Abstract
Rationale and Objectives.
A standard lung template could improve population-level analyses for computed tomography (CT) scans of the lung. We develop a fully-automated pre-processing pipeline for image analysis of the lungs using updated methodologies and R software that results in the creation of a standard lung template. We apply this pipeline to CT scans from a sarcoidosis population, exploring the influence of registration on radiomic analyses.
Materials and Methods.
Using 65 high-resolution CT scans from healthy adults, we create a standard lung template by segmenting the left and right lungs, non-linearly registering lung masks to an initial template mask, and using an unbiased, iterative procedure to converge to a standard lung shape (Dice similarity coefficient ≥0.99). We compare three-dimensional radiomic features between control and sarcoidosis patients, before and after registration to a study-specific lung template.
Results.
The final lung template had a right lung volume of 2967 cm3 and left lung volume of 2623 cm3, with a median HU = −862. Registration significantly affected radiomic features, shifting the HU distribution to the left, decreasing variability, and increasing smoothness (p<0.0001). The registration improved detective ability of radiomics; for contrast, autocorrelation, energy and homogeneity, the group effect was significant post-registration (p<0.05), but was not significant pre-registration.
Conclusion.
The final lung template and software used for its creation are publicly available via the lungct R package to facilitate its use in practice. This study advances lung imaging by developing tools to improve population-level analyses for various lung diseases.
Keywords: template creation, atlas, lung, computed tomography, R software
Introduction
In many analyses of imaging data, population-level inference is of interest. For example, in functional magnetic resonance imaging of the brain, there is a need to aggregate data across individuals to inform which areas of the brain activate during a task. To aggregate across individuals, the images are warped to a common space to maintain spatial alignment. This alignment is done by creating a reference image, commonly called a template or atlas and warping each individual’s image to the reference image.
Creation of templates has focused heavily on those of the brain and magnetic resonance imaging modalities1. However, there are various other organs and imaging modalities for which identification of regions where images differ between groups of individuals is of interest. Examples include lungs2, lymph nodes3, liver4, and spleen5, which are often imaged with computed tomography (CT). Thus, there is a need for development of templates for other organs and imaging modalities. The focus of this paper is to develop a lung template for CT.
A reference template, or atlas, for the lungs would provide a standardized coordinate system for lung imaging studies, which would make it possible to (1) perform whole-lung comparisons across individuals in a more objective and principled manner, (2) identify anatomical regions that differ between groups, and (3) compare findings across lung studies. In neuroimaging, the standard brain has enabled researchers to study the normal aging process of brains1, locate brain regions that differ between schizophrenic and healthy patients7, improve the diagnostic accuracy of Alzheimer’s disease8, and identify genes related to neurological disorders9, among many others. These studies use statistical techniques, such as voxel- and deformation-based morphometry10,11, and imaging-genetics9, that enable objective whole-brain comparisons within and across studies. With the establishment of a standard lung template, we believe that these neuroimaging techniques can be adapted for the lung to uncover many findings related to emphysema, sarcoidosis, idiopathic pulmonary fibrosis, and other lung diseases.
Additionally, current objective and automated techniques for population-level inference of the lungs include radiomic analyses12 and machine-learning13,14,15. These methods could benefit from a standard lung template. Specifically, radiomic analyses can be confounded by region of interest size16. Further, machine- or deep- learning methods typically require equal image resolution, or even equal image size for fully connected layers17. Registration of lung CT scans to a common lung template would standardize images to the same image dimensions prior to analysis, removing issues related to image size and spacing in both radiomic and machine-learning methods.
To our knowledge, only a single method for atlas construction for the lungs has been established2,18. This approach by Li et al. uses CT scans from twenty normal volunteers, in-house segmentation and registration methods, and transformation-averaging for the construction of the final lung atlas. This standard lung is not freely available for download limiting its use in practice. Additionally, updated methodologies for segmentation, registration, and template creation have been developed since, along with open-source software programs for their implementation19,20 (see Online Supplemental Material for details).
In this paper, we develop a fully-automated pre-processing pipeline for medical image analysis of lung CT scans using updated methodologies and software available in R that results in the creation of a publicly available, unbiased estimate of a standard lung from a healthy adult population. We apply this pre-processing methodology to CT scans from a diseased population of patients with sarcoidosis, whereby we explore the influence of registration on radiomic analyses as well as performing regional analyses.21.
Materials and Methods
Healthy Control Population
For the creation of a standard lung, data from N=108 non-smoking, healthy control patients between the ages of 45 and 80 years with no history of lung disease and normal post-bronchodilator spirometry was obtained from COPDGene, a retrospective cohort study with recruitment between October 2006 and January 201122,23. Research chest high-resolution CT scans were obtained with patients at full inspiration, a tube potential of 120 kVP, tube current of 400 mA and a variety of scanner manufacturers (General Electric Medical Systems, Siemens and Philips), which resulted in different reconstructed slice thicknesses (0.625, 0.75, or 0.9mm), slice intervals (0.625, 0.5, and 0.45mm), and convolution kernels (Standard, B31f, and B). Of the 108 controls, six patients were excluded due to missing CT scans (N=2) or inaccurate segmentations from VIDA Diagnostics (N=4), resulting in N=102 healthy patients.
Of the 102 healthy patients available for use in our study, 32 were males. Since sex, age, BMI, and lung volume affect the lung size, shape, and function24,25, three patients were chosen for the initial templates that varied according to these characteristics, including two females and one male. To create a balanced sample across sex for template creation, all 31 remaining male participants (mean age: 63.5 years, range: 46–78 years) and 31 randomly sampled scans from females (mean age: 61.7 years, range: 45–78 years) were selected, for a total of 62 participants.
Image Processing Pipeline for Lung CT
Pre-Processing.
As the first step in the pre-processing pipeline, we converted all images from raw DICOM (Digital Imaging and Communications in Medicine) to three-dimensional NIfTI (Neuroimaging Informatics Technology Initiative) using dcm2niix (https://github.com/rordenlab/dcm2niix) from the dcm2niir R package interface26. We reset image origins to zero and resampled the data to 1×1×1mm (or 1 mm3) format, to normalize scans to the same space and resolution.
Lung Segmentation.
COPDGene provided left and right lung segmentations from the proprietary Pulmonary Workstation 2 software (VIDA Diagnostics, Inc, Coralville, IA)23. However, to make a fully automated image processing pipeline, we also created a publicly available segment_lung_lr function in the lungct R package to segment the left and right lungs. As this is a newly developed image segmentation package, we provide details on the approach here.
The segment_lung_lr function identifies the left and right lungs from the CT scan using a combination of thresholding- and region-based segmentation methodology. First, the lung and airways are detected from the original CT scan by Hounsfield unit (HU) thresholding. Typically, normal lung tissue corresponds to radiodensity between −700 to −600 HU27. As diseased lung tissue can have variable ranges of radiodensity, segment_lung_lr allows the user to specify a maximum HU threshold (default −300), with the minimum HU threshold set at −1024 HU, which is the radiodensity of air. Second, the large airways (i.e. trachea and large bronchi) are detected by identifying the region near the mid-line that is less radiodense than normal lung tissue via histogram analysis. To detect the left and right lungs, the large airways are first removed from the lung and airway segmentation. Next, a connected components analysis is used to determine if the left and right lungs are separated in the segmentation. If the left and right lungs are not separated in the segmentation following the initial removal of the airways, the maximum HU threshold is lowered, followed by an erosion of the segmented mass. Once the left/right segmentations are identified, the segmentations are dilated to reverse the prior erosion necessary to discriminate the left from the right lung. Finally, the right lung is classified based on a center of gravity located to the left of the left lung; thus, if the original scan has left/right orientation flipped, it will remain that way through the segmentation process.
To confirm the reliability of segment_lung_lr, these segmentations were compared to the corresponding segmentations from VIDA Diagnostics using the Dice similarity coefficient (DSC), average symmetric surface distance (ASSD), and three-dimensional measures: volume, surface area, length, width, and depth. The three-dimensional measures were calculated on lung masks using labelGeometryMeasures function in the ANTsR R package28. To compare these measures across VIDA Diagnostics and lungct segmentations, Wilcoxon signed-rank tests were used due to small sample size and skewed data.
