Abstract
As cells encounter adverse environmental conditions, such as hypoxia, oxidative stress or nutrient deprivation, they trigger stress response pathways to protect themselves until transient stresses have passed. Inhibition of translation is a key component of such cellular stress responses and mounting evidence has revealed the importance of a class of tRNA-derived small RNAs called tiRNAs in this process. The most potent of these small RNAs are those with the capability of assembling into tetrameric G-quadruplex (G4) structures. However, the mechanism by which these small RNAs inhibit translation has yet to be elucidated. Here we show that eIF4G, the major scaffolding protein in the translation initiation complex, directly binds G4s and this activity is required for tiRNA-mediated translation repression. Targeting of eIF4G results in an impairment of 40S ribosome scanning on mRNAs leading to the formation of eIF2α-independent stress granules. Our data reveals the mechanism by which tiRNAs inhibit translation and demonstrates novel activity for eIF4G in the regulation of translation.
INTRODUCTION
Protein synthesis consumes a significant fraction of cellular energy expenditure. The initiation of this process is tightly regulated to ensure that metabolic conditions are adequate to support this energy intensive process [reviewed in (1)]. This regulatory control is particularly important in cells exposed to adverse conditions that tax energy stores and metabolic resources. Under these conditions, the cellular stress response acts to conserve energetic reserves and redirect resources towards the repair of stress-induced damage to enhance cell survival. Canonical cap-dependent translation occurs when eukaryotic initiation factor (eIF) 4F binds to the m7GTP mRNA cap. eIF4F is a heterotrimeric complex composed of eIF4E, the cap binding protein, eIF4A, a DEAD-box helicase, and eIF4G, a large scaffold protein that facilitates interactions with other eIFs, notably, eIF3 and poly(A) binding protein (PABP). Building upon the eIF4F–cap complex, the 48S pre-initiation complex scans through the 5′ untranslated region (5′ UTR), pausing at the AUG start codon. In response to stress, these processes are modified to reduce general cap-dependent translation, resulting in reduced energy expenditure, and favor non-canonical translation initiated on transcripts bearing upstream open reading frames or IRES elements, resulting in the production of proteins that help cells survive the stress. Mechanistically, phosphorylation of eIF2α by one of four eIF2α kinases or disruption of the eIF4F complex by mTOR-regulated 4E-BP, play major roles in this stress-induced regulatory process. We and others have discovered a stress-activated tRNA-derived non-coding RNA that similarly targets the translation initiation complex to re-program protein synthesis during stress. This process is initiated by the angiogenin-induced cleavage of the anti-codon loop of tRNA to produce 5′- and 3′-tRNA-derived stress-induced RNAs (tiRNAs) (2,3).
Selected tiRNAs that contain a stretch of guanosines at their extreme 5′ end, termed a terminal oligoguanine (TOG) motif, are potent inhibitors of translation (4). This activity is intrinsic to the molecules and not a result of bulk tRNA depletion as is true for other responses (5). Only ∼1% of all tRNAs are cleaved. Translation repression by TOG-containing tiRNAs is a result of displacement of eIF4F from the m7GTP mRNA cap. This results in the formation of stress granules (SGs), stored condensates of stalled pre-initiation complexes (6). Surprisingly, SG formation by tiRNAs is not a result of eIF2α phosphorylation, the canonical mechanism of triggering SG formation. Our initial results suggested that the cold-shock domain containing protein, YB-1, was necessary for tiRNA-mediated eIF4F displacement and translation repression (4); however, follow up work revealed that YB-1 was required for SG formation but not for eIF4F displacement or translation repression (7).
The TOG motif bestows translation inhibition activity by facilitating the formation of tetrameric G-quadruplex (G4) containing molecules (8,9). G4s are formed by stacking of planar structures called G-quartets that form by Hoogsteen base-pairing of four guanosines. G4s have multiple roles in RNA biology as they regulate transcription, mRNA splicing, mRNA localization, translation and more [reviewed in (10)]. In addition to their roles in normal biological processes, G4s have also been implicated in multiple pathological states, such as in C9ORF72-associated amyotrophic lateral sclerosis (ALS) (11). The tetrameric G4-containing form of TOG-containing tiRNAs (G4-tiRNAs) is the bioactive molecule. Ionic conditions that disfavour G4 formation or chemical modifications that prevent G4 formation abolish activity (9).
Here, we present data demonstrating that G4-tiRNAs inhibit translation by directly targeting the HEAT1 domain of eIF4G, the major scaffolding protein necessary for translation initiation. This activity is solely dependent upon eIF4G as all other human proteins are dispensable. We confirm that G4 structures are necessary for this activity, and reveal that eIF4G has G4 binding activity. This mechanism of translation inhibition results in a failure of the scanning step of translation initiation. Pharmaceutical inhibition of scanning has previously been shown to trigger the formation of SGs in a phospho-eIF2α independent manner (12,13). Thus, data presented here reveals new functionality for eIF4G, demonstrates how G4-tiRNAs inhibit translation and induce phospho-eIF2α independent SGs.
MATERIALS AND METHODS
Generation of plasmids
Human eIF4E cDNA was cloned between BamHI and XhoI of pET28a (EMD Millipore) generating pET28-eIF4E. Fragments of eIF4G were amplified with BglII and XhoI linkers by PCR and cloned into pET28a between BamHI and XhoI generating pET28-eIF4G (90–651) and pET28-eIF4G(90–1129). Poliovirus 2A protease was cloned into pET32a between BamHI and XhoI to generate the pET32a-2APro plasmid. pET21b (EMD Millipore) was modified encode a Smt3 tag to aid in solubility of target proteins. Smt3 with an N-terminal 10X His tag was cloned between the XbaI and XhoI sites of pET21b, generating the pET21–10XHis-Smt3 plasmid used in this publication. The HEAT1 domain of eIF4G was amplified and cloned between BamHI and XhoI into this vector generating pET21–10XHis-Smt3-eIF4G (HEAT1). Previously described bicistronic reporters were used as templates for the generation of monocistronic IRES reporters (14). In each case, the upstream firefly luciferase cistron was deleted by digesting with NheI and XbaI. The plasmid backbone was re-ligated to generate the pT7-IRES-NLuc-A50 series of plasmids. Then, NanoLuc was removed by digestion with NdeI and XhoI and replaced with firefly luciferase to generate the pT7-IRES-FLuc-A50 series of plasmids.
