Abstract
Adenosine-to-inosine (A-to-I) RNA editing is a common post transcriptional modification. It has a critical role in protecting against false activation of innate immunity by endogenous double stranded RNAs and has been associated with various regulatory processes and diseases such as autoimmune and cardiovascular diseases as well as cancer. In addition, the endogenous A-to-I editing machinery has been recently harnessed for RNA engineering. The study of RNA editing in humans relies heavily on the usage of cell lines as an important and commonly-used research tool. In particular, manipulations of the editing enzymes and their targets are often developed using cell line platforms. However, RNA editing in cell lines behaves very differently than in normal and diseased tissues, and most cell lines exhibit low editing levels, requiring over-expression of the enzymes. Here, we explore the A-to-I RNA editing landscape across over 1000 human cell lines types and show that for almost every editing target of interest a suitable cell line that mimics normal tissue condition may be found. We provide CLAIRE, a searchable catalogue of RNA editing levels across cell lines available at http://srv00.recas.ba.infn.it/atlas/claire.html, to facilitate rational choice of appropriate cell lines for future work on A-to-I RNA editing.
INTRODUCTION
Adenosine-to-inosine (A-to-I) RNA editing is one of the most common post-transcriptional modifications in metazoan (1–3). It is catalyzed by the adenosine deaminase that acts on RNA (ADAR) family of enzymes. In humans, this family consists of three members: ADAR1 (ADAR) and ADAR2 (ADARB1) two catalytically active dsRNA-binding proteins (4–7) and ADAR3 (ADARB2), which contains the dsRNA binding domains but lacks catalytic activity. While ADAR3 does not perform A-to-I editing it is believed to act as dominant negative regulator of editing (8,9). Inosines can pair with cytosine, and they are recognized by the ribosome during translation as guanosines, leading to a modified protein product (recoding) (10,11). However, most editing activity occurs in non-coding regions (12). In primates, editing mostly occurs in the Alu repetitive elements (13–16), mainly by ADAR1 (17,18), while ADAR2 is associated with most recoding sites (17).
RNA editing is believed to have a critical role in protecting against false activation of innate immunity by endogenous double stranded transcripts (19–21). It also plays an important role in various regulatory processes such as splicing (22–24), microRNA processing (25,26), microRNA targeting (27–31) and mRNA stability (32,33). Altered editing may lead to various diseases (34,35) such as autoimmune (36–39), cardiovascular (40,41) and neurological (42–48) diseases, and cancer development (49–53). Finally, ADAR enzymes are utilized for newly developed RNA engineering approaches (54–58).
Cell lines are extensively utilized for RNA editing studies. Cancer derived cell lines are immortalized cells originating usually from tumor tissues. Their ease of growth and the ability to grow indefinitely have made them a mainstay of biological research (59). However, there is a large genomic and phenotypic variability within cell lines. It has been shown that using misidentified, over passaged or contaminated cell lines can result in a serious reduction of phenotypic quality which can hinder discovery and reproducibility (60,61). Thus, great care must be taken in choosing the appropriate cell lines, best suited to answer the research question.
The RNA editing landscape in cell lines has not yet been characterized systematically. Previous works on RNA editing in cell lines has mostly found a markedly lower level of editing compared to normal or diseased tissue samples (13,53,62). Thus, many cell line studies of RNA editing have relied on overexpression of ADAR to ensure a measurable level of editing that can then be manipulated. These disturbances can cause artifacts unaccounted for and hinder reproducibility.
Here, we analyzed editing across over one thousand unique cancer cell lines (63,64), and created a catalogue of RNA-editing levels in coding and non-coding regions, as well as ADAR1 and ADAR2 expression levels. This database allows for a rational choice of the most appropriate cell line for experimental research in RNA editing.
MATERIALS AND METHODS
Data
RNA-seq fastq files for 675 GCLB cell lines were downloaded from the European Genome-phenome Archive (GCLB; https://www.ebi.ac.uk/ega/datasets/EGAD00001000725) (64). Additional 933 CCLE cell lines RNA-seq BAM files were downloaded from the GDC legacy archive (https://portal.gdc.cancer.gov/legacy-archive/) (63), and transformed into fastq files using the bamtofastq command (samtools suite version 1.2; htslib 1.2.1). Libraries for both datasets were created using poly-A selection, Illumina TruSeq protocol and paired-end sequenced on either Illumina HiSeq 2000 or HiSeq 2500. Library preparation and sequencing protocols were similar for both datasets except for read length, which is 101 bp for CCLE and 75 bp for GCLB. Therefore, we trimmed 13 bp from each end of CCLE reads, to get 75 bp-long trimmed reads that match the GCLB read length. In addition, HEK293 fastq files were downloaded from the Cell Atlas (65) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA183192) and added to the cell line samples set. The ten pairs of duplicates of cancerous cell-lines available in Cell Atlas were used to further analyze reproducibility (see Results). Libraries for the Cell Atlas were also created using poly-A selection and the Illumina TruSeq protocol, and sequenced on the Illumina HiSeq 2500. Reads are paired-end, 101 bp long, and were trimmed to 75 bp. Cell lines appearing in both CCLE and GDC were analyzed and presented independently. We used the FastQC script (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) for quality control of the RNA-seq data.
Data analysis
We used the GRCh37/hg19 human reference genome for all analyses. Gene expression levels were calculated in transcript per million (TPMs) units using Salmon version 0.11.2 (66). Fastq files were aligned to the reference genome using STAR version 2.5.2b (67) and parameters –alignIntronMax 1000000 –alignMatesGapMax 1000000 –alignSJoverhangMin 8 –outFilterMismatchNmax 999 –outFilterMismatchNoverReadLmax 0.1 –outFilterMultimapNmax 1 –outSAMattributes All –outSAMtype BAM SortedByCoordinate –outWigType bedGraph –quantMode GeneCounts.