Registration.
Symmetric Normalization (SyN) registration29, chosen based on its success in the EMPIRE10 intra-subject thoracic CT registration challenge20, was used to transform the resampled lung masks to a common space (described in the template creation section below). The registration performs an affine transformation, followed by SyN deformable transformation, and was optimized via mutual information30. Left and right lung masks were registered separately to that of the initially selected template, due to differences in left and right lung size and shape.
Template Creation.
For template creation of the lung, we followed the iterative method presented in Avants et al.31 and implemented in buildTemplate (ANTsR) given to its success in the brain. Adapting for the lung, we created the publicly available get_template function in the lungct R package (Figure 1). In brief, a lung mask from a randomly selected patient is chosen as the initial template; then, all remaining masks are registered to the initial template, resulting in three-dimensional transformations along with transformed masks in common space; the average transformation is applied to the average transformed mask, resulting in a new template; this process is repeated until convergence. While the default in buildTemplate is to perform three iterations, we define convergence as having a DSC between successive iterations of at least 0.99.
Figure 1:
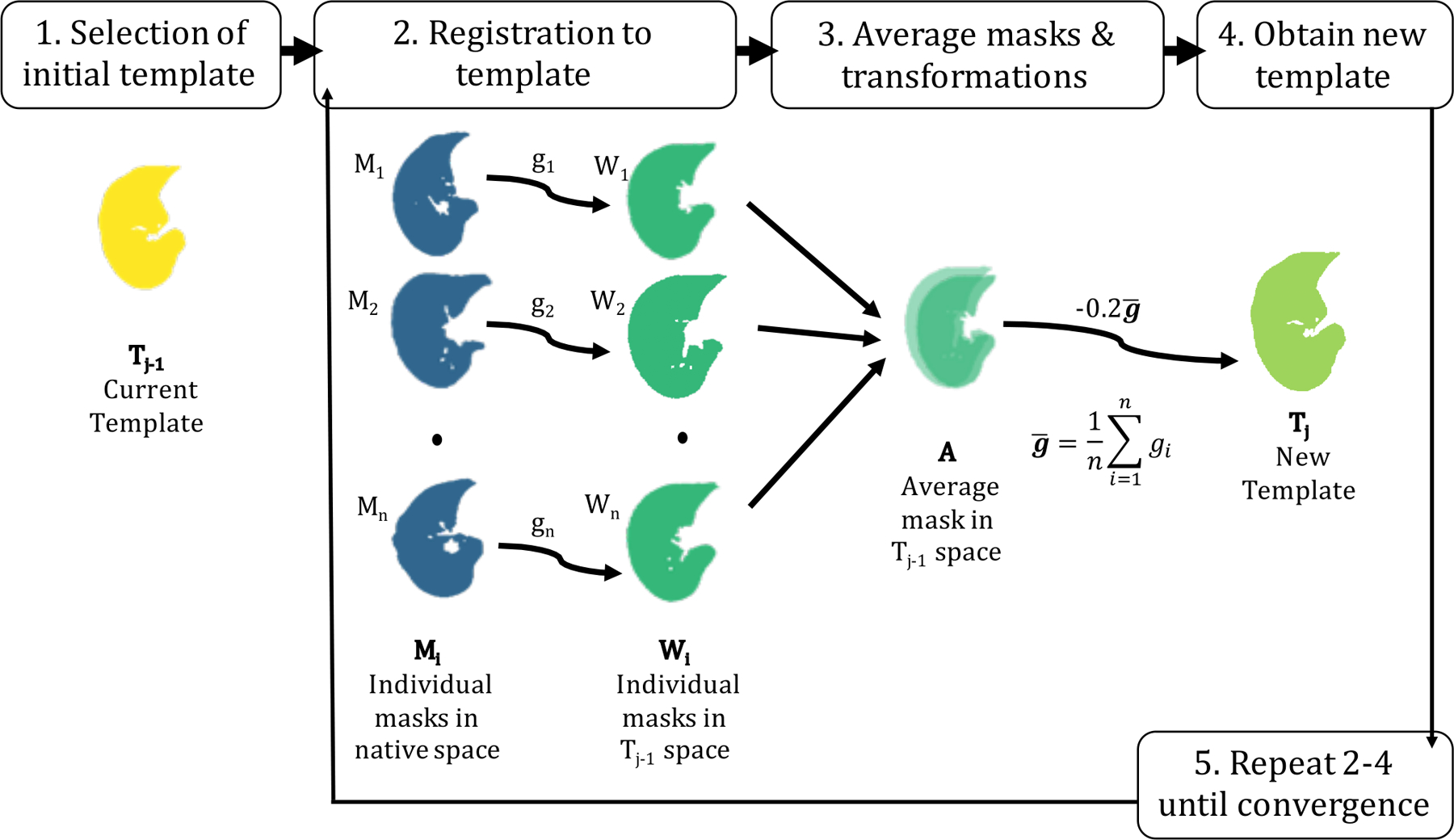
Template creation methodology. An initial template mask, T1, is randomly selected (Step 1), as this method does not depend on the choice of the initial template. Then, all remaining masks, Mi, where i∈1…n and n is the total number of masks, are registered to the template (Step 2), and interpolated using a linear interpolator. This is performed separately for the left and right lungs. All warped masks, Wi, now in template space, are averaged voxel-wise to obtain an average mask, A, in template space. The average template mask, A, was thresholded above a value of 0.5 to maintain average lung volume. The diffeomorphic transformations (i.e. the differentiable maps from the moving image to the fixed space), including the affine registration, were averaged to create an average transformation, ḡ (Step 3). Finally, the average transformation, ḡ is multiplied by a gradient step of −0.2 and applied to the average image, A, resulting in a new template, Tj (Step 4) where j∈1…J and J is the total number of iterations. Steps 2–4 are repeated until convergence.
Three healthy templates were created from the 62 healthy control patients, using three different initial template masks to investigate whether the resulting template mask could be dependent on the starting image. To confirm we had reached convergence, three dimensional measures were calculated for the template masks at every iteration. Once the final healthy template masks were obtained, final registrations were performed on all original masks in native space, resulting in a set of N=62 final transformations and masks. The final transformations on the lung segmentations were applied to their respective CT images, to obtain warped images in common space. Using the warped images, a final healthy template was constructed using voxel-wise mean and standard deviations HUs. Additionally, the final transformations were also applied to their respective lobe segmentations from VIDA Diagnostics. Using majority vote at each voxel in the warped lobe segmentations, an average lobe mask was created in template space.
Application of Image Processing Pipeline for Lung CT
To illustrate how our image processing pipeline can be used in common lung analyses for diseased populations, we conducted radiomic analyses between sarcoidosis patients and healthy controls.
Study Populations.
The population of healthy controls included all N=102 non-smoking healthy control patients from the COPDGene study as described in the Healthy Control Population section above. The population of sarcoidosis patients were recruited as part of the NHLBI funded Genomic Research in Alpha-1 Antitrypsin Deficiency and Sarcoidosis (GRADS) study. The GRADS study is a multi-center, observational cohort exploring the role of the microbiome and genome in patients with Alpha-1 Antitrypsin Deficiency and/or Sarcoidosis32. Patients were eligible for GRADS if they had a confirmed diagnosis of sarcoidosis via biopsy or manifestations consistent with acute sarcoidosis (Lofgren’s syndrome), met one of the nine study phenotypes and provided signed informed consent32. As part of GRADS, uniform clinical data was obtained including pulmonary function testing, a chest radiograph (for Scadding staging classification), and a research chest HRCT based on the COPDGene protocol22. Research chest HRCT scans used the same manufacturers and parameters as COPDGene above, with the exception of the effective tube current, which was based on BMI (range: 160–330 mA) for GRADS patients. Of the 330 patients with sarcoidosis, N=321 had a CT scan usable for quantitative analysis and were used in this study.
Study-Specific Template.