Purification of recombinant proteins
All proteins were expressed in Rosetta 2 pLysS Escherichia coli cells after transfection of specific plasmids. Cells were grown at 37°C in 2XYT media until the OD reached 0.6–0.8 and then induced with 1 mM IPTG for 4 h. Cells were pelleted by centrifugation and frozen at –80°C for later use.
eIF4G (HEAT1) was purified by disrupting cells by sonication in 20 mM Tris [pH 8.0], 150 mM NaCl. Lysate was clarified by centrifugation (20 min at 21 000 × g). Lysate was incubated with 2 ml of Ni-NTA agarose (Invitrogen) for 2 h at 4°C. Beads were washed 1× with 20 mM Tris (pH 8.0), 150 mM NaCl, 10 mM Imidazole then 1x with 20 mM Tris (pH 8.0), 500 mM NaCl, 10 mM Imidazole and then 1× with 20 mM Tris (pH 8.0), 150 mM NaCl, 10 mM Imidazole before elution with 20 mM Tris (pH 8.0), 150 mM NaCl, 250 mM Imidazole. Eluted protein was dialyzed overnight into 10 mM Sodium Phosphate (pH 7.5), 300 mM NaCl, 2 mM β-Mercaptoethanol, 5 mM MgCl2, 5% glycerol. The following day, the protein was dialyzed against fresh buffer for an additional 4 h. Precipitate was cleared by centrifugation and supernatant was concentrated with Amicon Ultra-15 Centrifugal Filter Unit (NMWL 30 kDa). Finally, ATP was added to a final concentration of 1 mM. eIF4E was purified by the same manner except it was dialyzed against 20 mM Tris [pH 8.0], 150 mM NaCl, 0.1 mM EDTA, 10% glycerol. Poliovirus 2A protease was purified under denaturing conditions and resolubilized as previously described (15). Purified 2A protease was concentrated with Amicon Ultra-15 Centrifugal Filter Unit (NMWL 10 kDa) and stored at –20 in TMO.1 buffer (50 mM Tris [pH 7.4], 20% glycerol, 1 mM EDTA, 1 mM DTT, 12.5 MgCl2, 100 mM KCl).
Purification of eIF4F and cap binding assay
Highly purified eIF4F was purified from rabbit reticulocytes as previously described (16). A graphical representation of the purification schema is shown in Figure 1A. For purification of eIF4E:eIF4G complexes from E. coli, pET28-eIF4E, pET28-eIF4G (90–651), or pET28-eIF4G (90–1129) were transformed into Rosetta2 pLysS cells, grown in 2XYT media until OD reached 0.6 and then induced with 1 mM IPTG for 4 h. Aliquots of bacteria were spun down (1 ml for eIF4E and 10 ml for eIF4G) and frozen at –80°C. For purification, frozen aliquots were thawed on ice and resuspended in 20 mM Tris [pH 8.0], 150 mM NaCl, 0.5 mg/ml lysozyme. Cells were further disrupted by sonication and lysate was clarified by centrifugation. 10% NP-40 was added to supernatant to a final concentration of 0.5%. Individual lysates or combined lysates were incubated with m7GTP-agarose (Jena Biosciences) for 2 h and washed 3× with 20 mM Tris [pH 8.0], 150 mM NaCl, 0.5% NP-40. Cap competition assays were performed with small RNAs as described previously (4).
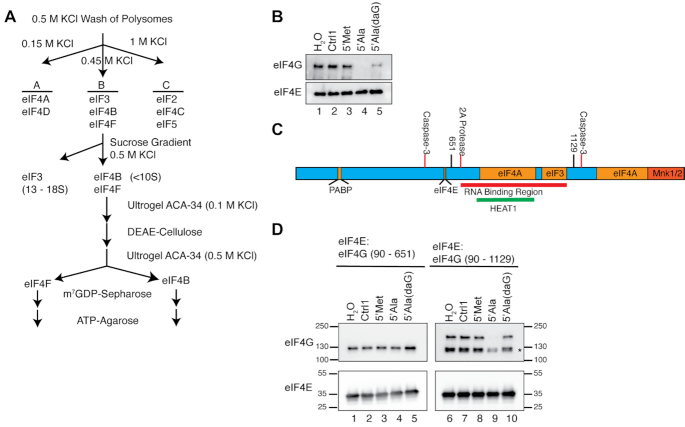
Figure 1.
G4-tiRNAs directly target eIF4F. (A) Strategy of purification of eIF4F adapted from (16). (B) Purified human eIF4F is sensitive to TOG containing tiRNAs in a G-quadruplex dependent manner. Purified eIF4F was bound to m7GTP-agarose and challenged with indicated small RNAs. 5′tiRNAAla efficiently disrupted the eIF4F complex, but 5′tiRNAAla(daG), which cannot form a G4 efficiently had reduced activity. (C) Schematic of eIF4G indicating domains required for interaction with other proteins (orange), protease cleavage sites (red lines), sites of truncations (black lines). RNA Binding region (red bar) and HEAT1 repeat (green bar) are indicated. (D) Recombinant eIF4G containing the RNA binding region is sensitive to 5′tiRNAAla. eIF4E and truncation of eIF4G were expressed and purified from E. coli, bound to m7GTP-agarose and challenged with indicated RNAs. 5′tiRNAAla could disrupt eIF4F when eIF4G retained the RNA binding regions (90–1129). This disruption was dependent upon G4 formation as 5′tiRNAAla(daG) is incapable of disrupting eIF4F.
Protease cleavage reactions and cleavage product capture
One 10-cm2 plate of U2OS cells were lysed in 900 μl of NP-40 Lysis Buffer (20 mM Tris [pH 8.0], 150 mM NaCl, 0.5% NP-40) to which the optimal amount of recombinant 2A protease needed for cleavage was determined. Cleavage reactions were carried out in lysis buffer at room temperature for 2 hours while tumbling. Cleaved eIF4G was captured using eIF4E-saturated m7GTP-agarose (Jena Biosciences). eIF4E-saturated m7GTP-agarose was generated by adding 20 μl of purified eIF4E (100 pmol) to 100 μl of m7GTP-agarose and tumbled for 2 hours at 4°C. Beads were washed extensively with NP-40 lysis buffer. For generation of Casp3 cleaved eIF4G, U2OS cells were treated for 16 hours with 500 μM CisPt to induce apoptosis. Cell lysates were prepared with NP-40 Lysis Buffer (20 mM Tris [pH 8.0], 150 mM NaCl, 0.5% NP-40). Cleaved eIF4G fragments were captured from lysate with eIF4E saturated m7GTP-agarose (Jena Bioscience). Cap competition assays were performed with small RNAs as described previously (4).
Electrophoretic mobility shift assays
Synthetic RNA was end labeled with ATP, [γ-32P]-3000 Ci/mmol (Perkin Elmer) using T4 Polynucleotide Kinase (New England Biolabs). RNA was purified by gel filtration with Illustra MicroSpin G-25 Columns (GE Healthcare). 1 pmol of purified RNA was used per reaction and indicated amounts of recombinant protein. RNA and proteins were mixed in 10 mM sodium phosphate (pH 7.5), 300 mM NaCl, 2 mM β-mercaptoethanol, 5 mM MgCl2, 5% glycerol, 1 mM ATP and incubated at room temperature for 10 min before loading onto 5% acrylamide gel (29:1 Acrylamide:Bis-acrylamide, 0.5× TBE, 5% glycerol). Gels were dried and visualized by autoradiography. For cold competition assays, after 10 min of incubation at room temperature, 1 μl of RNA at indicated concentration was added and complexes were incubated for a further 10 min at room temperature before separating complexes by electrophoresis.