The global editing level was assessed by the Alu Editing Index (AEI) (18,68), following the protocol detailed in Roth et al. (18), with default parameters. In addition, we quantified editing at specific coding sites (editing sites within the coding sequence, both synonymous and non-synonymous) using Reditools (69) Known script with parameters -v 2 -n 0.01 -r 1 –T [6–6]. These sites were compiled from a previously published list of editing sites observed in normal GTEx tissues (17). From this previously published list, we discarded sites that are not located in RefSeq coding sequences, as downloaded from UCSC genome browser website (70). We also discarded sites for which the reported average editing level was <1% in both brain-cerebellum and heart-artery GTEx samples (17). We added to the list ten conserved coding editing sites that were found in (71) (Supplementary Table S1), but missing from the larger list. This resulted in a list of 314 coding sites (Supplementary Table S2).
For comparison, AEI and editing levels for each coding site were calculated for all GTEx samples (72), using the same methodology. GTEx tissues were aggregated to match cell line tissues of origin and are presented in Supplementary Table S3.
To calculate gene ranked-expression, expression level (TPM) of all genes was calculated using Salmon, as described above. The gene list was then sorted by expression level, and each gene was assigned a rank, which is its location in the list relative to the full list length. That is, the most highly-expressed gene is assigned a ranked-expression value (or expression quantile) of 100, reflecting the fact its expression is equal or higher than 100% of the genes. Genes expressed at a level that equals the median over all genes are assigned a value of 50, etc.
Linear regressions and multiple linear regressions were computed using the gene expression levels as calculated above. The R lm function was used with default parameters (i.e. linear regression using QR decomposition) to fit the AEI data to a linear model including the expression of each of the ADAR genes separately (simple regression), or to a model including ADAR1 the and one other gene (two-variable regression). For the two-variable regression, all genes were tested, and P-values were Bonferroni-corrected.
Plotting and statistics were done using R and python in-house scripts
Growing and transfecting cells
Cells were ordered from the ATCC and grown in the following mediums: Cama-1: DMEM + 10% FBS, ZR-75-1: RPMI-1640 + 10% FBS, NCI-H1573: RPMI-1640 + 5% FBS. Cells were kept at 37°C and passaged once reaching confluence. DNA and RNA were extracted using Norgen RNA/DNA Purification Kit (Cat. 48700). cDNA was prepared using iScript advanced reverse transcriptase. PCRs for AZIN1 were done using Invitrogen Platinum SuperFi PCR master mix with the following primers.
RNA
FP: TCGCAGTTAATATCATAGC
RP: AAGGCACAAAGAAGAAGT
DNA
FP: GTTCTTCTGGTGGAGTCCCT
RP: ACAGTTCAAATATCCCATCTGCC
Editing levels of sanger sequences were calculated using EditR (73).
RESULTS
We analyzed the editing landscape in 1610 cell-line samples: 933 cell-line samples available in The Cancer Cell Line Encyclopedia (CCLE) (63) database, 675 samples from Genentech Cell Line Bank (GCLB) (64), as well as two additional samples of the non-cancer-derived cell line HEK293 obtained from the Cell Atlas (65). Of these, 460 cell lines are found in both the CCLE and GCLB datasets (Figure 1A). These cell-lines cover a wide range of source tissues, mostly lung cancers and hematopoietic and lymphoid cancers (226 and 193 unique cell lines, respectively) (Figure 1B, Supplementary Table S4).
Figure 1.
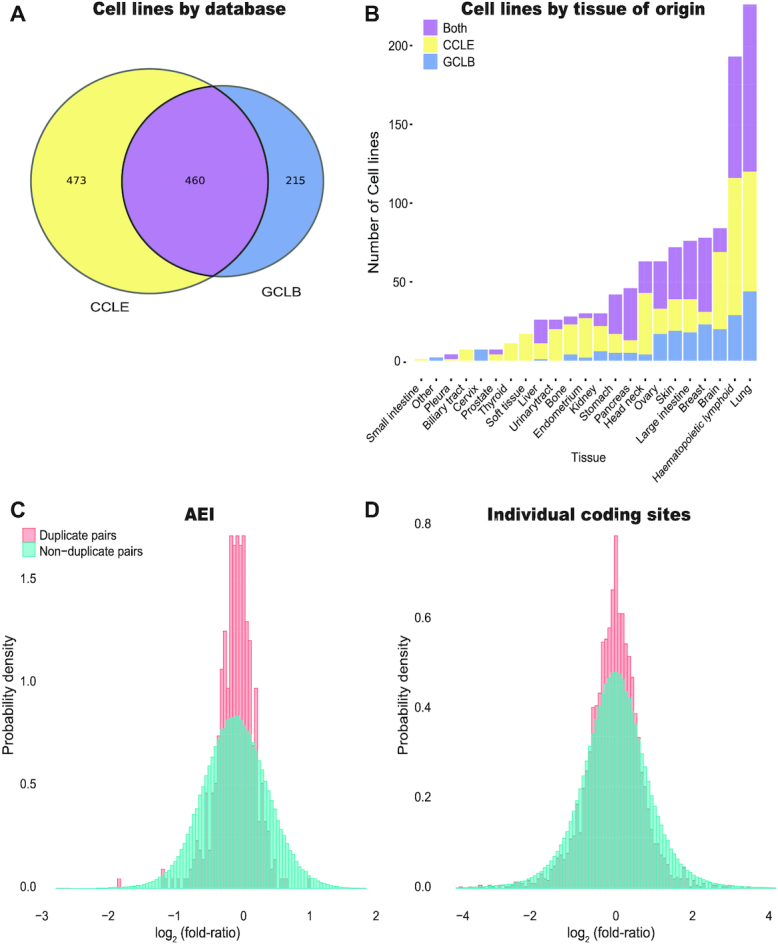
Variability of editing in Cell-lines. (A) We study 933 cell line samples from the CCLE dataset (63) and 675 samples from the GCLB dataset (64). Data for 460 cell lines is found in both datasets. (B) The cell lines analyzed originate from a variety of tissues, mostly from lung, and hematopoietic and lymphoid tissues. (C) Ratios of AEI values calculated for two biological replicated of each of 460 cell lines (same cell line type appearing in both datasets, cultured and sequenced in different labs) follow a log-normal distribution. The distribution is compared with that of 211,140 comparisons of non-duplicate cell lines (ratios of AEI values for different cell-lines from a different dataset, cultured and sequenced in different labs). Since the ratios follow a log-normal distribution, we plot the distribution for the logarithm of the ratio, log2(editing of cell line from CCLE/editing of cell line from GCLB), which is approximately normal. The half width at half maximum (HWHM) of the distribution of log2(ratio) for duplicates is 0.26, compared with a HWHM of 0.56 for pairs of non-duplicate cell lines. (D) Ratios of editing levels at specific sites calculated for two biological replicated of each of 460 cell lines (same cell line type appearing in both datasets, cultured and sequenced in different labs), and compared with the distribution of ratios obtained for specific sites in each of the 211,140 non-duplicate cell line pairs. For each sample, only well covered sites were considered (minimum coverage of 20 reads with at least 10 counts of ‘G’ in each of the duplicates; median number of sites analyzed per sample is 6), and for each pair ratios were calculated only for sites that are well covered in both pair mates. Altogether, we considered 3170 ratios between duplicates, and 788,484 for non-duplicate cell line pairs. The HWHM of the log2(ratio) for duplicates is 0.59, compared to a HWHM of 0.82 for non-duplicate cell lines.
