Abstract
Tyrosyl-DNA phosphodiesterase 2 (TDP2) reverses Topoisomerase 2 DNA–protein crosslinks (TOP2-DPCs) in a direct-reversal pathway licensed by ZATTZNF451 SUMO2 E3 ligase and SUMOylation of TOP2. TDP2 also binds ubiquitin (Ub), but how Ub regulates TDP2 functions is unknown. Here, we show that TDP2 co-purifies with K63 and K27 poly-Ubiquitinated cellular proteins independently of, and separately from SUMOylated TOP2 complexes. Poly-ubiquitin chains of ≥ Ub3 stimulate TDP2 catalytic activity in nuclear extracts and enhance TDP2 binding of DNA–protein crosslinks in vitro. X-ray crystal structures and small-angle X-ray scattering analysis of TDP2-Ub complexes reveal that the TDP2 UBA domain binds K63-Ub3 in a 1:1 stoichiometric complex that relieves a UBA-regulated autoinhibitory state of TDP2. Our data indicates that that poly-Ub regulates TDP2-catalyzed TOP2-DPC removal, and TDP2 single nucleotide polymorphisms can disrupt the TDP2-Ubiquitin interface.
INTRODUCTION
Tyrosyl-DNA phosphodiesterase 2 (TDP2, a.k.a TTRAP, EAPII) is a multifunctional protein, with roles in HIV viral genome integration (1), cell proliferation (2,3), ribosome biogenesis (4), viral RNA processing (5), the RNA–DNA damage response (6–8), and is a key modulator of resistance to chemotherapeutic drugs. Knockout or RNAi knockdown of TDP2 sensitizes cells to cancer chemotherapeutics such as etoposide that target Topoisomerase 2 (TOP2) (9,10) and increases the frequency of genome rearrangements caused by inaccurate repair of the resulting DNA damage (11,12). TDP2 inactivating mutations are linked to spinocerebellar ataxia, autosomal recessive 23 (SCAR23), which manifests as intellectual disability, seizures, and ataxia, highlighting the importance of TDP2 for normal neuronal development (11,12).
Mammalian TOP2 (TOP2α and TOP2β) relieves DNA topological strain by introducing transient double-strand breaks (DSBs) through which another DNA duplex is passed, then resealing the DNA break to restore genome integrity. The strand passage reaction catalyzed by TOP2 is both essential and dangerous, as failure to complete the re-ligation step generates DNA DSBs that are covalently linked to the active-site tyrosine of TOP2 through a 5′-phosphotyrosine (5′-Y) linkage (13), which results in a DNA lesion called a TOP2 DNA–protein crosslink (TOP2-DPC). Chemotherapeutic drugs such as etoposide or doxorubicin that poison the TOP2 re-ligation step cause accumulation of TOP2-DPCs and apoptotic cell death (14). Environmental toxicants and DNA damage also alter the TOP2 cleavage-religation cycle causing accumulation of TOP2-DPCs (15–17). A robust DNA damage response (DDR) to TOP2-DPCs is essential to maintain genome integrity, and TDP2 is a rapid responder to TOP2-DPCs (10). TDP2 displays high specificity for cleaving 5′-Y linkages, and this activity generates unadducted 5′-phosphate DNA ends that can be rejoined by the cellular non-homologous end joining (NHEJ) machinery (9,10,18–20).
Like many DDR processes (21), TOP2-DPC repair is modulated by signaling with Ubiquitin family of post-translational modifications such as SUMO2 (Small Ubiquitin-like Modifier 2) (10,22). The SUMO E3/E4 ligase ZATTZNF451 (23,24) (poly-Zinc finger Associated with TDP2 and TOP2) binds and catalyzes the modification of TOP2-DPCs with SUMO2/3. In turn, TDP2 binds SUMO2/3 through a split SUMO Interacting Motif (split-SIM) to recruit TDP2 to DNA damage. ZATT also alters the conformation of TOP2-DPCs so TDP2 can access and hydrolyze the 5′-Y, thereby ‘licensing’ TOP2-DPCs for repair (10). TDP2 is unique amongst the EEP (endonuclease/exonuclease/phosphatase) family of phosphodiesterases in that it contains an N-terminal Ub-binding ubiquitin-associated (UBA) domain (Figure 1A). Poly-Ub chains are formed by linking a Ub to any of seven lysines on another Ub, yielding seven possible types of poly-Ub, each with different signaling consequences for DNA repair (21). Caenorhabditis elegans TDP2 reportedly binds K48 and K63 linked di-Ub (25), and biochemical analysis of both human and C. elegans enzymes indicate that the N-terminal UBA domain inhibits TDP2 catalytic activity (18,20). However, whether poly-Ub regulates human TDP2 activity, the type of poly-Ub that is bound by TDP2, and how poly-Ub binding is related to TDP2 interactions with SUMO2 modified TOP2-DPC resolution is unknown.
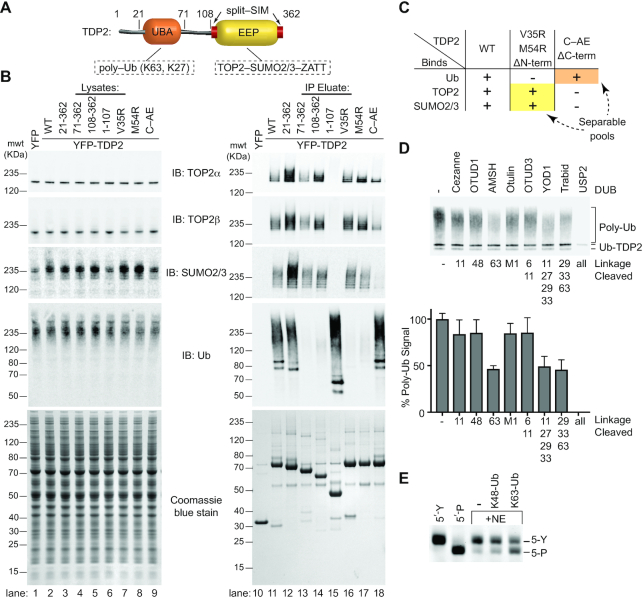
Figure 1.
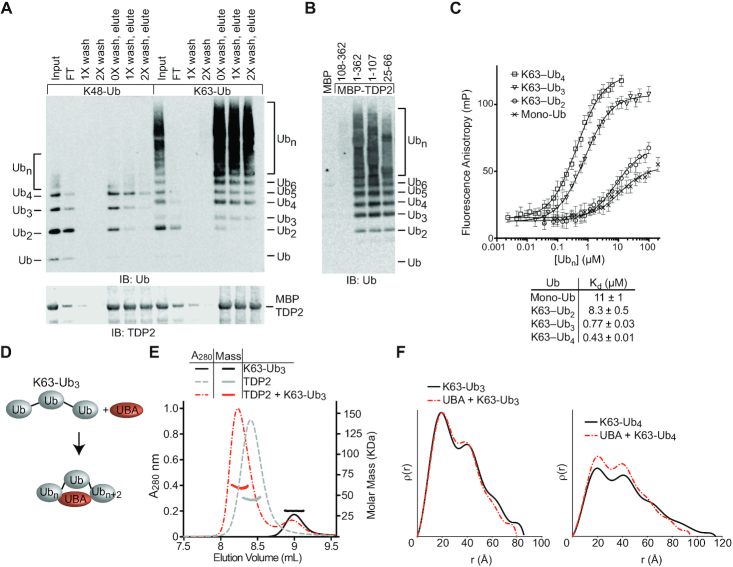
TDP2 binds poly-ubiquitin. (A) Domain architecture of TDP2. TDP2 contains an N-terminal ubiquitin-associated (UBA) domain and a C-terminal endonuclease/exonuclease/phosphatase (EEP) catalytic domain. (B) YFP-TDP2 lysates and immunoprecipitates were separated by SDS-PAGE and probed with the indicated antibodies. (C) TDP2 binds poly-Ubiquitinated proteins independently of SUMOylated Topoisomerase 2. (D) YFP-TDP2 immunoprecipitates were treated with de-Ubiquitinases (DUBs) that cleave poly-Ub linked through the indicated lysines, then (upper) analyzed by western blotting for Ub. (lower) the poly-Ub signal intensity from the western blots was quantified and normalized to samples without DUB. N = 6, error bars ± s.d. DUBs that hydrolyze K63- or K27- linked poly-Ub decrease the poly-Ub signal, showing that TDP2 associates with both K27 and K63-linked poly-Ub. (E) Nuclear extracts (NE) containing TDP2 hydrolyze a phosphotyrosyl-DNA (5′-Y) substrate to a 5′-phosphate (5′-P) DNA product, assayed by denaturing PAGE. Addition of K63-linked, but not K48-linked poly-Ub stimulates this activity.
In this work, we examined immunoprecipitated TDP2 variants with impaired SUMO2/3 or Ub-binding, and found that TDP2 interacts with separable pools of Ubiquitinated or SUMOylated proteins, with TOP2 and ZATT present only in the SUMOylated fraction. We find that TDP2 also associates with poly-Ub containing K63 and K27 Ub-chain linkages, but not K48 poly-Ub, which is usually associated with proteasomal degradation (26). The TDP2 UBA domain binds K63-Ub chains of three or more Ub in length, and TDP2 tyrosyl–DNA phosphodiesterase activity is stimulated by K63-linked poly-Ub. To determine the molecular basis for the TDP2-Ub interactions, we solved two ultra-high resolution (0.85Å) X-ray crystal structures of the human TDP2-UBA domain bound to Ub. Combined results from small-angle X-ray scattering analysis and mutagenesis of TDP2-Ub3 complexes reveal that two Ub-interacting surfaces on the UBA domain interact with the first and third Ub of a poly-ubiquitin chain. Collectively, these results underscore a role for Ub binding in modulating TDP2 activity and interaction with its DNA-protein crosslink reversal pathway components.
MATERIALS AND METHODS
All chemicals were purchased from Sigma unless stated otherwise.
