Abstract
The chromatin organizer SATB1 is highly enriched in thymocytes and is essential for T-cell development. Although SATB1 regulates a large number of genes important for T-cell development, the mechanism(s) regulating expression of SATB1 during this process remain elusive. Using chromatin immune precipitation-seq-based occupancy profiles of H3K4me3 and H3Kme1 at Satb1 gene locus, we predicted four different alternative promoters of Satb1 in mouse thymocytes and characterized them. The expression of Satb1 transcript variants with distinct 5′ UTRs occurs in a stage-specific manner during T-cell development and is dependent on TCR signaling. The observed discrepancy between the expression levels of SATB1 mRNA and protein in developing thymocytes can be explained by the differential translatability of Satb1 transcript variants as confirmed by polysome profiling and in vitro translation assay. We show that Satb1 alternative promoters exhibit lineage-specific chromatin accessibility during T-cell development from progenitors. Furthermore, TCF1 regulates the Satb1 P2 promoter switch during CD4SP development, via direct binding to the Satb1 P2 promoter. CD4SP T cells from TCF1 KO mice exhibit downregulation of P2 transcript variant expression as well as low levels of SATB1 protein. Collectively, these results provide unequivocal evidence toward alternative promoter switch-mediated developmental stage-specific regulation of SATB1 in thymocytes.
INTRODUCTION
The development of thymocytes begins in thymus soon after a common lymphoid progenitor from the bone marrow migrates to it (1–3). Thymic developmental stages are typically characterized based on the surface expression of CD4 and CD8 co-receptors into CD4−CD8− double negative (DN), CD4+CD8+ double positive (DP) and single positive (SP) -either CD4+CD8− (CD4SP) or CD4−CD8+ (CD8SP) (4). DP thymocytes that are T-cell receptor (TCR) re-arranged undergo TCR signal mediated positive and negative selection and differentiate into either CD4+SP or CD8+SP thymocytes (5–7). Thus, TCR signaling plays a pivotal role during thymocyte development, during which it activates a plethora of transcription factors (TFs) leading to the selection of functional T cells in thymus. SATB1 (Special AT-rich binding protein 1), a thymocyte enriched regulator, is indispensable for thymocyte development (8,9). SATB1 is a higher-order chromatin organizer and a lineage-specific TF (10,11). SATB1 forms an unusual ‘cage-like’ three-dimensional structure in mouse thymocytes and presumably circumscribes the heterochromatin (11,12). SATB1 tethers specialized AT-rich genomic regions and thereby causes the looping of the chromatin (10,13), thus regulating the chromatin ‘loopscape’ (12). Further, SATB1 regulates the target gene expression by acting as the docking site for a number of chromatin modifiers and nucleosome remodelers (14,15). SATB1 is a member of the SATB family proteins that are implicated in chromatin looping, chromatin dynamics and transciptional regulation (12,16). The other SATB family member is SATB2, which along with SATB1 has been studied in various cancer models, ascribing them as characteristic markers for disease progression (17).
Studies using Satb1 knockout (KO) mice revealed that thymocytes fail to develop beyond DP stage in the absence of SATB1 (8). SATB1 is essential for positive and negative selection of thymocytes, and for the establishment of immune tolerance (9). SATB1 is also essential for the development of thymic regulatory CD4+ cells (Tregs) from their precursors in the thymus, thereby playing an important role in immune tolerance (18). Considering the diversity of functions assigned to SATB1, studying its regulation is crucial for understanding the cell type-specific functional outcome. Post-translational modifications, such as phosphorylation and acetylation of SATB1, have contrasting effects on the transcriptional activity of SATB1 and also on its propensity for the recruitment of its interaction partners (19,20). Further, SATB1 is negatively regulated by FOXP3 induced micro-RNAs miR-7 and miR-155, which specifically target 3′ UTR of Satb1 (21,22). Interestingly, in T cells, SATB1 is under the control of TCR signaling (23). However, the precise molecular mechanism of transcriptional regulation of SATB1 via TCR signaling is still unclear.
Alternative promoter usage along with other alternative processing events contributes toward generation of diversity in the transcriptome and the proteome, thereby increasing the complexity of an organism. Utilization of alternative promoters increases the frequency of alternative splicing events in the genome (24). Alternative promoters modulate gene expression in a tissue or developmental stage-specific manner (25,26). Additionally, they provide complexity to gene regulation by modulating the sub-cellular localization of mRNA, generating transcript isoforms with alternative first exons and affect the translation efficiency by harboring regulatory elements in the alternative first exons (27).
Here, we investigate the regulation of Satb1 during thymic T-cell development via alternative promoter usage, and how SATB1 expression is stringently controlled via these promoters. We demonstrate that the differential translatability of Satb1 transcript variants contributes to the regulation of SATB1 protein levels during T-cell development. We show that the combinatorial expression of Satb1 transcript variants regulates SATB1 protein levels during the development of T cells. Furthermore, the dynamic accessibility of the Satb1 alternative promoters is regulated in a lineage-specific manner during the development of T cells from their progenitors. Our study reveals that the Wnt-responsive TF TCF1 mediates Satb1 alternative promoter switch during development of CD4SP from DP cells. Collectively, the findings reported here demonstrate that TCR signaling induces a switch in the usage of Satb1 alternative promoters during the development of T cells in the thymus.
MATERIALS AND METHODS
Mice
Three-week-old C57BL/6 mice were used for sorting of thymocyte subpopulations in different T-cell developmental stages. Six-week-old C57BL/6 mice were used to isolate naïve CD4+ T cells from spleen. TCF1 knockout (TCF1 KO) mice were obtained from H. Clevers. All mice were bred and maintained under specific pathogen free environment and experimental procedures were performed according to the guidelines of the National Facility for Gene Function in Health and Disease, IISER Pune, Experimental Animal Facility at NCCS Pune and National Institute of Aging, Baltimore, USA.
T-cell isolation and culture
Single cell suspension of thymocytes was prepared by macerating thymii from three-week-old mice and was used for the surface staining. Prior to the surface staining of total thymocytes, Fc receptor blocking was performed using the purified rat anti-mouse CD16/CD32 (Cat no. 55314, Clone 2.4G2, BD Biosciences). Thymocytes were then surface stained using the following fluorochrome-tagged antibodies: eFluor 450 anti-mouse CD4 (Cat no. 48-0041-82, Clone GK1.5, eBioscience); APC anti-mouse CD4 (Cat no. 17-0041-82, Clone GK1.5, eBioscience); FITC anti-mouse CD4 (Cat no. 53729, Clone GK1.5, BD Biosciences); PE anti-mouse CD8a (Cat no. 553032 Clone 53-6.7, BD Biosciences); eFluor 450 anti-mouse CD24 (Cat no. 48-0242-82, Clone M1/69, eBioscience); PerCP Cy5.5 anti-mouse CD24 (Cat no. 45-0242-82, Clone M1/69, eBioscience); PE-Cy7 anti-mouse TCRβ (Cat no. 25-5961-82, Clone H57-597, eBioscience); PE-Cy7 anti-mouse CD69 (Cat no. 552879, Clone H1.2F3, BD Biosciences); PE anti-mouse CD25 (Cat no. 553866, Clone PC61, BD Biosciences) and BV510 anti-mouse CD25 (Cat no. 563037, Clone PC61, BD Biosciences). CD4−CD8− double negative (DN), CD4+CD8+ DP, CD4+ single positive (CD4SP), immature CD4SP (CD4+CD24+), mature CD4SP (CD4+CD24−) and CD8+ single positive (CD8SP) thymocytes were sorted using FACS Aria III SORP (BD Biosciences). Spleen from six-week-old mice were used for enrichment of naïve CD4+ T cells by magnetic sorting using the mouse CD4+ T-cell enrichment kit (BD Biosciences) followed by FACS sorting. For TCR activation experiments, naïve CD4+ T cells were stimulated with 0.5 μg/ml of plate bound anti-mouse CD3 (Cat no. 14-0032-82, Clone 17A2, eBioscience) and 1.5 μg/ml of soluble anti-mouse CD28 (Cat no. 14-0281-82, Clone 37.51, eBioscience) for 48 h. Cells were harvested for RNA extraction and for intracellular staining. Intracellular staining was performed using Alexa Fluor 647 anti-SATB1 (Cat no. 562378, Clone 14/SATB1, BD Biosciences) and PE anti-mouse Nur77 (Cat no. 12-5965-82, Clone 12.14, eBioscience) by using Foxp3/Transcription Factor staining kit (eBioscience). Flow cytometry analyses were performed using BD FACS Canto II (BD Biosciences).
5′. Rapid amplification of cDNA ends (5′ RACE)
Single cell suspension of thymocytes was prepared using thymii from three week-old C57BL/6 mice. Total thymocytes were used for RNA extraction using Trizol (Invitrogen) method and cDNA synthesis was performed using SMARTer RACE 5′/3′ kit (Clontech) according to the manufacturer's protocol. 5′ RACE polymerase chain reaction (PCR) was carried out using the forward universal primer mix provided with the kit, and the reverse primer 5′-TGCTCCCAAGCCTTCCTCTTCCTAGAG-3′ specific to the exon-2 of Satb1. The resulting PCR products were subjected to gel electrophoresis and DNA bands were gel purified using Nucleospin gel extraction kit (MACHEREY-NAGEL). Nested PCR was performed in case of E1b and E1d by using the following Satb1 exon-2 specific reverse primer 5′-CTGTCTTACAGATCACCTGCCAG-3′. The amplified DNA fragments were cloned into linearized pRACE vector provided with the kit, and then propagated by transformation of DH5α strain of Escherichia coli (Promega). Recombinant plasmid DNAs were isolated from an individual bacterial clones by alkaline lysis method and were subjected to sequencing by Sanger sequencing method.
Quantitative real-time PCR analysis (qRT-PCR)
Isolation of total RNA from sorted thymocyte subpopulations and from peripheral CD4+ T cells was performed using Qiagen RNeasy mini kit (Qiagen). Following DNase I (Promega) digestion, RNA was subjected to cDNA synthesis using high capacity cDNA synthesis kit (Applied Biosystems). Quantitative RT-PCR analyses were performed using SYBR green qPCR master mix (Roche) at the following PCR conditions: step 1, 95°C, 5 min; step 2, 95°C, 45 s, 60°C, 45 s, 72°C, 1 min for 40 cycles. The change in gene expression was calculated using the formula ΔCt = Ct Target − Ct Control. Normalized transcript expression was calculated using the equation 2−(ΔCt), where ΔCt = Ct Target − Ct Control. The oligonucleotide primer sequences used for qRT-PCR analyses are listed in the Supplementary Table S1.