Microscale thermophoresis assay
MicroScale Thermophoresis (MST) experiments were performed according to the instrumental protocol in a Monolith NT.115 (red/blue) instrument (NanoTemper Technologies). Serial dilutions of eIF4G-HEAT1 were made using a buffer containing 10 mM sodium phosphate (pH 7.5), 300 mM NaCl, 2 mM β-mercaptoethanol, 5 mM MgCl2, 5% glycerol, and 1 mM ATP. 3′-FAM labeled RNA oligos (IDT) were used as targets for the binding experiments. RNA solutions were heated at 95°C in presence of 100 mM KCl and allowed to cool down to room temperature to facilitate G4 formation. RNA concentration was kept constant at 20 nM throughout the experiments. The RNA-protein mixture was incubated at room temperature for 15 min before running the binding efficiency test experiments. MST experiments were performed using 40% and 60% MST power and between 20–80% LED power at 24 °C. The MST traces were recorded using the standard parameters: 5 s MST power off, 30 s MST power on and 5 s MST power off.
The data presented here are the average of 3 independent experiments. Average normalized fluorescence (%) was plotted against HEAT1 concentration to determine the binding constant (Kd). Ligand depletion model with one binding site was used (GraphPad Prism 8) to fit the binding which follows the following model: Y = Bmax*X / (Kd + X)
Crosslinking assays
In vitro transcription was preformed using synthetic oligonucleotides containing the T7 RNA polymerase promoter and the 5′ UTR of pT7-FLuc-A50 by the method originally described by (17) with modifications described in (18). The sequence of the transcribed RNA was: 5′ – GGGAAUUCACCGGUACUACUGUCAGCGCUAGC – 3′. Transcribed RNAs were PAGE purified overnight, ethanol precipitated and resuspened in H2O. RNAs were capped using vaccinia capping enzyme (New England Biolabs) according to manufactures instructions. 5 pmol of capped, radiolabelled RNA was incubated with 5 μl of rabbit reticulocyte lysate (Promega) and 100 pmol of indicated tiRNAs. Reactions were incubated on ice for 10 min and then crosslinked in Stratolinker (1.6 J). Crosslinked complexes were denatured in SDS-loading dye and heated to 100°C for 10 min before running on 4–20% Novex gel. Gels were dried and complexes were visualized by autoradiography.
In vitro translation assays
pT7-FLuc-A50, pT7-PV-FLuc-A50, and pT7-EMCV-FLuc-A50 were linearized with NotI and transcribed with HiScribe T7 High yield RNA Synthesis Kit according to manufacturer's instructions (New England Biolabs). For NanoLuc reporters with differing 5′ UTR lengths, pNL1.1 (Promega) was used as a PCR template. Forward primers contained a T7 promoter for transcription and were positioned to generate the indicated 5′ UTR length. Reverse primer added a synthetic poly(A) tail. RNAs was transcribed with HiScribe T7 High yield RNA Synthesis Kit according to manufacturer's instructions (New England Biolabs). In vitro translation assays were performed as previously described (4).
RESULTS
G4-tiRNAs directly act upon eIF4F components
We began our investigation of whether G4-tiRNAs directly targeted eIF4F by using eIF4F purified from rabbit reticulocytes. Previously, we had purified eIF4F from cellular lysates using a single step m7GTP-agarose purification. However, this approach co-purifies hundreds of cap-associated proteins (19,20). Therefore, we used a more stringent purification strategy to purify eIF4F away from other cellular proteins (Figure 1A). We chose 5′tiRNAAla as the prototypical G4-tiRNA as we have previously characterized this RNA species structurally and functionally (4,7–9). We bound purified eIF4F to m7GTP-agarose and challenged eIF4F with various control RNAs or 5′tiRNAAla. We used the inactive 5′tiRNAMet as a control as it does not repress translation, contain a TOG motif or form a G-quadruplex (4,9). Control RNAs (Ctrl1 and 5′tiRNAMet) did not affect the association of eIF4F with m7GTP-agarose, but 5′tiRNAAla efficiently displaced eIF4G as has been previously shown with eIF4F from 1-step m7GTP-agarose purification (4). Our previous work showed that 5′tiRNAAla-mediated displacement of eIF4F requires the G4 structure as this function is attenuated by 7-deazaguanosine substitutions that prevent G4 assembly (9). In line with this data, 7-deazaguonisine substituted 5′tiRNAAla is reduced in its ability to repress translation or trigger the formation of stress granules. In a similar fashion, 7-deazaguanosine (daG) substituted 5′tiRNAAla is reduced in its ability to displace purified eIF4F complexes from m7GTP-agarose (Figure 1B, ln 5). The purification schema used here has been shown to reliably purify eIF4F from other cap associated proteins (16); however, to ensure that 5′tiRNAAla does not target contaminating human proteins, we assembled complexes composed of Escherichia coli-derived recombinant proteins. A full biochemical interrogation of eIF4G has been stymied by the inability to recombinantly express and purify the full-length protein. For our purposes, we used the ability of properly folded eIF4G to interact with eIF4E as a basis for purification. Properly folded, active eIF4E was purified from E. coli using m7GTP-agarose and properly folded, active eIF4G was purified based on its ability to interact with this protein (Supplementary Figure S1A, B). We used this system to interrogate domains that might be necessary for these functional activities. eIF4G contains a poorly defined RNA binding region roughly spanning amino acids Glycine-682 to Arginine-1115 (Figure 1C). We expressed and purified two truncations of eIF4G, one spanning amino acids 90–651 that lacks the RNA binding region (RBR-) and a second spanning amino acids 90–1129 that includes the RNA binding region (RBR+). By challenging these complexes with 5′tiRNAAla or control RNAs, we showed that 5′tiRNAAla is incapable of displacing the RBR– fragment (Figure 1D, left panel). However, the RBR+ fragment is sensitive to 5′tiRNAAla (Figure 1D, right panel). Fortuitously, the RBR+ fragment co-purified with a C-terminal truncation of the approximate size of the RBR– fragment (*). The location of the antibody epitope confirms that this is a C-terminal and not an N-terminal truncation (Supplementary Figure S2A). This co-purifying fragment, which must lack much of the RBR, is less sensitive to 5′tiRNAAla, thus serving as an ideal internal control for the requirements of displacement. Further, neither full length RBR+ fragment, nor the truncation, is sensitive to 7-deazaguanosine substituted 5′tiRNAAla. Thus, these data show that displacement of eIF4G from m7GTP caps by G4-tiRNAs is independent of other human proteins, requires the RNA binding region of eIF4G, and the G-quadruplex structure of tiRNAs.
