Abstract
S100A12 is a member of the Ca2+ binding S100 family of proteins that functions within the human innate immune system. Zinc sequestration by S100A12 confers antimicrobial activity when the protein is secreted by neutrophils. Here, we demonstrate that Ca2+ binding to S100A12’s EF-hand motifs and Zn2+ binding to its dimeric interface cooperate to induce reversible self-assembly of the protein. Solution and magic angle spinning nuclear magnetic resonance spectroscopy on apo-, Ca2+-, Zn2+-, and Ca2+,Zn2+-S100A12 shows that significant metal binding-induced chemical shift perturbations, indicative of conformational changes, occur throughout the polypeptide chain. These perturbations do not originate from changes in the secondary structure of the protein, which remains largely preserved. While the overall structure of S100A12 is dominated by Ca2+ binding, Zn2+ binding to Ca2+-S100A12 introduces additional structural changes to helix II and the hinge domain (residues 38–53). The hinge domain of S100A12 is involved in the molecular interactions that promote chemotaxis for human monocyte, acute inflammatory responses and generates edema. In Ca2+-S100A12, helix II and the hinge domain participate in binding with the C-type immunoglobulin domain of the receptor for advanced glycation products (RAGE). We discuss how the additional conformational changes introduced to these domains upon Zn2+ binding may also impact the interaction of S100A12 and target proteins such as RAGE.
Graphical Abstract
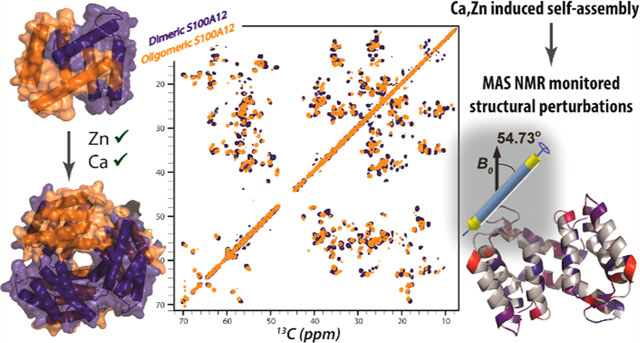
The antimicrobial activity by the human innate immune system halts pathogen growth and proliferation by sequestering critical nutrients. This “nutritional immunity” is a first level of defense against microbial invasion.1,2 Invading microbes require transition metal ions such as Fe2+, Mn2+, and Zn2+ for many of their proteins involved in cellular metabolism. By confiscating these metal ions, immune cells limit their access to pathogens. This limitation leads to a “fight over metals” at the host pathogen interface.2,3 In humans, several members of the S100 family of proteins sequester transition metal ions following secretion into the extracellular space during infection.2,4 S100A8 and S100A9 sequester Fe2+, Mn2+, Zn2+, and Ni2+,5–10 while S100A7 and S100A12 bind Zn2+.2,4,11–13
Expressed exclusively in vertebrates, S100 proteins are Ca2+ binding small polypeptides that are responsible for cell proliferation, signal transduction, differentiation, cell cycle regulation, and transcription.14,15 S100A12 (also known as Calgranulin C) is a 92-amino acid member of this family that performs Zn2+ binding-mediated antimicrobial functions.2,4 In vitro, S100A12 has been shown to exhibit antimicrobial activities against Brugia malayi,16 to remove Zn2+ from standard microbial growth media, and to inhibit the growth of several pathogenic fungi and human gastrointestinal pathogen Listeria monocytogenes.13 Elevated levels of this protein have been detected in gastric biopsies of patients colonized with Helicobacter pylori.17 These findings have led to the conclusion that the antimicrobial properties of S100A12 are exerted by sequestration of Zn2+. Once excreted, S100A12 initiates a pro-inflammatory signaling cascade by interacting with pattern recognition receptors such as the receptor for advanced glycation end products (RAGE) and Toll-like receptor 4.4,18–21 Increased serum levels of S100A12 have been reported in patients suffering from neurodegenerative, metabolic, and neoplastic disorders.22 S100A12 has been suggested to be used as a marker for disease and infection.23–25 These findings show that S100A12 is a key component of the innate immune system mediating both antimicrobial and proinflammatory activities.
S100A12 is composed of four α-helices (helices I–IV) and two Ca2+ binding EF-hand loops [G9–T37 and I54–A85 (Figure 1)].26–28 The motif connecting helices II and III is named the “hinge” (K38–V53). Although members of the S100 family share a high degree of sequence similarity, variations responsible for functional divergence are observed in the hinge region. In human S100A12, the hinge domain is responsible for initiating chemotactic activity for human monocytes in vitro and generating proinflammatory responses and edema in vivo.29 Apo-S100A12 is dimeric; structural studies infer that the dimer is predominantly stabilized by hydrophobic interactions mediated by helix IV and portions of helix I.26 Zn2+ and Cu2+ bind to the same site at the S100A12 dimer interface.26,30 The transition metal ions bind via a His3Asp motif composed of H85 and H89 from one subunit and H15 and D25 from the second subunit (Figure 1), resulting in two metal binding sites per dimer. Although Zn2+-mediated S100A12 antimicrobial activities have been established, the biological role of binding of Cu2+ to S100A12 is unknown.
Figure 1.
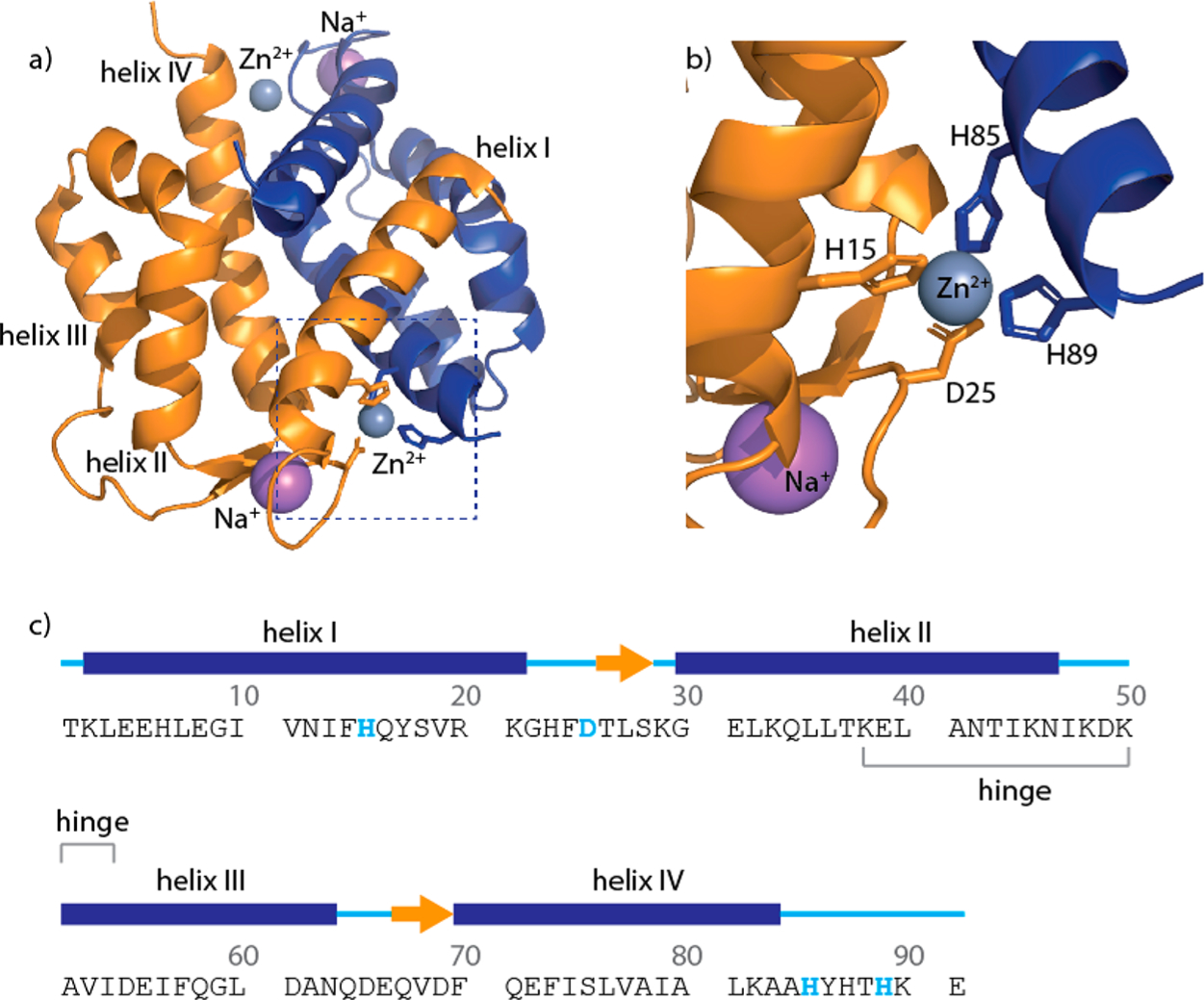
Amino acid sequence and structure of zinc-bound S100A12. (a) Crystal structure of Zn2+-S100A12 (PDB entry 2WCB). (b) Close-up showing the zinc binding motif with residues H15 and D25 (orange) from one monomer and H85 and H89 (blue) from the second monomer. (c) Polypeptide sequence and secondary structure of the apoprotein based on the crystal structure (PDB entry 2WCF). Gray and purple spheres denote zinc and sodium ions, respectively.
Many S100 proteins self-assemble. The S100A8/S100A9 heterodimer forms tetramers.31 Metal binding initiates self-assembly of S100A12. S100A12 hexamers are observed in structures crystallized in the presence of excess calcium,28 while tetramers are observed in structures crystallized in the presence of Zn2+.32 Self-assembly of S100 proteins is functionally relevant;33,34 higher-order S100A12 assemblies are present in blood serum and human tissue.35,36 Additionally, hexameric S100A12 interacts with RAGE and Toll-like receptor 4 (TLR-4),21,37,38 suggesting that the binding of oligomeric S100A12 to membrane receptors may trigger cellular signaling and inflammation. Clearly, a structural understanding of oligomeric S100A12 is necessary to evaluate its biological role.
Although insight into the structure of S100A12 has been obtained for its apo and Ca2+-, Ca2+-, and Cu2+,Zn2+-bound forms by nuclear magnetic resonance (NMR)39 and X-ray crystallography,26,27,30 a structure of S100A12 simultaneously bound to Zn2+ and Ca2+ is not available. The current belief is that Ca2+ binding dominates the architecture of S100A12 and binding of Zn2+ to Ca2+-S100A12 introduces only minor perturbations into the structure. The absence of a dual divalent cation-bound structure is important as the Ca2+ concentration in serum is high; Zn2+ sequestration-mediated antimicrobial activity by S100A12 clearly must involve the calcium-bound protein. Additionally, both divalent cations are necessary for S100A12 induction of Toll-like receptor 4 signaling.21 These findings underscore the unexplored importance of synergistic cation-mediated activation of S100A12 in the immune response and the importance of structural characterization of this state of the protein.