Cycloheximide and MG132 chase assay
Three-week-old C57BL/6 mice were used for isolation of thymus. Thymi were used for the preparation of single cell suspension and subjected to Fc receptor blocking using the purified anti-CD16/CD32 (Clone 2.4G2, BD Biosciences). Thymocytes were then surface stained using the following fluorochrome tagged antibodies: FITC anti-mouse CD4 (Clone GK1.5, BD Biosciences); PE anti-mouse CD8a (Clone 53–6.7, BD Biosciences). CD4+CD8+ DP thymocytes were FACS sorted using FACS Aria III SORP (BD biosciences). Sorted DP thymocytes were cultured in the presence or absence of different concentrations of MG-132 (Calbiochem), and Cycloheximide (Sigma-Aldrich) in RPMI-1640 media supplemented with 10% FBS and penicillin-streptomycin for 4 h. After incubation, cells were harvested and used for western blotting.
Western blotting
Cells were lysed using RIPA lysis buffer (50 mM Tris–HCl at pH 7.4, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1% NP40, 0.25% sodium deoxycholate, 1 mM PMSF and 1× protease inhibitor cocktail (Roche) and the protein quantification was performed using bicinchoninic acid (BCA) protein assay kit (Thermo Scientific). Total protein was separated on 10% or 12.5% sodium dodecyl sulphate (SDS) polyacrylamide gel and then transferred to polyvinylidene fluoride (PVDF) membrane. The PVDF membrane was then blocked with 5% non-fat milk and probed with the following antibodies; anti-SATB1 (dilutions 1:1000 for polyclonal antibody against the N-terminal region of SATB1 prepared in-house, 1:500 for monoclonal antibody from Santa Cruz Biotechnology, Cat no. sc-376096), anti-GAPDH (1:4000, Abcam Cat no. ab8245), anti-ubiquitin (1:1000, Millipore clone P4D1-A11, Cat no. 05-944), anti-β-ACTIN (1:4000, Sigma, clone-AC 74, cat no. A2228), anti-TCF1 (1:500, Santa Cruz Biotechnology, Cat no. sc-271453) and anti-γ-tubulin (1:4000, Sigma, Cat no. T5326). The signals were visualized using ECL luminescence detection reagent (Bio-Rad) on ImageQuant LAS 4000 system (GE Healthcare Life Sciences).
In vitro transcription, in vitro translation and luciferase reporter assays
Full length Satb1 5′ UTRs such as P1L, P1S, P2, P3 and P4 were amplified from thymocyte cDNA and then cloned upstream of firefly luciferase gene in pGL3 basic vector (Promega). The sequences of an individual clones were confirmed by Sanger sequencing. A template for in vitro transcription was prepared by PCR amplification of firefly luciferase coding sequences (CDS) along with distinct Satb1 5′ UTRs by using a pair of primers which include T7 promoter in the sense primer specific to the 5′ UTR and an antisense primer specific to the firefly luciferase gene. The resulting PCR amplified products were gel extracted and quantified using spectrophotometer (NanoDrop 2000c, Thermo Scientific). Equimolar concentrations of DNA normalized to their base pair lengths were used for in vitro transcription reaction using T7 RNA polymerase, according to the manufacturer's protocol of mMESSAGE mMACHINE T7 kit (Invitrogen). In vitro transcribed RNA was purified by LiCl2 precipitation method and equimolar concentrations of RNA normalized to their base pair sizes were used for in vitro translation reaction using rabbit reticulocyte lysate (Invitrogen). Luciferase activity was measured in 5 μl of in vitro translated product by using the luciferase assay kit (Promega). The sequences of DNA oligonucleotide primers used for the cloning in this experiment are included in Supplementary Table S2.
Polysome profiling analysis
Thymi from six 3-week-old C57BL/6 mice were immediately snap frozen as soon as mice were dissected and were pulverized under liquid nitrogen. The powder was then lysed in the lysis buffer (5 mM Tris–HCl at pH 7.5, 2.5 mM MgCl2, 1.5 mM KCl, 100 μg/ml cycloheximide (Sigma-Aldrich), 0.1% Triton-X-100, 2 mM dithiothreitol (DTT), 500 U/ml RNase inhibitor (Applied Biosystems), 1× EDTA free protease inhibitor cocktail (Roche). After lysis, the nuclei were pelleted down by centrifugation at 14 000 rpm for 15 min, at 4°C, without brake during deceleration using a bench top centrifuge (Rotor IL-053, Eppendorf). The supernatant was collected into pre-chilled 1.5 ml tubes and OD260nm was measured using spectrophotometer (NanoDrop 2000c, Thermo Scientific). The supernatant with OD260nm of 40 was layered onto 10 ml linear sucrose gradient (10–50% sucrose (w/v)), which was prepared in 1× gradient buffer (20 mM HEPES at pH 7.6, 100 mM KCl, 5 mM MgCl2, 2 mM DTT, 100 μg/ml cycloheximide (Sigma-Aldrich), 500 U/ml RNase inhibitor (Applied Biosystems), 1× EDTA free protease inhibitor cocktail (Roche) and centrifuged in a SW40Ti rotor (Beckman Coulter) for 3 h at 35 000 rpm at 4°C, with no brake applied during deceleration. Sucrose gradient was then subjected to fractionation using a gradient fractionation system (ISCO Model 160 gradient former). RNA was extracted from these fractions using Trizol (Invitrogen) method. Following DNase I (Promega) digestion, RNA was subjected to cDNA synthesis which was performed using iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time PCR (qRT-PCR) analyses were performed using SYBR green qPCR master mix (Roche) and by using Satb1 alternative transcript-specific amplification primers listed in the Supplementary Table S1.
ChIP-qPCR assay
Chromatin immune precipitation (ChIP) was performed using chromatin isolated from DP and CD4SP thymocytes. Approximately 107 DP and CD4SP thymocytes were crosslinked with 1% formaldehyde at room temperature for 10 min. After formaldehyde crosslinking, 125 mM glycine was used for neutralization. Crosslinked cells were subjected to the nuclei isolation using the hypotonic buffer (25 mM Tris–HCl at pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.1% NP40, 1 mM DTT, 0.5 mM PMSF and 1× protease inhibitor cocktail (Roche). Isolated nuclei were lysed using the lysis buffer (50 mM Tris–HCl at pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Sodium deoxycholate, 0.1% SDS, 0.5 mM PMSF and 1× protease inhibitor cocktail (Roche) and chromatin was subjected to the sonication using Bioruptor Twin sonicator (Diagenode, Belgium) for 20–30 cycles with 30 s ‘on’ and 30 s ‘off‘ to obtain the chromatin fragment size of 200–500 bp. Preclearing of the sonicated chromatin was performed using a cocktail containing 50% protein A/G beads slurry (Thermo scientific) and the precleared chromatin was subjected to the immune precipitation with anti-TCF1 antibody (Santa Cruz Biotechnology, sc-271453, Cell Signaling Technology C46C7) overnight at 4°C. Similarly, normal IgG was used as an isotype control. Immunoprecipitated complexes were pulled down by adding Protein A/G beads cocktail which was pre-saturated with 10 mg/ml tRNA and 1% bovine serum albumin and incubated at 4°C for 4 h. The immunoprecipitated bead bound chromatin was washed thoroughly, and subjected to elution by using the elution buffer (1% SDS, 0.1M NaHCO3). The eluted chromatin was de-crosslinked and the protein and RNA contamination was removed by treating with proteinase K (Sigma-Aldrich), and RNase A (Sigma-Aldrich), respectively. Further, the immunoprecipitated chromatin was purified and subjected to the quantitative PCR analysis using the formula ΔCt = Ct Target − Ct Input. Fold differences in enrichment were calculated using the equation 2−(ΔCt), where ΔCt = Ct Target − Ct Input, for both IgG and TCF1 immunoprecipitated DNA. The primer sequences used for ChIP-PCR analysis are listed in the Supplementary Table S1.
Transfections and luciferase activity assay
Satb1 P2 promoter region was amplified from the genomic DNA of mouse thymocytes and was cloned upstream to the firefly luciferase gene in pGL3 basic vector (Promega). Similarly TCF1 gene was amplified from the cDNA of mouse thymocytes, and then cloned into p3X-CMV-FLAG10 vector (Sigma). The P2 promoter containing pGL3 basic vector was overexpressed in HEK293T cells in the presence or absence of TCF1 overexpression using Lipofectamine 3000 transfection reagent (Invitrogen). After 48 h of transfection, cells were harvested for immunoblotting and luciferase activity assay. The P2 promoter activity was measured by subjecting the cell lysates to luciferase activity assay using dual luciferase assay kit using the manufacturer's protocol (Promega). Renilla luciferase activity was used as endogenous control. The sequences of cloning primers used in this experiment are listed in Supplementary Table S2.
Statistical analysis
The represented data were the mean ± SEM from three independent experiments. Statistical analyses were performed using GraphPad. Unpaired two-tailed Student's t-test and one-way ANOVA were used as indicated to compare the data points. P -value of <0.05 was considered as significant and P -values (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001) have been included in the figure legends. The label ‘ns’ indicates the change relative to the control sample was not statistically significant.
Motif analysis
TCF1 and input sequence (GSE46662) reads were aligned to the reference mouse genome mm10 using Bowtie2 (v2.4.1). The sorted bam files were used for peak calling to identify TCF1 binding cites using MACS2. Motif identification and analysis was performed using MEME-Suite (28). The top 500 peaks were shortlisted and a 100 bp region around the summit were subjected to MEME-ChIP analysis (http://meme-suite.org/tools/meme-chip). TCF1 motif was primarily enriched in these sequences. This motif was used to identify genome-wide TCF1 sites using FIMO (http://meme-suite.org/tools/fimo). The resulting GFF file was uploaded onto the IGV for visualization. Similar strategy was used for NFkB and STAT6 binding site analysis of Satb1 promoters.
Data base availability, accession numbers and NGS analysis
GSE77695 dataset (29) was used for analysis of ATAC-seq performed in hematopoietic stem cells (HSCs), multipotent progenitors (MPPs), common lymphoid precursors (CLPs), B cells, CD4 T cells and CD8 T cells. GSE100738 (30) dataset was used for ATAC-seq analyses performed in thymocytes in the different stages of T-cell development. GSE20898 (31) dataset was used for ChIP-seq analyses of genome-wide occupancy of H3K4me3, H3K27me3 and H3K4me1 performed in mouse DP thymocytes. GSE46662 (32) dataset was used for TCF1 ChIP-seq analysis. The raw reads were aligned using Bowtie2 (33) and peak calling was performed using MACS2 (34). The peaks were visualized using IGV genome browser (35). RNA-seq analysis of DP and CD4SP thymocytes was performed using the dataset GSE48138 (36). RNA-seq read alignment was performed using TopHat2 (37,38). Cuffdiff was used for further differential gene expression analysis (38).