The RNA binding region of eIF4G is necessary and sufficient for G4-tiRNA-mediated displacement
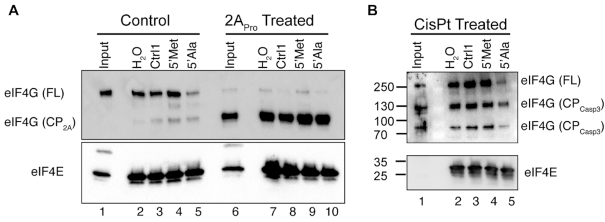
eIF4G interacts with multiple protein binding partners and has numerous post-translational modifications, all of which would be absent from the E. coli-derived recombinant protein. To confirm that the insensitivity of the RBR- fragment is due to the lack of the RNA binding region and not an artifact due to expression in E. coli, we returned to human cells. Owing to its central role in regulating translation, eIF4G is a target of multiple cellular and viral proteases. Following infection by poliovirus, eIF4G is cleaved by virally expressed 2A protease (2APro) between Arginine-681 and Glycine-682, separating the eIF4E binding site from the RNA binding region (Supplementary Figure S2A). We treated U2OS cell lysate with recombinantly expressed 2APro and determined the amount of protease needed to fully cleave endogenous eIF4G (Supplementary Figure S2B, C). The eIF4G cleavage product resulting from 2APro cleavage (CP2A) was purified by interaction with recombinant eIF4E bound to m7GTP-agarose. Pull-downs from untreated and treated lysates were challenged with 5′tiRNAAla or control RNAs. As before, untreated eIF4G remained sensitive to 5′tiRNAAla despite being captured by recombinant eIF4E (Figure 2A). However, eIF4G (CP2A), which lacks the RNA binding region, was insensitive to 5′tiRNAAla, confirming data from eIF4G truncations expressed in E. coli. eIF4G is also targeted by cleaved Caspase-3 (Casp3) upon the induction of apoptosis. In contrast to 2APro, Casp3 targets two different sites on eIF4G, generating a central cleavage product between amino acids 532 and 1176 (Supplementary Figure S2A). The cleavage product that is purifiable by its interaction with eIF4E on m7GTP-agarose lacks the N- and C-termini of endogenous eIF4G. We triggered Casp3-mediated eIF4G cleavage by treating cells with cisplatin (CsPt) (Supplementary Figure S2D), a potent apoptosis inducing chemotherapeutic agent that results in eIF4G cleavage (21). We confirmed that the central cleavage product was affinity purified based on its interaction with eIF4E on m7GTP-agarose, but the N-terminal cleavage product was not (Supplementary Figure S2E, F). We tested if the resultant cleavage product (CPCasp3) remained sensitive to tiRNAs as before. Indeed, owing to the fact that this cleavage product retains the RNA binding regions, it remained sensitive to 5′tiRNAAla (Figure 2B). These data support our previous data that G4-tiRNAs directly target eIF4F (Figure 1) and reveal that the central RNA binding region is both necessary and sufficient for targeting of endogenous eIF4G.
Figure 2.
Endogenously expressed eIF4G requires the RNA binding region for G4-tiRNA activity. (A) eIF4G was cleaved with poliovirus 2A protease which separated the eIF4E binding site from the RNA binding region. This fragment was purified on m7GTP-agarose based on its ability to interact with eIF4E. Full length (FL) eIF4G is sensitive to 5′tiRNAAla but the 2A protease cleavage product (cp2A), which lacks the RNA binding region, is insensitive. (B) eIF4G was cleaved by activated caspase3 after induction of apoptosis by cisplatin. The cleavage product containing the eIF4E binding site retains the RNA binding region but lacks the N- and C-termini. This cleavage product (cpcasp3) was purified on m7GTP-agarose and challenged with different small RNAs. It remained sensitive to 5′tiRNAAla.
G4-tiRNAs directly interact with the HEAT1 domain of eIF4G
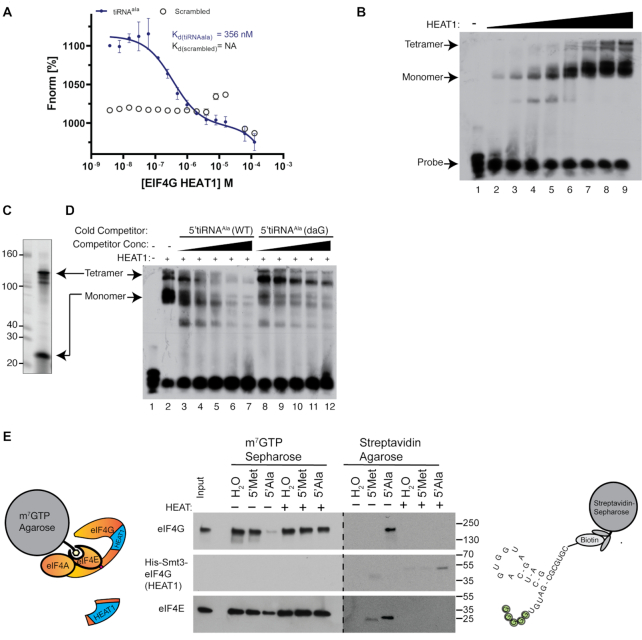
Within the RBR is a conserved structural domain known as a HEAT repeat. HEAT repeats are found in a wide variety of proteins and play a variety of roles. They are composed of alternatively stacked α-helices (22). Throughout eIF4G, there are 3 distinct HEAT repeats and thus, this initial repeat is referred to as HEAT1. This domain has been shown to have RNA binding activity and thus we hypothesized that G4-tiRNAs may directly interact with the HEAT1 domain. Thus, we expressed and purified the HEAT1 repeat in E. coli (Supplementary Figure S3). We performed microscale thermophoresis (MST) and RNA electrophoretic mobility shift assays (rEMSA) to quantify the affinity of G4-tiRNAs for the eIF4G HEAT1 domain (Figure 3A, B). RNA binding analysis by MST revealed that the HEAT1 repeat of eIF4G interacted with 5′tiRNAAla but not with a scrambled G-rich control RNA (Figure 3A). Notably, the affinity of HEAT1 for 5′tiRNAAla is higher than the reported affinity of HEAT1 for the EMCV IRES (Kd = 0.41 μM versus 1.3 μM, respectively) (23). MST analysis using a scrambled control revealed no interaction (Figure 3A). By performing rEMSA analysis, we confirmed that the HEAT1 domain interacts with 5′tiRNAAla (Figure 3B). Unexpectedly, HEAT1 shifted 5′tiRNAAla as two distinct complexes. This was reminiscent of the two conformers that 5′tiRNAAla adopts in isolation: a monomeric conformer and a G-quadruplex containing tetrameric conformation (Figure 3C). To confirm that these complexes reflect monomeric single stranded (ss)RNA-bound HEAT1 and tetrameric G-quadruplex (G4)-bound HEAT1, we performed cold competition assays (Figure 3D). HEAT1 was bound to radiolabeled 5′tiRNAAla and competed with cold unmodified 5′tiRNAAla (WT) or 7-deazaguanosine modified 5′tiRNAAla [5′Ala(daG)] (9). As expected, competition of radiolabeled 5′tiRNAAla with cold 5′tiRNAAla competed both complexes. However, competition of radiolabeled 5′tiRNAAla with cold 5′tiRNAAla (daG) competed the faster moving complex with the same efficiency as 5′tiRNAAla (WT). However, competition of the more slowly moving complex was severely impaired. These results indicate that HEAT1 is capable of binding to ssRNA, as has previously been shown (24,25), but also to G4-containing RNA. Finally, we determined whether purified recombinant HEAT1 could block the ability of 5′tiRNAAla to disrupt the eIF4F complex. We used m7GTP-agarose to pull down eIF4F complexes from cell lysates, then challenged with tiRNAs or control RNAs in the absence or presence of recombinant HEAT1 (Figure 3E). The ability of 5′tiRNAAla to displace eIF4G (and a portion of eIF4E) was abolished in the presence of recombinant HEAT1 (Figure 3E, lane 7). As the RNAs used in this experiment contained a 3′ biotin moiety, we were able to recover them and the proteins from the supernatant using streptavidin-sepharose. In the absence of HEAT1, a portion of displaced eIF4G and eIF4E became associated with biotin-5′tiRNAAla (Figure 3E, lane 10). In the presence of HEAT1, biotin-5′tiRNAAla pulled down His-HEAT1, but not eIF4G or eIF4E (Figure 3E, lane 13). Thus, the interaction between 5′tiRNAAla and the HEAT1 repeat of eIF4G is necessary for 5′tiRNAAla activity.
