Abstract
OncomiRs are microRNAs that are associated with early onset of specific cancers. To identify microRNAs involved in pediatric acute lymphoblastic leukemia (ALL) subtypes T-ALL and B-ALL, peripheral blood and bone marrow samples were independently subjected to microarray analysis using two different high-fidelity array platforms. The unique and common gene signatures from both arrays were validated by TaqMan individual assays in 100 pediatric ALL samples. Survival studies were carried out in the test set and validation set with 50 randomly selected samples in each set. MicroRNA expression profile revealed characteristic signatures for distinguishing T and B lineages and identified 51 novel microRNAs in pediatric ALL. Interestingly, the present study also revealed endogenous similarities and differences between blood and bone marrow within each ALL subtype. When Cox regression analysis was carried out with these identified microRNAs, 11 of them exhibited expression levels significantly correlated with survival. Validation of some of the common and relevant microRNAs from both arrays showed that their targets are involved in key oncogenic signaling pathways. Thus, this study suggests that microRNAs have the potential to become important diagnostic tools for identification and monitoring clinical outcomes in ALL patients.
Introduction
Acute lymphoblastic leukemia (ALL) represents a heterogeneous disease characterized by various underlying genetic abnormalities that block B- or T-cell differentiation and abets abnormal cell proliferation and survival [1]. Among acute leukemias in children, about 60% is ALL, of which 80%-85% cases are of B-lineage and the rest are of T-lineage [[2], [3], [4]]. Current diagnostic regimen involves the use of flow cytometric immunotyping on peripheral blood (PB) or bone marrow (BM) specimens which is cumbersome [5]. Thus, there is an imminent need for the development of molecular markers for diagnostic and prognostic purpose. MicroRNAs are small noncoding RNAs that play an important role in the regulation of gene expression. Abnormal gene expression by microRNAs is correlated with the initiation and progression of many cancer types [6,7] and is referred to as OncomiRs. They have been shown to regulate both lymphoid and myeloid lineages in the hematopoietic system [8]. Distinct signatures of microRNA expression are so fine-tuned in hematopoietic cell differentiation that their deregulation might contribute to leukemogenesis [9,10]. Studies revealed that overexpression of miR-155 in transgenic mouse showed enhanced B-cell proliferation and eventually developed ALL [11]. MicroRNA expression signatures differentiate ALL from Acute Myeloid Leukemia (AML), and combinations could discriminate both cases with an accuracy of 97%-99% [12]. Such studies have also revealed the involvement of microRNAs in hematopoietic stem cell and leukemic stem cell functions [13]. Several microRNAs were also correlated with drug resistance in ALL, implicating another major role of microRNAs in tumorigenesis [14]. Such tangible evidence of microRNAs acting as regulators in shaping tumor pathophysiology and its progression provided the impetus for us to profile the entire complement of microRNAs associated with ALL. Therefore, we aimed to identify novel signatures that could be used to classify the T and B subtypes and establish blood-based microRNAs for potential diagnostic and prognostic markers for childhood ALL in future.
We identified novel microRNAs which can distinguish the two subtypes, T-ALL and B-ALL, either by PB or BM samples. The other interesting outcome of this study was the common and unique signatures of microRNAs of PB and BM within the same subtype. The expression pattern of 11 microRNAs was associated with patient survival in both test and validation data sets. We opine that potential miRNA biomarkers identified in this study would further help in classification of subtypes and determination of clinical outcome of ALL.
Material and Methods
Patient Database and Sample Collection
A total of 50 Ph-negative ALL patients having more than 90% blast cells as per May-Grünwald-Giemsa staining of children aged between 3 and 14 years reported to Regional Cancer Centre (RCC), Trivandrum, were selected for the study. Equal number of peripheral blood and bone marrow samples of same patients was included in all experiments. Controls consisted of 20 samples, 10 PB (healthy volunteers), and 10 BM reported to RCC for various other reasons. Patients were treated in a risk adaptive manner based on age, WBC count, and prednisone response. For collection of RBC free clear pellet, BM and PB samples were mixed with RBC lysis buffer for 15 minutes at room temperature followed by 10-minute centrifugation at 2500 rpm. Pellet was washed in phosphate-buffered saline, suspended in RNAlater (Invitrogen, USA), and stored at −80°C until RNA isolation.
Morphological Evaluation and Sorting by Flow Cytometry
These patients were Ph negative and had undergone similar treatments according to Berlin-Frankfurt-Münster (BFM)-based chemotherapy regimen. To distinguish the two immunophenotypes of ALL, i.e., B-ALL and T-ALL, morphological characterization of BM and PB samples was performed by flow cytometry [15] using a panel of fluorochrome conjugated monoclonal antibodies. PB and BM smears were stained with Giemsa and myeloperoxidase stain for morphologic evaluation. Cells (1 × 104) were stained within 24 hours of collection using whole blood lyse wash technique. Ten milliliters of diluted antibodies was added to each sample followed by addition of leukocyte common antigen and incubated for 10 minutes. Two milliliters (1:10) of FACSLYSE solution (BD, Biosciences, San Jose, CA) was added to all samples, vortexed, incubated again for 10 minutes, and centrifuged carefully at 1500 rpm for 5 minutes, and supernatant was discarded. Sheath fluid was added and vortexed well. Six parameters and four-color immunophenotyping of the samples were performed using FACS Calibur flow cytometer (Becton Dickinson, San Jose, CA). Data were analyzed using Cell Quest pro software (Version 6.0; Supplementary Figure S1).
RNA Extraction and cDNA Synthesis
Total RNA was isolated from a total of 100 ALL samples (50 PB and 50 BM) along with 10 healthy controls using mirVana Kit (ThermoFisher Scientific, USA) as per manufacturer's instructions. Purity and quantity of RNA were measured using NanoDrop ND 1000 spectrophotometer (NanoDrop Technologies, USA), and integrity was checked by determining RNA integrity number using an Agilent Bioanalyzer (2100). Only those RNA samples with RNA integrity number ≥ 7 were taken for this study. First-strand cDNA was synthesized from total RNA (1 μg) using high-capacity cDNA synthesis reverse transcription kit (Applied Biosystems, USA) according to manufacturer's instructions.