RESULTS
Identification of alternative promoter signature at Satb1 gene locus in murine thymocytes
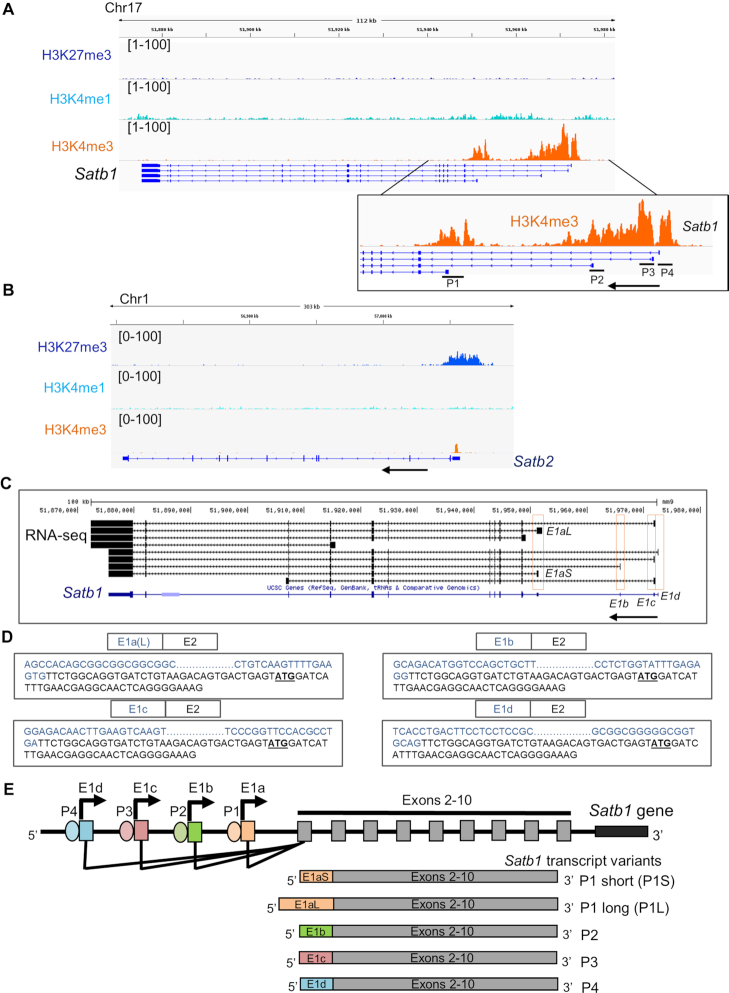
To unravel the molecular mechanisms of SATB1 regulation during T-cell development, we characterized the regulatory elements of Satb1 gene. As a first step toward this, we chose to map the epigenetic landscape of the Satb1 locus, especially the upstream region. We profiled key histone modification marks namely, H3K4me3, H3K4me1 and H3K27me3 at the Satb1 gene locus (Figure 1A) using publicly available ChIP-seq datasets of mouse DP thymocytes (31). Analysis of these histone marks revealed multiple regions upstream of Satb1 transcription start site (TSS) that were enriched only in the H3K4me3 mark, but not H3K4me1 (Figure 1A); the characteristic chromatin modification feature of a promoter element (39,40). The overlay of the ChIP-seq analyses with UCSC annotated Ref-seq data for mouse Satb1 revealed that the H3K4me3 peaks corresponded to different first exonic regions of Satb1 (Figure 1A), indicative of multiple transcription initiation events occurring at Satb1 gene locus. Therefore, the indicated regions at the Satb1 locus with H3K4me3 marks might act as putative promoters of the Satb1 gene. Interestingly, unlike Satb1, another SATB family TF Satb2 harbors a single region upstream of the TSS with occupancy of H3K4me3, but not that of H3K4me1 (Figure 1B), suggesting that the presence of alternative promoter-like regulatory elements is exclusive to Satb1 among the SATB family genes.
Figure 1.
Identification of alternative promoters of Satb1 in mouse thymocytes. (A) Publicly available ChIP-seq datasets of key histone modifications namely H3K4me3, H3K4me1 and H3K27me3 were analyzed for their occupancy profile at the Satb1 locus. Inset shows a magnified plot for the H3K4me3 modifications at the Satb1 alternative promoters P1–P4. Arrow depicts the direction of the sense transcription. (B) Occupancies of these histone modifications were also mapped at the Satb2 locus. (C) RNA-seq analyses were performed using publicly available datasets for mouse thymocyte subpopulations CD3low DP and CD4SP. The transcript sequences were aligned with the Satb1 gene locus, first exons of which are labeled E1a through E1d (the first exons are boxed). (D) 5′ RACE PCR was performed using total RNA from mouse thymocytes and the Satb1 exon-2-specific reverse primer. The characterized sequences of distinct first exons of Satb1 transcript variants, such as E1a(L), E1b, E1c and E1d are shown as confirmed by Sanger sequencing. Also, see Supplementary Figure S1. (E) Schematic depiction of the genomic locations of alternative promoters such as P1, P2, P3 and P4 at the Satb1 locus. Satb1 alternative promoter usage leads to alternative splicing resulting in the generation of Satb1 transcript variants (below) with alternative first exons such as E1aS, E1aL, E1b, E1c and E1d (Genomic distances are not drawn to scale).
To evaluate the usage and transcriptional activity of the predicted Satb1 alternative promoters, we performed 5′ rapid amplification of cDNA ends (5′ RACE) (41) using RNA extracted from mouse thymocytes. The analysis identified multiple Satb1 transcript variants with alternative first exons; E1a(L), E1b, E1c and E1d (Figure 1D and Supplementary Figure S1). In case of E1b and E1d, nested PCR was performed for their characterization as mentioned in the methods. The E1a(L) exon is located 2.48 Kb upstream of the E2 exon and similarly, E1b, E1c and E1d exons are located 17.14, 23.18 and 23.875 Kb upstream, respectively, from the E2 exon. Hence, these alternative first exons are alternatively spliced to the E2 exon, resulting in the generation of multiple Satb1 transcript variants with different 5′ end sequences (Figure 1C and Supplementary Figure S1). Further, we analyzed publicly available RNA-seq data from mouse thymocytes (CD3low DP and CD4+ SP) (36) and confirmed that indeed Satb1 gene exhibits alternative transcript expression in mouse thymocytes (Figure 1C). RNA-seq analysis also identified Satb1 transcript variants with E1a, E1b, E1c and E1d exons (Figure 1C). The E1a containing transcript variant generates two isoforms with short (E1aS) and long (E1aL) alternative first exons, the latter being detected in the 5′ RACE analysis. The alternative first exons of Satb1 mRNA variants do not contribute to making of SATB1 protein and therefore the protein CDS of Satb1 begins from its second exon (Supplementary Figure S1). Therefore, the alternative first exons along with a part of second exon upstream to the protein coding mRNA sequence of Satb1, act as 5′ UTR of Satb1 mRNA. Mapping the sequences of Satb1 alternative first exons to the mouse genome revealed that their TSSs reside within the above identified distinct H3K4me3 ChIP-seq peaks (Supplementary Figure S2), indicating that distinct alternative promoters (hereafter named P1, P2, P3 and P4) are used for the generation of Satb1 transcript variants with alternative 5′ UTRs, named P1 (E1a)—both P1S (E1aS) and P1L (E1aL), P2 (E1b), P3 (E1c) and P4 (E1d), respectively (Figure 1E).
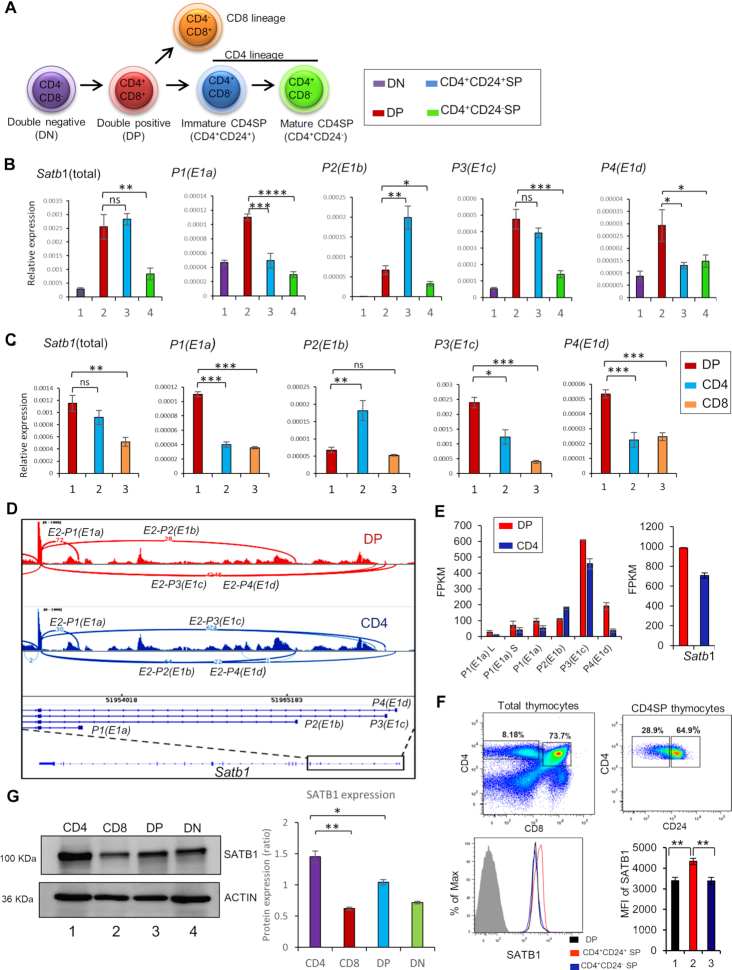
Cell type-specific expression of Satb1 transcript variants during development of T cells
To explore the importance of Satb1 transcript variants expression, we first characterized their expression pattern during T-cell development (Figure 2A). We performed quantitative RT-PCR analysis of Satb1 transcript variants in various developmental stages of thymocytes namely, CD4−CD8− double negative (DN), CD4+CD8+ DP, total CD4−CD8+SP (CD8SP), total CD4+CD8−SP (CD4SP), CD4+CD24+SP (immature CD4SP) and CD4+CD24−SP (mature CD4SP) thymocytes. The expression analysis revealed a differential expression pattern of Satb1 transcript variants in a cell type-specific manner during T-cell development. For instance, DP and immature CD4SP thymocytes exhibit higher levels of Satb1 mRNA compared to other developmental stages, such as DN, mature CD4SP and CD8SP (Figure 2B and C). We then evaluated the expression pattern of Satb1 transcript variants in these thymic T-cell subtypes. The expression of P1 and P4 transcript variants was higher in DP thymocytes compared to other cell types (Figure 2B). P3 transcript variant expression was higher in DP as well as in immature CD4SP compared to DN, mature CD4SP and CD8SP thymocytes (Figure 2B and C). Interestingly, unlike other transcript variants, P2 variant has shown distinct expression pattern, specific to CD4+ T cells. During development, P2 transcript expression begins during the transition from DN to DP and its levels further increase in CD4+ SP cells, but not in the CD8+ T cells which exhibit reduction in the P2 transcript (Figure 2B and C). Specifically among the CD4+ T cells, P2 is highly expressed in immature CD4SP compared to mature CD4SP (Figure 2B). Altogether, these analyses indicated that Satb1 mRNA in DP stage majorly consists of the P1, P3 and P4 transcript variants, but less of the P2 variant. P2 was predominantly expressed in immature CD4SP thymocytes along with the P3 transcript variant (Figure 2C). These findings revealed cell type-specific expression of a combination of Satb1 transcript variants during T-cell development. Further, RNA-seq data from CD3low DP and CD4SP thymocytes (36) was analyzed and differential splicing of Satb1 alternative first exons in these two thymocyte population was plotted (Figure 2D) with their expression levels indicated as FPKM. The analysis corroborates the presence of Satb1 alternative splice variants in thymic T cells (Figure 2E). Altogether, the expression analysis along with the RNA-seq analysis revealed that P3 transcript variant is expressed throughout development, and has a high abundance among Satb1 transcript variants. Along with P3 variant, the alternative transcript variants of Satb1 such as P1L, P1S, P2 and P4 exhibit developmental stage-specific expression in a combinatorial manner during the development of T cells.