Most of human RNA editing activity occurs in Alu repetitive elements (13–16). Thus, the editing levels in Alu can serve as an indication for the overall editing of a sample. In addition, there is much interest in specific editing sites, mainly recoding sites, and many studies of these sites rely on using cell lines. Accordingly, for each sample we evaluated the global editing through the Alu Editing Index (AEI) (18) (Supplementary Table S4), as well as the editing level in each of 314 known specific sites within coding sequences (Supplementary Table S2, Methods). The full information is available as Supplementary Tables S2–S4. A dedicated website, CLAIRE (Cell Line A-to-I RNA Editing) facilitates on-line searches of the database, and is available as part of REDIportal (74), at http://srv00.recas.ba.infn.it/atlas/claire.html.
Editing levels are reproducible for cells lines of the same type
Differences in the editing profile across cell line types exceed the ones caused by stochastic biological noise within the sample, inherent genomic variations within cell lines of the same type, batch effects and varying biological conditions in different labs. To show that, we looked at 460 cell lines which appear in both the CCLE and GCLB datasets. We compared the editing levels between the two biological replicates of these 460 cell-lines (one from the CCLE dataset and the other from the GCLB dataset). In addition, we compared the editing levels of each cell line from the CCLE dataset against the editing levels of all cell lines in the GCLB dataset excluding its own duplicate. First, we looked at the AEI, and calculated the log2 fold-ratio. The distribution of the fold-ratios is log-normal, and the half width at half maximum (HWHM) of the log2(ratio) is 0.26 for duplicates (equivalent to a typical fold ratio of 1.20), compared with HWHM = 0.56 for random pairs of cells (typical fold ratio of 1.48) (F-test, P-value <2.2e–16) (Figure 1C). For coding sites, we calculated the editing level per site and looked again at the distribution of log2(ratio) for well-covered, well-edited, sites (minimum coverage of 20 reads with at least 10 counts of ‘G’). The HWHM of the log2(ratio) for duplicates is 0.59 (typical fold ratio of 1.50), compared to HWHM = 0.82 for non-duplicate pairs (typical fold ratio of 1.76) (F-test, P-value <2.2e–16) (Figure 1D).
We further analyzed an additional dataset from the Cell Atlas (65) that contains ten pairs of technical duplicates. For each of the ten cell line types, two replicates were cultured and sequenced under the same conditions. Again, we looked at the ratios of editing levels (AEI, and levels at specific sites) in each of these ten pairs of samples. As expected, the similarity between pairs of technical duplicates is much higher than the biological replicates studies above. For the AEI, HWHM = 0.07 for duplicates compared to 0.5 for non-duplicate pairs, meaning a typical fold ratio of 1.05 and 1.41 respectively (F-test, P-value = 5.72e–06). For the coding sites, HWHM = 0.36 for duplicates and 1.21 for non-duplicate pairs, and the typical fold ratios are 1.28 and 2.31, respectively (F-test, P-value = 0.030).
We thus conclude that while some variability is expected for samples originated from the same cell line origin but cultured and sequenced in different labs, this variability is much lower than that between different types of cancerous cell lines. Therefore, knowing in advance the expected editing levels in each cell line type can assist rational design of RNA editing experiments, by choosing the cell line with the desired editing profile.
ADAR expression level is not a good predictor of editing levels
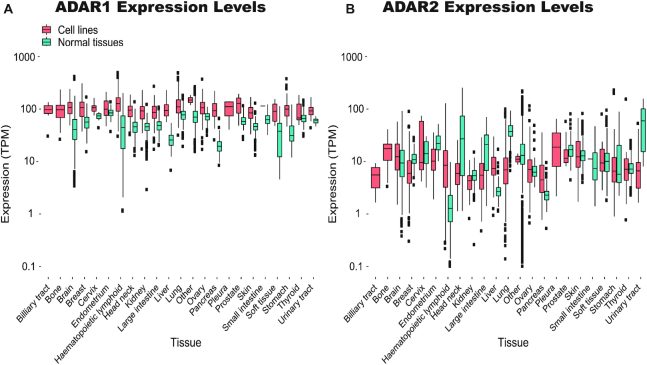
One may assume that choosing cell lines with high expression levels of the ADAR enzymes is sufficient to ensure editing levels that mimic normal tissues. Notably, while editing in cell lines is lower, the median ADAR1 mRNA levels in cell lines is even somewhat higher than in normal tissues (103.1 TPM compared to 50.0 TPM) (Figure 2A and Supplementary Table S4). To verify that this high relative level of ADAR1 expression does not follow from loss of expression of other genes, we looked at the ranked expression of ADAR1 (Methods), and verified that the ranks of ADAR1 (expression quantiles) are higher in cell lines as well (Supplementary Figure S1A). ADAR2 expression levels are generally much lower than ADAR1, and the median ADAR2 expression in cell lines is 7.4 TPM, comparable to normal tissues (Figure 2B, Supplementary Figure S1B and Supplementary Table S4).