Cell culture, western blots, and antibodies
HEK293F cells (Invitrogen) that stably express YFP, YFP-TDP2 1–362, and YFP-TDP2 C-AE mutant were described previously (10). HEK293F cell lines that stably express other variants of YFP-tagged TDP2 were generated as described, and immunoprecipitations were performed as described (10). T121 transformed wildtype and Tdp2−/− MEFs were previously described (10,54). The absence of mycoplasma contamination was confirmed using Universal Mycoplasma Detection Kit (ATCC). Protein samples were separated by SDS-PAGE, transferred to PVDF membrane (Millipore), and probed with antibodies using the iBind system (Invitrogen). Antibodies used: rabbit polyclonal anti-TDP2: Santa Cruz Biotech (sc-135214 lot# d1212), mouse monoclonal anti-Ub: Cell Signaling Technologies (3936, ref# 10/2014), mouse monoclonal anti-MBP: New England Biolabs (E8032 Lot# 0101804), rabbit monoclonal anti-TOP2α: Abcam (ab52934 lot# GR241638-22), rabbit monoclonal anti-TOP2β: Abcam (ab109524, lot# GR42963-15), mouse monoclonal anti SUMO2/3: Abcam (ab81371 lot# GR3233360-1), mouse anti-GFP/YFP: Roche (27575600, lot# 11814460001), and mouse monoclonal anti-V5: Invitrogen (46-0705 lot# 1517583). Secondary antibodies: IRDYE680 anti-rabbit: LICOR (926-68021 C51007-05) and IRDYE800 anti-mouse: LICOR (926-32210 lot# C30109-03) were visualized with an Odyssey Fc imager and ImageStudio software (LICOR) according to manufacturer's protocol.
Recombinant proteins
MBP-TDP2 (amino acids 1–362, 21–362 and 108–362) and untagged TDP2 (amino acids 1–362 and 108–362) were purified as described (18). TDP2 N-terminus (amino acids 1–107) was cloned into pDest566 and expressed and purified similarly to wildtype TDP2, except the final butyl-sepharose purification was replaced with anion exchange purification on a HiTrap Q-Sepharose column (GE Life Sciences). TDP2-UBA domain (amino acids 25–66) was cloned into vector pMCSG9 (27), and all TDP2-UBA mutants were generated with the Quickchange Mutagenesis kit (Stratagene). Protein was expressed in BL21 Rosetta2 Escherichia coli (EMD Biosciences), with 50μM IPTG at 16°C for 16 hours. Cells were lysed by sonication in GF100 buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 1 mM Tris(2-carboxyethyl)phosphine (TCEP)) with the addition of 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mg/ml (cheetah) of lysozyme. MBP-UBA proteins were affinity purified on amylose resin (New England Biolabs) and eluted in GF100 supplemented with 10 mM Maltose, followed by size-exclusion chromatography on a 16/60 S-200 column (GE Life Sciences) in GF100 buffer. For crystallization and fluorescein-labeling, the MBP tag was removed by TEV protease digestion and the UBA domain protein was purified sequentially on a 4.6/10 Source 15Q column (GE Life Sciences) and a 16/60 S-75 column (GE Life Sciences) in GF100 buffer. Protein was concentrated to 2 mM using a 3K cutoff centrifugal filter (Millipore). Bovine Ub protein (identical sequence to human protein) was purchased from Sigma for crystallization trials or expressed from Addgene plasmid 12647 and purified as described (28). Ub chains were synthesized by in vitro polymerization and purified as described (29).
Fluorescein labeling of TDP2-UBA domain protein
All reactions were done at 4°C. 100 μM TDP2-UBA domain protein was dialyzed overnight into phosphate-buffered saline (PBS) with 0.5 mM TCEP. 10 mM NHS-Fluorescein in DMSO was added to UBA protein to a final concentration of 1 mM, and the coupling reaction was allowed to proceed for 2 h at 22°C in the dark. The reaction was quenched by the addition of 100 mM Tris pH 9 and 10 mM BME for 2 hours, and unincorporated fluorescein was removed by dialysis in GF100 for 16 h. Fluorescein labeled UBA protein was purified on a 4.6/10 Source15Q column (GE Life Sciences). The UBA domain does not contain any lysine residues, therefore the only primary amine group is the N-terminus, and this chemistry will add a single fluorescein to the N-terminus of the protein. Attachment of a single fluorescein to the UBA protein was confirmed by mass spectrometry.
In vitro pull-down assays
To 500 μl reactions in binding buffer (10 mM HEPES pH 7.5, 50 mM NaCl, 0.5 mM TCEP, 2.5% (v/v) glycerol, 0.1 mg ml−1 BSA, and 0.1% (v/v) Tween-20) containing 1 μM of indicated Maltose Binding Protein (MBP) fusion or MBP protein were added 1 μg Ub chain mixtures (Enzo Life Sciences) for experiments in Figure 1, 1 μg in vitro-synthesized Ub chains for experiments in Figures 5 and 6, or 1 μM TDP2 V5-tagged TDP2108–362 for Figure 7, followed by 20 μl of amylose resin (NEB). Reactions were mixed on a nutator at 4°C for 2 h, transferred to a micro-spin column (Bio-Rad), centrifuged at 1000 g for 1 min, washed with 100 μl binding buffer, eluted with 100 mM maltose, and run on a NuPage (ThermoFisher) 4–12% Bis–Tris SDS-PAGE, followed by immunoblotting with probes for the bait protein.
Figure 5.
The UBA domain contains two Ubiquitin binding surfaces. (A) Alignment of the UBA domain and two Ub molecules that are common to the asymmetric units from the two crystal forms shows two interfaces are present in all cases. (B) Interface 1 consists of several hydrophobic TDP2 residues that bind to Ub on the hydrophobic patch. (C) A network of polar hydrogen bonding residues surrounds the hydrophobic core of Interface 1. (D) A comparison of TDP2-UBA residues in helix α1 and α3 that interact with the hydrophobic patch of ubiquitin to those commonly found in UBA domains, as shown for the DSK2 UBA-Ub structure (PDB ID: 4UN2). (E) A potassium ion (purple) bridges Ub and the hydroxyl group of the conserved UBA domain residue Y61. (F) TDP2 Y61F removes the crystallographically observed K+ ion ligand and destabilizes UBA domain binding to K63-Ub3 binding. N = 4, average ± s.d. *P < 0.05, **P < 0.01, two-tailed t-test. (G) Interface 2 consists of a hydrophobic pocket that cradles Ub T9. (H) Interface 2 contains a water-mediated hydrogen bond between Ub residue T9 and UBA residue E49.
Figure 6.
Mutational analysis of the UBA-Ub interfaces. (A) Five mutant TDP2-UBA proteins containing a mutation in a solvent-exposed residue (pink) were engineered. (B) Four mutant UBA-Ub proteins containing a mutation corresponding to a SNP (teal) were engineered. (C) Western blot analysis of K63 poly-Ub chains bound to mutant MBP-UBA proteins in an in vitro pull-down assay. (D) Modeling of Interface 1 mutations V35R and M54R shows they would clash with Ub residues. Black dotted outline and arrow indicate steric clashes that are incompatible with the interfaces observed in crystal forms 1 and 2 (E) TDP2 R56 forms several hydrogen bonding interactions with neighboring residues. (F) Modeling of C45L and C45W shows these residues should cause a small clash in Interface 2. E49A would remove part of the Ub T9 binding pocket and through-water hydrogen bond.
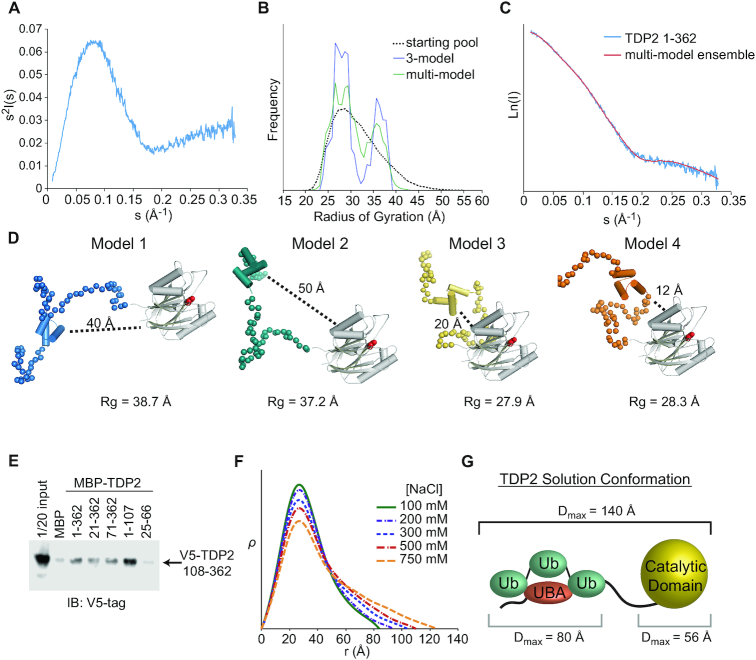
Figure 7.
The N-terminus of TDP2 interacts with the catalytic domain. (A) Kratky plot for TDP2 is typical of a predominantly folded protein with some disordered regions. (B) EOM using a 3-model or multi-model ensemble selects a pool of models containing both extended (Rg ∼ 37 Å) and compact (Rg ∼ 28 Å). (C) Comparison of scattering calculated from a multi-ensemble model is a good fit to the experimentally determined scattering intensity. (D) Example models selected by EOM contain two classes - those with the UBA domain distal (40–50 Å) or proximal (10–20 Å) to the catalytic domain. (E) in vitro pulldown assay with MBP-tagged TDP2 constructs and V5-tagged TDP2 catalytic domain (108–362) shows an interaction between the flexible N-terminus of TDP2 and catalytic domain in trans. (F) ρ(r) plots calculated from SAXS data collected from TDP2 in buffers with the indicated concentration of NaCl. Higher salt concentrations disrupt protein-protein interactions and weakens the UBA-catalytic domain interaction, resulting in a more extended conformation of TDP2. (G) Maximum particle dimension (Dmax) determined by SAXS are consistent with an extended conformation of the TDP2-Ub3 complex.
Size-exclusion chromatography with multiangle light scattering (SEC-MALS)
100 μl of protein: 10 μM TDP2, 15 μM K63-Ub3, or a mixture of the two proteins were run on a 7.8 × 300 mm size-exclusion chromatography column with 300 Å pores and 5 μm particle size (Wyatt WTC-030S5) in buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 1 mM TCEP) on a HPLC (Agilent) with a μDAWN light-scattering-diode array. Data were analyzed with ASTRA 7.1.4.8 (Wyatt Technology) to calculate molar mass, and plots were generated with GraphPad Prism.
Synthesis of 5′-toluidylated DNA oligonucleotide
5′-[Phosphate]-AATCCGGATCCG-[Cy5]-3′ DNA oligonucleotide was purchased from Integrated DNA Technologies and coupled to Toluidine using EDC (Sigma) as described (19). The oligonucleotide was gel-purified to remove unmodified precursor, and purified on a C18 sep-pak (Waters) before use in DNA binding assays.
