Abstract
Active herbal or natural compounds have high chemical diversity and specificity than synthetic drugs. Recently, we have validated the hypoglycemic salutation of Oncocalyx glabratus in rodent model, and demonstrated the activation of PPARα/γ by its newly ioslated flavan derivative Oncoglabrinol C (5,3′-Dihydroxyflavan 7-4′-O-digallate) in liver cells (HepG2). Here we evaluated the potential of Oncoglabrinol C against Dichlorofluorescin (DCFH) and Methylglyoxal (MGO) induced endothelial cells (HUVEC) oxidative and apoptotic damage, including activation of PXR-mediated hepatic CYP3A4. Our MTT assay showed protection of ~57% and ~63.5% HUVEC cells by 10 and 20 μg/ml doses of Oncoglabrinol C, respectively through attenuating DCFH triggered free-radicals. Also, the two doses effectively protected ~53% and ~65.5% cells, respectively by reversing MGO toxicity. In DCFH and MGO treated cells, Oncoglabrinol C (20 μg/ml) effectively downregulated caspase 3/7 activity by ~33% and ~43.5%, respectively. Moreover, in reporter gene (dual-luciferase) assay, Oncoglabrinol C (20 μg/ml) moderately activated hepatic CYP3A4. Molecular docking of Oncoglabrinol C indicated its strong interactions with cellular caspase 3/7, PPARα/γ and PXR proteins, which supported its anti-apoptotic (antagonistic) as well as pro-hypoglycemic and PXR/CYP activating (agonistic) activities. Taken together, our findings demonstrated the potential of Oncoglabrinol C in reversing the endothelial oxidative and apoptotic damage as well as in the activation of hepatic CYP3A4. This warrants further evaluations of Oncoglabrinol C and related compounds towards developing effective and safe drugs against diabetes associated cardiovascular disorders.
Keywords: Oncocalyx glabratus, Oncoglabrinol C, Apoptosis, Caspase, CYP3A4
1. Introduction
In recent times, herbal medications have gained much popularity due to the global recognition of complementary and alternative medicine system. Plants derived natural compounds have high chemical diversity and biochemical specificity that often act more effectively than synthetic drugs (Clardy and Walsh, 2004, Ganesan, 2008). Oncocalyx glabratus belongs to Loranthaceae, the largest family of flowering parasitic plants commonly known as mistletoes (Calvin and Wilson, 2006), Mistletoes are regarded as ‘cure all’ medicinal plants worldwide (Adodo, 2004), including Saudi Arabia (Collenette, 1999). Phytochemical analysis of O. glabratus has evinced presence of flavonoids, terpenoids, steroids, tannis and reducing sugars (Waly et al., 2012). Endorsing the traditional use O. glabratus in diabetic patients (Obatomi et al., 1994), we have shown its hypoglycemic salutation in rodent model (Ahmed et al., 2015).
The peroxisome proliferator-activated receptor (PPAR) activator isoforms, PPARα and PPARγ are known to effectively lower the levels of blood sugar and lipids, respectively (Mirza et al., 2019). In addition, PPARα and PPARγ also are reported to be involved in the anti-inflammatory actions of several nonsteroidal anti-inflammatory drugs (Desvergne and Wahli, 1999, Flevét et al., 2006). Therefore, PPAR is considered as important targets towards developing effective drugs, including natural products against diabetes and associated cardiovascular disorders. In line with this, we have recently reported isolation of Oncoglabrinol C (5,3′-Dihydroxyflavan 7-4′-O-digallate), a new flavan derivative from O. glabratus that showed marked activation of both PPARα and PPARγ in cultured human liver cells (Ahmed et al., 2017).
Moreover, the in vivo cytotoxic effect of endogenous methylglyoxal (MGO) is known to mediate via oxidative stress and apoptosis (Kalapos, 2008). In the clinical cases of type 2 diabetes, elevation in plasma MGO is considered as one of the causative factors in hyperglycemia-associated macrovascular diseases (Sena et al., 2012). The endothelial cells (EC), the inner linings of blood vessels play an important role in modulating cardiovascular function and homeostasis (Choy et al., 2001). MGO has been shown to trigger hyperglycemia and apoptosis in cultured human EC, suggesting its prominent role in diabetic cardiovascular complications (Bourajjaj et al., 2003). In line with this, we have very recently reported a new proanthocyanidin from Rhus tripartita that ameliorated MGO-induced apoptosis of EC in vitro (Alqahtani et al., 2019).
The cytochrome P450 family enzyme CYP3A4 is crucial in metabolizing several known drugs, xenobiotics and bioactive natural or herbal products via activation of nuclear pregnane X receptor (PXR) (Al-Dosari and Parvez, 2018). Owing to the herb/drug associated adverse effects or organ toxicity, a good understanding of CYP3A4 modulatory activity of a herbal product is necessary (Parvez and Rishi, 2019). Taken together in the present study, we have extended the anti-glycemic analysis of O. glabratus derived Oncoglabrinol C, and assessed its in vitro therapeutic potential against oxidative and apoptotic damages in endothelial cells, including cytochrome 450 (CYP3A4) modulating activity in liver cells.
2. Materials and methods
2.1. Extraction, isolation and structure elucidation of the compounds
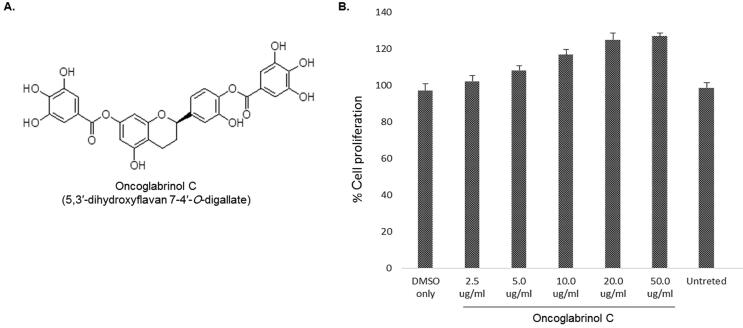
The extraction and isolation of the compound Oncoglabrinol C (C29H22O13) from the aerial parts of O. Glabratus along with structure elucidation (Fig. 1A) have been reported by us previously (Ahmed et al., 2017).
Fig. 1.