Figure 2.
Expression levels of the ADAR enzymes in cell lines and normal tissues. ADAR expression levels (TPM) were calculated for all cell lines in the two datasets. Normal levels were calculated for matching GTEx normal tissue samples (72) when available. (A) ADAR1 expression levels are overall higher in cell lines than in normal tissues (B) but ADAR2 levels mostly resemble those of normal tissues. Note the logarithmic scale.
However, high expression of ADAR1 does not ensure high levels of editing. To demonstrate this, we selected 10% of cell lines with the highest levels of ADAR1 expression and found that even in these cells AEI values may be much lower than normal tissues (Supplementary Figure S2A). Consistently, while ADAR1 levels in cell lines are moderately correlated with the AEI (Spearman, R2 = 0.28, P-value < 2.2e–16), this correlation is too weak to allow a reliable prediction of high AEI based on ADAR1 levels (Supplementary Figure S2B). No significant correlation was found between AEI and either ADAR2 or ADAR3. Taken together, these results show that ADAR levels are not a good enough proxy for choosing a cell line with appreciable editing.
There is much current interest in ADAR regulators. Correlation studies may supply some initial leads into such regulators. Following previous studies (17,18), we used a linear regression model to correlate AEI values with expression levels of ADAR1 and one additional gene (each time), searching for an appreciable increase in the adjusted R-squared. We found 105 genes for which the adjusted R-squared significantly increases by more than 0.1 (compared to the single-variable correlation of AEI and ADAR1, where the adjusted R-squared equals 0.27; Bonferroni ≤ 0.05, Supplementary Table S5). Interestingly, almost a third of these (32/105) are RNA binding genes (75) (marked with a star in Supplementary Table S5). These may be used as a starting point for future analyses. Notably, ADAR3 was not identified.
Editing in cell lines is reduced compared to normal tissues
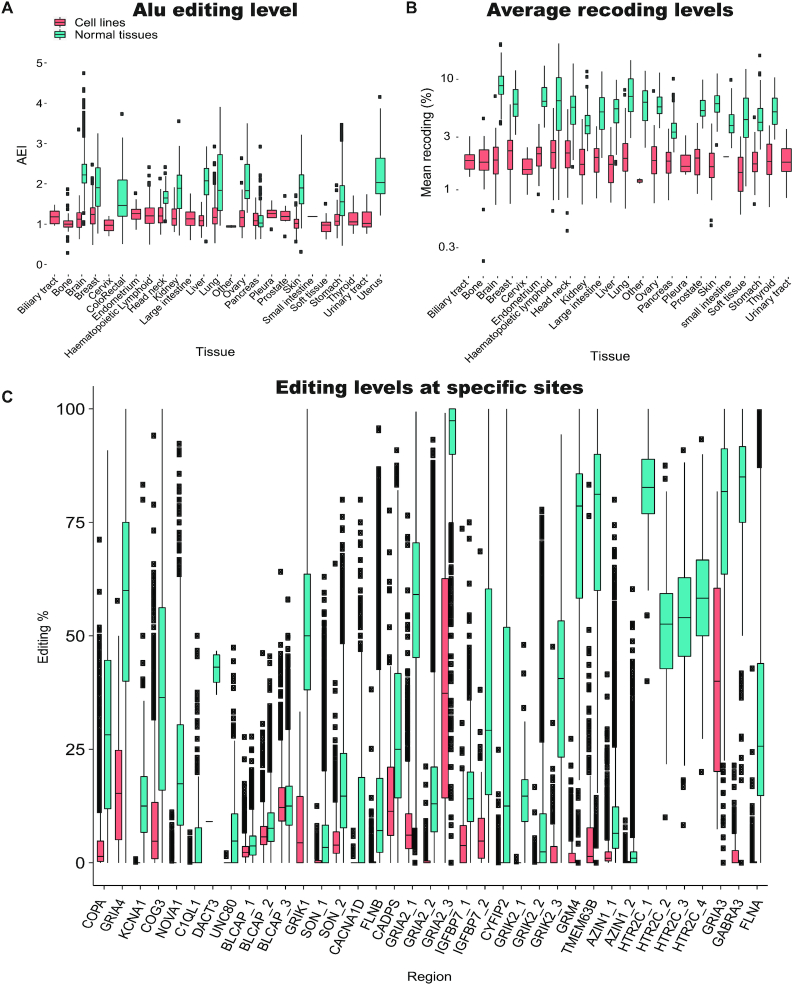
The AEI, which is calculated over all Alu sequences in the genome, provides a suitable measure for global editing activity (18). Its median value across normal tissues is 1.86. In comparison, the median AEI in cell lines is 1.14, and only 52 of the 1610 cell line samples exceed the normal median value.
Dividing the cell lines based on the tissue of origin, we compared the distribution of AEI values in cell lines and normal samples. In all tissues but pancreas the AEI in cell lines is markedly lower (Figure 3A). Furthermore, the differences between tissues, clearly seen in the normal tissues data, disappear to a large extent when these tissues are transformed into cell lines. However, the range of editing levels in cell lines within each tissue is large, and for each tissue one may find some cell lines that mimic the global editing levels seen in normal tissues.
Figure 3.
