Abstract
Nitrogen fixation is an important metabolic process carried out by microorganisms, which converts molecular nitrogen into inorganic nitrogenous compounds such as ammonia (NH3). These nitrogenous compounds are crucial for biogeochemical cycles and for the synthesis of essential biomolecules, i.e. nucleic acids, amino acids and proteins. Azotobacter vinelandii is a bacterial non-photosynthetic model organism to study aerobic nitrogen fixation (diazotrophy) and hydrogen production. Moreover, the diazotroph can produce biopolymers like alginate and polyhydroxybutyrate (PHB) that have important industrial applications. However, many metabolic processes such as partitioning of carbon and nitrogen metabolism in A. vinelandii remain unknown to date.
Genome-scale metabolic models (M-models) represent reliable tools to unravel and optimize metabolic functions at genome-scale. M-models are mathematical representations that contain information about genes, reactions, metabolites and their associations. M-models can simulate optimal reaction fluxes under a wide variety of conditions using experimentally determined constraints. Here we report on the development of a M-model of the wild type bacterium A. vinelandii DJ (iDT1278) which consists of 2,003 metabolites, 2,469 reactions, and 1,278 genes. We validated the model using high-throughput phenotypic and physiological data, testing 180 carbon sources and 95 nitrogen sources. iDT1278 was able to achieve an accuracy of 89% and 91% for growth with carbon sources and nitrogen source, respectively. This comprehensive M-model will help to comprehend metabolic processes associated with nitrogen fixation, ammonium assimilation, and production of organic nitrogen in an environmentally important microorganism.
Highlights
-
•
Genome-scale metabolic model of Azotobacter vinelandii DJ achives over 90% accuracy.
-
•
iDT1278 is the most comprehensive model to simulate diazotrophy.
-
•
Determining the most suitable culture conditions to produce polymers A. vinelandii.
-
•
Constraint-based modeling unravels links among nitrogen fixation and production of organic nitrogen.
1. Introduction
Azotobacter vinelandii is a gram-negative soil bacterium capable of converting atmospheric nitrogen gas (N2) into soluble ammonia (NH3) as well as into other important nitrogenous compounds (Gyurján et al., 1995; Howard and Rees, 1996). Azotobacter and related Azospirillium are estimated to fix up to 10–30% of the total nitrogen in the rhizosphere (Cleveland et al., 1999). Nitrogen fixation can be carried out under ambient conditions by any of the three highly specialized metal-dependent nitrogenases, referred to as molybdenum nitrogenase, vanadium nitrogenase, and iron-only nitrogenase (Setubal and Almeida, 2015; Sippel et al., 2017). Nitrogenases produce high concentrations of fixed ammonium, which is excreted and serves as essential nutrient for other organisms (Ambrosio et al., 2017; Tan et al., 2015). However, the activity of these enzymes is highly sensitive to molecular oxygen and energetically costly. Diazotrophs, such as A. vinelandii have developed specific strategies to protect the nitrogenase complex in diazotrophic conditions (Setubal and Almeida, 2015). One of the most studied mechanisms for nitrogenase protection is alginate biosynthesis. Alginate is transported to the extracellular space where it works as a barrier that decreases oxygen diffusion into the cytoplasm and thus maintains high functionality of oxygen-sensitive nitrogenases in anoxic environments (García et al., 2018; Pacheco-Leyva et al., 2016).
Alginate is of great industrial value because of its use as biocompatible and biodegradable exopolysaccharide. This polymer is used as gel-film-stabilizing, -thickening, or -forming agent in the food and pharmaceutical industry (Remminghorst and Rehm, 2006). Besides alginate bioproduction, A. vinelandii produces another attractive commercial polymer, i.e. polyhydroxybutyrate (PHB) (Yoneyama et al., 2015). PHB is synthetized by this microorganism under high carbon/nitrogen ratios as a carbon and energy reserve in the form of cysts (Stevenson and Socolofsky, 1966; Zúñiga et al., 2011). Both biopolymers can be produced in elevated concentrations, representing 30–70% of the dry biomass (Pacheco-Leyva et al., 2016; Yoneyama et al., 2015).
A. vinelandii has been shown to grow under a broad range of heterotrophic conditions and is able to metabolize, different sugars, alcohols, and organic acids as well as nitrogen-containing compounds (Nagai et al., 1972; Quiroz-Rocha et al., 2017; Sahoo et al., 2014; Shawky et al., 1987). Despite this metabolic versatility to use different carbon and nitrogen sources, several of the internal metabolic processes regarding carbon and nitrogen partitioning (division and distribution of an element into metabolic, structural or storage pools) in A. vinelandii remain unknown. Today there are five fully sequenced genomes available for A. vinelandii strains (e.g. A. vinelandii CA, DJ, CA6, DSM 279, and NBRC13581) (Noar et al., 2015; Setubal et al., 2009; Setubal and Almeida, 2015), enabling a comprehensive functional characterization of Azotobacter metabolism at genome-scale.
To comprehend the metabolic capabilities of Azotobacter vinelandii DJ we used a systems biology approach, which offers tools to predict the organism behavior based on mathematical representations of biological data. M-models can be reconstructed using semi-automated tools that generate a draft model. This draft model is further curated manually to increase its quality. To date, only two core M-models of Azotobacter vinelandii are available that contain a reduced number of metabolic reactions. These core reactions are in general related to nitrogen fixation or PHB and alginate production, disregarding most of the central metabolism of the microorganism (e.g. TCA cycle, lipid metabolism and some amino acids synthesis) (García et al., 2018; Inomura et al., 2018). Here we have developed a M-model for Azotobacter vinelandii DJ to contextualize metabolic processes associated with nitrogen fixation, ammonium assimilation, and production of organic nitrogen on genome-scale. Our model was successfully validated using high-throughput phenotypic data and physiological data.
2. Material and methods
2.1. Draft model generation
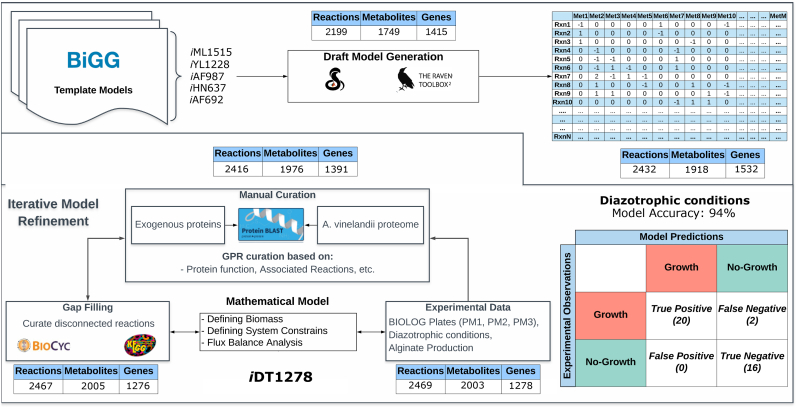
The draft model of A. vinelandii DJ was generated using The COBRA (Heirendt et al., 2019) and The RAVEN (Agren et al., 2013) Toolboxes. The proteome sequence was obtained from PATRIC database (Genome ID: 322710.5) and was used as input sequence to reconstruct the draft model based on protein homology. We selected five reference models as templates after alignment of the complete genome sequences of A. vinelandii DJ with all bacteria with available models in the BiGG Database (King et al., 2016). Templates included Escherichia coli str. K-12 substr. MG1655, model iML1515 (Monk et al., 2017), Klebsiella pneumoniae subsp. pneumoniae MGH 78578, model iYL1228 (Liao et al., 2011), Geobacter metallireducens GS-15, model iAF987 (Feist et al., 2014), Clostridium ljungdahlii DSM 13528, model iHN637 (Nagarajan et al., 2013), and Methanosarcina barkeri str. Fusaro, model iAF692 (Feist et al., 2006). Template models contained reactions associated with nitrogen fixation, H2 production, acetate consumption, amino acids catabolism and sugar degradation (Fig. 2). The generated draft model also contained genes (exogenous genes) from template models, which were later removed during the manual curation step.
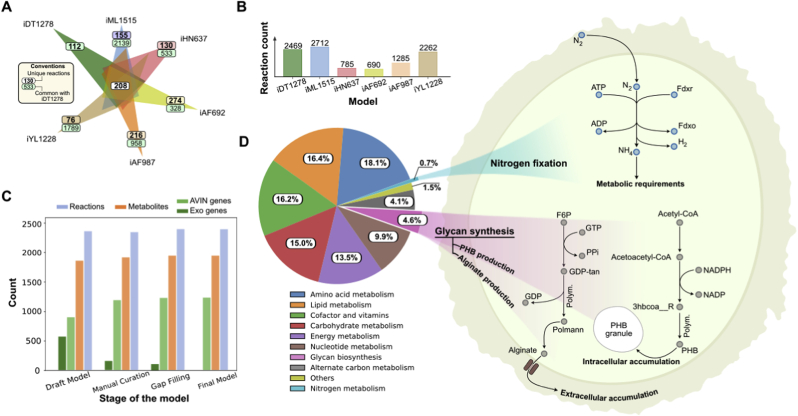
Fig. 2.
