SUMMARY
Typhoidal and nontyphoidal Salmonellae (NTS) cause typhoid fever and gastroenteritis respectively in humans. Salmonella Typhoid toxin contributes to typhoid disease progression and chronic infection, but little is known about the role of its NTS orthologue. We found that typhoid toxin and its NTS orthologue induce different clinical presentations. The PltB subunit of each toxin exhibits different glycan-binding preferences that correlate with glycan expression profiles of host cells targeted by each bacteria at the primary infection or intoxication sites. Through co-crystal structures of PltB subunits bound to specific glycan receptor moieties, we show that they induce markedly different glycan-binding preferences and virulence outcomes. Furthermore, immunization with the NTS S. Javiana or its toxin offers cross-reactive protection against lethal-dose typhoid toxin challenge. Cumulatively, these results offer insights into the evolution of host adaptations in Salmonella AB toxins, their cell and tissue tropisms, and the design for improved typhoid vaccines and therapeutics.
In Brief
Through structural and functional analyses of the typhoid fever-causing Salmonella’s Typhoid toxin and its orthologue in nontyphoidal/self-limiting gastroenteritis-causing Salmonella, Lee et al. offer insights into the evolution of host adaptations in Salmonella AB toxins, as well as the design for improved typhoid fever vaccines and therapeutics.
Graphical Abstract
INTRODUCTION
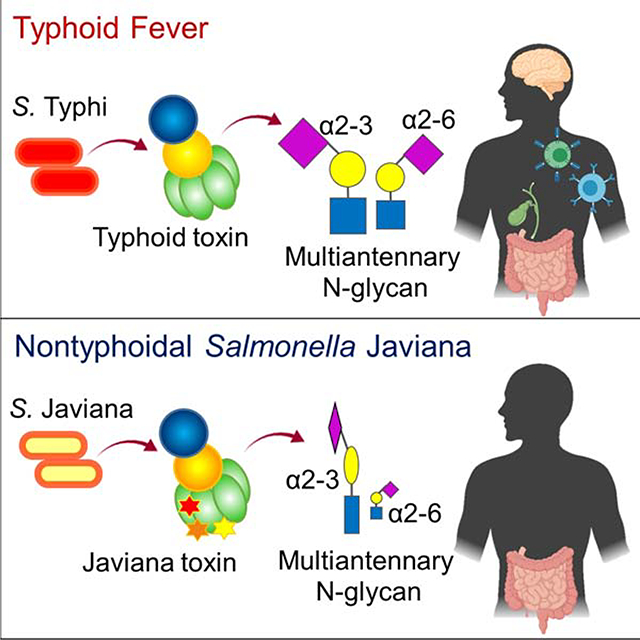
Salmonella infection is a major public health problem due to the global spread of multidrug-resistant and extensively drug-resistant strains (Feasey et al., 2015; Klemm et al., 2018; Parkhill et al., 2001). Salmonella enterica serovars can be divided into two groups based on disease outcomes in humans: typhoidal Salmonella and nontyphoidal Salmonella (NTS) (Gal-Mor et al., 2014; Johnson et al., 2018). Salmonella enterica serovar Typhi (S. Typhi) and S. Paratyphi are typhoidal Salmonellae causing life-threatening systemic infectious disease typhoid and paratyphoid fever respectively in humans. S. Enteritidis and S. Typhimurium are the most common NTS responsible for self-limiting gastroenteritis in healthy people globally, while S. Javiana is among the common NTS causing gastroenteritis in developed countries (Gilchrist and MacLennan, 2019; Jackson et al., 2013; Singletary et al., 2016). In addition to these cell and tissue adaptations in the same host, typhoidal Salmonella is adapted to exclusively infect humans, whereas NTS can infect both humans and animals (Ohl and Miller, 2001; Raffatellu et al., 2008; Tsolis et al., 2011).
In line with the severe disease outcomes and strict host adaptation, S. Typhi encodes few unique virulence factors, including typhoid toxin. Like S. Typhi, typhoid toxin appears to evolve to adapt in humans, in which its receptor binding subunit PltB plays a crucial role (Deng et al., 2014). Typhoid toxin consists of two enzymatic ‘A’ subunits, CdtB (nuclease) and PltA (mono ADP-ribosyltransferase), linked to a homopentamer of PltB (receptor-binding ‘B’ subunit). Due to its distinct A2B5 stoichiometry with two enzymes in a single toxin complex, typhoid toxin is unique in the bacterial AB toxin family (Song et al., 2013). CdtB subunit is a nuclease that induces host cell DNA damage and cell cycle arrest (Haghjoo and Galan, 2004; Spano et al., 2008). If this intoxication process is not intercepted, cell death can result. PltA is a mono ADP-ribosyltransferase that has amino acid sequence and structural similarities to pertussis toxin S1 (Spano et al., 2008). PltB recognizes the specific trisaccharide motif, N-acetylneuraminic acid (Neu5Ac)-α2–3/α2–6-galactose (Gal)-β1–3/β1–4-N-acetylglucosamine (GlcNAc), as host cell receptor for endocytosis, a required process for AB toxins to deliver their A subunits into the site of action in host cells (Song et al., 2013). Likewise, the interaction between PltB and its host cell surface glycan receptor determines the host, tissue, and cell tropism of bacterial AB toxins. For example, despite the ubiquity of the trisaccharide motif for PltB binding, Neu5Ac-α2–3/α2–6-Gal-β1–3/β1–4-GlcNAc, the pentameric configuration of PltB allows for preferential binding to this glycan motif displayed as part of multiantennary N-linked glycoproteins, resulting in high-affinity multivalent interactions (Yang et al., 2018b). Consistently, typhoid toxin has tropism to cells displaying multiantennary N-linked glycoproteins containing multiple Neu5Acs such as immune cells and endothelial cells of arterioles in the brain (Yang et al., 2018b). O-acetyl modification on the terminal sialic acid Neu5Ac of the glycan receptor moiety appears to also contribute to the tropism of typhoid toxin since the toxin exhibits the increased binding affinity to the O-acetylated glycan receptor motif (Nguyen et al., 2020). Its tropism to immune cells is anticipated to play a large role in altering innate and adaptive immune responses, which correspond to the suggested roles of this toxin in typhoid pathogenesis during both acute and chronic infections (Song et al., 2013; Song et al., 2010; Yang et al., 2018a; Yang et al., 2018b). However, we found that its NTS orthologue Javiana toxin does not cause the same observed toxicity despite the high amino acid sequence similarity between typhoid toxin and Javiana toxin. Here, we performed structure and function analysis to determine whether the amino acid sequence variations observed in typhoid and Javiana toxins reflect their evolution to adapt to specific host cells and tissues in humans that correspond to the primary infection or intoxication sites of the bacteria secreting these toxins.
RESULTS
Typhoid Toxin and Its NTS Orthologue Javiana Toxin Induce Different Clinical Presentations where Their Receptor Binding PltB Subunits Play a Crucial Role
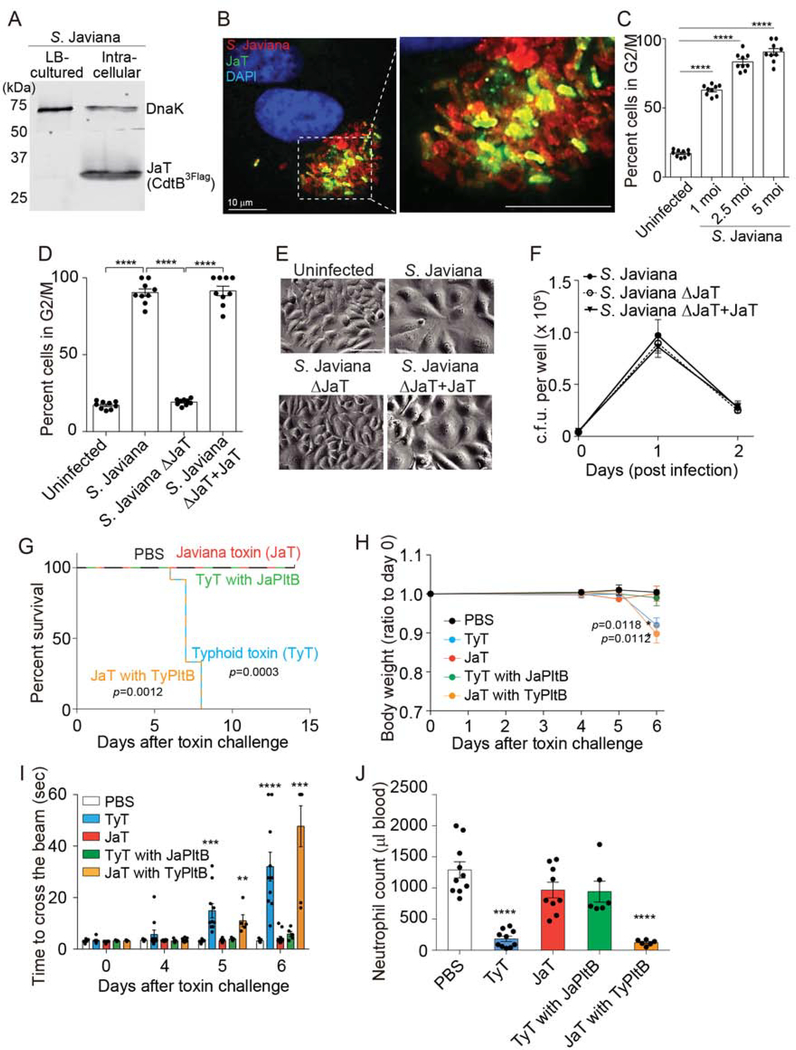
We investigated whether NTS S. Javiana secretes functional Javiana toxin during infection. Similar to the regulation mechanism of the typhoid toxin gene expression in S. Typhi, which senses and responds to environmental cues present within the Salmonella-containing vacuole (SCV) (Chang et al., 2016; Spano and Galan, 2008), we found that Javiana toxin is expressed exclusively by intracellular S. Javiana. Western blot analysis detecting Javiana toxin indicates that S. Javiana harvested from infected Henle-407 cells highly expressed Javiana toxin, but S. Javiana cultured in LB broth did not (Figure 1A). Consistently, fluorescent microscopy results indicate that Javiana toxin was produced by intracellular S. Javiana (Figure 1B). Javiana toxin secreted during infection was functional since this toxin was capable of intoxicating host cells measured by host cell cycle profiles via flow cytometry. The percentage of host cells arrested in G2/M was proportional to the multiplicity of infection (m.o.i.) of S. Javiana, determined by colony-forming unit (c.f.u.) assays, demonstrating the sensitivity of this assay in evaluating Javiana toxin produced during infection (Figure 1C). Approximately twenty percent of total cells were in G2/M stage in both uninfected Henle-407 cells and S. Javiana ΔJaT lacking the toxin genes (Figure 1D). In contrast, cells infected with either wild-type (WT) S. Javiana or the chromosomally complemented S. Javiana ΔJaT (S. Javiana ΔJaT + JaT) showed ~90% cells arrested in G2/M stage (Figure 1D). Consistent results were observed in host cell distension morphology analysis, another hallmark of CdtB-mediated toxicity in Henle-407 cells (Figure 1E). Data shown in Figure 1F demonstrate comparable c.f.u. numbers among the strains tested, indicating that Javiana toxin was the cause of the observed intoxication results. Taken together, these results demonstrate that (1) S. Javiana can express functional Javiana toxin and (2) infection into host cells is required for the expression of Javiana toxin.