Editing in cell lines is lower compared to normal tissues (A) Global level of editing, measured by the AEI, for cell lines and matched normal tissues. (B) The distribution of the average editing level at the evolutionarily conserved coding sites (Supplementary Table S1) (71) is presented for cell lines and normal tissue. Note the logarithmic scale. (C) Distributions of editing levels at specific evolutionarily conserved coding sites in cell lines and normal tissues. Note that for each site, a number of cell lines exhibit editing levels comparable to those found in normal tissues. The distribution of editing levels was calculated only for samples with at least 10 reads supporting the site.
It should be stressed, though, that specific Alu regions may behave differently than the global AEI trend (Supplementary Figure 3). Presumably, this could be due to the interplay between ADAR editing and other RNA binding proteins (75). If one is interested in a specific Alu region, ensuring a high global AEI may not be sufficient, and screening of several high-AEI cell lines would be required.
Most coding editing sites have a unique set of preferable cell lines
Similarly, editing at the 314 coding sites (Methods) is lower in cell lines. First, for each cell line and normal tissue sample we looked at the editing level averaged over all well covered (≥10 reads) coding sites. Then we averaged the results over all cell lines and normal samples for all 314 sites and, in particular, for the 37 evolutionarily conserved coding sites in mammalians (71) that are of special interest. The average level at the 314 sites over all cell lines was only 2.09%, compared to 6.85% for normal GTEx tissues. For the evolutionarily conserved sites the levels were 3.88% and 20.27%, respectively. This lower level of editing is observed in cell lines derived from all tissue types (Figure 3B). Looking at specific sites, most of them show either no editing or very low levels of editing in the majority of cell lines (Figure 3C). No significant correlation was observed between the editing levels per site and the expression of the target gene (tested for 285 coding sites for which editing was observed in at least 100 cell lines; Benjamini–Hochberg FDR = 0.05).
Nevertheless, for almost each of the 314 sites one may find a number of cell lines that do show editing levels comparable to, or even exceeding, those found in normal tissues. These cell lines are potential candidates for studying editing in these individual editing sites. For example, editing of the GRIA3 site chrX:122598962 is undetectable due to low or even no reads coverage in >1500 cell line samples analyzed. For this site, editing levels above 1% are seen in only 33 cell lines. However, there are a few cell lines showing editing levels above 70% at this site, resembling the high levels found in highly edited normal tissues such as the brain. Whereas editing levels are typically consistent across normal tissues (76), editing in cell lines shows an appreciable variance. For example, editing of the NEIL1 K>R recoding site chr15:75646086 is detectable in most cell lines, but the levels are widely distributed (average ± std: 37.1% ± 25.5 for cell lines, compared with 81.9%±14.4 for normal tissues).
The full list of cell lines and their editing levels in coding sites is provided as Supplementary Table S2. Only 14 of the 314 coding sites and 6 of the 37 conserved sites show a significantly lower editing level in all cell lines, compared to normal GTEx tissues (all cell line levels below 95% of normal samples). These sites include 4 sites of the serotonin receptor HTR2C for which we found no coverage in all cell line samples, a site in the DACT3 transcript (chr19:47152854) where the highest measured editing level in cell lines is only 9%, and a site in GABRA3 (chrX:151358319) for which the highest editing level observed in cell lines is 20–21% (in three cell lines), far lower than what is found in normal tissues.
Notably, strong editing in specific coding sites does not indicate overall strong editing. For example, the cell line KALS-1 exhibits one of the highest levels of editing in GRIA3 sites (72% and 68% for the two sites above) but very low editing in other, generally well-edited, sites (e.g. 8/5533 and 32/736 reads show editing at the well-edited FLNA and AZIN1 sites, respectively). Some cell lines even show appreciable (>10%) editing for multiple targets (e.g. OV56, MHH-NB-11, SCLC-21H, NCI-H1385 and CAMA-1 with >10 sites each) but there is no cell line that shows strong editing for all sites of interest, suitable for studying coding sites in general. Therefore, selection of the most appropriate cell line should be done based on the specific set of coding sites of interest.
To further demonstrate this point, we collected cell-lines which are among the top 10 mostly edited for at least 10 of the 314 sites, as well as the widely used HeLa and HEK293. For these cell lines, we quantified the editing level at the evolutionarily conserved sites (Figure 4A). Clearly, even these cell lines are not suitable for all coding sites. While they exhibit high levels of editing in multiple sites, for each cell line there are some sites that are poorly expressed or poorly edited.
Figure 4.
Choosing the right cell line for the target of interest. (A) A heatmap of editing levels in cell lines that are edited most strongly at evolutionarily conserved coding sites (Supplementary Table S1) (71), as well as the commonly used HEK293 and HeLa cells, present a non-uniform pattern of editing. Yellow indicates low coverage (<10 reads). Even the cell lines showing strong editing in multiple coding sites are not suitable for all sites. For each of these, there are many conserved coding sites that are poorly expressed or poorly edited. (B) Using our dataset, we have selected three cell lines predicted to yield appreciable editing levels at the AZIN1 recoding site, grown and sequenced to verify the editing levels. Chromatograms of DNA and RNA sanger sequencing for the cell lines CAMA-1, NCI-H1573 and ZR-75-1 show RNA editing levels of 42%, 36% and 67% respectively. DNA sequencing of the same cells confirm no variability at the genomic level.
Taken together, these results emphasize the importance of carefully choosing the proper cell line to study, so that it exhibits editing levels that resemble the physiological state at the specific sites relevant for the experiment, or globally. Making this choice is readily facilitated by the CLAIRE database and the on-line search form, available at http://srv00.recas.ba.infn.it/atlas/claire.html.
Sequencing of Azin1 cell lines show editing levels similar to the catalogue
To demonstrate the validity of our approach, we searched the database for cell lines appropriate for a study of editing in AZIN1. Editing of AZIN1 is of much interest due to its contribution to certain types of cancer (53,77). A recent study has looked into the role of AZIN1 editing in cancer using PLC8024 cells (53). These cells express ADAR1 strongly (93 TPM), but exhibit negligible levels of editing in AZIN1 (∼2% editing), the gene of interest. In another study (62), nine cell lines were cultured and sequenced to measure editing levels in AZIN1 and FLNB and discover the most appropriate cell lines for experimental use. Here we demonstrate how a simple search throughout our dataset supplies a variety of apt options, and may save researchers time and resources.