Figure 2.
Cell type-specific expression of Satb1 transcript variants during thymocyte development. (A) Schematic diagram depicting the various developmental stages of thymocytes. The relevant cell surface markers are indicated. The color code of various subpopulations of the cells is the same as the bars of relative expression values in (B). (B and C) Quantitative RT-qPCR analyses were performed to measure the expression levels of total Satb1 mRNA and Satb1 transcript variants in various developmental stages of thymocytes such as DN, DP, immature CD4SP and mature CD4SP, and DP, total CD4SP and total CD8SP. 18S rRNA expression was used as endogenous control for normalization. The presented data are from three independent experiments and is shown as means of ± SEM. P-values were calculated using ANOVA or unpaired two-tailed Student's t-test. *P < 0.05; **P < 0.01; ***P < 0.001. (D) RNA-seq analysis of CD3low DP and CD4 SP thymocytes was performed using publicly available datasets. Sashimi plot analysis represents the alternative splicing events of Satb1 alternative first exons between DP and CD4SP thymocytes. (E) RNA-seq based gene expression values (FPKM) corresponding to expression of the alternatively spliced variants of Satb1 and total SATB1 in DP and CD4 SP thymocytes are presented in the two histograms (F) Expression of SATB1 protein in CD4+CD8+ DP, immature CD4+CD24+ SP and mature CD4+CD24− SP thymocytes was analyzed using flow cytometry and the mean florescence intensities (MFI) of SATB1 in these three populations are shown; n = 3, P < 0.01. The percentage change from DP to CD4 immature is calculated using the formula: 100*[MFI(CD4imm)-MFI(DP)]/MFI(DP). Similarly, it was calculated for the other populations. (G) Western blotting to compare SATB1 protein levels in isolated populations of DN, DP, CD4 and CD8 thymocytes (panel on the left). β-ACTIN was used as an endogenous control. Densitrometric analysis was performed using Fiji (ImageJ v.1.52a), and normalization was achieved using respective ACTIN signals (histogram on the right); Statistical analysis was performed using ANOVA (Graphpad v8) n = 3, *P < 0.05, **P < 0.01.
Since we observed the presence of distinct combinations of Satb1 transcript variants in DP and CD4SP thymocytes, we also quantitated the levels of SATB1 protein in these developmental stages. We observed that SATB1 levels were higher with a modest 27.75% in the immature CD4SP thymocytes compared to the DP thymocytes (Figure 2F), as shown earlier (23). Next, to ascertain the protein expression diversity of Satb1 in major thymic T-cell stages, we carried out FACS based isolation of DN, DP, CD4 and CD8 populations followed by western blotting and densitometric analysis. The protein quantitation corroborates the discrepancy in SATB1 levels in DP and CD4 thymic lineages (Figure 2G). During T-cell development from DP to immature CD4SP, interestingly, we observed that the Satb1 transcript levels do not differ, but a significant difference was observed in SATB1 protein levels between these two populations. We therefore asked that what leads to such discrepancy at RNA and protein levels, and how does the combinatorial expression of Satb1 variants in DP and CD4SP thymocytes might play a role in differential SATB1 protein expression in thymocyte development? As shown in Figure 2B, majority of Satb1 transcripts expressed in DP consist of all Satb1 transcript variants except the P2, which is highly expressed only in the immature CD4SP cells. Before further dissecting the role of transcript variants in maintaining differential protein levels, we monitored the stability of SATB1 protein in DP thymocytes to ascertain that the observed phenomenon is due to differential translatability of these transcript variants and not degradation of SATB1 protein in DP thymocytes.
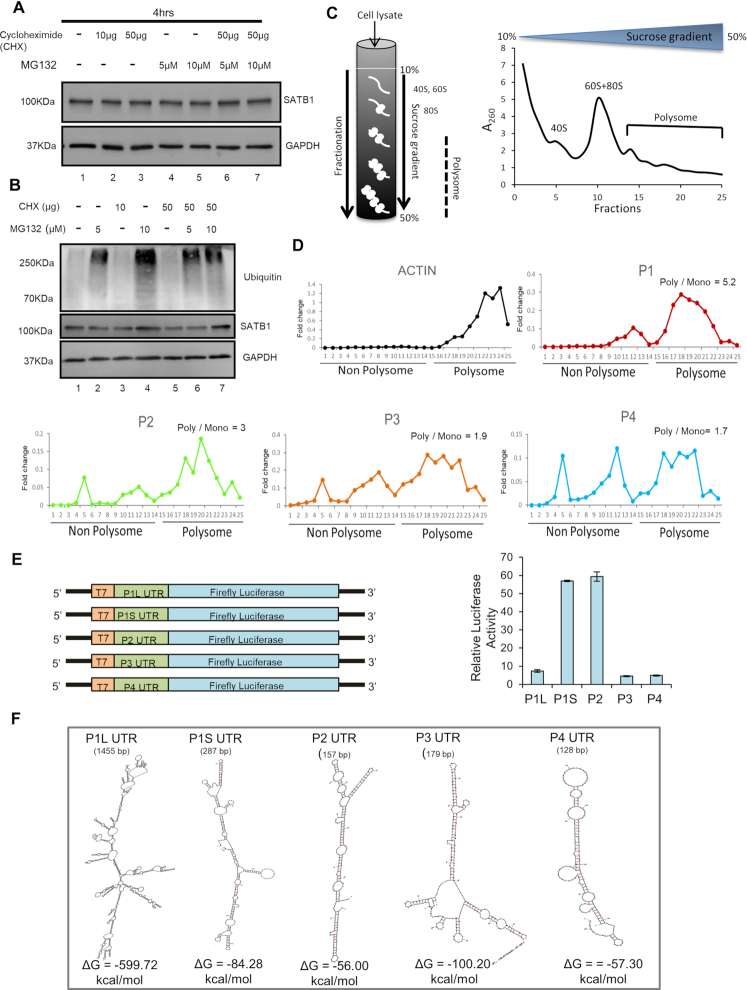
Satb1 transcript variants differ in translation efficiencies
To evaluate that the significantly lower level of SATB1 protein in DP thymocytes compared to immature CD4SP is not due to proteasomal degradation of SATB1 in DP, we performed MG132 and cycloheximide chase assay in DP thymocytes. DP thymocytes were treated with MG132, a proteasome inhibitor, in the presence of the translation inhibitor cycloheximide for 4 h. SATB1 protein levels in DP cells were not altered upon MG132 treatment in the presence or absence of cycloheximide (Figure 3A), although a slight increase in SATB1 was observed in total thymocytes upon the treatment (Figure 3B). Ubiquitination in total lysates was also monitored to confirm the efficacy of the MG132 treatment (Figure 3B). These results confirm that the decreased SATB1 protein levels in the DP compared to the immature CD4SP thymocytes are not due to proteasomal degradation. Although these findings suggest that the proteasomal machinery is not required, we cannot rule out the possibility of other mechanisms of active protein degradation. Next, to test whether Satb1 transcript variants regulate the levels of SATB1 protein, we performed polysome profiling of total thymocytes to monitor their partitioning in the ribosome-free and ribosome-bound states (Figure 3C). Interestingly, we found that all Satb1 transcript variants were present in the subunit, monosome and in the polysome bound states indicating that all Satb1 variants are translatable (Figure 3D). However, we observed that P1 and P2 transcript variants were comparatively more translatable than P3 and P4 as indicated by the ratio of polysome to monosome bound states (Figure 3D). Nevertheless, we cannot rule out the possibility that P3, being the most abundant transcript variant of Satb1, can contribute substantially to the SATB1 protein level despite its comparatively low translatability. Next, to ascertain whether Satb1 alternative transcripts can have differential translatability, we performed in vitro transcription and translation assays. Satb1 alternative 5′ UTRs were cloned upstream of firefly luciferase CDS in the pGL3 basic vector and used for in vitro transcription (Figure 3E). Equimolar ratios of in vitro transcribed RNAs normalized to their base-pair lengths were used for in vitro translation. The translated product was subjected to luciferase activity measurement. The luciferase assay showed that the P2 and P1S 5′ UTR sequences are most efficiently translatable (Figure 3E), validating the results of the polysome profiling. Luciferase activity was reduced in case of P1L, P3 and P4 5′UTR sequences, indicating that these Satb1 5′ UTRs exhibit lower translation efficiency in vitro (Figure 3E). We then performed secondary structure analysis of these 5′ UTR sequences by using the mfold web server (42), and found that the 5′ UTR sequences of P2 and P4 form less stable secondary structures followed by P1S, whereas 5′ UTRs of P1L and P3 can form more stable secondary structures, as indicated by their lower ΔG values (Figure 3F). Thus, secondary structures of the 5′ UTRs might explain the observed poor translatability of the P1L and P3 5′ UTRs in vitro. Although the P4 5 ’UTR sequence forms less stable secondary structure, we could not explain its low translatability in vitro. As there is a distinct combination of Satb1 transcript variants in DP thymocytes which in turn have distinct 5′ UTR sequences compared to immature CD4SP, we find that the comparatively lower SATB1 protein level in DP cells is due to lower translatability of the 5′ UTRs of Satb1. Therefore we next investigated how these alternative transcript variants were regulated in the thymic micro-environment.
Figure 3.
