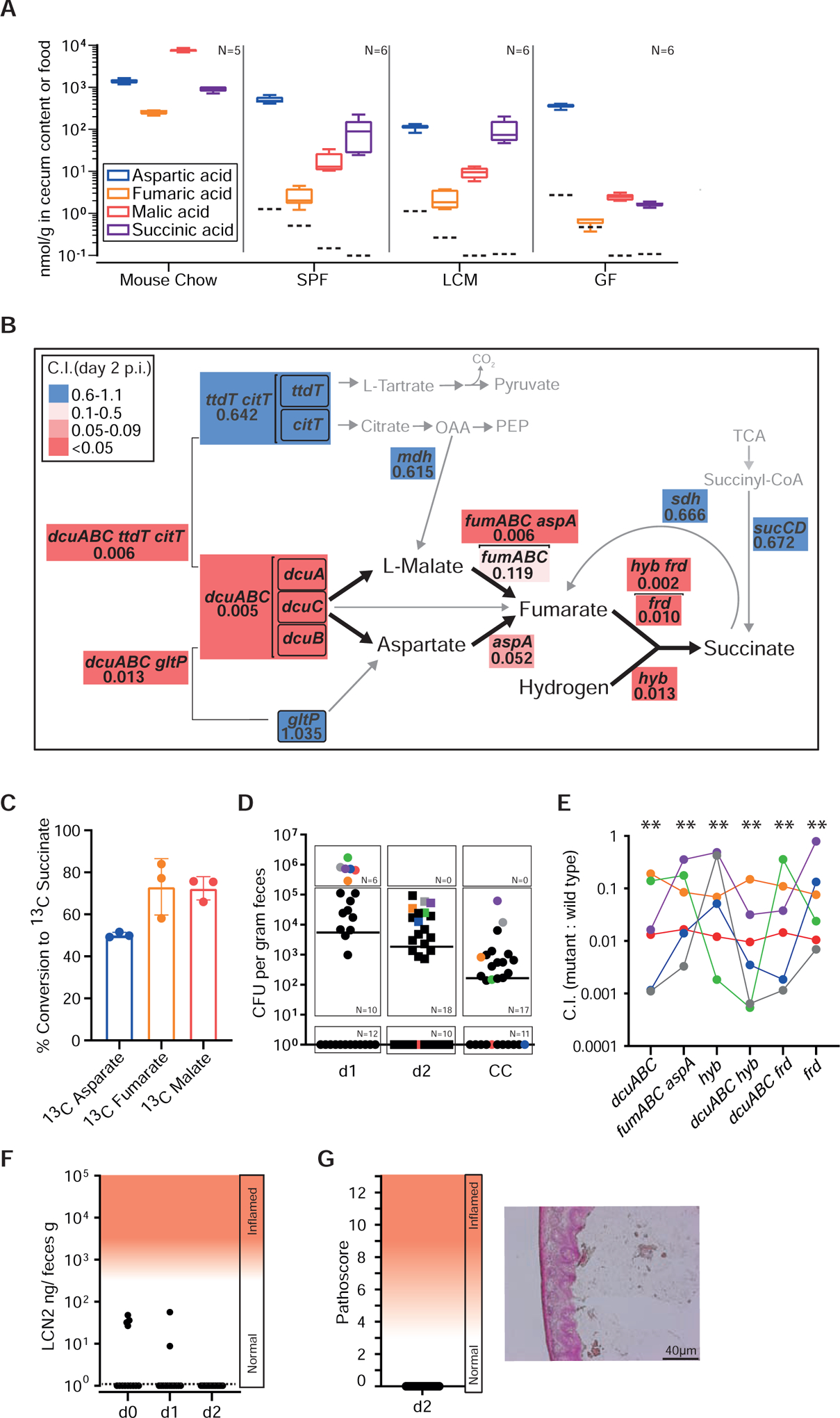
A. C4-Dicarboxlyate levels in the cecum lumen. Cecum contents (without particulate matter or cells) was recovered from mouse chow, germ free (GF), specific pathogen free (CONE), and LCM mice and analyzed by liquid chromatography mass spectrometry (LCMS). Independent measurements of n = 5 (mouse chow) and n = 6 (cecum samples) biological replicates. Dashed lines indicate the limits of detection; box plots extend from the 25th to 75th percentiles with whiskers going down to the smallest value and up to the largest. Center lines indicate median values. B. Competitive indices of site-directed mutants vs. wild type S. Typhimurium. We infected LCM mice with WITS-barcoded S. Typhimurium strain mixtures (5×106 cfu in total, by gavage; at least 6 independent experiments for each mutant) and analyzed the C.I. at days 1–4 pi by qPCR (Materials and Methods). All data are summarized in Tab. S3. Panel B shows data for day 2 pi. Gene symbols denote the mutants, analyzed. Arrows: metabolic conversions; Brackets indicate pathways that were collectively deleted. Numbers: median C.I. at day 2 pi; Colored boxes indicate the degree of attenuation, as indicated in the legend. Grey: Genes that are not essential for C4-dicarboxylate driven H2/fumarate respiration (mutant C.I. values 0.6–1.1). C. Conversion of 13C C4-dicarboxylates by S. Typhimurium. Wild type S. Typhimurium was grown anaerobically (37°C, 4% H2, 10% CO2, 86% N2) in minimal medium containing 45 mM 12C pyruvate and 10 mM of 13C aspartate, 13C fumarate or 13C L-malate (Star Methods). After reaching stationary phase, cell free culture supernatants were harvested and the amounts of released 13C succinate were analyzed by LC-MS (n = 3 independent cultures). D-G. Competitive infection experiment in CONE mice. N = 30 animals were infected for two days with wild type S. Typhimurium, S.TmdcuABC, S.TmfumAC fumB aspA, S.Tmhyb, S.TmdcuABC hyb, S.TmdcuABC frd, or S.Tmfrd (1:1:1:1:1:1:1 mix; 5×107 cfu in total, by gavage). D. Total fecal Salmonella loads as determined by plating. E. Competitive index, as determined by qPCR analysis of the strains’ barcodes. F. Gut inflammation as measured in feces by Lipocalin-2 ELISA. G. Histopathology of the infected cecum tissue at day 2 pi.
In contrast to fumarate, our metabolomics measurements revealed that aspartate (100–500 nmol/g cecum content) and malate (10–20 nmol/g cecum content) were highly abundant in LCM and CONE mice (Fig. 4A). Assuming that particulate matter and cells occupy 90% of the cecum lumen, the aspartate and malate concentrations in the extracellular space (1–5 mM or 100–200 μM respectively) are close to or above the reported KM-values of DcuA and DcuB (KM = 30–50 μM; (Engel et al., 1994, Six et al., 1994)). A second approach confirmed the abundance of malate and aspartate and established that free citrate is below the detection limit (Fig. S4A). Our assays did not provide data for free L/D-tartrate. The amounts of free succinate in the cecum lumen of LCM and CONE mice ranged between those of aspartate and malate (≈ 100 nmol/g cecum content; Fig. 4A). These succinate values are 6–30 fold lower than previously reported for gut contents sampled from humans and mice (Spiga et al., 2017, Cummings et al., 1987) (Meijer-Severs and van Santen, 1987). We speculate that these differences are attributable to differences in food, intestinal resorption rates, microbiota composition or bacterial lysis during sample preparation (as in Fig. S4B). Regardless, because succinate is the reduction end-product of fumarate respiration, it cannot serve as substrate for further anaerobic respiration during initial growth and must be secreted by the DcuABC antiporters (Unden et al., 2016). Collectively, these findings show that aspartate and malate are present in the cecum lumen and that their concentrations suffice to support H2/fumarate respiration during initial growth in the murine gut.
L-malate and L-aspartate serve as important nutrients for gut luminal H2/fumarate respiration.
To further deduce the role of malate and aspartate metabolism during the initial growth, we performed a series of competitive infection experiments in LCM mice and profiled aspA, fumA, fumB and fumC mutants for colonization defects. The aspA gene encodes for aspartate-ammonia lyase, which converts aspartate to fumarate and ammonia (Fig. 4B; (Williams and Lartigue, 1967)). The genome of S. Typhimurium also encodes three fumarases (encoded by fumA, fumB, or fumC), which are hydro-lyases catalyzing inter-conversion of L-malate and fumarate (Mercado-Lubo et al., 2009). In competitive infection assays, S.TmfumAC fumB and S.TmaspA both exhibited impaired initial growth as compared to wild type S. Typhimurium (C.I. = 0.119 or 0.052, respectively; day 2 pi; Fig. 4B; Tab. S3; for day 1 pi data see Fig. S4C) suggesting that catabolism of malate and aspartate is important for fueling gut luminal H2/fumarate respiration. Accordingly, the quadruple mutant S.TmfumAC fumB aspA, which can neither convert L-malate nor L-aspartate, showed severe attenuation (C.I. = 0.006 at day 2 pi; Fig. 4B; Tab. S3) to a similar level as observed for S.Tmfrd, S.Tmhyb, or S.Tmhyb frd (C.I. = 0.010, 0.013, or 0.002 respectively; day 2 pi; Fig. 4B; Tab. S3). This genetic evidence suggests that malate and aspartate are the main sources to drive H2/fumarate respiration during the initial growth of S. Typhimurium. By inference, we conclude that the import of fumarate from the gut lumen does not contribute significantly as initial growth in the absence of aspA, fumA, fumB and fumC was severely impaired. This is in line with our metabolomics measurements establishing low availability of free fumarate in the cecum lumen (Fig. 4A).
While anaerobic H2/fumarate respiration is well established for E. coli growth in vitro (Unden et al., 2016, Zientz et al., 1999, Zientz et al., 1996), it remained to be verified for S. Typhimurium. We performed anaerobic (4% H2, 10% CO2, 86% N2) S. Typhimurium growth experiments to verify the conversion of L-malate and aspartate to succinate. In the presence of C4-dicarboxylates, wild type S. Typhimurium grew anaerobically in minimal medium containing pyruvate, while much slower growth was observed in the absence of C4-dicarboxylates (Fig. S4D). Supplementing the minimal medium with 13C C4-dicarboxylates resulted in the consumption of all the available C4-dicarboxylate and the release of 50–70% mole equivalents of 13C succinate, as measured by mass spectrometry of the spent medium (Fig. 4C). Thus, similar to E. coli (Unden et al., 2016, Zientz et al., 1999, Zientz et al., 1996), S. Typhimurium can grow by anaerobic H2/fumarate respiration, if C4-dicarboxylates are available.
Besides DcuABC-mediated uptake, S. Typhimurium encodes several alternative transporters and enzymes which may replenish cellular malate and aspartate pools under anaerobic conditions (Fig. 4B). However, these do not appear to affect initial growth in the gut, as shown by a series of competitive infection experiments, which are summarized in Fig. 4B, Fig. S4C and Tab. S3 and discussed in the supplement (Supplemental Discussion).