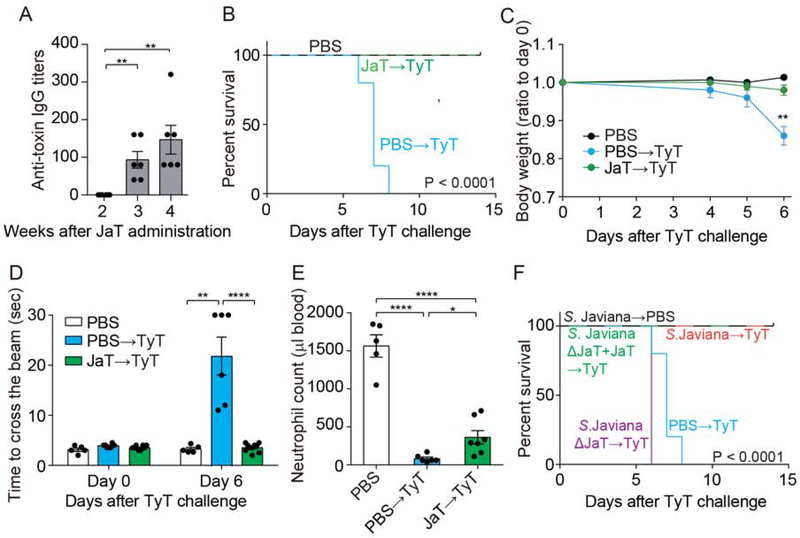
Figure 1. Typhoid Toxin and Its NTS Orthologue Javiana Toxin Induce Different Clinical Presentations where Their Receptor Binding PltB Subunits Play a Crucial Role.
A, Western blot analysis using anti-Flag antibody that detects Javiana toxin CdtB. DnaK was used as a loading control. B, Fluorescent microscopy images of Henle-407 cells infected with S. Javiana (red). Javiana toxin, green; DAPI (DNA), blue. Scale bar, 10 μm. C, Percentage of host cells arrested in G2/M stage. Henle-407 cells were left uninfected or infected with the indicated multiplicity of infection (m.o.i) of S. Javiana for 48 hrs. D, Cell cycle analysis profiles of Henle-407 cells that were left uninfected or infected (5 m.o.i.) for 2 days with indicated S. Javiana strains. E, Representative light microscope images that correspond to results shown in D. Scale bar, 100 μm. F, Colony-forming unit (c.f.u.) analysis of indicated S. Javiana strains in Henle-407 cells. G, Survival of animals receiving different toxin preparations. H, Bodyweight changes after intravenous administration of indicated toxin preparations. I, Measurements of time required for crossing the beam for animals treated as indicated. J, Circulating neutrophil numbers in peripheral blood of animals treated as indicated were measured 5 days after toxin challenge. At least three independent experiments were performed. Bars represent average ± standard deviation. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001. n=6–18 per group. Two-tailed unpaired t-tests were used for C, D, F, H-J, while low-rank test was used for G.
We asked whether both typhoid and Javiana toxins cause similar clinical presentations. To address this question, we administered endotoxin-free ultrapure toxin preparations into cytidine monophospho-N-acetylneuraminic acid hydroxylase (CMAH) null mice. CMAH converts Neu5Ac to N-glycolylneuraminic acid (Neu5Gc) in many mammals. However, humans do not produce a functional CMAH enzyme because humans encode a pseudogene for CMAH, therefore predominantly expressing Neu5Ac (Chou et al., 2002). Like humans, CMAH null mice exclusively express Neu5Ac. Furthermore, we previously demonstrated that typhoid toxin secreted by the human-adapted pathogen S. Typhi binds to glycan moieties terminated in Neu5Ac but not to the otherwise same glycan moieties terminated in Neu5Gc (Deng et al., 2014).
Consistent with previous observations (Song et al., 2013; Yang et al., 2018b), mice systemically administered typhoid toxin preparations showed many of the characteristic signs and symptoms of typhoid fever, lost weight, and eventually died (Figure 1G–J). Motor function impairment and leukopenia were objectively quantified via methods previously established (Figure 1I–J) (Yang et al., 2018b). Strikingly, considering the small amino acid variation between the two toxins, mice administered intravenously with an equal amount of Javiana toxin showed little to no disease (Figure 1G–J). These two toxins differ in six amino acids, three in PltA and three in PltB, while containing identical CdtB subunits. Because PltB (receptor-binding) and CdtB (nuclease) activities are essential for toxin endocytosis and intoxication in host cells respectively, but PltA does not (Song et al., 2013; Yang et al., 2018b), we hypothesized that the three amino acid variation on PltB is primarily responsible for this striking in vivo phenotype difference that typhoid and Javiana toxins induce. To test the hypothesis, two chimera toxins, Javiana toxin whose PltB subunits were replaced with the typhoid toxin counterpart (JaT with TyPltB) and typhoid toxin whose PltB subunits were replaced with the Javiana toxin counterpart (TyT with JaPltB), were generated and evaluated in mice. Mice systemically administered JaT with TyPltB phenocopied typhoid toxin, whereas TyT with JaPltB phenocopied Javiana toxin (Figure 1G–J), indicating that the amino acid variation found in these two toxins’ PltBs is responsible for different virulence outcomes.
Glycan Binding Preferences of Typhoid and Javiana Toxins Are Similar in a Broader Context, yet Markedly Different in Specific Details
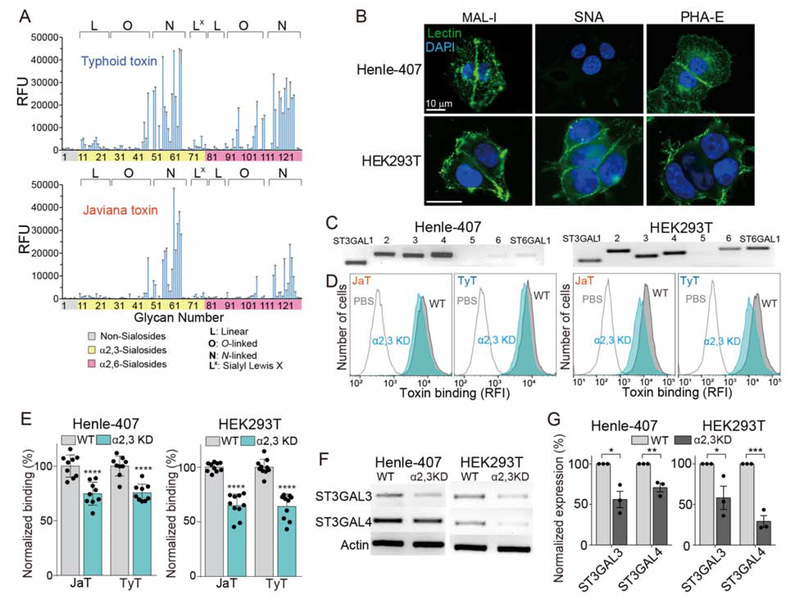
To understand the underlying molecular mechanism, we examined glycan-binding preferences of these two toxins. Alexa Fluor-555 conjugated toxin preparations were probed for 130 glycans on glycan array chips displaying sialylated multiantennary glycans (Peng et al., 2017; Yang et al., 2018b). Javiana toxin exhibits a narrower glycan-binding profile, as compared to typhoid toxin (Figure 2A). This difference is largely due to how the terminal Neu5Ac is linked to the underlying disaccharide, Gal-β1–3/β1–4-GlcNAc. In N-linked glycans, this configuration is either α2–3 or α2–6. Typhoid toxin binds well to both α2–3 and α2–6 sialosides, whereas Javiana toxin binds to α2–3 sialosides with a narrower specificity toward sialylated, extended, multiantennary N-linked glycans (Figure 2A and Table S1). Notably, Javiana toxin shows some yet much decreased binding to α2–6 sialosides (Figure 2A).
Figure 2. Glycan Binding Preferences of Typhoid and Javiana Toxins Are Similar in a Broader Context, yet Markedly Different in Specific Details.
A, Glycan array analyzing glycan-binding preferences of typhoid and Javiana toxins. RFU, relative fluorescent unit, represents toxin bindings to glycans in the array. B, Unpermeabilized Henle-407 and HEK293T cells stained with MAL-1-FITC, SNA-FITC, and DAPI. Scale bar, 10 μm. C, qRT-PCR analysis of enzymes involved in the biosynthesis of α2–3 and α2–6 sialosides. D-E, Toxin bindings to ST3GAL3- and ST3GAL4-depleted Henle-407 and HEK293T cells. KD, knockdown. Shown are representative histograms (D) and quantification results (E). F-G, qRT-PCR results of ST3GAL3- and ST3GAL4-depleted cells used in D and E. Shown are representative gel images (F) and quantification (G). At least three independent experiments were performed. Bars represent average ± standard deviation. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001. Two-tailed unpaired t-tests were used for E and G. See also Table S1.
To validate the glycan array results, we exploited two cell lines. Henle-407 cells almost exclusively express α2–3 sialosides whereas HEK293T cells express both α2–3 and α2–6 sialosides (Figure 2B). Specifically, Henle-407 cells bind to Maackia amurensis lectin I (MAL-1), a lectin specific to Neu5Ac-α2–3-Gal glycans but not to Sambucus nigra bark lectin (SNA), a lectin specific to Neu5Ac-α2–6-Gal glycans (Figure 2B). Furthermore, Henle-407 cells also bind to Phaseolus vulgaris (PHA)-E, a lectin specific to multiantennary N-glycans, indicating that Neu5Ac-α2–3-Gal glycans are displayed in the context of multiantennary N-linked glycans on Henle-407 cells (Figure 2B). HEK293T cells bind to MAL-1, SNA, and PHA-E, indicating that HEK293T cells express multiantennary α2–3 and α2–6 sialosides (Figure 2B). To determine the manipulation strategy for specific glycans in these cells, we quantified the expression of relevant enzymes involved in the biosynthesis of α2–3 and α2–6 sialosides. Six isotypes of ST3 β-galactoside α2,3-sialyltransferase (ST3GAL), ST3GAL1, 2, 3, 4, 5, and 6, can synthesize N-, O-linked glycoproteins and glycolipids terminated in the α2–3 configuration in cells, but three isotypes, ST3GAL3, 4, and 6, are known to be involved in the synthesis of N-linked glycans terminated in Neu5Ac-α2–3-Gal-β1–3/β1–4-GlcNAc (Varki et al., 2017) and therefore these are potentially relevant to the synthesis of glycan receptors for typhoid and Javiana toxins. ST6 β-galactoside α2,6-sialyltransferase 1 (ST6GAL1) and 2 can synthesize α2–6 sialosides, although ST6GAL1 is more relevant in this study as it is known to be expressed in various cells and tissues, whereas ST6GAL2 is brain tissue-specific (Varki et al., 2017). Semi-quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) on these enzymes using primer sets specific for detecting each glycosyltransferase isotype revealed that human intestinal epithelial Henle-407 cells express ST3GAL1, 2, 3, and 4, along with detectable, but low ST6GAL1 (Figure 2C left). HEK293T cells express ST3GAL1, 2, 3, 4, and 6 and ST6GAL1 (Figure 2C right). These results are consistent with the MAL-1 and SNA binding to glycans on the surface of Henle-407 and HEK293T (Figure 2B). Small hairpin RNA (shRNA) specific for ST3GAL3 and 4 resulted in a significant decrease in the expression levels of ST3GAL3 and ST3GAL4, as well as the binding of both toxins in Henle-407 and HEK293T cells (Figure 2D–G). Knockdown of α2–6 sialosides in HEK293T cells was unsuccessful, likely due to the low efficiency of the target oligonucleotides that we used in interfering with the expression of ST6GAL1, as well as the complex glycan biosynthesis mechanism that is intertwined with many enzyme isoforms for each step of glycan synthesis in host cells. Taken together, these results indicate that typhoid toxin binds a broader group of multiantennary N-linked α2–3 and α2–6 glycans, while Javiana toxin has a glycan-binding preference toward a narrower group of multiantennary N-linked α2–3 sialosides along with some binding to α2–6 sialosides.