(A) Chemical structure of O. glabratus derived Oncoglabrinol C (C29H22O13) and (B) MTT assay showing dose-dependent cell proliferative/growth stimulatory activity of Oncoglabrinol C on cultured human endothelial cell (HUVEC). Values on Y-axis: means of three determinations.
2.2. Cell culture, reagents and drugs
The human primary umbilical vein endothelial cells (HUVEC 16549; Cat# PCS-100-010; ATTCC, USA) and hepatoblastoma cells (HepG2; Cat# HB-8065; ATTCC, USA) were maintained in DMEM-GlutMax media (Cat# 41966-029; Gibco, USA), supplemented with bovine serum (10%; Cat# 10270; Gibco, USA) and penicillin-streptomycin mix (×1; Cat# 15240-062; Invitrogen,; USA) at 37 °C with 5% CO2 supply. For all experiments, cells (0.5 × 105/well) were grown overnight in 96-well flat-bottom culture plates. Dichlorofluorescin (DCFH), the inducer of oxidative cell damage, Methylglyoxal (MGO; Cat# M0252-25ML; Sigma-Alderich, Germany), the standard pro-apoptotic agent and Aminoguanidine (AG; Cat# 1937-19-5; Sigma-Alderich, Germany), the anti-apoptotic drug and Rifampicin (RMP; Cat# R3501; Sigma-Alderich, Germany), the PXR agonist were purchased.
2.3. Compounds preparations
Stock of Oncoglabrinol C was prepared by first dissolving in 50 μl dimethyl sulfoxide (DMSO; Cat# 67-68-5; Sigma, Germany), and then in complete DMEM-GlutMax media (1 mg/ml, final). Further working concentrations (50, 20, 10, 5, and 2.5 μg/ml) were reconstituted in complete media. Similarly prepared AG (0.05 mM) (Alqahtani et al., 2019) and RMP (10 μM) (Al-Dosari and Parvez, 2018) served as positive controls whereas DCFH (CC50 = 50 μg/ml) and MGO (0.5 mM) (Alqahtani et al., 2019) acted as negative controls. DMSO (0.1%) was included as a vehicle control or untreated control.
2.4. Cell proliferation or growth stimulation assay
The cell proliferative/growth stimulatory efficacy of Oncoglabrinol C was tested on cultured HUVEC cells, using MTT assay (TACS MTT Cell Proliferation Assay Kit, Cat# 4890-25-K; Tervigen, USA). Briefly, cells were treated with the triplicated doses of Oncoglabrinol C, including DMSO and MGO controls for 72 h. The MTT reagent (10 μl/well) was added and cells were incubated at RT for 4 h in dark, followed by treating with detergent solution (100 μl/well) after appearance of purple color. The culture was further incubated at 37 °C for 1.5 h, and the absorbance (λ = 570) was measured (Microplate reader ELx800; BioTek, USA). Data were analyzed for in relation to the untreated control, using equation [% Cell proliferation= (As − Ab)/(Ac − Ab)x 100, where As, Ab and Ac represented the absorbance of sample, blank and negative control, respectively], and presented (Excel software, Microsoft, USA). The experiment was repeated twice for reproducibility.
2.5. Cell protective assay
Assessment of cytoprotective activities of the selected doses of Oncoglabrinol C (5.0, 10.0 and 20.0 μg/ml) against DCFH and MGO induced injury was performed. In a duplicated set of HUVEC culture, the first set was co-treated with DCFH (50 μg/ml) and a dose of Oncoglabrinol C or AG (0.05 mM), including all controls in triplicate. The second set was co-treated with MGO (0.5 mM) and a dose of Oncoglabrinol C or AG (0.05 mM), including all controls in triplicate. After 2 days of incubation, MTT assay (TACS MTT Cell Proliferation Assay Kit, Tervigen, USA) was performed to determine the cell survival (%), as above. The assay was repeated twice for reproducibility.
2.6. Cellular anti-apoptotic signaling (caspase-3/7) assay
HUVEC cells were co-treated with DCFH (50 μg/ml) or MGO (0.5 mM) and the maximally active dose of Oncoglabrinol C (20.0 μg/ml), including all controls in triplicate. On day 2 post-incubation, caspase 3/7 activity (Apo-ONE-cas3/7 Assay Kit; Cat# G7792; Promega, USA) was measured as per the manual. Briefly, caspase-3/7 reagent was added (100 μl/well) to the cultures and mixed by gentle rocking, following incubation for ~6 hr at RT in dark. The data was analyzed in relation to the untreated control and presented. The assay was repeated twice for reproducibility.
2.7. PXR-mediated hepatic CYP3A4 activation assay
The PXR-dependent CYP3A4 activation property of Oncoglabrinol C was assessed in PXR-luciferase (pCDG-hPXR), CYP3A4-luciferase (pGL3-CYP3A4-XREM) as well as control (pRL) plasmids co-transfected HepG2 cells as described previously (Al-Dosari and Parvez, 2018). Briefly, 24 h post-transfection, cells were treated with two doses (10 and 20 μg; in triplicate) of Oncoglabrinol C or RMP (10 μM; positive control) or DMSO (0.1%; vehicle or negative control), and incubated for another 24 h. Cell lysates were prepared and subjected to luminescence measurement (Dual-Luciferase Reporter Assay System; Cat# E1910; Promega, USA) and presented as fold-expression of CYP3A4 gene in relation to negative control.
2.8. Computational analysis
2.8.1. Preparation of ligands
For molecular docking, the structure of Oncoglabrinol C (ligand) was drawn on 2D SKETCHER (Schrodinger-2018, LLC, NY, USA) and optimized along with control ligands: N-[(2S)-4-chloro-3-oxo-1-phenyl-butan-2-yl]-4-methyl-benzenesulfonamide (TQ8); 5-fluoro-1 h-indole-2-carboxylic acid-(2-mercapto-ethyl)-amide (FICA); (2S)-2-ethoxy-3-[4-(2-{4-[(methylsulfonyl)oxy] phenyl}ethoxy)phenyl] propanoic acid (AZ242); (2S)-(4-isopropylphenyl)[(2-methyl-3-oxo-5,7-dipropyl-2,3-dihydro-1,2-benzisoxazol-6-yl)oxy] acetate (C01) and 3-[(4-Methyl-1-piperazinyl)imino]methyl- rifamycin (RMP), using LIGPREP (Schrodinger-2018, LLC, NY, USA). The bond orders and angles were assigned to Oncoglabrinol C and the 2D structure was converted to a 3D structure followed by energy minimization, using OPLS3e (Optimized potential for Liquid Simulations) forcefield. Further, EPIK (Schrodinger-2018, LLC, NY, USA) was used to generate its ionization states (pH 7.0 ± 2.0) and a maximum of 32 conformations were generated.