In addition to transport of aspartate by DcuABC, the genome of S. Typhimurium encodes a H+/glutamate-aspartate transporter GltP (Deguchi et al., 1989). However, S.TmgltP colonized the gut about as efficiently as wild type S. Typhimurium (C.I. = 1.2 at day 2 pi, Fig. 4B) and disrupting gltP did not further attenuate the dcuA dcuB dcuC mutant (compare S.TmdcuABC vs. S.TmdcuABC gltP) suggesting that the DcuABC is the main import route for aspartate. Collectively, these findings suggest that Dcu transporters promote initial growth in the gut lumen by importing mainly malate and aspartate to drive H2/fumarate respiration.
A fully operating TCA cycle is not required for initial gut lumen colonization.
Next, we asked whether the observed phenotypes (Fig. 4B, Fig. S4C, Tab. S3) are attributed solely to defects in fumarate respiration or, alternatively, were due to other disturbances of the TCA cycle. The latter was conceivable, as several recent studies have reported full expression of the TCA cycle enzymes in antibiotic pretreated or inflamed guts (Ng et al., 2013) (Ali et al., 2014) (Wilson et al., 2019, Rivera-Chavez et al., 2016, Spiga et al., 2017). Some of the TCA enzymes and substrates are shared with H2/fumarate respiration. However, deletions in genes exclusively involved in the TCA cycle but not required for H2/fumarate respiration exhibited only minor effects on initial growth compared to frd, hyb or dcuABC mutants. Among them was an sdh mutant lacking succinate dehydrogenase activity, which colonized LCM mice almost as well as wild type S. Typhimurium (S.Tmsdh C.I. = 0.66, day 2 pi; Fig. 4B, Tab. S3). Similarly, deletion of sucCD lacking succinyl-CoA synthetase activity only mildly affected initial growth (S.TmsucCD C.I. = 0.68, day 2 pi; Fig. 4B, Tab. S3). This was in line with earlier work analyzing the gut-luminal growth of a S. Typhimurium TTSS-1/2 mutant incapable of eliciting mucosal inflammation (Spiga et al., 2017). In sum, our findings suggested that a functioning TCA cycle is not a prerequisite for initial growth in the murine gut.
DcuABC promotes S. Typhimurium growth in the face of a complex microbiota
To assess the role of dcuABC-mediated C4-dicarboxylate transport for initial growth in face of a complex microbiota, we performed competitive infection experiments with barcoded strains in CONE mice (Fig. 4D–G; Star Methods; no pretreatment). In line with the pronounced colonization resistance conferred by complex microbiotas (Barthel et al., 2003, Stecher et al., 2010, Wotzka et al., 2019), we detected wild type and/or Salmonella cells in feces of only 16/30 animals by day 1 pi (Fig. 4D). The pathogen loads declined further by day 2 pi, and we observed no signs of overt gut inflammation (Fig. 4F,G). The low colonization efficiency in CONE mice explains why we only detected robust signals for the barcodes of all 7 strains in the 6 mice carrying the highest fecal pathogen loads at day 1 pi (Fig. 4D,E; highlighted with colors; >105 cfu/g). This data is noisier than that from LCM mice. Nevertheless, in these 6 mice S.Tmfrd, S.Tmhyb, S.TmdcuABC frd, S.TmdcuABC, S.TmdcuABC hyb and S.TmfumAC fumB aspA were significantly attenuated compared to wild type S. Typhimurium (median attenuation: 10–100-fold). Subsequent experiments further substantiated this (see below; unperturbed CONE controls in Fig. S5C). In sum, these findings suggest that dcuABC-mediated C4-dicarboxylate transport promotes initial gut luminal growth in mice harboring different microbiotas.
Aspartate and Malate supplementation promotes gut luminal growth of S. Typhimurium in face of a complex microbiota.
Our data suggested that gut luminal levels of malate and aspartate are critical determinants promoting gut colonization. To assess whether colonization resistance is affected by malate or aspartate availability, we supplemented CONE mice with water (= negative control) or L-aspartate and L-malate (= experimental group) and infected the animals for 4 days with an equal mix of WITS-barcoded wild type S. Tm, S. TmdcuABC, S. TmfumABC aspA, S. Tmhyb, S. TmaspA, S. TmfumABC, and S. Tmfrd. We found that the total fecal S. Typhimurium load was 10–100 fold higher and more animals developed mucosal inflammation when aspartate and malate was supplemented (3/9 mice) as compared to controls (0/10 mice; Fig. 5A,B; Fig. S5A). Systemic colonization might also be slightly elevated in the L-aspartate/L-malate treated group. However, this effect was not significant (p≥0.05; Fig. S5B). These data indicated that aspartate and L-malate promote initial colonization and progression to disease.
In line with the data presented above (Fig. 4E, Fig. 2B, Tab. S3), S. TmdcuABC, S. TmfumABC aspA, S. Tmhyb, S. TmaspA, and S. Tmfrd were attenuated in the control CONE mice upon aspartate and malate supplementation (Fig. S5C). However, as total pathogen densities were low, these data were again quite noisy. This may also explain why we could not detect significant growth defects for some of the strains like S. TmfumABC in this experiment.
We also assessed if malate- and aspartate-inflicted changes in microbiota composition might contribute to pathogen growth. Malate and aspartate supplementation shifted the microbiota composition (Fig. S6A,B). However, this shift was mild compared to those inflicted by the onset of gut inflammation (LCM mice, Fig. 3D) or by high fat diet exposure of CONE mice (Fig. S6A,B, right side), a stimulus which can boost S. Typhimurium gut colonization by ≥1000-fold (Wotzka et al., 2019). Fumarate supplementation had an even milder effect on the microbiota composition than malate and aspartate supplementation (Fig. S6A,B). We speculate that this may be attributable to fumarate resorption by the host, as food contains much more fumarate than the cecum lumen of germ-free mice (Fig. 4A). Importantly, the fumarate supplementation did not enhance initial pathogen growth in the gut lumen or enteropathy (Fig. S6C,D). Thus, the observed small changes in microbiota composition alone appear insufficient for boosting initial growth of S. Typhimurium. However, we cannot exclude that they contribute in malate and aspartate supplemented mice.
Pathogen blooms in the aspartate and malate supplemented CONE mice still required dcuABC, aspA, hyb and frd (Fig. S5C). This supported the idea that pathogen growth still relied on H2/fumarate respiration. We cannot exclude that additional effects attributable to altered microbiota metabolism or community structure might further promote the pathogen’s growth. Collectively, our findings underscore the importance of C4-dicarboxylic acid levels in the gut lumen to promote initial growth of S. Typhimurium in CONE mice.
E. coli 8178 requires DcuAB for competitive gut luminal growth
Genome analyses indicated that DcuABC homologs and genes implicated in H2/fumarate respiration are encoded by several other enteric pathogens and by commensal E. coli strains (Tab. S4), including E. coli 8178. E. coli 8178 had been isolated from mice and is capable of limiting gut luminal S. Typhimurium growth after a shift to Western-type diet and in the inflamed gut (Stecher et al., 2012) (Wotzka et al., 2019). We hypothesized that Dcu-mediated C4-dicarboxylate import might contribute to the competitive capacity of E. coli 8178. Based on our S. Typhimurium data, we focused on DcuA and DcuB, as these should show the strongest phenotypes. To test our hypothesis, we performed a competitive infection experiment in mice harboring a complex microbiota that lacks commensal E. coli. We used CONX mice (Star Methods), as these animals allow sufficient E. coli and S. Typhimurium densities for our experiment (approx. 105-107 cfu/g stool at day 1 p.i.). The animals were inoculated with a mix of wild type S. Typhimurium, wild type E. coli 8178 and an isogenic mutant lacking DcuAB (E. colidcuAB; Key Resources Table). Control mice were inoculated with wild type S. Typhimurium alone (Fig. 6). Lipocalin-2 levels of CONX mice were slightly higher than those of uninfected CONE mice (Fig. 6D; compare to Fig. 3B, Fig. S1D). However, tissue sections revealed no enteropathy and Lipocalin-2 levels did not change over time (Fig. 6D,E), suggesting that initial growth lasted throughout the course of the 4 days of infection. In presence of E. coli, S. Typhimurium had a reduced initial growth in the gut lumen (Fig. 6A). We also observed a trend towards reduced spread to systemic sites (Fig. 6C). However, this trend was not significant and more animals would be necessary to verify this effect. E. colidcuAB was 10–100-fold attenuated in colonization compared to wild type E. coli (Fig. 6A,B). Wild type E. coli diminished initial S. Typhimurium growth in CONX mice more efficiently than E. colidcuAB (Fig. 6F) and E. colifrd was attenuated in competitive infections (Fig. 6G). These data confirm that commensal E. coli can fortify colonization resistance against S. Typhimurium(Velazquez et al., 2019) and suggest that DcuAB-dependent fumarate respiration promotes its’ competitive capacity in the gut lumen.
Figure 6. Competitive growth of E. coli 8178 requires DcuAB.