Glycan Binding Preferences of Typhoid and Javiana Toxins Are Correlated with Glycan Expression Profiles of Target Cells at the Primary Infection or Intoxication Sites of Two Salmonella that Produce the Toxins
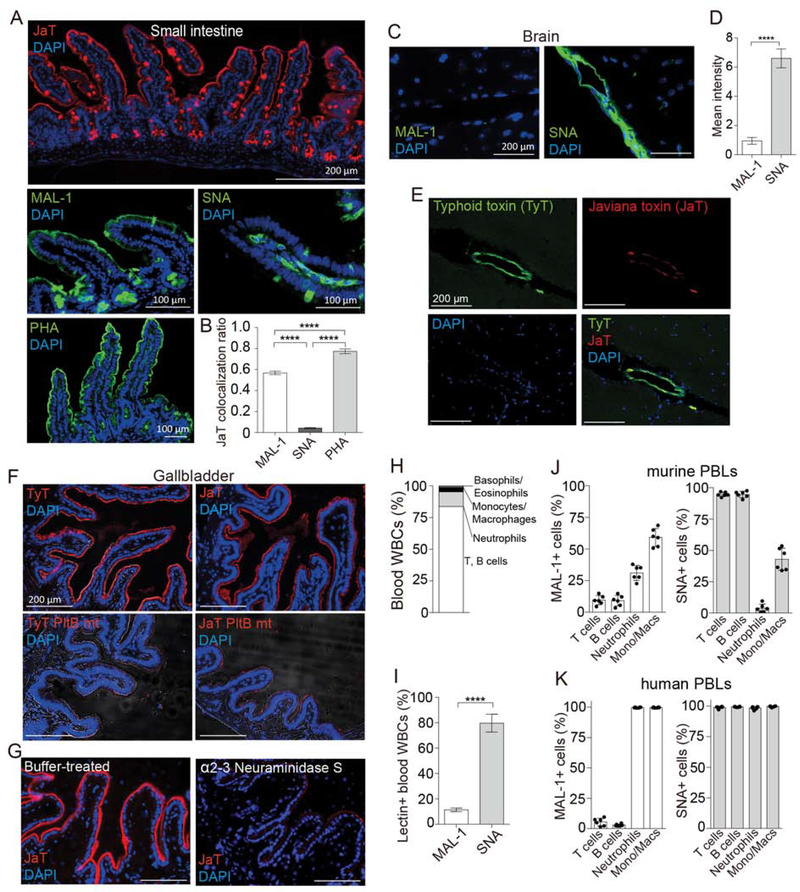
We investigated whether the three amino acid variation N29K, S50G, and T65I of two toxins’ PltB subunits is the evolutionary adaptation to host cells and tissues important for serovar specific infection and intoxication. The primary infection site of NTS is the intestinal epithelium, while typhoidal Salmonellae infect cells at both local and systemic sites and manipulate some of those cells for the pathogen’s benefits. These led us to hypothesize that intestinal and gallbladder epithelial cells predominantly express multiantennary α2–3 sialosides, while immune cells and endothelial cells of arterioles in the brain predominantly express multiantennary α2–6 sialosides. Perfusion-sacrificed and optimal cutting temperature compound (OCT)-embedded frozen intestine, brain, and gallbladder tissue sections were prepared from CMAH null mice for JaT, TyT, MAL-1, SNA, and PHA-L (Figure 3A–G). Primary peripheral blood leukocytes (PBLs) obtained from CMAH null mice and human volunteers were analyzed for their α2–3 and α2–6 sialoside expressions (Figure 3H–K). Similar to PHA-E, PHA-L is a lectin specific for detecting multiantennary N-linked glycans. Lectin-assisted glycan expression profile analysis indicates that small intestinal epithelial cells predominantly express multiantennary N-linked α2–3 sialosides (Figure 3A and B). In contrast, the α2–6 counterparts are the predominant form on endothelial cells of arterioles in the brain (Figure 3C and D), as well as immune cells of both mice and humans (Figure 3H–K). Consistent with data shown in Figure 1G–J and 2A, typhoid toxin bound well to brain endothelial cells while Javiana toxin binding to these cells was weak (Figure 3E). Furthermore, glycan expression analysis using glycan-binding specificities of typhoid and Javiana toxins and α2–3 neuraminidase S, an enzyme specific to remove α2–3 sialosides, indicates that the predominant glycan types displayed on the surface of gallbladder epithelial cells are multiantennary N-linked α2–3 sialosides (Figure 3F–G). Gallbladder colonization of S. Typhi is known to be important for chronic infection, shedding and transmission of S. Typhi. Overall, these results are in support of the hypothesis that the amino acid variation found in typhoid and Javiana toxins is the outcomes of host tissue and cell adaptations evolved in the context of the interaction between the host and Salmonella in the same host. Evolution of host adaptations in Salmonella AB toxins is further supported by the observation that Javiana toxin binds well to bovine and swine intestinal epithelial cells that predominantly express α2–3 sialosides in the context of multiantennary N-linked glycans (Figure S1).
Figure 3. Glycan Binding Preferences of Typhoid and Javiana Toxins Are Correlated with Glycan Expression Profiles of Target Cells at the Primary Infection or Intoxication Sites of Two Salmonella that Produce the Toxins.
A, Shown are representative small intestine tissue sections of CMAH null mice stained with Javiana toxin-AF555, MAL-1-FITC, SNA-FITC, PHA-FITC, and DAPI. B, Colocalization of Javiana toxin with lectins indicated in A in intestinal epithelial cells. C, Glycan expression profiles of the endothelial cells of arterioles in the CMAH null mouse brain. Brain tissue sections of CMAH null mice were stained with MAL-1-FITC, SNA-FITC, and DAPI. D, Quantification results associated with C. E, Representative brain tissue sections of CMAH null mice that were intravenously administered a 1:1 mixture of typhoid toxin-AF488 and Javiana toxin-AF555. DAPI was used to counterstain the tissue sections. F, Shown are representative gallbladder tissue sections of CMAH null mice stained with typhoid toxin-AF555 (top left), Javiana toxin-AF555 (top right), typhoid toxin PltBS35A-AF555 (a glycan-binding defective mutant of typhoid toxin; bottom left), and Javiana toxin PltBS35A-AF555 (a glycan-binding defective mutant of Javiana toxin; bottom right). DAPI was used to counterstain the tissues. G, Gallbladder tissue sections of CMAH null mice treated for 1 hr with either buffer only (left) or α2–3 Neuraminidase S (right) followed by staining with Javiana toxin-AF555 and DAPI. H, Subtypes of circulating white blood cells (WBCs) of CMAH null mice were counted in a hematology analyzer. I, Murine PBLs predominantly express α2–6 form of sialylated multiantennary N-linked glycans. J, α2–3 and α2–6 sialosides on the cell surface of murine PBLs. K, α2–3 and α2–6 sialosides on the cell surface of human PBLs. At least three independent experiments were performed. Bars represent average ± standard deviation. ** p<0.01. **** p<0.0001. n=6–9 per group. Two-tailed unpaired t-tests. Scale bar in A (top panel), C, E, F and G, 200 μm; scale bar in the middle and bottom panels, 100 μm. See also Figure S1.
The Role of PltB Amino Acid Sequence Variations in Glycan Binding Preferences and Virulence Outcomes
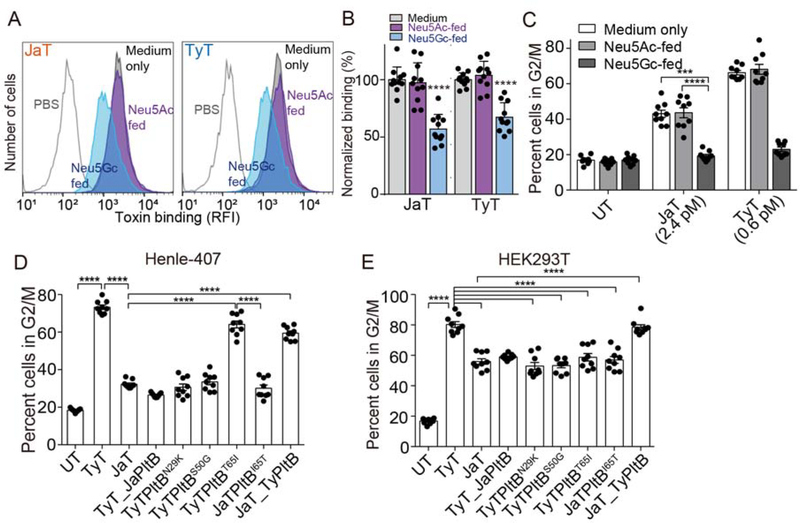
Understanding whether Javiana toxin has also evolved to use glycan receptor moieties terminated in Neu5Ac but not the counterparts terminated in Neu5Gc offers insights into their evolutionary timeline. For instance, typhoid toxin has evolved to the glycan moieties terminated in Neu5Ac (“human-type”) and does not bind to the otherwise same glycan moieties terminated in Neu5Gc (other mammals). In contrast, ArtAB toxin, an AB5 toxin (a probable ancestor of the A2B5 toxin family) secreted by NTS S. Typhimurium DT104 that infects both humans and animals, binds both Neu5Ac- and Neu5Gc-terminated sialosides (Gao et al., 2017). Moreover, subtilase toxin (also an AB5 toxin) from E. coli binds exclusively Neu5Gc-terminated glycans, which therefore is not toxic to humans (Byres et al., 2008). To determine the potential selectivity of Javiana toxin toward specific sialic acid type, Neu5Ac and/or Neu5Gc, we supplemented the media for Henle-407 cells with Neu5Ac or Neu5Gc for metabolic incorporation, resulting in isogenic Henle-407 cells expressing 99% Neu5Ac-terminated-glycans and up to 60% Neu5Gc-terminated-glycans, respectively (Deng et al., 2014). Cells displaying Neu5Ac were susceptible to both typhoid and Javiana toxins, whereas cells predominantly displaying Neu5Gc became resistant to both toxins (Figure 4A–C), supporting the concept that Javiana toxin was likely derived from typhoid toxin after its adaptation to human type sialic acid Neu5Ac. Furthermore, the phylogenetic tree obtained by analyzing S. Typhi pltB and its NTS orthologues also supports this evolutionary timeline (Figure S2–S3 and Table S2–S3). Notably, all S. Typhi identified thus far encode pltB and their sequences for typhoid toxin subunits including pltB are identical, while only some NTS serovars encode typhoid toxin orthologues and their pltB sequences contain some sequence variations (Table S2 and S3).