Figure 3.
The HEAT1 repeat of eIF4G interacts with 5′tiRNAAla and is required for activity. (A) eIF4G(HEAT1) was expressed and purified from E. coli. Microscale thermophoresis demonstrate that this domain interacted with 5′tiRNAAla (closed circles), but not a scrambled G-rich control (open circles). (B) RNA electrophoresis mobility shift assays (rEMSAs) confirm the interaction between eIF4G(HEAT1) and 5′tiRNAAla and also show that two different complexes are shifted. Lanes 1–9 were shifted with 0, 1.5, 3, 6, 12.5, 25, 50, 100 and 125 pmol of protein. (C) 5′tiRNAAla in the absence of proteins exists as two conformers as previously shown (9). (D) In a cold competition assay, the upper tetrameric complex is efficiently competed by unmodified 5′tiRNAAla. However, the upper band is less sensitive to 5′tiRNAAla(daG), which cannot assemble into G-quadruplex structures. Lanes 3 & 8, 4 & 9, 5 & 10, 6 & 11, 7 & 12 were competed with 6.25, 12.5, 25, 50 and 100 pmol of cold competitors, respectively. (E) 5′tiRNAAla’s interaction with the eIF4G(HEAT1) domain is required for tiRNA activity. m7GTP binding assays were performed with or without supplementation with recombinant eIF4G(HEAT1). Supplementation with this recombinant protein blocked 5′tiRNAAla activity against eIF4F. Upon recovery of tiRNA bound proteins by streptavidin-sepharose, eIF4G was purified without supplementation and eIF4G(HEAT1) was purified with supplementation.
G4-tiRNAs inhibit the scanning step of translation initiation
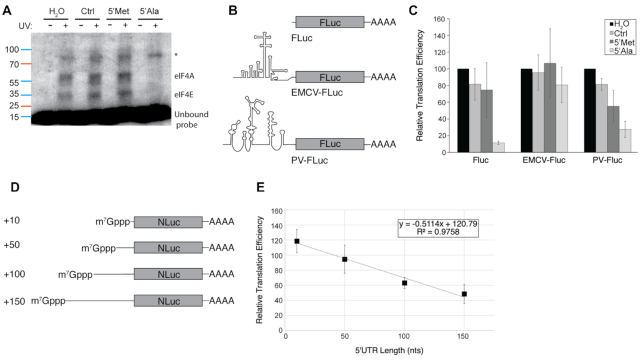
The heterotrimeric eIF4F complex is necessary for cap-dependent translation initiation, with each individual subunit playing distinct roles in the process. The HEAT1 domain of eIF4G is required for the scanning step of translation initiation (26). Additionally, eIF4A plays an outsized role in promoting efficient scanning. We confirmed that eIF4A was displaced from the 5′ UTRs of mRNAs through UV crosslinking. Rabbit reticulocyte lysates were incubated with capped radiolabled RNAs that comprised the 5′ UTR of a luciferase reporter and challenged with control RNAs or tiRNAs. Complexes were crosslinked, resolved on an SDS-PAGE gel and visualized by autoradiography. We note loss of crosslinks corresponding to proteins the size of eIF4A and eIF4E (Figure 4A). This corroborates our earlier data that demonstrated that eIF4A was displaced along with eIF4G in cap binding assays (4). Surprisingly, there is an additional, unidentified crosslink that is unaltered by 5′tiRNAAla. As tiRNAs target the HEAT1 domain and displace eIF4A, we sought to determine whether they specifically inhibited scanning. To this end, we assayed the ability of tiRNAs to repress the translation of an uncapped reporter or an uncapped reporter harbouring EMCV-A6 or Poliovirus (PV) IRES elements (Figure 4B). Our previous work showed that translation driven from the EMCV-A7, but not the EMCV-A6, IRES is inhibited by tiRNAs (4). The A7 variant binds with lower affinity binding to the eIF4G HEAT1 domain, suggesting a possible mechanism for the differential sensitivity to tiRNAs. Here we compared the ability of tiRNAs to inhibit translation initiated at PV or EMCV-A6 IRESes because both are picornavirus sequences that require a similar set of trans acting factors (TAFs) to initiate cap-independent translation [reviewed in (27)]. However, the biggest difference between the two is the location of the start codon relative to the assembly point of the ribosome and TAFs. EMCV assembles these factors immediately adjacent to the start codon such that minimal scanning of the ribosome is necessary prior to start codon recognition. In contrast, PV IRES is located greater than 400 nt upstream of the start codon requiring the pre-initiation complex to scan the 5′ UTR to reach the start codon. We quantified translation using firefly luciferase reporters assayed for luminescence (Figure 4C). Using this system, we confirmed that 5′tiRNAAla potently inhibits translation from uncapped transcripts, but not from EMCV-A6 driven transcripts. In contrast, despite requiring many of the same factors, PV IRES is potently inhibited by 5′tiRNAAla, suggesting, but not proving, that 5′tiRNAAla specifically effects the scanning step of translation initiation. Interestingly, 5′tiRNAMet, a tiRNA previously categorized as inactive, was also able to repress translation initiation from the poliovirus IRES suggesting that it may possess activity in a context-dependent manner.
Figure 4.