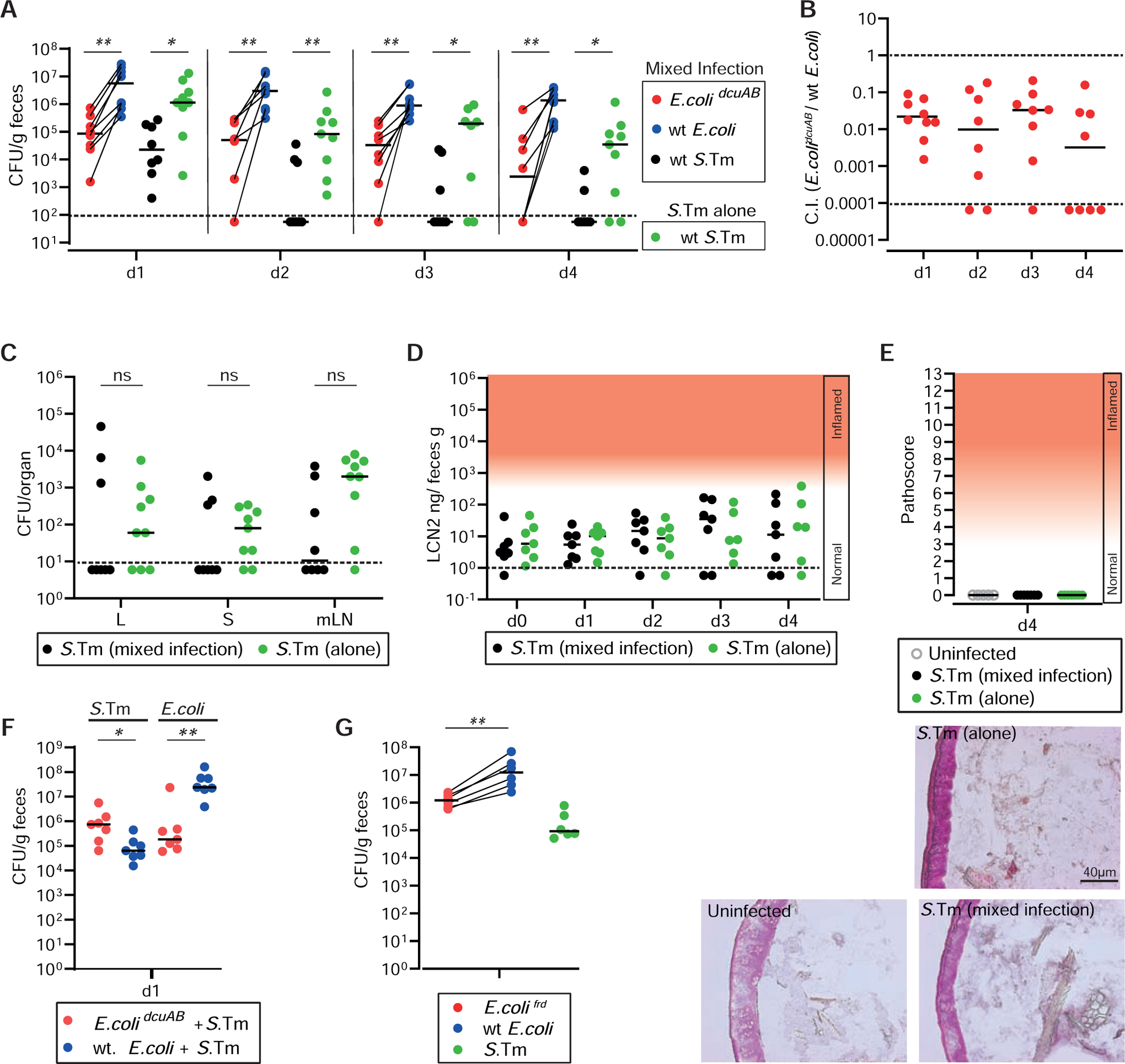
A. CONX mice were infected for 4 days with a 1:1:1 mix of wild type E. coli (CmR), E. colidcuAB (KmR) and wild type S. Typhimurium (SmR; N = 8) or with S. Typhimurium alone (N = 9; 108 cfu each; by gavage). A. Fecal loads of the indicated E. coli and S. Typhimurium strains as determined by plating on MacConkey agar. B. C.I. values for E. colidcuAB vs. wild type E. coli. C. S. Typhimurium loads in livers, spleens and mesenteric lymph nodes. D. Gut inflammation as measured by Lipocalin-2 ELISA. E. Representative images of hematoxylin and eosin (H&E) stained cecum tissue from uninfected mice or animals infected for 4 days with the mix, or wild type S. Typhimurium, alone. F. CONX mice were infected for 1 day with a 1:1 mix of wild type E. coli (CmR) and wild type S. Typhimurium (SmR) or with a 1:1 mix of E. colidcuAB (KmR) and wild type S. Typhimurium (N = 7; 108 cfu each; by gavage). We determined fecal loads of the indicated E. coli and S. Typhimurium strains by plating on MacConkey agar. G. CONX mice were infected for 1 day with a 1:1:1 mix of wt E. coli (CmR), E. colifrd (KmR) and wild type S. Typhimurium (SmR; N = 6). Fecal loads of the indicated E. coli and S. Typhimurium strains were determined by plating on MacConkey agar.
Discussion
The initial growth of S. Typhimurium in the anaerobic milieu of the microbiota-colonized gut is fueled in part by H2/fumarate respiration where fumarate serves as an alternative electron acceptor for electrons derived from the hyb H2-hydrogenase reaction to sustain anaerobic growth in the gut lumen (Maier et al., 2013). Here, we found that fumarate is obtained via conversion of L-aspartate and/or L-malate, which are imported in exchange for succinate, the reduction product of fumarate. This DcuABC-dependent antiporter process allows S. Typhimurium to maintain respiration in the absence of suitable alternative electron acceptors. This metabolic strategy employs just one or two steps of the reverse TCA cycle, i.e. the fumarate reductase (frd) and fumarase (fumABC).
What is the source of the L-aspartate and the L-malate? Aspartate is a typical food constituent. Our mouse diet contains about 1–2 μmol/g (≈0.01% w/w; Fig. 4A) free aspartate, and much additional aspartate is bound in proteins (18.5% w/w protein content). Most proteins are hydrolyzed, but most aspartate is resorbed before the digesta leave the small intestine. In pigs, aspartate resorption occurs with apparent transporter affinities of Km=5–20 mM (Buddington et al., 2001). If this holds true for mice, much of the free aspartate detected in the cecum lumen (100–500 nmol/g cecum content; corresponding to ≈1–5 mM, as discussed above) may originate directly from the food. This free aspartate concentration is sufficient for DcuABC-mediated C4-dicarboxylate uptake by S. Typhimurium, as the pathogen’s transporters feature 20–100 fold higher substrate affinity than the host’s transporters (DcuAB, Km(fumarate)≤0.1 mM; (Six et al., 1994, Janausch et al., 2002, Engel et al., 1994)). As germ-free mice, LCM mice and CONE mice have similar amounts of free aspartate in the cecum lumen (0.1–0.4 mM), the microbiota may not contribute much. Thus, most of the aspartate fueling S. Typhimurium growth in the cecum lumen may originate from food.
Malate is present in mouse food at 8 μmol/g (≈0.1% w/w). Rat feeding experiments with 14C-labled D/L-malic acid suggest that ≈0.3% of this malate will be excreted with the feces (Daniel, 1969). Based on these data, we would expect about 24 nmol/g free malate in the cecum lumen. While germ-free mice had only 2 nmol/g, we observed 10–20 nmol/g free malate in the cecum content of LCM and CONE mice. Therefore, this malate may derive from both, food and the microbiota. Our data indicates that the malate and the aspartate concentrations are in the same range as the C4-dicarboxylate Km values of the DcuABC transporters. This supports our hypothesis, that Dcu-mediated C4-dicarboxylate uptake fuels S. Typhimurium’s initial growth by H2/fumarate respiration. Interestingly, the substrates fueling this initial growth are co-fed from two distinct sources, the food (L-aspartate, some L-malate) and the microbiota (H2 (Maier et al., 2013), some L-malate).
In addition to the use of C4-dicarboxylates for H2/fumarate respiration, E. coli and S. Typhimurium can also use C4-dicarboxylates during aerobic growth (Unden et al., 2016). In this case, C4-dicarboxylates are mainly imported via DctA and serve as electron donors, which are oxidized to CO2. This occurs during a later phase of S. Typhimurium gut colonization, i.e. when gut inflammation commences and creates an oxidative gut-luminal milieu (Spiga et al., 2017). In most LCM animals, this occurs at days 3–4 pi when mucosal responses are induced by 10–100 fold (e.g. Il6, Il1b, nos2; Fig. S3D,E). The central carbon metabolism of S. Typhimurium adapts accordingly by switching away from H2/fumarate respiration to utilizing inflammation-elicited terminal electron acceptors (O2, NO −3, tetrathionate; (Winter et al., 2010) (Rivera-Chavez et al., 2016) (Spiga et al., 2017); see also Fig. S3) and diverse electron donors, including microbiota-released carbohydrates, lactate released by enterocytes (Gillis et al., 2018), 1,2-propanediol (Faber et al., 2017), galactarate and glucarate (Faber et al., 2016), formate (Hughes et al., 2017), ethanolamine (Thiennimitr et al., 2011) and succinate (Spiga et al., 2017). As a consequence, the full TCA cycle is engaged in the forward direction. This is confirmed by our data for days 3 and 4 pi in LCM mice (C.I. of S.Tmsdh = 0.28; C.I. of S.TmsucCD = 0.11; Tab. S3). In this phase of the infection, C4-dicarboxylate importers like DctA support S. Typhimurium blooms during gut inflammation by importing microbiota-derived succinate (Spiga et al., 2017). Thus, S. Typhimurium utilizes C4-dicarboxylates in distinct ways during initial growth vs. inflammation-promoted pathogen blooms.
Gut luminal growth of commensal E. coli 8178 and its competitive growth vs. S. Typhimurium also relies on DcuAB and fumarate reductase. This is in line with our S. Typhimurium data and earlier work on the growth of E. coli frd mutants in mice treated with low doses of streptomycin (Jones et al., 2011). Sequence data hints that other Enterobacteriaceae might also employ aspartate or malate to gain energy from H2/fumarate respiration. For example, orthologs of dcuABC, fumABC, aspA, frd and diverse H2-hydrogenases are observed in additional Salmonella enterica spp., commensal and pathogenic E. coli strains, Helicobacter pylori, Shigella flexneri and Yersinia enterocolitica, but not in other commensal microbiota strains analyzed (Tab. S4). Along these lines, it is also tempting to speculate that some microbiota members may inhibit infections by depleting, or fuel infections by releasing L-aspartate, L-malate or other C4-dicarboxylates, as suggested in the Restaurant hypothesis (Conway and Cohen, 2015) (Maltby et al., 2013). It would be interesting for future work to test these hypotheses.
Star Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Wolf-Dietrich Hardt (hardt@micro.biol.ethz.ch). Mice can be obtained from Jackson laboratories. Gnotobiotic mice are available upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse lines.
We used male and female 8–12 week old mice and assigned animals of either sex randomly to the experimental groups. All lines lack E. coli, as determined by plating on MacConkey agar. All mice originate from C57BL/6 or 129SvEv breeders originally obtained from Jackson laboratories. The mice with a normal complex microbiota (CONE, CONX) were specific pathogen-free and bred under full barrier conditions in individually ventilated cage systems in the EPIC mouse facility of ETH Zurich, Switzerland. CONE mice have a C57BL/6 genetic background, CONX mice have the 129SvEv genetic background. LCM mice (Stecher et al., 2010) (Maier et al., 2013) are ex-germ free C57BL/6 animals that have been associated with the strains of the altered Schädler flora. They are bred in flexible film isolators under strict exclusion of microbial contamination at the isolator facility of the EPIC mouse facility of ETH Zurich, Switzerland. Germ-free C57BL/6 mice were bred in flexible film isolators under strict exclusion of microbial contamination at the isolator facility of the EPIC mouse facility of ETH Zurich, Switzerland.
All studies were performed in accordance with ethical and legal requirements and were reviewed and approved by the Kantonales Veterinäramt Zürich under the licenses 222/2013, 193/2016 and ZH158/19.
Bacteria strains and culture conditions
All Salmonella strains are isogenic to S. Tm SB300, a re-isolate of SL1344. The E. coli strains are derivatives of E. coli 8178, an isolate from the stool of a SPF C57BL/6 mouse colony. They are listed in the KEY RESOURCES TABLE. All strains were routinely grown overnight at 37°C in Lysogeny broth (LB) with agitation. Strains were stored at − 80°C in peptone glycerol broth.
METHOD DETAILS
Construction of bacteria strains and plasmids
Transposon Mutagenesis.