We followed the same pre-processing, segmentation, registration, and template creation methodology as above. A study-specific template was created from 42 lung segmentations (10% of total sample size), of which half were randomly selected from the healthy controls and half randomly selected from sarcoidosis patients. Equal proportion of sarcoidosis and healthy patients were used to ensure that the resulting template would not be biased to group type. The standard healthy lung template could also have been used here, but we chose to create a study-specific template to illustrate how this could be done; furthermore, study-specific templates may increase detecting power for a study1. Once the final study-specific template was created, the final transformations were applied to their respective CT scans, to obtain warped scans in common space.
Effect of Registration on Radiomics.
To understand the influence of registration on radiomics, we performed a radiomic analysis21 between patients with sarcoidosis and healthy controls. Three-dimensional radiomic features were calculated on both healthy and diseased patients, before and after registration to the study-specific template. Radiomic features21,33 were calculated separately on the left and right lungs, using the RIA_lung function from the lungct R package. Specifically, eight radiomic features were calculated, including four first-order features (mean, standard deviation, skewness and kurtosis) and four grey-level co-occurrence matrix (GLCM) features (contrast, autocorrelation, energy, and homogeneity). GLCM features were calculated using 16 gray levels, equal probability bins, and a distance of 1 voxel, averaged over all directions. Linear mixed-effects models with a random intercept for subject were used to evaluate whether registration influences the group effect (sarcoidosis v. control) on radiomic features. These models were adjusted for lung, sex, age, race, height, BMI, and lung volume.
Differences in Radiomics by Lobe.
We did not have lobe segmentations on the sarcoidosis population. However, using our standard lobe template, we were able to perform regional lobe analyses without individual lobe masks. To do this, we transformed the standard lobe template into the study-specific space using SyN registration, creating a study-specific lobe template. Next, using the warped CT scans in the common study-specific space, we calculated the same eight radiomic features as above on the five different lobes (top left, bottom left, top right, middle right, and bottom right) using the study-specific lobe template as our mask for each scan. To evaluate whether the group effect (sarcoidosis v. control) on radiomic features changes across the lung lobes, we fit linear mixed-effects models with a random intercept for subject, adjusted for sex, age, race, height, BMI, and lung volume.
All results were considered significant at p<0.05. Data used in this study was approved by the local institutional review boards.
Results
All figures of the lung are in “radiological” convention, where the left side of the image is the right lung.
Patient Characteristics
In total, 62 patients were used for the template creation, including 50% female (N=31), 5% non-white (N=3), and 100% non-Hispanic. Additionally, patients had an average age of 62.3 (SD = 9.0) years, body mass index (BMI) of 28.5 (SD = 4.5), percent-predicted Forced Expiratory Volume in 1 second of 104.3 (SD = 14.3) and percent-predicted forced vital capacity of 99.6 (SD = 11.9). The primary patient whose lung segmentation was used as the final initial template (Patient A), was a white, non-Hispanic, female with 56 years and a BMI of 30.6.
Validation of lungct Segmentation
In general, segmentations from lungct were slightly more conservative than those from VIDA software, removing more airway and exterior edges (Figure 2). The mean DSC across lungct and VIDA software lung segmentations was 0.989 (SD = 0.007, minimum = 0.943, median = 0.990, maximum = 0.996), and the mean ASSD was 0.567 mm (SD = 0.263 mm, minimum = 0.12 mm, maximum = 1.97 mm). This indicates a high level of overlap and minimal amount of distance between the segmentation borders. For the right lung alone, the mean DSC was 0.989 (SD = 0.007) and the mean ASSD was 0.602 mm (SD = 0.288 mm); the right lung volume and surface area were significantly lower in lungct segmentations compared to those of VIDA (p=0.041 and p=0.046, respectively), although the relative differences were not biologically meaningful (0.58% and −0.34%, respectively). There were no significant differences between segmentation methods for length, depth, and width (p≥0.362) (Table 1). For the left lung alone, the mean DSC was 0.989 (SD = 0.008) and the mean ASSD was 0.537 mm (SD = 0.251 mm); the left lung volume and surface area was significantly lower in the lungct segmentation compared to the VIDA segmentation (p=0.033 and p<0.001), although, as in the right lung, the relative differences were not biologically meaningful for the left lung (0.62% and −0.97%, respectively). There were no significant differences between segmentation methods for length, depth and width (p≥0.108) (Table 1). Overall, the lungct segmentations were very similar in practice to the well-validated VIDA results, indicating this automated pipeline should work well for similar scans.
Figure 2.
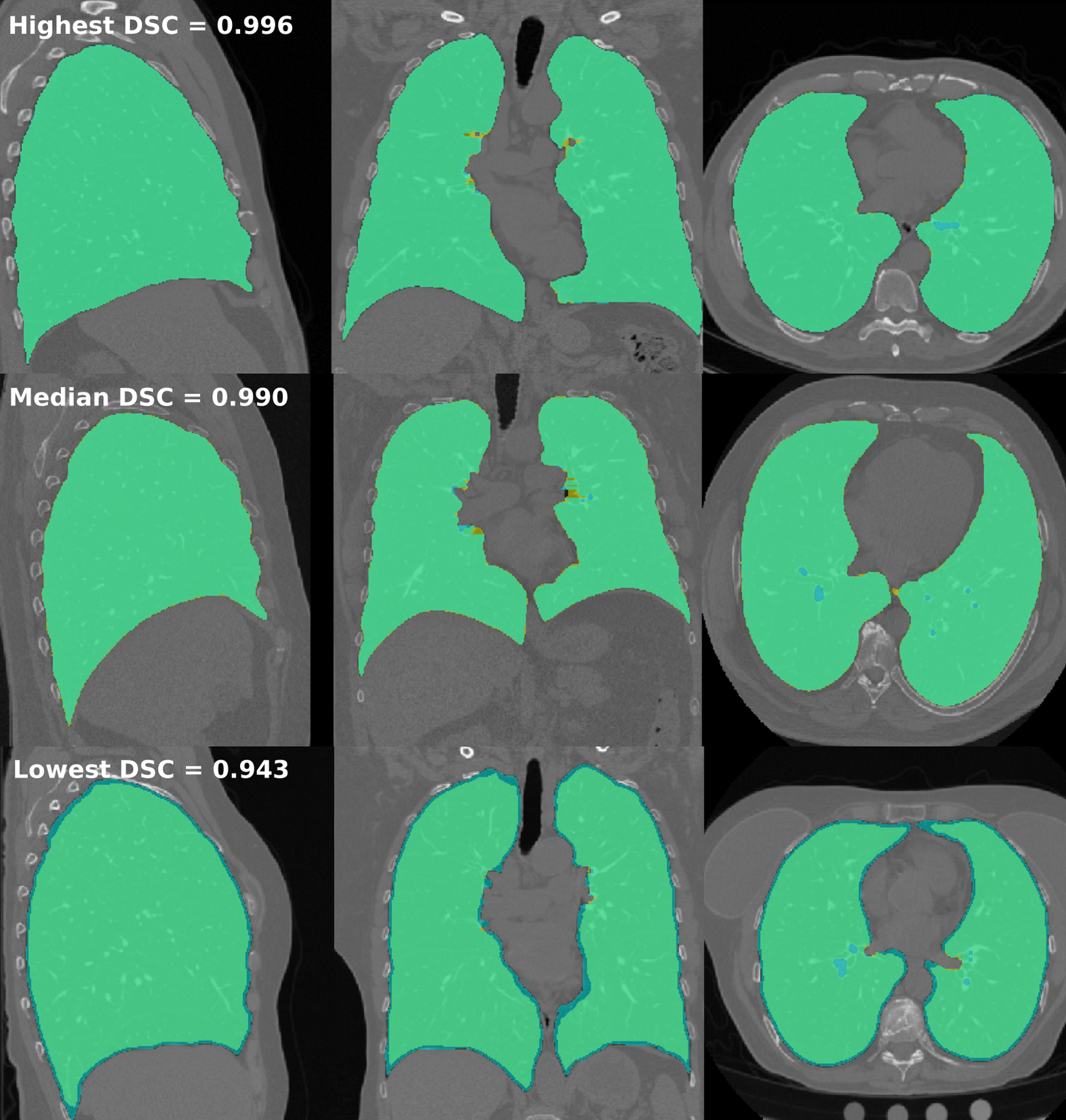
Comparison of lung segmentations between lungct and VIDA Diagnostics. Masks with the highest (row 1), median (row 2), and lowest (row 3) DSC between lungct and VIDA are shown in the sagittal, coronal, and axial planes (columns 1 – 3). Segmentations with lungct segmentations (yellow) are overlaid on top of VIDA segmentations (blue), with overlap shown in green. All figures of the lung are in “radiological” convention, where the left side of the image is the right lung.