Characteristics of iDT1278. A) Comparison among the template models (Escherichia coli str. K-12 substr. MG1655, Klebsiella pneumoniae subsp. pneumoniae MGH 78578, Geobacter metallireducens GS-15, Clostridium ljungdahlii DSM 13528 and Methanosarcina barkeri str. Fusaro) and iDT1278 reactions. The six models share 208 core metabolic reactions. A. vinelandii DJ model contains 112 unique reactions related to aromatic compounds metabolism, alginate and PHB production, etc. B) Number of reactions in the template models and A. vinelandii DJ. C) Change in the number of reactions, metabolites and genes at the different stages of the reconstruction process/manual curation of A. vinelandii DJ. D) Reactions distribution through the subsystems in the genome-scale model; subsystems were grouped into 11 groups summarizing the complete metabolism of A. vinelandii DJ. Nitrogen fixation, PHB, and alginate production are highlighted in the condensed pathway diagrams. Alginate accumulation occurs in the extracellular space meanwhile PHB storage happens in the cytoplasm compartment. Nitrogen fixation in A. vinelandii can be performed by different specialized nitrogenases.
2.2. Model refinement
2.2.1. Manual curation
We used PATRIC (Wattam et al., 2017) to identify essential genes for A. vinelandii DJ in the final model. We only extracted those genes that had a given Enzyme Commission (EC) number that could be used to obtain the GPR (gene-protein reaction) associations. The final list of reactions with EC number and gene association not previously present in the model were balanced and added to the model before analyzing GPR associations.
Model refinement included two major steps: manual curation/review of the GPR associations and gap-filling by adding new metabolic reactions in the model. In the first step of manual curation, we determined sequence similarity among A. vinelandii DJ proteins and the exogenous proteins in the GPRs to identify A. vinelandii (AVIN) genes closely related to the exogenous proteins. We identified proteins based on BLASTp criteria of ≥ 40% identity, e-value ≤ 1e−4, and query coverage ≥ 85%. A second step of manual curation was performed based on protein function, type of metabolic reaction, and GPR associations. Then all the GPR associations were manually curated to catalyze biological reactions that they were associated with. PATRIC essential genes previously identified were added in this step of manual curation. Remaining reactions with mixed AVIN and exogenous genes in the GPR association were manually curated in order to remove the genes that did not belong to A. vinelandii. Reactions with exclusively exogenous GPR associations were identified through previous manual curation steps. Afterwards, Flux Balance Analysis (FBA) was performed to identify which of these reactions carry any flux under experimental conditions (Orth et al., 2010). From these evaluated reactions, those with no flux and exogenous GPR associations were removed from the model.
2.2.2. Gap-filling
Gap-filling analysis was performed to identify the metabolites disconnected in the model. These metabolites were classified depending on the number of reactions present in the model or their capability to be consumed, produced, or both. Disconnected reactions were manually curated using information from different bioinformatic databases (e.g. KEGG, Biocyc). From these results, gap-filling was used to connect pathways through the data retrieved. A second step of gap-filling was accomplished to connect the metabolites from the medium conditions retrieved using literature information (Wong and Maier, 1985) through algorithms to identify the reactions involved in the carbon source assimilation. A total of 38 carbon sources were used under nitrogen fixation and ammonium assimilation conditions. Complementary, experimental data were generated using Biolog plates to test different carbon and nitrogen sources. This was employed to improve the quality of model predictions under a wide variety of conditions. A set of 190 carbon sources and 95 nitrogen sources were used to connect the networks properly. Subsequently, the GPR associations were verified for each reaction added during the gap-filling to maintain the quality of the model. Those reactions with no gene information and literature validation were conserved as orphan reactions.
2.2.3. Final quality control and quality analysis
Final quality check was performed by a person who did not perform the manual curation to assess the quality of the data. We performed in-silico GPR simulations to verify if the GPR associations are correctly assigned using the COBRA Toolbox algorithms. Next, we performed Mass Balance simulations on the model to check for unbalanced reactions added during the model refinement. Ultimately, the final model was tested looking for ATP, NADH, and NADPH free energy production, removing exchange reactions, and calculating their accumulation.
2.3. Constraints and growth simulations
Experimental data from the literature were retrieved to calculate the initial medium constraints. For each growth condition, the carbon, nitrogen, and hydrogen fluxes were initially determined depending on every value obtained from the literature. The constraints related to mineral compounds and exchange reactions are summarized in Table S1. Initially, a set of six different conditions were used to measure the accuracy of the model. The carbon sources verified in this stage of validation were carbohydrates under nitrogen fixation or ammonium assimilation conditions. The simulation results were compared to this set of experimental values to identify the quality in the model predictions. Subsequently, 38 carbon sources under nitrogen fixation and H2 consumption conditions from the literature (Wong and Maier, 1985) were used to test and increase the quality of the model. The uptake rates were estimated from the experimental conditions and set for all the carbon sources; nitrogen and H2 uptake rates were not fixed to a specific value according to the experimental conditions. Finally, model benchmarking was performed for 190 carbon sources (Biolog plates PM1 and PM2) and 95 nitrogen sources (Biolog plates PM3) to validate model predictions. Biolog microplates experiments to test carbon and nitrogen assimilation were performed in the present work, measuring the growth rate values in the plate reader for 96 h. For carbon sources evaluation, ammonium assimilation was not fixed to a specific value (non-diazotroph conditions). The experimental results from Biolog plates were matched with data retrieved from the literature to determine and evaluate model precision during the simulations. During the nitrogen condition simulations, pyruvate was used as the unique carbon source. Statistical parameters were calculated according to the comparison between the metabolic predictions and the experimental values. The model accuracy from the Biolog plates results was compared with the in-silico predictions of the A. vinelandii model from CarveMe to identify the quality of the model simulations of the present work. The alginate production capability of the model was tested using four different carbon sources (carbohydrates) from the literature (Revin et al., 2018). The carbon compound uptake rates were calculated according to the experimental values. The simulations were performed initially setting ammonium as unique nitrogen source and subsequently molecular nitrogen was established as the unique nitrogen source. Furthermore, the predicted values were compared to determine which conditions allow a higher alginate production rate. For polyhydroxybutyrate (PHB) production, the metabolic internal fluxes for the principal pathways related to the PHB synthesis were calculated (glycolysis, pentose phosphate pathway, Entner-Doudorrff pathway, and TCA cycle) and compared with fluxomic data determined by Wu et al. (2019). In silico predictions were performed through FBA, using The COBRA Toolbox and the Gurobi Optimizer v.8.0.1 solver (Gurobi Optimization) for MATLAB (MathWorks). Percent error between experimental values and in silico results were calculated to obtain model accuracy.
2.4. Carbon and nitrogen partitioning analysis
Experimentally determined growth phenotypes using Biolog plates were used to validate predicted carbon and nitrogen flux distributions. The internal fluxes for all the reactions of the model were calculated in-silico for all the carbon sources (PM1 and PM2) experiments. The reactions were grouped in general subsystems that represented the complete metabolism of A. vinelandii DJ. Subsequently, an average flux per subsystem was calculated using the flux values of all the reactions belonging to the subsystem. This procedure was performed to calculate the carbon and nitrogen distribution in each general subsystem under diazotrophic and non-diazotrophic conditions. Ultimately, the carbon and nitrogen distributions (the grouped average fluxes per subsystem) were compared through a linear correlation analysis in order to determine how the fluxes change through the experimental conditions for carbon sources set in the Biolog plates. High linear correlation coefficients (r) indicate a similar distribution of a specific subsystem (above 0.9), while low coefficients suggest different carbon or nitrogen distributions when comparing diazotroph and non-diazotroph conditions.
3. Results
3.1. Metabolic network reconstruction of A. vinelandii DJ
We used a semiautomatic approach to reconstruct the M-model of A. vinelandii DJ (Fig. 1). This approach has been previously applied for the reconstruction of M-models (Zuñiga et al., 2016). First, a draft model of A. vinelandii DJ was reconstructed using the genome annotation from PATRIC (Genome ID: 322710.5). Five manually curated and validated M-Models were used as protein homology templates: Escherichia coli str. K-12 substr. MG1655 (Monk et al., 2017), Klebsiella pneumoniae subsp. pneumoniae MGH 78578 (Liao et al., 2011), Geobacter metallireducens GS-15 (Feist et al., 2014), Clostridium ljungdahlii DSM 13528 (Nagarajan et al., 2013), and Methanosarcina barkeri str. Fusaro (Feist et al., 2006). The RAVEN and COBRA Toolboxes (Agren et al., 2013; Heirendt et al., 2019) were used to generate the draft reconstruction. Each reaction in the draft model was evaluated for energy (ATP, NADH and NADPH accumulation) and mass balances as part of the quality control tests to guarantee model functionality and accuracy. Reactions associated with template genes were conserved in the first draft model to ensure model connectivity as well as the model’s capability to perform simulations. Nitrogen fixation and hydrogen consumption reactions were imported from the M-model templates. The resulting draft model contained 2,432 metabolic reactions and 1,918 metabolites divided into three different compartments (cytoplasm, periplasm, and extracellular space).