We constructed the DNA-barcoded EZ:Tn5transposome by PCR from a linearized plasmid pZ922 template using primers that contained a random 17 nucleotide sequence flanking the RB-TnSeq PCR priming sites and the terminal inverted repeats of Tn5 (Fig. S1A; KEY RESOURCES TABLE). Specifically, we used primer tn5+ME+17N-F3 and primer tn5+ME-R2 to amplify a kanamycin resistance cassette from pZ922, which has an Illumina primer binding sequence at the 5’ end and a transcriptional terminator and a T7 promotor downstream. We performed a PCR in 100 μl total volume with 0.1 μmol of each primer, 25 ng of pZ922 template (NotI digested), and 0.5 μl Phusion DNA polymerase (NEB 1.0U/μl) with the following cycling program: initial denaturation step at 94 °C for 1 min followed by 30 cycles of 94 °C for 30 sec, 55 °C for 30 sec, 72 °C for 1 min, and a terminal extension step at 72 °C for 5 min. The PCR product was purified by gel electrophoresis in the absence of DNA intercalating dyes, excised from the gel for column-based DNA extraction (Macherey&Nagel), and eluted in 10 mM Tris/HCl, 1 mM EDTA (TE buffer). We prepared the transposome by adding 1 μl of EZ-Tn5 Transposon DNA (100μg/ml in TE) (Epicentre; 1.0 U/μl), and 1 μl of glycerol, mixed by vortexing, and incubated for 30 min at room temperature. Excess salts were removed by drop dialysis on a Type-VS Millipore membrane (0.025 μm pore size, Millipore). We electroporated 1 μl of the DNA-barcoded EZ:Tn5 transposome into 50 μl of electrocompetent wild type S. Typhimurium (prepared from mid log cultures; ~1010-1011 cfu/ml). Cells were immediately recovered in SOC medium, incubated for 45 min with agitation at 37 °C, and plated in batches on LB agar (50 μg/ml kanamycin). The transposon mutants were recovered and aliquots of a pool of 2000 colonies were stored in peptone glycerol at −80 °C.
RB-TnSEq Control Strains.
We constructed the RB-TnSEq control strains using the lamda red recombination system (Datsenko and Wanner, 2000) in order to insert the barcoded transposon into a fitness neutral locus. 40-nucleotide long sequences that are homologous to the intergenic region between the pseudogenes, malX and malY were attached to the ends of the barcoded transposons (as generated from above) by PCR using the primers, ydgA tn5-aphT-R2 and ydgA tn5-aphT-F2. PCR products were electroporated into a wild type S. Typhimurium carrying the pKD46 plasmid. Recombinant strains were selected on LB agar (50 μg/ml kanamycin). The transposon insertion was confirmed by PCR (primers ydgAtn5aphTcheckR and ydgAtn5aphTcheckF) and the associated 17 nucleotide sequence was determined by Sanger sequencing. To generate RB-TnSeq barcoded control strains with known DNA barcodes, we transduced from these strains five barcoded transposons each via P22 phage into the mutant strains, hybABChypO, cheY, and ssaV invG as well as wild type S. Typhimurium (KEY RESOURCES TABLE).
Mouse infection experiments.
8 to 12 week old mice were inoculated with S. Typhimurium cells prepared from a culture grown for 12 h in LB medium containing the appropriate antibiotic(s), diluted 1:20 and sub-cultured for 4 h in the same medium w/o antibiotics. Bacteria were washed twice with PBS and mice were infected with bacteria by gavage (5·106 cfu in 50 μl, if not stated otherwise). Mice were kept in grid-flor cages to avoid transmission between mice. Animals were sacrificed at day 4 pi by cervical dislocation. Fresh fecal pellets, whole cecum content, a piece of the liver, the mesenteric lymph nodes and the spleens were harvested and suspended in PBS (500 μl) using a Tissue Lyser device (Qiagen) before plating to determine the total bacterial population size. HE-staining of cyro-embedded tissues and subsequent pathoscoring for granulocyte infiltration was performed as described previously (Barthel et al., 2003).
Lipocalin-2 ELISA.
Lipocalin 2 was detected in feces homogenized in 500 μl sterile PBS by ELISA (DuoSet Lipocalin ELISA kit (DY1857, R&D Systems, Minneapolis, MN, USA).
Sequencing of transposon insertion sites and their associated barcodes.
Sequencing library preparation.
Transposon sequencing and DNA library preparation was performed as previously described (Yung et al., 2015, Christen et al., 2011). In brief, we used semi-arbitrarily primed (semi-AP) and nested PCR reactions to amplify the transposon junctions bearing the unique DNA barcodes as well as the adjacent genomic DNA and to add terminal adapter sequences compatible to the Illumina sequencing platform (Fig. S1B). To prepare the PCR template, we diluted the transposon library in sterile water to an OD of 1.0 and subjected the diluted sample to six freeze-thaw-heat cycles (submersion in liquid nitrogen for 1 min, followed by incubation at room temperature for 1 min and at 95 °C for 1 min). We then performed separate semi-AP PCRs with the crude bacterial lysate as template and the transposon specific primer (ARB-Tn5-R1) and one of the indexed semi-arbitrary primers (ARB3A_PE, ARB3C_PE, ARB3D_PE, ARB3F_PE; KEY RESOURCES TABLE), using the following program: (1) 94 °C for 3 min, (2) 94 °C for 30 s, (3) 42 °C for 30 s, slope −1 °C per cycle, (4) 72 °C for 1 min, (5) go 6 times to step 2; (6) 94 °C for 30 s, (7) 58 °C for 30 s, (8) 72 °C for 1 min, (9) go 25 times to step 6; (10) 72 °C for 3 min. We enriched and prepared the PCR products for Illumina sequencing by performing a second PCR with nested primers that carry terminal Illumina sequencing adapters (PE_PCR_Primer_1.0, PE_PCR_Primer_2.0; KEY RESOURCES TABLE) and the following program: (1) 94 °C for 3 min, (2) 94 °C for 30 s, (3) 64 °C for 30 s, (4) 72 °C for 1 min, (5) go 30 times to step 2; (6) 72 °C for 3 min. We size-selected 200 −500 bp fragments by gel electrophoresis (2% agarose) prior to Illumina sequencing.
Illumina sequencing.
Cluster-generation was performed within standard Illumina paired- end flow-cells (Illumina cluster chemistry v2) and settings according to the Illumina cluster generation protocol. The DNA sample concentration (25 ng/μl) of each transposon junction library was titrated to produce approximately 1.3 × 105 clusters per tile on the flow cell. Paired-end sequencing with 150 bp read length from both ends of a given DNA fragment was performed on an Illumina HiSeq instrument using sequencing chemistry (v4) and standard paired-end sequencing primers (PE 1.0 and PE 2.0, Illumina).
Mapping of the transposon insertion sites.
Raw transposon read processing and mapping of genomic insertion sites was performed using previously described data analysis routines (Christen et al., 2016, Christen et al., 2011). In brief, reads with at least a 15 bp long perfect match to the Tn5 transposon end sequence [‘GTGTATAAGAGACAG’] and bearing genomic inserts larger than 15 base pairs were selected. Transposon internal RB-TnSeq-barcode sequences were extracted and sequencing strings corresponding to the illumina adapters, transposon end or arbitrary PCR primers sequences were trimmed. Using bwa-07.12 (Li and Durbin, 2010), filtered transposon reads were aligned onto the S. Typhimurium NCBI reference genome (FQ312003). Reads with correct paired-end alignment, no mismatches within the first 15 bases of both paired-end reads, and with an insert size smaller than 700 bases, were selected for subsequent analysis. Transposon insertion sites were defined according to the first reference base detected adjacent to the transposon end sequence.
RB-TnSeq barcode assignment to genomic insertion sites.
Barcode sequences where unambiguously assigned to genomic insertion locations selecting the most abundant barcode sequence associated with a specific insertion site and excluding secondary barcode occurrences at alternative insertion sites with lower read counts. (Fig. S1B; Fig. S4). Cumulatively, we could unambiguously associate 1305 unique RB-TnSeq-barcodes with specific transposon insertion sites, corresponding to 732 genes.
Barcode annotation.
Each barcode was assigned to specific gene if the insertion locations occurred inside the protein coding sequence according to the reference genome coordinates (Tab. S3).
Analysis of RB-TnSeq mutant gut colonization phenotypes.
Sample preparation.
Fecal S. Typhimurium cells were enriched in 10 ml LB supplemented with 50 μg/ml of streptomycin or 3.0 ml LB supplemented with 12.5 μg/ml of tetracycline for 4 hours to select and enrich for living salmonallae. Bacterial cells were pelleted, resuspended in TE buffer, and stored at −20 °C. DNA was extracted from thawed pellets using commercial kits (Qiagen Mini DNA) according to manufacturer’s instructions. To PCR-amplify the barcode sequences, we added 200 ng of extracted DNA, 100 nmol of each primer (PE PCR Primer 1.0 and indexed PE2.0-ME, Table S1), 10 μmol dNTPs and standard amounts of reaction buffer and Taq (0.2 μl, 1 U/μL) in a 50 μl total volume. We ran the reaction using the following cycling program: initial denaturation step at 94 °C for 3.0 min followed by 23 cycles of 94 °C for 30 sec, 60 °C for 30 sec, 72 °C for 20 sec, and a terminal extension step at 72 °C for 2.0 min. PCR products were column-purified, measured for DNA concentration, pooled (450 ng per sample), and then gel purified. Purified PCR products were diluted to a final concentration 25 ng/μl in TE buffer.
Illumina sequencing.
Sequencing was performed as described, above (see sequencing of RB-TnSeq transposon insertion sites).
Barcode counting.
In order to analyze barcode sequence abundance in the inoculum and the output pools, we extracted random barcode sequences and index sequences used to track individual sample and experimental conditions from raw read files. Barcodes were demultiplexed and corresponding occurrences were computed using shell scripts (Fig. S3B). We only considered 100% matches.
Sample QC.
For each sample, a linear regression is made with the dilution series of wild type control strains to verify that r2 > 0.8. If this is not the case, two more successive linear regressions are made using only 4 or 3 control wild type RB-TnSeq strains with the higher titers. If in neither of these cases the sample maintains linearity r 2> 0.8, the sample is discarded completely. In order to avoid noise originating from RB-TnSeq mutants that were poorly represented in the inoculum pool, we only retained RB-TnSeq mutants with equal or higher number of reads than the lowest wild type RB-TnSeq strain that maintains linearity.
Rarefaction.
All of the samples were rarefied to the same value, equivalent to the shallowest sample that passed QC. This was done with the vegan R package (v 2.5-1)
Set detection limit.
Linear regressions were calculated for all the samples in a similar procedure to the Sample QC. All barcodes less abundant than the lowest linear wt control are deemed below detection limit and set to the corresponding value
Calculate fitness.
Fitness values were calculated as the number of observed barcode reads at a particular time point divided by the number of barcode reads observed in the inoculum. In the cases where there is more than one barcode per gene, the median value was selected. To test for significance, a Wilcox unpaired exact test was performed by comparing each RB-TnSeq gene fitness value and the corresponding mean fitness for the fitness values of the 3/2000, the 1/2000 and the 1/6000 dilutions of the RB-TnSeq-barcoded ssaV invG control strains, and then FDR adjusted.