Satb1 transcript variants exhibit differential translatability. (A) DP thymocytes were cultured in the presence or absence of the indicated concentrations of MG132 and cycloheximide for 4 h. Cells were harvested and used for immunoblotting which was performed by probing with anti-GAPDH and anti-SATB1 antibodies (N = 3). (B) Thymocytes were cultured in the presence or absence of the indicated concentrations of MG132 and/or cycloheximide. Ubiquitination and SATB1 levels were monitored in total thymocyte lysates as described in ‘Materials and Methods’ section. (C) Polysome profiling of total thymocytes were performed and OD at 260 nm was measured from the collected fractions as described in ‘Materials and Methods’ section. (D) Fractions from the gradient were used for RNA extraction and cDNA synthesis. Further, quantitative PCR (qPCR) analyses were performed to measure the levels of Satb1 transcript variants present in each fraction. The ratios of polysome to monosome bound Satb1 transcript variants are shown. (E) Translation efficiencies of Satb1 5′ UTRs were quantified using luciferase assay. The schematic illustration on the left depicts the cloning strategy for 5′ UTR sequences of Satb1 transcripts upstream of firefly luciferase CDS in pGL3 basic vector. Invitro transcription was performed using T7 polymerase. In vitro translation was performed using equimolar concentrations of in vitro transcribed RNA. Luciferase activity assay was performed with the in vitro translated products (N = 3) and the relative luciferase activity in the presence of each Satb1 5′ UTR was measured. (F) Secondary structure analysis of Satb1 5′ UTR sequences was performed using mfold web server. –ΔG values, which indicate the stability of secondary structures, were obtained for each 5′ UTR of Satb1 transcript variants.
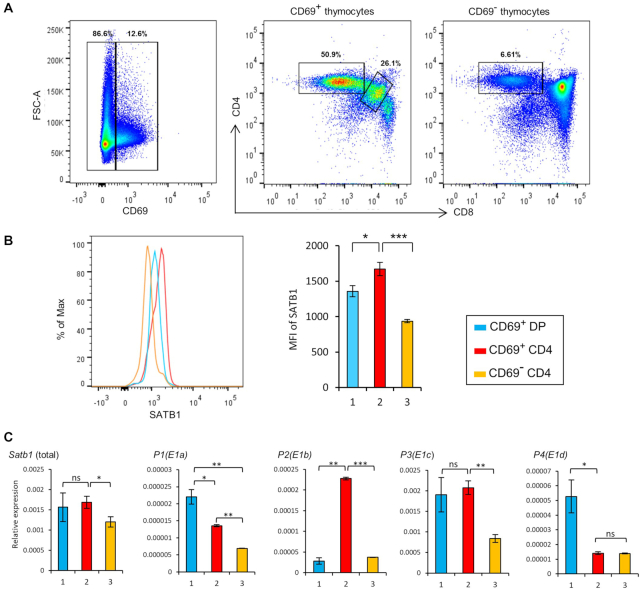
TCR-mediated activation leads to Satb1 promoter switch during T-cell development
TCR signaling plays an essential role in the differentiation and selection of developing thymocytes. SATB1 levels, as shown previously (23), are elevated during TCR-mediated positive selection of thymocytes and SATB1 is abundantly expressed in activated CD69+TCRhi population (Supplementary Figure S7). Interestingly, we had earlier observed that there is a switch from P1 to P2 in CD4+SP thymocytes (Figure 2B), which are mature CD4 thymocytes, hence positively selected. Therefore, we investigated whether TCR signal brings about Satb1 alternative promoter switch during the differentiation of DP into CD4SP. We first assessed the expression of SATB1 protein in TCR engaged DP and CD4SP thymocyte populations that express CD69 on their surface, which corresponds to the activated cells (Figure 4A, left and middle plots), in contrast with the CD69− thymocytes (Figure 4A, plot on the right), which comprise cells with no or very low active TCR signaling (also see Supplementary Figure S7). We observed that SATB1 protein levels were significantly but modestly higher (23.11%) in CD69+CD4SP than CD69+DP cells, but are comparatively low in CD69−CD4SP (45.08% and 31.01%) compared to CD69+CD4SP and CD69+DP cells, respectively (Figure 4B), suggesting that SATB1 expression is more responsive to TCR signal in CD4+ cells. Further, CD69+CD4SP thymocytes show higher expression of the P2 transcript variant along with P3 compared to the CD69+DP thymocytes (Figure 4C), indicating that the P2 promoter switch occurs only in the TCR engaged CD4SP thymocytes but not in the TCR-engaged DP thymocytes. This also suggests that P3 transcript remains abundant irrespective of the major signaling cascades occurring, and presumably plays a crucial role in maintaining SATB1 levels throughout all the stages of thymic T-cell development. These observations suggest that during thymic T-cell development, TCR signaling in the CD4SP thymocytes induces a switch in Satb1 promoter usage, and this promoter switch might lead to the observed higher SATB1 protein levels.
Figure 4.
TCR signal induces Satb1 promoter switch in CD4SP thymocytes. (A) Flow cytometry analysis of SATB1 expression in TCR signaling active DP and CD4SP thymocytes. Thymocytes were stained with anti-CD4, anti-CD8, anti-CD69 and anti-SATB1 antibodies and FACS analyses were performed using BD FACS canto II. (B) SATB1 expression in CD69+DP, CD69+CD4SP and CD69−CD4SP thymocytes is shown by histogram analysis using FlowJo V10.5.2. (C) CD69+DP, CD69+CD4SP, CD69−CD4SP thymocyte populations were FACS sorted and cells were used for RNA extraction and cDNA synthesis. qRT-PCR was performed to estimate the levels of total Satb1 transcripts as well as its transcript variants. The color key is same as used in (B). The represented data are from three independent experiments and is shown as means of ± SEM. P-values were calculated using one-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001.
Lineage-specific chromatin accessibility of Satb1 P2 promoter during T-cell development
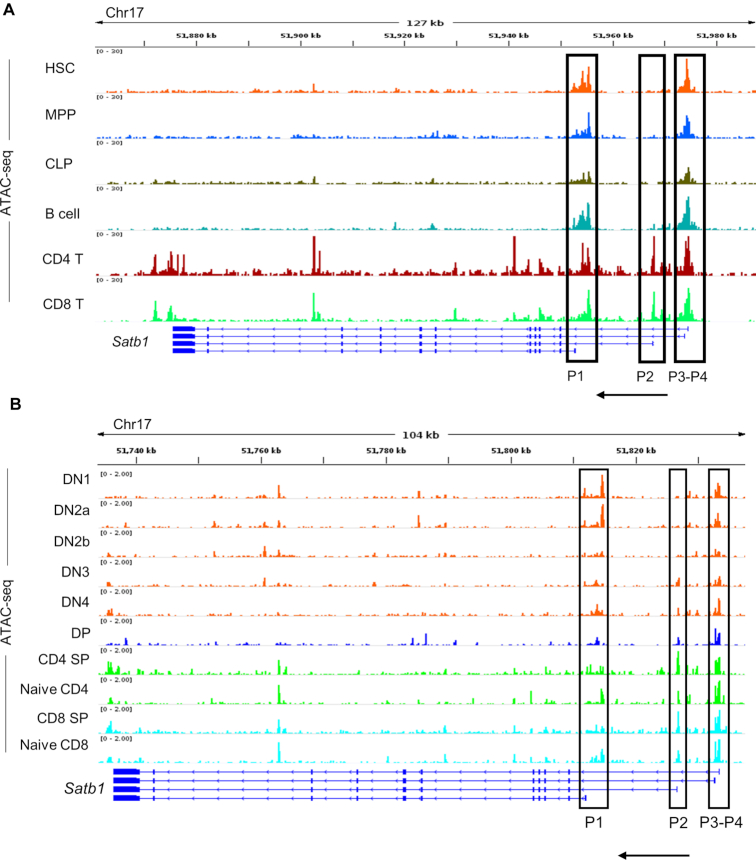
Since the expression of Satb1 transcript variants is cell type-specific during T-cell development (Figures 2B and 4C), we hypothesized that the transcriptional activity of Satb1 alternative promoters might rely on the availability of cell type-specific TFs. Therefore, we posited that Satb1 upstream locus might exhibit cell type-specific chromatin accessibility to achieve alternative transcript expression in a developmental stage-specific manner. To investigate this, we analyzed publicly available ATAC-seq chromatin accessibility data from the immunological genome project (https://www.immgen.org/) and investigated the dynamics of chromatin at the alternative promoters of Satb1 gene locus (29,30). We first assessed the chromatin openness of Satb1 alternative promoters during T-cell development from their progenitors (Figure 5A). During lymphocyte development, a precursor HSC differentiates into an MPP which then differentiates into a CLP (4). CLPs being bi-potential differentiate either into a B-cell lineage in the bone marrow or into a T-cell lineage once they enter the thymus (3). The ATAC-seq analyses revealed that the chromatin region around P1, P3 and P4 promoters exists in an open state throughout the development from the precursor HSC to MPP, CLP, B cells and T cells (Figure 5A). Interestingly, the P2 promoter was found to be in a closed chromatin configuration in the precursors namely, HSC, MPP and CLP. Furthermore, unlike other promoters, P2 promoter region was not accessible in the differentiated B-cell lineage as well (B-cells exhibit poor expression of SATB1), indicating its specificity toward the T-cell lineage (Figure 5A). Since chromatin accessibility of Satb1 alternative promoters revealed lineage specificity during development from precursor HSCs, we monitored the effect on chromatin dynamics of the Satb1 alternative promoters during T-cell development in the thymus. Among Satb1 alternative promoters, P1, P3 and P4 promoter regions exhibit accessibility throughout the T-cell developmental stages in the thymus. The chromatin accessibility at the P2 promoter region begins only at the DN3 stage onward, correlating with the occurrence of pre-TCR signaling. Its accessibility further increases in the later SP stages (Figure 5B). Among the various T-cell developmental stages, only CD4SP stage exhibited comparatively increased P2 promoter accessibility. Surprisingly, although the P2 promoter region displays chromatin accessibility in the CD8SP T cells, the expression of P2 transcripts is found predominantly in CD4+ T cells and not in CD8+ T cells (Figure 5B). It is possible that despite the open chromatin state at the P2 promoter, a variety of mechanisms could be responsible for its inactive state. We also observe an overall dramatic reduction in SATB1 levels in CD8 thymocytes compared to CD4 T cells (Figure 2G). Further studies are required to understand the mechanisms of SATB1 downregulation during the CD8 lineage differentiation.
Figure 5.
Lineage-specific chromatin dynamics of Satb1 alternative promoter regions during thymic T-cell development from precursor cells. (A) We used publicly available ATAC-seq data from HSCs, MPPs, CLPs, B cells and T cells to analyze the state of chromatin accessibility at the alternative promoters of the Satb1 gene locus in these cell types. Satb1 alternative promoter regions (P1, P2, P3 and P4) are marked by rectangular boxes at the Satb1 gene locus. (B) We analyzed the publicly available ATAC-seq datasets performed in various T-cell developmental stages; DN1, DN2a, DN2b, DN3, DN4, DP, CD4SP and CD8SP to assess the chromatin dynamics at the alternative promoters of the Satb1 gene locus. We also used ATAC-seq data from naïve CD4 and CD8 populations alongside the developmental stages. The distinct Satb1 alternative promoter regions (P1, P2, P3 and P4) are marked by rectangular boxes at the Satb1 gene locus.