Fig. 1.
Workflow used to reconstruct a metabolic model of A. vinelandii DJ. A draft model was created from five template models present in BiGG (Escherichia coli str. K-12 substr. MG1655, Klebsiella pneumoniae subsp. pneumoniae MGH 78578, Geobacter metallireducens GS-15, Clostridium ljungdahlii DSM 13528 and Methanosarcina barkeri str. Fusaro). The RAVEN toolbox Agren et al., 2013 for MATLAB was used to create the draft model from stoichiometric data. The initial draft model contained 2,432 reactions, 1,918 metabolites, and 1,532 genes. The iterative process of model refinement included manual curation, gap-filling and curation using experimental data. The resultant final model contained A. vinelandii specific metabolic processes such as nitrogen fixation, and production of alginate and PHB. The final model, containing 2,469 reactions, 2,003 metabolites, and 1,278 genes, predicted with 94% accuracy.
3.1.1. Model refinement
Model refinement was performed using two principal steps: manual curation and gap filling. Every gene-protein-reaction (GPR) association was verified using multiple databases (e.g. KEGG, Biocyc, BRENDA, and MetaNetX) and available information from the literature. Manual curation was based on protein sequence similarity. The genes annotated in the GPR associations were aligned to protein sequences of A. vinelandii. Sequences, which passed the BLASTp parameters (see Methods), were assigned functionality based on information in the bioinformatics databases. The assigned genes in the GPR rules were replaced with the A. vinelandii genes (AVIN).
The original draft model consisted of 1,532 genes (Fig. 2A) corresponding to 934 AVIN genes and 598 genes from the template. At the end of the second manual curation, the AVIN genes increased to 1,233 and the total number of template genes was 102. Intuitively, as the level of curation increases the number of genes from template models decreases. When the genes in the model were curated by functionality, the number of genes from template models was zero.
3.1.2. Gap filling
After the manual curation, the total number of reactions and metabolites in the model was 2,416 and 1,976, respectively. We used literature information and experimental data from the Biolog plates results (PM1 and PM2 for carbon sources and PM3 for nitrogen sources) to add or remove reactions in the model. Each reaction added to the model in this step was manually reviewed to maintain concordance in the GPR associations. Overall, a total of 51 reactions and 29 metabolites (mostly reactions related to carbohydrate and amino acids catabolism) were added to the model during the gap filling process (Fig. 1, Fig. 2C). The reactions added to the model were mainly transport and interconversion reactions to connect the carbon or nitrogen sources with intracellular metabolites from the model.
3.1.3. Model properties
The Azotobacter vinelandii DJ metabolic model (iDT1278) consists of 2,003 metabolites, 2,469 reactions and 1,278 genes (around 26% of all annotated coding genes in the genome). Specific details about the reactions and metabolites from the model are summarized in Table S1 iDT1278 was validated using experimental data under nitrogen fixation (diazotroph) and ammonium assimilation (non-diazotroph) conditions. iDT1278 contains all the reactions and genes involved in nitrogen fixation, PHB, and alginate biosynthesis (Fig. 2B).
The properties of iDT1278 are shown in Fig. 2. Most of the reactions in the model belong to amino acid metabolism, lipid metabolism, and cofactor and vitamins metabolism (60% of total reactions of the model). Specific metabolic capabilities of A. vinelandii DJ such as nitrogen fixation (nitrogen metabolism), PHB and alginate production (glycan and secondary metabolites biosynthesis) represent around 3% of the metabolic reactions. Template models used during the reconstruction share 208 reactions with iDT1278. Most of the reactions (a total of 2,139 reactions) were taken from the first template (Escherichia coli K12 substr. MG1555, iML1515) (Fig. 2A). Nitrogen fixation and H2 consumption pathways were obtained from the templates iHN637(Clostridium ljungdahlii DSM 13528) and iAF987 (Geobacter metallireducens GS-15). iDT1278 shares 208 reactions among all the template models (Fig. 2C) which are related to core metabolic pathways (TCA cycle, oxidative phosphorylation, amino acids metabolism, etc.). Table 1 shows a comparison of the properties of the different metabolic models reconstructed for A. vinelandii. As a result, iDT1278 represents, to our knowledge, the most comprehensive M-model of the diazotroph A. vinelandii available to date. However, the A. vinelandii metabolic model from CarveMe (Machado et al., 2018) contains the closest number of reactions, metabolites and genes using the BiGG database information.
Table 1.
Comparison of the principal model properties (reactions, metabolites and genes) available for A. vinelandii.
| Model provenience | Reactions | Metabolites | Genes | Reference |
|---|---|---|---|---|
| Present work | 2,469 | 2,003 | 1,278 | Present work |
| Model SEED | 1,570 | 1,416 | 903 | Henry et al. (2010) |
| CarveMe | 2,422 | 1,978 | 1,395 | Machado et al. (2018) |
| García (Alginate and PHB production) | 46 | 39 | 0 | García et al. (2018) |
| Inomura (Nitrogen fixation) | 33 | 17 | 0 | Inomura et al. (2018) |
3.1.4. Biomass objective function
The biomass objective function (BOF) contains the principal constituents and the abundance of each metabolite involved in biomass production. The proportion of each metabolite participating in the BOF composition is determined per gram of biomass. iDT1278 includes two biomass reactions: 1) An initial BOF was obtained from the first template (Escherichia coli K12 substr. MG1555, iML1515) based on their physiological similarity (Gram-negative bacteria); the stoichiometric coefficients of the amino acids present in the BOF were calculated based on the theoretical amino acid abundance in the genome, using 55% of the biomass composition from amino acids. 2) A second BOF was determined from the first reaction to predict the alginate production since A. vinelandii DJ produces alginate only under specific metabolic conditions (Noar et al., 2015). The second BOF contains the same constituents present in the first BOF plus periplasmic alginate in order to simulate the complete metabolism and alginate production of A. vinelandii.
3.2. iDT1278 predicts accurately phenotypic experimental data
3.2.1. Growth rates validation in carbon and nitrogen sources
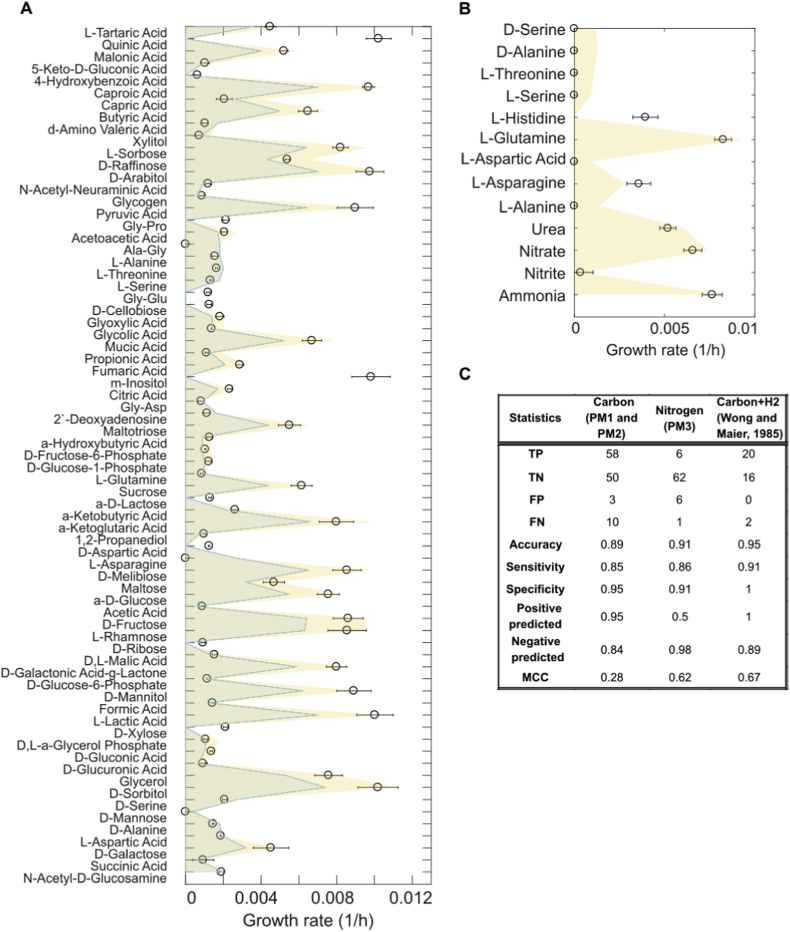
The model was validated under a wide range of different growth conditions (diazotrophic and non-diazotrophic growth), using high-throughput phenotypic data as well as literature information. Initially, iDT1278 was tested under six different experimental conditions, specifically, carbohydrates under diazotrophic and non-diazotrophic conditions (Table 2). The M-model predicted precisely the growth rates for all the carbon sources using ammonium or molecular nitrogen as nitrogen sources. For the carbon sources in non-diazotrophic conditions (sucrose, mannitol and glucose), the predicted growth rates are consistent with experimental values obtained from the literature, resulting in an average accuracy close to 95%. For example, the predicted growth using mannitol (uptake rate of 0.83 mmol/gDW/h) as sole carbon source was 0.0472 1/h, agreeing with the experimental data (0.045 ± 0.003 1/h). Average precision under nitrogen fixation conditions decreased significantly to 83%. Table 2 shows the comparison between experimental data from literature and predicted values for A. vinelandii DJ. Initial results showed higher model accuracy (12% more) when predicting growth rates using ammonium as nitrogen source compared to N2. Subsequently, flux balance analysis (FBA, Orth et al., 2010) was performed for a group of 38 carbon sources in diazotrophic and H2-consuming conditions (Wong and Maier, 1985). Statistical results show for the subset of 38 carbon sources (Fig. 3C) an accuracy of 95%, with 20 true positive predictions (100% positive predicted) and 16 true negative predicted results (89% negative predicted). Matthews correlation coefficient (MCC) was calculated under the conditions previously mentioned, obtaining a value of 0.67 (Fig. 3C). The false negative predictions obtained during the validation of aconitate and lactose are related to the absence of literature information about the enzymes which metabolize the carbon sources into familiar metabolites for the microorganism (e.g. the model lacks enzymes to convert lactose into glucose and fructose).