2.8.2. Preparation of cellular proteins
The molecular docking of Oncoglabrinol C with caspase 3, caspase 7, PPARα, PPARγ and PXR were performed using different modules of Schrodinger suite in MAESTRO interface (Schrodinger-2018, LLC, NY, USA) as described previously (Rehman et al., 2019). Briefly, the 3D coordinates of caspase 3 (PDB: 2XYG) (Rajkumaret al., 2011), caspase 7 (PDB: 1SHL) (Hardy et al., 2004), PPARα (PDB: 1I7G) (Cronet et al., 2001), PPARγ (PDB: 1ZEO) (Shi et al., 2005) and PXR (PDB: 1SKX) (Chrencik et al., 2005) were retrieved (www.rcsb.org). All four cellular proteins were pre-processed using PROTEIN PREPARATION WIZARD (Schrodinger-2018, LLC, NY, USA) by adding hydrogen atoms, assigning bond orders, and removing water molecules as well as any other heterogeneous molecules. The proteins structures were further refined by addition of any missing side-chains or loops (PRIME; Schrodinger-2018, LLC, NY, USA), and creating hydrogen bond networks (pH 7.4), followed by energy minimization (OPLS3e forcefield).
2.8.3. Grid generation and molecular docking
For docking, the grids were generated by selecting the bound ligand of each protein as the centroid of the grid box (caspase 3: 64 × 64 × 64 Å, caspase 7: 64 × 64 × 64 Å, PPARα: 72 × 72 × 72 Å, PPARγ: 72 × 72 × 72 Å and PXR: 72 × 72 × 72 Å), using GRID GENERATION TOOL (Schrodinger-2018, LLC, NY, USA). Docking of caspase, PPAR and PXR proteins with Oncoglabrinol C and control ligands were performed with extra-precision (XP) docking algorithm, using GLIDE (Schrodinger-2018, LLC, NY, USA). The ligand’s docking affinity (Kb) for each protein was estimated from docking energy (ΔG) using the relation: , where R and T were Boltzmann gas constant (1.987 cal/mol/K) and temperature (298 K), respectively (Rehamn et al., 2016).
2.9. Statistical analysis
All data (in triplicate) were presented as the mean ± SE and analyzed (one-way ANOVA), using SPSS software V. 25 (IBM, USA). The statistical differences between the control and treated samples were carried out using Tukey-Kramer post hoc analysis.
3. Results
3.1. Endothelial cell proliferative activity of Oncoglabrinol C
Five concentrations (2.5, 5, 10, 20 and 50 μg/ml) of Oncoglabrinol C were tested for their effects on HUVEC cells viability and proliferation. Compared to the insignificant effect of 2.5 μg/ml dose, the 5 μg/ml had growth stimulation activity by ~8% (Fig. 1B). On the other hand, while the 10 μg/ml dose moderately enhanced (~17%) the cell growth, 20 μg/ml dose showed the maximal cell proliferative activity by ~25%. However, treatment with 50 μg/ml dose did not contribute significantly (Fig. 1B).
3.2. Oncoglabrinol C protects HUVEC cells from oxidative damages
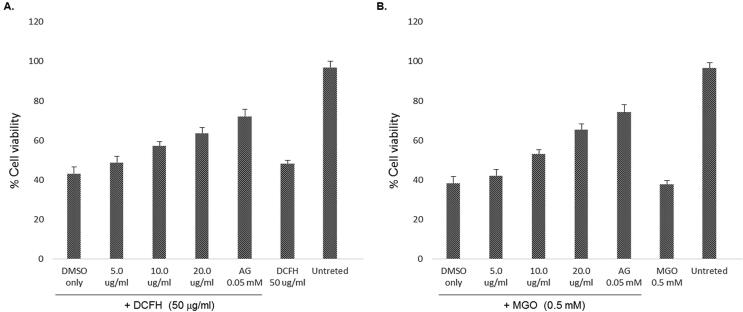
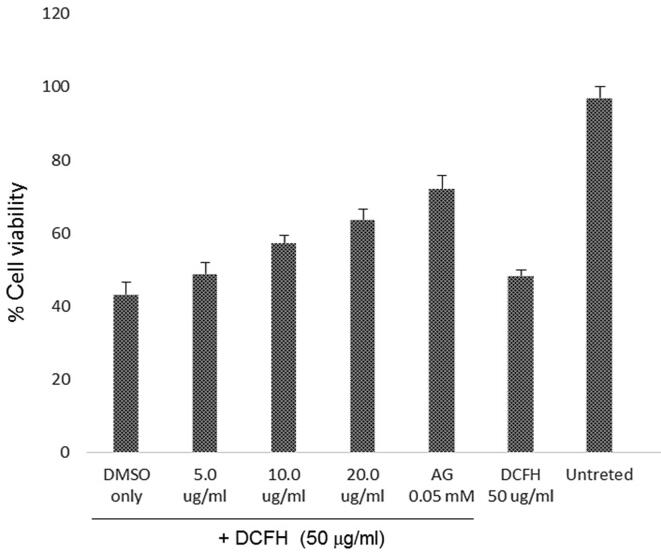
Based on the growth stimulatory activities, three doses (5, 10 and 20 μg/ml) of Oncoglabrinol C were further tested for cytoprotective potential against DCFH toxicity. Our MTT assay showed DCFH induced 49% cell death that was attenuated by Oncoglabrinol C treatment in a dose-dependent manner (Fig. 2A). While the 5 μg/ml dose had insignificant effect on 51% cell survival, treatment with 10 and 20 μg/ml doses of Oncoglabrinol C, protected ~57% and ~63.5% cells, respectively. Compared to these, AG protected the cell proliferation by ~72% against DCFH injury (Fig. 2A).
Fig. 2.
The MTT assay showing cytoprotective efficacy of Oncoglabrinol C against (A) Dichlorofluorescin (DCFH) induced oxidative stress and (B) Methylglyoxal (MGO) triggered apoptosis in cultured human endothelial cells (HUVEC). Values on Y-axis: means of three determinations.