Fluorescent polarization anisotropy (FP) assays were performed essentially as described (30). Briefly, fluorescein–UBA protein (30 nM) was incubated with the indicated concentrations of Ub or Ub chains in quadruplicate (N = 4) in FP buffer (20 mM HEPES pH 7.5, 100 mM NaCl, 1 mM TCEP, 0.2 mg ml−1 BSA and 5% (v/v) glycerol) in black, flat-bottomed 96-well plates for 10 min at 22°C, then each replicate was measured four times in a POLARstar Omega microplate reader (BMG Labtech) using excitation and emission wavelengths of 485 and 520 nm, respectively. TDP2 DNA binding experiments used MBP-TDP2 and K63-Ub3, and 10 nM DNA oligo (5′-[toluidyl]-AATCCGGATCCG-[Cy5]-3′) with excitation and emission wavelengths of 620 and 670 nm. Kd values were calculated by fitting FP data to a one-site binding model in GraphPad Prism.
TDP2 DNA Binding EMSA
DNA oligonucleotides (5′-[phosphate]-CACCACGGTGCCGAGG ATGACGATGAGCGCATTGTTAGATTTCA-[Cy5]-3′ and 5′-TGAAATCTAACAATGCGCTCATCG TCATCCTCGGCACCGT-3′) were annealed in 10 mM Tris pH 7.5 by heating to 75°C and slowly cooling to room temperature. MBP-TDP2 (50 mM HEPES, 200 mM NaCl, 10% glycerol) and K63-Ub3 (10 mM HEPES pH 7.5 100 mM NaCl, 1 mM TCEP, 25% glycerol) were with 4 nM oligonucleotides in a final reaction condition of 40 mM Tris, 6 mM HEPES pH 7.5, 0.08 mM EDTA, 3.5% (v/v) glycerol, 0.08 mM TCEP, 0.08 mg/ml BSA, and 30 mM NaCl for 30 min at 22°C. DNA/protein complexes were resolved on 0.9% agarose gels in 0.5× Tris/borate running buffer at 4°C for 1 h (10 V/cm), and scanned on a Typhoon RGB (GE) to visualize the Cy5 label.
Small-angle X-ray scattering (SAXS) analysis
Small angle X-ray scattering data were collected at the SIBYLS beamline (12.3.1) at the Advanced Light Source (Berkeley, CA). Data were processed and analyzed as described (10). RANCH (31) was used to generate 10 000 models of the TDP2 catalytic domain (PDB entry 4GZ1, chain A, residues 118-370 (18)) and UBA domain (crystal form 1, chain B, residues 25–66) connected by dummy atoms to represent the remainder of the protein in a random coil conformation. A genetic algorithm implemented in GAJOE (31) was used to select and evaluate models pools against the SAXS curves. SAXS analysis plots were generated using Microsoft Excel.
Crystallization and structure determination of UBA-Ub
Crystals of UBA-Ub protein complex were grown using the sitting-drop vapor diffusion method by mixing 200 nl of precipitant with 200 nl of protein mixture (7.5 mg ml−1 UBA protein and 13 mg ml−1 Ub in GF100 buffer). Crystal form 1 was obtained in 200 mM potassium formate and 14% (w/v) PEG3350, and crystal form 2 was obtained in 150 mM magnesium formate, 200 mM potassium formate, and 18% (w/v) PEG3350. Crystals grew over a period of two weeks at 4°C, and were transferred to a cryoprotectant containing crystallization condition supplemented with PEG3350 to 40% (w/v) for crystal form one and PEG3350 to 25% (w/v) and 16% (v/v) ethylene glycol for crystal form 2, then flash frozen in liquid nitrogen. X-ray diffraction datasets for sulfur-SAD phasing were collected at 105 K on the NIEHS in-house HF007 rotating anode X-ray source with VariMax HF mirrors and a Saturn944 CCD detector (Rigaku) for anomalous diffraction datasets. High-resolution datasets were collected at the Advanced Photon Source on beamline 22-ID. X-ray diffraction data were processed and scaled using the HKL2000 suite. Initial structures were solved using sulfur-SAD phasing with the HKL3000 suite (32,33), and iterative rounds of model building in COOT (34) and refinement against the high-resolution datasets with PHENIX (35) were used to produce the final models. Hydrogen atoms that did not show peaks in an Fo – Fc map calculated from a model lacking hydrogen atoms were removed for the final model. Crystal form 2 contained electron density between chains C and D that did not correspond to any buffer components, and was modeled as a benzoate that was likely present in the bovine Ub protein that was used to grow this crystal.
TDP2 5′-DNA phosphotyrosyl hydrolase activity assays
TDP2 5′-Y phosphotyrosyl hydrolase assays were performed essentially as described (18), with the addition of 1 mg ml−1 BSA and Ub or poly-Ub chains where indicated, yielding a final NaCl concentration of 110 mM. Reactions using recombinant TDP2 used 1 μM DNA oligonucleotide substrate and were performed at 22°C for 60 s. For detyrosylation experiments in HEK293F cell nuclear extract, 10 μl reactions contained 85 mM NaCl, 25 mM KCl, 2.5 μl of 1/40 diluted nuclear extract, Ub chains where indicated, and 50 nM DNA oligonucleotide substrate. Reactions were performed at 22°C for 15 min.
Processing of Top2-DPC reactions essentially as previously performed (10), but briefly, reactions conditions for TDP2-catalyzed hydrolysis of TOP2cc contain buffer A (50 mM Tris pH 8, 10 mM MgCl2, 30 ug/ul BSA, and 0.5 mM DTT) supplemented with 2 mM ATP, ZATT (100 nM), TOP2β-DPC (purification previously described, 10) and either 10 uM K63Ub3 or buffer (10 mM HEPES pH 7.5, 100 mM NaCl, 1 mM TCEP, 25% (v/v) glycerol, 2.5 mg/ml BSA). The addition of 100 nM TDP2 (residues 112–362 stored in 10 mM HEPES pH 7.5, 500 mM NaCl, 1 mM TCEP, 25% (v/v) glycerol, 1 mg/ml BSA starts 60 s reaction at 37°C. Protein bands were separated on NuPAGE 4–12% Bis–Tris SDS-PAGE and scanned using a Typhoon9500 imager (GE). Gel bands were quantified using Image Lab Software (Bio-Rad) and analyzed using Graphpad Prism. Quantification graphs were produced with data from 2 replicates and data were fit to a 4-parameter agonist vs response model average ± s.d.
Ub chain linkage analysis
To YFP-TDP2 immunoprecipitation eluates (containing 0.1 M glycine pH 2) 0.1 M Tris base was added to adjust the pH to near neutral. The UbiCrest kit (Boston Biochem) was used to selectively degrade poly-Ub containing the targeted linkage type according to the manufacturer's protocol. 10 μl reactions contained: 1 μl 10× DUB buffer, 4 μl neutralized YFP-TDP2 IP eluate, 3 μl ddH2O, and 2 μl DUB. Reactions were incubated at 37° for 30 min, followed by SDS-PAGE and immunoblotting with an anti-Ubiquitin antibody. The Ub signal intensity was quantified using ImageStudio software (LICOR) and normalized to control reactions that had no DUB added and USP2-treated samples where all cleavable Ub has been removed. Graphs were generated using GraphPad Prism. A decrease in anti-Ubiquitin signal following DUB treatment corresponds to the fraction of poly-Ub containing the linkage type(s) targeted by the DUB.
Clonogenic survival assays
Survival assays were carried out by seeding T121 transformed MEFs infected with the indicated lentiviral vector in 100 mm dishes in duplicate for each experimental condition. After 6 h, the indicated concentration of etoposide was added and cells were incubated for 10–12 days. Dishes were fixed and stained for manual colony counting in Crystal Violet solution (0.5% (w/v) Crystal violet in 20% (v/v) ethanol). The surviving fraction at each dose was calculated by dividing the average number of visible colonies in treated versus untreated dishes.
γH2AX foci analysis
Primary MEFs were infected with the indicated lentiviral vector and grown on coverslips for 7–10 days until confluency arrested. Cells were treated with 30 μM etoposide for 30 min, washed with fresh medium and allowed to repair for the indicated time. At each repair time point cells were fixed in ice-cold methanol for 10 min at –20°C and immunofluorescence was carried out as described (10, 54). Briefly, cells were permeabilized (2 min in PBS–0.2% (v/v) Triton X-100), blocked (30 min in PBS–5% (w/v) BSA) and incubated with the primary antibody for 1 h in PBS–1% (w/v) BSA. Cells were then washed (three times in PBS–0.1% (v/v) Tween 20), incubated with the secondary antibody for 30 min in 1% (w/v) BSA–PBS, washed again as described above, counterstained with DAPI (Sigma), and mounted in Vectashield (Vector Labs). γH2AX foci were manually counted (double-blind) in 40 cells for each experimental condition.
RESULTS
TDP2 associates with K27- and K63-linked poly-Ub chains
To examine the interplay between TDP2, TOP2, Ub and SUMO2/3, we generated a panel of HEK293F cell lines that stably express YFP-TDP2 constructs and analyzed co-immunoprecipitated proteins by western blot (Figure 1A, B and Supplementary Figure S1A). TDP2 associates with a high molecular weight population of Ubiquitinated proteins (Figure 1B). There are also two prominent Ubiquitinated protein bands at ∼75 and 85 kDa (Figure 1B, lane 11), which correspond to Ubiquitinated YFP-TDP2, as shown by co-staining of these bands with anti-Ub and anti-TDP2, as well as an antibody against GFP/YFP (Supplementary Figure S1B). TDP2 N-terminal truncations that lack the UBA domain (TDP2 aa 71–362 and 108–362) (Figure 1B, lanes 13 and 14) are nearly devoid of bound high-molecular weight Ub, but retain binding to SUMO2/3 and SUMOylated TOP2. By comparison, two TDP2 mutants (aa 1–107 or the split-SIM ‘C-AE’ mutant (10)) that do not bind SUMO2/3 both retain binding to cellular poly-Ubiquitinated proteins (Figure 1B, compare lane 11 versus 15 or 18). Furthermore, tandem affinity pulldowns with cells that co-express His6-Ub or His6-SUMO2 with YFP-TDP2 shows an absence of Ub-TOP2β or mixed Ub-SUMO2 chains (Supplementary Figure S1C). Thus, TDP2 interacts with poly-Ub independent of TOP2-SUMO2/3 (Figure 1C). Under the conditions evaluated here, TDP2 is not associated with Ub-TOP2.