Table 2.
Predicted and experimental growth rates reported for A. vinelandii DJ under different carbon and nitrogen sources.
| Carbon source | Nitrogen source | Experimental value (1/h) | Predicted growth (1/h) | Reference |
|---|---|---|---|---|
| Glucose | Ammonium | 0.0505 | 0.0486 | Clementi (1997) |
| Mannitol | Ammonium | 0.045 ± 0.003 | 0.0472 | Revin et al. (2018) |
| Sucrose | Ammonium | 0.076 ± 0.004 | 0.07 | Díaz-Barrera et al. (2016) |
| Glucose | Nitrogen | 0.06 ± 0.0002 | 0.09 | Wong (1988) |
| Fructose | Nitrogen | 0.048 ± 0.002 | 0.0517 | Wong (1988) |
| Galactose | Nitrogen | 0.074 ± 0.007 | 0.065 | Wong (1988) |
Fig. 3.
Model validation using high-throughput phenotypic data for different carbon and nitrogen sources. A) Predicted growth values of A. vinelandii DJ in nitrogen fixation (N2 + Carbon source) or ammonium assimilation (NH4 + Carbon source) conditions using 121 carbon sources (PM1 and PM2 from Biolog Plates). However, only 71 carbon sources are shown in Fig. 3. B) Estimated growth for 75 nitrogen sources, nevertheless just 12 nitrogen compounds are presented in the graph (PM3 from Biolog Plates). Pyruvate was used to constrain the model as the unique carbon source during the nitrogen validation. C) statistical parameters and Matthews correlation coefficient of the predictions for the carbon (first column) and nitrogen sources (second column) using the Biolog Plates information; true positive (TP), true negative (TN), false positive (FP) and false negative (FN). Statistical analysis of the estimations for 38 carbon sources under nitrogen fixation and H2 consumption conditions (third column). The data used during the simulations were obtained from Wong and Maier (1985).
Additional experimental validation was performed using Biolog plates for a set of carbon (PM1 and PM2) and nitrogen (PM3) sources to determine the growth rate values of A. vinelandii DJ. Out of 190 carbon sources from the Biolog plates, 123 compounds were identified in the model; the simulations were performed under two specific conditions: diazotrophic and non-diazotrophic simulation conditions. However, experimental results were obtained only under growth with ammonium as the unique nitrogen source. The same procedure used in PM1 and PM2 experiments was followed to estimate the growth rates with 75 different nitrogen sources. For this case, simulations were performed using pyruvate as the carbon source. Fig. 3 shows the complete analysis of the experimental and predicted data for all carbon (Fig. 3A) and nitrogen sources (Fig. 3B); statistical parameters (accuracy, sensitivity, specificity, positive predicted, negative predicted and Matthews correlation coefficient) were calculated for non-diazotrophic conditions. Subsequently, the same experimental results from the Biolog plates were used to calculate the statistical parameters of the CarveMe model to establish a comparison between iDT1278 and the CarveMe metabolic model accuracy. Details of CarveMe predictions and statistical parameters using Biolog plates data are presented in Table S3.
For carbon sources validation, an accuracy of 89% was achieved with 58 true positive predictions (95% positive predicted) and 50 true negative estimations (84% negative predicted). For this subset of compounds, accuracy decreased significantly in negative predictions (10 false negative estimations). These negative prediction disagreements involve carbohydrates and some amino acids as carbon sources. Some false negatives appear to be related to the lack of evidence about required transporters. Simulated growth rate values in ammonium assimilation conditions were significantly higher than in the nitrogen fixation conditions (>26%). Higher accuracy was observed in both positive and negative predictions in iDT1278 compared to CarveMe simulations (global accuracy of 61%, 72% positive predicted and 54% negative predicted). These statistical results support the clear differences between the quality predictions of iDT1278 and other metabolic models available for A. vinelandii. Significant changes in the growth rate values were not observed when using amino acids as carbon sources since these organic molecules contain nitrogen, which provides an indirect supply of this element to the microbe. For the nitrogen source validation, an overall accuracy of 91% was accomplished, nonetheless, the number of non-growing conditions increased considerably in comparison with the carbon condition experiments (almost 91% of nitrogen experiments resulted in no growth), indicating a reduced capability to grow in a wide range of nitrogen sources.
3.3. Carbon and nitrogen partitioning analysis
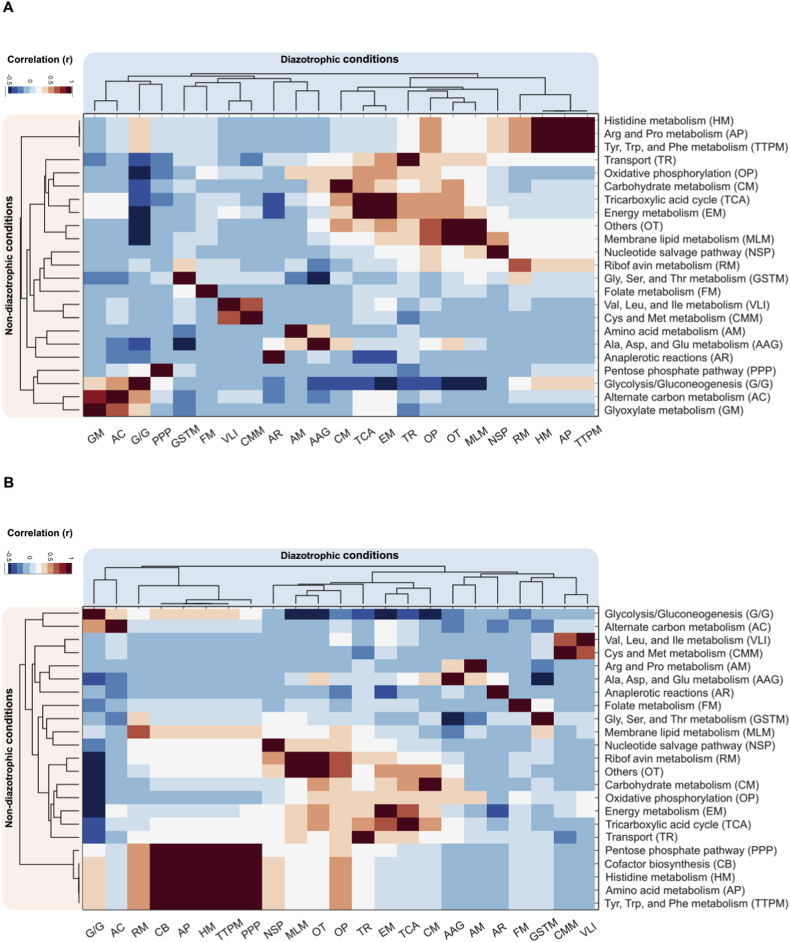
The average metabolic fluxes of carbon and nitrogen elements were determined for all carbon sources with growth rate values greater than 0.001 from Biolog plates data (61 total). The complete dataset with the experimental data is summarized in Table S2. The metabolic fluxes for both elements were grouped in 43 specific subsystems to identify the activity of the main pathways for all the experimental conditions. The highest average carbon fluxes (see Methods) were obtained from energy metabolism (82 mmol/gDW/h), oxidative phosphorylation (67.5 mmol/gDW/h), biomass and maintenance functions (36 mmol/gDW/h), and TCA cycle (25 mmol/gDW/h) for both nitrogen (N2 and NH4) conditions (disregarding transport fluxes). Similar tendencies were observed for the internal nitrogen flux distributions across the subsystems for diazotrophic and non-diazotrophic growth. However, the average flux distributions of carbon and nitrogen decline under diazotrophic conditions. Regarding global metabolic fluxes, the global carbon flux drops 4.9% and the nitrogen global flux value decreases 5.5%. The pathways with higher variation between carbon and nitrogen fluxes in diazotrophic and non-diazotrophic conditions were riboflavin metabolism (45%), glycine, serine and threonine metabolism (22%), alanine, aspartate and glutamate metabolism (26%), and nucleotide synthesis (10%). These subsystems are well-known for containing metabolites with high nitrogen content. The decline in the carbon and nitrogen global flux values of these pathways can be related to the low available nitrogen under diazotroph conditions due to the high energy cost of nitrogen fixation.