Using our database, we chose the widely used (78) CAMA-1, ZR-75-1 and NCI-H1573 cell lines, for which AZIN1 editing levels are 41%, 22% and 23% respectively. After growing the cells and extracting DNA and RNA, we found indeed appreciable editing levels of 42%, 36% and 67%, respectively (Figure 4B). The deviations between the levels reported in the database (based on the available RNA-seq data) and the ones we measured for cells grown in our lab are consistent with our a-priori estimates of the variability (Figure 1D). These results demonstrate the utility of our database to find appropriate cell lines that can be efficiently put into use.
DISCUSSION
Cell lines are a vital tool to study RNA editing. Generally, editing levels in cell lines are found to be lower when compared to normal tissues, and one often needs to overexpress ADAR in order to facilitate the study. Here, we point out the large variability of editing and ADAR expression levels within cell lines, even if they originate from the same tissue. Most of the variability may be traced to the cell line type, rather than the inherent biological and technical noise. It is therefore possible, and recommended, to choose a cell line for which editing at the target(s) of interest, or the AEI (if global editing is the focus of the study) are known in advance to be within the desired range.
Notably, while the analyzed cell lines are derived from cancer cells where editing is often globally elevated (50), they mostly exhibit reduced overall editing. Furthermore, we found that the tissue variability in terms of the editing profile largely disappears in cell lines. Thus, as is often the case, cell lines do not necessarily mimic all aspects of the behavior of their tissue of origin. Possibly, part of the difference is due to cell lines missing environmental and physiological signals mediated by the surroundings tissues and the interaction with the immune system that may play a critical role in the activation and modulation of RNA editing.
The dataset presented here includes ADAR1 and ADAR2 expression levels, RNA editing levels in specific sites, the AEI as a measure for global editing, for over 1000 unique cancer cell lines. We believe this cell line editome catalogue will facilitate rational selection of appropriate cell lines for RNA editing research, and promote understanding patterns of editing in different cell lines.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Udi Ben-Zvi for help with Python, Shalom Hillel Roth for help with statistics, and Ofir Shlifer for advice regarding the online search platform. We also thank Jenny Shapiro for help with the tissue cultures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Israel Science Foundation [2673/17, 1945/18 to E.E.]; International Collaboration Grant from the Jacki and Bruce Barron Cancer Research Scholars’ Program, a partnership of the Israel Cancer Research Fund and City of Hope, as supported by The Harvey L. Miller Family Foundation [205467 to E.Y.L.]. Funding for open access charge: Israel Cancer Research Fund [205467].
Conflict of interest statement. None declared.
REFERENCES
- 1. Eisenberg E., Levanon E.Y.. A-to-I RNA editing — immune protector and transcriptome diversifier. Nat. Rev. Genet. 2018; 19:473–490. [DOI] [PubMed] [Google Scholar]
- 2. Porath H.T., Knisbacher B.A., Eisenberg E., Levanon E.Y.. Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance. Genome Biol. 2017; 18:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002; 71:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bass B.L., Weintraub H.. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987; 48:607–613. [DOI] [PubMed] [Google Scholar]
- 5. Grice L.F., Degnan B.M.. The origin of the ADAR gene family and animal RNA editing. BMC Evol. Biol. 2015; 15:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Savva Y.a., Rieder L.E., Reenan R.a.. The ADAR protein family. Genome Biol. 2012; 13:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nishikura K. Functions and regulation of RNA editing by ADAR Deaminases. Annu. Rev. Biochem. 2010; 79:321–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen C.X., Cho D.S., Wang Q., Lai F., Carter K.C., Nishikura K.. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000; 6:755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Oakes E., Anderson A., Cohen-Gadol A., Hundley H.A.. Adenosine deaminase that Acts on RNA 3 (ADAR3) Binding to glutamate receptor subunit B Pre-mRNA inhibits RNA editing in glioblastoma. J. Biol. Chem. 2017; 292:4326–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Basilio C., Wahba A.J., Lengyel P., Speyer J.F., Ochoa S.. Synthetic polynucleotides and the amino acid code. V. PNAS. 1962; 48:613–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Licht K., Hartl M., Amman F., Anrather D., Janisiw M.P., Jantsch M.F.. Inosine induces context-dependent recoding and translational stalling. Nucleic Acids Res. 2019; 47:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morse D.P., Aruscavage P.J., Bass B.L.. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. PNAS. 2002; 99:7906–7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levanon E.Y., Eisenberg E., Yelin R., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D. et al.. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004; 22:1001–1005. [DOI] [PubMed] [Google Scholar]
- 14. Athanasiadis A., Rich A., Maas S.. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004; 2:e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blow M., Futreal A.P., Wooster R., Stratton M.R.. A survey of RNA editing in human brain. Genome Res. 2004; 14:2379–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim D.D.Y., Kim T.T.Y., Walsh T., Kobayashi Y., Matise T.C., Buyske S., Gabriel A.. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res. 2004; 14:1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan M.H., Li Q., Shanmugam R., Piskol R., Kohler J., Young A.N., Liu K.I., Zhang R., Ramaswami G., Ariyoshi K. et al.. Dynamic landscape and regulation of RNA editing in mammals. Nature. 2017; 550:249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roth S.H., Levanon E.Y., Eisenberg E.. Genome-wide quantification of ADAR adenosine-to-inosine RNA editing activity. Nat. Methods. 2019; 16:1131–1138. [DOI] [PubMed] [Google Scholar]