Next, to delineate the types of poly-Ub linkages bound to TDP2, we treated YFP-TDP2 immunoprecipitates with linkage-specific DeUBiquitinases (DUBs) and quantified poly-Ub by immunoblotting (36) (Figure 1D and Supplementary Figure S1D,E). Digestion with Cezanne, OTUD1, Otulin or OTUD3 did not significantly reduce the poly-Ub signal. Therefore, TDP2 does not bind significant amounts of linear, K6, K11 or K48 poly-Ub. The K63-specific AMSH DUB reduced the Ub signal by ∼50%, indicating that approximately half of the TDP2 bound poly-Ub contains K63 linkages. Trabid, which hydrolyzes K63, K29 and K33 linkages, decreased the Ub signal comparable to AMSH, suggesting that K29 and K33 linkages are not abundant in TDP2 immunoprecipitates. YOD1 hydrolyzes K11, K27, K29 and K33 linked poly-Ub, and this DUB reduced the poly-Ub signal by ∼50%. As K29 and K33 linkages do not appear to be present in TDP2-associated poly-Ub, and K11 poly-Ub comprises approximately 10% of TDP2-associated poly-Ub, we estimate the remaining 40% of TDP2-associated poly-Ub is K27-linked. Altogether, we conclude that TDP2 is associated with both K27-Ub and K63-Ub in HEK293F extracts.
To test if poly-Ub regulates endogenous TDP2 activity, we monitored the cellular 5′ tyrosyl-DNA phosphodiesterase activity in nuclear extracts using a 5-Y substrate (Supplementary Figure S2) in the presence or absence of purified poly-Ub chains (Figure 1E). K48 chains, which do not stably bind to TDP2 (Figure 1D), had little impact on TDP2 activity. Conversely, the addition recombinant K63 poly-Ub significantly stimulated endogenous 5′ tyrosyl-DNA phosphodiesterase activity (Figure 1E). As TDP2 is the sole source of 5′ tyrosyl-DNA phosphodiesterase activity in mammalian extracts (54), these data suggest that K63 poly-Ub regulates TDP2 catalytic activity. Notably, we were unable to examine the interaction between TDP2 and K27 poly-Ub as K27 poly-Ub is not currently commercially available and enzymatic methods for its synthesis have not been described.
TDP2 preferentially binds K63-linked Ub3
To further dissect the determinants of TDP2-Ub interactions, we assessed binding of recombinant K48 and K63-linked poly-Ub to Maltose-Binding Protein (MBP)-tagged human TDP2 (Figure 2A). While mono-Ub did not stably bind to TDP2, K63 poly-Ub bound more efficiently than K48 poly-Ub, as evidenced by an excess of unbound K48 poly-Ub in the column flow-through and depletion of bound K48 poly-Ub by washing with buffer. We next investigated which domains of TDP2 are required for binding poly-Ub using a panel of TDP2 deletion mutants. While the MBP control and the isolated EEP domain (residues 108–362) of TDP2 displayed no detectable binding, all constructs that contained the UBA domain (residues 25–66) bound to K63-Ub (Figure 2B), indicating that the UBA domain is necessary and sufficient to bind K63-Ub. To quantify TDP2-Ub interactions, we developed a fluorescence polarization anisotropy (FP) assay using N-terminal fluorescein-labeled UBA domain to measure solution dissociation constants (Kd) for K63-Ub chains of defined length. The UBA domain binds mono-Ub weakly (Kd 11 μM), while K63-Ub2 binds only slightly better (Kd 8.3 μM) (Figure 2C). Strikingly, K63-Ub3 (Kd 0.78 μM) binds TDP2 significantly better than either mono-Ub or K63-Ub2. Likewise, K63-Ub4 bound tightly to TDP2 in the nanomolar range (Kd 0.43 μM). We also observed trend for K48-Ub and K11-Ub, with an increase in binding affinity for chains ≥3 Ub in length (Supplementary Figure S3). Interestingly, while the UBA domain exhibited weaker binding to K11-Ub3, K48-Ub3 actually bound slightly better than K63-Ub chains showing that the UBA domain of TDP2 capable of tight binding to K48-Ub, even though the full-length protein does not (Figure 2A). High affinity poly-Ub≥3 chain interactions are likely mediated by TDP2 UBA binding to one Ub protomer (Ubn), and a second Ub spaced 2 protomers away (Ubn+2).
Figure 2.
TDP2-UBA domain binds K63Ub3 with a 1:1 stoichiometry. (A) TDP2 binds K48 and K63 poly-Ub chains. Poly-Ub interaction was assessed in binding reactions containing amylose resin, MBP-TDP2 fusion protein, and poly-Ub chains. Proteins present in the input, flow through (FT), washes, and elution were detected by SDS-PAGE and western blotting. Ubn indicates longer chains that are not resolved in the gel. (B) Domain analysis of TDP2. MBP-TDP2 fusion protein constructs were assayed for binding to K63-Ub chains as in panel A. (C) Fluorescence polarization anisotropy assay. Fluorescein-UBA protein was incubated with increasing amounts of the indicated Ub or K63-Ub chain and the FP value was measured at each concentration. Error bars, s.d. N = 4 replicates. Data were fit to single binding-site model (solid line) to calculate Kd values ± s.e.m. (D) Model for compaction of K63-Ub3 upon binding the UBA domain. (E) Molar mass determination of proteins using size-exclusion chromatography with multi-angle light scattering (SEC-MALS) indicates TDP2 binds Ub3 with a 1:1 stoichiometry. (F) ρ(r) plots calculated from SAXS scattering data of the indicated protein or protein complexes.
To define the stoichiometry of the TDP2:K63–Ub3 complex we used size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) (Figure 2E). Both TDP2 (MALS-MW = 48 KDa, calculated-MW = 41 kDa), and K63-Ub3 (MALS-MW = 30 kDa, calculated-MW = 26 kDa) were monomeric, and the TDP2:K63-Ub3 complex formed with 1:1 stoichiometry (MALS-MW = 66 kDa, calculated-MW = 67 kDa). We reasoned that a K63-Ub3 chain bound to TDP2 via the first and third Ub molecules might be structurally distinct from previously observed extended conformations of K63-Ub chains (37). To test this, we used Small Angle X-ray Scattering (SAXS) to compare the solution conformations of K63-Ub3 to TDP2-UBA bound complexes. Despite the addition of extra mass from the UBA domain, the K63-Ub3:UBA (radius of gyration, Rg = 25.0 Å, maximum particle dimension, Dmax = 80 Å) and K63-Ub4:UBA (Rg = 29.7 Å, Dmax = 95 Å) complexes displayed slightly compacted solution conformations when compared to free K63-Ub3 (Rg = 26.7 Å, Dmax = 85 Å) and K63-Ub4 (Rg = 32.4 Å, Dmax = 115 Å) (Figure 2F and Supplementary Figure S4A–D; Supplementary Table S1). These observations are consistent with the UBA domain binding and bridging distal Ub protomers, and suggest Ub chain binding avidity is provided by two Ub binding sites on a single TDP2 UBA domain.
K63-linked Ub3 regulates TDP2 substrate interactions
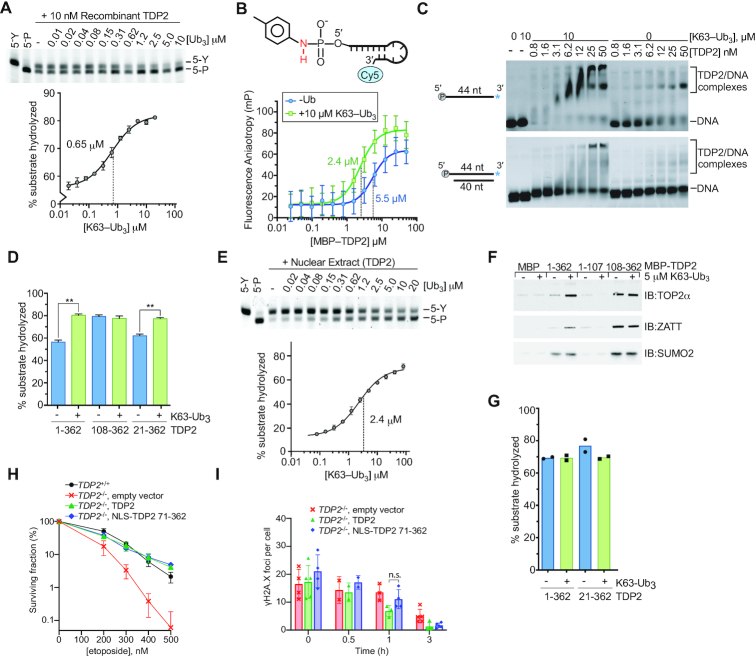
Given the specific high-affinity interaction between TDP2 and poly-Ub, we hypothesized that K63-Ub3 regulates TDP2 interactions with DNA–protein crosslink substrates. Consistent with the Ub-stimulated TDP2 tyrosyl-DNA phosphodiesterase activity in extracts (Figure 1E), K63-Ub3 also stimulated recombinant TDP2 tyrosyl-DNA phosphodiesterase activity ∼1.5-fold on a 5′-Y adducted oligonucleotide with a midpoint of stimulation at 0.65 μM (Figure 3A), which is similar to the Kd for K63-Ub3 of 0.77 μM (Figure 2C). In contrast, compared to K63-Ub3, stimulation by K48-Ub3 required relatively high triubiquitin concentrations, with a midpoint of 4 μM to increase TDP2 tyrosyl-DNA phosphodiesterase activity on a 5′-Y oligonucleotide (Supplementary Figure S4E), which is consistent with weaker binding to K48-polyUb observed for full-length TDP2 (Figure 2A).
Figure 3.
K63-Ub3 stimulates TDP2 activity. (A) Stimulation of TDP2 catalytic activity by K63-Ub3 chains. (upper) Recombinant TDP2 protein and the indicated [K63-Ub3] chains were incubated with 5′-phosphotyrosyl DNA substrate. Detyrosylation of the DNA was monitored by denaturing PAGE. (lower) Percent of product DNA formation on PAGE gels was plotted as a function of Ubiquitin concentration, and data were fit to a dose-response model (solid line) to determine EC50 values. Error bars, s.d. N = 3 replicates. (B) TDP2 binding to a non-hydrolyzable 5′-toluidyl, Cy5-labeled DNA oligonucleotide was measured using fluorescence polarization anisotropy. K63-Ub3 enhances binding of TDP2 to DNA. Error bars, s.d. N = 4 replicates. Data were fit to single binding-site model (solid line) to calculate Kd values ± s.e.m. (C) The effect of K63-Ub3 on TDP2 binding to DNA was assayed by EMSA using the indicated DNA substrates. (D) 5′-Y DNA hydrolysis catalyzed by TDP2 or TDP2 truncation mutants with 10 μM K63-Ub3 where indicated was quantified. N = 3, error bars s.d., **P < 0.001 two-tailed t-test. (E) Stimulation of TDP2 activity in nuclear extracts of HEK293F cells by K63-Ub3. (upper) Nuclear extract and K63-Ub3 were incubated with 5′-phosphotyrosyl DNA substrate. Detyrosylation of the DNA was monitored by denaturing PAGE. (lower) Product DNA formation on PAGE gels was plotted as a function of Ubiquitin concentration, and data were fit to a dose-response model (solid line) to determine EC50 values. K63-Ub3 stimulates TDP2 activity ∼5-fold in nuclear extract. Error bars, s.d. N = 3 replicates. (F) K63-Ub3 regulates the interaction between TDP2 and TOP2α and ZATT. The effect of K63-Ub3 on the interaction between TOP2α, ZATT, or SUMO2 and MBP-TDP2 fusion proteins was assayed using an in vitro pulldown assay. The indicated bait proteins were detected by SDS-PAGE and western blotting with the corresponding antibodies. (G) Effect of K63-Ub3 on TOP2-DPC hydrolysis by the indicated TDP2 variants. (H) Clonogenic survival of T121-transformed Tdp2−/− MEFs complemented with the indicated vectors shows that TDP2 that lacks the UBA domain (71–362) rescues knockout cells as efficiently as wildtype TDP2. Error bars, average ± s.d. (I) Repair of DSBs measured as disappearance of γH2A.X foci following treatment with 30μM etoposide for 30min and the indicated repair times in primary Tdp2−/− MEFs complemented with the indicated vectors shows that TDP2 lacking the UBA domain (71–362) rescues knockout cells as efficiently as wildtype TDP2. Average ± s.d, n.s. P = 0.12, two-tailed t-test.