TCF1 directly regulates the expression of SATB1 in CD4SP thymocytes
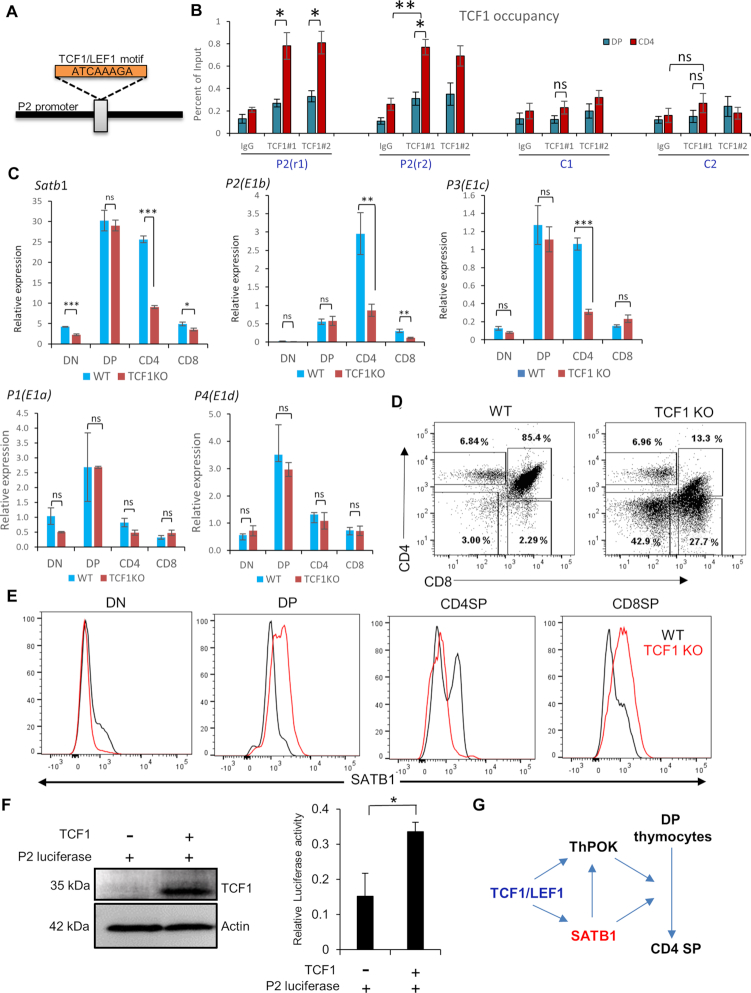
Since P2 promoter switch was identified during differentiation of DP into CD4SP T cells, we then asked whether any specific TF regulates this promoter switch in developing CD4SP thymocytes. CD4+ T-cell development from DP is influenced by multiple key TFs, such as Th-POK, GATA3, c-Myb and Tox (43–46). Recent reports also suggest that the loss of TCF1 and LEF1 results in the impairment of CD4SP from DP, and causes redirection of CD4SP into CD8SP cells (47). We hypothesized that either of these CD4SP T lineage specifying TFs might regulate Satb1 P2 promoter switch and hence the expression of CD4SP-specific Satb1 transcript variants. In silico motif search analysis yielded a canonical TCF1/LEF1 DNA-binding motif in the P2 promoter of Satb1 (Figure 6A), but not the others (Supplementary Figure S5). Interestingly, TCF1/LEF1 motif is well conserved at the P2 promoter of Satb1 gene locus in both human and mouse (Supplementary Figure S3), indicating that TCF1 might similarly regulate the Satb1 P2 promoter switch in human T cells as well. We then performed ChIP-qPCR analysis using anti-TCF1 antibody in DP as well as CD4SP thymocytes and found that TCF1 strongly binds to the P2 promoter of Satb1 in CD4SP thymocytes (Figure 6B), but shows a comparatively lower enrichment in DP thymocytes (Figure 6B). For the TCF1 genomic site-specificity on the Satb1 promoter, we used different control upstream regions for each promoter (abbreviated as cold regions; C1–C5 in Figure 6B and Supplementary Figure S6). Next, we analyzed the publicly available TCF1 ChIP-seq data from total thymocytes (32) and observed the occupancy of TCF1 on the P2 promoter, wherein TCF1/LEF1 binding motif was identified (Supplementary Figures S3 and 5). Concordantly, both ChIP-qPCR and ATAC-seq analysis show that TCF1 occupancy and chromatin accessibility on P2 promoter increases from DP to CD4 thymocytes (Figures 6B and 5B). We also utilized the TCF1 knockout (KO) mice model, which shows that the expression of both P2 as well as total Satb1 transcripts were significantly downregulated in TCF1 KO CD4SP thymocytes (Figure 6C–E). Although there is no cannonical TCF1/LEF1 binding site on P3 promoter and spanning region on either side (Supplementary Figure S5), there seems to be a substantial effect of TCF1 deficiency on P3 transcript expression only in CD4 thymocytes (Figure 6C), suggesting its TCF1 dependency. We also observed an appreciable binding of TCF1 on the P3 promoter in our ChIP-seq and ChIP-qPCR analyses (Supplementary Figures S4 and 6), which could possibly occur via a non-cannonical binding site of TCF1/LEF1 or through another partner DNA-binding factor (48). In line with the absence of TCF1 binding (Supplementary Figures S5 and 6), TCF1 KO led to no significant difference in P1 and P4 variants’ expression among different thymic populations (Figure 6C). There was a significant reduction in SATB1 protein levels in CD4SP cells from TCF1 KO mice, compared to the wildtype (Figure 6D and E). Surprisingly, although we did not observe a change in Satb1 or P2 transcripts in DPs upon TCF1 KO (which corroborates with both ChIP and ATAC seq data), but we observed an increase in SATB1 protein levels in DP thymocytes from TCF1 KO mice (Figure 6E). This might be due to the contrasting roles of TCF-1 in the form of an activator or a repressor in different thymocyte development stages (49) as we find in the case of CD8 thymic T cells wherein these two HMG-TFs TCF1/LEF1 repress the CD4 lineage-specific genes such as Cd4 and Foxp3 through their intrinsic HDAC activity (49). Satb1 could be one of the genes regulated in such a manner via TCF1/LEF1 factors. Further studies are needed to probe into the stage-specific regulation via TCF1. We also assessed the role of TCF1 in the transcriptional activity of P2 promoter by monitoring the P2 promoter activity in the presence or absence of TCF1 overexpression. We observed significant increase in the promoter activity of P2 in the presence of TCF1 as shown by the luciferase activity assay (Figure 6F). Collectively, these experiments provide an unequivocal evidence that TCF1 is important for P2 and P3 promoters’ expression in CD4SP thymocytes, thereby regulating the expression of SATB1 during T-cell development (Figure 6G). Since many TFs are induced upon T cell activation such as NFkB, STAT1/3, STAT6, ThPok, GATA3, Runt family proteins, etc. (7), multiple factors co-regulate gene expression post-TCR signal. We searched for NFkB and STAT6 motifs apart from TCF1 for their occurrence in Satb1 alternative promoters. We find that a consensus motif of STAT6 is present 300 bp downstream of the P2 promoter region, thus indicating a regulatory role of STAT6 toward SATB1 expression. Furthermore, we did not find any NFkB consensus binding site in or around the Satb1 promoter regions (Supplementary Figure S5). Binding of these TFs can occur at non-cannonical sequences as well and is facilitated by other binding partners. Therefore, it is plausible that these TFs in combination with other factors might be involved in the regulation of Satb1, and thus warrant further studies.
Figure 6.
TCF1 directly regulates the SATB1 expression in CD4SP thymocytes during T-cell development. (A) A TCF1/LEF1 canonical DNA-binding motif was identified in the P2 promoter region of Satb1 using MEME-Suite (Version 5.1.1, http://meme-suite.org/tools/meme). (B) TCF1 ChIP was performed in sorted DP and CD4SP thymocytes, followed by qRT-PCR analysis using primers spanning the TCF1/LEF1 binding site on P2 promoter. Two different antibody clones were used (depicted as #1 and #2 in the figure axis) to assess the antibody-factor specificity. Different control regions were used to ascertain whether TCF1 specifically binds to the Satb1 promoter regions, labeled C1 and C2, at 1000 and 1500 bp upstream of P2, respectively. (C) qRT-PCR analysis of expression profile of total Satb1, P1, P2, P3 and P4 variants in CD4SP thymocytes along with other sorted T-cell developmental stages from wild-type and TCF1 KO mice. Hprt gene expression was used as endogenous control. The represented data are from three independent experiments and is shown as means of ± SEM. P-values were calculated using one-way ANOVA or. *P < 0.05; **P < 0.01; ***P < 0.001. (D) Total thymocytes were isolated from either WT or TCF1 KO mice and stained with anti-CD4, anti-CD8 and anti-SATB1 antibodies. FACS analyses were performed using BD FACS canto II with the indicated gating. (E) Flow cytometry analysis shows the expression of SATB1 in thymocyte developmental stages; DN, DP, CDSP and CD8SP from wild-type and TCF1 KO mice as indicated by black and red color curved lines respectively. Solid gray peaks indicate the staining controls. (F) Satb1 P2 promoter was cloned in pGL3 basic vector and subjected to dual luciferase activity assay in the background of TCF1 overexpression in HEK293 cells. Renilla was used as an endogenous control. The represented data are from three independent experiments and is shown as means of ± SEM. P-values were calculated using the unpaired two-tailed Student's t-test. *P < 0.05. (G) A schematic representation depicting how TCF1 acts upstream of SATB1 during differentiation of CD4SP from DP in thymus.
TCR signaling induces Satb1 promoter switch in peripheral CD4+ T cells
In Figure 4B, we observed that the expression of Satb1 P2 transcript variant was TCR signal-dependent during T-cell development. We therefore evaluated whether the same regulatory mechanism exists in peripheral CD4+ T cells. Toward this, we performed in vitro TCR stimulation of purified naïve CD4+ T cells from spleen using anti-CD3 and anti-CD28. As expected, SATB1 protein levels increased upon TCR activation, which is evidenced by immunoblotting and flow cytometry analysis (Figure 7B). Expression analysis revealed that total Satb1 mRNA levels were also increased in the TCR activated CD4+ in comparison with the naïve CD4+ T cells (Figure 7C). When tested for the expression of Satb1 transcript variants upon activation, the expression of P2 transcript along with P3 was induced in the TCR activated CD4+ T cells compared to naïve T cells (Figure 7C). In contrast, TCR activation led to the downregulation of the P1 variant, and no significant increase was observed in case of the P4 transcript upon the treatment (Figure 7C). These results again confirm that the expression of P2 transcript variant along with P3 is required to upregulate SATB1 protein levels upon T-cell activation.
Figure 7.