- 19. Liddicoat B.J., Piskol R., Chalk A.M., Ramaswami G., Higuchi M., Hartner J.C., Li J.B., Seeburg P.H., Walkley C.R.. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015; 349:1115–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mannion N.M., Greenwood S.M., Young R., Cox S., Brindle J., Read D., Nellåker C., Vesely C., Ponting C.P., McLaughlin P.J. et al.. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014; 9:1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pestal K., Funk C.C., Snyder J.M., Price N.D., Treuting P.M., Stetson D.B.. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity. 2015; 43:933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones A.K., Buckingham S.D., Papadaki M., Yokota M., Sattelle B.M., Matsuda K., Sattelle D.B.. Splice-variant- and stage-specific RNA editing of the Drosophila GABA receptor modulates agonist potency. J. Neurosci. 2009; 29:4287–4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lev-Maor G., Ram O., Kim E., Sela N., Goren A., Levanon E.Y., Ast G.. Intronic Alus influence alternative splicing. PLoS Genet. 2008; 4:e1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rueter S.M., Dawson T.R., Emeson R.B.. Regulation of alternative splicing by RNA editing. Nature. 1999; 399:75–80. [DOI] [PubMed] [Google Scholar]
- 25. Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K.. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007; 315:1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alon S., Mor E., Vigneault F., Church G.M., Locatelli F., Galeano F., Gallo A., Shomron N., Eisenberg E.. Systematic identification of edited microRNAs in the human brain. Genome Res. 2012; 22:1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li L., Song Y., Shi X., Liu J., Xiong S., Chen W., Fu Q., Huang Z., Gu N., Zhang R.. The landscape of miRNA editing in animals and its impact on miRNA biogenesis and targeting. Genome Res. 2018; 28:132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K.. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006; 13:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pinto Y., Buchumenski I., Levanon E.Y., Eisenberg E.. Human cancer tissues exhibit reduced A-to-I editing of miRNAs coupled with elevated editing of their targets. Nucleic Acids Res. 2017; 46:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Altaf F., Vesely C., Sheikh A.M., Munir R., Shah S.T.A., Tariq A.. Modulation of ADAR mRNA expression in patients with congenital heart defects. PLoS One. 2019; 14:e0200968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vesely C., Tauber S., Sedlazeck F.J., von Haeseler A., Jantsch M.F.. Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic miRNAs. Genome Res. 2012; 22:1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang I.X., So E., Devlin J.L., Zhao Y., Wu M., Cheung V.G.. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 2013; 5:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scadden A.D.J. Inosine-containing dsRNA binds a stress-granule-like complex and downregulates gene expression in trans. Mol. Cell. 2007; 28:491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gatsiou A., Vlachogiannis N., Lunella F.F., Sachse M., Stellos K.. Adenosine-to-inosine RNA editing in health and disease. Antioxid. Redox Signal. 2018; 29:846–863. [DOI] [PubMed] [Google Scholar]
- 35. Gallo A., Vukic D., Michalík D., O’Connell M.A., Keegan L.P.. ADAR RNA editing in human disease; more to it than meets the I. Hum. Genet. 2017; 136:1265–1278. [DOI] [PubMed] [Google Scholar]
- 36. Rice G.I., Kasher P.R., Forte G.M.A., Mannion N.M., Greenwood S.M., Szynkiewicz M., Dickerson J.E., Bhaskar S.S., Zampini M., Briggs T.A. et al.. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 2012; 44:1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roth S.H., Danan-Gotthold M., Ben-Izhak M., Rechavi G., Cohen C.J., Louzoun Y., Levanon E.Y.. Increased RNA editing may provide a source for autoantigens in systemic lupus erythematosus. Cell Rep. 2018; 23:50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu J., Chen Z.J.. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014; 32:461–488. [DOI] [PubMed] [Google Scholar]
- 39. Shallev L., Kopel E., Feiglin A., Leichner G.S., Avni D., Sidi Y., Eisenberg E., Barzilai A., Levanon E.Y., Greenberger S.. Decreased A-to-I RNA editing as a source of keratinocytes’ dsRNA in psoriasis. RNA. 2018; 24:828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jain M., Mann T.D., Stulić M., Rao S.P., Kirsch A., Pullirsch D., Strobl X., Rath C., Reissig L., Moreth K. et al.. RNA editing of filamin A pre- mRNA regulates vascular contraction and diastolic blood pressure. EMBO J. 2018; 37:e94813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stellos K., Gatsiou A., Stamatelopoulos K., Perisic Matic L., John D., Lunella F.F., Jaé N., Rossbach O., Amrhein C., Sigala F. et al.. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016; 22:1140–1150. [DOI] [PubMed] [Google Scholar]
- 42. Kawahara Y., Ito K., Sun H., Aizawa H., Kanazawa I., Kwak S.. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004; 427:801. [DOI] [PubMed] [Google Scholar]
- 43. Srivastava P.K., Bagnati M., Delahaye-Duriez A., Ko J.-H., Rotival M., Langley S.R., Shkura K., Mazzuferi M., Danis B., van Eyll J. et al.. Genome-wide analysis of differential RNA editing in epilepsy. Genome Res. 2017; 27:440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gurevich I., Tamir H., Arango V., Dwork A.J., Mann J.J., Schmauss C.. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002; 34:349–356. [DOI] [PubMed] [Google Scholar]
- 45. Hwang T., Park C.-K., Leung A.K.L., Gao Y., Hyde T.M., Kleinman J.E., Rajpurohit A., Tao R., Shin J.H., Weinberger D.R.. Dynamic regulation of RNA editing in human brain development and disease. Nat. Neurosci. 2016; 19:1093–1099. [DOI] [PubMed] [Google Scholar]