In the presence of saturating amounts of K63-Ub3, less TDP2 is required (1.8 nM versus 4.8 nM) to hydrolyze 50% of the 5′-Y substrate (Supplementary Figure S5A), suggesting that K63-Ub3 enhances substrate binding by TDP2. To test this directly we generated a non-hydrolysable 5′-Y substrate analog by coupling toluidine to the 5′-phosphate of a Cy5-labeled DNA oligonucleotide, yielding a 5′-Toluidilated DNA (5′-Tol) with a phosphoramidate bond (Supplementary Figure S2) that is resistant to hydrolysis by TDP2 (Supplementary Figure S5B). Fluorescence polarization anisotropy binding analysis (Figure 3B) shows that MBP-TDP2 binds 5′-Tol DNA in the presence or absence of K63-Ub3, but has >2-fold enhanced affinity (Kd 5.5 versus 2.4 μM) in the presence of K63-Ub3. K63 poly-Ub was reported to bind double-strand DNA over 40 bp in length (38), so we used EMSA to evaluate TDP2 binding to a 40 bp DNA containing a phosphorylated 4nt 5′-overhang that characteristic of the DNA structure in a TOP2-DPC (Figure 3C). Under these conditions, we find that K63-Ub3 does not bind DNA, but does enhance binding of TDP2 to both single-strand and double-strand DNA.
K63-linked Ub3 regulates TDP2 activity and interactions with TOP2 and ZATT via the UBA domain
TDP2 binds to Poly-Ub and DNA substrates through different domains, which raises the question of how Ub binding to the UBA domain activates EEP domain catalytic activity. We compared the effect of K63-Ub3 on the activity of full-length TDP2 (TDP2FL) and a construct that contained only the EEP domain (TDP2108-362) using a 5′-Y DNA substrate (Figure 3D and Supplementary Figure S5C). K63-Ub3 stimulated the activity of TDP2FL to a level comparable to that of the EEP domain, which was unaffected by Ub. This suggests that the UBA domain of TDP2 negatively regulates the catalytic activity, and Ub binding stimulates activity. Rao et al. previously reported that acidic residues N-terminal to the UBA domain bind in the active-site of TDP2 to regulate activity of the C. elegans homolog (25). We tested a human TDP2 truncation that lacks the equivalent acidic N-terminus of human (TDP221-362), and found this fragment displays comparable stimulation by K63-Ub3, indicating that the acidic residues N-terminal to the UBA domain are dispensable for Ub-mediated regulation of human TDP2. We then extended our analysis to test the regulation of endogenous TDP2 in nuclear extract (Figure 3E), and found that K63-Ub3 stimulated TDP2 5′-Y hydrolase activity to an even greater extent than that observed for the purified recombinant protein (∼5-fold versus ∼1.5-fold). This difference suggests that additional cellular proteins or post-translational modification of TDP2 either stabilize the inhibited state or further activate the Ub-bound state to enhance the regulatory effect of K63-Ub3.
TDP2 also hydrolyzes intact TOP2-DPCs, in a direct reversal pathway that is promoted through interactions with TOP2 and SUMO2/3, and licensed by ZATT (10). We tested whether K63-Ub3 influences interactions with these proteins in a pull-down assay. TOP2α, ZATT and SUMO2 bind to both TDP2FL and TDP2108-362 (Figure 3F and Supplementary Figure S5D). K63-Ub3 did not alter binding to SUMO2, but increased the amount of TOP2α and ZATT that bound to TDP2FL. TDP2108-362 binds TOP2α and ZATT stronger than TDP2FL and binding is unaffected by K63-Ub3. Thus the K63-Ub3 also regulates binding to TOP2 and ZATT and this effect is mediated through the UBA domain of TDP2. However, K63-Ub3 does not appear to cause a further increase in activity of ZATT-licensed TOP2-DPCs (Figure 3G).
A previous report found that TDP2 deficient DT40 cells could not be complemented by overexpression of an N-terminal TDP2 truncation that removed the UBA domain (25). Given the proposed regulatory role for Ub binding, we evaluated the functional role of the UBA domain in the cellular response to etoposide in mouse embryonic fibroblasts (MEFs). We found that Tdp2−/− mouse embryonic fibroblasts MEFs cells were similarly complemented by expression of WT or an UBA-deleted human transgene, both in a colony-forming assay for etoposide sensitivity and in the disappearance of γH2AX foci after acute etoposide treatment (Figures 3H and I). Overall, these results are consistent with ubiquitin binding serving to tune and regulate mammalian TDP2 activity, rather than being prerequisite for its function.
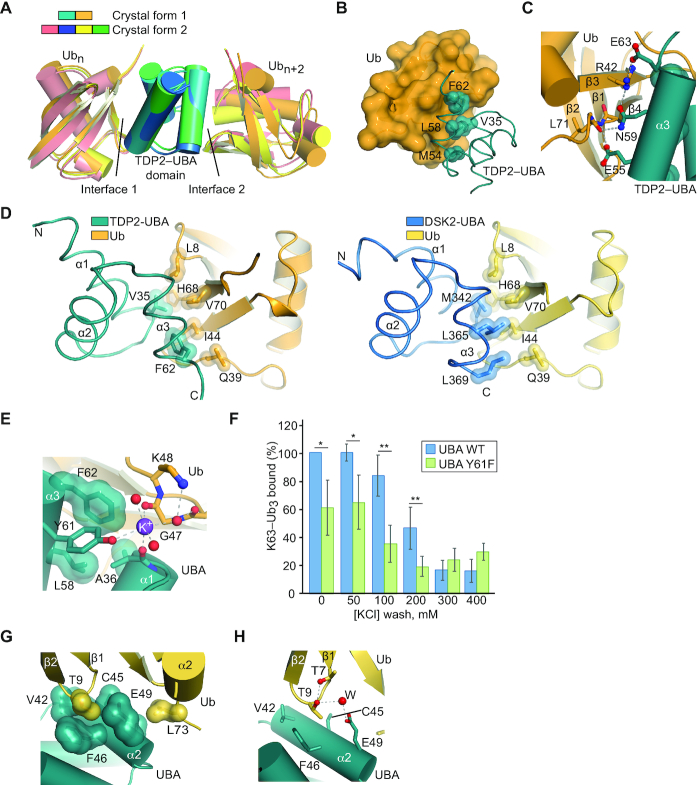
Molecular architecture of the TDP2-UBA:poly-Ub complex
To define the molecular architecture of the TDP2-UBA:poly-Ub complex, we crystallized the human TDP2 UBA domain in complex with mono-Ub in two crystal forms. Crystal form 1 contains one UBA domain and one Ub in the asymmetric unit (ASU) (Figure 4A) whereas crystal form 2 has two UBA domains and two Ub molecules per ASU (Figure 4B). Both crystal forms diffracted X-rays to sub-Angstrom (0.85 Å) resolution (Supplementary Table S2), which provides a detailed view of the molecular interfaces and allows individual atoms to be visualized. Hydrogen atoms from well-ordered regions can be observed as positive peaks in a Fo-Fc map calculated in the absence of hydrogens (Figure 4C), and K+ ions produce large peaks in an anomalous difference Fourier map (Figure 4D).
Figure 4.
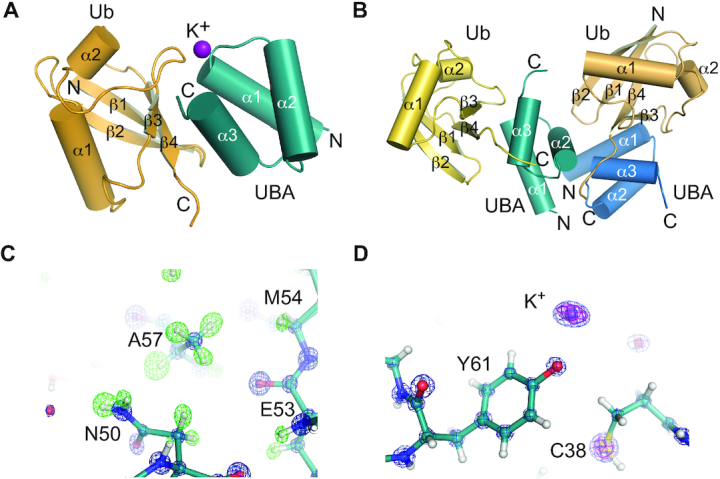
Structures of human TDP2-UBA domain bound to Ubiquitin. (A) Structure of TDP2 UBA-Ubiquitin in crystal form 1. The asymmetric unit contains 1 Ub and 1 UBA protein. (B) Structure of TDP2 UBA-Ubiquitin in crystal form 2. The asymmetric unit contains 2 Ub and 2 UBA protein. (C) Electron density from crystal form 2 shown for a 2mFo – dFc map contoured at 5σ (blue) and a mFo – dFc map calculated in the absence of hydrogens contoured at +3σ (green). (D) 2mFo – dFc map contoured at 5σ (blue) and anomalous difference Fourier contoured at 5σ (magenta).