We present in this paper structural and assembly studies of S100A12 designed to distinguish the effects that are unique to Ca2+ or Zn2+ binding and the physiologically relevant condition of simultaneous binding of these cations. Although Zn2+ much more potently stimulates S100A12 self-assembly than does Ca2+, the two cations cooperate to facilitate higher-order assembly. Solution-state and magic angle spinning (MAS) NMR characterization of Zn2+- and Zn2+,Ca2+-bound oligomeric S100A12 reveals that although Ca2+ binding is known to introduce major structural rearrangements into the apoprotein and the structure of Ca2+,Zn2+-S100A12 is expected to be dominated by calcium binding, significant conformational changes are induced by the binding of zinc to Ca2+-S100A12. These structural perturbations may promote self-assembly by affording energetically favorable oligomers and assist in the interaction of S100A12 with membrane receptors. On the basis of the observed reversible self-assembly of S100A12 in the presence of Zn2+ and/or Ca2+ and the structural changes introduced due to zinc binding, we propose a cellular model of S100A12 function in the extracellular space. Our hypothesis suggests that the overexpression and secretion of S100A12 at disease sites and subsequent binding of Zn2+ and Ca2+ lead to its self-assembly. The structural changes introduced upon binding of divalent metal ions to assembled S100A12, as reported herein, are hypothesized to induce cellular signaling specific to inflammation and disease activities. In our model, these signaling pathways are not initiated in healthy cells with normal S100A12 expression where dimeric protein is expected to be largely present.
MATERIALS AND METHODS
Purification of S100A12 and Sample Preparation for NMR Measurements.
The cDNA of human S100A12 (UniProt entry P80511) was synthesized and subcloned into the pET-41a(+) vector by GenScript. The cloned plasmid was transformed into BL21(DE3) Escherichia coli cells. The cell culture was incubated at 37 °C in Luria broth or minimal medium containing [U-15N]NH4Cl and/or [U-13C]-D-glucose. Upon induction by 1 mM isopropyl β-D-1-thiogalactopyrano-side (IPTG), the cell culture was grown at 18 °C for 20 h and harvested by centrifugation at 4000 rpm for 30 min. The cell pellet was resuspended in lysis buffer [50 mM Tris-HCl (pH 7.4)] and sonicated, and the lysate was centrifuged at 15000 rpm and 4 °C for 40 min. The supernatant was loaded onto a diethylaminoethanol (DEAE) anion exchange column preequilibrated with loading buffer [50 mM Tris-HCl (pH 7.4)]. Eluted fractions containing S100A12 were identified by SDS-PAGE and pooled together. Five millimolar ethylenediaminetetraacetic acid (EDTA) was added to remove divalent metal ions followed by dialysis to remove excess EDTA. The protein was further purified with a size exclusion column (GE Healthcare Life Science, HiLoad 16/600 Superdex 75 prep grade) with 10 mM HEPES (pH 7.0), 150 mM NaCl running buffer. Size exclusion chromatography was performed twice to completely remove any trace amounts of EDTA remaining after dialysis. The amino acid sequence of purified S100A12 was verified by liquid chromatography–electrospray ionization tandem mass spectrometry (LC–ESI-MS/MS). Two equivalents of Zn2+ and 20 equiv Ca2+ were added for Zn2+-S100A12 and Ca2+,Zn2+-S100A12 NMR sample preparation. For the preparation of NMR samples containing zinc, 200 μM Zn2+ was added to 100 μM protein and the solution was then concentrated to the desirable protein concentration. The samples for solution NMR measurements were prepared at a S100A12 concentration of 1 mM in 20 mM HEPES buffer (pH 7.5) containing 150 mM NaCl, 5% D2O, and 200 μM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS). MAS NMR samples were prepared by gradual addition of a 30% (w/v) PEG-3350 aqueous solution to 2 mM protein samples in 20 mM HEPES buffer (pH 7.5) containing 150 mM NaCl, using a protocol similar to that reported previously.40
Mass Spectrometry.
S100A12 sequence analysis and determination of the purity of the protein after purification were conducted by LC-MS and LC-MS/MS. The sample preparation protocol and mass spectrometry analysis are reported in the Supporting Information.
Analytical Ultracentrifugation (AUC).
AUC studies were conducted in a Beckman XL-I analytical ultracentrifuge using the An60-Ti rotor with the absorption optics set to either 230 or 280 nm to expand the accessible protein concentration range. The protein absorbance was tracked using double-sector cell assemblies with the sample blanked against buffer. All experiments were performed at 20 °C in 50 mM Tris-HCl and 150 mM NaCl (pH 7.5) to which the indicated amount of divalent cation had been added. For AUC sample preparation, stock metal ion solutions were added to 0.1 mM apoprotein to achieve concentrations of 0.2 mM Zn2+ and/or 20 mM Ca2+. These concentrations of the divalent metal ions are sufficient for complete metal binding based on published Kd values and our NMR measurements. The protein solution was dialyzed against buffer containing 0.2 mM Zn2+ and/or 20 mM Ca2+ and concentrated to 2.5 mM. Additional dilutions conducted for AUC studies used buffers containing the respective metal ions at the concentrations mentioned above. The partial specific volume of the protein and the solute density and viscosity were calculated using Sednterp (B. Hayes, T. Laue, and J. Philo, Sedimentation Interpretation Program, University of New Hampshire, Durham, NH) from the amino acid and buffer compositions, respectively. Using these values, resolved sedimentation parameters were corrected to standard conditions (20,W). To facilitate comparison with published crystal structures, sedimentation and diffusion coefficients were calculated from PDB entries 2wcf, 2wc8, and 1GQM using HYDROPRO.41
Sedimentation velocity (SV) experiments were performed at 55000 rpm. Approximately 100 scans were acquired over the course of a SV experiment. A subset of these scans were analyzed by the time-derivative (dc/dt) method using DCDT+ version 2.4.2.42,43 The dc/dt method removes time-invariant noise from the data by subtracting pairs of scans. Normalizing and averaging multiple scan pairs improves the precision of resolved sedimentation parameters. Sedimentation equilibrium (EQ) experiments were conducted using six channel centerpieces in the Ti-60 rotor; samples were equilibrated for 24 h at each of three rotor speeds and globally analyzed using HeteroAnalysis version 1.0.114 (J. L. Cole and J. W. Lary, Analytical Ultracentrifugation Facility, Biotechnology Services Center, University of Connecticut, Storrs, CT).
NMR Spectroscopy.
1H–15N HSQC44 and three-dimensional (3D) 1H–15N TOCSY-HSQC,45 NOESY-HSQC,46,47 and 1H–15N–13C CBCACONH48 solution NMR experiments were performed at 14.1 T (600 MHz) on a Varian NMR spectrometer outfitted with an HCN cryoprobe. The Larmor frequencies were 599.93, 150.87, and 60.79 MHz for 1H, 13C, and 15N, respectively, and the sample temperature was maintained at 25 °C. A 3D 1H–15N–13C HNCACB49 experiment was conducted at the City University of New York’s Advanced Science Research Center at 16.4 T on a standard bore Bruker Avance III HD spectrometer equipped with a 5 mm quadruple resonance inverse QCI-F cryoprobe. Ca2+-S100A12 solution NMR measurements were performed under conditions reported in the literature at 14.1 T.39 13C–13C dipolar assisted rotational resonance (DARR)50 MAS NMR experiments were performed at 14.1 T on a Varian NMR spectrometer using a fast MAS PhoenixNMR 1.6 mm HXY probe. The spectra were acquired at a MAS frequency of 12 kHz controlled by a Tecmag MAS controller. The temperature was maintained at 4.0 ± 0.1 °C. The Larmor frequencies were 599.93 and 150.87 MHz for 1H and 13C, respectively. The typical pulse lengths were 2.4 μs (1H) and 2.2 μs (13C). 1H–13C cross-polarization (CP) was performed with a linear amplitude ramp (80–100%); the CP contact time was 1.5 ms, and the DARR mixing time was 15 ms. A two-pulse phase modulation (TPPM)51 decoupling was applied during the acquisition. 3D NCACX and NCOCX spectra were acquired at 21.15 and 17.6 T, respectively, on a Bruker Avance II spectrometer outfitted with a 3.2 mm HCN EFree probe at the New York Structural Biology Institute. All 3D spectra were acquired at a MAS frequency of 14 kHz. These experiments utilized SPECIFIC-CP52 heteronuclear magnetization transfer from 15N to 13Cα/13CO using a 4 ms tangent amplitude ramp on the 15N channel, followed by the DARR mixing sequence. The contact time for 1H–15N CP was 1.5 ms, and a recycle delay of 2.0 s was used for all experiments.
NMR Data Processing and Secondary Structure Propensity (SSP) Calculations.
All NMR spectra were processed with NMRpipe53 and analyzed with Sparky.54 A forward linear prediction to twice the number of original data points followed by zero filling to twice the total number of points was used for all experiments. A 30°- and 60°-shifted sine bell apodization with water suppression function were used for both dimensions on solution NMR spectra. All solid-state spectra were processed using 30°-, 45°-, 60°-, or 90°-shifted sine bell apodization followed by a Lorentzian-to-Gaussian transformation in all dimensions.
SSPs were calculated using 13Cα and 13Cβ chemical shifts by a method described by Marsh et al.55 The SSP program (provided by the authors) calculates a single score representing the fraction of α and β structure propensities. An SSP score of 1 represents a fully formed α structure, while a score of −1 represents a fully formed β structure. Positive and negative SSP values between 1 and −1 indicate α and β structures, respectively, and the magnitude of the score gives the propensity of the secondary structure.
RESULTS
Characterization of Apo-S100A12 and Metal-Bound S100A12.
As the first step in determining the structural changes in S100A12 that occur during its divalent cation-mediated oligomerization, we characterized the dimeric apoprotein by solution NMR spectroscopy. As expected, apo-S100A12 is a stable dimer under the experimental conditions used in this study in the absence of any divalent cations. The results of SV analysis are consistent with a monodisperse dimer at S100A12 concentrations from 6 to 233 μM. The time-derivative profiles are described well by the normal distribution, and the measured values of S increase with a decrease in protein concentration. For a monodisperse particle, the ratio of the sedimentation and diffusion coefficients provides an accurate estimate of the molecular weight (Figure 2a and Table 1). The resolved coefficients are consistent with those of the S100A12 dimer calculated by the program HydroPro from the crystal structure of PDB entry 2WCF (Table 1).
Figure 2.