TaqMan Low-Density Array (TLDA) and Locked Nucleic Acid (LNA) Array Experiments
TLDA (version 2.0) which contains 667 human miRNAs covering Sanger miRBase (ver10.0) was performed as per manufacturer’s instructions. A total number of 48 samples (equal number of B-ALL (24) and T-ALL (24) lineages, each group consisting of PB (12) and BM (12) samples from the same patient, were taken for TLDA experiments along with 20 normal (10 PB and 10 BM) samples. The experiment was repeated with LNA arrays (version 11.0) containing 1372 miRNAs from hmr miRbase 14.0 + miRPlus from Exiqon, Denmark, following manufacturer's instructions.
TaqMan Individual Validation Assays
Common microRNAs that showed significant up/downregulation from TLDA and LNA arrays were subjected to double-blind TaqMan individual validation assays in 100 samples (including array samples) following manufacturer's instructions and MIQE guidelines.
MicroRNA Target Validation by Quantitative Polymerase Chain Reaction (qPCR) and Immunoblotting
Genes reported to be involved in ALL were retrieved from different databases such as KEGG [16], LeGenD (https://www.bioinformatics.org/legend/legend.htm), LGB (http://lgb.adibiosolutions.com), and COSMIC [17]. Target prediction of the identified miRNAs was performed with miRanda program [18], TargetScan [19], and PicTar [20] and LeukmiR database [21]. Interaction network between the miRNAs and their targets in various signaling pathways was generated using sna package in “R” [22]. Relevant and significant 11 microRNAs were selected for target validation studies. AntimiRs (miR-136, miR-137, miR-143, miR-466) and mimics (miR-649, miR-200c, miR-105, miR-432, miR-659, miR-662, miR-921) for these miRNAs, targeting ALL genes involved in key pathways, were purchased from Exiqon, Denmark, and evaluated on their oncogenic targets such as caspase 3, caspase 8, c-Myc, p53, PAX5, and STAT 3 obtained from in silico studies through qPCR and immunoblotting techniques. The quantification and densitometric analyses were done using “Image J” and presented.
Survival Analysis
A total of 397 differentially expressed and significant microRNAs were used for this study. One hundred samples from 50 ALL patients were divided into two groups. Fifty patient samples assigned to test set were used in TLDA/LNA array analysis, and 50 patient samples assigned to validation set were used for TaqMan individual assays. Cox regression hazard ratio (HR) was assessed between miRNA expression and overall survival time in each patient using “survival” package [23] in R (version 3.1.2). MiRNAs at log-rank P value < .01 and HR > 1 were categorized as risky, while those with HR < 1 were categorized as protective. The risk score for each protective and risky microRNA was calculated for each patient belonging to the test and validation group using the following formula:
Risk score = (regression coefficient of miR-1 × miR-1 expression) + regression coefficient of miR-nth × miR-nth expression). Further, patients with risk score greater than 60th percentile were grouped as high risk, and those with risk score less than 60th percentile were considered as the low-risk group. Overall survival was estimated by the Kaplan-Meier curve [24] for these two groups, and log-rank test was used to evaluate the significance of the difference of overall survival (P value ≤ .05) between high-risk and low-risk group.
MS/LTQ-Orbitrap Analysis
Proteome analyses of ALL versus normal samples were carried out as described in Saxena et al. [25].
Statistical Analysis
Statistical analysis of TLDA data was performed using StatMiner (spotfire) software from Integromics. Differentially expressed microRNAs were considered as significant and valid for those with FDR-adjusted P value ≤ .05.
Results
MicroRNA Signatures Differentiate the Two Hematopoietic Lineages of ALL
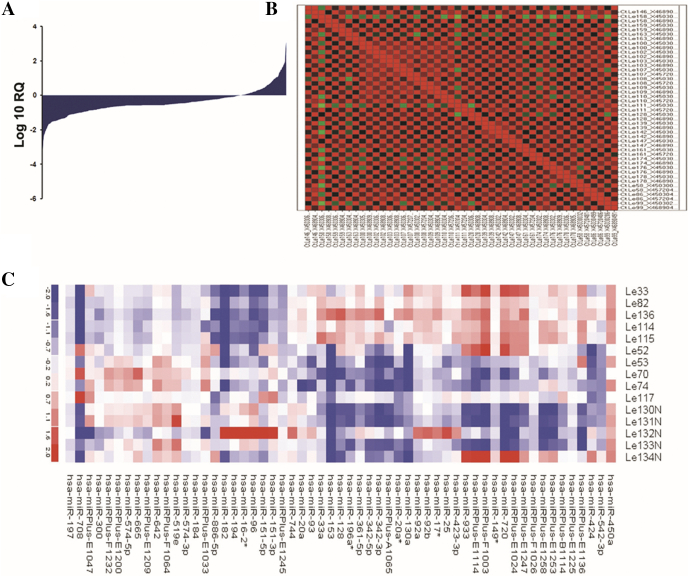
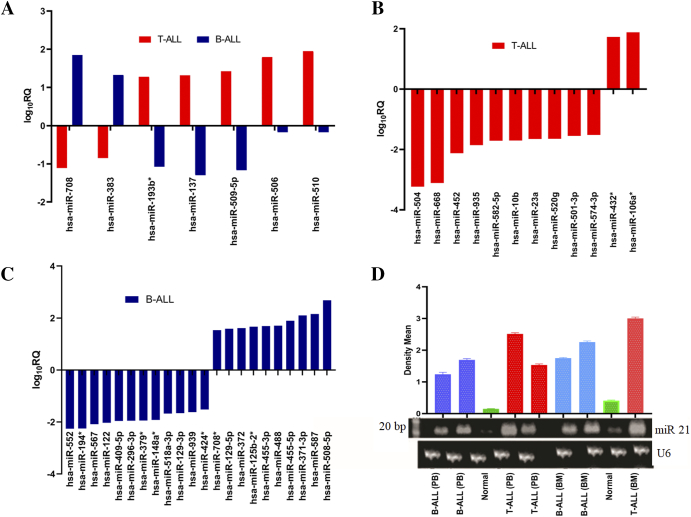
Out of a total 667 microRNAs in Taqman low-density arrays, 397 were detected as valid and significant in ALL and hence taken for further analysis. Of these, 337 (85%) microRNAs were downregulated and 60 (15%) microRNAs were upregulated (Supplementary Figure S2). Though the microRNA expression profile indicated a downregulation trend in ALL patients (Figure 1A, Supplementary Table S1), there were marked differences in the microRNA composition and degree of expression between T-ALL and B-ALL subtypes compared to healthy controls (Figure 1, B and C, Supplementary Figure S3). Validation of significant microRNAs from each individual group was carried out in 100 samples using blind TaqMan individual assays (Supplementary Figure S4). This reconfirmed and validated the results from both array analyses. MicroRNA expression profile further revealed the common and unique signatures between T-ALL and B-ALL subtypes (Supplementary Figure S5A, Supplementary Table S2, A and B). Interestingly, there were sets of microRNAs that were common to these subtypes but exhibited opposite trends in expression (Figure 2A). Twelve microRNAs in T-ALL and 22 microRNAs in B-ALL were uniquely and differentially expressed (Figure 2, B and C). The respective fold up- and downregulation of the microRNAs along with corresponding P values are shown in Supplementary Tables S3, A and B. Moreover, upregulation of miR-21 in both PB and BM of T-ALL and B-ALL subtypes was confirmed by northern blotting (Figure 2D). Our in-silico studies (in press) supported microarray data that classified 51 of them as novel ALL microRNAs, which were not reported earlier.