Calculate CI.
The CI (Competitive index) is calculated by dividing the fitness values of each RB-TnSeq mutant to the mean fitness value of the 3/2000, the 1/2000 and the 1/6000 dilutions of the RB-TnSeq-barcoded wild type S. Typhimurium control strains. To test for significance, a Wilcox unpaired exact test is performed for each gene CI and the corresponding mean CI for the 3/2000, the 1/2000 and the 1/6000 dilutions of the RB-TnSeq-barcoded ssaV invG control strains, and then FDR adjusted. The decision to use the RB-TnSeq-barcoded ssaV invG strains for comparison (and not the RB-TnSeq-barcoded wild type strains) is that the the RB-TnSeq-barcoded wild type strains had already been used for calculating the CI. During co-infections with wild type S. Typhimurium, the ssaV invG strains are known to colonize the gut lumen just as well as the isogenic wild type strain (Stecher et al., 2007). Therefore, we could use the RB-TnSeq-barcoded ssaV invG strains for comparison (Fig. S3A)
Analysis of WITS mutant gut colonization phenotypes.
Inoculum mixtures were prepared by mixing equal ratios of the indicated WITS-barcoded strains including a WITS21-barcoded wild type S. Typhimurium strain (KEY RESOURCES TABLE). An equivalent proportion (50% of the total cfu) of non-barcoded wild type S. Typhimurium was added to ensure equivalent disease kinetics between different experiments. If two independent clones were available from a given mutant (KEY RESOURCES TABLE), we used the different clones in the different independent replicates of each experiment and verified that both clones yielded equivalent results. This strategy avoided misinterpretation of our data that might be caused by non-neutral second-site mutations.
Sample preparation for WITS-distribution analysis.
Samples were prepared and DNA was recovered, as described above (Analysis of RB-TnSeq mutant phenotypes). rtqPCR analysis was performed using the FastStart Universal SYBR Green Master Mix (Roche) and primers listed in KEY RESOURCES TABLE. The temperature profiles were described previously (Maier et al., 2014b) (Grant et al., 2008) (Kaiser et al., 2013). The population size of each tagged strain was calculated by multiplying the number of kanamycin resistant colonies/g feces with the ratio of WITS determined by rtqPCR.
To calculate Shannon Evenness (EH), we used the formula: , where ln, with H=Shannon’s diversity index, S=total number of strains, pi=proportion of S made up of the ith strain.
16S rRNA gene sequencing
Microbial DNA was extracted from feces as described(Wotzka et al., 2019). Feces were collected before the infection (day 0) or at days 1, 2, 3, or 4 as indicated. The samples were immediately frozen in liquid nitrogen for storage at −80°C. The AllPrep DNA/RNA Kit (Qiagen) was used to extract DNA, using 600 ml of RLT buffer. One unit of 3 mm metal beads (Berani Kugellager AG) and 600 mg of 0.1 mm zirconia beads (OPS Diagnostics) were added to the tube containing the feces, which werebead-beaten two times at 30 Hz for 5 min with a 5 minute incubation time in between, using a Retsch M400 apparatus. The lysates were transferred into the corresponding columns which were then washed as described in the manufacturer’s protocol. Finally, the bacterial DNA was eluted in 100 μl elution buffer (EB).
The NEXTflex® 16S V4 Amplicon-Seq Kit 2.0 (Barcodes 1–96; Bio Scientific) was employed to produce the 16S libraries. Genomic DNA (30 ng/μl) was used as template and the library preparation was performed according to the manufacturer’s protocol (25 μl reaction volumes). A modified primer pair was used for the first PCR reaction, replacing the primer pair 515f-806r from the kit with the degenerate primers 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806rB (5’-GGACTACNVGGGTWTCTAAT-3’) as described52,53. Q5 High-Fidelity DNA Polymerase (BioConcept (NEB)) was used for the first PCR reaction: 1) first denaturation: 95°C for 4 min; 2) denaturation in each PCR cycle: 95°C for 30 sec; 3) annealing (each PCR cycle): 56°C for 30 sec; 4) extension (each PCR cycle): 72°C for 90 sec; 5) final extension at the end of the reaction: 72°C for 4 min. The cycles 2–4 were repeated 8 times. The PCR products were purified using AMPure XP magnetic beads (Beckman Coulter SA), eluted in 20 μl of resuspension buffer and served as templates in the next PCR reaction, which employed barcoded primers for multiplexing. The quantity of the amplicons was measured using a Qubit fluorometer and quality was verified using a Fragment Analyzer (Advanced Analytical). DNA concentrations were measured using a a Qubit fluorometer. PCR products (approx. 450 bp length) were diluted to a final concentration of 60 ng DNA per 20 μl multiplexed sample. Illumina MiSeq paired end read sequencing was performed at the Functional Genomics Center Zürich.
The raw sequencing data were processed with custom scripts to execute the following commands of the USEARCH software (v9.1.13): the fastq_mergepairs command was used with default settings to merge and quality-filter paired reads. The merged reads were subsequently filtered using the fastq_filter command (-fastq_maxee 0.1). We only merged reads with perfect primer matches and we only selected reads of ≥100 bp length. The fastx_uniques command was used for de-replication. Then, data were clustered into operational taxonomic units (OTUs; 97% threshold) using the cluster_otus command (-minsize 2). Thereby, chimeric sequences were also removed. The usearch_global command (-strand both; -id 0.97) was used to quantify the OTU abundances for each sample. Taxonomic annotation was done by querying OTU sequences against the SILVA database (v 128). For this purpose, the usearch_global command (-id 0.90; -maxaccepts 20; -maxrejects 500; -strand both; -top_hits_only) was performed. The scripts used for these steps are available in https://github.com/SushiLab/Amplicon_Recipes.git and the raw data is available from ENA with the accession number PRJEB35643
Downstream analyses were done in RStudio (version 1.0.143 based on [R] version 3.3.3) using the libraries vegan and ggplot2. Read counts were rarefied to 55,000 (CONE experiments) or 100,000 (LCM experiments) reads per sample. To analyze changes in the microbiota composition between samples, time points or treatments, the Bray-Curtis dissimilarity was analyzed.
Anaerobic culture of S. Typhimurium.
S. Typhimurium cells were cultured in minimal M9 medium (12.8 g/L Na2HPO4∙7H2O, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 100 μM CaCl2, 2 mM MgSO4) supplemented with Wolin’s trace elements (13.4 μM EDTA, 3.1 μM FeCl3-6H2O, 0.62 μM ZnCl2, 76 nM CuCl2-2H2O, 42 nM CoCl2-2H2O, 162 nM H3BO3, 8.1 nM MnCl2-4H2O), 250 mg/L 12C Histidine-HCl and 45 mM 12C pyruvate. 10 mM 13C C4-dicarboxylate (aspartate, fumarate, malate) was added, as indicated. Growth was analyzed under anaerobic conditions (4% H2, 10% CO2, 86% N2) at 37 °C. Supernatant was sampled at the transition from exponential to stationary phase and centrifuged for 2.0 min at 11,000 × g at 4 °C to pellet and remove bacterial cells. Samples were diluted (1:1000 v/v) in a buffer corresponding to the starting conditions of the mobile phase used for liquid chromatography (see below: mass spectrometry, metabolite measurements) containing unlabeled standards.
Mass spectrometry.
Materials.
Stable isotope labeled (SIL) standards [13C4]-fumaric acid (99%), [13C4, 15N]-L-aspartic acid (99%), [1, 4-13C2]-succinic acid (99%) and [13C4]-L-malic acid (98%) were purchased from Cambridge Isotope Laboratories.
Sample preparation.
Cecum contents were recovered in pre-weighed 2 ml microfuge tubes. 0.5 ml of ice cold PBS was added per 1 g cecum content. Samples were homogenized using a tissuelyzer (Qiagen) at 25 Hz for 2.0 min, chilled for 1 min and homogenized again for another 2 min. Samples were centrifuged for 5.0 min at 20,000 × g at 4 °C to pellet cells. Supernatants were collected and centrifuged for 30 min to pellet remaining suspended particulate matter. Clear supernatants were syringe-filtered (0.22 μm). Samples were centrifuged for 2 min at 20,000 × g. Mouse chow (MC) pellets (1 pellet per sample) were coarsely grounded and collected in 2 ml microfuge tubes. MilliQ water was added to achieve a concentration of 0.033 g mouse chow /ml and cleared supernatants were prepared, as described above. Samples were diluted (1:10 v/v for cecum content samples, 1:1 v/v for extracted mouse chow) in dH2O containing a mix of SIL standards (aspartic acid, fumaric acid, malic acid, succinic acid). Prior to acquisition, samples were further diluted 1:10 (v/v) in a buffer corresponding to the starting conditions of the mobile phase used for liquid chromatography (see below).
Metabolite measurements.
Microscale ultrahigh performance liquid chromatography mass spectrometry (UPLC-MS) was performed with a Dionex Ultimate 3000 (Thermo Scientific) hyphenated to a Q Exactive Plus mass spectrometer (Thermo Scientific) with a heated electrospray ionization (HESI) source. Separation of metabolites was achieved with a C18 column (Kinetex®, 2.6 μm Polar C18, with dimensions 100×2.1 mm) as the stationary phase, and LC-MS grade solvents A/B/C/D as a mobile phase. Solvent A (1.5% formic acid in dH2O), and C (methanol) were kept constant at 10% and 25% respectively. For elution, solvent B (acetonitrile) was used against solvent D (dH2O) with the following gradient: 0 min, 0%; 0.4 min, 0%; 1.4 min, 50%; 2.4 min, 50%; 2.9 min, 0%; 7.0 min, 0%. Flow rate was 500 μl min−1; the injection volume was 2 μl. The mass spectrometer was operating in negative FTMS mode (m/z range 80 to 1000); resolution was set to 70000 (at m/z 200). Ionization was performed using a HESI probe with a sheath and aux gas flow rate of 60 and 20 respectively. Spray voltage was 3.0 kV, the S-lens RF level 50.0, capillary temperature 275°C and the aux gas heater was set to 400°C. Between samples, blank measurements were included.
Metabolites were identified based on exact monoisotopic mass (±0.002 Da), matching retention time and predicted mass shifts with corresponding SIL analytical standards.
Metabolites were quantified using unlabeled or SIL compounds as internal standards and custom written scripts employing the Python-based platform eMZed2 (Kiefer et al., 2013) (Hartl et al., 2017) (http://emzed.ethz.ch/). Briefly, corresponding monoisotopic (unlabeled) and respective (labeled) SIL peaks for each compound were determined by targeted extraction. Peak areas were calculated by integration using the trapezoidal rule. Background signals as determined by the median response in blank measurements were subtracted. For Fig. 4A, a linear response was confirmed by calibration curves of isotope labeled standards spiked into a mix (n=6) of the respective sample matrices (i.e. cecum content from CONE, LCM, or GF mice). Average metabolite concentrations were estimated for each sample type, and approximately equimolar concentrations of respective SIL standards were spiked into individual samples. As the obtained peak areas from SIL standards and analyzed samples were accordingly in the same range, metabolite concentrations were directly determined by calculating the ratio of peak areas Aunlabeled (sample) over Alabeled (SIL standard) and multiplication with the known concentration of the corresponding spiked SIL standards.