The grouped average fluxes of carbon and nitrogen elements per subsystem calculated for diazotrophic and non-diazotrophic conditions were compared through a linear correlation analysis to determine how the subsystem flux values behave under both nitrogen (N2 and NH4) conditions. Fig. 4 presents the linear correlations values between diazotrophic and non-diazotrophic conditions of the active subsystems. For carbon and nitrogen partitioning analysis, the highest correlation coefficients (Pearson correlation >0.95, p-value <1 × 10−30) were observed in all the amino acids pathways, biomass and maintenance functions, energy metabolism, carbohydrate metabolism, and some subsystems related to lipid metabolism (lipopolysaccharide biosynthesis and glycerophospholipid metabolism), demonstrating similar metabolic flux distributions through these specific subsystems when comparing diazotrophic and non-diazotrophic conditions. However, weak correlation values were obtained in riboflavin metabolism (r = 0.66), oxidative phosphorylation (r = 0.32 in carbon fluxes and r = 0.37 in nitrogen distributions) and transport to the inner membrane (r = 0.12 and r = 0.31, respectively), showing an average flux distribution decrease of 45% for these specific subsystems when comparing carbon and nitrogen activity in diazotrophic against non-diazotrophic conditions. Additionally, strong correlation values were observed in specific subsystem clusters for both nitrogen conditions.
Fig. 4.
Carbon and nitrogen partitioning distribution from Biolog plates experimental data. A) Average carbon flux correlation coefficients through all subsystems in diazotrophic and non-diazotrophic conditions; B) average nitrogen flux correlation coefficients through all subsystems in diazotrophic and non-diazotrophic conditions.
3.4. Alginate and PHB production estimated through in-silico experiments
A. vinelandii DJ contains specific mechanisms to produce and secrete alginate into the extracellular space. In iDT1278 we manually curated six specific reactions related to alginate biosynthesis including 16 different genes. One of the most important reactions involved in this pathway is the alginate epimerase (EC 5.1.3.37), which encompass a complex protein system to synthetize the alginate polymer (Pacheco-Leyva et al., 2016).
We evaluated the model accuracy to growth and alginate production using four carbon sources under diazotrophic and non-diazotrophic conditions. Experimental data retrieved from the literature (Revin et al., 2018) was only available for growth with ammonium. Simulations were confirmed to accurately predict (true positive predictions) alginate production rates with three carbon sources (glucose, mannitol, and sucrose). For example, the predicted growth using glucose (uptake rate of 0.33 mmol/gDW/h) as the sole carbon source was 0.1478 1/h and an alginate production rate of 0.25 mmol/gDW/h, agreeing with the experimental data (growth rate of 0.152 ± 0.012 1/h and alginate production rate of 0.265 ± 0.018 mmol/gDW/h). Lactose was the only compound with a mismatch between reported values and in-silico outputs (false negative). The disagreement appears to be associated to the lack of information in the literature and bioinformatic databases about the essential enzymes in the metabolism of this disaccharide in A. vinelandii DJ.
We compared the growth rates determined under diazotrophic and non-diazotrophic conditions for alginate production. The simulation analysis showed a significant increase in the growth rates (close to 28%) when the A. vinelandii consumes ammonium as a nitrogen source instead of molecular nitrogen. Additionally, a second comparison of alginate production rates between ammonium and molecular nitrogen exhibited the same increasing tendency when ammonium is used as a unique nitrogen source instead of molecular nitrogen (around 27%).
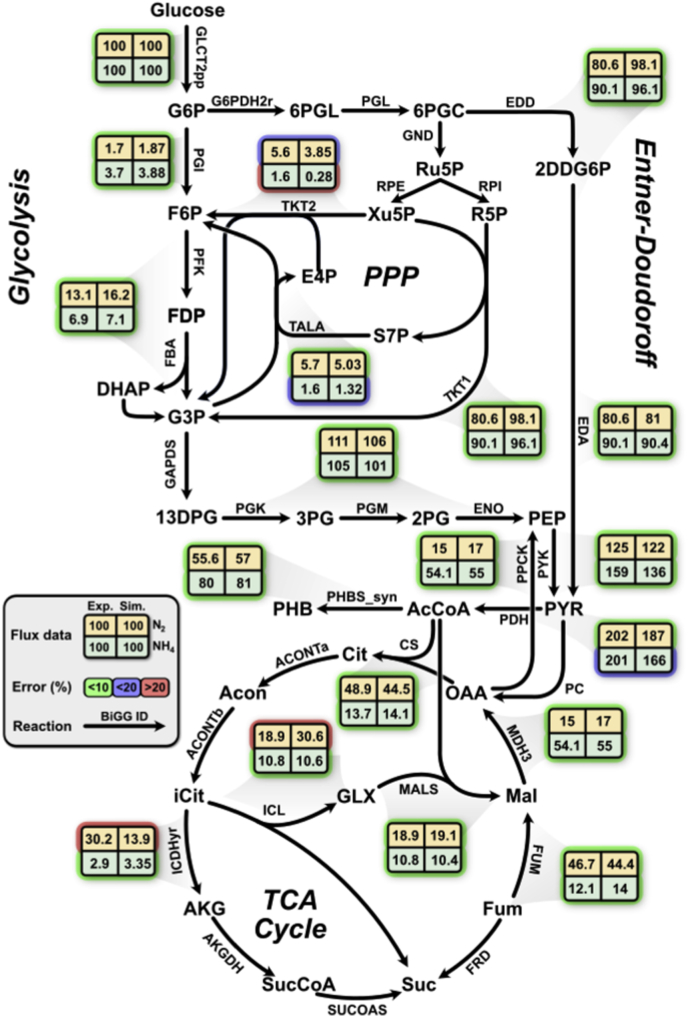
While alginate is transported to the extracellular space, PHB is intracellularly stored (Yoneyama et al., 2015). To simulate PHB accumulation in A. vinelandii we incorporated a sink reaction to the model iDT1278. Simulated flux distributions about PHB production were validated using fluxomic data retrieved from Wu et al. (2019). The metabolic fluxes of the reactions involved in the PHB synthesis and related pathways (glycolysis, pentose phosphate pathway, the Entner-Deundoroff pathway, and the TCA cycle) were calculated through FBA for diazotrophic and non-diazotrophic conditions. The simulation results were compared with the experimental measured fluxes (Wu et al., 2019) and the percent error was estimated by reaction (Fig. 5). A general agreement in the reaction fluxes was observed under both nitrogen (N2 and NH4) conditions. A total of 16 out of 19 reaction flux estimations presented a global accuracy above 90% for diazotrophic and non-diazotrophic conditions. Disagreements were detected in three specific reactions: the transketolase (TKT2), aconitase (ACONTa/ACONTb) and isocitrate dehydrogenase (ICDHyr) in which the percent errors were above 20%. According to the predicted and experimental results, a higher PHB production is obtained under non-diazotroph conditions due to higher energy cost for nitrogen fixation. Additionally, most of the carbon coming from the glucose is metabolized through the Entner-Doudoroff (ED) pathway, generating less energy than glycolysis, but producing pyruvate (PHB precursor) in fewer steps. The available pyruvate generated though the ED pathway is mostly used in the TCA cycle and the PHB synthesis (Castillo et al., 2013; Wu et al., 2019), allowing the growth of the microorganism and the production of this biopolymer.
Fig. 5.
Metabolic flux map distribution of A. vinelandii under diazotrophic and non-diazotrophic conditions. The map displays major metabolic pathways involved in the PHB production. Values of metabolic flux are normalized to a glucose uptake rate of 100. Predicted metabolic fluxes were compared against fluxomic data determined by Wu et al., in 2019. The reactions were labeled according to their percent error (green, blue and red) and nitrogen source (green and yellow). Abbreviations: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; FDP, fructose-1,6-bisphosphate; G3P, 3-phosphoglycerate; DHAP, dihydroxyacetone phosphate; 13DPG, 3-phosphoglyceroil phosphate; 2 PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; PYR, pyruvate; Cit, citrate; Acon, aconitate; iCit, isocitrate; AKG, a-ketoglutarate; sucCoA, succinyl coenzyme A; Suc, succinate; Fum, fumarate; Mal, malate; OAA, oxaloacetate; GLX, glyoxylate; PHB, polyhydroxybutyrate; 6PGL, 6-phospho-glucono-1,5-lactone; 6PGC, 6-phospho-gluconate; 2DDG6P, 2-Dehydro-3-deoxygluconate 6-phosphate; Ru5P, ribulose-5-phosphate; R5P, ribose-5-phosphate; S7P, sedoheptulose-7-phosphate; X5P, xylulose-5-phosphate; E4P, erythrose-4-phosphate. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
4.1. Model reconstruction
Here we have created the first comprehensive genome-scale metabolic model for A. vinelandii DJ (iDT1278) that focuses on nitrogen assimilation, nitrogen fixation, as well as on alginate and PHB production. The model consists of 1,278 genes involved in 2,469 reactions. Compared to the first microorganism template used in the present work (Escherichia coli str. K-12 substr. MG1655, iML1515), the percentage of metabolic genes per genome decreases from 31% (iML1515) to 26% (iDT1278). However, a higher percentage of metabolic genes (from 26% of A. vinelandii to 12% of the average genes in the photosynthetic models) was observed when iDT1278 was compared to three photosynthetic organisms in the BiGG database: C. vulgaris UTEX 395, iCZ843 (Zuñiga et al., 2016), Synechocystis sp. PCC 6803, iJN678 (Nogales et al., 2012) and C. reinhardtii, iCR1080 (Chang et al., 2011). The M-model is accurate to 89% for all the carbon sources and 91% for nitrogen sources. The model was validated using a wide variety of carbon (159 compounds) and nitrogen (75 metabolites) sources. iDT1278 shown a significant higher accuracy (27% upper) in the predictions compared to the CarveMe model simulations. Additionally, iDT1278 predicted accurately the growth ratio and production values of alginate and PHB production under diazotrophic and non-diazotrophic conditions. To our knowledge, this is the first M-model at genome-scale capable to simulate several carbon and nitrogen conditions (close to 250 conditions) with a high precision even when comparing internal metabolic fluxes.