Figure 1.
Expression trend of microRNAs in ALL (A) by TLDA represented as log10RQ and (B) TLDA heat map depicting gene expression profiles of microRNAs. Green color in the heat map depicts downregulated microRNAs, and red color represents upregulated microRNAs. (C) Heat map illustrating unsupervised hierarchical clustering of microRNA expression using LNA arrays.
Figure 2.
MicroRNA expression profile of T-ALL and B-ALL reveals common and unique signatures. (A) Common microRNAs with reverse expression trend (at log10RQ > 1) between T-ALL and B-ALL (B and C) unique microRNAs at log10RQ > 1.5 in T-ALL and B-ALL, respectively. (D) Northern blot analysis showing an overexpression of miR-21 in PB and BM of both subtypes compared to healthy individuals.
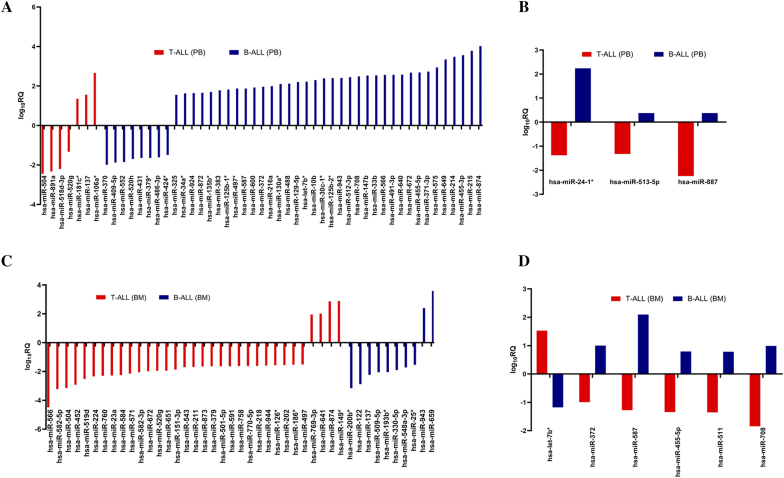
Peripheral Blood Profile of T-ALL and B-ALL Revealed Common and Unique MicroRNAs
Comparative analysis of PB in both T- and B-cell subtypes showed marked differences in their microRNA expression profiles revealing unique and common microRNAs expressed in the two subtypes (Supplementary Figure S5B, Supplementary Table S4, A and B). Analysis of PB of both subtypes showed that a set of 7 microRNAs and 45 microRNAs was uniquely expressed in T-ALL and B-ALL, respectively (Figure 3A). Significantly affected microRNAs included hsa-miR-181c*, miR-137, miR-106a*, miR-504, miR-891a, and miR-518d-3p in T-ALL and hsa-miR-455-5p, miR-566, miR-708, miR-372, miR-34a*, and miR-424*in B-ALL (Supplementary Table S5). Interestingly, among the common ones at same cutoff value, miR-24-1*, miR-513-5p, and miR-887 showed opposite expression patterns in PB of both subtypes (Figure 3B).
Figure 3.
Differential microRNA expression signature of PB and BM distinguishes T-ALL and B-ALL. (A), common significant microRNAs in PB with opposite expression between T-ALL and B-ALL (B), unique microRNAs in BM of T-ALL and B-ALL (C) and common microRNA in BM with opposite expression between T-ALL and B-ALL (D); log10RQ > 1.5.
Distinct MicroRNA Signatures Found in T-ALL and B-ALL Bone Marrow Samples
Differences in the microRNA signatures were also observed in the BM samples of both T- and B-cell types (Supplementary Figure S5C). A set of 33 microRNAs expressed was unique to T-ALL BM, while 10 were specific to BM of B-ALL (Figure 3C). The above-mentioned unique expression of microRNAs in BM of both subtypes was characterized by the most significant downregulation and upregulation exclusively to T-ALL and B-ALL subtype as compared to control (Supplementary Table S6). Further analysis revealed an inverse expression trend of a set of microRNAs between BM of T-ALL and B-ALL subtypes. Hsa-miR-372, miR-455-5p, miR-511, miR-587, and miR-708 were downregulated in T-ALL BM while they were upregulated in B-ALL BM, whereas hsa-let-7b* showed an inverse trend (Figure 3D). These microRNAs with their respective fold change and corresponding adjusted P values are shown in Supplementary Table S7, A and B, respectively.