Limits of detection (LODs) were determined for the data shown in Fig. 4A using a statistical method (Armbruster and Pry, 2008). Briefly, samples were prepared as described above, and low concentration SIL standards were spiked into a mix (n=6) of respective matrices and repeatedly measured (n=8 per matrix). LODs were then calculated based on the obtained standard deviation (SD) between repeatedly measured samples.
The Z-score was based on a confidence interval of 95% and corrected for low sample number with the corresponding t-distribution. Obtained LODs were subsequently confirmed by manual inspection of calibration curves.
Additional quantifications of C4-dicarboxylates and other free metabolites in the mouse cecum in Fig. S4A
Chromatographic separation was performed via hydrophilic interaction (HILIC) on a AdvanceBio MS Spent Media column (100 × 2.1 mm, Agilent technologies) using a mobile phase A: H2O, 10 mM ammonium acetate titrated at pH 9.0 and B: Acetonitrile, 10 mM ammonium acetate titrated at pH 9.0. Samples were prepared as above and diluted 10 times in B phase. 2 μl of sample were injected at 100% B and 0.250 ml/min followed by the following gradient: 0.0 min 100%, 2.0 min 100%, 10.0 min 60%, 10.50 min 100%, 18.0 min 100%. The qTOF (Agilent 6520) was operated in negative mode at 2 GHz extended dynamic range and with a mass/ charge (m/z) range of 50–1000 and the following source parameters: VCap: 3500 C, nozzle voltage: 2000 V, gas temp: 325°C m drying gas 5 l/min, nebulizer 30 psig. Online mass calibration was performed using a second ionization source and a constant flow (5 ul/min) of reference solution (119.0363, 301.9981 and 980.0163). Compounds were identified based on the retention time of chemical standards and their accurate mass (tolerance 20 ppm) The MassHunter Quantitative Analysis Software (Agilent, version 7.0) was used for peak integration. Quantification was based on dilution series of chemical standards. Limits of detection for each metabolite was determined as mean +3.3x standard deviation of the signal detected in the lowest detected standard concentration. Obtained LODs were subsequently confirmed by manual inspection of calibration curves.
DNA sequence analyses for Tab. S4.
We identified Salmonella enterica Typhimurium SL1344 genes of interest (dcuABC, aspA, fumABC, frdABCD, sdh and H2-hydrogenases) in genomes from representative enteropathogens and microbiota using a reciprocal best BLAST hit approach. We used parameters previously recommended for the most sensitive and precise detection of orthologous genes in pairwise comparisions of genomes (Moreno-Hagelsieb and Latimer, 2008). Specifically, we used BLASTp of the NCBI BLAST+ (version 2.8.1) suite (Camacho et al., 2009) with soft filtering and Smith-Waterman alignment (“-seg yes - soft_masking true -use_sw_tback”) to reciprocally align the protein sequences of S. enterica Typhimurium with those of other genomes. We subsequently parsed the results for the best reciprocal hits using a custom python script and considered two genes as orthologs if the respective alignment score exceeded 60 bits. Protein sequences were annotated to Clusters of Orthologous Groups (Galperin et al., 2015) with EggNOG-mapper 2.0.0 (Huerta-Cepas et al., 2017) [4] in diamond mode (-m diamond), targetting the ‘bact’ database version 5.0.0 (Huerta-Cepas et al., 2019) (-d bact), using 8 threads per annotation (--cpu 8).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
The Mann Whitney test was performed using Graphpad Prism Version 6.0 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com). P values of < 0.05 (two-tailed) were considered as statistically significant. * P < 0.05, ** P < 0.01, *** P < 0.001.
Downstream analysis of the RB-TnSeq and 16s data were performed with custom R scripts including the packages vegan (2.5–5), ggplot2 (2.2.1), dplyr (0.8.0), tidyr (0.8.2), ggrepel (0.7.0) and readr (1.1.1).
For analyzing mass spectrometry data, we used the MassHunter Quantitative Analysis Software (Agilent, version 7.0), as detailed, above.
For analyzing the C.I. data in Fig. S3B, the Multiple Comparisons Test with Sidak`s correction was used.
For analyzing the attenuation of E. colidcuAB (Fig. 6A), Wilcoxon’s matched-pairs signed rank test (nonparametric, paired) was performed.
DATA AND CODE AVAILABILITY
The published article includes all datasets generated or analyzed during this study. All code is freely available upon request (except for the commercial data analysis programs listed). The RB-TnSeq data are listed in Tab. S1, the gut colonization phenotypes of the analyzed mutants are listed in Tab. S2. The 16S sequencing data is available at http://www.ebi.ac.uk/ena/data/view/PRJEB35643.
Supplementary Material
Table S1. Phenotypes of RB-TnSEq mutants at days 1–4 pi (related to Fig. 1).
Table S2. Gut colonization phenotypes of the 29 S. Typhimurium in-frame deletion mutants included in the confirmation experiment (related to Fig. 1).
Table S3. Median C.I. values of the indicated S. Typhimurium mutants in LCM mice (related to Fig. 2B, Fig. 4B and Fig. S4C).
Table S4. Distribution of orthologous genes implicated in C4-dicarboxylate fueled H2/fumarate respiration (related to Fig.6).
Table S5. Bacterial Strains (related to Key Resource Table).
Table S6. Oligonucleotides (related to Key Resource Table).
Highlights.
A high-throughput screen identifies key Salmonella genes used in initial gut colonization Host food and microbiota provide luminal aspartate and malate for Salmonella respiration The DcuABC transporters pump aspartate and malate into the Salmonella cell Aspartate and malate conversion into fumarate fuels growth by H2/fumarate respiration
Acknowledgements
We are grateful to Gottfried Unden, the members of the Hardt, Sunagawa, Christen, Sauer and the Vorholt lab for helpful scientific discussions. The RCHCI staff (especially Katharina Holzinger, Dennis Mollenhauer & Sven Nowok) for excellent support of our animal work is gratefully acknowledged. We thank Chris Field for help with computing Table S4. W.-D. H. is supported by the SNF (310030_53074 and 310030B_173338/1; Sinergia CRSII_154414/1), ETH Zurich (ETH-33 12-2), the Promedica Foundation, the Helmut Horten Foundation and the Monique Dornonville de la Cour Stiftung. L.L. receives grant LA 4572/1-1 from the Deutsche Forschungsgemeinschaft. J.A.V. is supported by the Swiss National Science Foundation (31003A-173094). B.C. is supported by the Swiss National Science Foundation, (310030_184664 and CRSII5_177164). SS is supported by the ETH and Helmut Horten Foundation. MM and SP are supported, in part, by NIH grants R03 AI139557, R01 AI136520, R01 AI044198, USDA 2017-67015-26085 and 2017-67017-26180.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors have no conflicting financial interests.
References
- ALI MM, NEWSOM DL, GONZALEZ JF, SABAG-DAIGLE A, STAHL C, STEIDLEY B, DUBENA J, DYSZEL JL, SMITH JN, DIEYE Y, ARSENESCU R, BOYAKA PN, KRAKOWKA S, ROMEO T, BEHRMAN EJ, WHITE P & AHMER BMM 2014. Fructose-Asparagine Is a Primary Nutrient during Growth of Salmonella in the Inflamed Intestine. Plos Pathogens, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARMBRUSTER DA & PRY T 2008. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev, 29 Suppl 1, S49–52. [PMC free article] [PubMed] [Google Scholar]
- BAKKEREN E, DOLOWSCHIAK T & D. M,RJ 2017. Detection of Mutations Affecting Heterogeneously Expressed Phenotypes by Colony Immunoblot and Dedicated Semi-Automated Image Analysis Pipeline. Front Microbiol, 8, 2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARTHEL M, HAPFELMEIER S, QUINTANILLA-MARTINEZ L, KREMER M, ROHDE M, HOGARDT M, PFEFFER K, RUSSMANN H & HARDT WD 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar typhimurium colitis model that allows analysis of both pathogen and host. Infection and Immunity, 71, 2839–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIN KIM O, LUX S & UNDEN G 2007. Anaerobic growth of Escherichia coli on D-tartrate depends on the fumarate carrier DcuB and fumarase, rather than the L-tartrate carrier TtdT and L-tartrate dehydratase. Archives of Microbiology, 188, 583–589. [DOI] [PubMed] [Google Scholar]
- BOTT M 1997. Anaerobic citrate metabolism and its regulation in enterobacteria. Archives of Microbiology, 167, 78–88. [PubMed] [Google Scholar]
- BUDDINGTON RK, ELNIF J, PUCHAL-GARDINER AA & SANGILD PT 2001. Intestinal apical amino acid absorption during development of the pig. American Journal of Physiology-Regulatory Integrative and Comparative Physiology, 280, R241–R247. [DOI] [PubMed] [Google Scholar]
- BUFFIE CG & PAMER EG 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol, 13, 790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BYNDLOSS MX, OLSAN EE, RIVERA-CHAVEZ F, TIFFANY CR, CEVALLOS SA, LOKKEN KL, TORRES TP, BYNDLOSS AJ, FABER F, GAO Y, LITVAK Y, LOPEZ CA, XU G, NAPOLI E, GIULIVI C, TSOLIS RM, REVZIN A, LEBRILLA CB & BAUMLER AJ 2017. Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science, 357, 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAMACHO C, COULOURIS G, AVAGYAN V, MA N, PAPADOPOULOS J, BEALER K & MADDEN TL 2009. BLAST+: architecture and applications. BMC Bioinformatics, 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CANALS R, XIA XQ, FRONICK C, CLIFTON SW, AHMER BMM, ANDREWS-POLYMENIS HL, PORWOLLIK S & MCCLELLAND M 2012. High-throughput comparison of gene fitness among related bacteria. Bmc Genomics, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHRISTEN B, ABELIUK E, COLLIER JM, KALOGERAKI VS, PASSARELLI B, COLLER JA, FERO MJ, MCADAMS HH & SHAPIRO L 2011. The essential genome of a bacterium. Molecular Systems Biology, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHRISTEN M, BEUSCH C, BOSCH Y, CERLETTI D, FLORES-TINOCO CE, DEL MEDICO L, TSCHAN F & CHRISTEN B 2016. Quantitative Selection Analysis of Bacteriophage phi CbK Susceptibility in Caulobacter crescentus. Journal of Molecular Biology, 428, 419–430. [DOI] [PubMed] [Google Scholar]