Figure 4. The Role of PltB Amino Acid Sequence Variations in Glycan Binding Preferences and Virulence Outcomes.
A-B, Toxin binding analysis of Henle-407 cells fed with either Neu5Gc or Neu5Ac. Shown are representative flow histograms (A) and quantification (B). C, Toxicity assays of cells that correspond to A and B. D-E, Cell cycle profile analysis of Henle-407 cells (D) and HEK293T (E) treated with indicated toxin preparations. UT, untreated; TyT, typhoid toxin; JaT, Javiana toxin; TyT_JaPltB, a typhoid toxin mutant containing Javiana toxin PltB subunits; TyTPltBN29K, TyTPltBS50G, and TyTPltBT651; typhoid toxin mutants containing singly substitution to amino acids found on Javiana toxin PltB; JaTPltBI65T, a Javiana toxin mutant containing I65T substitution; JaT_TyPltB, a Javiana toxin mutant containing Typhoid toxin PltB subunits. At least three independent experiments were performed. Bars represent average ± standard deviation. ***, p<0.001, ****, p<0.0001. Two-tailed unpaired t-tests were used for B-E. See also Figure S2–S3 and Table S2–S3.
To understand the role of PltB N29K, S50G, and T65I substitutions in glycan-binding preferences toward α2–3 and/or α2–6 sialosides (linkage between the terminal Neu5Ac and the underlying disaccharide) and virulence outcomes, we generated typhoid toxin mutants containing either singly substitution to amino acids found in Javiana toxin PltB or all three substitutions, followed by the evaluation of those toxin variants in Henle-407 (predominantly α2–3 sialosides) and HEK293T cells (both α2–3 and α2–6 sialosides) (Figure 2B). Javiana toxin mutants containing either I65T substitution or typhoid toxin PltB were also included. Overall, we found that both N29K and S50G substitutions play large roles for the narrower binding specificity of Javiana toxin to α2–3 sialosides because typhoid toxin containing PltBN29K or PltBS50G resulted in a marked decrease of cell cycle arrest in G2/M in both Henle-407 and HEK293T cells (Figure 4D–E). Typhoid toxin containing PltBT651 did not cause a decrease of G2/M cell cycle arrest in Henle-407 cells but caused a significant decrease of G2/M cell cycle arrest in HEK293T cells, indicating that T65I plays a large role in inhibiting the binding of Javiana toxin to α2–6 sialosides (Figure 4D–E).
Structure and Function Analysis of S. Typhi PltB Subunits Bound to α2–3 and α2–6 Sialosides
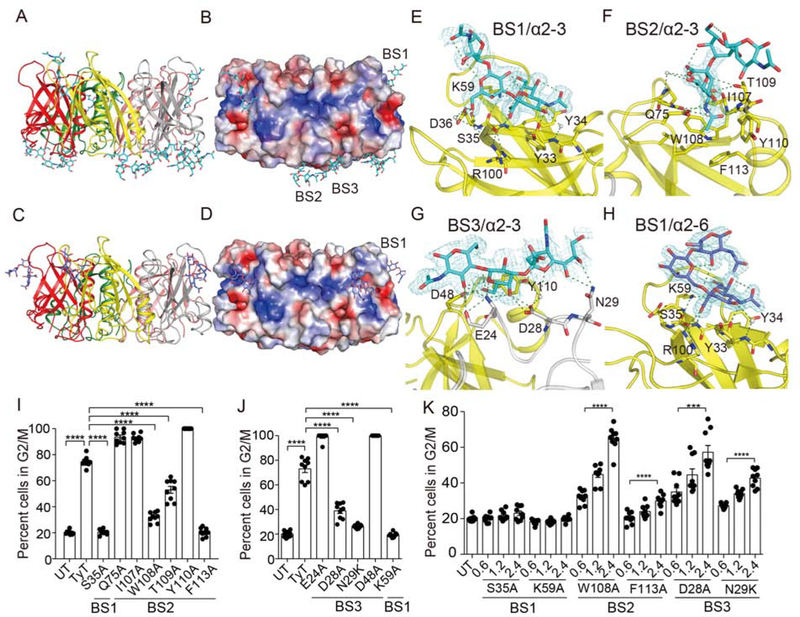
To reveal the atomic interface between PltB and α2–3/α2–6 sialosides, we solved X-ray crystal structures of typhoid toxin PltB homopentamers bound to Neu5Ac-α2–3-Gal-β1–4-GlcNAc or Neu5Ac-α2–6-Gal-β1–4-GlcNAc, respectively (Figure 5A–H and S4, and Table S4 and S5). We reveal three glycan-binding sites on PltB, one lateral binding site and two binding sites in the bottom of PltB, which we named BS1–3, totaling 15 binding sites per PltB pentamer in one combined toxin (Figure 5A–H). N29 is on the BS3, S50 is near the BS2 and BS3, and T65 is close to the BS1 (Figure 5B, 5D, 6A–C, and S4–S6). We found that Neu5Ac-α2–3-Gal-β1–4-GlcNAc (α2–3 sialosides) can use all three binding sites BS1–3, whereas, like GD2 glycolipid glycan (Deng et al., 2014), Neu5Ac-α2–6-Gal-β1–4-GlcNAc (α2–6 sialosides) use only BS1 that is laterally located where S35 and K59 are key residues (Figure 5A–H). W108, T109, and F113 on the BS2 play a critical role in causing Henle-407 cell cycle arrest in G2/M, while D28 and N29 in the BS3 are important in intoxicating Henle-407 cells (predominantly α2–3 sialosides), indicating the active roles of the BS2 and BS3 in typhoid toxin binding to α2–3 sialosides (Figure 5I–J). To understand whether certain binding sites are preferred sites for toxin binding to α2–3 sialosides, Henle-407 cells were treated with various doses of typhoid toxin containing S35A and K59A for BS1, W108A and F113A for BS2, or D28A and N29K for BS3 and their intoxication profiles were analyzed, revealing that the BS1 has greater accessibility to α2–3 sialosides than the BS2 and 3 (Figure 5K).
Figure 5. Structure and Function Analysis of S. Typhi PltB Subunits Bound to α2–3 and α2–6 Sialosides.
A, Crystal structure of typhoid toxin PltB pentamer in complex with Neu5Ac-α2–3-Gal-β1–4-GlcNAc (Cyan sticks, sugar carbon atoms; blue sticks, nitrogen atoms; red sticks, oxygen atoms) is shown as a ribbon cartoon with each protomer depicted in a different color. B, Surface charge distribution of the PltB pentamer structure and Neu5Ac-α2–3-Gal-β1–4-GlcNAc. BS1–3, binding sites 1–3. C, Ribbon diagram of crystal structure of typhoid toxin PltB pentamer in complex with Neu5Ac-α2–6-Gal-β1–4-GlcNAc (Purple sticks, sugar carbon atoms; blue sticks, nitrogen atoms; red sticks, oxygen atoms). D, Surface charge distribution of the PltB pentamer structure and Neu5Ac-α2–6-Gal-β1–4-GlcNAc. BS1, binding site 1. E-H, Close-up views of the binding sites with glycans indicated and their electron density maps. I, Structure and function analysis of the BS1 and BS2 of the PltB. S35 is on the BS1, while Q75, I107, W108, T109, Y110, and F113 are on the BS2. Cell cycle profile analysis of Henle-407 cells treated with indicated toxin preparations. J, Structure and function analysis of the BS1 (K59) and BS3 (E24, D28, N29, D48). K, Intoxication profiles of Henle-407 cells that were left untreated or treated for 66 hrs with indicated doses of typhoid toxin mutants (0–2.4 pM). At least three independent experiments were performed. Bars represent average ± standard deviation. *** p<0.001. **** p<0.0001. n=9 per group. Two-tailed unpaired t-tests. See also Figure S4 and Table S4–S5.
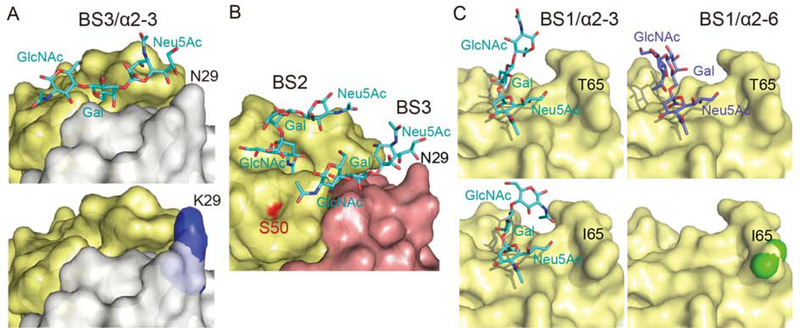
Figure 6. Structure Comparison Analysis of WT PltB, PltBN29K, and PltBT65I Bound to Specific Glycan Receptor Moieties.
A, Close-up views of the BS3 of PltB (top panel) and PltBN29K (bottom panel) soaked in a buffer containing Neu5Ac-α2–3-Gal-β1–4-GlcNAc. B, Close-up views of the binding sites BS2 and BS3 of WT PltB with Neu5Ac-α2–3-Gal-β1–4-GlcNAc. The location of the residue S50 is highlighted in red. We could not obtain crystal of PltBS50G. C, Close-up views of the BS1 of PltB (top right) and PltBT651 (bottom right) soaked in a buffer containing Neu5Ac-α2–6-Gal-β1–4-GlcNAc, as well as the BS1 of PltB (top left) and PltBT651 (bottom left) soaked in a buffer containing Neu5Ac-α2–3-Gal-β1–4-GlcNAc. See also Figure S5–S6 and Table S4–S5.
To determine the role of each substitution found in Javiana toxin PltB, we also solved crystal structures of PltBN29K native and PltBN29K bound Neu5Ac-α2–3-Gal-β1–4-GlcNAc or Neu5Ac-α2–6-Gal-β1–4-GlcNAc, respectively, as well as PltBT651 native and PltBT651 soaked for various times in a crystallization condition buffer containing Neu5Ac-α2–3-Gal-β1–4-GlcNAc or Neu5Ac-α2–6-Gal-β1–4-GlcNAc, respectively (Figure 6A and C, Figure S5–S6, and Table S4–S5). Crystal structure of PltBN29K soaked in a crystallization condition buffer containing Neu5Ac-α2–3-Gal-β1–4-GlcNAc reveals that this glycan is bound on the BS1 and BS2 of PltBN29K, but not on BS3 (Figure 6A and Figure S5). The BS3 of WT PltB containing N29 can accommodate this glycan but the K29 counterpart cannot because K29 hinders the binding of terminal Neu5Ac to this binding site (Figure 6A). These results are consistent with the narrower specificity of Javiana toxin to α2–3 sialosides than that of typhoid toxin (Figure 2A). Results shown in Figure 4D and Figure 6B indicate that PltBS50G contributes to the narrower binding ability of Javiana toxin to α2–3 sialosides, but we could not obtain crystal of PltBS50G.