- 46. Khermesh K., Erchia A.M.D., Barak M., Annese A., Wachtel C., Levanon E.Y., Picardi E., Eisenberg E.L.I.. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer's disease. RNA. 2016; 22:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Eran A., Li J.B., Vatalaro K., McCarthy J., Rahimov F., Collins C., Markianos K., Margulies D.M., Brown E.N., Calvo S.E. et al.. Comparative RNA editing in autistic and neurotypical cerebella. Mol. Psychiatry. 2013; 18:1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Breen M.S., Dobbyn A., Li Q., Roussos P., Hoffman G.E., Stahl E., Chess A., Sklar P., Li J.B., Devlin B. et al.. Global landscape and genetic regulation of RNA editing in cortical samples from individuals with schizophrenia. Nat. Neurosci. 2019; 22:1402–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cenci C., Barzotti R., Galeano F., Corbelli S., Rota R., Massimi L., Di Rocco C., O’Connell M.A., Gallo A.. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J. Biol. Chem. 2008; 283:7251–7260. [DOI] [PubMed] [Google Scholar]
- 50. Paz-Yaacov N., Bazak L., Buchumenski I., Porath H.T.T., Danan-Gotthold M., Knisbacher B.A.A., Eisenberg E., Levanon E.Y.Y.. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 2015; 13:267–276. [DOI] [PubMed] [Google Scholar]
- 51. Fumagalli D., Gacquer D., Campbell P.J., Sotiriou C., Detours V., Lefort A., Libert F., Brown D.. Principles governing A-to-I RNA editing in the breast cancer transcriptome article principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep. 2015; 13:277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Han L., Diao L., Yu S., Xu X., Li J., Zhang R., Yang Y., Werner H.M.J., Eterovic A.K., Yuan Y. et al.. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015; 28:515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen L., Li Y., Lin C.H., Chan T.H.M., Chow R.K.K., Song Y., Liu M., Yuan Y.-F., Fu L., Kong K.L. et al.. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013; 19:209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Montiel-Gonzalez M.F., Vallecillo-Viejo I., Yudowski G.A., Rosenthal J.J.C.. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. PNAS. 2013; 110:18285–18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Merkle T., Merz S., Reautschnig P., Blaha A., Li Q., Vogel P., Wettengel J., Li J.B., Stafforst T.. Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat. Biotechnol. 2019; 37:133–138. [DOI] [PubMed] [Google Scholar]
- 56. Katrekar D., Chen G., Meluzzi D., Ganesh A., Worlikar A., Shih Y.-R., Varghese S., Mali P.. In vivo RNA editing of point mutations via RNA-guided adenosine deaminases. Nat. Methods. 2019; 16:239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Qu L., Yi Z., Zhu S., Wang C., Cao Z., Zhou Z., Yuan P., Yu Y., Tian F., Liu Z. et al.. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol. 2019; 37:1059–1069. [DOI] [PubMed] [Google Scholar]
- 58. Abudayyeh O.O., Gootenberg J.S., Franklin B., Koob J., Kellner M.J., Ladha A., Joung J., Kirchgatterer P., Cox D.B.T., Zhang F.. A cytosine deaminase for programmable single-base RNA editing. Science. 2019; 365:382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Risbridger G.P. Human cell lines as tools of our trade: ‘laying it on the (cell) line’. Mol. Endocrinol. 2015; 29:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hughes P., Marshall D., Reid Y., Parkes H., Gelber C.. The costs of using unauthenticated, over-passaged cell lines: how much more data do we need. BioTechniques. 2007; 43:575–586. [DOI] [PubMed] [Google Scholar]
- 61. Geraghty R.J., Capes-Davis A., Davis J.M., Downward J., Freshney R.I., Knezevic I., Lovell-Badge R., Masters J.R.W., Meredith J., Stacey G.N. et al.. Guidelines for the use of cell lines in biomedical research. Br. J. Cancer. 2014; 111:1021–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Qin Y.R., Qiao J.J., Chan T.H.M., Zhu Y.H., Li F.F., Liu H., Fei J., Li Y., Guan X.Y., Chen L.. Adenosine-to-inosine RNA editing mediated by adars in esophageal squamous cell carcinoma. Cancer Res. 2014; 74:840–851. [DOI] [PubMed] [Google Scholar]
- 63. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G. V., Sonkin D. et al.. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z., Liu H., Degenhardt J., Mayba O., Gnad F., Liu J. et al.. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2014; 33:306–312. [DOI] [PubMed] [Google Scholar]
- 65. Thul P.J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Björk L., Breckels L.M. et al.. A subcellular map of the human proteome. Science. 2017; 356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 66. Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C.. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017; 14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bazak L., Levanon E.Y., Eisenberg E.. Genome-wide analysis of Alu editability. Nucleic Acids Res. 2014; 42:6876–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Picardi E., Pesole G.. REDItools: high-throughput RNA editing detection made easy. Bioinformatics. 2013; 29:1813–1814. [DOI] [PubMed] [Google Scholar]
- 70. Karolchik D. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004; 32:493D–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pinto Y., Cohen H.Y., Levanon E.Y.. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 2014; 15:R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N. et al.. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S.. EditR: a method to quantify base editing from sanger sequencing. CRISPR J. 2018; 1:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Picardi E., D’Erchia A.M., Lo Giudice C., Pesole G.. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2017; 45:D750–D757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Quinones-Valdez G., Tran S.S., Jun H.-I., Bahn J.H., Yang E.-W., Zhan L., Brümmer A., Wei X., Van Nostrand E.L., Pratt G.A. et al.. Regulation of RNA editing by RNA-binding proteins in human cells. Commun. Biol. 2019; 2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Greenberger S., Levanon E.Y., Paz-Yaacov N., Barzilai A., Safran M., Osenberg S., Amariglio N., Rechavi G., Eisenberg E.. Consistent levels of A-to-I RNA editing across individuals in coding sequences and non-conserved Alu repeats. BMC Genomics. 2010; 11:608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Okugawa Y., Toiyama Y., Shigeyasu K., Yamamoto A., Shigemori T., Yin C., Ichikawa T., Yasuda H., Fujikawa H., Yoshiyama S. et al.. Enhanced AZIN1 RNA editing and overexpression of its regulatory enzyme ADAR1 are important prognostic biomarkers in gastric cancer. J. Transl. Med. 2018; 16:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bairoch A. The cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 2018; 29:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.