Table 1.
Comparison of volumetric measures between healthy lung masks from lungct R package and VIDA Diagnostics. Results are presented as mean (95% CI). P-values are from a Wilcoxon Signed Rank test due to the relatively small (N=62) sample of paired, skewed data. R. Diff stands for the relative difference in measurements from lungct segmentations as compared to that of VIDA.
| Measure | lungct | VIDA | P-value | R. Diff (%) |
|---|---|---|---|---|
| Right | ||||
| Volume (cm3) | 3008 (2845, 3171) | 3026 (2863, 3189) | 0.041 | −0.58 |
| Surface Area (cm2) | 1542 (1486, 1597) | 1547 (1491, 1603) | 0.046 | −0.34 |
| Length (cm) | 23.56 (23.02, 24.10) | 23.55 (23.01, 24.09) | 0.795 | 0.05 |
| Depth (cm) | 17.98 (17.61, 18.35) | 17.99 (17.62, 18.36) | 0.736 | −0.06 |
| Width (cm) | 11.99 (11.72, 12.25) | 12.00 (11.73, 12.27) | 0.362 | −0.14 |
| Left | ||||
| Volume (cm3) | 2672 (2530, 2815) | 2689 (2546, 2831) | 0.033 | −0.62 |
| Surface Area (cm2) | 1466 (1415, 1517) | 1480 (1429, 1531) | 0.000 | −0.97 |
| Length (cm) | 24.40 (23.83, 24.97) | 24.41 (23.84, 24.99) | 0.758 | −0.06 |
| Depth (cm) | 17.74 (17.37, 18.12) | 17.75 (17.38, 18.13) | 0.705 | −0.06 |
| Width (cm) | 10.65 (10.44, 10.87) | 10.67 (10.46, 10.89) | 0.108 | −0.21 |
Healthy Lung Template Characteristics
Using segmentations from lungct and patient A as the initial template, the final template mask converged to an average lung shape after 14 iterations of get_template, as both the left and the right lung masks had a DSC ≥ 0.99 between the 13th and 14th iterations. The ASSD between the 13th and 14th iterations was also low, at 0.61 mm in the left lung and 0.48 mm in the right lung. The final template mask created for the right lung had an average lung volume of 2967 cm3, surface area of 1407 cm2, length of 23.0 cm, depth of 18.1 cm, and width of 11.9 cm. For the left lung, the final template had an average volume of 2623 cm3, surface area of 1352 cm2, length of 23.9 cm, depth of 17.8 cm, and width of 10.6 cm. These three-dimensional measures of the final lung template are consistent with the average three-dimensional measures from the 62 lung masks prior to registration (Table 1); for example, the 95% confidence interval for the right lung volume (and the lungct segmentation) was from 2845 to 3171 cm3 prior to registration, which includes the right lung volume of the final template (2967 cm3).
The final template, containing the average HU per voxel, can be seen in the first row of Figure 3, along with the standard deviation HU (bottom row). Across all voxels, the mean HU ranged from −961 to −425 HU (median: −862 HU), with the SD ranging from 19 to 2041 HU (median: 80 HU). As shown by more opacification on the scan, the HU was generally higher near the inner lungs, where the bronchi are located; this area also had larger variability across scans. The highest variability was found on the exterior edges of the lungs, most likely an artifact of imperfect segmentation and registration. Furthermore, the final lobe template, seen in Figure 4, consisted of five lobes, two in the left and three in the right. The lobe volumes were 1285 cm3 in the top-left, 1333 cm3 in the bottom-left, 1039 cm3 in the top-right, 458 cm3 in the middle-right, and 1464 cm3 in the bottom right.
Figure 3.
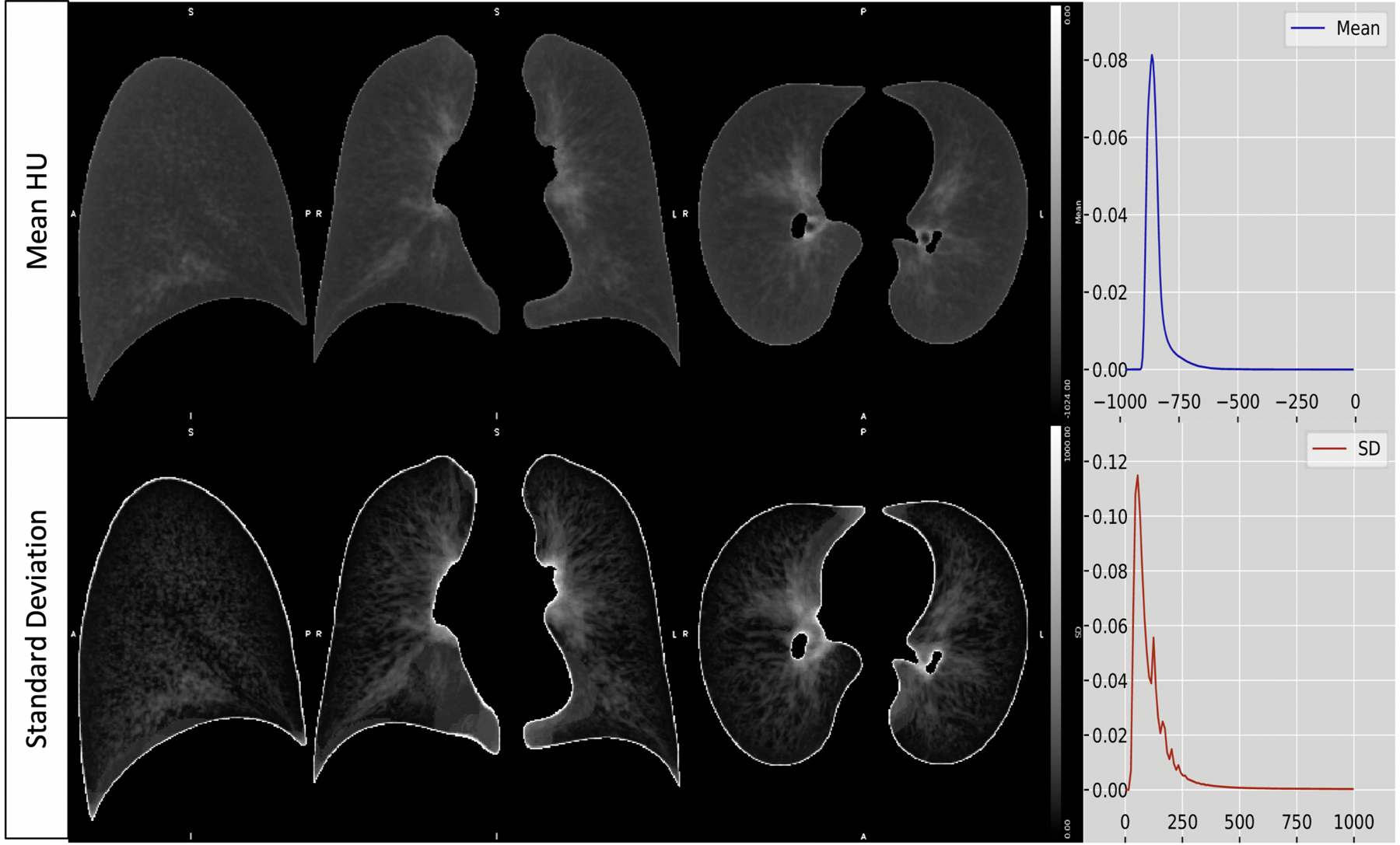
The final template, containing the mean HU per voxel (top row), along with the standard deviation HU (bottom row) in the sagittal, coronal, and axial planes (columns 1 – 3). The fourth column shows the right-skewed histogram HU densities for the mean and standard deviation templates. All figures of the lung are in “radiological” convention, where the left side of the image is the right lung.