PltBN29K shows the comparable binding to Neu5Ac-α2–6-Gal-β1–4-GlcNAc glycans, which occurs through the BS1 (Figure S5 right panels), further supporting our observation that α2–6 sialosides use only the lateral binding site BS1 of PltB. This, along with results shown in Figure 4E, led us to hypothesize that T65I plays a crucial role in the diminished glycan-binding preference of Javiana toxin to α2–6 sialosides. Co-crystal structures of PltBT651 with glycans are consistent with the findings shown in Figure 2A and 4E that T65I variation plays a large role in the markedly decreased binding ability of Javiana toxin to α2–6 sialosides. PltBT651 can bind to Neu5Ac-α2–3-Gal-β1–4-GlcNAc, but, unlike WT or PltBN29K that can bind to α2–6 sialosides (2 hr soaking), PltBT651 did not bind to Neu5Ac-α2–6-Gal-β1–4-GlcNAc glycan even after 20 hr soaking (Figure 6C, Figure S6, and Table S4). Overall, these structure and function studies reveal that typhoid toxin can use all three binding sites for its binding to α2–3 sialosides, whereas it uses only the lateral binding site BS1 for toxin binding to α2–6 sialosides. In contrast, its NTS orthologue Javiana toxin shows a narrower specificity to α2–3 sialosides and markedly decreased binding ability to α2–6 sialosides, which are primarily due to its amino acid variation of N29K and S50G for the former and T65I variation for the latter.
Immunization with Javiana Toxin or S. Javiana Offers Cross-Reactive Protection against Lethal-Dose Typhoid Toxin Challenge
Lastly, we asked whether Javiana toxin can offer cross-reactive protection against lethal-dose typhoid toxin challenge because two toxins are similar. Groups of CMAH null mice were administered with either PBS alone or Javiana toxin and evaluated their anti-toxin IgG titers at 2, 3, and 4 weeks after toxin administration (Figure 7A). After 4 weeks, when anti-toxin antibody titers were elevated at a significant level, these mice were challenged with LD100 of typhoid toxin. We found that exposure to Javiana toxin elicited a humoral immune response that offered cross-reactive protection against lethal-dose typhoid toxin challenge. Mice pre-exposed to Javiana toxin survived, maintained body weight, and showed normal beam-walking and less severe leukopenia (Figure 7B–E). Similarly, mice infected with either WT S. Javiana or S. Javiana ΔJaT + JaT offered cross-reactive protection against lethal-dose typhoid toxin challenge, whereas mice infected with S. Javiana ΔJaT died (Figure 7F). Overall, these results support the possibility of cross-reactive protection (or natural immunity) against typhoid toxin in NTS endemic regions if some NTS serovars are a producer of a typhoid toxin orthologue.
Figure 7. Immunization with Javiana Toxin or S. Javiana Offers Cross-Reactive Protection against Lethal- Dose Typhoid Toxin Challenge.
A, Anti-toxin IgG titers in peripheral blood of CMAH null mice intravenously administered Javiana toxin preparations. B, Survival of mice pre-exposed to Javiana toxin followed by lethal-dose typhoid toxin challenge. C, Body weight changes of mice after the typhoid toxin challenge. D, Measurements of time required for crossing the beam for animals treated as indicated. E, Circulating neutrophil numbers in peripheral blood of mice treated as indicated. F, Survival of animals infected with indicated S. Javiana strains followed by lethal-dose typhoid toxin challenge. Three independent experiments were performed. Bars represent average ± standard deviation. * p<0.05, ** p<0.01, **** p<0.0001. n=5–8 per group. Two-tailed unpaired t-tests were used for A, C-E, while low-rank test was used for B and F.
DISCUSSION
Typhoid toxin is unique among the bacterial AB toxin family members in that it has two enzymatic subunits in a single toxin complex, reflecting its evolution from two separate groups of bacterial toxins and its adaptation in the context of the host and S. Typhi interaction. Consistently, typhoid toxin is proposed to exacerbate typhoid disease progression and chronic infection (Song et al., 2013; Song et al., 2010; Yang et al., 2018b). When administered to laboratory animals, typhoid toxin recapitulates many of the characteristic signs and symptoms of typhoid fever, in particular leukopenia and neurologic complications (Song et al., 2013; Yang et al., 2018b). In humanized mice engrafted with human fetal liver stem and progenitor cells that partially supported S. Typhi survival and replication, typhoid toxin contributed significantly to the establishment of persistent infection by its tropism to immune cells (Song et al., 2010; Yang et al., 2018b). Several clinical reports have shown the abundant presence of anti-CdtB antibodies in the sera of recovered typhoid patients, indicating that S. Typhi produces typhoid toxin during infection in humans (Charles et al., 2014; Liang et al., 2013; Sharma et al., 2018; Tran Vu Thieu et al., 2017). Primary human cells and tissues relevant to typhoid signs express the specific glycan receptor moieties for typhoid toxin, implying that typhoid toxin can target these cells during human infection (Yang et al., 2018b). Consistently, considerable numbers of cases showed leukopenia and neurologic signs resembling the phenotype observed in the typhoid toxin intoxicated mice (Ali et al., 1997; Azmatullah et al., 2015; Pohan, 2004; Qamar et al., 2018; Sejvar et al., 2012).
Unlike S. Typhi that causes the life-threatening disease typhoid fever, NTS serovars such as S. Typhimurium, S. Enteritidis, and S. Javiana cause local infections in the gastrointestinal (GI) tract and subsequently self-limiting gastroenteritis. Unlike many other NTS serovars, S. Javiana encodes a typhoid toxin orthologue, Javiana toxin, in its chromosome. Our results support the hypothesis that Javiana toxin has evolved to adapt to epithelial cells of the GI tract which correspond to the primary infection site for NTS serovars, while typhoid toxin has evolved to adapt to various cell types including epithelial, endothelial, and immune cells that correlate to the dynamic infectious cycle of S. Typhi that involves both local and systemic infections. Likewise, the subsequent virulence outcomes were consistent with host adaptations of these two toxins to specific cells relevant to the infectious niches of two Salmonella serovars producing these A2B5 toxins. In support, unlike typhoid toxin, Javiana toxin was not toxic to CMAH null mice when administered systemically while a chimera Javiana toxin mutant containing Typhi PltB became toxic to mice, like typhoid toxin, highlighting the evolution of host adaptations in Salmonella A2B5 toxins and their tissue and cell tropism.
To gain insight into the molecular mechanism by which the three amino acid variations in PltBs of these two toxins resulted in different glycan-binding preferences, we solved X-ray co-crystal structures of Typhi PltB and its mutants resembling each amino acid variation found in Javiana PltB bound to either α2–3 sialosides or α2–6 sialosides. It is important to note that we overcame technical difficulties associated with solving co-crystal structures of PltB bound to different groups of sialosides, revealing the three binding sites on Typhi PltB and the flexible use of these binding sites in accommodating different groups of glycan receptor moieties. These results are consistent with a broader tissue and cell specificity of typhoid toxin and correlate to the dynamic infectious cycle of S. Typhi that involves many cell types during its local and systemic infections.
Our results obtained from multi-pronged approaches consisting of glycan array, cell intoxication assays, and co-crystal structure analysis followed by mutagenesis studies indicate that N29K and S50G variations are responsible for the narrower binding specificity of Javiana toxin to α2–3 sialosides, while T65I variation plays a large role for the diminished binding of Javiana toxin to α2–6 sialosides, as compared to Typhi PltB. This information not only explains how PltBs of the two toxins display markedly different glycan-binding preferences but also provides important insights into the virulence outcome prediction of emerging bacteria that may contain a typhoid toxin orthologue.
In this structure and function study, we chose S. Javiana toxin because this NTS serovar is among the common NTS causing food poisoning in developed countries and encodes a typhoid toxin orthologue. However, PltB amino acid variations that do not find in Javiana toxin are present in other NTS orthologues encoded by less commonly found Salmonella serovars (Figure S3). Similar structure and function studies on those amino acid variations would inform us about their tissue and host tropisms and their role in disease progression in humans and other animals. These investigations would also inform us regarding whether other NTS orthologues of typhoid toxin may offer cross-reactive protection for typhoid-related illness in some cases. We found that, in in vivo intoxication mouse model, Javiana toxin produced by S. Javiana can offer cross-reactive protection against lethal-dose typhoid toxin challenge.
In short, structure and function analysis of Salmonella Typhoid toxin and its NTS orthologue offers insights into the evolution of host adaptations in Salmonella A2B5 toxins that have evolved and that are continuously evolving in the context of the interaction between the host and Salmonella producing these toxins. We demonstrated that small amino acid variations in PltB subunits are sufficient to change their glycan-binding preferences to match with glycans predominantly expressed on target host cells. Information obtained from this study may also offer insights into the design for improved typhoid fever vaccines and therapeutics.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeongmin Song (ieongmin.song@cornell.edu).
Materials Availability
Plasmids and bacterial strains generated for this study will be made available to other researchers via the Institutional Material Transfer Agreements.
Data and Code Availability
The published article includes all datasets generated during this study. Coordinates for the atomic structures have been deposited in the RCSB Protein Data Bank under PDB IDs of 6P4M, 6P4N, 6P4P, 6P4Q, 6P4R, 6P4S and 6P4T.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Bacterial Strains, Plasmids, and Other Materials
WT Salmonella enterica serovar Javiana has been described previously (den Bakker et al., 2011).
Plasmids used in this study that were constructed using the Gibson assembly cloning strategy (Gibson et al., 2009) are listed in Table S6. S. Javiana mutant strains were generated as previously described (Song et al., 2013), which is also listed in Table S6. For infection experiments, S. Javiana strains were grown at 37 °C in 2 ml LB broth containing 0.3 M NaCl to an OD600nm of ~0.9 after inoculation from an overnight culture at a dilution of 1:133. Fluorescein-conjugated lectins, MAL-1, SNA, and PHA, were purchased from Vector Laboratories.
Mammalian Cell Culture Conditions
Human intestinal epithelial Henle-407 cells and human embryonic kidney HEK293T cells were cultured in DMEM high glucose (Invitrogen) supplemented with 10% FBS (Hyclone cat# SH30396.03, Lot# AD14962284). Sialic acid contents of the FBS used were validated, which was ~99% Neu5Ac and less than 1% Neu5Gc. Cells were kept at 37°C in a cell culture incubator with 5% CO2. Mycoplasma testing was conducted regularly as part of the cell maintenance practice. Metabolic incorporation of Neu5Ac or Neu5Gc was carried out as previously described (Deng et al., 2014). Briefly, cells were cultured in a standard medium supplemented with 10 mM Neu5Ac or 10 mM Neu5Gc (Inalco) as follows. Henle-407 (1.5 × 104) cells were seeded onto 12-well culture plates in 1 ml media with or without 10 mM of indicated sialic acids. A stock solution of 50 mM sialic acids was freshly prepared in a DMEM medium whose final pH was adjusted to neutral with NaOH. During the feeding period, the cells were continuously monitored and maintained below 80% confluence. After 3 days of growth, cells were split into 12-well plates at a cell density of 6 × 104 per well for Henle-407 cells, followed by overnight incubation. Then the cells were used for different assays as indicated in the figure legend.