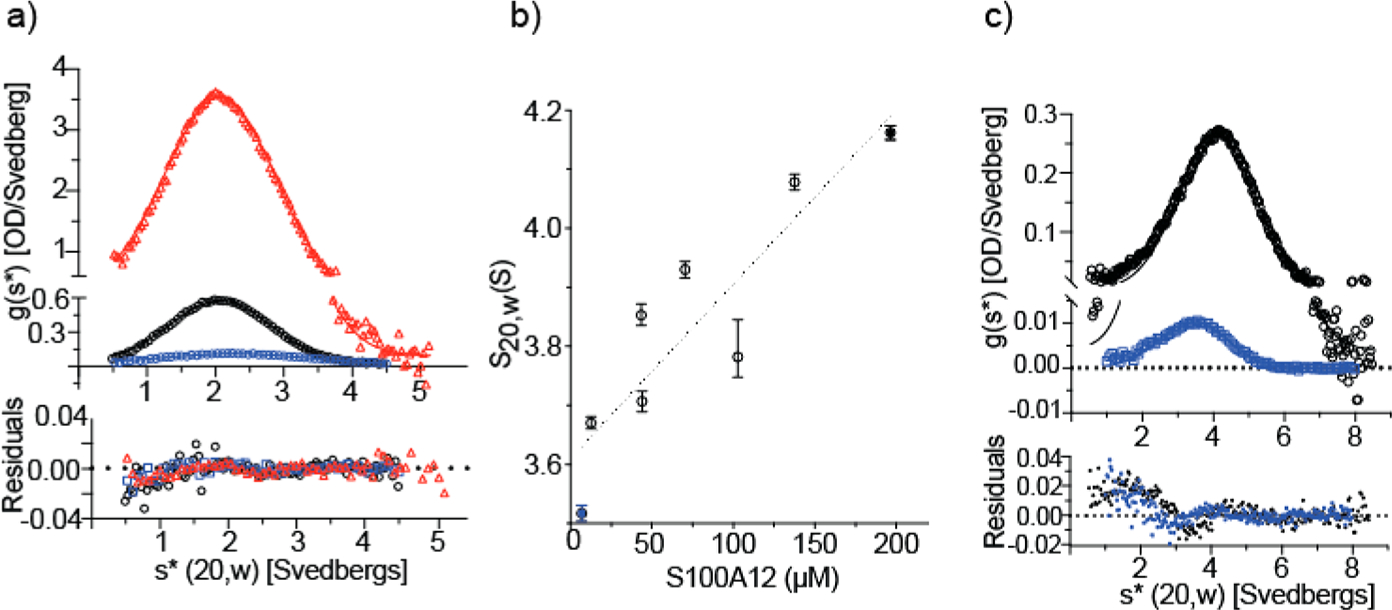
Sedimentation velocity (SV) analyses of (a) apo-S100A12 and (b and c) Zn2+-S100A12 with the cation present in 2-fold molar excess relative to the protein. Panels A and C show the time-derivative distributions and the best fits of the data to a single-component (normal) distribution (Table 1 for panel A). In panel A, apo-S100A12 was analyzed at 233 μM (red), 23 μM (black), and 6 μM (blue). Sedimentation of the first sample was tracked at 280 nm; the latter samples were tracked at 230 nm. Panel C shows two of the Zn2+-S100A12 samples that were analyzed: 196 μM (black) and 6.9 μM (blue) tracked at 280 and 230 nm, respectively. Panel B shows that S20,w increases with S100A12 concentration for the eight protein concentrations analyzed; the solid blue and black symbols represent the two distributions shown in panel C. The dotted line depicts a linear regression that highlights the upward trend of the data and does not represent a particular assembly model.
Table 1.
Summary of the Sedimentation Parameters Determined from Velocity Centrifugation of Apo-S100A12
| 233 ,μM S100A12a | 23 ,μM S100A12a | 6 μM S100A12a | PDB entry 2WCFb | |
|---|---|---|---|---|
| S (20,w) (S) | 2.195 (2.185, 2.205) | 2.165 (2.163, 2.167) | 2.483 (2.469, 2.497) | 2.4 |
| S/D (Mw) (kDa) | 19.90 (19.25, 20.55) | 20.75 (20.57, 20.93) | 18.15 (17.11, 19.12) | 20.3 |
| D (20,w) (F) | 10.53 (10.20, 10.86) | 9.96 (9.87, 10.04) | 13.05 (12.29, 13.89) | 9.7 |
The parameters were obtained from fitting the time-derivative distributions shown in Figure 2 to a single-component model as described in Materials and Methods.
The hydrodynamic parameters were calculated from the indicated crystal structure using the program HydroPro as described in Materials and Methods.
Figure 3a shows 14.1 T 1H–15N HSQC spectrum of the apoprotein. The spectrum shows 110 well-resolved peaks, 91 of which originate from the backbone amide nitrogen atoms. Additional 3D 1H–15N NOESY-HSQC, TOCSY-HSQC (Figure 3b), and 1H–13C–15N HNCACB, CBCACONH experiments allowed for the assignment of 80 of 92 backbone and 86 side chain (of 110) resonances. These assignments are presented on the 1H–15N HSQC spectrum in Figure 3a. Solution NMR studies and resonance assignments of calcium-bound S100A12 have been reported.39 To ensure our sample preparation methodology generates spectra identical to those reported in the literature, we performed 1H–13C–15N HNCACB, CBCACONH experiments on uniformly 13C- and 15N-labeled Ca2+-S100A12 (Figure S3). The chemical shifts obtained from our measurements were in complete agreement with the reported assignments39 and allowed for the identification of 73 backbone resonances.
Figure 3.
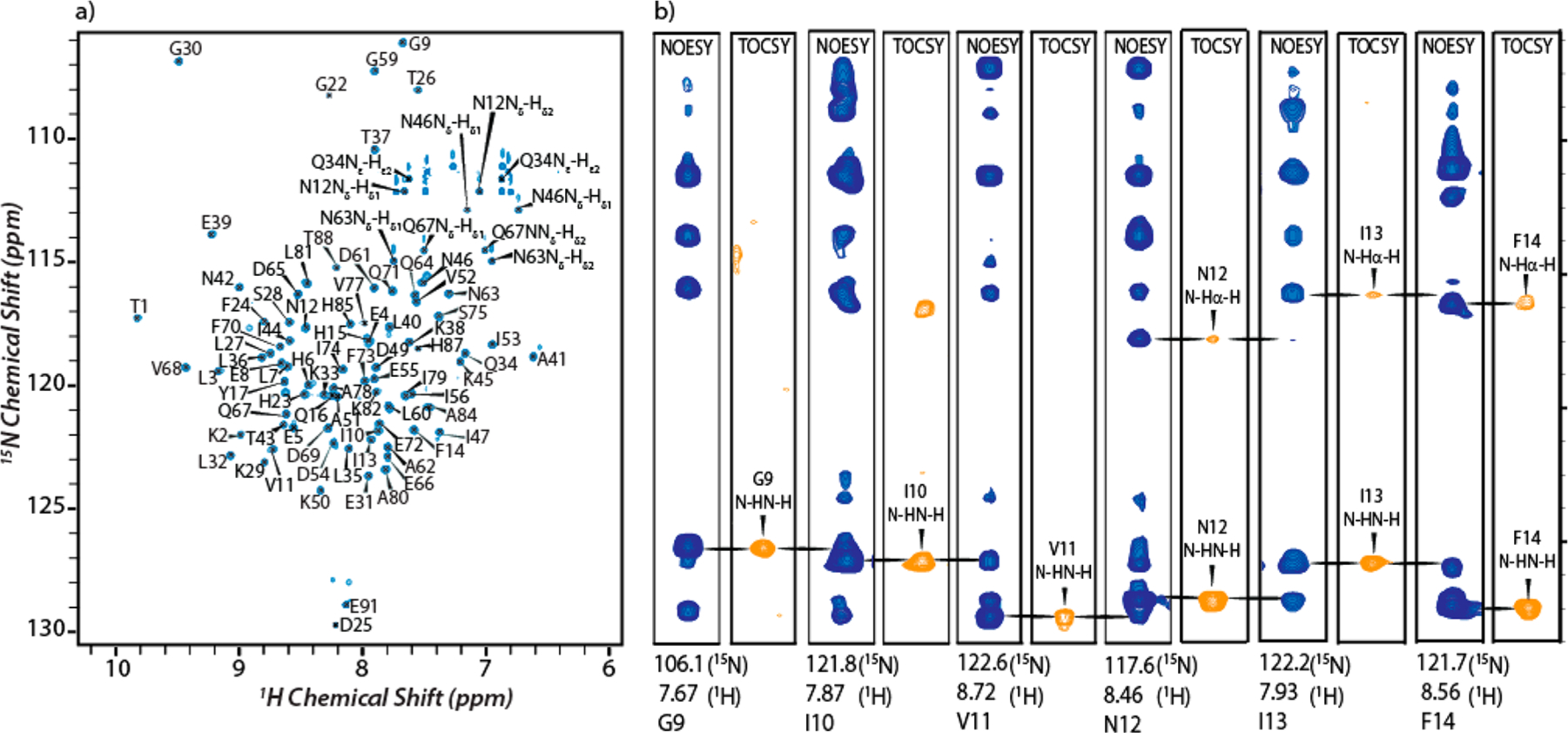
1H–15N HSQC spectrum and selected two-dimensional planes of the 3D 1H–15N NOESY-HSQC and TOCSY-HSQC spectra: (a) 14.1 T 1H–15N HSQC spectrum of apo-S100A12 with assignment of resonances and (b) strip plots from 1H–15N NOESY-HSQC (blue) and TOCSYHSQC (orange) spectra for residues G9–F14.
In contrast to the apoprotein, zinc-bound S100A12 exhibited a dramatic loss of NMR signal intensities. The two-dimensional (2D) 1H–15N HSQC spectrum of zinc-bound S100A12 showed only a few signals (Figure S4). Similar signal loss was observed for Ca2+,Zn2+-S100A12. These results are presumably because of the increased molecular weights of oligomeric S100A12 assemblies, which prohibit their detection in the solution state due to attenuated molecular tumbling. This conjecture was confirmed by AUC analysis. SV analyses were conducted as a function of S100A12 concentration in which Zn2+ was present in 2-fold molar excess. In contrast to that of the apoprotein, the level of sedimentation of Zn2+-S100A12 increases with an increase in protein concentration (Figure 2b). In addition, the time-derivative profiles systematically deviate from the normal distribution (Figure 2c). This behavior is diagnostic of a system undergoing reversible self-assembly; for such a system, the molecular weight is not accurately reported by the ratio of the sedimentation and diffusion constants. Because S20,w does not plateau with S100A12 concentration over the analyzed range, we are unable to estimate the size of the terminal oligomer of the assembly reaction from the SV experiments.