4.2. Model validation
4.2.1. Nitrogen fixation
iDT1278 accurately predicts the growth of A. vinelandii using different carbon sources under diazotrophic and non-diazotrophic conditions. The model contains all required reactions and constraints to successfully simulate the BOF representing the growth of the organism. Model predictions have been confirmed by experimental validation using Biolog plates (PM1, PM2 and PM3). With this information we elucidated the preferred mechanism used by the A. vinelandii DJ to fix nitrogen while growing with different carbon sources. The N2 uptake depends on the ammonium concentration and metal cofactor concentrations required for the nitrogenases (García et al., 2018; Inomura et al., 2018). In our model, a difference in growth rate can be observed under diazotrophic and non-diazotrophic conditions. The growth rate under different carbon sources is higher during NH4 assimilation than during N2 fixation. However, the growth rates are quite similar for growth with amino acids under diazotrophic and non-diazotrophic conditions. This can be explained by the fact that amino acids release ammonium when metabolized which then becomes readily available to the organism. N2 fixation on the other hand is an ATP-dependent process and the organism must employ more energy to convert the N2 into ammonium.
Growth rate values decrease considerably when A. vinelandii DJ grows diazotrophically but nitrogen and carbon flux distributions per subsystem (e.g. amino acid, lipid, and carbohydrate metabolism) behave very similar in both nitrogen conditions. These flux distribution correlations suggest that the major discrepancies in the metabolism of A. vinelandii fixing nitrogen are related to the cofactor and vitamin pathways (riboflavin metabolism) and oxidative phosphorylation (energy generation). According to the model predictions (iDT1278), the vitamin, cofactor and oxidative phosphorylation pathways in A. vinelandii are involved in the generation of the precursors for the BOF; when ammonium is used as a nitrogen source, the carbon and nitrogen fluxes in these pathways are significantly higher (30%) than the fluxes estimated using molecular nitrogen as nitrogen source.
4.3. Alginate production
Alginate represents an important exopolysaccharide for A. vinelandii and is synthetized to reduce oxygen availability and thus increase nitrogenase activity for enhanced nitrogen fixation (Galindo et al., 2007; Nosrati et al., 2012). Additionally, this polymer has industrial relevance to multiple fields such as in pharmaceutical (Azevedo et al., 2014), biotechnological (Tomida et al., 2010), and food industry applications (Kuda et al., 1998). Therefore, elucidating the mechanism of alginate production could potentially provide insights for increasing production of this valuable biopolymer. iDT1278 accurately predicts three out of four carbon sources capable of producing alginate and the accompanying nitrogen sources to maximize biosynthesis of this polymer. The model also successfully predicts the decline in the growth and alginate production when molecular nitrogen is used as a nitrogen source. Alginate metabolism has been studied using genetic and regulation approaches to explain the synthesis of this valuable biopolymer, since most of the genes involved in this metabolic pathway are regulated by the presence of oxygen (Ertesvåg et al., 1995; Lloret et al., 1996; Núñez et al., 2000). In the present work we show that alginate production can also be explained based exclusively on metabolic requirements using mathematical and metabolic representations.
4.4. PHB production
PHB, like alginate, is synthesized by A. vinelandii to reduce oxygen availability and promote nitrogen fixation. iDT1278 contains all the genes and specific reactions involved in the production of PHB. The polymer is a high value product used in the production of biodegradable plastics and other environmental friendly polymers (Galindo et al., 2007). iDT1278 accurately predicts most of the metabolic flux values in the reactions (85% of the reactions) involved in PHB synthesis and pathways related to the generation of PHB precursors and energy metabolism (glycolysis, pentose phosphate pathway, the Entner-Doudoroff pathway, and the TCA cycle) for diazotrophic and non-diazotrophic conditions. The model is capable of accurately simulating all the variations presented in the flux distribution values for every reaction involved in the PHB synthesis under different conditions (ammonium or molecular nitrogen as unique nitrogen sources) using only metabolic data (stoichiometry and biochemistry data).
As with alginate production, PHB metabolism in A. vinelandii has been well studied using exclusively genetic and regulation perspectives since PHB accumulation mainly occurs during oxygen limitations (Castillo et al., 2013; Vijayendra et al., 2007); however, PHB metabolic distribution and biosynthesis can also be described using metabolic data according to the predictions performed by iDT1278. Our model could as a result be deployed to potentially optimize PHB production in A. vinelandii.
4.5. Network properties
Most of the False Negative (6 out of 10 total) and False Positive (6 total) predictions from the model related to carbohydrates and amino acids substrates could not be confirmed to belong to a pathway or to an enzyme that converts these carbon sources to internal metabolites present in the model either from literature information or available genomic and metabolomic databases. Moreover, when using amino acids as growth substrates, the model predicts very similar growth rate values for diazotrophic and non-diazotrophic conditions since some amino acids can release ammonium further supporting growth. These discrepancies between experimental evidence and model predictions could be related to the limited information available concerning specific metabolic mechanisms for metabolizing carbon sources as carbohydrates, lipids and amino acids in A. vinelandii or processes related to genetic, transcriptional and regulation. Indeed, while nitrogen metabolism is tightly regulated in A. vinelandii (Hamilton et al., 2011; Toukdarian and Kennedy, 1986), transcriptional and translational regulation is not currently part of this M-model but could in part be recapitulated by including all macromolecular synthesis as part of a subsequent ME-models (Liu et al., 2019, 2014; O’Brien et al., 2013). Furthermore, nitrogenase activity and activity of other nitrogen-associated pathways are also regulated at the enzyme level (Pacheco-Leyva et al., 2016), potentially also contributing further to the lack of agreement between model predictions and experimental data.
4.6. Future aspects
Insight into the metabolic processes using the genome-scale model iDT1278 could benefit low-cost media optimization for A. vinelandii and to further improve production processes as PHB, alginate and other biopolymers with high industrial value. Although this environmentally important model organism has been isolated and studied for more than 100 years, its physiology and some metabolic pathways have yet to be fully understood. We believe the current model provides a valuable step along the path towards better characterization of this important microbe in isolation as part of a microbial community (Tan et al., 2015; Zuñiga et al., 2019). Unraveling this knowledge for A. vinelandii could improve its use in industrial applications (Noar and Bruno-Bárcena, 2018), and the systems biology approaches presented here may provide a tool to help in achieving that goal.
Author contributions
CZ, AZ, and KZ conceived the study. DT and CZ reconstructed the model and performed all simulations. DT and AP performed manual curation. JDT performed growth experiments. CZ, DT, AP, JDT, and MJB discussed the results. DT, AP, CZ, and KZ wrote the manuscript with input from all co-authors.
Funding
This material is based upon work supported by the National Science Foundation under Grant No. 1332344 and CBET-1804187, and the U.S. Department of Energy (DOE), Office of Science, Office of Biological & Environmental Research under Awards DE-SC0012658 and DE-SC0019388. D.T. was in part supported by Mexican National Research Council, CONACYT, fellowship No. 932962.
CRediT authorship contribution statement
Diego Tec Campos: Investigation, Methodology, Writing - original draft. Cristal Zuñiga: Conceptualization, Supervision, Project administration. Anurag Passi: Data curation, Writing - review & editing. John Del Toro: Investigation. Juan D. Tibocha-Bonilla: Validation. Alejandro Zepeda: Validation, Funding acquisition. Michael J. Betenbaugh: Investigation, Funding acquisition. Karsten Zengler: Conceptualization, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We acknowledge Manish Kumar and Erick Armingol (UC San Diego) for providing input and assistance throughout all stages of this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2020.e00132.