3.3. Oncoglabrinol C protects HUVEC cells from apoptotic damages
Oncoglabrinol C tested at 5, 10 and 20 μg/ml doses showed a dose-dependent cytoprotection against MGO induced 52% apoptosis (Fig. 2B). While the 5 μg/ml dose of Oncoglabrinol C showed insignificant effect on ~42% surviving cells, treatment with 10 and 20 μg/ml doses protected ~53% and ~65.5% cells, respectively. Comparatively, AG restored ~74% cell proliferation via attenuation of MGO (Fig. 2B).
3.4. Oncoglabrinol C downregulates cellular caspases
To further investigate the possible anti-apoptotic mechanism of Oncoglabrinol C towards HUVEC cells protection, its maximally active dose (20 μg/ml) was tested against DCFH and MGO, individually. Our data showed ~74% DCFH-induced and ~88% MGO-induced caspase 3/7 activity that were effectively downregulated to ~41% and ~44.5%, respectively by Oncoglabrinol C (Fig. 3). AG treatment suppressed caspase 3/7 to 53% and 48.5% against DCFH and MGO, respectively.
Fig. 3.
The anti-apoptotic assay showing inhibition of Dichlorofluorescin (DCFH) and Methylglyoxal (MGO) induced cellular caspase-3/7 expressions by Oncoglabrinol C (Onco C; 20 μg/ml) in cultured human endothelial cells (HUVEC). Values on Y-axis: means of three determinations.
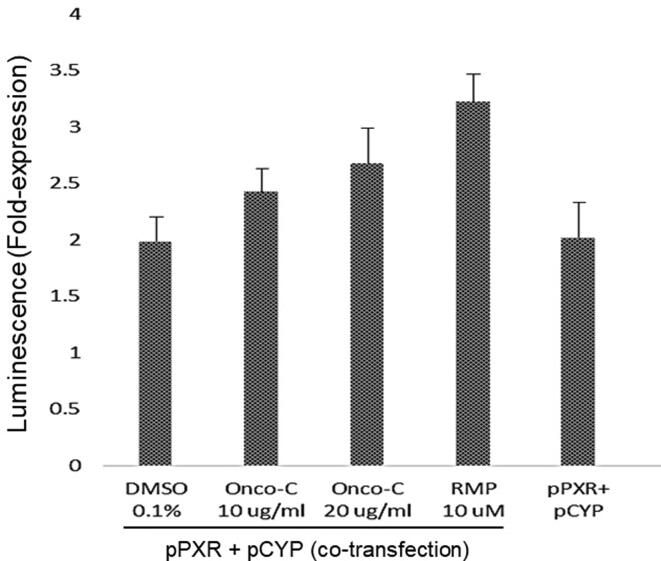
3.5. Oncoglabrinol C moderately activates hepatic CYP3A4
Our luciferase-reporter gene assay showed that Oncoglabrinol C at doses 10 and 20 μg/ml, moderately activated hepatic PXR-dependent CYP3A4 expression as compared RMP in HepG2 cells (Fig. 4). This is the first study on CYP3A4 stimulation by O. glabratus derived flavan derivative, suggesting its safe consumption in relation to drug metabolism.
Fig. 4.
The luciferase-reporter gene assay showing activation of hepatic CYP3A4 by Oncoglabrinol C (Onco C; 10 and 20 μg/ml) in cultured liver cells (HepG2). Positive control: Rifampicin (RMP; 10 μM). Vehicle control: DMSO (0.1%). Values on Y-axis: means of three determinations.
3.6. In silico molecular interactions of Oncoglabrinol C with cellular proteins
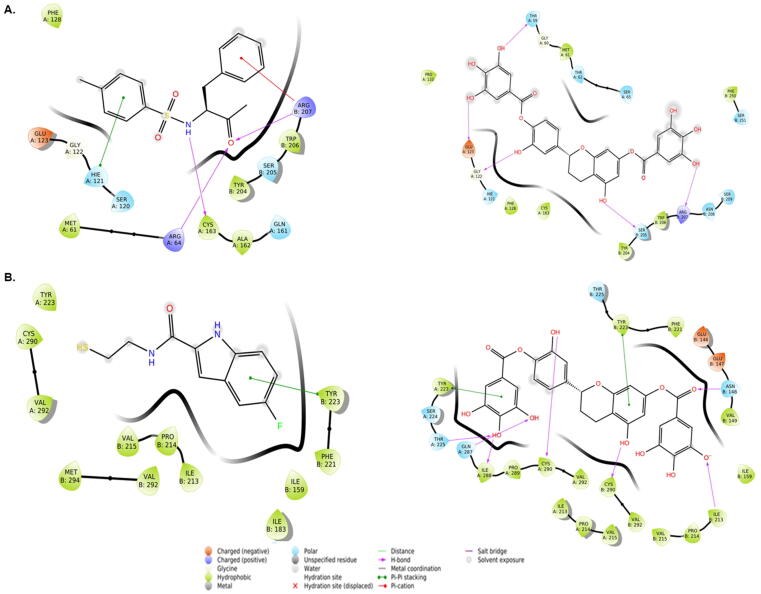
3.6.1. Interaction of caspase 3 with TQ8 (control antagonist) and Oncoglabrinol C
The docking of inhibitor ligand TQ8 revealed that the caspase 3-TQ8 complex was stabilized by hydrogen bonding with Arg64 and Cys163 of chain A, and Arg207 of chain B (Fig. 5A, left; Table 1). In addition, His121 of chain A and Arg207 of chain B were also involved in Pi-Pi stacking and Pi-cation interaction, respectively. The estimated docking energy and the corresponding docking affinity of TQ8 towards caspase 3 were −5.61 kcal mol−1 and 1.30 × 104 M−1 respectively. Likewise, Oncoglabrinol C was found to bind at the interface of chains A and B of caspase 3 through hydrogen bonding with Thr59, Gly122, and Glu123 of chain A, and Ser205 and Arg207 of chain B (Fig. 5A right, Table 1). In addition, some polar residues (Thr62, Ser65 and Hie121 of chain A, and Asn208, Ser209 and Ser251 of chain B) also participated in the interaction. The calculated docking energy and the corresponding docking affinity of Oncoglabrinol C towards caspase 3 were −6.90 kcal mol−1 and 1.15 × 105 M−1, respectively. Interestingly, some caspase 3 residues (Met61, Hie121, Gly122, Gln123, Phe128 and Cys163 of chain A, and Tyr204, Ser205, Trp206 and Arg208 of chain B) were found to interact with both TQ8 and Oncoglabrinol C.