An alternative AUC approach is sedimentation equilibrium (SE) that reports the weight-average molecular weight (Mw). S100A12 samples were analyzed in a buffer containing Ca2+, Zn2+, or both Zn2+ and Ca2+. The results of the SE analysis show that Zn2+ is a much more potent facilitator of S100A12 self-assembly than is Ca2+ and that, when bound simultaneously, the two cations cooperate to drive the self-assembly. When the data sets are fit to the single-component model, the Mw values determined do not correspond to multiples of the dimer [21146, 42292, and 63439 Da for the dimer, tetramer, and hexamer, respectively (Figure 4 and Table 2)]. The fits to the single-component model display nonrandom residuals (data not shown) demonstrating cation-mediated reversible self-assembly of S100A12. This result is consistent with the SV results obtained for Zn2+-S100A12 (Figure 2b,c).
Figure 4.
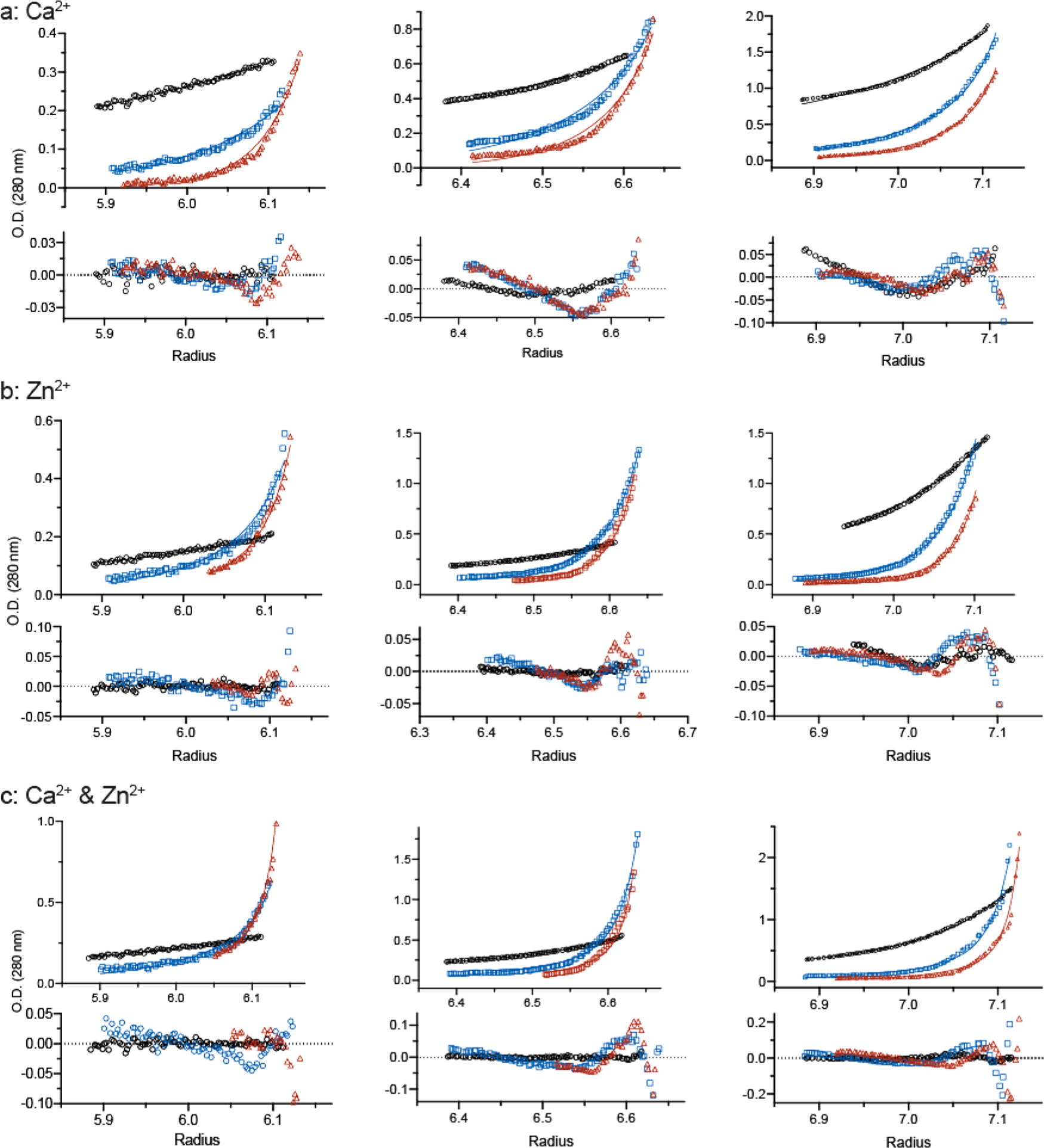
Sedimentation equilibrium analysis of S100A12 equilibrated in a buffer containing (a) 20 mM Ca2+, (b) 0.2 mM Zn2+, and (c) 20 mM Ca2+ and 0.2 mM Zn2+. The protein concentrations loaded into each sector of the six-channel centerpiece were 33 μM (left), 98 μM (middle), and 327 μM (right). The concentration gradients were determined using the absorption optics set at 280 nm. The best global fits of the dimer–octamer model to the three protein concentrations equilibrated at 12000 (black), 24000 (blue), and 30000 rpm (red) are shown along with the residuals; the resolved Kd values are cited in the text.
Table 2.
Summary of the Sedimentation Parameters Determined by Equilibrium Centrifugation of S100A12 Complexed with Ca2+, Zn2+, and Both Zn2+ and Ca2+
| metal ion | Mw (Da) | Kd(DX-O) (μM) |
|---|---|---|
| 20 mM Ca2+mM | 32750 (28610, 37489) | 4585.3 (4005.7, 5248.8) |
| 0.2 mM Zn2+ | 47320 (41005, 54607) | 56.7 (49.1, 65.4) |
| 0.2 mM Zn2+ and 20 Ca2+ | 68225 (59228, 78588) | 13.2 (11.5, 15.2) |
Attempts to fit each of these data sets to a variety of self-assembly models failed to yield a “clear winner” displaying random residuals. One issue is that sufficiently high protein concentrations cannot be analyzed to significantly populate terminal oligomers of each self-assembly reaction, if in fact assembly can reach saturation. To provide a consistent numerical comparison, we invoked Occam’s razor and report the Kd values from fits to the dimer-octamer assembly model that can embrace all three conditions (Table 2). This calculation highlights the observation that Zn2+ and Ca2+ cooperate to stimulate S100A12 to form high-order oligomers that are too large for solution NMR analysis. These data successfully explain the dramatic loss of NMR signal intensity for Zn2+- and Ca2+-bound S100A12 noted above. We therefore turned to solid-state NMR structural characterization of the oligomeric Zn2+- and Ca2+-bound S100A12 assemblies.
MAS NMR Spectra of Dimeric and Oligomeric S100A12.
Figure 5a shows 2D MAS NMR 13C–13C (12 kHz MAS; Bo = 14.1 T) and 13C–15N (12 kHz MAS; Bo = 16.4 T) correlation spectra of apo-S100A12, which exhibits highly resolved NMR spectra in the solid state. With the aid of 3D HNCACB and CBCACONH solution NMR experiments, assignments were made for 81 residues in the 2D 13C–13C correlation MAS NMR spectrum (Figure 5a). The near-perfect alignment of solid- and solution-state spectra shows that the structure of the apoprotein in the solid-state NMR sample is identical to that in the solution, demonstrating that the sample preparation protocol employed for MAS NMR measurements does not disrupt the overall structure of the protein while maintaining a hydrated environment that mimics solution-like conditions. A similar strategy was used for Ca2+-S100A12. The 2D MAS NMR 13C–13C (12 kHz MAS; Bo = 14.1 T) correlation spectrum of Ca2+-S100A12 was acquired, and Cα and Cβ resonances were assigned by comparison with 3D HNCACB and CBCACONH solution NMR data. The detection of Zn2+- and Ca2+,Zn2+-S100A12 assemblies required MAS NMR measurements. Well-resolved NMR spectra were observed for the zinc-bound S100A12 in the solid state, suggesting homogeneous S100A12 assemblies. An overlay of 13C–13C MAS correlation spectra of the apoprotein and zinc-bound protein revealed several chemical shift perturbations (CSPs) in the Cα and Cβ resonances, as shown in Figure 5b. In particular, significant CSPs (around 1 ppm) were observed for Cα atoms. On the basis of these perturbations and 3D NCACX and 3D NCOCX measurements, assignments of 57 of 92 amino acids (62% of the polypeptide chain acids) in the zinc-bound assemblies were made (Figure 5c–e and Figure S5).
Figure 5.