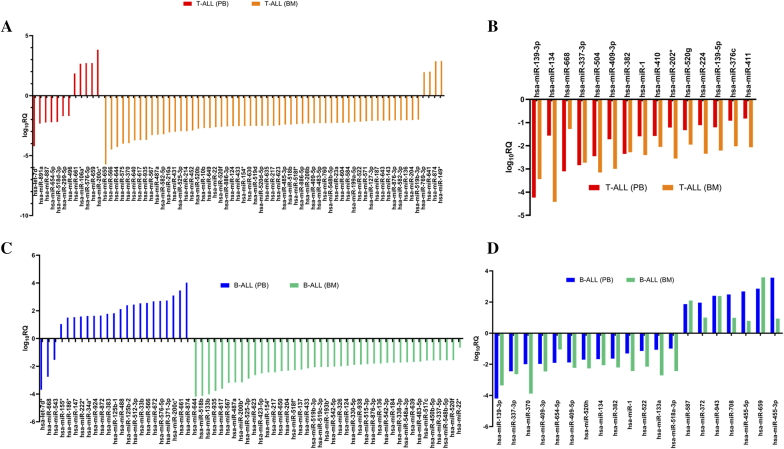
MicroRNA Profiling Unfolded Endogenous Differences Between Peripheral Blood and Bone Marrow Within ALL Subtypes
The most interesting finding of the current study was the endogenous differences in the expression pattern of a set of microRNAs between PB and BM samples within the same subtype of leukemia (Supplementary Figure S5, D and E). In T-ALL, 12 microRNAs of PB and 59 miRNAs of BM were identified to be differentially expressed (Figure 4A, Supplementary Table S8, A and B). Similarly, at the same cutoff value, 23 microRNAs were unique to PB, and 41 downregulated microRNAs were unique to BM of B-ALL subtype (Figure 4B, Supplementary Table S9, A and B). Among the microRNAs that showed similar expression profile in PB and BM of same subtypes, 15 microRNAs were downregulated in T-ALL subtype (Figure 4C, Supplementary Table S10A), while 20 microRNAs exhibited similar expression signature in B-ALL subtype (Figure 4D, Supplementary Table S10B).
Figure 4.
Unique and common microRNA expression signature discloses endogenous differences and similarities within T-ALL and B-ALL subtypes (A), unique microRNA expression signature between PB and BM distinguishes endogenous differences within T-ALL subtype (B) and B-ALL subtype (C), common microRNA signature reveals endogenous similarities between PB and BM subtype of T-ALL (D) and B-ALL subtype (log10RQ > 1.5).
MicroRNA Signatures Associated with Survival
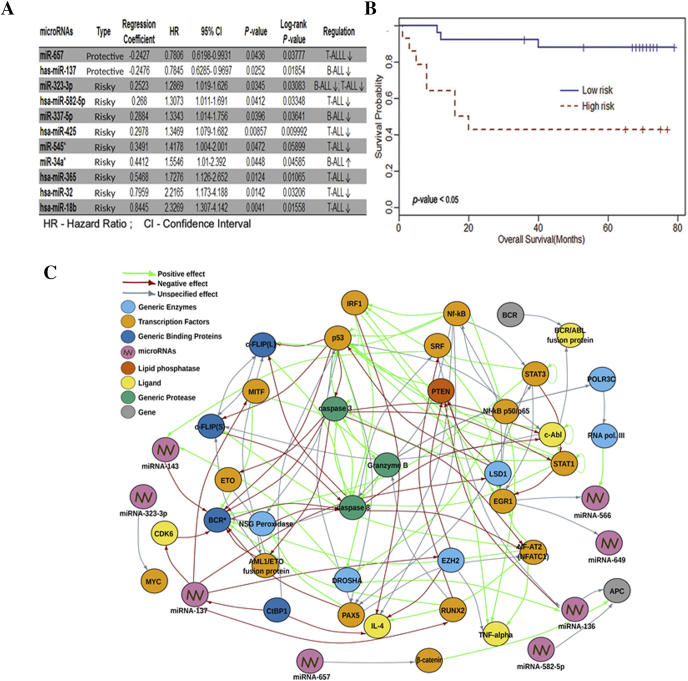
Cox proportional hazard regression analysis was carried out individually in 100 ALL samples (50 each in test and validation sets) to identify the correlation of expression signatures with patients' survival. Out of 397 microRNAs identified with ALL, 11 of them significantly correlated with overall survival. The risk score for each patient in the test set was calculated based on the correlation of the identified microRNA signatures to patient survival, and a score at 60th percentile divided patients into high- and low-risk groups. The patients in test set having risk score below cutoff value had low median survival of 18 months and were categorized as high-risk group, while the patients having risk score above the cutoff had better median survival of 69 months and thus categorized as low-risk group. The validation set confirmed our prediction and showed that patients in the low-risk group indeed had better survival rates (median survival: 69 months; HR: 0.76; 95% CI 0.64-0.91; P value = .0033) compared to high-risk group (median survival: 18 months; HR:1.09; 95% CI 1.03-1.17; P value = .0038). Among the 11 microRNAs, two of them were classified as protective and nine of them as risk indicative in patients' overall survival (P value ≤ .05) in ALL (Figure 5A). Kaplan-Meier curve was generated against the overall survival in these two risk groups (Figure 5B).
Figure 5.
Survival analysis of significant miRNAs. (A) Cox regression analyses using survival package indicating HR and respective P values. (B) Survival analysis depicted by Kaplan-Meier curve. (C) sna package of “R” program was used to find oncogenic targets of microRNAs.