Figure 4.

The final lobe template fit to the standard lung template space. All figures of the lung are in “radiological” convention, where the left side of the image is the right lung.
As part of a sensitivity analysis, convergence was also monitored by comparing three-dimensional metrics from three different initial templates across iterations (Figure 5 & Figure 6). On the initial templates from the three representative patients, patient A had intermediate volume, surface area, length, and depth in both the right and left lungs prior to registration; patient B had the highest volume, surface area, length, and depth in both the right and left lungs prior to registration, with patient C at the lowest values. However, as seen by the converging lines, by iteration 14, patient A, B and C had similar values across all metrics (Figure 5). Furthermore, at the initial iterations, the template masks show much variability in terms of shape, smoothness, and rotation between the three patients; however, by the 14th iteration, there are very minor visual differences among the template masks (Figure 6).
Figure 5.
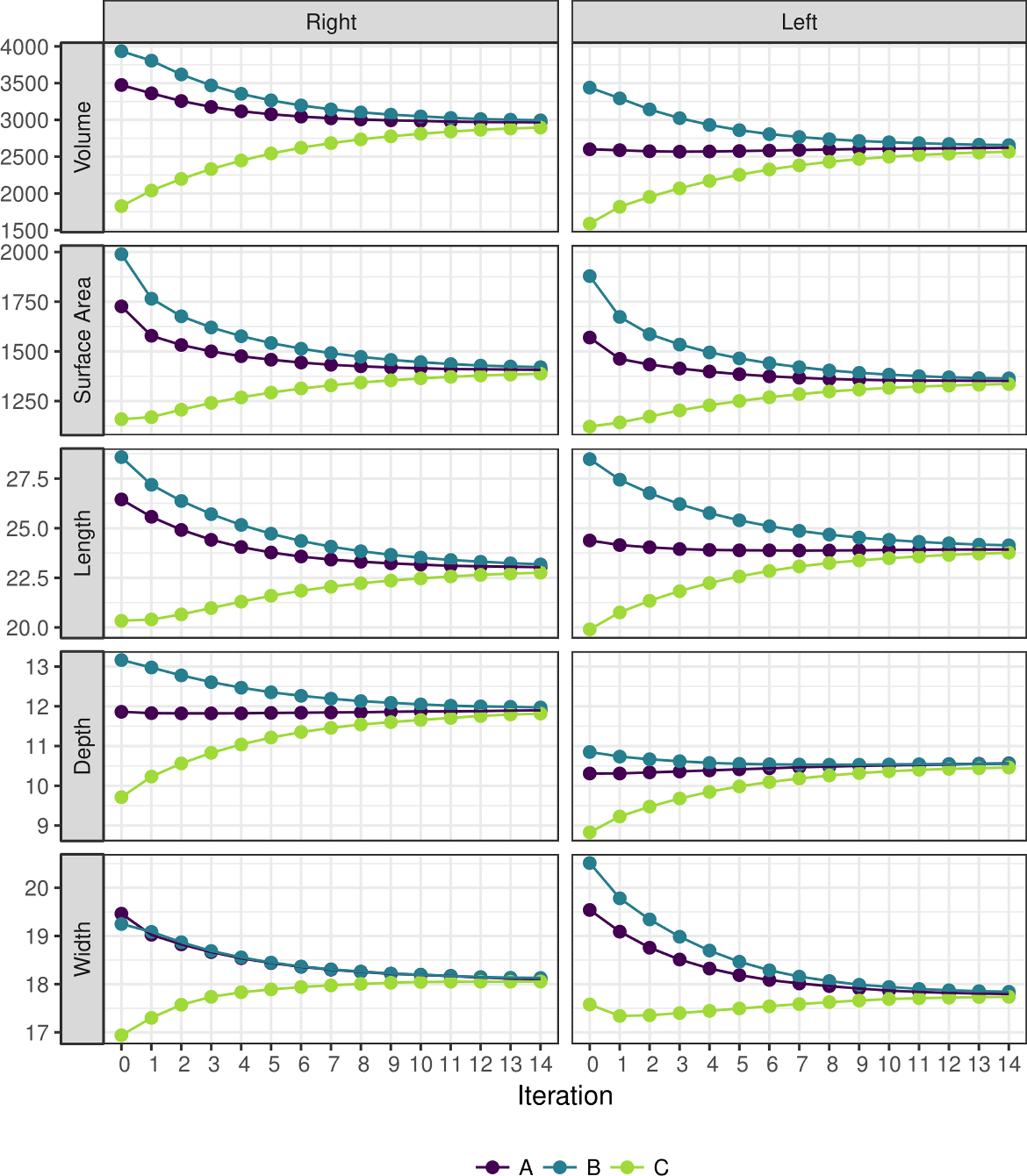
Three-dimensional measures for templates at each iteration, using lung masks from three different patients (A, B, and C) as the initial template for both the right and left lungs. Volume is given in cm3; surface area in cm2; length, width, and depth in cm. All figures of the lung are in “radiological” convention, where the left side of the image is the right lung.
Figure 6.
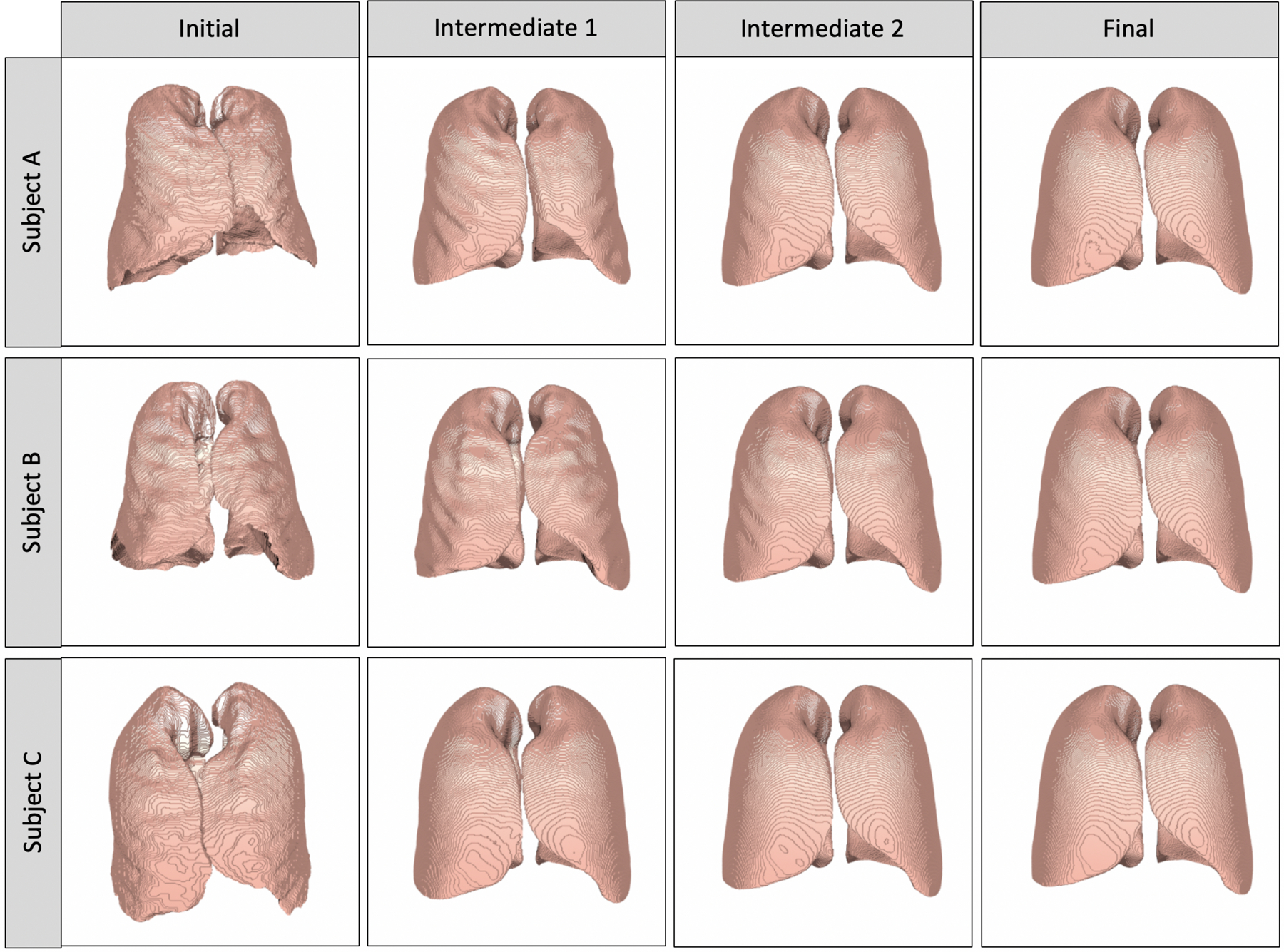
Three-dimensional contour plots of templates at the initial (0th), intermediate (5th, 10th) and final (14th) iterations, using different persons for the initial template (Patients A, B, C). All figures of the lung are in “radiological” convention, where the left side of the image is the right lung.