Fig. 5.
The in silico molecular docking analysis (Ligplot) showing interaction of Oncoglabrinol C with (A) control antagonist TQ8 (left) and caspase 3 (right), and (B) control antagonist FICA (left) and caspase 7 (right).
3.6.2. Interaction of caspase 7 with FICA (control antagonist) and Oncoglabrinol C
The docking of FICA with caspase 7 revealed that the caspase 7-FICA complex was stabilized by one Pi-Pi interaction with Tyr223 of chain B (Fig. 5B, left; Table 2). In addition to this, several other residues (Tyr223, Cys290 and Val292 of chain A, and Ile159, Ile183, Ile213, Pro214, Val215, Phe221, Val292, and Met294 of chain B) also participated in stabilizing caspase 7-FICA complex. The docking energy and the corresponding docking affinity of FICA towards caspase 7 were estimated to be −7.31 kcal mol−1 and 2.42 × 105 M−1, respectively. Similarly, Oncoglabrinol C was found to bind at the interface of chains A and B of caspase 7 through hydrogen bonding with Thr225, Gln287, Ile288 and Cys290 of chain A, and Asn148, Ile213 and Cys290 of chain B (Fig. 5B, right; Table 2). In addition, both Tyr223 of chain A and B participated in a Pi-Pi interaction with Oncoglabrinol C. The estimated docking energy and the corresponding docking affinity of Oncoglabrinol C towards caspase 7 were −8.79 kcal mol−1 and 2.80 × 106 M−1. Notably, there were some common residues of caspase 7 (Tyr223, Cys290 and Val292 of chain A, and Ile159, Ile213, Pro214, Val215, Phe221, Tyr223 and Val292 of chain B) that interacted with both FICA and Oncoglabrinol C.
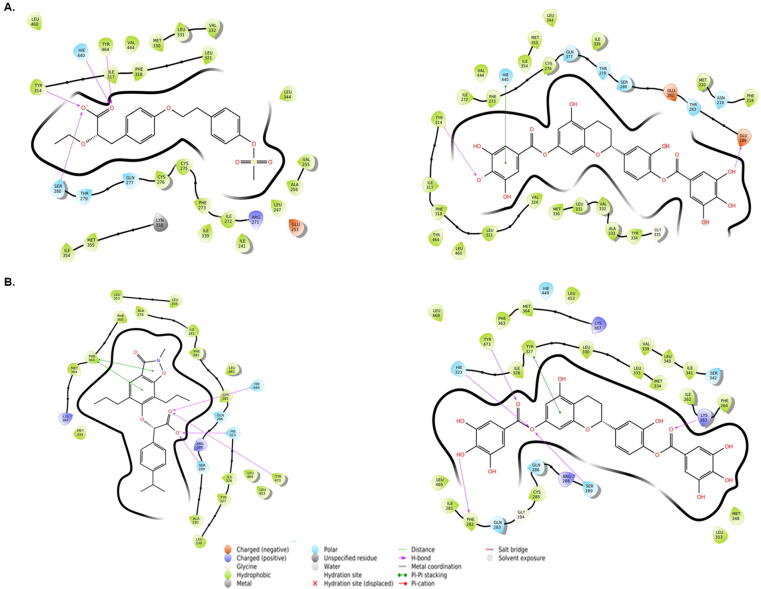
3.6.3. Interaction of PPARα with AZ242 (control agonist) and Oncoglabrinon-C
AZ242 is a dihydro-cinnamate derived agonist which binds LBD of PPARα. Our molecular docking results well collaborated with the AZ242-PPARα interaction X-ray crystallographic data on hydrogen bonding with Ser280, Tyr314, Hie440 and Tyr464 (Fig. 6A, left; Table 3). In addition, other charged and polar residues (Glu251, Arg271, Gln277 and Thr279) also participated in stabilizing AZ242-PPARα complex. The estimated docking energy and the corresponding docking affinity of AZA242 towards PPARα were −10.85 kcal mol−1 and 9.08 × 107 M−1, respectively. Similarly, Oncoglabiranol C was found to bind the central cavity of PPARα through hydrogen bonding with Glu286 and Tyr314 residues (Fig. 6B, right; Table 3). The residue His440 formed a Pi-Pi stacking interaction with Oncoglabiranol C while some charged and polar residues such as Asn219, Gln277, Thr279, Ser280, Glu282 and Thr283 also interact with Oncoglabrinol C. The calculated docking energy and the corresponding docking affinity of Oncoglabrinol C towards PPARα were −7.54 kcal mol−1 and 3.39 × 105 M−1, respectively. Interestingly, there were some common amino acid residues (Ile272, Phe273, Cys276, Ser280, Tyr314, Ile317, Phe318, Leu321, Met330, Leu331, Val332, Ile339, Leu344, Met355, Hie440, Val444 and Leu460) that interacted with both AZ242 and Oncoglabrinol C. Notably, in the X-ray crystal structure of PPARα-AZ242 complex, Ser280, Tyr314, His440 and Tyr464 residues were shown critical for the binding of the agonist (Cronet et al., 2001).
Fig. 6.
The in silico molecular docking analysis (Ligplot) showing interaction of Oncoglabrinol C with (A) control agonist AZ242 (left) and PPARα (right), and (B) control agonist C01 (left) and PPAR γ (right).
3.6.4. Interaction of PPARγ with C01 (control agonist) and Oncoglabrinol C
The docking analysis revealed that the C01-PPARγ complex was stabilized by hydrogen bonding with Ser289, Hie323, Hie449 and Tyr473 (Fig. 6B, left; Table 4). In addition, while other charged and polar residues (Gln286, Lys367 and Arg288) involved in the interaction, Phe363 formed two Pi-Pi stacked interactions with C01. The docking energy and the corresponding docking affinity of C01 towards PPARγ gamma were estimated to be −13.23 kcal mol−1 and 5.05 × 109 M−1 respectively. Similarly, Oncoglabrinol C was found to bind the central cavity of PPARγ Lys263, Phe282, Ser289, Hie323 and Tyr473 residues through hydrogen bonding (Fig. 6B, right; Table 4). Also, while Tyr327 formed a Pi-Pi stacking interaction, some charged and polar residues (Arg288, Lys367, Gln283, Gln286, Ser342 and His449 also interacted with Oncoglabrinol C. The estimated docking energy and the corresponding docking affinity of Oncoglabrinol C towards PPARγ were −14.76 kcal mol−1 and 6.69 × 1010 M−1, respectively. Interestingly, the were some common residues of PPARγ (Ile281, Cys285, Gln286, Ser289, Hie323, Ile326, Leu330, Met334, Leu353, Met364, Hie449, Leu453, Leu465, Leu469, and Tyr473) that interacted with both C01 and Oncoglabrinol C. Notably, in the X-ray crystal structure of PPARγ-C01 complex, Ser289, His323, His449 and Tyr473 were critical for the binding of the agonist (Cronet et al., 2001).