Mouse experiment
All animal experiments were conducted according to protocols approved by Cornell University’s Institutional Animal Care and Use Committee. Age- and sex-matched 5- to 8-week-old CMAH null mice were randomly allocated to each group. The mice used in this study were originally purchased from the Jackson Laboratory and bred in a vivarium in the animal facility at Cornell University. All the knockout mice used were genotyped before and after the experiments.
Isolation of Human Peripheral Blood Leukocytes
The inclusion of six human volunteers (3 male and 3 female healthy adult volunteers) was conducted according to protocols approved by Cornell University’s Institutional Review Board for Human Participants. Peripheral blood samples were collected by submandibular bleeding in Microtainer tubes coated with EDTA as an anticoagulant (BD Biosciences). PBLs were prepared by lysing erythrocytes through incubation of blood in ammonium-chloride-potassium (ACK) lysis buffer (Lonza).
METHODS DETAILS
Mammalian Cell Intoxication Assay
Host cell cycle profile quantification via flow cytometry and cell distension morphology analysis via microscopy were conducted as previously described (Deng et al., 2014; Song et al., 2013; Yang et al., 2018b). Briefly, after the treatment of cells with indicated toxins and bacteria, cells were trypsinized, harvested, washed, and fixed for 2 hours at −20 °C in a buffer containing 70% ethanol in PBS. Fixed cells were washed with PBS for 2 times and resuspended in 500 μl of PBS containing 50 μg ml−1 propidium iodide, 100 μg ml−1 RNase A, and 0.05% Triton X-100. After incubation for 40 min at 37 °C, cells were washed with PBS, resuspended in 200 μl PBS, filtered, and analyzed via flow cytometry. DNA contents of cells were determined using FlowJo software (Treestar).
Bacterial Infections and Colony-Forming Unit (C.F.U.) Assay
Human intestinal epithelial Henle-407 cells were infected with WT or mutant S. Javiana strains indicated (5 m.o.i) for 1 hr, followed by PBS-washing and a 30-minute incubation of the cells with 100 μg ml−1 gentamicin to remove all the extracellular bacteria. Then the cells were washed with PBS and lysed by incubating them in 1 ml of sterile PBS containing 0.1% sodium deoxycholate for 15 min at room temperature. The cells for the 24 hr or 48 hr infection were maintained in complete cell culture media containing 10 μg ml−1 gentamicin for an indicated duration by changing old media with new media containing 10 μg ml−1 gentamicin daily. Bacterial numbers were determined by plating 10-fold serial dilutions of homogenates on LB agar plates and incubated overnight at 37 °C. Colonies were counted and the number of total c.f.u. recovered was calculated. Randomly selected colonies from each independent experiment were validated by conducting a PCR-based genotyping assay using primer sets amplifying S. Javiana-specific genomic regions.
Recombinant Toxin Overexpression and Purification
Overexpression and purification of typhoid and Javiana toxins and their mutants were carried out as previously described (Song et al., 2013; Yang et al., 2018b). Briefly, Escherichia coli (E. coli) BL21(DE3)pLysS strains carrying the different plasmids were grown to an OD60o of 0.6–0.7 at 37 °C; expression of the toxin was induced by the addition of 0.5 mM IPTG, followed by overnight incubation at 29 °C. Bacterial cells were pelleted by centrifugation, resuspended in a buffer containing 15 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mg ml−1 lysozyme, 10 pg ml−1 DNase I and 1× protease inhibitor cocktail (Roche), lysed via sonication, pelleted, and affinity-purified using Ni-NTA resin (Qiagen). The eluates were diluted in a buffer containing 20 mM MES, pH 6.0 and loaded onto a Hi-trap SP ion-exchange column (GE Healthcare). Fractions from the ion-exchange chromatography were monitored by SDS-PAGE, concentrated, and further purified using a Superdex 200 column (GE Healthcare). Final fractions were examined for purity on 15% SDS-PAGE. In the case of N29K and T65I PltB homopentamers, the eluates were diluted in a buffer containing 15 mM Tris-HCl, pH 8.0 and loaded onto a Hi-trap Q column (GE Healthcare), followed by size-exclusion chromatography using a Superdex 200 column. Purified toxin preparations were further subjected to high-capacity endotoxin removal resin (Pierce, Thermo Fisher Scientific). LPS levels of all the toxin preparations were determined using a ToxinSensor chromogenic LAL endotoxin assay kit (GenScipt; <0.2 EU ml−1).
Alexa Fluor-555 Labeling of Javiana and Typhoid Toxins
Indicated toxin preparations were fluorescently labeled with Alexa Fluor-555 dye (Molecular Probes, Thermo Fisher Scientific) according to the vendor’s recommendation. Alexa Fluor-555 dye has a succinimidyl ester moiety that reacts with primary amines of proteins to form stable dye-protein conjugates. Purified toxin preparations (0.5 mg ml−1) were incubated with a reactive dye for 1 h at room temperature and applied to a size-exclusion chromatography column provided by the vendor to separate the dye-protein conjugates from the free dye. The degree of labeling was determined by measuring the absorbance of the conjugate solution at 280 nm and 555 nm, which yielded the toxin labeling degree with 4~6 dye/holotoxin ratios. The holotoxin’s predicted extinction coefficient is 191,400 M−1 cm−1.
Mouse Experiments
Groups of mice were injected with 100 pi solutions containing PBS buffer alone, or 2 pg of the indicated purified toxin preparations (endotoxin-free) via retro-orbital injections. Changes in the behavior, weight, and survival of the toxin-injected mice were closely monitored. Balance beam test was performed in a double-blind manner, as previously described (Yang et al., 2018b). When indicated, CMAH null mice were fasted for 4 hrs, orally injected with 100 μl of bicarbonate buffer to neutralize stomach acid, and then infected with 109 c.f.u. of S. Javiana, S. Javiana AJaviana toxin (JT) or S. Javiana AJT + JT (a chromosomally complemented strain to maintain the same copy number of Javiana toxin as that expressed by WT S. Javiana), using a gavage needle.
Peripheral Blood Leukocyte Counts
Blood samples were collected by submandibular bleeding in Microtainer tubes coated with EDTA as an anticoagulant (BD Biosciences), kept at room temperature, and analyzed within 2 hrs after blood collection using a Hemavet 950FS hematology analyzer (Drew Scientific).
Frozen Tissue Preparation
The mice were anesthetized with isoflurane, followed by sacrifice-perfusion, which was conducted by sequentially administering 10% sucrose and 4% paraformaldehyde using a pressure-controlled Perfusion One system (Leica Biosystems) at indicated time points. After perfusion, the mouse brain, spleen, and gallbladder were extracted and fixed in 4% paraformaldehyde for 24 h at 4 °C. Tissues were washed with PBS, immersed in a solution containing 30% sucrose, and incubated overnight for cryoprotection. The brain tissue was trimmed into coronal sections and placed in cassettes for Tissue-Tek OCT embedding. The embedded tissues were flash-frozen in isopentane cooled to −80 °C. Cryosections of frozen tissue samples were cut to be −8 μm thick and stored in −80 °C until staining.
Immunofluorescent Staining
The frozen tissue sections or cell lines were fixed with 4% paraformaldehyde (PFA) for 10 min, washed with PBS and blocked in 3% BSA/PBS for 30 min. The slides were incubated with Fluorescein-labeled lectins (Mal-1, SNA, PHA) or Alexa Fluor-555-labeled toxins for 1 h at room temperature or overnight at 4 °C. Then sections were washed with PBS and the nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI). The slides were mounted in ProLong antifade mounting solution (Molecular Probes, Thermo Fisher Scientific). Digital photomicrographs were taken using a Leica DMI6000B/DFC340 FX fluorescence microscope system. When indicated, the gallbladder tissue sections were treated with α2–3 Neuraminidase S (New England BioLabs, P0743) by carrying out the following procedures: fixation with 1% PFA/PBS for 15 min, washing with PBS, and incubation at 37°C for 1 hr with 30 μl of a mixture containing 50 mM sodium acetate, pH5.5, 5 mM CaCl2, and 24 units of α2–3 Neuraminidase S (per tissue section). The tissues treated with either the enzyme or buffer only were washed with PBS and blocked for 30 min in a PBS buffer containing 3% BSA, followed by incubation at 4 °C for overnight in a PBS/3%BSA containing Javiana toxin conjugated to Alexa Fluor 555. The tissues were then washed, counterstained with DAPI, mounted using ProLong antifade mounting solution (Molecular Probes, ThermoFisher Scientific), and viewed. In the case of other animal tissue experiments, the 5 μm tissue sections of formalin-fixed, paraffin-embedded bovine ileum (BP-309) and swine (PP-309) were obtained from Zyagen, followed by standard deparaffinization and rehydration steps with xylene and a descending concentration gradient of ethanol solutions (100, 95, 75 and 60%; 5 min for each solution), respectively, as well as an antigen retrieval step by boiling in citrate buffer, pH 6.0 for 20 min. The tissue sections were blocked with PBS/ 5% BSA, stained with indicated lectins for overnight at 4 °C, counterstained with DAPI, mounted, and viewed.
Image Acquisition, Equipment, and Settings
Immunofluorescent images were acquired with a Leica DMI6000B/DFC340 FX fluorescence microscope system. For imaging of Javiana and typhoid toxins and other lectin bindings on tissue sections, 1,600 * 1,200-pixel full-frame pictures of various channels were recorded as 16-bit TIFF files with *20 (numerical aperture (NA) 0.5) or *40 (NA 0.75) objectives. The filter wavelengths were as follows: Alexa Fluor-488 (plant lectins) – excitation filter 470/40 nm, emission filter 525/50 nm; Alexa Fluor-555 (Javiana and typhoid toxins) – excitation filter 545/30 nm, emission filter 610/75 nm; DAPI – excitation filter 340~380 nm, emission filter 425 nm. The fluorescent signal intensity of images was quantified using the measure function of ImageJ (National Institutes of Health, USA) after subtracting the background. The frequency of Mal-1, SNA, or PHA colocalization with Javiana or typhoid toxins was objectively determined using ImageJ with Manders’ coefficients within the Just Another Colocalization Plugin. Recorded images were merged using the ImageJ merge channels function and processed further with Adobe Photoshop to adjust the brightness and contrast equally for all recordings.