Influence of Registration on Radiomic Analyses
Table 2 shows the characteristics of the patient population used in the radiomic analysis. There were 423 patients, of which 102 were healthy and 321 had a confirmed case of sarcoidosis. Compared to the healthy population, the sarcoidosis population were significantly younger (52.9 vs. 62.4 years), taller (67.0 vs 65.5 inches), had increased BMI (30.6 vs 28.2) and smaller lung volumes (4.42 vs. 5.30 L), and higher proportion of males (45.8% vs. 31.4%) and lower proportion of whites (73.0% vs. 95.1%) (p≤0.001).
Table 2.
Patient characteristics used in the radiomic analyses. Results are presented as mean (SD) for continuous variables, and count (%) for categorical variables.
| Overall | Healthy | Sarcoidosis | P-value | |
|---|---|---|---|---|
| Sample Size | 423 | 102 | 321 | |
| Male | 179 (42.3) | 32 (31.4) | 147 (45.8) | 0.014 |
| White | 330 (78.4) | 97 (95.1) | 233 (73.0) | <0.001 |
| Age at enroll | 55.2 (10.55) | 62.4 (9.2) | 52.9 (9.9) | <0.001 |
| Height (in) | 66.6 (4.1) | 65.5 (3.6) | 67.0 (4.2) | 0.001 |
| BMI | 30.0 (6.3) | 28.2 (5.1) | 30.6 (6.5) | 0.001 |
| Lung volume | 4.89 (1.27) | 5.30 (1.17) | 4.41 (1.25) | <0.001 |
Radiomic features changed significantly post-registration compared to pre-registration across all first-order and GLCM features for both healthy controls and sarcoidosis subjects (Table 3).The mean HU, standard deviation and contrast significantly decreased post-registration for both healthy controls and sarcoidosis patients. The skewness, kurtosis, autocorrelation, energy and homogeneity significantly increased for both healthy controls and sarcoidosis patients (Table 3). The changes in first-order radiomic features indicate that the registration procedure shifts the HU distribution slightly to the left, decreases variability across voxels, increases right skew and results in more peaked distributions. The changes in GLCM features indicate that the registration procedure increases smoothness on the CT scans.
Table 3.
Influence of registration and group on radiomic values. For the registration effect, negative values indicate lower average radiomic values post-registration. For the group effect, negative values indicate lower average radiomic values in healthy controls. Results are presented as mean (95% CI). Bolded results are significant at a significance threshold of 0.05.
| Outcome | Registration Effect (Post- v. Pre-registration) | Group Effect (Control v. Sarcoidosis) | ||
|---|---|---|---|---|
| Controls | Sarcoidosis | Pre-Registration | Post-Registration | |
| Mean | −6.22 (−7.97, −4.47) | −8.69 (−9.70, −7.69) | −12.37 (−20.24, −4.51) | −9.90 (−17.76, −2.04) |
| SD | −18.44 (−19.74, −17.13) | −21.93 (−22.68, −21.17) | −15.94 (−20.38, −11.49) | −12.45 (−16.89, −8.01) |
| Skew | 0.51 (0.46, 0.55) | 0.46 (0.44, 0.49) | 0.49 (0.34, 0.64) | 0.53 (0.38, 0.68) |
| Kurtosis | 5.93 (5.52, 6.34) | 4.49 (4.26, 4.73) | 4.33 (3.01, 5.65) | 5.77 (4.45, 7.08) |
| Contrast | −4.46 (−4.70, −4.22) | −5.17 (−5.31, −5.03) | 0.36 (−0.51, 1.23) | 1.07 (0.20, 1.94) |
| Autocorrelation | 2.15 (2.00, 2.30) | 3.14 (3.05, 3.23) | 0.09 (−0.34, 0.52) | −0.90 (−1.33, −0.47) |
| Energy (x1000) | 1.37 (1.28, 1.45) | 1.72 (1.67, 1.77) | −0.05 (−0.26, 0.17) | −0.40 (−0.61, −0.18) |
| Homogeneity (x100) | 4.44 (4.22, 4.66) | 5.21 (5.09, 5.34) | −0.15 (−0.89, 0.60) | −0.92 (−1.67, −0.17) |
While the significant influence of registration on radiomics may be concerning initially, we found that the registration procedure does not deter our ability to find differences in radiomics between sarcoidosis and healthy control subjects (Table 3). Both pre- and post-registration, the mean HU and standard deviation were significantly lower, and skewness and kurtosis were significantly higher in the sarcoidosis population as compared to the controls. For all GLCM features (contrast, autocorrelation, energy, and homogeneity), the group effect was not significant pre-registration (p>0.05), but was significant post-registration (p<0.05). This indicates that registration to a standard lung template can improve our ability to find significant effects in radiomics, since we reduce noise, thereby enhancing signal, in our registered images.
Effect of Lobe Region on Radiomic Analyses
Significant differences between sarcoidosis and controls were observed for nearly all radiomic features and lung lobes (Table 4). Further, these group effects significantly differed across lobe regions for the standard deviation (p<0.0001), skewness (p=0.0002), kurtosis (p<0.0001), contrast (p=0.0105) and autocorrelation (p=0.0086). There were no significant lobe effects for the mean (p=0.3943), energy (p=0.0873) or homogeneity (p=0.0559). For standard deviation and skewness, the magnitude of the group effect was largest in the upper lobe of the right lung, and smallest in the lower lobe of the left lung. For kurtosis, the magnitude of the group effect was largest in the upper lobes of the left and right lungs, and smallest in the lower lobe of the left lung. For contrast and autocorrelation, the magnitude of the group effect was largest in the middle lobe of the right lung, and smallest in the lower lobe of the left lung. These results indicate that there are regional lobe differences in radiomics between sarcoidosis and healthy controls.
Table 4.
Differences in radiomic values between control and sarcoidosis subjects by lobe. Negative values indicate lower average radiomic values in the healthy controls as compared to the sarcoidosis population. Results are presented as mean (95% CI). P-values indicate whether the group effect changed significantly across lobes, with significant results bolded.