Research data for this article
iDT1278 is available in. mat and. xml format as well as the carbon and nitrogen partitioning analysis in the repository from Zengler Lab: https://github.com/cristalzucsd/AvinelandiiDJ.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Agren R., Liu L., Shoaie S., Vongsangnak W., Nookaew I., Nielsen J. The RAVEN toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio R., Ortiz-Marquez J.C.F., Curatti L. Metabolic engineering of a diazotrophic bacterium improves ammonium release and biofertilization of plants and microalgae. Metab. Eng. 2017;40:59–68. doi: 10.1016/j.ymben.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Azevedo M.A., Bourbon A.I., Vicente A.A., Cerqueira M.A. Alginate/chitosan nanoparticles for encapsulation and controlled release of vitamin B2. Int. J. Biol. Macromol. 2014;71:141–146. doi: 10.1016/j.ijbiomac.2014.05.036. [DOI] [PubMed] [Google Scholar]
- Castillo T., Heinzle E., Peifer S., Schneider K., Peña M.C.F. Oxygen supply strongly influences metabolic fluxes, the production of poly(3-hydroxybutyrate) and alginate, and the degree of acetylation of alginate in Azotobacter vinelandii. Process Biochem. 2013;48:995–1003. doi: 10.1016/j.procbio.2013.04.014. [DOI] [Google Scholar]
- Chang R.L., Ghamsari L., Manichaikul A., Hom E.F.Y., Balaji S., Fu W., Shen Y., Hao T., Palsson B.Ø., Salehi-Ashtiani K., Papin J.A. Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism. Mol. Syst. Biol. 2011;7:518. doi: 10.1038/msb.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi F. Alginate production by Azotobacter vinelandii. Crit. Rev. Biotechnol. 1997;17:327–361. doi: 10.3109/07388559709146618. [DOI] [PubMed] [Google Scholar]
- Cleveland C.C., Townsend A.R., Schimel D.S., Fisher H., Howarth R.W., Hedin L.O., Perakis S.S., Latty E.F., Von Fischer J.C., Elseroad A., Wasson M.F. Global patterns of terrestrial biological nitrogen (N2) fixation in natural ecosystems. Global Biogeochem. Cycles. 1999;13:623–645. doi: 10.1029/1999GB900014. [DOI] [Google Scholar]
- Díaz-Barrera A., Andler R., Martínez I., Peña C. Poly-3-hydroxybutyrate production by Azotobacter vinelandii strains in batch cultures at different oxygen transfer rates. J. Chem. Technol. Biotechnol. 2016;91:1063–1071. doi: 10.1002/jctb.4684. [DOI] [Google Scholar]
- Ertesvåg H., Høidal H.K., Hals I.K., Rian A., Doseth B., Valla S. A family of modular type mannuronan C-5-epimerase genes controls alginate structure in Azotobacter vinelandii. Mol. Microbiol. 1995;16:719–731. doi: 10.1111/j.1365-2958.1995.tb02433.x. [DOI] [PubMed] [Google Scholar]
- Feist A.M., Nagarajan H., Rotaru A.-E., Tremblay P.-L., Zhang T., Nevin K.P., Lovley D.R., Zengler K. Constraint-based modeling of carbon fixation and the energetics of electron transfer in Geobacter metallireducens. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist A.M., Scholten J.C.M., Palsson B.Ø., Brockman F.J., Ideker T. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo E., Peña C., Núñez C., Segura D., Espín G. Molecular and bioengineering strategies to improve alginate and polydydroxyalkanoate production by Azotobacter vinelandii. Microb. Cell Factories. 2007;6:1–16. doi: 10.1186/1475-2859-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A., Ferrer P., Albiol J., Castillo T., Segura D., Peña C. Metabolic flux analysis and the NAD(P)H/NAD(P)+ ratios in chemostat cultures of Azotobacter vinelandii. Microb. Cell Factories. 2018;17:10. doi: 10.1186/s12934-018-0860-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurján I., Korányi P., Preininger É., Varga S.S., Paless G. Azospirillum VI and Related Microorganisms. Springer Berlin Heidelberg; Berlin, Heidelberg: 1995. Artificial plant-azotobacter symbiosis for atmospheric nitrogen fixation; pp. 401–413. [DOI] [Google Scholar]
- Hamilton T.L., Ludwig M., Dixon R., Boyd E.S., Dos Santos P.C., Setubal J.C., Bryant D.A., Dean D.R., Peters J.W. Transcriptional profiling of nitrogen fixation in Azotobacter vinelandii. J. Bacteriol. 2011;193:4477–4486. doi: 10.1128/JB.05099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heirendt L., Arreckx S., Pfau T., Mendoza S.N., Richelle A., Heinken A., Haraldsdóttir H.S., Wachowiak J., Keating S.M., Vlasov V., Magnusdóttir S., Ng C.Y., Preciat G., Žagare A., Chan S.H.J., Aurich M.K., Clancy C.M., Modamio J., Sauls J.T., Noronha A., Bordbar A., Cousins B., El Assal D.C., Valcarcel L.V., Apaolaza I., Ghaderi S., Ahookhosh M., Ben Guebila M., Kostromins A., Sompairac N., Le H.M., Ma D., Sun Y., Wang L., Yurkovich J.T., Oliveira M.A.P., Vuong P.T., El Assal L.P., Kuperstein I., Zinovyev A., Hinton H.S., Bryant W.A., Aragón Artacho F.J., Planes F.J., Stalidzans E., Maass A., Vempala S., Hucka M., Saunders M.A., Maranas C.D., Lewis N.E., Sauter T., Palsson B.Ø., Thiele I., Fleming R.M.T. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat. Protoc. 2019;14:639–702. doi: 10.1038/s41596-018-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C.S., DeJongh M., Best A.A., Frybarger P.M., Linsay B., Stevens R.L. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 2010;28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- Howard J.B., Rees D.C. Structural basis of biological nitrogen fixation. Chem. Rev. 1996;96:2965–2982. doi: 10.1021/cr9500545. [DOI] [PubMed] [Google Scholar]
- Inomura K., Bragg J., Riemann L., Follows M.J. A quantitative model of nitrogen fixation in the presence of ammonium. PloS One. 2018;13 doi: 10.1371/journal.pone.0208282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King Z.A., Lu J., Dräger A., Miller P., Federowicz S., Lerman J.A., Ebrahim A., Palsson B.O., Lewis N.E. BiGG Models: a platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016;44:D515–D522. doi: 10.1093/nar/gkv1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuda T., Goto H., Yokoyama M., Fujii T. Effects of dietary concentration of laminaran and depolymerised alginate on rat cecal microflora and plasma lipids. Fish. Sci. 1998;64:589–593. doi: 10.2331/fishsci.64.589. [DOI] [Google Scholar]
- Liao Y.-C., Huang T.-W., Chen F.-C., Charusanti P., Hong J.S.J., Chang H.-Y., Tsai S.-F., Palsson B.O., Hsiung C.A. An experimentally validated genome-scale metabolic reconstruction of Klebsiella pneumoniae MGH 78578, iYL1228. J. Bacteriol. 2011;193:1710–1717. doi: 10.1128/JB.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.K., Lloyd C., Al-Bassam M.M., Ebrahim A., Kim J.-N., Olson C., Aksenov A., Dorrestein P., Zengler K. Predicting proteome allocation, overflow metabolism, and metal requirements in a model acetogen. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1006848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.K., O’Brien E.J., Lerman J.A., Zengler K., Palsson B.O., Feist A.M. Reconstruction and modeling protein translocation and compartmentalization in Escherichia coli at the genome-scale. BMC Syst. Biol. 2014;8:110. doi: 10.1186/s12918-014-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret L., Barreto R., León R., Moreno S., MartÍnez-Salazar J., Espín G., Soberón-Chávez G. Genetic analysis of the transcriptional arrangement of Azotobacter vinelandii alginate biosynthetic genes: identification of two independent promoters. Mol. Microbiol. 1996;21:449–457. doi: 10.1111/j.1365-2958.1996.tb02554.x. [DOI] [PubMed] [Google Scholar]
- Machado D., Andrejev S., Tramontano M., Patil K.R. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018;46:7542–7553. doi: 10.1093/nar/gky537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk J.M., Lloyd C.J., Brunk E., Mih N., Sastry A., King Z., Takeuchi R., Nomura W., Zhang Z., Mori H., Feist A.M., Palsson B.O. iML1515, a knowledgebase that computes Escherichia coli traits. Nat. Biotechnol. 2017;35:904–908. doi: 10.1038/nbt.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Nishizawa Y., Doin P.A., Aiba S. Energy difference in metabolism between glucose and acetate in chemostat culture of Azotobacter vinelandii. J. Gen. Appl. Microbiol. 1972;18:201–208. doi: 10.2323/jgam.18.201. [DOI] [Google Scholar]
- Nagarajan H., Sahin M., Nogales J., Latif H., Lovley D.R., Ebrahim A., Zengler K. Characterizing acetogenic metabolism using a genome-scale metabolic reconstruction of Clostridium ljungdahlii. Microb. Cell Factories. 2013;12:118. doi: 10.1186/1475-2859-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noar J.D., Bruno-Bárcena J.M. Azotobacter vinelandii: the source of 100 years of discoveries and many more to come. Microbiology. 2018;164:421–436. doi: 10.1099/mic.0.000643. [DOI] [PubMed] [Google Scholar]