5′tiRNAAla blocks the scanning step of translation initiation. (A) 5′tiRNAAla abolishes eIF4E and eIF4A crosslinks to a synthetic 5′UTR. A capped radiolabelled RNA was in incubated in rabbit reticulocyte lysate with indicated unlabelled RNAs. Lystates were crosslinked, run on an SDS-PAGE gel and visualized by autoradiography. Results indicate that eIF4A is efficiently displaced from the RNA along with eIF4E. An unidentified protein (*) is unaffected by 5′tiRNAAla. (B) Firefly luciferase reporter constructs were generated without an IRES, with the EMCV IRES or with the poliovirus IRES. (C) 5′tiRNAAla blocks uncapped RNA translation but EMCV IRES is refractory to 5′tiRNAAla activity as monitored by luciferase activity. However, poliovirus IRES driven translation is sensitive to 5′tiRNAAla suggesting that scanning is the sensitive step. (D) Capped NanoLuc reporters were generated with differing 5′ UTR lengths to increase the requirement for scanning (E) As 5′ UTR length and requirement for scanning increases, so does the translation inhibitory activity of 5′tiRNAAla as measured by luciferase activity. The effect on each individual 5′UTR reporter was normalized to an in vitro translation assay completed without treatment with 5′tiRNAAla.
We previously showed that capped mRNAs are less sensitive to tiRNA-mediated repression than uncapped mRNAs. In light of data presented above, we thought that perhaps the reason for the reduced sensitivity of capped mRNA reporters was the relatively short (∼25 nt) 5′ UTR of these constructs, thereby reducing the necessity of scanning. If true, by increasing the 5′ UTR and thus scanning dependency, we might increase the sensitivity of capped reporters to 5′tiRNAAla. Genome wide studies using Silvestrol showed that mRNAs with longer 5′ UTRs were more sensitive to inhibition of eIF4A (28). Thus, we generated a series of NanoLuc reporters with 5′ UTRs of 10, 50, 100 or 150 nt (Figure 4D). Indeed, the translation repression activity of 5′tiRNAAla increased as a function of the 5′ UTR length (Figure 4E), strongly implicating the scanning step as the inhibited step of translation repression.
DISCUSSION
Under optimal cellular conditions tRNAs serve their role by delivering amino acids to an elongating polypeptide chain. However, they also can take on various non-canonical roles, one of which is through stress-dependent inhibition of translation initiation. In humans and in response to stress, ANG or other members of the RNase A superfamily target the anticodon loop of tRNAs (2,3,29). Cleavage at this site generates two smaller ncRNAs that we termed tRNA-derived stress-induced RNAs (tiRNAs), but have also been referred to as tRNA halves or more generally as tRNA fragments. Those 5′tiRNAs that have a stretch of 5 guanosines at their 5′ end are potent inhibitors of translation (4). We termed this sequence motif the terminal oligoguanine (TOG) motif. We have shown that this sequence is important because it allows for the formation of tetrameric TOG-containing tiRNAs that assemble via a G-quadruplex (9). Additionally, the assembly of these tiRNA structures may be neuroprotective (8). The manner in which they inhibit translation initiation results in the formation of stress granules; however, unlike canonical stress granule formation these form independently of eIF2α phosphorylation (6). Their formation does depend upon the RNA binding protein YB-1 (7). Despite all of this work, we have not been able to elucidate the mechanism through which tiRNAs inhibit translation until now. Here we show that the G-quadruplex structures of TOG-containing tiRNAs interact with the HEAT1 repeat of eIF4G. This interaction disrupts the stability of eIF4F, of which eIF4G is a major component, on the m7GTP mRNA cap. Failure to assemble eIF4F on the m7GTP cap specifically inhibits the scanning of the 48S ribosome. These data now explain how tiRNAs can trigger the formation of stress granules in a phospho-eIF2α independent manner. Pharmacological inhibition of scanning by rocaglamide A or pateamine A similarly results in stress granule formation without phosphorylation of eIF2α.
The RNA binding region of eIF4G has been vaguely defined, but is conserved amongst eukaryotes. In yeast, there are three regions capable of binding RNA and this activity is necessary for survival (24). Yeast eIF4G has an affinity for poly(U) stretches and eIF4G binding to these stretches may regulate their translation (25). Mammalian eIF4G can bind cellular mRNA and its RNA binding activity is required for efficient interaction of eIF4E with the cap structure (30). However, the RNA binding properties of eIF4G have been best studied in the context of viral internal ribosome entry sites (IRESes). In plants, the Barley Yellow Dwarf Virus IRES binds eIF4G to drive cap-independent translation initiation (31). In humans, the poliovirus (PV) IRES recruits eIF4G to drive cap-independent translation initiation although the precise amino acids required for IRES binding are not known (32). Similarly, the encephalomyocarditis virus (EMCV) IRES also directly interacts with eIF4G and has the best described structural data (23). The HEAT1 repeat of eIF4G directly interacts with the J/K domain of the EMCV IRES.
This finding does suggest a level of selectivity in tiRNA-mediated translational inhibition. This was first suggested by the finding that upon transfection into cells, tiRNAs inhibit 10–20% of global protein synthesis, not 100%. Our present findings suggest that mRNAs that are most dependent upon eIF4G are most sensitive to tiRNA-mediated translation repression. In yeast, knockout of both eIF4G paralogs is lethal (33), but ablation of eIF4G does not result in complete translational arrest (34). In fact, despite its presumed central role in assembling the pre-initiation complex, degron-mediated depletion of eIF4G only inhibits the translation of 94 mRNAs (1.6%), whereas the translation efficiency of 99 mRNAs is increased. Similar effects were seen in normal immortalized human cells (MCF10A) upon siRNA-mediated knockdown of eIF4GI (35). Thus, it is possible that rather than globally altering mRNA translation, inactivation of eIF4G by tiRNAs may alter the translation of a subset of mRNAs. Amongst the most likely targets are mRNAs that harbor terminal oligopyrimidine (TOP) tracts. These mRNAs largely encode components of the translation machinery, notably, all mRNAs encoding ribosomal proteins. Their translation is highly sensitive to environmental conditions. In fact, the translation of these mRNAs is largely dependent upon eIF4G. Pharmacological inhibition of mTOR leads to the dephosphorylation of 4E-BP proteins which then competes with eIF4G for interaction with eIF4E. Under these conditions, the most sensitive mRNAs are those encoded by TOP-mRNAs (36). Additionally, this result is phenocopied by knockdown of eIF4G in human cells (35). Moreover, eIF4G aids in stabilizing eIF4E on the m7GTP cap (37). Thus, disruption of eIF4G could have an additive effect by reducing the interaction of eIF4E with mRNAs as has been seen in our experiments. Like eIF4G, certain mRNAs are more dependent upon eIF4E. Whereas 50% reduction in the levels of eIF4E has little effect on global translation rates, translation of a subset of mRNAs is significantly reduced (38). Similar results have been seen in cultured cell as well (39). Similarly, overexpression of eIF4E has the ability to transform NIH3T3 and Rat-2 fibroblasts (40) and these data have been confirmed in mice (41).