| Outcome | Left Upper Lobe | Left Lower Lobe | Right Upper Lobe | Right Middle Lobe | Right Lower Lobe | P-value |
|---|---|---|---|---|---|---|
| Mean | −11.39 (−19.66, −3.12) | −11.08 (−19.35, −2.81) | −12.53 (−20.80, −4.26) | −7.10 (−15.37, 1.18) | −10.55 (−18.82, −2.28) | 0.3943 |
| SD | −15.58 (−20.08, −11.09) | −9.14 (−13.64, −4.65) | −17.03 (−21.52, −12.53) | −13.45 (−17.95, −8.96) | −11.38 (−15.87, −6.88) | <0.0001 |
| Skew | 0.63 (0.46, 0.79) | 0.42 (0.26, 0.59) | 0.64 (0.48, 0.81) | 0.60 (0.44, 0.77) | 0.49 (0.32, 0.65) | 0.0002 |
| Kurtosis | 6.67 (5.19, 8.15) | 3.90 (2.41, 5.38) | 6.62 (5.14, 8.11) | 6.64 (5.15, 8.12) | 4.95 (3.46, 6.43) | <0.0001 |
| Contrast | 1.06 (0.30, 1.83) | 0.69 (−0.07, 1.46) | 0.91 (0.15, 1.68) | 1.41 (0.64, 2.18) | 0.79 (0.03, 1.56) | 0.0105 |
| Autocorrelation | −0.87 (−1.26, −0.48) | −0.61 (−1.00, −0.22) | −0.74 (−1.13, −0.35) | −0.97 (−1.36, −0.58) | −0.62 (−1.01, −0.23) | 0.0086 |
| Energy (x1000) | −0.44 (−0.67, −0.20) | −0.25 (−0.49, −0.01) | −0.39 (−0.62, −0.15) | −0.43 (−0.66, −0.19) | −0.32 (−0.55, −0.08) | 0.0873 |
| Homogeneity (x100) | −1.01 (−1.76, −0.26) | −0.60 (−1.35, 0.15) | −0.83 (−1.58, −0.08) | −1.22 (−1.97, −0.47) | −0.72 (−1.47, 0.03) | 0.0559 |
Discussion
In this study, we introduced a straightforward open-source pipeline for processing lung CT data in R that does not require visual reads. We applied this pipeline to 62 CT scans to create a publicly available, unbiased template of the lung from a healthy, non-Hispanic adult population in the United States. We also explored the influence of registration on the behavior of radiomic features, as well as performing a regional lobe radiomic analysis using a population of sarcoidosis patients and healthy controls.
For our pre-processing pipeline, we chose to implement all steps in R statistical software to provide an open-source platform that is easily accessible to a broad-analytic group. Our results indicate that our lungct segmentation methodology in R performs as well as that from the VIDA software for healthy patients. We also show that the symmetric normalization registration from ANTsR is flexible enough to register lung masks between people. Further, this automated pre-processing pipeline (segmentation, registration, and template creation) was robust to a diseased population of patients with sarcoidosis.
To ensure we reached an unbiased template of the lung, we used an iterative algorithm until convergence to a common shape. Rather than relying on the previous recommendation of a fixed number of iterations31, we defined convergence by a DSC between successive iterations of 0.99 or greater. We found that a DSC of 0.99 or greater corresponds to an ASSD of <1 mm; as all images were resampled to 1 mm3, this suggests the average error in boundary identification is a sub-voxel distance. Additionally, similar three-dimensional measures (volume, surface area, length, depth, and width) were observed for all initial templates chosen, resulting in a less biased template. If a small and fixed number of iterations were used (<14), the resulting final template could have been markedly different, and would be dependent on the initial template chosen. Further, the volumetric measures of the final left and right lung template were consistent with the average volumetric measures prior to analysis, indicating our template creation approach preserves volume, surface area, depth, width, and height across the left and right lungs.
By applying the final transformations obtained from the registration of lung segmentations to the original CT scans, we were able to obtain individual CT scans in a common space. Since the intensity and texture on lung CT scans are important in studies regarding lung diseases, we wanted to understand the impact of registration on radiomic features of the lung. We found that registration significantly affects first-order and GLCM radiomic features, by shifting the distribution of HUs slightly to the left, decreasing variability across voxels, and changing the HU patterns on the CT scan to appear smoother. The differences in both the first-order and GLCM radiomics can be explained by the non-linear transformation coupled with the linear interpolation from the registration procedure. For original scans that are larger in volume than that of the template, the HU across voxels are concatenated down to a smaller number of voxels by linear interpolation, following the non-linear transformation. Conversely, for original scans that are smaller in volume than that of the template, the HU of individual voxels are expanded into multiple voxels to be similar to their surroundings. In both cases, the linear interpolation following the non-linear registration results in updated HU at each voxel, which are more similar to the mode of the distribution. Since the HU distribution on CT scans of the lung is right-skewed, this registration procedure will result in a more right-skewed distribution as values are pulled to the mode, increasing the peakedness (i.e. increases kurtosis) of the distribution as well as shifting the HU distribution to the left (i.e. decreasing the mean HU). Furthermore, since this interpolation averages our voxels, the HU patterns appear smoother on the registered CT, which explains the changes in the GLCM features.
However, these registration effects did not deter, but rather enhanced, our ability to find group effects in radiomics. Specifically, for all GLCM features, the group effect was not significant pre-registration, but was significant post-registration. By transforming all scans to a common space during the registration procedure, our results suggest that we reduce noise and enhance signal in the registered images, resulting in more sensitivity to detect differences.
Another advantage of having a standard lung template is our ability to perform regional level analyses. While we did not have, or create, lobe segmentations for our sarcoidosis population, we were able to perform radiomic analyses between sarcoidosis patients and healthy controls at each lobe by using our created lobe template as a mask for each scan in study-specific space. In this application, we found that there are regional differences in radiomics between sarcoidosis patients and healthy controls, with the largest differences found in the upper and middle lobes of the right lung, matching existing literature34. Similar regional analyses could be performed for other lung diseases using our lobe mask, or other regional masks that may be of interest, such as those identifying vessels, airways and/or parenchymal tissues.
While we developed an unbiased template of the lung, a standard lung atlas using different methodology than shown here has previously been developed by Li et al2. Our method differs from Li’s by (1) obtaining a larger population of healthy control patients (from N=20 to N=62), (2) using updated, fully-automatic, open-source software for segmentation, registration29, and template creation31, which allows the creation of study-specific lung templates1, and (3) freely providing our resulting standard lung template for public download to facilitate its use in practice (BLINDED URL).
Our study is limited by sample size. We used as many CT scans from healthy individuals as were available to us; however, we recognize that our sample size for template creation of N=62 is modest, and is comprised of mostly white, non-Hispanic persons. To generalize to more diverse populations, we encourage researchers to use our pipeline to create lung templates specific to the population under study. Further, our pipeline uses a simple methodology for lung segmentation that works well for healthy and sarcoidosis scans; however, we recognize that there are many lung diseases with unique pathology, making segmentation difficult. We are investigating an R interface with the Chest Imaging Platform35 to provide more segmentation methods in our lungct package. Also, our registration method is not anchored on any specific anatomy, which may affect the quality of registration. Since anatomic anchoring requires a visual-read, we opted against it in order to develop a fully-automated and time-efficient pipeline. However, we provided an average lobe template which can be used in future analyses to align lung fissures across individuals, thereby removing potential variation in HU due to misalignment of internal structures. Registration could also be performed by registering HU, which may result in better alignment, but at the cost of masking biological variability.
All results in this paper were obtained utilizing parallel processing on the Joint High-Performance Computing Exchange at Johns Hopkins Bloomberg School of Public Health. However, all code contained herein can also be implemented efficiently on standard personal computers with or without parallelization. For instance, on a MacBook Pro 2.9 GHz Intel Core i7 with 16 GB RAM, left/right lung segmentation via lungct R package is performed in approximately one minute, and SyN registration via ANTsR R package is performed on both the left and right lung masks in approximately ten minutes for each scan without parallelization. Times are for high-resolution CT scans with original dimension of roughly 512×512×500 voxels.
To conclude, we developed a fully-automated, open-source pipeline for processing lung CT data, that resulted in a publicly available, unbiased template of the lung. We also showed that this pipeline can improve our ability to find differences in radiomics, and be used to perform regional-level analyses. We believe that the standard lung template will enable researchers to perform whole-lung, population level analyses in a more objective and sensitive manner, resulting in a better understanding of lung diseases. The R package that implements our methodology, including the template data in NIfTI file format, is located at BLINDED URL and published on Neuroconductor at https://neuroconductor.org/package/lungct.
Supplementary Material
Acknowledgments
The project described was supported by Award Number U01 HL089897 and Award Number U01 HL089856 from the National Heart, Lung, and Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health. The COPDGene® project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Novartis, Pfizer, Siemens and Sunovion.
Additional data from the GRADS study was supported by Award Numbers: U01 HL112707, U01 HL112707, U01 HL112694, U01 HL112695, U01 HL112696, U01 HL112702, U01 HL112708, U01 HL112711, U01 HL112712.