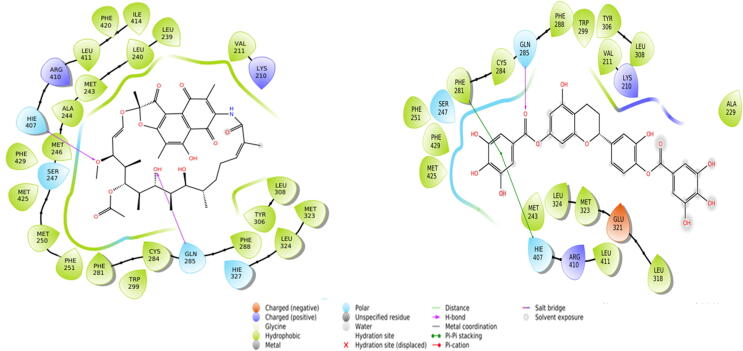
3.6.5. Interaction of PXR with RMP (control agonist) and Oncoglabrinol C
RMP, a macrolide antibiotic is a known ligand activator of PXR. The docking nalysis revealed that the RMP-PXR complex was stabilized by hydrogen bonding with Gln285 (Fig. 7, left; Table 5). In addition, while other charged and polar residues (Lys210, Ser247, His327 and Arg410) were also involved in the RMP-PXR interaction, RMP also formed one Pi-Pi interaction with PXR His407 residue. Several other hydrophobic residues also participated in stabilizing the ligand-protein complex. The estimated docking energy and affinity of RMP towards PXR were −6.84 kcal mol−1 and 1.04 × 105 M−1, respectively. Similarly, Oncoglabrinol C was found to bind the agonist site of PXR through hydrogen bonding with Gln285 (Fig. 7, right; Table 5). While PXR residues Phe281 and Hie407 formed Pi-Pi stacking interactions, some charged and polar residues (Lys210, Ser247, Glu321 and Arg410) also interacted with Oncoglabrinol C. Several other hydrophobic residues also stabilized the complex. The estimated docking energy and affinity of the stabilized complex were −5.31 kcal mol−1 and 7.86 × 104 M−1, respectively. Interestingly, some PXR residues (Val211, Met243, Phe251, Cys284, Phe288, Trp299, Tyr306, Leu308, Met323, Leu324, Leu411, Met425, Phe429) were commonly involved in interactions with both RMP and Oncoglabrinol C.
Fig. 7.
The in silico molecular docking analysis (Ligplot) showing interaction of Oncoglabrinol C with control agonist RMP (left) and PXR (right).
4. Discussion
Isolated natural compounds often perform better than randomly synthesized drugs that originate from a limited number of parent molecules with a much lower steric complexity (Clardy and Walsh, 2004, Ganesan, 2008). In addition, since natural active compounds have high chemical diversity and biochemical specificity, they offer great promise as potentially effective new drugs. O. glabratus, globally known as ‘cure all’ medicinal plant is also used to treat diabetic patients (Obatomi et al., 1994). Recently we have experimentally validated its hypoglycemic potential in rodent model of diabetes [8] as well as reported isolation of a new flavan Oncoglabrinol C as a potent activator of PPARα and PPARγ in liver cells (Ahmed et al., 2017). Moreover, in hypergycemic conditions, elevated level of MGO is known to induce oxidative stress and apoptosis of EC and associated macrovascular diseases (Sena et al., 2012, Bourajjaj et al., 2003). In the present study, we have therefore, assessed the in vitro anti-oxidative and anti-apoptotic potential of Oncoglabrinol C in human endothelial cells.
The cellular reactive oxygen species (ROS) are the highly toxic products of redox reactions of endogenous or exogenous sources (Opara, 2006) The accumulating excess of cellular ROS can damage lipids, proteins or nucleic acids, and inhibit their normal growth and function and promote tissue damages. In experimental settings, DCFH is generally used to estimate in vitro free-radicals induced oxidative stress through the oxidation of DCFH into the fluorescent DCF (Rota et al., 1999). We therefore, used DCFH to induce oxidative cytotoxicity in HUVEC cells and assessed the anti-oxidative activity of different concentrations of Oncoglabrinol C. Our data showed a dose-depend salutation of Oncoglabrinol C where the 10 and 20 μg/ml doses protected ~57% and ~63.5% cells, respectively through attenuating DCFH triggered ROS. Further, owing to the diabetic cardiovascular complications, we assessed Oncoglabrinol C activity toward reversing the MGO induced HUVEC apoptosis. Of the tested doses, 10 and 20 μg/ml of Oncoglabrinol C effectively protected ~53% and ~65.5% cells, respectively via amelioration of MGO associated cell damage.