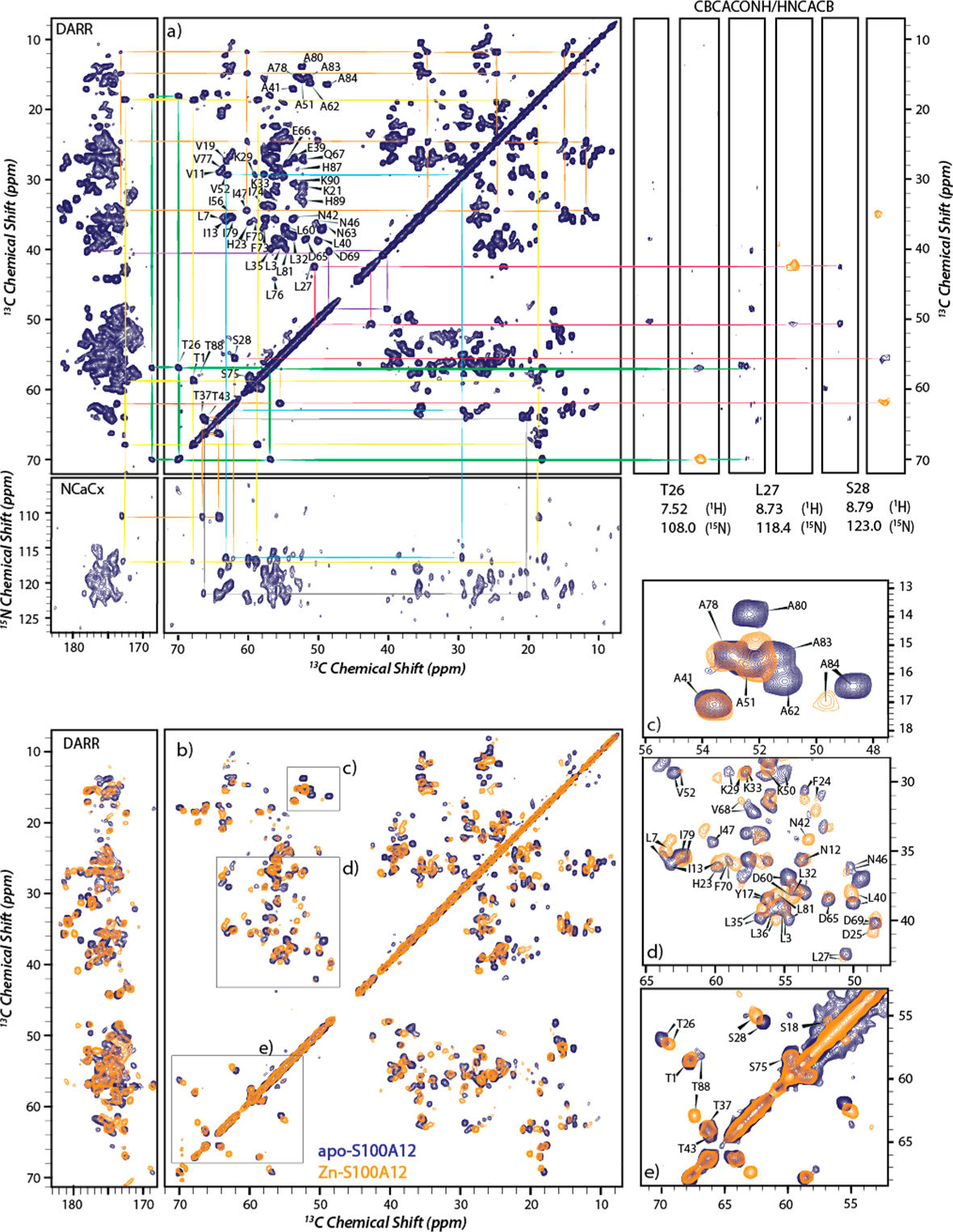
MAS NMR spectra of apo- and Zn2+-S100A12. (a) 13C–13C DARR (14.1 T) and 13C–15N NCACX (16.4 T) 2D correlation spectra of apo-S100A12 with resonance assignments (left) and strip plots from 1H–13C–15N CBCACONH (blue) and HNCACB (orange) solution NMR spectra for residues T26–S28 (right). Signals belonging to an assigned residue in MAS and solution spectra are connected with the same color. (b) Overlay of Zn2+-S100A12 (orange) and apo-S100A12 (blue) 13C–13C DARR MAS spectra acquired at 14.1 T. (c–e) Close-ups of panel b with assignments of the perturbed residues. All solid-state NMR spectra were acquired at 12 kHz MAS.
2D (13C–13C DARR; Bo = 14.1 T) and 3D (15N–13C, NCACX and NCOCX; Bo = 16.4 T; Figure S6) MAS NMR spectra of Ca2+,Zn2+-S100A12 allowed for the assignment of 66 of 92 amino acids in the polypeptide chain. Narrow line widths of approximately 0.46–0.62 ppm were observed, indicating a high degree of conformational homogeneity. Because Ca2+ is known to introduce major structural rearrangements into the apoprotein, the protein bound to both Ca2+ and Zn2+ was compared with Ca2+-S100A12 to monitor structural perturbations upon Zn2+ binding. The 13C–13C MAS NMR spectrum of Ca2+,Zn2+-S100A12 overlays well on the spectrum of Ca2+-bound S100A12 (Figure 6), suggesting that the structure remains largely unaltered upon binding of Zn2+ to Ca2+-S100A12. As expected, CSPs were observed for the metal binding residues. However, a closer inspection reveals several major perturbations were also observed in residues that do not belong to the metal binding scaffold. Some of these residues are shown in the close-ups of the 13C–13C overlay in Figure 6, and a detailed description of these perturbations is presented below.
Figure 6.
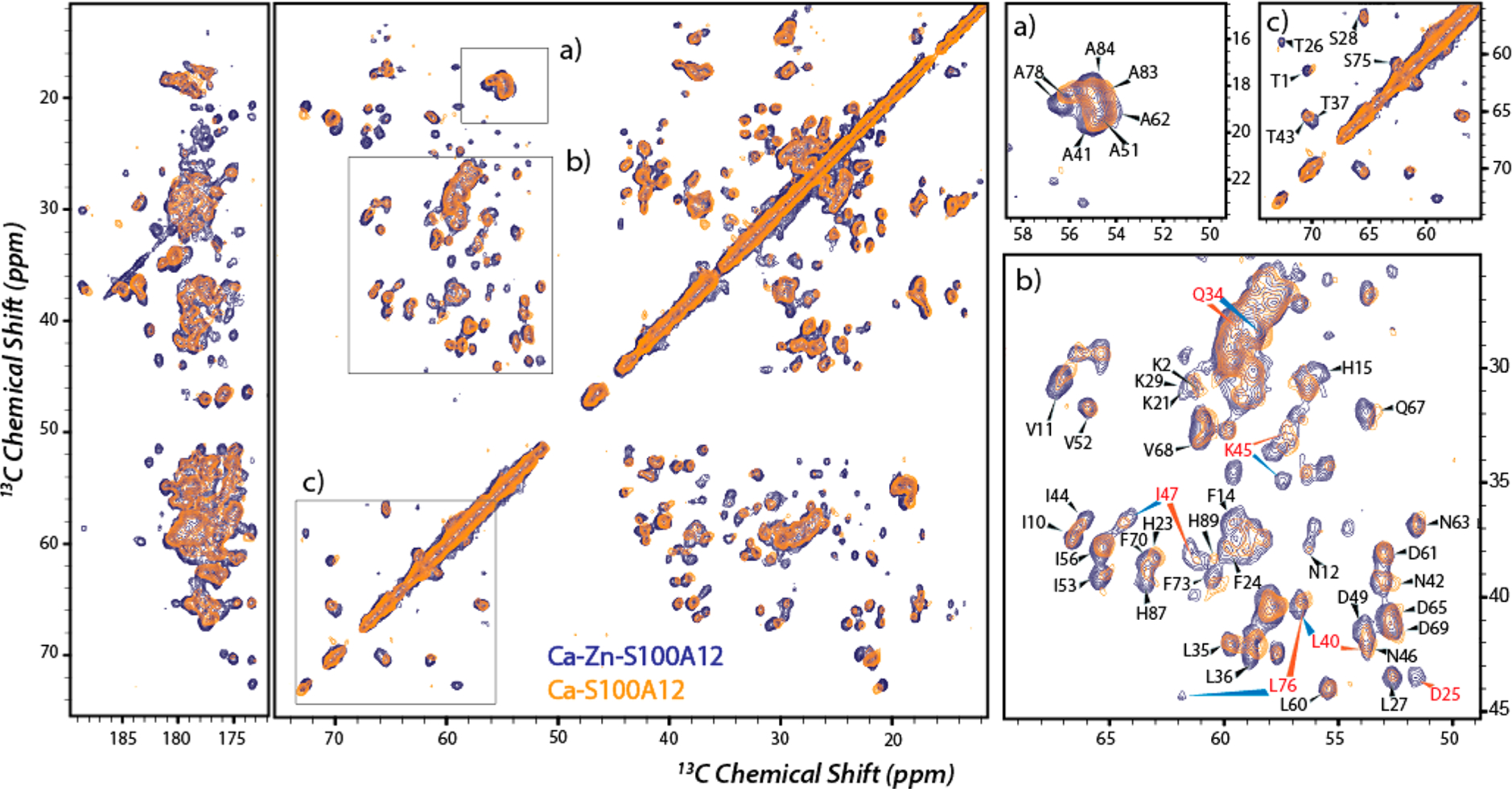
Overlay of 13C–13C correlation MAS NMR spectra of Ca2+,Zn2+-S100A12 (blue) and Ca2+-S100A12 (orange). Panels a–c are close-ups with assignments showing perturbed residues. Residues marked in red show significant CSP upon zinc binding. The experimental conditions were the same as those described for Figure 5.
Chemical Shift Perturbations upon Binding of Zinc to Apo- and Ca2+-S100A12.
Several CSPs can be observed for both Cα and Cβ nuclei in 13C–13C correlation spectra upon binding of zinc to apo- and Ca2+-S100A12. These observed CSPs, derived from 2D and 3D spectra, were plotted against the residue numbers for Cα, Cβ, and NH nuclei, as shown in Figure 7. On the basis of the line widths of the 13C–13C MAS NMR signals, a perturbation in the backbone Cα chemical shift of >0.3 ppm was considered to reflect changes in the local structure of the protein. Any perturbation of <0.15 ppm was considered to denote residues with conserved structural domains. The crystal structures of the apoprotein and the Ca2+-bound protein show four helices (helices I–IV) connected by loop regions. Binding of Zn2+ to the apoprotein induced changes in the entire polypeptide chain as monitored by the CSPs. While most residues in helix II exhibited CSPs of ~0.3 ppm, large CSPs were observed for helices I and IV, indicating major structural perturbations in these helices (Figure 7, left). The hinge region (residues K38–V53), which has been shown to be responsible for target recognition,29,56,57 exhibited CSPs of >0.3 ppm for few residues. CSPs for backbone 15N amide nitrogen nuclei (NH), deduced from 3D measurements, are also presented in Figure 7. Most residues with large perturbations in Cα chemical shifts also showed significant perturbations in NH and Cβ chemical shifts.
Figure 7.

Site-specific CSPs for binding of zinc to apo-S100A12 (left) and Ca2+-S100A12 (right). Cα, NH, and Cβ perturbations are colored pink, yellow, and blue, respectively. The secondary structure of the apoprotein determined from the crystal structure (PDB entry 2WCF) is displayed at the top: α-helices (blue bars), β-strands (yellow arrows), and loop regions (cyan lines). The dashed horizontal lines represent CSPs of 0.15 and 0.3 ppm for Cα and Cβ and 0.50 and 1.00 ppm for NH.
Contrary to the zinc-induced conformational changes that were observed in all domains of the apoprotein, structural perturbations in Ca2+-S100A12 are more specific as shown in the right panel of Figure 7. As expected, zinc binding residues in helices I and IV (H15, D25, H85, and H89) and the amino acids in their vicinity exhibit perturbations. A majority of the residues in helix III showed Cα CSPs of <0.3 ppm; however, major CSPs were observed for most of the residues in helix II and the hinge region denoting structural variation in this motif of the protein between the dimeric and oligomeric assemblies. This suggests the conformation of helix II and the hinge region are modulated by the binding of zinc to Ca2+-S100A12.
Secondary Structure Propensities.