The translation of a subset of mRNAs may actually be increased upon destabilization of eIF4F. Here the most likely transcripts are those containing upstream open reading frames (uORFs). These mRNAs are typically translationally silenced as the uORF prevents the ribosome from decoding the true ORF. However, in certain cellular conditions, notably under stress, leaky scanning allows for bypassing of the uORF and recognition of the true ORF. Under conditions in which scanning is disrupted by tiRNAs, it is conceivable that uORF containing transcripts would be upregulated. Further we should note that that while G4-tiRNAs displace components of eIF4F from reporter RNAs (Figure 4A), there is at least one unidentified protein that is insensitive. As of yet, we have not confirmed the identity of this protein, but, based on its molecular weight, two of the most likely candidates are eIF4B or eIF3D. The fact that eIF4B is recruited to mRNAs in a cap-dependent manner would argue against it, but it does not rule it out. Alternatively, eIF3D has been shown to be an alternative cap binding protein that can drive an eIF3-dependent eIF4F-independent mode of translation initiation (42). It is tempting to speculate that these mRNAs, such as c-Jun, may be insensitive to G4-tiRNA mediated translation inhibition. More recent data suggests that translation that is eIF4F-independent, but still cap-dependent, is widespread in cells (43). tiRNAs could shift the balance from eIF4F-depentent translation to eIF3D-DAP5 dependent translation. Another study demonstrates that threonyl-tRNA synthetase (TRS) can drive translation in an eIF4F-independent manner (44). TRS can function in a similar manner to eIF4G by binding eIF4E homolog protein (4EHP/eIF4E2), which binds the mRNA cap, and eIF3, PABP and eIF4A.
Our work also raised another possibility regarding the wider role of tiRNAs in cellular biology. We have previously characterized G4-tiRNAs as the most potent inhibitors of cellular translation. This was based on the use of a single mRNA reporter. However, results presented here (Figure 4), suggest that, depending upon the mRNA used, other tiRNAs, such as 5′tiRNAMet may function as translational repressors. Given the wide variety of mechanisms by which translation is regulated, it would not be surprising if other tiRNAs emerge as inhibitors of specific subsets of RNAs. In fact, the differential generation of tiRNAs in response to different stresses suggests that there may be selectivity in this process (45).
Perhaps the most surprising finding of our current study is that eIF4G possesses G-quadruplex binding ability. This is particularly relevant with regards to non-canonical translation initiation. As discussed above, viruses have adopted several mechanisms of recruiting eIF4G to mRNAs to promote cap-independent translation initiation. However, there are many ways that cellular mRNAs can also promote translation via cap-independent mechanisms. Systematic approaches have suggested that over 500 mRNAs may host elements that promote cap-independent translation activity (46). Some mRNAs may accomplish this through Watson-Crick base-pairing with the 18S rRNA (47,48) or by affinity to ribosomal proteins (49,50). Some mRNAs may harbor cap-independent translational enhancers (CITE) that help them compete for the translation machinery and may aid in promoting translation when cap recognition is suppressed (51,52). Finally, and most controversially, some cellular mRNAs may contain IRES elements that are analogous to those found in viral RNAs [reviewed in (53–55)]. The utilization of cellular IRESes seems to be particularly relevant during stress, when cap-dependent translation is inhibited [e.g. (56–58)]. Surprisingly, many of the reported cellular IRESes, such as VEGF, NRF2, FGF2 and SNCA, have been shown to contain G4s and the formation of G4s is required for cap-independent translation (59–62). Other cellular G-quadruplexes are associated with non-canonical translation processes known as repeat associated non-ATG (RAN) translation. The best studied instance of this is in the GGGGCC repeat expansion of C9ORF72 that is causative for Amyotrophic Lateral Sclerosis (ALS). Here, non-canonical RAN translation produces toxic dipeptide repeats (DPRs) that contribute to neuronal cell death [reviewed in (63)]. In light of the data presented here, it is possible that upon inhibition of cap-dependent translation, eIF4G could be directly recruited to certain G-quadruplexes contained within human IRES-like structures. Since eIF4G is the scaffold upon which the rest of the translation initiation machinery is built, its direct recruitment of it to an mRNA could facilitate cap-independent translation. In fact, we have previously presented data that tiRNAs may protect cells against C9ORF72-mediated cell toxicity (8). Further work exploring this possibility should provide many insights in the future.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM124458 to S.M.L.,GM128981 to W.C.M., GM126150 to P.I., GM126901 to P.A.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Holcik M., Sonenberg N.. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005; 6:318–327. [DOI] [PubMed] [Google Scholar]
- 2. Yamasaki S., Ivanov P., Hu G.F., Anderson P.. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009; 185:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akiyama Y., Lyons S.M., Fay M.M., Abe T., Anderson P., Ivanov P.. Multiple ribonuclease A family members cleave transfer RNAs in response to stress. 2019; bioRxiv doi:21 October 2019, preprint: not peer reviewed 10.1101/811174. [DOI]
- 4. Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P.. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011; 43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Donovan J., Rath S., Kolet-Mandrikov D., Korennykh A.. Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA. 2017; 23:1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Emara M., Ivanov P., Hickman T., Dawra N., Tisdale S., Kedersha N., Hu G., Anderson P.. Angiogenin-induced tiRNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010; 285:10959–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lyons S.M., Achorn C., Kedersha N.L., Anderson P.J., Ivanov P.. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res. 2016; 44:6949–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ivanov P., O’Day E., Emara M.M., Wagner G., Lieberman J., Anderson P.. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:18201–18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lyons S.M., Gudanis D., Coyne S.M., Gdaniec Z., Ivanov P.. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat Commun. 2017; 8:1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fay M.M., Lyons S.M., Ivanov P.. RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J. Mol. Biol. 2017; 429:2127–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reddy K., Zamiri B., Stanley S.Y., Macgregor R.B. Jr., Pearson C.E.. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 2013; 288:9860–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dang Y., Kedersha N., Low W.K., Romo D., Gorospe M., Kaufman R., Anderson P., Liu J.O.. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 2006; 281:32870–32878. [DOI] [PubMed] [Google Scholar]
- 13. Sadlish H., Galicia-Vazquez G., Paris C.G., Aust T., Bhullar B., Chang L., Helliwell S.B., Hoepfner D., Knapp B., Riedl R et al.. Evidence for a functionally relevant rocaglamide binding site on the eIF4A-RNA complex. ACS Chem. Biol. 2013; 8:1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aulas A., Fay M.M., Lyons S.M., Achorn C.A., Kedersha N., Anderson P., Ivanov P.. Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 2017; 130:927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yalamanchili P., Banerjee R., Dasgupta A.. Poliovirus-encoded protease 2APro cleaves the TATA-binding protein but does not inhibit host cell RNA polymerase II transcription in vitro. J. Virol. 1997; 71:6881–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grifo J.A., Tahara S.M., Morgan M.A., Shatkin A.J., Merrick W.C.. New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 1983; 258:5804–5810. [PubMed] [Google Scholar]