The TDP2 UBA domain is composed of the expected three-helix bundle UBA architecture (39), but lacks the fourth α-helix observed in the C. elegans TDP2 variant UBA domain (25). A comparison of the molecular interfaces found in our two structures reveals that two UBA-Ub interfaces (interfaces 1 and 2, Figure 5A and Supplementary Figure S6A) are common between these unrelated crystal forms, and other crystal-lattice interactions are not (Supplementary Figure S6B–G). Interface 1 consists of an interaction between helices α1 and α3 of the UBA domain and the hydrophobic patch of Ub, centered on strands β3 and β4 (Figure 5B, C). UBA domains often bind Ub via a methionine-glycine motif at the end of α1 and leucine(s) near the end of α3 (39,40), but these residues are absent in human TDP2 and appear to be substituted by a valine (V35) and a phenylalanine (F62) that protrudes from α3 (Figure 5D). These residues, along with M54 and L58 form a surface that recognizes the hydrophobic residues L8, I44, and V70 on Ub (Figure 5B). A network of hydrogen bonds and a salt bridge between UBA E63 and Ub R42 flanks the hydrophobic interface (Figure 5C). Unexpectedly, we also observe an ion coordinated between the UBA-Ub interface 1 in crystal form 1, as indicated a peak of 14σ on the 2Fo – Fc map, 13 σ on the anomalous scatter map. The CheckMyMetal (41) program validates the octahedral coordination geometry and oxygen ligand bond length (average of 2.7 Å) as being most consistent with binding of potassium (K+) ion at this site (Figures 4D and 5E). The K+ ion bridges the UBA domain Y61 hydroxyl and A36 peptide carbonyl with the Ub peptide carbonyl of E47 (Figure 5E). Consistent with a role for a monovalent ion in stabilization of the UBA–Ub complex, a Y61F mutation destabilizes binding to poly-Ub at moderate salt concentration (Figure 5F).
Interface 2 consists of an interaction between UBA helix α2 residues and the β1–β2 turn of Ub (Figure 5G). The side chain methyl group of a Ub Threonine residue (T9) occupies a pocket formed by UBA C45, F46 and E49, and additional Van der Waals interactions occur between Ub L73 and UBA A48 and E49. A water-mediated hydrogen bond between Ub T9 and UBA E49 comprises the remainder of interface 2 (Figure 5H). With the orientation of Ubs positioned at interface 1 and 2, the closest distance between any lysine side chain and C-terminus of the other Ub is greater than 20 Å, suggesting that these two Ubs represent the Ubn and Ubn+2 of a bound Ub3, and the Ub monomer that would bridge these (Ubn+1) is not present in the crystal lattices.
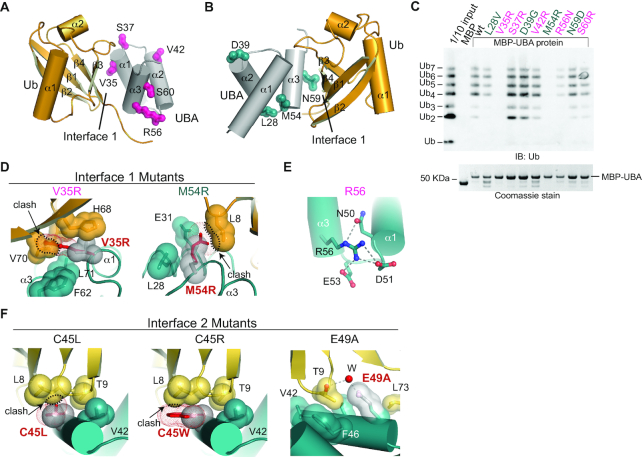
Mutational analysis of TDP2 UBA–Ub interfaces.
We probed the contribution of interfaces 1 and 2 to poly-Ub binding by assaying binding of K63-Ub chains to MBP-tagged mutant UBA proteins (Figure 6), or cellular Ubiquitinated proteins (Figure 1B). A V35R mutation designed to disrupt Interface 1 (Figure 6D), ablates K63-Ub binding to MBP-UBA (Figure 6C) and also blocks binding to cellular poly-Ub (Figure 1B, lane 16). Conversely, mutation of residues involved in crystal contacts but not interfaces 1 or 2 (S37R, V42R, and S60R) do not significantly alter binding to poly-Ub. By comparison, an R56N substitution which is predicted to disrupt folding of the UBA domain (Figure 6E) showed reduced Ub-binding in MBP-pulldowns (Figure 6C), and was impaired for binding all lengths of Ub chains tested (Supplementary Figure S7B).
Several non-synonymous single-nucleotide polymorphisms (SNPs) of TDP2 have been identified that result in amino-acid substitutions in the both the EEP (19) and UBA domain of TDP2 (42). We analyzed the impacts of four SNPs: L28V, D39G, M54R and N59D. In the UBA-Ub structure L28 is solvent exposed, D39 forms a hydrogen bond that caps the N-terminus of helix α2, and neither of these residues form part of interface 1 or 2. M54 forms part of the UBA surface that recognizes the hydrophobic patch of Ub in Interface 1, and N59 forms 2 hydrogen bonds to Ub in interface 1 (Figure 5C). L28V, D39G, and N59D do not have major impacts on poly-Ub chain binding (Figure 6C). However, M54R completely abolishes binding to recombinant poly-Ub (Figure 6C) as well as cellular poly-Ub (Figure 1B, lane 17). Modeling of M54R in the UBA-Ub structure shows that even the best accommodated rotamers result in a steric clash with Ub in Interface 1 (Figure 6D). Together, these results suggest Interface 1 is a major determinant of Ub binding, and show that a human TDP2 SNP encoding an M54R variant disrupts the TDP2–ubiquitin interaction.
To examine the contribution of Ub Interface 2 to TDP2-poly-Ub interactions (Supplementary Figure S7A), we designed additional UBA mutations to target Interface 2. C45W and C45L create steric clashes with Ub L8, and E49A disrupts through-water hydrogen bonds to Ub T9 (Figure 6F). In the FP Ub binding assay, these three Interface 2 mutants do not alter binding to mono-Ub or K63-Ub2, but do impair binding to K63-Ub3, (Supplementary Figure S7B). Thus Interface 2 contributes to the enhanced affinity for K63-Ub≥3 over Ub2, and we conclude it is an important determinant of K63 poly-Ub binding.
Solution conformation of TDP2
We used Small-Angle X-ray Scattering (SAXS) to probe the solution conformation of TDP2 and evaluate whether the N-terminal domain interacts with the catalytic domain of TDP2. Kratky analysis of solution scattering from TDP2FL reveals a bell-curve that peaks but does not return to baseline at sI2 > 0.2, and is characteristic proteins with both ordered and disordered regions. (Figure 7A). These observations are consistent with the TDP2 UBA domain being linked to the protein via a proteolytically labile linker peptide (18). Given the flexibility of the system, we used an ensemble approach in Ensembles of Mixtures (EOM) (31) to model the disordered segments of the N-terminal domain of TDP2 that are not observed in the UBA domain structure (residues 26–65 of crystal form 1) or catalytic domain structure (residues 110–362) (19). We systematically varied the test pool size, and found that pools of 3 or more models were required for an optimal fit to the data (Figure 7B and Supplementary Figure S8A; Supplementary Table S3). Interestingly, a comparison of the distribution of Rg values from the models in the whole pool with those selected by the genetic algorithm shows a bimodal distribution of models (Figure 7B, C), suggesting that the N-terminal domain of TDP2 may adopt two different conformations. Alignment of the four most frequently selected TDP2 models shows that the N-terminus is variable in position and thus quite flexible (Figure 7D). Interestingly, for two of the models the UBA domain is found in close proximity to the catalytic domain (12–20 Å), while the other two models position the UBA domain further away from the catalytic domain (40–50 Å). This suggests that the flexibility of the N-terminus enables TDP2 to exist in two conformations—one in which the UBA domain is close to or interacting with the catalytic domain, and the other in which the UBA domain is distal from the catalytic domain.
We posited that direct interactions between the N-terminal and catalytic domains could promote a more compact TDP2 conformation. Therefore, we examined whether these domains associate in trans using a V5-tagged catalytic domain construct and MBP-tagged truncation mutants of TDP2 (Figure 7E). Compared to an MBP control, the N-terminal domain of TDP2 (aa 1–107) bound to the TDP2 catalytic domain, while a construct encompassing a minimal UBA domain (aa 25–66) did not, suggesting residues flanking the UBA domain (1–24 and/or 67–107) are required for this interaction. We hypothesized that higher salt could disrupt the interaction between the N-terminal and EEP domains and destabilize the compacted conformation of TDP2. Electron pair distribution functions calculated from SAXS of TDP2 in buffers ranging from 100 to 750 mM NaCl show that higher salt promotes a more extended conformation of TDP2 (Figure 7F), consistent with salt disrupting the physical interactions that stabilize the compact conformation. We then evaluated the solution conformation of TDP2 bound to K63-Ub3 using SAXS and found that it adopts an extended conformation 140 Å in length (Figure 7G and Supplementary Figure S8B-D). This is consistent with Ub3 interacting with and sequestering the UBA away from the catalytic domain, which relieves the inhibitory effect of the N-terminal domain.
DISCUSSION
The DDR is a highly coordinated response that is regulated by post-translational modification with both Ub and SUMO. Understanding how TDP2 binds both of these modifiers is critical for understanding how TDP2 is regulated in response to DNA damage. MRE11 nuclease can cleave DNA adjacent to a TOP2-DPC (43), the proteasome or SPARTN protease can degrade the protein component of TOP2-DPCs followed by TDP2-catalyzed 5′-Y removal (6,44,45), or ZATT and TDP2 can directly reverse TOP2-DPCs (10). Cells undoubtedly use post-translational modifiers including poly-Ub and SUMOylation to regulate repair pathway selection (21), which may be a function of cell type, cell cycle stage, or chemotherapeutic drug dosage. For example, proteasomal degradation of TOP2-DPCs has only been observed at high concentrations of etoposide (25-250 μM) (44,46–48), whereas cell death occurs at ∼200 nM (10,47). It is unclear whether proteasome-mediated TOP2-DPC removal occurs to a meaningful extent in unchallenged cells or those exposed to a therapeutic dosage of etoposide. Therefore, in this work we examined how TDP2 interacts with poly-Ub in unchallenged cells that are repairing basal levels of TOP2-DPCs.