- CONWAY T & COHEN PS 2015. Commensal and Pathogenic Escherichia coli Metabolism in the Gut. Microbiology Spectrum, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUMMINGS JH, POMARE EW, BRANCH WJ, NAYLOR CPE & MACFARLANE GT 1987. Short Chain Fatty-Acids in Human Large-Intestine, Portal, Hepatic and Venous-Blood. Gut, 28, 1221–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DANIEL JW 1969. Metabolism of L- and Dl-Malic Acids by Rats. Food and Cosmetics Toxicology, 7, 103–&. [DOI] [PubMed] [Google Scholar]
- DATSENKO KA & WANNER BL 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America, 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEGUCHI Y, YAMATO I & ANRAKU Y 1989. Molecular-Cloning of Glts and Gltp, Which Encode Glutamate Carriers of Escherichia-Coli-B. Journal of Bacteriology, 171, 1314–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIARD M, GARCIA V, MAIER L, REMUS-EMSERMANN MNP, REGOES RR, ACKERMANN M & HARDT WD 2013. Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature, 494, 353–356. [DOI] [PubMed] [Google Scholar]
- DUBINI A, PYE RL, JACK RL, PALMER T & SARGENT F 2002. How bacteria get energy from hydrogen: a genetic analysis of periplasmic hydrogen oxidation in Escherichia coli. International Journal of Hydrogen Energy, 27, 1413–1420. [Google Scholar]
- ENGEL P, KRAMER R & UNDEN G 1992. Anaerobic Fumarate Transport in Escherichia-Coli by an Fnr-Dependent Dicarboxylate Uptake System Which Is Different from the Aerobic Dicarboxylate Uptake System. Journal of Bacteriology, 174, 5533–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENGEL P, KRAMER R & UNDEN G 1994. Transport of C-4-Dicarboxylates by Anaerobically Grown Escherichia-Coli - Energetics and Mechanism of Exchange, Uptake and Efflux. European Journal of Biochemistry, 222, 605–614. [DOI] [PubMed] [Google Scholar]
- FABER F, THIENNIMITR P, SPIGA L, BYNDLOSS MX, LITVAK Y, LAWHON S, ANDREWS-POLYMENIS HL, WINTER SE & BAUMLER AJ 2017. Respiration of Microbiota-Derived 1,2-propanediol Drives Salmonella Expansion during Colitis. Plos Pathogens, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FABER F, TRAN L, BYNDLOSS MX, LOPEZ CA, VELAZQUEZ EM, KERRINNES T, NUCCIO SP, WANGDI T, FIEHN O, TSOLIS RM & BAUMLER AJ 2016. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature, 534, 697–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FABICH AJ, JONES SA, CHOWDHURY FZ, CERNOSEK A, ANDERSON A, SMALLEY D, MCHARGUE JW, HIGHTOWER GA, SMITH JT, AUTIERI SM, LEATHAM MP, LINS JJ, ALLEN RL, LAUX DC, COHEN PS & CONWAY T 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun, 76, 1143–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRETER R, BRICKNER H, BOTNEY M, CLEVEN D & ARANKI A 1983. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect Immun, 39, 676–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GALPERIN MY, MAKAROVA KS, WOLF YI & KOONIN EV 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res, 43, D261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GILLIS CC, HUGHES ER, SPIGA L, WINTER MG, ZHU WH, DE CARVALHO TF, CHANIN RB, BEHRENDT CL, HOOPER LV, SANTOS RL & WINTER SE 2018. Dysbiosis-Associated Change in Host Metabolism Generates Lactate to Support Salmonella Growth (vol 23, pg 54, 2018). Cell Host & Microbe, 23, 570–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOLBY P, DAVIES S, KELLY DJ, GUEST JR & ANDREWS SC 1999. Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C-4-dicarboxylates in Escherichia coli. Journal of Bacteriology, 181, 1238–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRANT AJ, RESTIF O, MCKINLEY TJ, SHEPPARD M, MASKELL DJ & MASTROENI P 2008. Modelling within-host spatiotemporal dynamics of invasive bacterial disease. Plos Biology, 6, 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARTL J, KIEFER P, MEYER F & VORHOLT JA 2017. Longevity of major coenzymes allows minimal de novo synthesis in microorganisms. Nature Microbiology, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HECHT AL, CASTERLINE BW, EARLEY ZM, GOO YA, GOODLETT DR & BUBECK WARDENBURG J 2016. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep, 17, 1281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOISETH SK & STOCKER BAD 1981. Aromatic-Dependent Salmonella-Typhimurium Are Non-Virulent and Effective as Live Vaccines. Nature, 291, 238–239. [DOI] [PubMed] [Google Scholar]
- HUERTA-CEPAS J, FORSLUND K, COELHO LP, SZKLARCZYK D, JENSEN LJ, VON MERING C & BORK P 2017. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol Biol Evol, 34, 2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUERTA-CEPAS J, SZKLARCZYK D, HELLER D, HERNANDEZ-PLAZA A, FORSLUND SK, COOK H, MENDE DR, LETUNIC I, RATTEI T, JENSEN LJ, VON MERING C & BORK P 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res, 47, D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGHES ER, WINTER MG, DUERKOP BA, SPIGA L, FURTADO DE CARVALHO T, ZHU W, GILLIS CC, BUTTNER L, SMOOT MP, BEHRENDT CL, CHERRY S, SANTOS RL, HOOPER LV & WINTER SE 2017. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe, 21, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ILG K, ENDT K, MISSELWITZ B, STECHER B, AEBI M & HARDT WD 2009. O-Antigen-Negative Salmonella enterica Serovar Typhimurium Is Attenuated in Intestinal Colonization but Elicits Colitis in Streptomycin-Treated Mice. Infection and Immunity, 77, 2568–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JANAUSCH IG, ZIENTZ E, TRAN QH, KROGER A & UNDEN G 2002. C-4-dicarboxylate carriers and sensors in bacteria. Biochimica Et Biophysica Acta-Bioenergetics, 1553, 39–56. [DOI] [PubMed] [Google Scholar]
- JONES SA, GIBSON T, MALTBY RC, CHOWDHURY FZ, STEWART V, COHEN PS & CONWAY T 2011. Anaerobic respiration of Escherichia coli in the mouse intestine. Infect Immun, 79, 4218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAISER P, SLACK E, GRANT AJ, HARDT WD & REGOES RR 2013. Lymph Node Colonization Dynamics after Oral Salmonella Typhimurium Infection in Mice. Plos Pathogens, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIEFER P, SCHMITT U & VORHOLT JA 2013. eMZed: an open source framework in Python for rapid and interactive development of LC/MS data analysis workflows. Bioinformatics, 29, 963–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLEEFELD A, ACKERMANN B, BAUER J, KRAMER J & UNDEN G 2009. The Fumarate/Succinate Antiporter DcuB of Escherichia coli Is a Bifunctional Protein with Sites for Regulation of DcuS-dependent Gene Expression. Journal of Biological Chemistry, 284, 265–275. [DOI] [PubMed] [Google Scholar]
- KROGER C, COLGAN A, SRIKUMAR S, HANDLER K, SIVASANKARAN SK, HAMMARLOF DL, CANALS R, GRISSOM JE, CONWAY T, HOKAMP K & HINTON JCD 2013. An Infection-Relevant Transcriptomic Compendium for Salmonella enterica Serovar Typhimurium. Cell Host & Microbe, 14, 683–695. [DOI] [PubMed] [Google Scholar]
- LANGRIDGE GC, PHAN MD, TURNER DJ, PERKINS TT, PARTS L, HAASE J, CHARLES I, MASKELL DJ, PETERS SE, DOUGAN G, WAIN J, PARKHILL J & TURNER AK 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Research, 19, 2308–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI H & DURBIN R 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics, 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOPEZ CA, RIVERA-CHAVEZ F, BYNDLOSS MX & BAUMLER AJ 2015. The Periplasmic Nitrate Reductase NapABC Supports Luminal Growth of Salmonella enterica Serovar Typhimurium during Colitis. Infect Immun, 83, 3470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUTGENS M & GOTTSCHALK G 1980. Why a Co-Substrate Is Required for Anaerobic Growth of Escherichia-Coli on Citrate. Journal of General Microbiology, 119, 63–70. [DOI] [PubMed] [Google Scholar]
- MAIER L, BARTHEL M, STECHER B, MAIER RJ, GUNN JS & HARDT WD 2014a. Salmonella Typhimurium strain ATCC14028 requires H2-hydrogenases for growth in the gut, but not at systemic sites. PLoS One, 9, e110187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAIER L, DIARD M, SELLIN ME, CHOUFFANE ES, TRAUTWEIN-WEIDNER K, PERIASWAMY B, SLACK E, DOLOWSCHIAK T, STECHER B, LOVERDO C, REGOES RR & HARDT WD 2014b. Granulocytes Impose a Tight Bottleneck upon the Gut Luminal Pathogen Population during Salmonella Typhimurium Colitis. Plos Pathogens, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAIER L, VYAS R, CORDOVA CD, LINDSAY H, SCHMIDT TS, BRUGIROUX S, PERIASWAMY B, BAUER R, STURM A, SCHREIBER F, VON MERING C, ROBINSON MD, STECHER B & HARDT WD 2013. Microbiota-derived hydrogen fuels Salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe, 14, 641–51. [DOI] [PubMed] [Google Scholar]
- MAIER RJ 2005. Use of molecular hydrogen as an energy substrate by human pathogenic bacteria. Biochemical Society Transactions, 33, 83–85. [DOI] [PubMed] [Google Scholar]
- MALTBY R, LEATHAM-JENSEN MP, GIBSON T, COHEN PS & CONWAY T 2013. Nutritional Basis for Colonization Resistance by Human Commensal Escherichia coli Strains HS and Nissle 1917 against E. coli O157:H7 in the Mouse Intestine. Plos One, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEIJER-SEVERS GJ & VAN SANTEN E 1987. Short-chain fatty acids and succinate in feces of healthy human volunteers and their correlation with anaerobe cultural counts. Scand J Gastroenterol, 22, 672–6. [DOI] [PubMed] [Google Scholar]
- MERCADO-LUBO R, LEATHAM MP, CONWAY T & COHEN PS 2009. Salmonella enterica serovar Typhimurium mutants unable to convert malate to pyruvate and oxaloacetate are avirulent and immunogenic in BALB/c mice. Infect Immun, 77, 1397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIKI T, GOTO R, FUJIMOTO M, OKADA N & HARDT WD 2017. The Bactericidal Lectin RegIIIbeta Prolongs Gut Colonization and Enteropathy in the Streptomycin Mouse Model for Salmonella Diarrhea. Cell Host Microbe. [DOI] [PubMed] [Google Scholar]