The SSP for the apoprotein and the three metal-bound states of S100A12 were calculated using Cα and Cβ chemical shifts (see Materials and Methods) and are plotted versus residue number in Figure 8. The SSP values for the apoprotein predicted four helical domains and β-sheet propensity for several loop residues, in good agreement with the secondary structure determined by X-ray crystallography (Figure 8, left).26 These findings indicate that MAS NMR results are consistent with the predicted secondary structure of the apoprotein and that the overall conformation of protein is not affected by sample preparation or during spinning. A comparison of SSPs for apo- and Zn2+-S100A12 (Figure 8, left) shows Zn2+ binding does not disrupt the overall domainwise secondary structure of apo-S100A12. The helical domains in the apoprotein and the loop regions with β-sheet propensity (such as residues 24–27) retain their conformation upon self-assembly. However, compared to those of the apoprotein, the residues at the C-terminus of the oligomeric assemblies exhibit significantly increased α-helical propensity, suggesting extension of helix IV upon zinc binding. Similarly, the overall secondary structure of Ca2+-S100A12 is preserved upon Zn2+ binding, as shown in the right panel of Figure 8. The extension of helix IV upon binding of zinc to the apoprotein is also observed for Ca2+-S100A12.
Figure 8.
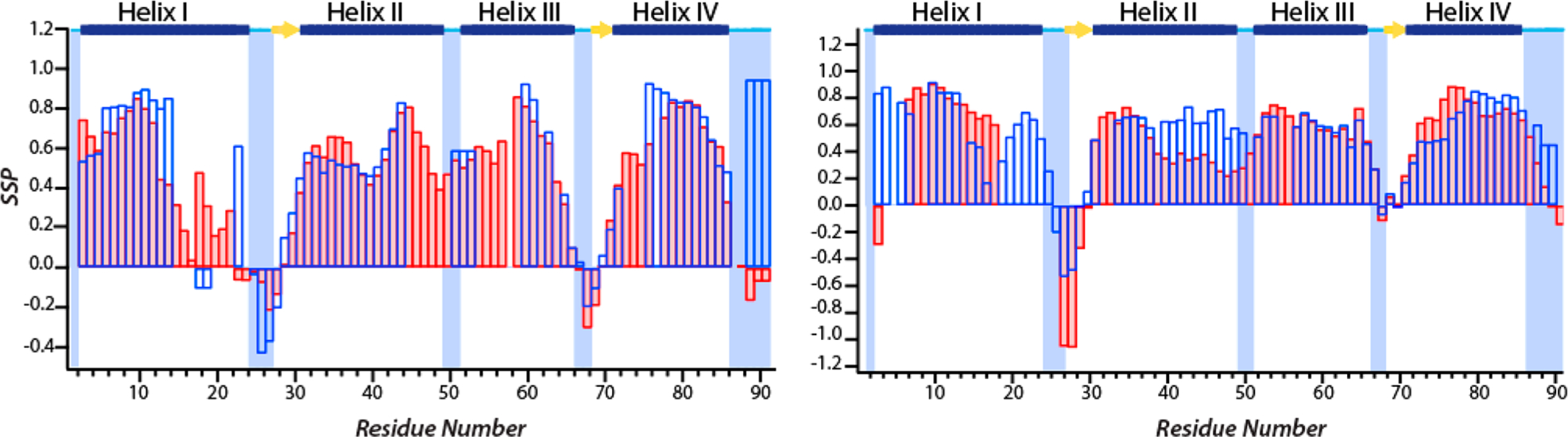
Comparison of the SSPs predicted from Cα and Cβ chemical shifts: apo-S100A12 (pink bars) and Zn2+-S100A12 (empty blue bars) (left) and Ca2+-S100A12 (pink bars) and Ca2+,Zn2+-S100A12 (empty blue bars) (right). The secondary structure of the apoprotein and Ca2+-bound protein determined from the crystal structure (PDB entries 2WCF and 1E8A) is displayed at the top: α-helices (blue bars), β-strands (yellow arrows), and loop regions (cyan lines).
■ DISCUSSION
Oligomeric S100A12 has been observed both in vivo and in vitro, and binding of these higher-order oligomers has been implicated in mechanisms of cellular signaling.32,35,36,38 While divalent cations clearly facilitate S100A12 self-assembly, the literature is inconsistent with regard to the precise conditions that induce S100A12 self-assembly. For instance, hexameric assemblies were observed when S100A12 was crystallized in the presence of Ca2+,28 although both Zn2+ and Ca2+ have been proposed to be necessary for oligomerization.32 The extreme peak broadening that we observed in our NMR analysis of 0.2–2 mM S100A12 in solutions containing Ca2+ and Zn2+ suggested that the protein forms higher-order assemblies in the presence of both divalent cations.
Although discrete oligomers and assembly models could not be resolved from our AUC analyses, it is clear that Zn2+ is 2 orders of magnitude more effective than Ca2+ in stimulating S100A12 self-assembly. In addition, the two cations cooperate to enhance oligomerization. The cooperation of the two cations makes structural sense because each cation binds to discrete crystallographically identified sites. While our results did not determine the terminal oligomer formed by Ca2+- and Zn2+-bound S100A12, they do clearly demonstrate that divalent cations stimulate reversible S100A12 self-assembly. It is this propensity that is likely to be relevant to selective cellular regulation.
As shown by our AUC results, binding of zinc to Ca2+-S100A12 affords increased propensity for self-assembly; however, structural changes introduced by zinc binding are not known. We performed NMR investigation of the apoprotein and Ca2+- and Zn2+-bound protein. Using 2D and 3D 1H–13C–15N correlation solution NMR spectroscopy, nearly complete (88%) backbone peak assignments were made for the apoprotein and 81% of amino acids were identified for Ca2+-S100A12 on the basis of the reported assignments.39 Zn2+- and Ca2+,Zn2+-bound assemblies prohibited their characterization by solution NMR because of their high molecular weight, consistent with the results of AUC experiments. In the solid state, MAS NMR spectra of Zn2+-and Ca2+,Zn2+-S100A12 gave well-resolved NMR spectra that allowed for the structural comparison with the apo and Ca2+-bound forms. Although complete backbone assignments for Zn2+- and Ca2+,Zn2+-S100A12 are not available, it is note-worthy that the current assignments are made throughout the polypeptide backbone, and consequently, the analysis presented in this report is not biased toward a specific domain of S100A12.
Comparison of Apo- and Zn2+-S100A12.
The observed Cα CSPs are mapped on the crystal structure of apo-S100A12 and displayed in Figure 9. According to the crystal structure (PDB entry 2WCF), four main helices in apo-S100A12 are linked by loops and short β-strands.26 Large perturbations were measured for helices I and IV, suggesting conformational changes in these domains upon zinc binding. Zinc binding introduces CSPs to several residues in Ca2+ binding EF-hand loop I near D25 and in the C-terminal residues near H85 and H89, consistent with the His3Asp binding motif, which is composed of H15, D25, H85, and H89. Our results indicate that all four helices undergo conformational changes upon zinc binding. A comparison of SSPs of apo- and zinc-S100A12 revealed that the helical regions retain their secondary structure upon oligomerization. This also suggests that the observed CSPs do not represent a change in the secondary structure of these domains. Similarly, albeit CSPs were observed for few residues, the SSP of the hinge region remained unperturbed and this motif exists primarily in its α-helical conformation in the two states. SSPs predicted by MAS NMR measurements reveal that the loop residues following helix IV in the apoprotein with β-sheet propensity acquire an α-helical conformation in Zn2+-bound assemblies. This increased helical propensity indicating extension of helix IV was also observed in the Zn2+-bound S100A12 crystal structure and can be introduced due to binding of Zn2+ to H85 and H89 at the C-terminus.26
Figure 9.

Observed Cα CSPs mapped onto the crystal structure of apo-S100A12 (PDB entry 2WCF). Color scheme: gray for unassigned residues, blue for residues with CSPs between 0 and 0.15 ppm, cyan for residues with CSPs between 0.15 and 0.3 ppm, gradient from orange to red for residues with CSPs between 0.30 and 2.00 ppm, and red for residues with CSPs of >2.00 ppm. The zinc binding residues are shown as sticks and labeled in red.
The crystal structure of Zn2+-bound S100A12 has been reported.26 In this work, close contacts between two dimeric Zn2+-S100A12 units arranged in a plausible tetrameric form were identified, which allowed the authors to propose a tetramerization interface. On the basis of this observation, helix I was suggested to participate in the tetramer interface, which was also proposed to be stabilized by extension of helix IV. Minor variations in helix I are observed between the crystal structures of apo- and Zn2+-S100A12 (PDB entries 2WCB and 2WCF, respectively), as indicated by the root-mean-square deviation of 1.18 Å and shown in the overlay of the two structures in Figure S7. The observed CSPs in helix I in our studies, which contains metal binding H15, may originate from the perturbations introduced upon metal binding.
Comparison of Ca2+- and Ca2+,Zn2+-S100A12.
Binding of zinc to the apoprotein introduces structural perturbations, as shown by crystallographic studies and the results presented here. However, binding of calcium to apo-S100A12 imposes major structural rearrangement on helices II and III as shown in an overlay of the two structures in Figure S8. Therefore, the overall structure of Ca2+,Zn2+-S100A12 is expected to be dominated by calcium-induced perturbations to the apoprotein. Although a structure of Ca2+,Zn2+-S100A12 is not available, S100A12 bound to calcium and copper has been investigated by X-ray crystallography.30 Surprisingly, an overlay of Ca2+- and Ca2+,Cu2+-S100A12 crystal structures reveals no major variations in these structures (Figure S9). Because Zn2+ and Cu2+ share the same metal binding site, the structure of Ca2+,Cu2+-S100A12 is expected to be similar to that of Ca2+,Zn2+-S100A12, and currently, this structure is being used as a surrogate structure of Zn2+,Ca2+-S100A12.
Our MAS NMR studies show that the overall secondary structure is conserved between Ca2+- and Ca2+,Zn2+-S100A12. The 13C–13C correlation spectra of Ca2+-S100A12 and Ca2+,Zn2+-S100A12 overlay well, with most regions of the polypeptide chain exhibiting minor perturbations and suggesting that the overall architecture of the protein is modulated by calcium binding, consistent with the similarities between the Ca2+- and Ca2+,Cu2+-S100A12 crystal structures. As expected, zinc binding introduces CSPs into helices I and IV at the metal binding scaffold and neighboring residues. However, a closer look revealed major CSPs for helix II and the hinge region (Figure 10). Because this region of the polypeptide chain does not contain zinc-coordinating residues, these CSPs indicate zinc binding modulated long-range conformational changes introduced into the protein. Therefore, our findings demonstrate that the Ca2+,Cu2+-S100A12 crystal structure, which does not show these conformational changes, should not be equated to the structure of the Zn2+- and Ca2+-bound protein.
Figure 10.