Here, we have shown that TDP2 binds to both poly-Ubiquitinated and SUMOylated proteins, and these are two independent and separate protein populations. While it is known that TDP2 binding to SUMO2/3 promotes recruitment of TDP2 to TOP2-DPCs (10), the role of Ub-signaling in TDP2 functions was poorly defined. It has been speculated that TDP2 interacts with K48-Ub chains to promote recruitment to TOP2-DPCs that are repaired by a proteasome-mediated TOP2 degradation pathway (25). Contrary to this hypothesis, we did not observe Ub-TOP2 in TDP2 IP samples, and find that TDP2 does not interact strongly with K48 poly-Ub in cellular extracts, suggesting that Ubiquitination of TOP2 is not involved in the recruitment of TDP2 to TOP2-DPCs. Instead, we find that TDP2 interacts with K27 and K63 poly-Ub, which are signaling molecules that regulate the DDR (21,22). K63-Ub≥3 enhances TDP2 binding to TOP2, ZATT, and DNA, and promotes catalytic activity on 5′-Y DNA (Figure 8). Overall, our data are congruent with the main function of poly-Ub binding by TDP2 being a regulatory one that promotes repair of TOP2-generated DNA damage and could function as a check on TDP2 hydrolase activity. Therefore, TDP2 might require both SUMO2/3 and poly-Ub signals, and this dual post-translational modifier regulatory mechanism may also be important to prevent aberrant TDP2 hydrolase activity.
Figure 8.
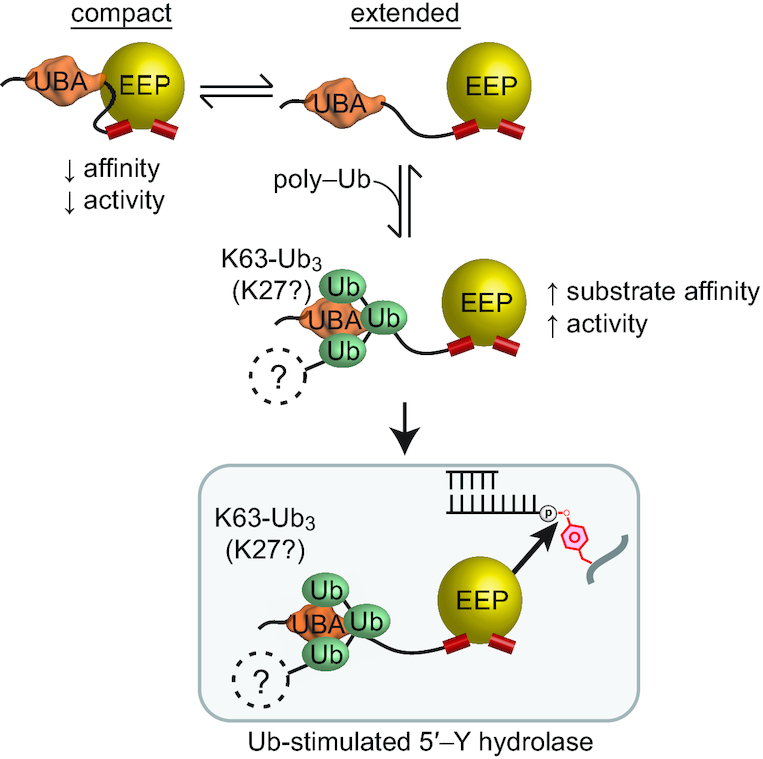
Proposed model for regulation of TDP2 by Ubiquitin and SUMO2/3. (A) TDP2 forms a compact state where the UBA domain decreases catalytic activity, or an extended conformation with increased catalytic activity. Binding to poly-Ubiquitin stabilizes the extended and active conformation. Proteins that are K63 (or K27) poly-ubiquitinated can function as regulatory signals that increase TDP2 hydrolase activity on 5′-Y DNA.
TDP2 homologs have a conserved domain structure, with an N-terminal UBA domain connected by a flexible linker to the C-terminal catalytic domain. The linker region appears to be relatively conserved in length as well, suggesting that in addition to the presence of an N-terminal UBA domain, the spatial arrangement and interdomain flexibility is critical for proper regulatory control of TDP2. The regulatory function of the UBA domain described in this work provides a rationale for this domain arrangement (18,20). The UBA domain and the span of flexible residues that link it to the catalytic domain allow sufficient mobility for the UBA domain to engage and regulate the catalytic domain as well as bind poly-Ub containing K27 and K63 linkages. The UBA domain uses two interfaces to interact with Ub protomers separated by one, or possibly more Ubs within a K63-linked poly-Ub chain, and therefore provides specificity for K63-Ub of three or more monomers in length. As K63 poly-Ub chains play important roles in DNA repair and non-homologous end joining (21,38,49), the UBA domain of TDP2 could integrate TDP2 into the K63-Ub signal network that directs repair of DSBs and TOP2-DPCs. Recently, it was reported that N-terminally truncated forms of TDP2 are expressed from alternative transcription start sites, resulting in proteins with altered cellular distribution and lacking core residues of the UBA domain (50,51). Our data suggests that in addition to being unable to bind poly-Ub, these truncated variants likely possess alternatively regulated tyrosyl DNA phophodiesterase activity.
K27-linked poly-Ub is a substantial component of the TDP2 interactome, yet K27 linkages are quite rare, representing <0.1% of the Ub isopeptide linkages in a cell (52). The enrichment of K27 poly-Ub in TDP2 immunoprecipitates suggests it is important for TDP2 functions such as recruitment to sites of DNA damage and/or activation of tyrosyl-DNA-phosphodiesterase activity. The identity of the K27 and/or K63 poly-Ubiquitinated proteins bound to TDP2 is not known, but we can speculate as to what they may be. A recent report (22) demonstrated that RNF168 catalyzes poly-Ubiquitination of histones H2A and H2AX with K63 and K27 linkages in response to DNA DSBs generated by etoposide treatment. Since TDP2 binds both K27 and K63 poly-Ub and repairs DNA damage caused by etoposide, RNF168 could be an important source of poly-Ub that directs TDP2-mediated repair of TOP2-DPCs.
A comparison of our findings of human TDP2 to those reported for Ub-binding by the C. elegans TDP2 homolog (ceTDP2) (25) reveals a number of features that distinguish hTDP2 from ceTDP2, which may be attributed to species-specific differences in TOP2-DPC repair. The UBA domain of ceTDP2 is a variant of the UBA domain with an additional fourth α-helix that is not conserved (Supplementary Figure S7A) in the human protein. ceTDP2 was reported to bind both K48-Ub2 (Kd 8 ± 4 μM) and K63-Ub2 (Kd 9 ± 4 μM) (25), whereas hTDP2 binds K63-Ub3 with higher affinity (0.77 μM; Figure 1E). Longer Ub chains were not tested for binding to ceTDP2, therefore, it is unknown if a similar linkage specificity exists for ceTDP2. It has been proposed that ceTDP2 is a dimeric protein, whereas hTDP2 is monomeric (Figure 2B) (18,25). Furthermore, hTDP2 can act in concert with human ZATT to directly reverse TOP2-DPCs without proteolytic processing, while C. elegans lacks a clear ZATT homolog (10). Collectively, these data suggest that although both species have a TDP2 homolog with an N-terminal UBA domain, there may be species-specific differences in Ub-mediated signaling and TDP2 function.
In addition to being a key effector of TOP2-DPC repair, TDP2 is a multifunctional protein that functions in several other non-DNA repair pathways (1–5,53,55). As an important step towards interpreting how TDP2 can participate in Ub-mediated signaling pathways and how truncation and mutation can alter Ub binding, we have described a comprehensive biochemical and biophysical analysis of the interaction between the UBA domain and Ub. This work provides a template by which to examine Ub-dependent links to TDP2 function in DNA repair, as well as other cellular pathways, and how it may be altered by SNPs or expression of mutated TDP2 variants.
DATA AVAILABILITY
Coordinates for TDP2 UBA-Ub crystal form 1 (6Q00) and crystal form 2 (6Q01) are available in the RCSB Protein Data Bank.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge Dr Jason Williams of the NIEHS Mass Spectrometry Research and Support Group for mass-spec analyses, Dr Lars Pedersen of the NIEHS Structure Function Group for X-ray facility support, Dr Jessica Wojtaszek for assistance with SEC-MALS, and Dr Monica Pillon for comments.
Author contributions: Conceptualization—R.S.W., M.J.S; Methodology—M.J.S., C.D.A., J.M.K., J.A.L., F.C.L., R.S.W.; Investigation—M.J.S., C.D.A., A.A.R., L.R.B., J.M.K., J.A.L.; Writing - original draft, M.J.S., R.S.W.; Writing - reviewing and editing, R.S.W., C.D.A., A.A.R., M.J.S., F.C.L.; Funding Acquisition R.S.W., F.C.L.; Supervision R.S.W., F.C.L. The authors declare no competing financial interests.