The authors were also supported by Award Numbers R01 HL114587 and R01 HL142049 from the National Institutes of Health.
The authors would like to thank Dr. Ciprian Crainiceanu from Johns Hopkins Bloomberg School of Public Health (Baltimore, Maryland) and Dr. Tasha Fingerlin from National Jewish Health (Denver, Colorado) for their input and guidance throughout the project.
References
- 1.Evans AC, Janke AL, Collins DL, Baillet S. Brain templates and atlases. NeuroImage 2012;62:911–922. [DOI] [PubMed] [Google Scholar]
- 2.Li B, Christensen GE, Hoffman EA, McLennan G, Reinhardt JM. Establishing a normative atlas of the human lung: computing the average transformation and atlas construction. Acad Radiol 2012;19:1368–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feuerstein M, Glocker B, Kitasaka T, Nakamura Y, Iwano S, Mori K. Mediastinal atlas creation from 3-D chest computed tomography images: application to automated detection and station mapping of lymph nodes. Med Image Anal 2012;16:63–74. [DOI] [PubMed] [Google Scholar]
- 4.Dura E, Domingo J, Ayala G, Marti-Bonmati L, Goceri E. Probabilistic liver atlas construction. Biomed Eng OnLine 2017;16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong C, Chen Y-W, Foruzan AH, Lin L, Han X-H, Tateyama T, Wu X, Xu G, Jiang H. Segmentation of liver and spleen based on computational anatomy models. Comput Biol Med 2015;67:146–160. [DOI] [PubMed] [Google Scholar]
- 6.Good CD, Johnsrude IS, Ashburner J, Henson RNA, Friston KJ, Frackowiak RSJ. A Voxel-Based Morphometric Study of Ageing in 465 Normal Adult Human Brains. NeuroImage 2001;14:21–36. [DOI] [PubMed] [Google Scholar]
- 7.Honea R, Crow TJ, Passingham D, Mackay CE. Regional Deficits in Brain Volume in Schizophrenia: A Meta-Analysis of Voxel-Based Morphometry Studies. Am J Psychiatry 2005;162:2233–2245. [DOI] [PubMed] [Google Scholar]
- 8.Busatto GF, Diniz BS, Zanetti MV. Voxel-based morphometry in Alzheimer’s disease. Expert Rev Neurother 2008;8:1691–1702. [DOI] [PubMed] [Google Scholar]
- 9.Bigos KL, Weinberger DR. Imaging genetics—days of future past. NeuroImage 2010;53:804–809. [DOI] [PubMed] [Google Scholar]
- 10.Ashburner J, Friston KJ. Voxel-based morphometry--the methods. NeuroImage 2000;11:805–821. [DOI] [PubMed] [Google Scholar]
- 11.Ashburner J, Hutton C, Frackowiak R, Johnsrude I, Price C, Friston K. Identifying global anatomical differences: deformation-based morphometry. Hum Brain Mapp 1998;6:348–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Royen FS, Moll SA, van Laar JM, van Montfrans JM, de Jong PA, Mohamed Hoesein FAA. Automated CT quantification methods for the assessment of interstitial lung disease in collagen vascular diseases: A systematic review. Eur J Radiol 2019;112:200–206. [DOI] [PubMed] [Google Scholar]
- 13.Hua K-L, Hsu C-H, Hidayati SC, Cheng W-H, Chen Y-J. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets Ther 2015;8:2015–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin H-C, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging 2016;35:1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88. [DOI] [PubMed] [Google Scholar]
- 16.Dercle L, Ammari S, Bateson M, Durand PB, Haspinger E, Massard C, Jaudet C, Varga A, Deutsch E, Soria J-C, Ferté C. Limits of radiomic-based entropy as a surrogate of tumor heterogeneity: ROI-area, acquisition protocol and tissue site exert substantial influence. Sci Rep 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krizhevsky A, Sutskever I, Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, eds. Advances in Neural Information Processing Systems 25 Curran Associates, Inc.; 2012. p. 1097–1105. [Google Scholar]
- 18.Li B, Christensen GE, Hoffman EA, McLennan G, Reinhardt JM. Establishing a normative atlas of the human lung: intersubject warping and registration of volumetric CT images. Acad Radiol 2003;10:255–265. [DOI] [PubMed] [Google Scholar]
- 19.Mansoor A, Bagci U, Foster B, Xu Z, Papadakis GZ, Folio LR, Udupa JK, Mollura DJ. Segmentation and image analysis of abnormal lungs at CT: current approaches, challenges, and future trends. RadioGraphics 2015;35:1056–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy K, Van Ginneken B, Reinhardt JM, Kabus S, Ding K, Deng X, Cao K, Du K, Christensen GE, Garcia V, others. Evaluation of registration methods on thoracic CT: the EMPIRE10 challenge. IEEE Trans Med Imaging 2011;30:1901–1920. [DOI] [PubMed] [Google Scholar]
- 21.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJWL, Dekker A, Fenstermacher D, Goldgof DB, Hall LO, Lambin P, Balagurunathan Y, Gatenby RA, Gillies RJ. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic Epidemiology of COPD (COPDGene) Study Design. COPD J Chronic Obstr Pulm Dis 2011;7:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zach JA, Newell JD Jr, Schroeder J, Murphy JR, Curran-Everett D, Hoffman EA, Westgate PM, Han MK, Silverman EK, Crapo JD, others. Quantitative CT of the lungs and airways in healthy non-smoking adults. Invest Radiol 2012;47:596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovalev V, Prus A, Vankevich P. Mining lung shape from x-ray images. International Workshop on Machine Learning and Data Mining in Pattern Recognition Springer; 2009. p. 554–568. [Google Scholar]
- 25.Melo LC, da Silva MAM, do N Calles AC. Obesity and lung function: a systematic review. Einstein Sao Paulo 2014;12:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muschelli J dcm2niir: Conversion of DICOM to NIfTI Imaging Files Through R. 2018.
- 27.Kazerooni EA, Gross BH. Cardiopulmonary imaging Lippincott Williams & Wilkins; 2004. [Google Scholar]
- 28.Avants BB, Tustison N, Song G. Advanced normalization tools (ANTs). Insight J 2009;2:1–35. [Google Scholar]
- 29.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 2008;12:26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattes D, Haynor DR, Vesselle H, Lewellen TK, Eubank W. PET-CT image registration in the chest using free-form deformations. IEEE Trans Med Imaging 2003;22:120–128. [DOI] [PubMed] [Google Scholar]
- 31.Avants BB, Yushkevich P, Pluta J, Minkoff D, Korczykowski M, Detre J, Gee JC. The optimal template effect in hippocampus studies of diseased populations. Neuroimage 2010;49:2457–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moller DR, Koth LL, Maier LA, Morris A, Drake W, Rossman M, Leader JK, Collman RG, Hamzeh N, Sweiss NJ, Zhang Y, O’Neal S, Senior RM, Becich M, Hochheiser HS, Kaminski N, Wisniewski SR, Gibson KF, GRADS Sarcoidosis Study Group. Rationale and Design of the Genomic Research in Alpha-1 Antitrypsin Deficiency and Sarcoidosis (GRADS) Study. Sarcoidosis Protocol. Ann Am Thorac Soc 2015;12:1561–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolossváry M, Kellermayer M, Merkely B, Maurovich-Horvat P. Cardiac Computed Tomography Radiomics: A Comprehensive Review on Radiomic Techniques. J Thorac Imaging 2018;33:26–34. [DOI] [PubMed] [Google Scholar]
- 34.Ryan SM, Fingerlin TE, Mroz M, Barkes B, Hamzeh N, Maier LA, Carlson NE. Radiomic Measures from Chest HRCT Associated with Lung Function in Sarcoidosis. Eur Respir J 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.San Jose Estepar R, Ross JC, Harmouche R, Onieva J, Diaz AA, Washko GR. Chest imaging platform: an open-source library and workstation for quantitative chest imaging C66. LUNG IMAGING II: NEW PROBES AND EMERGING TECHNOLOGIES American Thoracic Society; 2015. p. A4975–A4975. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