In Silico Identification and In Vitro Validation of MicroRNAs and Their Targets
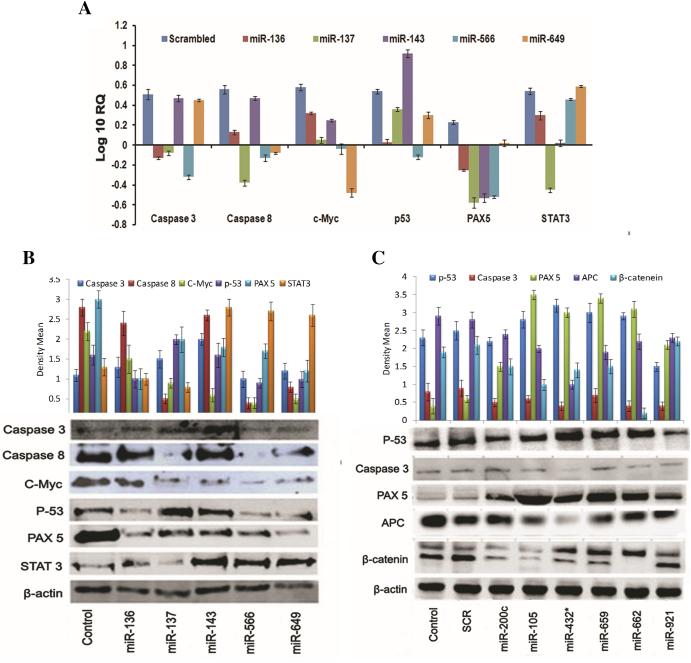
In silico pathway analysis using KEGG and GeneGo delineated the interaction of microRNAs and their targets in oncogenic pathways (Figure 5C, Supplementary Figure S6 and S7). Knockdown probes for miR-136, miR-137, miR-143, and miR-466 and mimic probes for miR-649, miR-200c, miR-105, miR-432, miR-659, miR-662, and miR-921 were used for in vitro validation to elucidate the functional role of microRNAs in leukemia cell lines. Plausible oncogenic pathway moieties such as caspase-3, caspase-8, and p53 were tested with knockdown probes, and STAT3, c-Myc, PAX-5, APC, and β-catenin were tested with mimics. qPCR results indicated a direct shutdown of caspase-3 by antisense oligonucleotides of miR-136, miR-137, miR-143, and miR-566 and by mimics of miR-649. Additionally, a similar regulation with respect to PAX-5, p53, and c-Myc was observed with all four antisense oligonucleotides and mimics of miR-649. These results clearly classified them as negative regulators of the respective targets (Figure 6A) which were further corroborated by immunoblotting studies (Figure 6B, Supplementary Figure S8). Mimics of miR-432 acted as negative regulators of APC and Caspase-3, while those of miR-200c, miR-105, miR-432*, miR-659, and miR-662 defined themselves as positive regulators of nonphosphorylated form of the oncogene β-catenin. This was further delineated using GeneGo pathway analysis that revealed the direct or indirect regulation of PAX-5 and BCR/ABL fusion protein by miRs-136, 137, 143, 566, and 649 as well as identified the involvement of diverse range of important generic enzymes and binding proteins in major oncogenic pathways (Figure 6C). Protein profiling of the ALL versus normal samples revealed differential expression of proteins found in ALL (Supplementary Figure S9 and S10). T-ALL proteome identified unique proteins specific to PB and BM samples highlighting endogenous differences between them. GeneGo MetaCore pathway analysis of the leukemic proteome identified alternative targets which are likely to be involved in the pathogenesis of ALL (Supplementary Figure S11 and S12).
Figure 6.
Effect of mimics and anti-miRs on their oncogenic targets tested in CCR5 cell lines. (A) Q-PCR analysis showing the knockdown and over expression of respective targets using selected microRNAs. (B) Immunoblot analysis showing the respective target knockdown and overexpression of caspase 3, caspase 8, c-Myc, p53, PAX5, and STAT 3. (C) p-53, caspase 3, PAX5, APC, and β-catenin. “Image J” was used for densitometeric analysis.
Discussion
We have performed a systematic method of genome-wide microRNA analysis in pediatric ALL using two different microRNA microarray platforms. Herein, we identified 51 novel microRNAs that have not been reported earlier to be associated with ALL. Unsupervised hierarchical clustering of microRNA profiles enabled stratification between T-ALL and B-ALL subtypes and revealed molecular differences between PB and BM within each subtype. Target validation unveiled direct and indirect interaction between OncomiRs: target networks suggesting a strong role of these microRNAs in leukemogenesis.
MicroRNA expression profiling exhibited a general trend of downregulation (85%) among the ALL patients. Common and altered expression of some microRNAs was detected in both T-ALL and B-ALL samples. miR-21, the first microRNA known to be expressed in almost all cancers [26], exhibited similar elevated expression in our study. The let-7 series, well known for their tumor suppressive attribute in different malignancies, were found to be downregulated. As observed here, upregulation of hsa-miR-496 has previously been linked with poor prognosis, while miR-134 expression is a favorable prognostic marker in ALL [27,28]. In support of our findings, upregulation of miR-708 and miR-128 has been associated with pathogenesis of high-risk common precursor B-ALL [29,30] and their subsequent downregulation with pediatric T-ALL [30,31]. The occurrence of these microRNAs that are common to both B-ALL and T-ALL, although with differential expression levels, helped in classifying the samples as ALL. In the present study, distinct microRNA expression in PB and BM samples was observed within the same subsets of ALL. In a similar study, Schotte et al. (2012) showed few of these microRNAs as significantly upregulated in B-ALL progenitor/stem cells [32], although not much has been reported about endogenous differences in these subtypes. Considering these observations, it seems likely that the molecular differences observed between PB and BM may reflect a key role of microRNAs involved in the hematopoietic cell differentiation depending upon cell types.
Further in our study, expression of 11 microRNAs was correlated with overall survival. Negative association of microRNA expression with survival identified two and nine of them as protective and risk indicative, respectively. We classified downregulation of mir-137 and miR-657 as protective in the present study; however, upregulation of these miRs has been reported with poor survival in lung cancer [33] and hepatocellular carcinoma [34]. The risk-indicative miR-582-5p reported here was observed in high-grade bladder cancer and in other ALL cases [[35], [36], [37]]. Further, Fulci et al. (2009) reported that miR-425-5p could discriminate B-ALL subgroups and its downregulation identified in BCR/ABL+ aberration [38]. Reduced expression of miR-18b and miR-32 was classified with shorter overall survival in various cancers [[39], [40], [41], [42]]. Recent studies from Wang et al. (2016) and Song et al. (2014) identified that lower expression of miR-323-3p and miR-545 was indicative of poor prognosis. Both these microRNAs were responsible for increased tumorigenesis with reduced survival in pancreatic ductal adenocarcinoma, respectively [43,44]. Downregulation of miR-337-5p and miR-365 was correlated with lymph node metastasis and poor survival in multiple cancers [[45], [46], [47], [48]]. Taken together, these studies have established an important role for 11 distinct microRNAs identified in our study that they could be used as predictive prognostic markers.
Target identification of miRNA genes is challenging because miRNAs bind to the target mRNA through incomplete sequence pairing, making it difficult to locate the complementary site. Network analysis between novel microRNAs and their targets revealed their potential role in regulation of various oncogenic signaling pathways. Pathway analyses with the deregulated microRNAs suggested that they were implicated in the functional regulation of various receptors, their cognate ligands, transcription factors, and apoptotic proteins found in major oncogenic pathways. Our pathway enrichment along with the qPCR and Western blot data analyses indicated a mechanistic interaction between differentially regulated microRNAs and their targets. This interdisciplinary approach, by studying in silico applications along with in vitro and in vivo validation methods, could help to characterize key pathways that impact cell survival and leukemogenesis leading to formulation of novel treatment regimens.