Notes
Present address: Matthew J. Schellenberg, Department of Biochemistry and Molecular Biology, Mayo Clinic, Rochester, MN 55905, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural research program of the US National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS) [1Z01ES102765 to R.S.W.]; Work in the F.C.L. lab is supported by Ministerio de Economía y Competitividad, Gobierno de España [SAF2017-89619-R, European Regional Development Fund]; European Research Council [ERC-CoG-2014-647359]; University of Seville Predoctoral Studentship [PIF-2011 to J.A.L.]; M.J.S. is supported by Mayo Clinic start-up funds and the Center for Biomedical Discovery new investigator funds; Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory; Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences [W-31-109-Eng-38]; SAXS data were collected at the Advanced Light Source (ALS), a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the Department of Energy, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies (IDAT) program, supported by DOE Office of Biological and Environmental Research. Additional support comes from the National Institute of Health project ALS-ENABLE [P30 GM124169]; High-End Instrumentation Grant [S10OD018483]. Funding for open access charge: US government, Intramural NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Zhang J.Q., Wang J.J., Li W.J., Huang L., Tian L., Xue J.L., Chen J.Z., Jia W.. Cellular protein TTRAP interacts with HIV-1 integrase to facilitate viral integration. Biochem. Biophys. Res. Commun. 2009; 387:256–260. [DOI] [PubMed] [Google Scholar]
- 2. Li C., Fan S., Owonikoko T.K., Khuri F.R., Sun S.Y., Li R.. Oncogenic role of EAPII in lung cancer development and its activation of the MAPK-ERK pathway. Oncogene. 2011; 30:3802–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou C., Shen Q., Xue J., Ji C., Chen J.. Overexpression of TTRAP inhibits cell growth and induces apoptosis in osteosarcoma cells. BMB Rep. 2013; 46:113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vilotti S., Biagioli M., Foti R., Dal Ferro M., Lavina Z.S., Collavin L., Del Sal G., Zucchelli S., Gustincich S.. The PML nuclear bodies-associated protein TTRAP regulates ribosome biogenesis in nucleolar cavities upon proteasome inhibition. Cell Death Differ. 2012; 19:488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Virgen-Slane R., Rozovics J.M., Fitzgerald K.D., Ngo T., Chou W., van der Heden van Noort G.J., Filippov D.V., Gershon P.D., Semler B.L.. An RNA virus hijacks an incognito function of a DNA repair enzyme. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:14634–14639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao R., Schellenberg M.J., Huang S.Y., Abdelmalak M., Marchand C., Nitiss K.C., Nitiss J.L., Williams R.S., Pommier Y.. Proteolytic degradation of topoisomerase II (Top2) enables the processing of Top2.DNA and Top2.RNA covalent complexes by tyrosyl-DNA-phosphodiesterase 2 (TDP2). J. Biol. Chem. 2014; 289:17960–17969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wallace B.D., Williams R.S.. Ribonucleotide triggered DNA damage and RNA-DNA damage responses. RNA Biol. 2014; 11:1340–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andres S.N., Schellenberg M.J., Wallace B.D., Tumbale P., Williams R.S.. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ. Mol. Mutagen. 2015; 56:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cortes Ledesma F., El Khamisy S.F., Zuma M.C., Osborn K., Caldecott K.W.. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009; 461:674–678. [DOI] [PubMed] [Google Scholar]
- 10. Schellenberg M.J., Lieberman J.A., Herrero-Ruiz A., Butler L.R., Williams J.G., Munoz-Cabello A.M., Mueller G.A., London R.E., Cortes-Ledesma F., Williams R.S.. ZATT (ZNF451)-mediated resolution of Topoisomerase 2 DNA-protein cross-links. Science. 2017; 357:1412–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gomez-Herreros F., Zagnoli-Vieira G., Ntai I., Martinez-Macias M.I., Anderson R.M., Herrero-Ruiz A., Caldecott K.W.. TDP2 suppresses chromosomal translocations induced by DNA topoisomerase II during gene transcription. Nat. Commun. 2017; 8:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zagnoli-Vieira G., Bruni F., Thompson K., He L., Walker S., de Brouwer A.P.M., Taylor R., Niyazov D., Caldecott K.W.. Confirming TDP2 mutation in spinocerebellar ataxia autosomal recessive 23 (SCAR23). Neurol Genet. 2018; 4:e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burden D.A., Osheroff N.. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998; 1400:139–154. [DOI] [PubMed] [Google Scholar]
- 14. Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem. Biol. 2013; 8:82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kingma P.S., Osheroff N.. The response of eukaryotic topoisomerases to DNA damage. Biochim. Biophys. Acta. 1998; 1400:223–232. [DOI] [PubMed] [Google Scholar]
- 16. Sabourin M., Osheroff N.. Sensitivity of human type II topoisomerases to DNA damage: stimulation of enzyme-mediated DNA cleavage by abasic, oxidized and alkylated lesions. Nucleic Acids Res. 2000; 28:1947–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deweese J.E., Osheroff N.. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 2009; 37:738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schellenberg M.J., Appel C.D., Adhikari S., Robertson P.D., Ramsden D.A., Williams R.S.. Mechanism of repair of 5′-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nat. Struct. Mol. Biol. 2012; 19:1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schellenberg M.J., Perera L., Strom C.N., Waters C.A., Monian B., Appel C.D., Vilas C.K., Williams J.G., Ramsden D.A., Williams R.S.. Reversal of DNA damage induced Topoisomerase 2 DNA-protein crosslinks by Tdp2. Nucleic Acids Res. 2016; 44:3829–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi K., Kurahashi K., Gao R., Tsutakawa S.E., Tainer J.A., Pommier Y., Aihara H.. Structural basis for recognition of 5′-phosphotyrosine adducts by Tdp2. Nat. Struct. Mol. Biol. 2012; 19:1372–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jackson S.P., Durocher D.. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 2013; 49:795–807. [DOI] [PubMed] [Google Scholar]
- 22. Gatti M., Pinato S., Maiolica A., Rocchio F., Prato M.G., Aebersold R., Penengo L.. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep. 2015; 10:226–238. [DOI] [PubMed] [Google Scholar]
- 23. Cappadocia L., Pichler A., Lima C.D.. Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat. Struct. Mol. Biol. 2015; 22:968–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eisenhardt N., Chaugule V.K., Koidl S., Droescher M., Dogan E., Rettich J., Sutinen P., Imanishi S.Y., Hofmann K., Palvimo J.J. et al.. A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat. Struct. Mol. Biol. 2015; 22:959–967. [DOI] [PubMed] [Google Scholar]
- 25. Rao T., Gao R., Takada S., Al Abo M., Chen X., Walters K.J., Pommier Y., Aihara H.. Novel TDP2-ubiquitin interactions and their importance for the repair of topoisomerase II-mediated DNA damage. Nucleic Acids Res. 2016; 44:10201–10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grice G.L., Nathan J.A.. The recognition of ubiquitinated proteins by the proteasome. Cell. Mol. Life Sci. 2016; 73:3497–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stols L., Gu M., Dieckman L., Raffen R., Collart F.R., Donnelly M.I.. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 2002; 25:8–15. [DOI] [PubMed] [Google Scholar]
- 28. Brzovic P.S., Lissounov A., Christensen D.E., Hoyt D.W., Klevit R.E.. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell. 2006; 21:873–880. [DOI] [PubMed] [Google Scholar]
- 29. Dong K.C., Helgason E., Yu C., Phu L., Arnott D.P., Bosanac I., Compaan D.M., Huang O.W., Fedorova A.V., Kirkpatrick D.S. et al.. Preparation of distinct ubiquitin chain reagents of high purity and yield. Structure. 2011; 19:1053–1063. [DOI] [PubMed] [Google Scholar]
- 30. Tumbale P., Appel C.D., Kraehenbuehl R., Robertson P.D., Williams J.S., Krahn J., Ahel I., Williams R.S.. Structure of an aprataxin-DNA complex with insights into AOA1 neurodegenerative disease. Nat. Struct. Mol. Biol. 2011; 18:1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bernado P., Mylonas E., Petoukhov M.V., Blackledge M., Svergun D.I.. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 2007; 129:5656–5664. [DOI] [PubMed] [Google Scholar]
- 32. Otwinowski Z., Minor W.. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 33. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hospenthal M.K., Mevissen T.E.T., Komander D.. Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest). Nat. Protoc. 2015; 10:349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Datta A.B., Hura G.L., Wolberger C.. The structure and conformation of Lys63-linked tetraubiquitin. J. Mol. Biol. 2009; 392:1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu P., Gan W., Su S., Hauenstein A.V., Fu T.M., Brasher B., Schwerdtfeger C., Liang A.C., Xu M., Wei W.. K63-linked polyubiquitin chains bind to DNA to facilitate DNA damage repair. Sci. Signal. 2018; 11:eaar8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hofmann K., Bucher P.. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 1996; 21:172–173. [PubMed] [Google Scholar]
- 40. Michielssens S., Peters J.H., Ban D., Pratihar S., Seeliger D., Sharma M., Giller K., Sabo T.M., Becker S., Lee D. et al.. A designed conformational shift to control protein binding specificity. Angew. Chem. Int. Ed. Engl. 2014; 53:10367–10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zheng H., Chordia M.D., Cooper D.R., Chruszcz M., Muller P., Sheldrick G.M., Minor W.. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat. Protoc. 2014; 9:156–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoa N.N., Shimizu T., Zhou Z.W., Wang Z.Q., Deshpande R.A., Paull T.T., Akter S., Tsuda M., Furuta R., Tsutsui K. et al.. Mre11 is essential for the removal of lethal Topoisomerase 2 covalent cleavage complexes. Mol. Cell. 2016; 64:580–592. [DOI] [PubMed] [Google Scholar]
- 44. Mao Y., Desai S.D., Ting C.Y., Hwang J., Liu L.F.. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J. Biol. Chem. 2001; 276:40652–40658. [DOI] [PubMed] [Google Scholar]
- 45. Vaz B., Popovic M., Newman J.A., Fielden J., Aitkenhead H., Halder S., Singh A.N., Vendrell I., Fischer R., Torrecilla I. et al.. Metalloprotease SPRTN/DVC1 orchestrates replication-coupled DNA-protein crosslink repair. Mol. Cell. 2016; 64:704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiao H., Mao Y., Desai S.D., Zhou N., Ting C.Y., Hwang J., Liu L.F.. The topoisomerase IIbeta circular clamp arrests transcription and signals a 26S proteasome pathway. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:3239–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee K.C., Bramley R.L., Cowell I.G., Jackson G.H., Austin C.A.. Proteasomal inhibition potentiates drugs targeting DNA topoisomerase II. Biochem. Pharmacol. 2016; 103:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ban Y., Ho C.W., Lin R.K., Lyu Y.L., Liu L.F.. Activation of a novel ubiquitin-independent proteasome pathway when RNA polymerase II encounters a protein roadblock. Mol. Cell. Biol. 2013; 33:4008–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nakada S. Opposing roles of RNF8/RNF168 and deubiquitinating enzymes in ubiquitination-dependent DNA double-strand break response signaling and DNA-repair pathway choice. J. Radiat. Res. 2016; 57:i33–i40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chou A.C., Aslanian A., Sun H., Hunter T.. An internal ribosome entry site in the coding region of tyrosyl-DNA phosphodiesterase 2 drives alternative translation start. J. Biol. Chem. 2019; 294:2665–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang S.N., Dalla Rosa I., Michaels S.A., Tulumello D.V., Agama K., Khiati S., Jean S.R., Baechler S.A., Factor V.M., Varma S. et al.. Mitochondrial tyrosyl-DNA phosphodiesterase 2 and its TDP2(S) short isoform. EMBO Rep. 2018; 19:e42139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ziv I., Matiuhin Y., Kirkpatrick D.S., Erpapazoglou Z., Leon S., Pantazopoulou M., Kim W., Gygi S.P., Haguenauer-Tsapis R., Reis N. et al.. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol. Cell. Proteomics. 2011; 10:M111.009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zucchelli S., Vilotti S., Calligaris R., Lavina Z.S., Biagioli M., Foti R., De Maso L., Pinto M., Gorza M., Speretta E. et al.. Aggresome-forming TTRAP mediates pro-apoptotic properties of Parkinson's disease-associated DJ-1 missense mutations. Cell Death Differ. 2009; 16:428–438. [DOI] [PubMed] [Google Scholar]
- 54. Gomez-Herreros F., Romero-Granados R., Zeng Z., Alvarez-Quilon A., Quintero C., Ju L., Umans L., Vermeire L., Huylebroeck D., Caldecott K.W. et al.. TDP2-dependent non-homologous end-joining protects against topoisomerase II-induced DNA breaks and genome instability in cells and in vivo. PLoS Genet. 2013; 9:e1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Riccio A.A., Schellenberg M.J., Williams R.S.. Molecular mechanisms of Topoisomerase 2 DNA-protein crosslink resolution. Cell. Mol. Life Sci. 2020; 77:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates for TDP2 UBA-Ub crystal form 1 (6Q00) and crystal form 2 (6Q01) are available in the RCSB Protein Data Bank.