TCR signal mediates Satb1 promoter switch in the peripheral CD4+ T cells. (A) Naïve CD4+ T cells were isolated from spleen of six-week-old C57BL/6 mice and were subjected to TCR activation by culturing in the presence of plate bound anti-CD3 and soluble anti-CD28 for 48 h. Cells were harvested and used for qRT-PCR, western blotting and FACS analysis. Naïve and TCR-activated cells were processed for western blotting which was performed using anti-SATB1 and anti-tubulin. Two replicates are shown (Lanes 1–2 and 3–4) to demonstrate the robustness of TCR signaling mediated upregulation of SATB1 in CD4 T cells. (B) FACS analysis were performed to generate the histograms of SATB1 and Nur77 by staining cells with anti-CD4, anti-CD8, anti-CD25, anti-SATB1 and anti-Nur77. (C) Naïve and TCR-activated cells were used for RNA extraction and cDNA synthesis. Next, quantitative RT-PCR (qRT-PCR) analyses were performed to measure the expression of total Satb1 and its transcript variants. Expression of 18S rRNA was used as an endogenous control. The represented data are from three independent experiments and is shown as means of ± SEM. P-values were calculated using unpaired two-tailed Student's t-test. *P < 0.05; **P < 0.01.
DISCUSSION
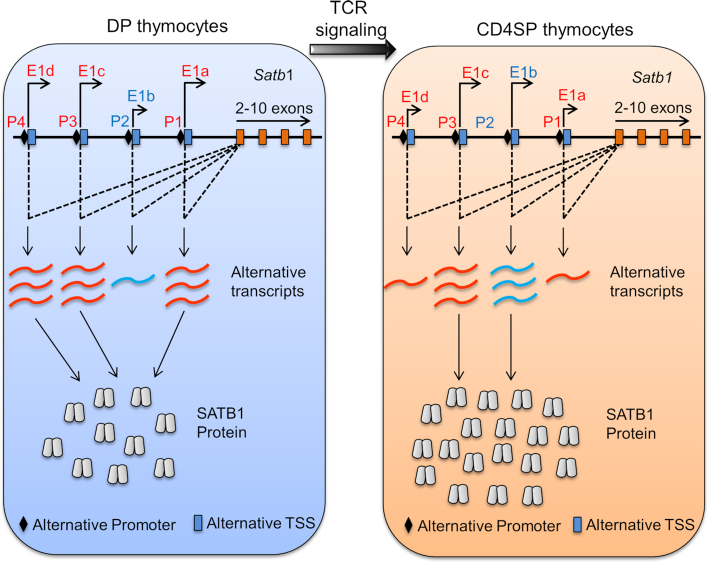
In the current study, we have uncovered a mechanism for SATB1 regulation by a TCR signal-mediated alternative promoter switch during T-cell development. We have identified four alternative promoters (P1, P2, P3 and P4) of Satb1 gene in mouse thymocytes. We show that Satb1 alternative promoter usage leads to alternative splicing of Satb1, thus generating different Satb1 transcript variants in mouse thymocytes. However, the Satb1 gene locus in human T cells exhibits three alternative promoters (50), some of which are similar to that of mouse indicating their evolutionary conservation. It has been well established that the usage of alternative promoters regulates the gene expression in a tissue or developmental stage-specific manner. Mammalian genomes utilize alternative promoters to increase the diversity in the transcriptome and proteome (24). The alternative splice variants of Satb1 differ in the non-overlapping first exon sequences, which we have shown to be 5′ UTRs.
The importance of Satb1 regulation comes from its diverse, context-specific functions; SATB1 is a chromatin organizer that is expressed predominantly in the thymus (51), and plays a pivotal role during thymocyte development by regulating the expression of multiple genes encoding T-cell-specific cytokines and surface receptors (8,9). SATB1 regulates the expression of RAG1 and RAG2 genes in the DP thymocytes, thus allowing the rearrangement of TCRα gene (52). During T-cell development, SATB1 protein exhibits differential expression pattern (23) and is TCR responsive. Here, we show that SATB1 is abundantly expressed in the immature CD4SP thymocytes compared to other developmental stages of thymocytes. In this study, we have focused on delineation of how Satb1 expression is tightly and coordinately regulated during T-cell development. Among thymocyte developmental stages, the DP and immature CD4SP thymocytes show higher expression of Satb1 mRNA. However, the transcript variants of Satb1 exhibit cell type-specific expression pattern in a combinatorial manner of different 5′ UTRs in these thymocyte populations during T-cell development. DP thymocytes show higher expression of P1, and P4 transcript variants along with constitutively expressed P3 which is the predominant transcript variant among the Satb1 transcript variants. Strikingly, P2 transcript variant is highly expressed specifically in CD4SP thymocytes but not in CD8SP thymocytes during their development from DP. Among CD4SP thymocytes, immature CD4SP exhibits higher expression of P2 transcript variant. Along with P2, immature CD4SP thymocytes also express high levels of P3 transcript variant, which can contribute significantly to SATB1 levels in CD4SP T cells despite its comparatively low translatablity. Our analyses identified the cell type and developmental stage-specific expression of combination of Satb1 mRNA variants during thymocyte development, and how combination of highly translatable and abundant transcript variants P2 and P3, respectively, can lead to higher SATB1 levels when DP cells develop into CD4 cells. In CD4 thymocytes, particularly, SATB1 is shown to be essential for the activation of genes encoding lineage specifying TFs, such as ThPOK, RUNX3 and FOXP3 (53).
More recently, Kanakura and colleagues showed that the differential expression of Satb1 regulates HSC heterogeneity (54) showing the importance of the stringent control of SATB1 expression in a lineage-dependent manner. We also showed that not just the transcripts’ expression, but the translatability of each variant is important in determining the protein levels of SATB1, both in vivo and in vitro. The combination of transcript variants expressed in DP are comparably less translatable than the ones used by immature CD4SP, resulting in the increased SATB1 protein levels in immature CD4SP than DP. Thus, during transition from DP to immature CD4SP, the usage of Satb1 alternative promoters switches from the combination of P1, P3 and P4 to that of P2 and P3. Notably, P3 remains the most abundant transcript for Satb1 which expresses constitutively pre- and post-TCR signal (Figure 4C), hinting that P3 expression is perpetually required for SATB1 expression. Further studies using a deletion of the P3 region from Satb1 locus in mouse model can provide definitive information on the role of P3. Since Satb1 transcript variants express in a combinatorial manner to maintain the differential levels of SATB1 protein in a stage-dependent manner, it would be interesting to study whether perturbations in any of the Satb1 alternative promoters affect the levels of SATB1 protein, thereby underscoring the stringency of SATB1 expression and consequently its functions in a stage-specific manner.
Connecting the promoter switch with an important signaling pathway, we further show that expression of the P2 variant in CD4 thymocytes is induced by TCR signaling. Ex vivo, upon anti-CD3/CD28 activation of peripheral naïve CD4+ T cells, we observe that expression of both P2 and P3 increases significantly, suggesting that Satb1 is regulated by TCR signal via selective alternative promoter usage. Furthermore, we observed that only the P2 promoter of Satb1 exhibits a characteristic cell type-specific chromatin accessibility during T-cell development from progenitors. We have identified that TCF-1 acts as a regulator of SATB1 by directly binding to the Satb1 P2 promoter and mediating the P2 promoter switch during CD4SP development from DP cells. We also observed TCF1 binding on P3 promoter despite the absence of its cannonical binding site in P3 region (Supplementary Figure S5). Concordantly, CD4SP thymocytes from TCF1 KO mice exhibited lower levels of SATB1 protein along with Satb1, P2 and P3 expression, indicating the importance of TCF1 in maintaining SATB1 protein levels via P2 promoter switch and a combination of P2 and P3 in CD4SP thymocytes. However, whether the P2 promoter switch is critical for CD4SP T-cell development by perturbation of the P2 promoter is not investigated in this study. Along with the TCR signaling, cytokine signaling is important in early gene regulation, including Satb1 (55). Various studies have shown the cross-talk of cytokine and TCR signals in gene regulation during Th2 differentiation (13,20,55,56). In a separate study, we recently showed that during human Th2 differentiation, Satb1 is regulated by NFkB and cytokine signaling-mediated P2 promoter switch (50), which also demonstrates the regulation of chromatin organizer SATB1 via alternate usage of three promoters.
In conclusion, we demonstrate that alternative promoter usage drives Satb1 expression during thymocyte development as well as peripheral T-cell activation. The selective expression of a combination of Satb1 transcript variants during T-cell development plays a crucial role toward the regulation of SATB1 protein levels as summarized in the schematic diagram (Figure 8). We hypothesize that this regulation is mediated by TCR signal-induced binding of TCF1 and transcriptional activation at P2 and P3 promoters specifically in the CD4SP thymocytes. SATB1 is an important regulator of T-cell development, and its expression is regulated differentially in a cell type/developmental stage-specific manner during T-cell development. The cell type-specific differential accessibility of the alternative promoters at the Satb1 locus warrants further studies to fully dissect the chromatin architecture based mechanism of Satb1 regulation via its multiple alternative promoters.
Figure 8.
A schematic model depicting TCR signal-mediated Satb1 alternative promoter switch in developing thymocytes. During thymic T-cell development, DP thymocytes are generated as a result of pre-TCR signaling at DN4 stage. At the DP stage of development, TCRα gene rearrangement takes place and DP thymocytes begin to receive TCR signaling. Persistent or stronger TCR signaling at the DP stage leads to their differentiation toward CD4+ T cells, whereas cessation or weaker TCR signaling leads to CD8+ T-cell differentiation. During T-cell development, DP thymocytes express P1 (P1S and P1L) and P4 transcript variants along with the constitutively active P3 variant. Upon TCR signal mediated differentiation of DP into CD4SP, CD4+ T cells switch to increased expression of P2 along with P3 variant, presumably due to direct binding by the TF TCF1. Since Satb1 transcript variants exhibit differential translatability, the combinatorial expression of Satb1 transcript variants plays a key role in maintaining SATB1 protein levels.
Supplementary Material
ACKNOWLEDGEMENTS
I.P. wishes to thank Dr Krishanpal Karmodiya for help with ChIP-seq analysis during the initial stages of the study. Authors wish to thank the staff at the Experimental animal facility at NCCS, the National Facility for Gene Function in Health and Disease at IISER Pune, and the flow cytometry facilities at IISER Pune and NCCS Pune.