Cellular caspases are a class of cysteine-aspartate proteases that play crucial roles in maintaining cellular homeostasis by via apoptosis and inflammation (Kumar, 2006). On stimulation, pro-caspases are activated by dimerization/oligomerization following cleavage into a small and a large sub-unit that interact with each other to form an active heterodimer caspase with two active sites located in close proximity to the dimer interface (Shi, 2004). Therefore, the inhibition of caspases presents a promising therapeutic intervention in apoptotic cell and tissue damage in acute and chronic diseases. In order to have an insight into the possible molecular mechanism involved in anti-oxidative and anti-apoptotic salutations of the most active dose (20 μg/ml) of Oncoglabrinol C, we therefore, measured the caspase 3/7 expressions in DFCH or MGO treated HUVEC cells. Our data showed that Oncoglabrinol C effectively downregulated caspase 3/7 activity to ~41% and ~44.5% in DCFH and MGO injured cells, respectively. This was in line with our previous observation on R. tripartita derived Rhuspartin, a new proanthocyanidin that ameliorated MGO-induced apoptosis of HUVEC cells (Alqahtani et al., 2019). To further strengthen our in vitro observation, we carried out in silico molecular docking of Oncoglabrinol C with caspase 3 and 7 along with their antagonistic ligand controls TQ8 and FICA, respectively. Alike TQ8, Oncoglabrinol C strongly interacted with caspase 3 chain A (Met61, Hie121, Gly122, Gln123, Phe128 and Cys163) and chain B (Tyr204, Ser205, Trp206 and Arg208) residues through hydrogen bonding. Similarly, like FICA, Oncoglabrinol C also made a stable complex with caspase 7 chain A (Tyr223, Cys290 and Val292) and chain B (Ile159, Ile213, Pro214, Val215, Phe221, Tyr223 and Val292) residues through hydrogen bonding. In addition, some other residues were also involved in Pi-Pi and Pi-cation interactions to further stabilize the Oncoglabrinol C and caspase 3 and 7 complexes.
PPARα and PPARγ, the ligand-activated transcription factors regulate energy metabolism, glucose homeostasis, cell proliferation as well as tissue inflammation (Mirza et al., 2019, Michalik et al., 2006). The 3D structure of PPAR consists of five distinguished regions/domains namely N-terminal region, DNA-binding domain (DBD), flexible hinge region, ligand binding domain (LBD) and C-terminal region. It is reported that the function of PPAR is controlled by the specific conformation of LBD, which in turn can be induced by the binding of its natural ligand and various agonists and antagonists (Zoete et al., 2007). We have recently shown marked hypoglycemic activity of Oncoglabrinol C through PPARα and PPARγ activations in cultured liver cells (Ahmed et al., 2017). To further support this in vitro observation, we also docked Oncoglabrinol C with PPARα and PPARγ along with their agonistic controls AZ242 and C01, respectively. In line with the control ligands, Oncoglabrinol C showed strong interactions with the LBD residues of both PPARα and PPARγ through hydrogen bonding. Notably, there were some common residues of PPAR that participated in both control ligands and Oncoglabrinol C interactions.
PXR up-regulates the expression of CYP3A4, which metabolizes more than 50% of drugs in response to xenobiotics (Quatrochi and Guzelian, 2001, Al-Dosari and Parvez, 2018). Since medicinal herbs or natural products are often orally consumed, their high phytoconstituents contents may potentially affect the CYP3A4 activity leading to herb/drug non-response or organ toxicity (Parvez and Rishi, 2019). In this report therefore, we also tested PXR-dependent CYP3A4 activation potential of Oncoglabrinol C using reporter gene assay. Our data revealed that Oncoglabrinol C moderately activated CYP3A4 in cultured liver cells, suggesting its safe consumption in relation to drug metabolism and efficacy. To support this in vitro observation, docking of Oncoglabrinol C with PXR along with its agonistic controls RMP was performed. Oncoglabrinol C showed strong interactions with the agonist site of PXR through hydrogen bonding. Notably, there were some common residues of PXR that participated in RMP and Oncoglabrinol C interactions.
5. Conclusions
Our findings demonstrated in vitro therapeutic salutation of Oncoglabrinol C, a novel flavan derivative from O. glabratus in the reversal of DCFH and MGO induced oxidative stress apoptosis in endothelial cells though downregulation of caspase 3/7 activity. This was further endorsed by in silico docking that showed strong interaction of Oncoglabrinol C with cellular caspase 3/7 and PPARα/γ proteins, indicating its antagonistic (anti-apoptotic) and agonistic (pro-hypoglycemic) activity, respectively.
This is consistent with the multiple known mechanisms involved in oxidative cell damaging and apoptosis. Also, this is the first report on hepatic CYP3A4 activation by O. glabratus derived Oncoglabrinol C, suggesting its efficacy and safe consumption. Nonetheless, this warrants further evaluations of chemically related natural compounds towards developing effective drugs against diabetes associated cardiovascular disorders.
Acknowledgments
Acknowledgement
The authors thankfully acknowledge the Deanship of Scientific Research, King Saud University, Riyadh for funding this work (Project No RG-1435-053).
Declaration of Competing Interest
All authors declare no conflict of interest.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2020.04.004.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Adodo, A., 2004. Nature Power: A Christian Approach to Herbal Medicine. third ed. Benedictine Publication Nigeria/Generation Press Ltd., Lagos/Surulere.
- Ahmed S., Al-Rehaily A.J., Ahmad M.S., Yousaf M., Nur-e-Alam M., Parvez M.K., Al-Dosari M.S., Noman O.M., Khan S.I., Khan I.A. Chemical constituents from Oncocalyx glabratus and their biological activities. Phytochem. Lett. 2017;20:128–132. [Google Scholar]
- Ahmed S., Mothana R.A., Al-Rehaily A.J. In-vivo hypoglycemic and anti-diabetic study of Oncocalyx glabratus. Bangladesh J. Pharmacol. 2015;10:830–835. [Google Scholar]
- Al-Dosari M.S., Parvez M.K. Novel plant inducers of PXR-dependent cytochrome P4503A4 expression in HepG2 cells. Saudi Pharm J. 2018;26:1069–1072. doi: 10.1016/j.jsps.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqahtani, A.S., Abdel-mageed., W,M., Shahat, A.A., Parvez, M.K., Al-Dosari, M.S., Malik, A., Abdel-Kader, M.S., Alsaid, M.S., 2019. Proanthocyanidins from the stem bark of Rhus tripartita and their amelioration of methylglyoxal-induced apoptosis of endothelial cells. J. Food Drug Anal. 27, 358–365. [DOI] [PMC free article] [PubMed]
- Bourajjaj M., Stehouwer C., Van Hinsbergh V., Schalkwijk C. Role of methylglyoxal adducts in the development of vascular complications in diabetes mellitus. Biochem. Soc. Trans. 2003;31:1400–1402. doi: 10.1042/bst0311400. [DOI] [PubMed] [Google Scholar]
- Calvin C.L., Wilson C.A. Comparative morphology of epicortical roots in old and new world Loranthaceae with reference to root types, origin, patterns of longitudinal extension and potential for clonal growth. Flora. 2006;20:51–64. [Google Scholar]
- Chrencik J.E., Orans J.O., Moore L.B., Xue Y., Peng L., Collins J.L., Wisely G.B., Lambert M.H., Kliewer S.A., Redinbo M.R. Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol. Endocrinol. 2005;19:1125–1134. doi: 10.1210/me.2004-0346. [DOI] [PubMed] [Google Scholar]
- Choy J.C., Granville D.J., Hunt D.W., McManus B.M. Endothelial cell apoptosis: biochemical characteristics and potential implications for atherosclerosis. J. Mol. Cell. Cardiol. 2001;33:1673–1690. doi: 10.1006/jmcc.2001.1419. [DOI] [PubMed] [Google Scholar]
- Clardy J., Walsh C. Lessons from natural molecules. Nature. 2004;432:829–837. doi: 10.1038/nature03194. [DOI] [PubMed] [Google Scholar]
- Collenette, S., 1999.Wild Flowers of Saudi Arabia. National Commission for Wild Life Conservation and Development, Kingdom of Saudi Arabia.