- Noar J., Loveless T., Navarro-Herrero J.L., Olson J.W., Bruno-Bárcena J.M. Aerobic hydrogen production via nitrogenase in Azotobacter vinelandii CA6. Appl. Environ. Microbiol. 2015;81:4507–4516. doi: 10.1128/AEM.00679-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales J., Gudmundsson S., Knight E.M., Palsson B.O., Thiele I. Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2678–2683. doi: 10.1073/pnas.1117907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosrati R., Owlia P., Saderi H., Olamaee M., Rasooli I., Akhavian T.A. Correlation between nitrogen fixation rate and alginate productivity of an indigenous Azotobacter vinelandii from Iran. Iran. J. Microbiol. 2012;4(3):153–159. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3465543/pdf/IJM-4-153.pdf [PMC free article] [PubMed] [Google Scholar]
- Núñez C., León R., Guzmán J., Espín G., Soberón-Chávez G. Role of Azotobacter vinelandii mucA and mucC gene products in alginate production. J. Bacteriol. 2000;182:6550–6556. doi: 10.1128/jb.182.23.6550-6556.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien E.J., Lerman J.A., Chang R.L., Hyduke D.R., Palsson B.Ø. Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol. Syst. Biol. 2013;9:693. doi: 10.1038/msb.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., Thiele I., Palsson B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco-Leyva I., Guevara Pezoa F., Díaz-Barrera A. Alginate biosynthesis in azotobacter vinelandii : overview of molecular mechanisms in connection with the oxygen availability. Int. J. Polym. Sci. 2016;1–12 doi: 10.1155/2016/2062360. 2016. [DOI] [Google Scholar]
- Quiroz-Rocha E., Moreno R., Hernández-Ortíz A., Fragoso-Jiménez J.C., Muriel-Millán L.F., Guzmán J., Espín G., Rojo F., Núñez C. Glucose uptake in Azotobacter vinelandii occurs through a GluP transporter that is under the control of the CbrA/CbrB and Hfq-Crc systems. Sci. Rep. 2017;7:858. doi: 10.1038/s41598-017-00980-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remminghorst U., Rehm B.H.A. Bacterial alginates: from biosynthesis to applications. Biotechnol. Lett. 2006;28:1701–1712. doi: 10.1007/s10529-006-9156-x. [DOI] [PubMed] [Google Scholar]
- Revin V.V., Kostina E.G., Revina N.V., Shutova V.V. Effect of nutrient sources on the alginate accumulation in the culture liquid of Azotobacter vinelandii D-05 and obtaining biocomposite materials. Braz. Arch. Biol. Technol. 2018;61 doi: 10.1590/1678-4324-2018160406. [DOI] [Google Scholar]
- Sahoo R.K., Ansari M.W., Dangar T.K., Mohanty S., Tuteja N. Phenotypic and molecular characterisation of efficient nitrogen-fixing Azotobacter strains from rice fields for crop improvement. Protoplasma. 2014;251:511–523. doi: 10.1007/s00709-013-0547-2. [DOI] [PubMed] [Google Scholar]
- Setubal J.C., Almeida N.F. Biological Nitrogen Fixation. John Wiley & Sons, Inc; Hoboken, NJ, USA: 2015. The Azotobacter vinelandii genome: an update; pp. 225–234. [Google Scholar]
- Setubal J.C., dos Santos P., Goldman B.S., Ertesvag H., Espin G., Rubio L.M., Valla S., Almeida N.F., Balasubramanian D., Cromes L., Curatti L., Du Z., Godsy E., Goodner B., Hellner-Burris K., Hernandez J.A., Houmiel K., Imperial J., Kennedy C., Larson T.J., Latreille P., Ligon L.S., Lu J., Maerk M., Miller N.M., Norton S., O’Carroll I.P., Paulsen I., Raulfs E.C., Roemer R., Rosser J., Segura D., Slater S., Stricklin S.L., Studholme D.J., Sun J., Viana C.J., Wallin E., Wang B., Wheeler C., Zhu H., Dean D.R., Dixon R., Wood D. Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J. Bacteriol. 2009;191:4534–4545. doi: 10.1128/JB.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawky B.T., Ghali Y., Ahmed F.A., Kahil T. Biochemical studies on the effect of various carbon sources on growth, nitrogen fixtion, and main cellular constituents of Azotobacter vinelandii, strain IV grown under various cultivation conditions. Acta Biotechnol. 1987;7:361–369. doi: 10.1002/abio.370070417. [DOI] [Google Scholar]
- Sippel D., Schlesier J., Rohde M., Trncik C., Decamps L., Djurdjevic I., Spatzal T., Andrade S.L.A., Einsle O. Production and isolation of vanadium nitrogenase from Azotobacter vinelandii by molybdenum depletion. JBIC J. Biol. Inorg. Chem. 2017;22:161–168. doi: 10.1007/s00775-016-1423-2. [DOI] [PubMed] [Google Scholar]
- Stevenson L.H., Socolofsky M.D. Cyst formation and poly-beta-hydroxybutyric acid accumulation in Azotobacter. J. Bacteriol. 1966;91:304–310. doi: 10.1128/jb.91.1.304-310.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Zuniga C., Zengler K. Unraveling interactions in microbial communities - from co-cultures to microbiomes. J. Microbiol. 2015;53:295–305. doi: 10.1007/s12275-015-5060-1. [DOI] [PubMed] [Google Scholar]
- Tomida H., Yasufuku T., Fujii T., Kondo Y., Kai T., Anraku M. Polysaccharides as potential antioxidative compounds for extended-release matrix tablets. Carbohydr. Res. 2010;345:82–86. doi: 10.1016/j.carres.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Toukdarian A., Kennedy C. Regulation of nitrogen metabolism in Azotobacter vinelandii: isolation of ntr and glnA genes and construction of ntr mutants. EMBO J. 1986;5:399–407. doi: 10.1002/j.1460-2075.1986.tb04225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayendra S.V.N., Rastogi N.K., Shamala T.R., Anil Kumar P.K., Kshama L., Joshi G.J. Optimization of polyhydroxybutyrate production by Bacillus sp. CFR 256 with corn steep liquor as a nitrogen source. Indian J. Microbiol. 2007;47:170–175. doi: 10.1007/s12088-007-0033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam A.R., Davis J.J., Assaf R., Boisvert S., Brettin T., Bun C., Conrad N., Dietrich E.M., Disz T., Gabbard J.L., Gerdes S., Henry C.S., Kenyon R.W., Machi D., Mao C., Nordberg E.K., Olsen G.J., Murphy-Olson D.E., Olson R., Overbeek R., Parrello B., Pusch G.D., Shukla M., Vonstein V., Warren A., Xia F., Yoo H., Stevens R.L. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017;45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T.Y. Effects of mannose on the growth of N(2)-fixing Azotobacter vinelandii. Appl. Environ. Microbiol. 1988;54:473–475. doi: 10.1128/aem.54.2.473-475.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T.Y., Maier R.J. H2-dependent mixotrophic growth of N2-fixing Azotobacter vinelandii. J. Bacteriol. 1985;163:528–533. doi: 10.1128/jb.163.2.528-533.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Herold R.A., Knoshaug E.P., Wang B., Xiong W., Laurens L.M.L. Fluxomic analysis reveals central carbon metabolism adaptation for diazotroph Azotobacter vinelandii ammonium excretion. Sci. Rep. 2019;9:13209. doi: 10.1038/s41598-019-49717-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama F., Yamamoto M., Hashimoto W., Murata K. Production of polyhydroxybutyrate and alginate from glycerol by Azotobacter vinelandii under nitrogen-free conditions. Bioengineered. 2015;6:209–217. doi: 10.1080/21655979.2015.1040209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuñiga C., Li C.-T., Huelsman T., Levering J., Zielinski D.C., McConnell B.O., Long C.P., Knoshaug E.P., Guarnieri M.T., Antoniewicz M.R., Betenbaugh M.J., Zengler K. Genome-scale metabolic model for the green alga Chlorella vulgaris UTEX 395 accurately predicts phenotypes under autotrophic, heterotrophic, and mixotrophic growth conditions. Plant Physiol. 2016;172:589–602. doi: 10.1104/pp.16.00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuñiga C., Li C.-T., Yu G., Al-Bassam M.M., Li T., Jiang L., Zaramela L.S., Guarnieri M., Betenbaugh M.J., Zengler K. Environmental stimuli drive a transition from cooperation to competition in synthetic phototrophic communities. Nat. Microbiol. 2019;4:2184–2191. doi: 10.1038/s41564-019-0567-6. [DOI] [PubMed] [Google Scholar]
- Zúñiga C., Morales M., Le Borgne S., Revah S. Production of poly-β-hydroxybutyrate (PHB) by Methylobacterium organophilum isolated from a methanotrophic consortium in a two-phase partition bioreactor. J. Hazard Mater. 2011;190:876–882. doi: 10.1016/j.jhazmat.2011.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.