Glycan Expression Profiling of Murine and Human Peripheral Blood Leukocytes (PBLs)
PBLs were stained using a premix antibody cocktail containing various human and mouse antibodies (Key Resources Table), along with either Fluorescein-MAL-1 (Vector Labs) or Fluorescein-SNA (Vector Labs). The stained cells were read using a BD FACS Canto II flow cytometry system (BD Biosciences) and analyzed using FlowJo software. Murine PBLs were gated based on their cell surface staining profiles, CD3+, CD19+, CD11b+Ly6G+ (mouse) or CD15+CD16+ (human), or CD11b+CD11c+ (mouse) or CD14+ (human) and the surface expressions of α2–3 and α2–6 sialosides on T cells, B cells, neutrophils, and monocytes were quantified by analyzing their co-staining profiles with MAL-1 or SNA.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Flag, Clone M2 | Sigma | Sigma-Aldrich Cat# F1804, RRID:AB_262044 |
| Anti-DnaK, Clone 8E2/2 | Abcam | Abcam Cat# ab69617, RRID:AB_1209209 |
| Goat anti-moue IgG DyLight800 | Thermo Fisher | Thermo Fisher Scientific Cat# SA5-10176, RRID:AB_2556756 |
| Anti-mouse CD45, 30-F11 | Biolegend | BioLegend Cat# 103102, RRID:AB_312967 |
| Mouse anti-human CD3 conjugated to PE-Cy7, Clone SK7 | BD Biosciences | BD Biosciences Cat# 557851, RRID:AB_396896 |
| Mouse anti-human CD19 conjugated to PE, Clone HIB19 | BD Biosciences | BD Biosciences Cat# 561741, RRID:AB_10893795 |
| Mouse anti-human CD14 conjugated to PerCP-Cy5.5, Cl one M5E2 | BD Biosciences | BD Biosciences Cat# 561116, RRID:AB_2033939 |
| Mouse anti-human CD15 conjugated to APC, Clone HI98 | BD Biosciences | BD Biosciences Cat# 551376, RRID:AB_398501 |
| Mouse anti-human CD16 conjugated to APC-Cy7, Clone 3G8 | BD Biosciences | BD Biosciences Cat# 557758, RRID:AB_396864 |
| Hamster anti-mouse CD3 conjugated to PE-Cy7, Clone 145-2C11 | BD Biosciences | BD Biosciences Cat# 552774, RRID:AB_394460 |
| Anti-mouse CD19 conjugated to APC-Cy7, Clone MB19-1 | eBioscience | Thermo Fisher Scientific Cat# A15391, RRID:AB_2534405 |
| Anti-mouse CD11c conjugated to PerCP-Cy5.5, Clone N418 | eBioscience | Thermo Fisher Scientific Cat# 45-0114-82, RRID:AB_925727 |
| Anti-mouse CD11b conjugated to PE, Clone M1/70 | eBioscience | Thermo Fisher Scientific Cat# 12-0112-82, RRID:AB_2734869 |
| Anti-mouse Ly6G conjugated to APC, Clone 1A8 | eBioscience | Thermo Fisher Scientific Cat# 17-9668-82, RRID:AB_2573307 |
| Bacterial and Virus Strains | ||
| See Table S6 for full list of Salmonella Strains | See Table S6 | See Table S6 |
| Biological Samples | ||
| N/A | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Salmonella A2B5 toxins (a.k.a., Typhoid Toxin, Javiana Toxin) and their pentameric B subunits | This manuscript | N/A |
| Critical Commercial Assays | ||
| Alexa Fluor 555 Protein Labeling Kit | ThermoFisher | A20174 |
| Lipofectamine 3000 | ThermoFisher | L3000008 |
| RNeasy mini kit | Qiagen | 74104 |
| iScript cDNA synthesis kit | Bio-Rad Laboratories | 1708891 |
| Green Tag Polymerase | Genscript | E00043 |
| JCSG | Qiagen | N/A |
| PEGRx | Hampton Research | N/A |
| Deposited Data | ||
| PltB Crystal Structures with Sialosides | RCSB Protein Data Bank | PDB IDs of 6P4M, 6P4N, 6P4P, 6P4Q, 6P4R, 6P4S, 6P4T |
| Glycanarray Results | This manuscript | See Table S1 |
| Experimental Models: Cell Lines | ||
| Henle-407 Cells | Originally obtained from ATCC | Validated using intestinal epithelial markers |
| HEK293T Cells | Originally obtained from ATCC | ATCC Cat# CRL-3216, RRID:CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| B6.129X1-Cmah tm1Avrk/J Mice (a.k.a. Cmah KO) | Jackson Lab | IMSR Cat# JAX:017588, RRID:IMSR_JAX:017588 |
| Oligonucleotides | ||
| See Table S7 for full list | IDT | See Table S7 |
| Recombinant DNA | ||
| See Table S6 for full list | See Table S6 | See Table S6 |
| Software and Algorithms | ||
| GraphPad Prism 6 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ RRID:SCR_002798 |
| Excel | Microsoft | Office 365 RRID:SCR_016137 |
| FlowJo V10 | TreeStar | https://www.flowjo.com/ RRID:SCR_008520 |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ RRID:SCR_003070 |
| Unicorn 6.3 | GE Healthcare Life Sciences | https://www.gelifesciences.com/en/us/shop/unicorn-6-3-p-01118 RRID:SCR_000004 |
| PyMol | Schrodinger | https://pymol.org/2/ RRID:SCR_000305 |
| PHENIX | (Adams et al., 2010) | https://www.phenix-online.org/ RRID:SCR_014224 |
| Coot | (Emsley et al., 2010) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ RRID:SCR_014222 |
| Other | ||
| Fluorescein Maackia aumurensis lectin (Mal-I) | Vector laboratories | Vector Laboratories Cat# FL-1311, RRID:AB_2336567 |
| Fluorescein Sambucus nigra bark lectin (SNA) | Vector laboratories | Vector Laboratories Cat# FL-1301, RRID:AB_2336719 |
| Fluorescein Phaseolus vulgaris erythroagglutinin (PHA-E) | Vector laboratories | Vector Laboratories Cat# FL-1121, RRID:AB_2336651 |
| Fluorescein Phaseolus vulgaris leucoagglutinin (PHA-L) | Vector laboratories | Vector Laboratories Cat# FL-1111, RRID:AB_2336655 |
Glycan Microarray
Glycan microarray analysis was performed in the laboratory of J.C. Paulson. Alexa Fluor-555-Javiana and typhoid toxin stocks were diluted in 1xPBS-Tween 20 (PBS-T) (50 pg ml−1 final) and incubated on the array surface for 1 h at room temperature. Following sequential washes in PBS-T, PBS, and sterile dH2O, the bound toxin was detected using an Innoscanl 100AL (Innopsys) fluorescent microarray scanner. Fluorescent signal intensity was measured using Mapix (Innopsys), and the mean intensity minus mean background of four replicate spots was calculated. A complete list of the glycans on the array is presented in Table S1.
Measurement of Anti-Toxin Antibody Responses Cross-Reacting to Typhoid Toxin
Antibody levels in the sera of immunized mice were examined using a direct enzyme-linked immunosorbent assay (ELISA). ELISA plates (Maxisorp, Nunc) were coated with 50 ng purified typhoid toxoid in 100 μl plate-coating buffer (50 mM carbonate-bicarbonate buffer, pH 9.6), and incubated overnight at 4 °C. Wells were washed with PBS containing 0.05% Tween 20 and blocked with PBS containing 1% BSA for 1 h at 37 °C. A twofold serial dilution of the sera contained in 50 μl PBS/0.05% Tween 20/0.5% BSA was added to each well and incubated for 2 h at 37 °C. After washing, bound antibodies were detected with horseradish peroxidase (HRP)-conjugated anti-mouse immunoglobulin IgG (Southern Biotech) at a 1:10,000 dilution in PBS/0.05% Tween/0.5% BSA. Wells were then incubated with an HRP substrate, tetramethylbenzidine (Sigma), for 10–30 min and the reaction was stopped by the addition of 100 μl of 1 M H3PO4. Relative serum antibody levels were determined by standard endpoint ELISA titration.
Typhoid Toxin Binding Assay
Cells were collected by trypsinization, washed with PBS, resuspended in 100 μl PBS containing 4 ng ml−1 Alexa Fluor-555-labelled purified toxin preparations. Cells were incubated in the presence of the labeled toxin preparations for 15 min at 4 □, washed with PBS, fixed with 1% paraformaldehyde, and analyzed by flow cytometry.
Semi-Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA from indicated cells were isolated using RNeasy mini kit (Qiagen) followed by cDNA synthesis using iScript cDNA synthesis kit (Bio-Rad Laboratories) and qRT-PCR using gene-specific primers (Table S7). Gene expression levels were normalized relative to the expression level of β-actin using Image J (NIH).
Knockdown of ST3GAL3 and ST3GAL4 in Cells
RNA interference vector pSUPER.retro.puro (OligoEngine) was used to generate plasmids expressing shRNA constructs that allowed to specifically knockdown the expression of ST3GAL3 and ST3GAL4 that are critical in synthesizing the terminal Neu5Ac-α2–3-Gal of N-linked glycoproteins. Oligonucleotides used are: 5’-
GATCCCCTCGCAAGTGGGCTAGAATCTTCAAGAGAGATTCTAGCCCACTTGCGATTTTTC-3’ and 5’-CGAGAAAAATCGCAAGTGGGCTAGAATCTCTCTTGAAGATTCTAGCCCACTTGCGAGGG for ST3GAL3, and 5’-GATCCCCGAGTGATAAGAAGCGGGTATTCAAGAGATACCCGCTTCTTATCACTCTTTTTC −3’ and 5’-TCGAGAAAAAGAGTGATAAGAAGCGGGTATCTCTTGAATACCCGCTTCTTATCACTCGGG for ST3GAL4. These oligonucleotides (Integrated DNA Technologies) were annealed to form double-stranded DNA and cloned into the BglII and XhoI sites of pSUPER.retro.puro vector. Henle-407 and HEK293T cells were transfected with these plasmids using Lipofectamine 3000 (Invitrogen). The knockdown of these enzymes was validated by conducting qRT-PCR using primer sets specific to ST3GAL3 and ST3GAL4 (Table S7).
Crystallization
Purified C-terminal 6x His-tagged PltB wildtype and mutants were prepared as described previously and used for crystallization (Deng et al., 2014). After crystal optimization trials, PltB, PltBN29K and PltBT65I crystals appeared in 1 to 2 days at room temperature using the hanging drop vapor diffusion method in a mixture of 2 μl of protein and 2 μl of reservoir solution. Conditions for crystallization are described in Table S4. Neu5Ac-α2–3-Gal-β1–4-GlcNAc (Carbosynth) and Neu5Ac-α2–6-Gal-β1–4-GlcNAc (Carbosynth) were introduced into PltB crystals via soaking. Hanging drop containing crystals were mixed with 1 μL of stock solution of α2–3 or α2–6 sialosides in reservoir buffers to yield a final concentration of 5 mM of sialosides. After 2 hours of incubation with sialosides, the crystals were cryoprotected and flash-frozen in liquid nitrogen.