To gain further insights about the functional role of miRNA signatures, target genes were extracted from GO and KEGG pathways. Validation of a few significant microRNAs using qPCR and immunoblotting suggested similar functional interactions. For example, knockdown of miRs such as miR-136, miR-137, miR-143, miR-566, and miR-649 led to increased expression of proapoptotic signaling molecules such as caspase 3 and 8, thereby suggesting interplay between the upregulation of these microRNAs and carcinogenesis. Similarly, mimics of miR-432 negatively regulated APC and caspase 3, while miR-200c, miR-105, miR-432*, miR-659, and miR-662 had profound positive regulatory effect on the nonphosphorylated form of β-catenin, an oncogene indicating a strong network interaction between these microRNAs and wnt pathway regulators. Increased expression levels of PAX5 with mimics indicated that these microRNAs might be involved in its negative regulation that plays a central role in B-cell development and differentiation. This observation is supported by an earlier report that PAX5 mRNA levels were high in B-ALL patients [49]. These insights offer possibility of developing novel therapeutic approaches based upon the targets identified. Comparison of the proteome from patient groups with the controls revealed differential expression of proteins commonly reported in different cancers. Recent studies suggest the involvement of prolactin inducible protein as survival factor in seminal plasma of ALL patients, which also correlated with our studies [50]. Detailed studies are needed to compare protein dynamics in the subtypes of ALL that might lead to an interesting outcome.
In conclusion, we have deciphered the microRNA complement of ALL with a proof of concept for generating potential biomarkers that could be used for diagnosis, prognosis, and identifying potential therapeutic targets of ALL. MicroRNA regulation is an essential component in assessing the clinical outcome of pediatric ALL. The putative microRNA:target interaction studies also suggested novel oncogenic targets for therapeutic intervention. Our extensive validation strategy generated potential biomarkers for diagnosis and prognosis which would aid in the clinical paradigm for ALL subtype classification.
Ethics Approval and Consent to Participate
This study was approved by institutional review board and ethics committee of RCC and Centre for Cellular and Molecular Biology (CCMB), and all procedures were performed following the Declaration of Helsinki, after obtaining written informed consent from the parents/guardians.
Availability of Data and Materials
The complete data generated from this study are available at Gene Expression Omnibus GSE60386 (for TLDA) and GSE59199 (for LNA arrays) at http://www.ncbi.nlm.nih.gov/geo/. All the protocols associated with this study can be made available by email request to the corresponding author.
Declaration of Competing Interest
Authors declare no competing interests.
Funding
This work was funded by Department of Biotechnology (BT/PR10021/AGR/36/28/2007), Ministry of Science and Technology, Government of India.
Authors’ Contributions
Author contributions: L. D. K. and R. A. N. designed the study. L. E. A. and R. A. N. collected samples. R. A. N. and P. K. K. performed diagnosis, treatment, and management of patients. R. A. N., V. K. V., S. S. B., E. R. P., and L. E. A. carried out experiments and acquired data. L. D. K., R. A. N., V. K. V., S. S. B., and A. R. performed analyses and interpretation of the data. L. D. K., R. A. N., A. R., V. K. V., S. S. B., and P. K. editing and reviewing the manuscript.
Acknowledgements
Authors are grateful to Dr. Alpana Razdan and Dr. Mainak Banerjee, Perkin Elmer/LABINDIA, for TLDA statistical analysis using StatMiner R program and Dr. Torben and Dr. Michael, Exiqon, for LNA microarray analysis. We also acknowledge the assistance provided by Mr. Challa Kiran for northern analysis, immunoblotting, and proteomics experiments; and Ms. Aparna Kumari for maintaining and updating “LeukmiR” database at CCMB server. Special thanks to Mr. Aviral Kumar for his help with making all figures; Drs. V. Dinesh Kumar, Priya Jacob, and Dr. Guruprasad for critical reading and their suggestions regarding the manuscript; and Mr. Velu Mani Selvaraj and Mr. S. Guruprasadh for help in editing the manuscript. Special thanks to late Prof. Lalji Singh, ex-Director, CCMB, for his encouragement and support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100800.