Author contributions’: Designed experiments: I.P., A.M., S.G.; performed experiments: I.P., A.M.; Sh.G. and G.L. provided help and expertise in mice experiments; ATAC-seq, ChIP-Seq and RNA-Seq analysis: I.P., A.M., S.K.; TCF KO experiments: K.P.G., A.V. and J.M.-S. provided expertise in TCF KO mice experiments, J.M.-S. supported the research involving TCF KO mice. Polysome profiling experiments: V.D., I.P.; V.S. provided expertise in polysome profiling; S.G., V.S., G.L. were involved in regular discussions and data interpretations. I.P., A.M. and S.G. wrote the manuscript. S.G. supervised and supported the research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology, Ministry of Science and Technology (DBT) [BT/01/COE/09/07, BT/MED/30/SP11288/2015]; National Institutes of Health, Intramural Research Program of the National Institute on Aging (in part); CSIR Fellowships (to I.P., A.M.); IISER Pune Post-Doctoral Fellowship (to S.K.); Science and Engineering Research Board (SERB) [JC Bose Fellowship, JCB/2019/000013]. Funding for open access charge: DBT; SERB.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wallis V.J., Leuchars E., Chwalinski S., Davies A.J.. On the sparse seeding of bone marrow and thymus in radiation chimaeras. Transplantation. 1975; 19:2–11. [DOI] [PubMed] [Google Scholar]
- 2. Donskoy E., Goldschneider I.. Thymocytopoiesis is maintained by blood-borne precursors throughout postnatal life. A study in parabiotic mice. J. Immunol. 1992; 148:1604–1612. [PubMed] [Google Scholar]
- 3. Mori S., Shortman K., Wu L.. Characterization of thymus-seeding precursor cells from mouse bone marrow. Blood. 2001; 98:696–704. [DOI] [PubMed] [Google Scholar]
- 4. Germain R.N. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2002; 2:309–322. [DOI] [PubMed] [Google Scholar]
- 5. von Boehmer H., Teh H.S., Kisielow P.. The thymus selects the useful, neglects the useless and destroys the harmful. Immunol. Today. 1989; 10:57–61. [DOI] [PubMed] [Google Scholar]
- 6. Robey E., Fowlkes B.J.. Selective events in T cell development. Annu. Rev. Immunol. 1994; 12:675–705. [DOI] [PubMed] [Google Scholar]
- 7. Starr T.K., Jameson S.C., Hogquist K.A.. Positive and negative selection of T cells. Annu. Rev. Immunol. 2003; 21:139–176. [DOI] [PubMed] [Google Scholar]
- 8. Alvarez J.D., Yasui D.H., Niida H., Joh T., Loh D.Y., Kohwi-Shigematsu T.. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000; 14:521–535. [PMC free article] [PubMed] [Google Scholar]
- 9. Kondo M., Tanaka Y., Kuwabara T., Naito T., Kohwi-Shigematsu T., Watanabe A.. SATB1 plays a critical role in establishment of immune tolerance. J. Immunol. 2016; 196:563–572. [DOI] [PubMed] [Google Scholar]
- 10. de Belle I., Cai S., Kohwi-Shigematsu T.. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J. Cell Biol. 1998; 141:335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cai S., Han H.J., Kohwi-Shigematsu T.. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 2003; 34:42–51. [DOI] [PubMed] [Google Scholar]
- 12. Galande S., Purbey P.K., Notani D., Kumar P.P.. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr. Opin. Genet. Dev. 2007; 17:408–414. [DOI] [PubMed] [Google Scholar]
- 13. Cai S., Lee C.C., Kohwi-Shigematsu T.. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006; 38:1278–1288. [DOI] [PubMed] [Google Scholar]
- 14. Kumar P.P., Bischof O., Purbey P.K., Notani D., Urlaub H., Dejean A., Galande S.. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 2007; 9:45–56. [DOI] [PubMed] [Google Scholar]
- 15. Yasui D., Miyano M., Cai S., Varga-Weisz P., Kohwi-Shigematsu T.. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002; 419:641–645. [DOI] [PubMed] [Google Scholar]
- 16. Zhou L.Q., Wu J., Wang W.T., Yu W., Zhao G.N., Zhang P., Xiong J., Li M., Xue Z., Wang X. et al.. The AT-rich DNA-binding protein SATB2 promotes expression and physical association of human (G)gamma- and (A)gamma-globin genes. J. Biol. Chem. 2012; 287:30641–30652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Naik R., Galande S.. SATB family chromatin organizers as master regulators of tumor progression. Oncogene. 2019; 38:1989–2004. [DOI] [PubMed] [Google Scholar]
- 18. Kitagawa Y., Ohkura N., Kidani Y., Vandenbon A., Hirota K., Kawakami R., Yasuda K., Motooka D., Nakamura S., Kondo M. et al.. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 2017; 18:173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pavan Kumar P., Purbey P.K., Sinha C.K., Notani D., Limaye A., Jayani R.S., Galande S.. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol. Cell. 2006; 22:231–243. [DOI] [PubMed] [Google Scholar]
- 20. Notani D., Gottimukkala K.P., Jayani R.S., Limaye A.S., Damle M.V., Mehta S., Purbey P.K., Joseph J., Galande S.. Global regulator SATB1 recruits beta-catenin and regulates T(H)2 differentiation in Wnt-dependent manner. PLoS Biol. 2010; 8:e1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beyer M., Thabet Y., Muller R.U., Sadlon T., Classen S., Lahl K., Basu S., Zhou X., Bailey-Bucktrout S.L., Krebs W. et al.. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat. Immunol. 2011; 12:898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McInnes N., Sadlon T.J., Brown C.Y., Pederson S., Beyer M., Schultze J.L., McColl S., Goodall G.J., Barry S.C.. FOXP3 and FOXP3-regulated microRNAs suppress SATB1 in breast cancer cells. Oncogene. 2012; 31:1045–1054. [DOI] [PubMed] [Google Scholar]
- 23. Gottimukkala K.P., Jangid R., Patta I., Sultana D.A., Sharma A., Misra-Sen J., Galande S.. Regulation of SATB1 during thymocyte development by TCR signaling. Mol. Immunol. 2016; 77:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Landry J.R., Mager D.L., Wilhelm B.T.. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 2003; 19:640–648. [DOI] [PubMed] [Google Scholar]
- 25. Asnagli H., Afkarian M., Murphy K.M.. Cutting edge: Identification of an alternative GATA-3 promoter directing tissue-specific gene expression in mouse and human. J. Immunol. 2002; 168:4268–4271. [DOI] [PubMed] [Google Scholar]
- 26. Bharti K., Liu W., Csermely T., Bertuzzi S., Arnheiter H.. Alternative promoter use in eye development: the complex role and regulation of the transcription factor MITF. Development. 2008; 135:1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davuluri R.V., Suzuki Y., Sugano S., Plass C., Huang T.H.. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008; 24:167–177. [DOI] [PubMed] [Google Scholar]
- 28. Bailey T.L., Johnson J., Grant C.E., Noble W.S.. The MEME Suite. Nucleic Acids Res. 2015; 43:W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shih H.Y., Sciume G., Mikami Y., Guo L., Sun H.W., Brooks S.R., Urban J.F. Jr, Davis F.P., Kanno Y., O'Shea J.J.. Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell. 2016; 165:1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yoshida H., Lareau C.A., Ramirez R.N., Rose S.A., Maier B., Wroblewska A., Desland F., Chudnovskiy A., Mortha A., Dominguez C. et al.. The cis-regulatory atlas of the mouse immune system. Cell. 2019; 176:897–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei G., Abraham B.J., Yagi R., Jothi R., Cui K., Sharma S., Narlikar L., Northrup D.L., Tang Q., Paul W.E. et al.. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011; 35:299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dose M., Emmanuel A.O., Chaumeil J., Zhang J., Sun T., Germar K., Aghajani K., Davis E.M., Keerthivasan S., Bredemeyer A.L. et al.. beta-Catenin induces T-cell transformation by promoting genomic instability. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thorvaldsdottir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu G., Tang Q., Sharma S., Yu F., Escobar T.M., Muljo S.A., Zhu J., Zhao K.. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 2013; 14:1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A. et al.. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007; 39:311–318. [DOI] [PubMed] [Google Scholar]
- 40. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al.. Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Frohman M.A. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 1993; 218:340–356. [DOI] [PubMed] [Google Scholar]
- 42. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. He X., He X., Dave V.P., Zhang Y., Hua X., Nicolas E., Xu W., Roe B.A., Kappes D.J.. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005; 433:826–833. [DOI] [PubMed] [Google Scholar]
- 44. Wang L., Wildt K.F., Zhu J., Zhang X., Feigenbaum L., Tessarollo L., Paul W.E., Fowlkes B.J., Bosselut R.. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat. Immunol. 2008; 9:1122–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maurice D., Hooper J., Lang G., Weston K.. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. 2007; 26:3629–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aliahmad P., Kaye J.. Development of all CD4 T lineages requires nuclear factor TOX. J. Exp. Med. 2008; 205:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Steinke F.C., Yu S., Zhou X., He B., Yang W., Zhou B., Kawamoto H., Zhu J., Tan K., Xue H.H.. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat. Immunol. 2014; 15:646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hrckulak D., Kolar M., Strnad H., Korinek V.. TCF/LEF transcription factors: an update from the internet resources. Cancers (Basel). 2016; 8:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xing S., Li F., Zeng Z., Zhao Y., Yu S., Shan Q., Li Y., Phillips F.C., Maina P.K., Qi H.H. et al.. Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nat. Immunol. 2016; 17:695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Khare S.P., Shetty A., Biradar R., Patta I., Chen Z.J., Sathe A.V., Reddy P.C., Lahesmaa R., Galande S.. NF-kappaB signaling and IL-4 signaling regulate SATB1 Expression via alternative promoter usage during Th2 differentiation. Front. Immunol. 2019; 10:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dickinson L.A., Joh T., Kohwi Y., Kohwi-Shigematsu T.. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992; 70:631–645. [DOI] [PubMed] [Google Scholar]
- 52. Hao B., Naik A.K., Watanabe A., Tanaka H., Chen L., Richards H.W., Kondo M., Taniuchi I., Kohwi Y., Kohwi-Shigematsu T. et al.. An anti-silencer- and SATB1-dependent chromatin hub regulates Rag1 and Rag2 gene expression during thymocyte development. J. Exp. Med. 2015; 212:809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kakugawa K., Kojo S., Tanaka H., Seo W., Endo T.A., Kitagawa Y., Muroi S., Tenno M., Yasmin N., Kohwi Y. et al.. Essential roles of SATB1 in specifying T lymphocyte subsets. Cell Rep. 2017; 19:1176–1188. [DOI] [PubMed] [Google Scholar]
- 54. Doi Y., Yokota T., Satoh Y., Okuzaki D., Tokunaga M., Ishibashi T., Sudo T., Ueda T., Shingai Y., Ichii M. et al.. Variable SATB1 levels regulate hematopoietic stem cell heterogeneity with distinct lineage fate. Cell Rep. 2018; 23:3223–3235. [DOI] [PubMed] [Google Scholar]
- 55. Lund R., Ahlfors H., Kainonen E., Lahesmaa A.M., Dixon C., Lahesmaa R.. Identification of genes involved in the initiation of human Th1 or Th2 cell commitment. Eur. J. Immunol. 2005; 35:3307–3319. [DOI] [PubMed] [Google Scholar]
- 56. Ahlfors H., Limaye A., Elo L.L., Tuomela S., Burute M., Gottimukkala K.V., Notani D., Rasool O., Galande S., Lahesmaa R.. SATB1 dictates expression of multiple genes including IL-5 involved in human T helper cell differentiation. Blood. 2010; 116:1443–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.