- Cronet P., Petersen J.F., Folmer R., Blomberg N., Sjoblom K., Karlsson U., Lindstedt E.L., Bamberg K. Structure of the PPAR-alpha and -gamma ligand binding domain in complex with AZ 242; ligand selectivity and agonist activation in the PPAR family. Structure. 2001;9:699–706. doi: 10.1016/s0969-2126(01)00634-7. [DOI] [PubMed] [Google Scholar]
- Desvergne B., Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- Flevét C., Fruchart J.C., Staels B. PPARα and PPARγ dual agonists for the treatment of type 2 diabetes and the metabolic syndrome. Curr. Opin. Pharmacol. 2006;6:606–614. doi: 10.1016/j.coph.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Ganesan A. The impact of natural products upon modern drug discovery. Curr. Opin. Chem. Biol. 2008;12:306–317. doi: 10.1016/j.cbpa.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Hardy J.A., Lam J., Nguyen J.T., O'Brien T., Wells J.A. Discovery of an allosteric site in the caspases. PNAS. 2004;101:12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalapos M.P. The tandem of free radicals and methylglyoxal. Chem. Biol. Interact. 2008;171(251–27):1. doi: 10.1016/j.cbi.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2006;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- Michalik, L., Auwerx, J., Berger, J.P., Chatterjee, V.K., Glass, C.K., Gonzalez, F,J., Grimaldi, .PA., Kadowaki, T., Lazar, M.A., O'Rahilly, S,, Palmer, C.N., Plutzky, J., Reddy, J.K., Spiegelman, B.M., Staels, B., Wahli, W., 2006. International union of pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol. Rev. 58,726–741. [DOI] [PubMed]
- Mirza A.Z., Althagafi I.I., Shamshad H. Role of PPAR receptor in different diseases and their ligands: Physiological importance and clinical implications. Eur. J. Med. Chem. 2019;166:502–513. doi: 10.1016/j.ejmech.2019.01.067. [DOI] [PubMed] [Google Scholar]
- Obatomi D.K., Bikomo E.O., Temple V.J. Anti-diabetic properties of the African mistletoe in streptozotocin-induced diabetic rats. J. Ethnopharmacol. 1994;3:13–17. doi: 10.1016/0378-8741(94)90111-2. [DOI] [PubMed] [Google Scholar]
- Opara E.C. Oxidative stress. Dis. Mon. 2006;2:183–198. doi: 10.1016/j.disamonth.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Parvez, M.K., Rishi, V., 2019. Herb-drug interactions and hepatotoxicity. Curr Drug Metab. 20, 275–282. [DOI] [PubMed]
- Quatrochi L.C., Guzelian P.S. CYP3A regulation: from pharmacology to nuclear receptors. Drug Met. Disposition. 2001;29:615–622. [PubMed] [Google Scholar]
- Rajkumar G., Stjepan J., Peer R.E.M., Amedeo C., Markus G.G. In silico identification and crystal structure validation of caspase-3 inhibitors without a P1 aspartic acid moiety. Acta Crystallogr. A. 2011;F67:842–850. doi: 10.1107/S1744309111018604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehamn M.T., Ahmed S., Khan A.U. Interaction of meropenem with ‘N’ and ‘B’ isoforms of human serum albumin: a apectroscopic and aolecular docking Study. J. Biomol. Struct. Dyn. 2016;34:1849–1864. doi: 10.1080/07391102.2015.1094411. [DOI] [PubMed] [Google Scholar]
- Rehman M.T., AlAjmi M.F., Hussain A., Rather G.M., Khan M.A. High-throughput virtual screening and Molecular dynamics simulation identified ZINC84525623 a potential inhibitor of NDM-1. Int. J. Mol. Sci. 2019;20:819. doi: 10.3390/ijms20040819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota C., Chignell C.F., Mason R.P. Evidence for free radical formation during the oxidation of 2’-7’-dichlorofluorescin to the fluorescent dye 2’-7’-dichlorofluorescein by horseradish peroxidase: possible implications for oxidative stress measurements. Free Radical Biol. Med. 1999;27:873–881. doi: 10.1016/s0891-5849(99)00137-9. [DOI] [PubMed] [Google Scholar]
- Sena C.M., Matafome P., Crisóstomo J., Rodrigues L., Fernandes R., Pereira P., Seiça R.M. Methylglyoxal promotes oxidative stress and endothelial dysfunction. Pharmacol. Res. 2012;65:497–506. doi: 10.1016/j.phrs.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Shi G.Q., Dropinski J.F., McKeever B.M., Xu S. Design and synthesis of alpha-aryloxyphenylacetic Acid Derivatives: a novel class of PPAR alpha/gamma dual agonists with potent antihyperglycemic and lipid modulating activity. J. Med. Chem. 2005;48:4457–4468. doi: 10.1021/jm0502135. [DOI] [PubMed] [Google Scholar]
- Shi Y. Caspase activation. Cell. 2004;117:855–858. doi: 10.1016/j.cell.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Waly N.M., Ali AE El-Din, Jrais R.N. Botanical and biological studies of six parasitic species of family Loranthaceae growing in Kingdom of Saudi Arabia. Int. J. Environ. Sci. 2012;4:196–205. [Google Scholar]
- Zoete V., Grosdidier A., Michielin O. Peroxisome proliferator-activated receptor structures: ligand specificity, molecular switch and interactions with regulators. BBA. 2007;1771:915–925. doi: 10.1016/j.bbalip.2007.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.