Appendix A. Supplementary Data
Supplementary Figures
Supplementary Tables
References
- 1.Mitelman F., Heim S. Chromosome abnormalities in cancer. Cancer Detect. Prev. 1990;14(5):527–537. [PubMed] [Google Scholar]
- 2.Chiaretti S., Li X., Gentleman R., Vitale A., Vignetti M., Mandelli F.…Foa R. Gene expression profile of adult T-cell acute lymphocytic leukemia identifies distinct subsets of patients with different response to therapy and survival. Blood. 2004;103(7):2771–2778. doi: 10.1182/blood-2003-09-3243. PMid:14684422. [DOI] [PubMed] [Google Scholar]
- 3.Linet M.S., Ries L.A., Smith M.A., Tarone R.E., Devesa S.S. Cancer surveillance series: recent trends in childhood cancer incidence and mortality in the United States. J. Natl. Cancer Inst. 1999;91(12):1051–1058. doi: 10.1093/jnci/91.12.1051. PMid:10379968. [DOI] [PubMed] [Google Scholar]
- 4.Belson M., Kingsley B., Holmes A. Risk factors for acute leukemia in children: a review. Environmental health perspectives. 2006;115(1):138–145. doi: 10.1289/ehp.9023. 17366834 PMC1817663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vardiman J.W., Thiele J., Arber D.A., Brunning R.D., Borowitz M.J., Porwit A.…Bloomfield C.D. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. 19357394 [DOI] [PubMed] [Google Scholar]
- 6.Blenkiron C., Goldstein L.D., Thorne N.P., Spiteri I., Chin S.F., Dunning M.J.…Tavaré S. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8(10):R214. doi: 10.1186/gb-2007-8-10-r214. 17922911 PMC2246288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Connell R.M., Rao D.S., Chaudhuri A.A., Boldin M.P., Taganov K.D., Nicoll J.…Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 2008;205(3):585–594. doi: 10.1084/jem.20072108. 18299402 PMC2275382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garzon R., Croce C.M. MicroRNAs in normal and malignant hematopoiesis. Curr. Opin. Hematol. 2008;15(4):352–358. doi: 10.1097/MOH.0b013e328303e15d. 18536574 [DOI] [PubMed] [Google Scholar]
- 9.Kluiver J., Kroesen B.J., Poppema S., Van den Berg A. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia. 2006;20(11):1931. doi: 10.1038/sj.leu.2404387. 16990772 [DOI] [PubMed] [Google Scholar]
- 10.Schotte D., Chau J.C.K., Sylvester G., Liu G., Chen C., Van Der Velden V.H.J.…Den Boer M.L. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia. 2009;23(2):313. doi: 10.1038/leu.2008.286. 18923441 [DOI] [PubMed] [Google Scholar]
- 11.de Yébenes Virginia G. Regulation of B-cell development and function by microRNAs. Immunol. Rev. 2013;253(1):25–39. doi: 10.1111/imr.12046. 23550636 https://doi.org/10.1111/imr.12046. PMC3686225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mi S., Lu J., Sun M., Li Z., Zhang H., Neilly M.B.…Bohlander S.K. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl. Acad. Sci. 2007;104(50):19971–19976. doi: 10.1073/pnas.0709313104. 18056805 PMC2148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung S.S., Hu W., Park C.Y. The role of microRNAs in hematopoietic stem cell and leukemic stem cell function. Ther. Adv. Hematol. 2011;2(5):317–334. doi: 10.1177/2040620711410772. 23556099 PMC3573414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Si W., Shen J., Zheng H. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics. 2019;11(25) doi: 10.1186/s13148-018-0587-8. 30744689 PMC6371621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Björklund E., Mazur J., Söderhäll S., Porwit-MacDonald A. Flow cytometric follow-up of minimal residual disease in bone marrow gives prognostic information in children with acute lymphoblastic leukemia. Leukemia. 2003;17(1):138. doi: 10.1038/sj.leu.2402736. 12529671 [DOI] [PubMed] [Google Scholar]
- 16.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. 9847135 PMC148090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forbes S.A., Bindal N., Bamford S., Cole C., Kok C.Y., Beare D.…Teague J.W. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2010;39(suppl_1):D945–D950. doi: 10.1093/nar/gkp995. 19906727 PMC2808858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. https://doi.org/10.1186/gb-2003-5-1-p1 PMid:14709173 PMCid:PMC395733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. 15652477 [DOI] [PubMed] [Google Scholar]
- 20.Krek A., Grün D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J.…Rajewsky N. Combinatorial microRNA target predictions. Nat. Genet. 2005;37(5):495. doi: 10.1038/ng1536. 15806104 [DOI] [PubMed] [Google Scholar]
- 21.Rawoof A., Swaminathan G., Tiwari S., Nair R.A., Dinesh Kumar L. LeukmiR: a database for miRNAs and their targets in acute lymphoblastic leukemia. Database, 2020. 2020 doi: 10.1093/database/baz151. 32128558 PMC7054207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butts C.T. Social network analysis with sna. J. Stat. Softw. 2008;24(6):1–51. doi: 10.18637/jss.v024.i06. 18612375 PMC2443947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Therneau T.M., Grambsch P.M. Modeling survival data: extending the Cox model. Springer; New York, NY: 2000. Expected survival; pp. 261–287.10696153 [DOI] [Google Scholar]
- 24.Srinivasan S., Patric I.R.P., Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6(3):e17438. doi: 10.1371/journal.pone.0017438. 21483847 PMC3069027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxena S., Dupont S., Meghah V., Meena Lakshmi M.G., Singh S.K., Brahmendra Swamy C.V., Idris M.M. Proteome map of the neural complex of the tunicate C iona intestinalis, the closest living relative to vertebrates. Proteomics. 2013;13(5):860–865. doi: 10.1002/pmic.201200148. 23300126 [DOI] [PubMed] [Google Scholar]
- 26.Medina P.P., Nolde M., Slack F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86. doi: 10.1038/nature09284. 20693987 [DOI] [PubMed] [Google Scholar]
- 27.Schotte D., Pieters R., Den Boer M.L. MicroRNAs in acute leukemia: from biological players to clinical contributors. Leukemia. 2012;26(1):1–12. doi: 10.1038/leu.2011.151. 21701489 [DOI] [PubMed] [Google Scholar]
- 28.Mi S., Lu J., Sun M., Li Z., Zhang H., Neilly M.B.…Bohlander S.K. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl. Acad. Sci. 2007;104(50):19971–19976. doi: 10.1073/pnas.0709313104. 18056805 PMC2148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X., Li D., Zhuang Y., Shi Q., Wei W., Ju X. Retracted: overexpression of miR-708 and its targets in the childhood common precursor B-cell ALL. Pediatr. Blood Cancer. 2013;60(12):2060–2067. doi: 10.1002/pbc.24583. 23970374 [DOI] [PubMed] [Google Scholar]