X-Ray Data Collection and Structure Determination
PltB crystals with sialosides were submitted to data collection at GMCA APS (www.gmca.aps.anl.gov) 23-ID-B beamline at an incident beam of 1.87 A in wavelength and 100K. PltB diffraction data from single crystals were processed using HKL-2000 (Otwinowski and Minor, 1997). PltBN29K and PltBT651 crystals with and without sialosides were submitted to data collection with Rigaku XtaLAB Synergy diffractometer using an incident beam of 1.54184 A from a Cu Ka PhotonJet microfocus source and 100K. PltBN29K and PltBT651 diffraction data from single crystals were processed using CrysAlisPro (Rigaku). All structures were phased using the molecular replacement method in PHENIX (Adams et al., 2010) using previously published PltB apo structure (PDB 4RHR) as a search model (Deng et al., 2014). The bound sialosides were fitted into density within the soaked crystal structures. Rebuilding and real-space refinement were performed with Coot (Emsley et al., 2010) together with reciprocal space refinements in PHENIX (Adams et al., 2010). Figures were prepared using PyMol (Schrodinger Inc). The data collection and refinement statistics are summarized in Table S5.
PltB DNA and Amino Acid Sequence Comparison Analysis
FastME 2.0 phylogeny inference program (Lefort et al., 2015) with the F84 DNA evolution model (Felsenstein and Churchill, 1996) was used to analyze S. Typhi pltB and pltB orthologues from NTS serovars (Figure S2). Salmonella serovars and their pltB sequences used in this analysis are summarized in Table S2 and S3. Amino acid sequence analysis shown in Figure S3 was performed using CLUSTALW.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were tested for statistical significance with GraphPad Prism software. The number of replicates for each experiment and the statistical test performed are indicated in the figure legends. Image analysis and quantification and cell cycle profile analysis and quantification were performed using ImageJ and FlowJo software, respectively.
Supplementary Material
Highlights.
Typhoid toxin and non-typhoid Javiana toxin induce different clinical presentations.
Typhoid toxin PltB uses both α2–3 and α2–6 sialosides to enter target cells.
Amino acid variations in Javiana PltB render different glycan-binding preference.
Javiana toxin offers cross-reactive protection against typhoid toxin challenge.
ACKNOWLEDGMENTS
We thank Jun Zhao and Lingquan Deng for glycan pdb files, and Samantha N. MacMillan and the X-ray Diffraction Facility of the Department of Chemistry and Chemical Biology for the assistance of acquisition of protein X-ray diffraction data from the Rigaku home-source. This work was supported in part by NIH R01 AI137345, AI139625, AI141514, R03 AI135767, and a grant from the Mizutani Foundation for Glycoscience to J.S.; NIH R01 AI114730 to J.C.P.; NIH ACB-12002, AGM-12006, 1S10D012289, and DOE DE-AC02-06CH11357 to GMCA@APS, the Argonne National Laboratory. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali G, Rashid S, Kamli MA, Shah PA, and Allaqaband GQ (1997). Spectrum of neuropsychiatric complications in 791 cases of typhoid fever. Trop Med Int Health 2, 314–318. [DOI] [PubMed] [Google Scholar]
- Azmatullah A, Qamar FN, Thaver D, Zaidi AK, and Bhutta ZA (2015). Systematic review of the global epidemiology, clinical and laboratory profile of enteric fever. J Glob Health 5, 020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byres E, Paton AW, Paton JC, Lofling JC, Smith DF, Wilce MC, Talbot UM, Chong DC, Yu H, Huang Sv et al. (2008). Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature 456, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SJ, Song J, and Galan JE (2016). Receptor-Mediated Sorting of Typhoid Toxin during Its Export from Salmonella Typhi-Infected Cells. Cell Host Microbe 20, 682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles RC, Liang L, Khanam F, Sayeed MA, Hung C, Leung DT, Baker S, Ludwig A, Harris JB, Larocque RCV et al. (2014). Immunoproteomic analysis of antibody in lymphocyte supernatant in patients with typhoid fever in Bangladesh. Clin Vaccine Immunol 21, 280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Hayakawa T, Diaz S, Krings M, Indriati E, Leakey M, Paabo S, Satta Y, Takahata N, and Varki A (2002). Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc Natl Acad Sci U S A 99, 11736–11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Bakker HC, Moreno Switt AI, Govoni G, Cummings CA, Ranieri ML, Degoricija L, Hoelzer K, Rodriguez-Rivera LD, Brown S, Bolchacova Ev et al. (2011). Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genomics 12, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Song J, Gao X, Wang J, Yu H, Chen X, Varki N, Naito-Matsui Y, Galan JE, and Varki A (2014). Host adaptation of a bacterial toxin from the human pathogen salmonella typhi. Cell 159, 1290–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feasey NA, Gaskell K, Wong V, Msefula C, Selemani G, Kumwenda S, Allain TJ, Mallewa J, Kennedy N, Bennett A, et al. (2015). Rapid emergence of multidrug resistant, H58-lineage Salmonella typhi in Blantyre, Malawi. PLoS Negl Trop Dis 9, e0003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J, and Churchill GA (1996). A Hidden Markov Model approach to variation among sites in rate of evolution. Mol Biol Evol 13, 93–104. [DOI] [PubMed] [Google Scholar]
- Gal-Mor O, Boyle EC, and Grassl GA (2014). Same species, different diseases: how and why typhoidal and nontyphoidal Salmonella enterica serovars differ. Front Microbiol 5, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Deng L, Stack G, Yu H, Chen X, Naito-Matsui Y, Varki A, and Galan JE (2017). Evolution of host adaptation in the Salmonella typhoid toxin. Nat Microbiol 2, 1592–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Gilchrist JJ, and MacLennan CA (2019). Invasive Nontyphoidal Salmonella Disease in Africa. EcoSal Plus 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghjoo E, and Galan JE (2004). Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci U S A 101, 4614–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson BR, Griffin PM, Cole D, Walsh KA, and Chai SJ (2013). Outbreak-associated Salmonella enterica serotypes and food Commodities, United States, 1998–2008. Emerg Infect Dis 19, 1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Mylona E, and Frankel G (2018). Typhoidal Salmonella: Distinctive virulence factors and pathogenesis. Cell Microbiol 20, e12939. [DOI] [PubMed] [Google Scholar]
- Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, Wong VK, Dallman TJ, Nair S, Baker, Sv et al. (2018). Emergence of an Extensively Drug-Resistant Salmonella enterica Serovar Typhi Clone Harboring a Promiscuous Plasmid Encoding Resistance to Fluoroquinolones and Third-Generation Cephalosporins. MBio 9, e00105–00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort V, Desper R, and Gascuel O (2015). FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol Biol Evol 32, 2798–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Juarez S, Nga TV, Dunstan S, Nakajima-Sasaki R, Davies DH, McSorley S, Baker S, and Felgner PL (2013). Immune profiling with a Salmonella Typhi antigen microarray identifies new diagnostic biomarkers of human typhoid. Sci Rep 3, 1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Lee S, Yang YA, Ahn C, Sim JH, Kei TG, Barnard KN, Yu H, Millano SK, Chen X, et al. (2020). The role of 9-O-acetylated glycan receptor moieties in the typhoid toxin binding and intoxication. PLoS Pathog 16, e1008336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohl ME, and Miller SI (2001). Salmonella: a model for bacterial pathogenesis. Annu Rev Med 52, 259–274. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, et al. (2001). Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413, 848–852. [DOI] [PubMed] [Google Scholar]
- Peng W, de Vries RP, Grant OC, Thompson AJ, McBride R, Tsogtbaatar B, Lee PS, Razi N, Wilson IA, Woods RJ, et al. (2017). Recent H3N2 Viruses Have Evolved Specificity for Extended, Branched Human-type Receptors, Conferring Potential for Increased Avidity. Cell Host Microbe 21, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohan HT (2004). Clinical and laboratory manifestations of typhoid fever at Persahabatan Hospital, Jakarta. Acta Med Indones 36, 78–83. [PubMed] [Google Scholar]
- Qamar FN, Yousafzai MT, Sultana S, Baig A, Shakoor S, Hirani F, Wassay A, Khushboo S, Mehmood J, Freeman A, et al. (2018). A Retrospective Study of Laboratory-Based Enteric Fever Surveillance, Pakistan, 2012–2014. J Infect Dis 218, S201–S205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Wilson RP, Winter SE, and Baumler AJ (2008). Clinical pathogenesis of typhoid fever. J Infect Dev Ctries 2, 260–266. [DOI] [PubMed] [Google Scholar]
- Sejvar J, Lutterloh E, Naiene J, Likaka A, Manda R, Nygren B, Monroe S, Khaila T, Lowther SA, Capewell L, et al. (2012). Neurologic manifestations associated with an outbreak of typhoid fever, Malawi--Mozambique, 2009: an epidemiologic investigation. PLoS One 7, e46099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma T, Sharma C, Sankhyan A, Bedi SP, Bhatnagar S, Khanna N, Gautam V, Sethi S, Vrati S, and Tiwari A (2018). Serodiagnostic evaluation of recombinant CdtB of S. Typhi as a potential candidate for acute typhoid. Immunol Res 66, 503–512. [DOI] [PubMed] [Google Scholar]
- Singletary LA, Karlinsey JE, Libby SJ, Mooney JP, Lokken KL, Tsolis RM, Byndloss MX, Hirao LA, Gaulke CA, Crawford RW, et al. (2016). Loss of Multicellular Behavior in Epidemic African Nontyphoidal Salmonella enterica Serovar Typhimurium ST313 Strain D23580. MBio 7, e02265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Gao X, and Galan JE (2013). Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin. Nature 499, 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Willinger T, Rongvaux A, Eynon EE, Stevens S, Manz MG, Flavell RA, and Galan JE (2010). A mouse model for the human pathogen Salmonella typhi. Cell Host Microbe 8, 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano S, and Galan JE (2008). A novel pathway for exotoxin delivery by an intracellular pathogen. Curr Opin Microbiol 11, 15–20. Epub 2008 Feb 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano S, Ugalde JE, and Galan JE (2008). Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 3, 30–38. [DOI] [PubMed] [Google Scholar]
- Tran Vu Thieu N, Trinh Van T, Tran Tuan A, Klemm EJ, Nguyen Ngoc Minh C, Voong Vinh P, Pham Thanh D, Ho Ngoc Dan T, Pham Duc T, Langat P, et al. (2017). An evaluation of purified Salmonella Typhi protein antigens for the serological diagnosis of acute typhoid fever. J Infect 75, 104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis RM, Xavier MN, Santos RL, and Baumler AJ (2011). How to become a top model: impact of animal experimentation on human Salmonella disease research. Infect Immun 79, 1806–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JHV et al. (2017). Essentials of Glycobiology, 3rd edition. Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Yang YA, Chong A, and Song J (2018a). Why Is Eradicating Typhoid Fever So Challenging: Implications for Vaccine and Therapeutic Design. Vaccines (Basel) 6, E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YA, Lee S, Zhao J, Thompson AJ, McBride R, Tsogtbaatar B, Paulson JC, Nussinov R, Deng L, and Song J (2018b). In vivo tropism of Salmonella Typhi toxin to cells expressing a multiantennal glycan receptor. Nat Microbiol 3, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated during this study. Coordinates for the atomic structures have been deposited in the RCSB Protein Data Bank under PDB IDs of 6P4M, 6P4N, 6P4P, 6P4Q, 6P4R, 6P4S and 6P4T.