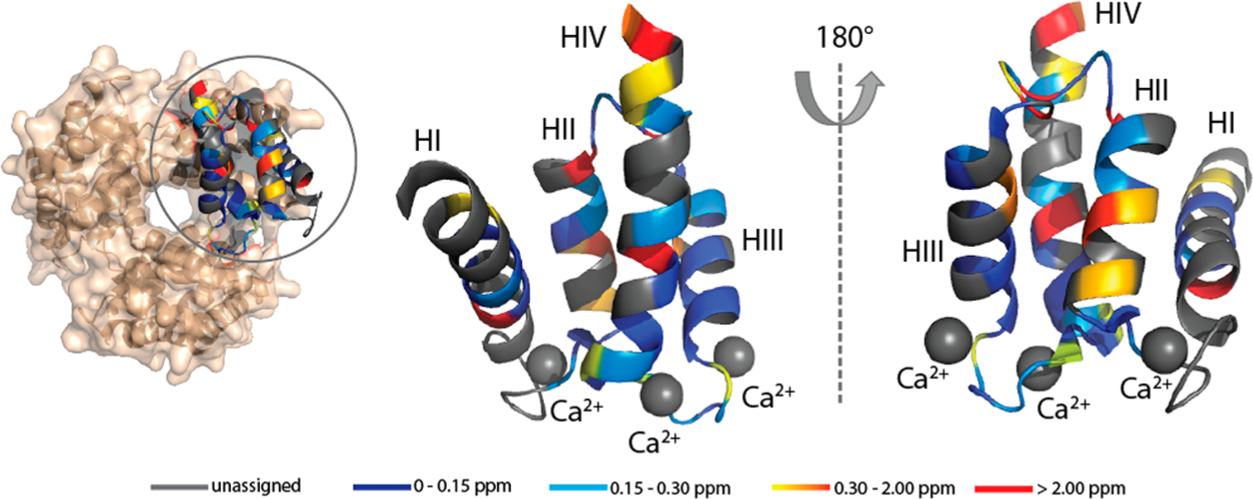
Observed Cα CSPs observed upon binding of Zn2+ to Ca2+-S100A12 mapped onto the crystal structure of hexameric S100A12 (PDB entry 1GQM). The CSPs are mapped on the monomeric chain for the sake of clarity. Color scheme: gray for unassigned residues, blue for residues with CSPs between 0 and 0.15 ppm, cyan for residues with CSPs between 0.15 and 0.3 ppm, gradient from orange to red for residues with CSPs between 0.30 and 2.00 ppm, and red for residues with CSPs of >2.00 ppm. The calcium ions are denoted by gray spheres.
Many S100 proteins such as S100B, S100A1, S100A2, S100A3, S100A5, S100A6, S100A7, S100A8/A9, S100A12, and S100A16 bind to Zn2+. On the basis of structural studies, the overall architecture upon Ca2+ binding is retained after Zn2+ ligation.58 A more methodical comparison can be made for S100A7 for which structures of both Ca2+- and Ca2+,Zn2+-bound states are available.59 In these structures, Zn2+ binding does not appear to affect the conformation of helix II. In contrast, binding of Zn2+ to Ca2+-S100B induces more pronounced changes in helices III and IV.60,61 These comparisons suggest that although the architecture of most S100 proteins is dominated by Ca2+ binding, the extent and the nature of Zn2+-mediated structural modulation may not be similar for all S100 proteins.
The C-type immunoglobulin domain of RAGE binds to Ca2+-S100A12 via an interface composed of helix II and the hinge domain.37 The perturbations in these regions observed in our MAS NMR studies may represent biologically pertinent Zn2+-modulated structural changes in S100A12 that could dictate its interactions with membrane receptors such as RAGE and TLR-4. Leu40, Ile44, Ile47, and Ile53 of the hinge domain have been shown to play an important role in S100A12-induced chemotaxis and edema.29 Interestingly, in our NMR studies, we observed large CSPs for these residues upon binding of Zn2+ to Ca2+-S100A12. The observed perturbations, denoting conformational changes, could also affect the aforementioned functions of the hinge domain of S100A12, which may be dependent on metal binding.
Due to the high calcium concentration in serum, it is reasonable to suggest that upon secretion into the extracellular space, S100A12 is bound to calcium. Therefore, during its antimicrobial activity in the serum afforded by zinc sequestration, our results indicate that S100A12 is likely to exist in oligomeric form. In the immune response, S100A12 also interacts with membrane receptors to initiate the proinflammatory signaling cascade.4,19 The self-assembly of S100A12 during the immune response may also suggest that oligomeric protein (as opposed to dimers) could be responsible for triggering the inflammatory pathways in human bodies.
We close with some thoughts about how the observed reversible S100A12 self-assembly and the variation in the assembly formation facilitate a feedback loop in the immune system that allows the cells to regulate inflammation. During infection, overexpression and excretion of S100A12 into the extracellular space result in the formation of oligomers upon zinc binding, which may interact with the membrane receptors to initiate inflammation and induce cellular signaling. We suggest that dimeric S100A12, which may not lead to inflammation, is present in a healthy cell under normal levels of S100A12 expression. The ability of the protein to reversibly assemble may also have ramifications in the context of its interactions with membrane receptors. Our results indicate that S100A12 should not be considered as discrete tetrameric or hexameric oligomers but rather as a protein concentration-dependent continuum. Binding of S100A12 to different physiological targets such as RAGE and induction of Toll-like receptor 4 signaling may be facilitated by structurally distinct forms of the protein.21 Lastly, the long-range conformational changes to helix II and the hinge region induced by zinc binding suggest that studies in the future focusing on the role of these domains in facilitating receptor interactions may provide insights into the mechanism of S100A12-induced cellular signaling.
■ CONCLUSION
S100A12 self-assembles upon zinc binding, indicating that this member of the innate immune response is likely to exist in oligomeric form during its antimicrobial functions. We demonstrate that although calcium alone can initiate the self-assembly of S100A12, zinc and calcium cooperate to increase the propensity of the protein to self-assemble. Magic angle NMR studies demonstrate that binding of zinc to Ca2+-S100A12 induces major chemical shift perturbations to helix II and the hinge region. These perturbations in NMR chemical shifts may indicate functionally relevant structural changes, which could influence the interactions of S100A12 with membrane receptors, affecting cellular signaling. The conformational changes introduced in the hinge region may, in turn, impact the S100A12-mediated chemotactic and inflammatory responses. Taken together, this work demonstrates that zinc binding-driven structural changes should be considered as a contributor to the mechanism of action of S100A12 in human immune response and inflammatory pathways.
Supplementary Material
■ ACKNOWLEDGMENTS
The authors thank Dr. Boris Itin at the New York Structural Biological Center and Dr. James Aramini of the Advanced Science Research Center, City University of New York, for assistance with NMR measurements.
Funding
This work was supported by the City University of New York and Research Foundation startup funds to R.G. and the New York State Office for People with Developmental Disabilities. M.B.’s contribution to this project is supported by the National Institutes of Health (1R01-GM129350).
■ ABBREVIATIONS
- CSP
chemical shift perturbation
- TLR-4
Toll-like receptor 4
- DSS, 4
4-dimethyl-4-silapentane-1-sulfonic acid
- ESI-MS
electrospray ionization mass spectrometry
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- MALDI-TOF
matrix-assisted laser desorption ionization time-of-flight
- PDB
Protein Data Bank
- PEG-3350
polyethylene glycol 3350
- RAGE
receptor for advanced glycation end product
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.9b00123.
Mass spectrometry of apo-S100A12, 1H–13C–15N NMR spectra of Ca2+-S100A12, 1H–15N HSQC spectra of Zn2+-S100A12, sequential MAS NMR assignments for Zn2+- and Ca2+,Zn2+-S100A12, and overlays of available crystal structures of S100A12 (PDF)
Accession Codes
UniProt entry P80511.
The authors declare no competing financial interest.