- 30.Li M., Fu W., Wo L., Shu X., Liu F., Li C. miR-128 and its target genes in tumorigenesis and metastasis. Exp. Cell Res. 2013;319(20):3059–3064. doi: 10.1016/j.yexcr.2013.07.031. 23958464 [DOI] [PubMed] [Google Scholar]
- 31.de Oliveira J.C., Brassesco M.S., Scrideli C.A., Tone L.G., Narendran A. MicroRNA expression and activity in pediatric acute lymphoblastic leukemia (ALL) Pediatr. Blood Cancer. 2012;59(4):599–604. doi: 10.1002/pbc.24167. 22492670 [DOI] [PubMed] [Google Scholar]
- 32.Schotte D., De Menezes R.X., Moqadam F.A., Khankahdani L.M., Lange-Turenhout E., Chen C.…Den Boer M.L. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica. 2011;96(5):703–711. doi: 10.3324/haematol.2010.026138. 212421864 PMC3084917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu S.L., Chen H.Y., Chang G.C., Chen C.Y., Chen H.W., Singh S.…Su T.J. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13(1):48–57. doi: 10.1016/j.ccr.2007.12.008. 18167339 [DOI] [PubMed] [Google Scholar]
- 34.Zhang L., Yang L., Liu X., Chen W., Chang L., Chen L.…Yen Y. MicroRNA-657 promotes tumorigenesis in hepatocellular carcinoma by targeting transducin-like enhancer protein 1 through nuclear factor kappa B pathways. Hepatology. 2013;57(5):1919–1930. doi: 10.1002/hep.26162. 23175432 [DOI] [PubMed] [Google Scholar]
- 35.Uchino K., Takeshita F., Takahashi R.U., Kosaka N., Fujiwara K., Naruoka H.…Yoshiike M. Therapeutic effects of microRNA-582-5p and-3p on the inhibition of bladder cancer progression. Mol. Ther. 2013;21(3):610–619. doi: 10.1038/mt.2012.269. 23295946 PMC3589153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lionetti M., Biasiolo M., Agnelli L., Todoerti K., Mosca L., Fabris S.…Bortoluzzi S. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myelomas. Blood. 2009;114(25):e20–e26. doi: 10.1182/blood-2009-08-237495. https://doi.org/10.1182/blood.V114.22.2824.2824. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H., Yang J.H., Zheng Y.S., Zhang P., Chen X., Wu J.…Qu L.H. Genome-wide analysis of small RNA and novel microRNA discovery in human acute lymphoblastic leukemia based on extensive sequencing approach. PLoS One. 2009;4(9):e6849. doi: 10.1371/journal.pone.0006849. PMid:19724645 PMCid:PMC2731166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fulci V., Colombo T., Chiaretti S., Messina M., Citarella F., Tavolaro S.…Macino G. Characterization of B-and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosom. Cancer. 2009;48(12):1069–1082. doi: 10.1002/gcc.20709. 19760605 [DOI] [PubMed] [Google Scholar]
- 39.Fonseca-Sanchéz M.A., Pérez-Plasencia C., Fernández-Retana J., Arechaga-Ocampo E., Marchat L.A., Rodríguez-Cuevas S.…López-Camarillo C. microRNA-18b is upregulated in breast cancer and modulates genes involved in cell migration. Oncol. Rep. 2013;30(5):2399–2410. doi: 10.3892/or.2013.2691. 23970382 [DOI] [PubMed] [Google Scholar]
- 40.Zhu D., Chen H., Yang X., Chen W., Wang L., Xu J., Yu L. miR-32 functions as a tumor suppressor and directly targets SOX9 in human non–small cell lung cancer. OncoTargets Ther. 2015;8:1773. doi: 10.2147/OTT.S72457. 26229485 PMC4516199. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Zhang D., Ni Z., Xu X., Xiao J. MiR-32 functions as a tumor suppressor and directly targets EZH2 in human oral squamous cell carcinoma. Med. Sci. Monit. 2014;20:2527. doi: 10.12659/MSM.892636. 25472588 PMC4266205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J., Kuai X., Song M., Chen X., Yu Z., Zhang H., Mao Z. microRNA-32 inhibits the proliferation and invasion of the SGC-7901 gastric cancer cell line in vitro. Oncol. Lett. 2014;7(1):270–274. doi: 10.3892/ol.2013.1667. 24348862 PMC3861597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C., Liu P., Wu H., Cui P., Li Y., Liu Y.…Gou S. MicroRNA-323-3p inhibits cell invasion and metastasis in pancreatic ductal adenocarcinoma via direct suppression of SMAD2 and SMAD3. Oncotarget. 2016;7(12):14912. doi: 10.18632/oncotarget.7482. 26908446 PMC4924761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song B., Ji W., Guo S., Liu A., Jing W., Shao C.…Jin G. miR-545 inhibited pancreatic ductal adenocarcinoma growth by targeting RIG-I. FEBS Lett. 2014;588(23):4375–4381. doi: 10.1016/j.febslet.2014.10.004. 25315416 [DOI] [PubMed] [Google Scholar]
- 45.Hamdi K., Goerlitz D., Stambouli N., Islam M., Baroudi O., Neili B.…Elgaaied A.B. miRNAs in sera of Tunisian patients discriminate between inflammatory breast cancer and non-inflammatory breast cancer. Springerplus. 2014;3(1):636. doi: 10.1186/2193-1801-3-636. 26034677 PMC4447743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu C., Wang C., Guan X., Liu Y., Li D., Zhou X.…Zhang C.Y. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS One. 2014;9(3):e92292. doi: 10.1371/journal.pone.0092292. 24651474 PMC3961321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nie J., Liu L., Zheng W., Chen L., Wu X., Xu Y.…Han W. microRNA-365, down-regulated in colon cancer, inhibits cell cycle progression and promotes apoptosis of colon cancer cells by probably targeting Cyclin D1 and Bcl-2. Carcinogenesis. 2011;33(1):220–225. doi: 10.1093/carcin/bgr245. 22072615 [DOI] [PubMed] [Google Scholar]
- 48.Bai J., Zhang Z., Li X., Liu H. MicroRNA-365 inhibits growth, invasion and metastasis of malignant melanoma by targeting NRP1 expression. Cancer Biomark. 2015;15(5):599–608. doi: 10.3233/CBM-150500. 26406949 [DOI] [PubMed] [Google Scholar]
- 49.Firtina S., Sayitoglu M., Hatirnaz O., Erbilgin Y., Oztunc C., Cinar S.…Devecioglu O. Evaluation of PAX5 gene in the early stages of leukemic B cells in the childhood B cell acute lymphoblastic leukemia. Leuk. Res. 2012;36(1):87–92. doi: 10.1016/j.leukres.2011.07.017. 21813177 [DOI] [PubMed] [Google Scholar]
- 50.Jain P., Ojha S.K., Kumar V., Bakhshi S., Singh S., Yadav S. Differential seminal plasma proteome signatures of acute lymphoblastic leukemia survivors. Reprod. Biol. 2019 doi: 10.1016/j.repbio.2019.11.002. 31711845 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Tables
Data Availability Statement
The complete data generated from this study are available at Gene Expression Omnibus GSE60386 (for TLDA) and GSE59199 (for LNA arrays) at http://www.ncbi.nlm.nih.gov/geo/. All the protocols associated with this study can be made available by email request to the corresponding author.