- MORENO-HAGELSIEB G & LATIMER K 2008. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics, 24, 319–24. [DOI] [PubMed] [Google Scholar]
- NG KM, FERREYRA JA, HIGGINBOTTOM SK, LYNCH JB, KASHYAP PC, GOPINATH S, NAIDU N, CHOUDHURY B, WEIMER BC, MONACK DM & SONNENBURG JL 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature, 502, 96–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ORGANIZATION, W. H. & NATIONS, F. A. A. O. O. T. U. 2002. Risk Assessments of Salmonella in Eggs and Broiler Chickens. Microbiological Risk Assessment Series. [Google Scholar]
- POS KM, DIMROTH P & BOTT M 1998. The Escherichia coli citrate carrier CitT: a member of a novel eubacterial transporter family related to the 2-oxoglutarate/malate translocator from spinach chloroplasts. Journal of Bacteriology, 180, 4160–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRICE MN, WETMORE KM, WATERS RJ, CALLAGHAN M, RAY J, LIU HL, KUEHL JV, MELNYK RA, LAMSON JS, SUH Y, CARLSON HK, ESQUIVEL Z, SADEESHKUMAR H, CHAKRABORTY R, ZANE GM, RUBIN BE, WALL JD, VISEL A, BRISTOW J, BLOW MJ, ARKIN AP & DEUTSCHBAUER AM 2018. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature, 557, 503–+. [DOI] [PubMed] [Google Scholar]
- RIVERA-CHAVEZ F, WINTER SE, LOPEZ CA, XAVIER MN, WINTER MG, NUCCIO SP, RUSSELL JM, LAUGHLIN RC, LAWHON SD, STERZENBACH T, BEVINS CL, TSOLIS RM, HARSHEY R, ADAMS LG & BAUMLER AJ 2013. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog, 9, e1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIVERA-CHAVEZ F, ZHANG LF, FABER F, LOPEZ CA, BYNDLOSS MX, OLSAN EE, XU G, VELAZQUEZ EM, LEBRILLA CB, WINTER SE & BAUMLER AJ 2016. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe, 19, 443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANA TG, FLAUGNATTI N, LUGO KA, LAM LH, JACOBSON A, BAYLOT V, DURAND E, JOURNET L, CASCALES E & MONACK DM 2016. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A, 113, E5044–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMONSEN J, MOLBAK K, FALKENHORST G, KROGFELT KA, LINNEBERG A & TEUNIS PF 2009. Estimation of incidences of infectious diseases based on antibody measurements. Stat Med, 28, 1882–95. [DOI] [PubMed] [Google Scholar]
- SIX S, ANDREWS SC, UNDEN G & GUEST JR 1994. Escherichia coli possesses two homologous anaerobic C4-dicarboxylate membrane transporters (DcuA and DcuB) distinct from the aerobic dicarboxylate transport system (Dct). J Bacteriol, 176, 6470–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SONNENBURG JL, XU J, LEIP DD, CHEN CH, WESTOVER BP, WEATHERFORD J, BUHLER JD & GORDON JI 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science, 307, 1955–9. [DOI] [PubMed] [Google Scholar]
- SPIGA L, WINTER MG, FURTADO DE CARVALHO T, ZHU W, HUGHES ER, GILLIS CC, BEHRENDT CL, KIM J, CHESSA D, ANDREWS-POLYMENIS HL, BEITING DP, SANTOS RL, HOOPER LV & WINTER SE 2017. An Oxidative Central Metabolism Enables Salmonella to Utilize Microbiota-Derived Succinate. Cell Host Microbe, 22, 291–301 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STECHER B 2015. The Roles of Inflammation, Nutrient Availability and the Commensal Microbiota in Enteric Pathogen Infection. Microbiol Spectr, 3. [DOI] [PubMed] [Google Scholar]
- STECHER B, BARTHEL M, SCHLUMBERGER MC, HABERLI L, RABSCH W, KREMER M & HARDT WD 2008. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol, 10, 1166–80. [DOI] [PubMed] [Google Scholar]
- STECHER B, BERRY D & LOY A 2013. Colonization resistance and microbial ecophysiology: using gnotobiotic mouse models and single-cell technology to explore the intestinal jungle. FEMS Microbiol Rev, 37, 793–829. [DOI] [PubMed] [Google Scholar]
- STECHER B, CHAFFRON S, KAPPELI R, HAPFELMEIER S, FREEDRICH S, WEBER TC, KIRUNDI J, SUAR M, MCCOY KD, VON MERING C, MACPHERSON AJ & HARDT WD 2010. Like Will to Like: Abundances of Closely Related Species Can Predict Susceptibility to Intestinal Colonization by Pathogenic and Commensal Bacteria. Plos Pathogens, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STECHER B, DENZLER R, MAIER L, BERNET F, SANDERS MJ, PICKARD DJ, BARTHEL M, WESTENDORF AM, KROGFELT KA, WALKER AW, ACKERMANN M, DOBRINDT U, THOMSON NR & HARDT WD 2012. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci U S A, 109, 1269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STECHER B & HARDT WD 2011. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol, 14, 82–91. [DOI] [PubMed] [Google Scholar]
- STECHER B, ROBBIANI R, WALKER AW, WESTENDORF AM, BARTHEL M, KREMER M, CHAFFRON S, MACPHERSON AJ, BUER J, PARKHILL J, DOUGAN G, VON MERING C & HARDT WD 2007. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. Plos Biology, 5, 2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRIPP ST, SOBOH B, LINDENSTRAUSS U, BRAUSSEMANN M, HERZBERG M, NIES DH, SAWERS RG & HEBERLE J 2013. HypD is the scaffold protein for Fe-(CN)2CO cofactor assembly in [NiFe]-hydrogenase maturation. Biochemistry, 52, 3289–96. [DOI] [PubMed] [Google Scholar]
- TCHAWA YIMGA M, LEATHAM MP, ALLEN JH, LAUX DC, CONWAY T & COHEN PS 2006. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect Immun, 74, 1130–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THIENNIMITR P, WINTER SE, WINTER MG, XAVIER MN, TOLSTIKOV V, HUSEBY DL, STERZENBACH T, TSOLIS RM, ROTH JR & BAUMLER AJ 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proceedings of the National Academy of Sciences of the United States of America, 108, 17480–17485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNDEN G, STRECKER A, KLEEFELD A & KIM OB 2016. C4-Dicarboxylate Utilization in Aerobic and Anaerobic Growth. EcoSal Plus, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VELAZQUEZ EM, NGUYEN H, HEASLEY KT, SAECHAO CH, GIL LM, ROGERS AWL, MILLER BM, ROLSTON MR, LOPEZ CA, LITVAK Y, LIOU MJ, FABER F, BRONNER DN, TIFFANY CR, BYNDLOSS MX, BYNDLOSS AJ & BAUMLER AJ 2019. Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nat Microbiol, 4, 1057–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WETMORE KM, PRICE MN, WATERS RJ, LAMSON JS, HE J, HOOVER CA, BLOW MJ, BRISTOW J, BUTLAND G, ARKIN AP & DEUTSCHBAUER A 2015. Rapid Quantification of Mutant Fitness in Diverse Bacteria by Sequencing Randomly Bar-Coded Transposons. Mbio, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMS VR & LARTIGUE DJ 1967. Quaternary structure and certain allosteric properties of aspartase. J Biol Chem, 242, 2973–8. [PubMed] [Google Scholar]
- WILSON RP, TURSI SA, RAPSINSKI GJ, MEDEIROS NJ, LE LS, KOTREDES KP, PATEL S, LIVERANI E, SUN S, ZHU W, KILPATRICK L, WINTER SE, GAMERO AM & TUKEL C 2019. STAT2 dependent Type I Interferon response promotes dysbiosis and luminal expansion of the enteric pathogen Salmonella Typhimurium. PLoS Pathog, 15, e1007745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WINTER SE, THIENNIMITR P, WINTER MG, BUTLER BP, HUSEBY DL, CRAWFORD RW, RUSSELL JM, BEVINS CL, ADAMS LG, TSOLIS RM, ROTH JR & BAUMLER AJ 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature, 467, 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOEHLKE G & DIMROTH P 1994. Anaerobic Growth of Salmonella-Typhimurium on L(+)-Tartrate and D(−)-Tartrate Involves an Oxaloacetate Decarboxylase Na+ Pump. Archives of Microbiology, 162, 233–237 [DOI] [PubMed] [Google Scholar]
- WOTZKA SY, KREUZER M, MAIER L, ARNOLDINI M, NGUYEN BD, BRACHMANN AO, BERTHOLD DL, ZUND M, HAUSMANN A, BAKKEREN E, HOCES D, GUL E, BEUTLER M, DOLOWSCHIAK T, ZIMMERMANN M, FUHRER T, MOOR K, SAUER U, TYPAS A, PIEL J, DIARD M, MACPHERSON AJ, STECHER B, SUNAGAWA S, SLACK E & HARDT WD 2019. Escherichia coli limits Salmonella Typhimurium infections after diet shifts and fat-mediated microbiota perturbation in mice. Nat Microbiol, 4, 2164–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOTZKA SY, NGUYEN BD & HARDT WD 2017. Salmonella Typhimurium Diarrhea Reveals Basic Principles of Enteropathogen Infection and Disease-Promoted DNA Exchange. Cell Host Microbe, 21, 443–454. [DOI] [PubMed] [Google Scholar]
- YUNG MMC, PARK DM, OVERTON KW, BLOW MJ, HOOVER CA, SMIT J, MURRAY SR, RICCI DP, CHRISTEN B, BOWMAN GR & JIAO YQ 2015. Transposon Mutagenesis Paired with Deep Sequencing of Caulobacter crescentus under Uranium Stress Reveals Genes Essential for Detoxification and Stress Tolerance. Journal of Bacteriology, 197, 3160–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZIENTZ E, JANAUSCH IG, SIX S & UNDEN G 1999. Functioning of DcuC as the C4-dicarboxylate carrier during glucose fermentation by Escherichia coli. J Bacteriol, 181, 3716–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZIENTZ E, SIX S & UNDEN G 1996. Identification of a third secondary carrier (DcuC) for anaerobic C4-dicarboxylate transport in Escherichia coli: roles of the three Dcu carriers in uptake and exchange. J Bacteriol, 178, 7241–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Phenotypes of RB-TnSEq mutants at days 1–4 pi (related to Fig. 1).
Table S2. Gut colonization phenotypes of the 29 S. Typhimurium in-frame deletion mutants included in the confirmation experiment (related to Fig. 1).
Table S3. Median C.I. values of the indicated S. Typhimurium mutants in LCM mice (related to Fig. 2B, Fig. 4B and Fig. S4C).
Table S4. Distribution of orthologous genes implicated in C4-dicarboxylate fueled H2/fumarate respiration (related to Fig.6).
Table S5. Bacterial Strains (related to Key Resource Table).
Table S6. Oligonucleotides (related to Key Resource Table).
Data Availability Statement
The published article includes all datasets generated or analyzed during this study. All code is freely available upon request (except for the commercial data analysis programs listed). The RB-TnSeq data are listed in Tab. S1, the gut colonization phenotypes of the analyzed mutants are listed in Tab. S2. The 16S sequencing data is available at http://www.ebi.ac.uk/ena/data/view/PRJEB35643.