■ REFERENCES
- (1).Hood MI, and Skaar EP (2012) Nutritional immunity: transition metals at the pathogen–host interface. Nat. Rev. Microbiol 10, 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Kehl-Fie TE, and Skaar EP (2010) Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol 14, 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Weinberg ED (1975) Nutritional immunity: host’s attempt to withhold iron from microbial invaders. JAMA, J. Am. Med. Assoc 231, 39–41. [DOI] [PubMed] [Google Scholar]
- (4).Zackular JP, Chazin WJ, and Skaar EP (2015) Nutritional immunity: S100 proteins at the host-pathogen interface. J. Biol. Chem 290, 18991–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Nakashige TG, Zygiel EM, Drennan CL, and Nolan EM (2017) Nickel sequestration by the host-defense protein human calprotectin. J. Am. Chem. Soc 139, 8828–8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, and Skaar EP (2008) Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319, 962–965. [DOI] [PubMed] [Google Scholar]
- (7).Brophy MB, Hayden JA, and Nolan EM (2012) Calcium ion gradients modulate the zinc affinity and antibacterial activity of human calprotectin. J. Am. Chem. Soc 134, 18089–18100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hayden JA, Brophy MB, Cunden LS, and Nolan EM (2013) High-affinity manganese coordination by human calprotectin is calcium-dependent and requires the histidine-rich site formed at the dimer interface. J. Am. Chem. Soc 135, 775–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Nakashige TG, Zhang B, Krebs C, and Nolan EM (2015) Human calprotectin is an iron-sequestering host-defense protein. Nat. Chem. Biol 11, 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hayden JA, Brophy MB, Cunden LS, and Nolan EM (2013) High-affinity manganese coordination by human calprotectin is calcium-dependent and requires the histidine-rich site formed at the dimer interface. J. Am. Chem. Soc 135, 775–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lee KC, and Eckert RL (2007) S100A7 (Psoriasin)- mechanism of antibacterial action in wounds. J. Invest. Dermatol 127, 945–957. [DOI] [PubMed] [Google Scholar]
- (12).Gläser R, Harder J, Lange H, Bartels J, Christophers E, and Schröder J-M (2005) Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat. Immunol 6, 57. [DOI] [PubMed] [Google Scholar]
- (13).Cunden LS, Gaillard A, and Nolan EM (2016) Calcium ions tune the zinc-sequestering properties and antimicrobial activity of human S100A12. Chem. Sci 7, 1338–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, and Geczy CL (2013) Functions of S100 proteins. Curr. Mol. Med 13, 24–57. [PMC free article] [PubMed] [Google Scholar]
- (15).Donato R (2003) Intracellular and extracellular roles of S100 proteins. Microsc. Res. Tech 60, 540–551. [DOI] [PubMed] [Google Scholar]
- (16).Gottsch JD, Eisinger SW, Liu SH, and Scott AL (1999) Calgranulin C has filariacidal and filariastatic activity. Infect. Immun 67, 6631–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Haley KP, Delgado AG, Piazuelo MB, Mortensen BL, Correa P, Damo SM, Chazin WJ, Skaar EP, and Gaddy JA (2015) The human antimicrobial protein calgranulin C (S100A12) participates in control of Helicobacter pylori growth and regulation of virulence. Infect. Immun 83, 2944–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Foell D, Wittkowski H, Vogl T, and Roth J (2007) S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J. Leukocyte Biol 81, 28–37. [DOI] [PubMed] [Google Scholar]
- (19).Leclerc E, Fritz G, Vetter SW, and Heizmann CW (2009) Binding of S100 proteins to RAGE: an update. Biochim. Biophys. Acta, Mol. Cell Res 1793, 993–1007. [DOI] [PubMed] [Google Scholar]
- (20).Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, et al. (1999) RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell 97, 889–901. [DOI] [PubMed] [Google Scholar]
- (21).Kessel C, Fuehner S, Zell J, Zimmermann B, Drewianka S, Brockmeyer S, Holzinger D, Hinze C, Wittkowski H, and Foell D (2018) Calcium and zinc tune autoinflammatory Toll-like receptor 4 signaling by S100A12. J. Allergy Clin. Immunol 142, 1370–1373.e8. [DOI] [PubMed] [Google Scholar]
- (22).Pietzsch J, and Hoppmann S (2009) Human S100A12: a novel key player in inflammation? Amino Acids 36, 381–389. [DOI] [PubMed] [Google Scholar]
- (23).Wittkowski H, Frosch M, Wulffraat N, Goldbach-Mansky R, Kallinich T, Kuemmerle-Deschner J, Frühwald MC, Dassmann S, Pham T-H, Roth J, and Foell D (2008) S100A12 is a novel molecular marker differentiating systemic-onset juvenile idiopathic arthritis from other causes of fever of unknown origin. Arthritis Rheum. 58, 3924–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Manolakis AC, Kapsoritakis AN, Tiaka EK, and Potamianos SP (2011) Calprotectin, calgranulin C, and other members of the s100 protein family in inflammatory bowel disease. Dig. Dis. Sci 56, 1601–1611. [DOI] [PubMed] [Google Scholar]
- (25).Oesterle A, and Hofmann Bowman MA (2015) S100A12 and the S100/calgranulins: Emerging biomarkers for atherosclerosis and possibly therapeutic targets. Arterioscler., Thromb., Vasc. Biol 35, 2496–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Moroz OV, Blagova EV, Wilkinson AJ, Wilson KS, and Bronstein IB (2009) The crystal structures of human S100A12 in apo form and in complex with zinc: new insights into S100A12 oligomerisation. J. Mol. Biol 391, 536–551. [DOI] [PubMed] [Google Scholar]
- (27).Moroz OV, Antson AA, Murshudov GN, Maitland NJ, Dodson GG, Wilson KS, Skibshøj I, Lukanidin EM, and Bronstein IB (2001) The three dimensional structure of human S100A12. Acta Crystallogr., Sect. D: Biol. Crystallogr 57, 20–29. [DOI] [PubMed] [Google Scholar]
- (28).Moroz O, Antson A, Dodson E, Burrell H, Grist S, Lloyd R, Maitland N, Dodson G, Wilson K, Lukanidin E, and Bronstein IB (2002) The structure of S100A12 in a hexameric form and its proposed role in receptor signalling. Acta Crystallogr., Sect. D: Biol. Crystallogr 58, 407–413. [DOI] [PubMed] [Google Scholar]
- (29).Yan WX, Armishaw C, Goyette J, Yang Z, Cai H, Alewood P, and Geczy CL (2008) Mast cell and monocyte recruitment by S100A12 and its hinge domain. J. Biol. Chem 283, 13035–13043. [DOI] [PubMed] [Google Scholar]
- (30).Moroz O, Antson A, Grist S, Maitland N, Dodson G, Wilson K, Lukanidin E, and Bronstein I (2003) Structure of the human S100A12–copper complex: implications for host-parasite defence. Acta Crystallogr., Sect. D: Biol. Crystallogr 59, 859–867. [DOI] [PubMed] [Google Scholar]
- (31).Korndorfer IP, Brueckner F, and Skerra A (2007) The crystal structure of the human (S100A8/S100A9)2 heterotetramer, calprotectin, illustrates how conformational changes of interacting alpha-helices can determine specific association of two EF-hand proteins. J. Mol. Biol 370, 887–98. [DOI] [PubMed] [Google Scholar]
- (32).Moroz OV, Burkitt W, Wittkowski H, He W, Ianoul A, Novitskaya V, Xie J, Polyakova O, Lednev IK, Shekhtman A, Derrick PJ, Bjoerk P, Foell D, and Bronstein IB (2009) Both Ca2+ and Zn2+ are essential for S100A12 protein oligomerization and function. BMC Biochem. 10, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Vogl T, Leukert N, Barczyk K, Strupat K, and Roth J (2006) Biophysical characterization of S100A8 and S100A9 in the absence and presence of bivalent cations. Biochim. Biophys. Acta, Mol. Cell Res 1763, 1298–306. [DOI] [PubMed] [Google Scholar]
- (34).Ostendorp T, Leclerc E, Galichet A, Koch M, Demling N, Weigle B, Heizmann CW, Kroneck PMH, and Fritz G (2007) Structural and functional insights into RAGE activation by multimeric S100B. EMBO J. 26, 3868–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Shepherd CE, Goyette J, Utter V, Rahimi F, Yang Z, Geczy CL, and Halliday GM (2006) Inflammatory S100A9 and S100A12 proteins in Alzheimer’s disease. Neurobiol. Aging 27, 1554–1563. [DOI] [PubMed] [Google Scholar]
- (36).Larsen A, Bronstein IB, Dahl O, Wentzel-Larsen T, Kristoffersen EK, and Fagerhol MK (2007) Quantification of S100A12 (EN-RAGE) in blood varies with sampling method, calcium and heparin. Scand. J. Immunol 65, 192–201. [DOI] [PubMed] [Google Scholar]
- (37).Xie J, Burz DS, He W, Bronstein IB, Lednev I, and Shekhtman A (2006) Hexameric calgranulin C (S100A12) binds to the receptor for advanced glycated end products (RAGE) using symmetric hydrophobic target-binding patches. J. Biol. Chem 282, 4218–4231. [DOI] [PubMed] [Google Scholar]
- (38).Kessel C, Fuhner S, Brockmeyer S, Wittkowski H, and Föll D (2015) OP0194 Hexameric S100A12 is Required for Pro-Inflammatory TLR4-Signalling. Ann. Rheum. Dis 74, 144.4. [Google Scholar]
- (39).Hung K-W, Hsu C-C, and Yu C (2013) Solution structure of human Ca2+-bound S100A12. J. Biomol. NMR 57, 313–318. [DOI] [PubMed] [Google Scholar]
- (40).Sun S, Han Y, Paramasivam S, Yan S, Siglin AE, Williams JC, Byeon I-JL, Ahn J, Gronenborn AM, and Polenova T (2012) Solid-state NMR spectroscopy of protein complexes. In Protein NMR Techniques, pp 303–331, Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Ortega A, Amoros D, and Garcia de la Torre J (2011) Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J 101, 892–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Philo JS (2006) Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem 354, 238–46. [DOI] [PubMed] [Google Scholar]
- (43).Stafford WF 3rd (1992) Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem 203, 295–301. [DOI] [PubMed] [Google Scholar]
- (44).Cavanagh J, Fairbrother WJ, Palmer AG III, Rance M, and Skelton NJ (2007) Protein NMR Spectroscopy: Principles and Practice, Academic Press. [Google Scholar]
- (45).Marion D, Driscoll PC, Kay LE, Wingfield PT, Bax A, Gronenborn AM, and Clore GM (1989) Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: Application to interleukin 1b. Biochemistry 28, 6150–6156. [DOI] [PubMed] [Google Scholar]
- (46).Marion D, Kay LE, Sparks SW, Torchia DA, and Bax A (1989) Three-dimensional heteronuclear NMR of 15N-labeled proteins. J. Am. Chem. Soc 111, 1515–1517. [Google Scholar]
- (47).Zuiderweg ERP, and Fesik SW (1989) Heteronuclear three-dimensional NMR spectroscopy of the inflammatory protein C5a. Biochemistry 28, 2387–2391. [DOI] [PubMed] [Google Scholar]
- (48).Grzesiek S, and Bax A (1992) Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc 114, 6291–6293. [Google Scholar]
- (49).Grzesiek S, and Bax A (1992) An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J. Magn. Reson 99, 201–207. [Google Scholar]
- (50).Takegoshi K, Nakamura S, and Terao T (2001) 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett 344, 631–637. [Google Scholar]
- (51).Bennett AE, Rienstra CM, Auger M, Lakshmi KV, and Griffin RG (1995) Heteronuclear decoupling in rotating solids. J. Chem. Phys 103, 6951–6958. [Google Scholar]
- (52).Baldus M, Petkova AT, Herzfeld J, and Griffin RG (1998) Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol. Phys 95, 1197–1207. [Google Scholar]
- (53).Delaglio F, Grzesiek S, Vuister GW, et al. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277. [DOI] [PubMed] [Google Scholar]
- (54).Goddard T, and Kneller D (2006) Sparky: NMR assignment and integration software, University of California, San Francisco. [Google Scholar]
- (55).Marsh JA, Singh VK, Jia Z, and Forman-Kay JD (2006) Sensitivity of secondary structure propensities to sequence differences between α- and g-synuclein: Implications for fibrillation. Protein Sci. 15, 2795–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Goyette J, and Geczy CL (2011) Inflammation-associated S100 proteins: new mechanisms that regulate function. Amino Acids 41, 821–842. [DOI] [PubMed] [Google Scholar]
- (57).Kligman D, and Hilt DC (1988) The S100 protein family. Trends Biochem. Sci 13, 437–443. [DOI] [PubMed] [Google Scholar]
- (58).Gilston BA, Skaar EP, and Chazin WJ (2016) Binding of transition metals to S100 proteins. Science China. Sci. China: Life Sci 59, 792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Brodersen DE, Nyborg J, and Kjeldgaard M (1999) Zinc-Binding Site of an S100 Protein Revealed. Two Crystal Structures of Ca2+-Bound Human Psoriasin (S100A7) in the Zn2+-Loaded and Zn2+-Free States. Biochemistry 38, 1695–1704. [DOI] [PubMed] [Google Scholar]
- (60).Wright NT, Inman KG, Levine JA, Cannon BR, Varney KM, and Weber DJ (2008) Refinement of the solution structure and dynamic properties of Ca2+-bound rat S100B. J. Biomol. NMR 42, 279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Wilder PT, Varney KM, Weiss MB, Gitti RK, and Weber DJ (2005) Solution Structure of Zinc- and Calcium-Bound Rat S100B as Determined by Nuclear Magnetic Resonance Spectroscopy. Biochemistry 44, 5690–5702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