- 17. Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C.. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987; 15:8783–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lyons S.M., Ricciardi A.S., Guo A.Y., Kambach C., Marzluff W.F.. The C-terminal extension of Lsm4 interacts directly with the 3′ end of the histone mRNP and is required for efficient histone mRNA degradation. RNA. 2014; 20:88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tcherkezian J., Cargnello M., Romeo Y., Huttlin E.L., Lavoie G., Gygi S.P., Roux P.P.. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev. 2014; 28:357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Webb N.R., Chari R.V., DePillis G., Kozarich J.W., Rhoads R.E.. Purification of the messenger RNA cap-binding protein using a new affinity medium. Biochemistry. 1984; 23:177–181. [DOI] [PubMed] [Google Scholar]
- 21. Marissen W.E., Lloyd R.E.. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 1998; 18:7565–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marcotrigiano J., Lomakin I.B., Sonenberg N., Pestova T.V., Hellen C.U., Burley S.K.. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell. 2001; 7:193–203. [DOI] [PubMed] [Google Scholar]
- 23. Imai S., Kumar P., Hellen C.U., D'Souza V.M., Wagner G.. An accurately preorganized IRES RNA structure enables eIF4G capture for initiation of viral translation. Nat. Struct. Mol. Biol. 2016; 23:859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berset C., Zurbriggen A., Djafarzadeh S., Altmann M., Trachsel H.. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. RNA. 2003; 9:871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zinshteyn B., Rojas-Duran M.F., Gilbert W.V.. Translation initiation factor eIF4G1 preferentially binds yeast transcript leaders containing conserved oligo-uridine motifs. RNA. 2017; 23:1365–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pestova T.V., Kolupaeva V.G.. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002; 16:2906–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thompson S.R. Tricks an IRES uses to enslave ribosomes. Trends Microbiol. 2012; 20:558–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rubio C.A., Weisburd B., Holderfield M., Arias C., Fang E., DeRisi J.L., Fanidi A.. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014; 15:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu H., Feng J., Liu Q., Sun F., Tie Y., Zhu J., Xing R., Sun Z., Zheng X.. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009; 583:437–442. [DOI] [PubMed] [Google Scholar]
- 30. Yanagiya A., Svitkin Y.V., Shibata S., Mikami S., Imataka H., Sonenberg N.. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol. Cell Biol. 2009; 29:1661–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Banerjee B., Goss D.J.. Eukaryotic initiation factor (eIF) 4F binding to barley yellow dwarf virus (BYDV) 3′-untranslated region correlates with translation efficiency. J. Biol. Chem. 2014; 289:4286–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Breyne S., Yu Y., Unbehaun A., Pestova T.V., Hellen C.U.. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:9197–9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goyer C., Altmann M., Lee H.S., Blanc A., Deshmukh M., Woolford J.L. Jr., Trachsel H., Sonenberg N.. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell Biol. 1993; 13:4860–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park E.H., Zhang F., Warringer J., Sunnerhagen P., Hinnebusch A.G.. Depletion of eIF4G from yeast cells narrows the range of translational efficiencies genome-wide. BMC Genomics. 2011; 12:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramirez-Valle F., Braunstein S., Zavadil J., Formenti S.C., Schneider R.J.. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J. Cell Biol. 2008; 181:293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thoreen C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., Sabatini D.M.. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012; 485:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Haghighat A., Sonenberg N.. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 1997; 272:21677–21680. [DOI] [PubMed] [Google Scholar]
- 38. Truitt M.L., Conn C.S., Shi Z., Pang X., Tokuyasu T., Coady A.M., Seo Y., Barna M., Ruggero D.. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell. 2015; 162:59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yanagiya A., Suyama E., Adachi H., Svitkin Y.V., Aza-Blanc P., Imataka H., Mikami S., Martineau Y., Ronai Z.A., Sonenberg N.. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol. Cell. 2012; 46:847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lazaris-Karatzas A., Montine K.S., Sonenberg N.. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990; 345:544–547. [DOI] [PubMed] [Google Scholar]
- 41. Ruggero D., Montanaro L., Ma L., Xu W., Londei P., Cordon-Cardo C., Pandolfi P.P.. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 2004; 10:484–486. [DOI] [PubMed] [Google Scholar]
- 42. Lee A.S., Kranzusch P.J., Doudna J.A., Cate J.H.. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature. 2016; 536:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de la Parra C., Ernlund A., Alard A., Ruggles K., Ueberheide B., Schneider R.J.. A widespread alternate form of cap-dependent mRNA translation initiation. Nat Commun. 2018; 9:3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jeong S.J., Park S., Nguyen L.T., Hwang J., Lee E.Y., Giong H.K., Lee J.S., Yoon I., Lee J.H., Kim J.H et al.. A threonyl-tRNA synthetase-mediated translation initiation machinery. Nat. Commun. 2019; 10:1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Saikia M., Krokowski D., Guan B.J., Ivanov P., Parisien M., Hu G.F., Anderson P., Pan T., Hatzoglou M.. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J. Biol. Chem. 2012; 287:42708–42725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weingarten-Gabbay S., Elias-Kirma S., Nir R., Gritsenko A.A., Stern-Ginossar N., Yakhini Z., Weinberger A., Segal E.. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 2016; 351:aad4939. [DOI] [PubMed] [Google Scholar]
- 47. Dresios J., Chappell S.A., Zhou W., Mauro V.P.. An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2006; 13:30–34. [DOI] [PubMed] [Google Scholar]
- 48. Vanderhaeghen R., De Clercq R., Karimi M., Van Montagu M., Hilson P., Van Lijsebettens M.. Leader sequence of a plant ribosomal protein gene with complementarity to the 18S rRNA triggers in vitro cap-independent translation. FEBS Lett. 2006; 580:2630–2636. [DOI] [PubMed] [Google Scholar]
- 49. Xue S., Tian S., Fujii K., Kladwang W., Das R., Barna M.. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015; 517:33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hertz M.I., Landry D.M., Willis A.E., Luo G., Thompson S.R.. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol. Cell Biol. 2013; 33:1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Andreev D.E., Dmitriev S.E., Terenin I.M., Shatsky I.N.. Cap-independent translation initiation of apaf-1 mRNA based on a scanning mechanism is determined by some features of the secondary structure of its 5′ untranslated region. Biochemistry (Mosc.). 2013; 78:157–165. [DOI] [PubMed] [Google Scholar]
- 52. Shatsky I.N., Terenin I.M., Smirnova V.V., Andreev D.E.. Cap-Independent Translation: What's in a Name. Trends Biochem. Sci. 2018; 43:882–895. [DOI] [PubMed] [Google Scholar]
- 53. Gilbert W.V. Alternative ways to think about cellular internal ribosome entry. J. Biol. Chem. 2010; 285:29033–29038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jackson R.J. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb. Perspect. Biol. 2013; 5:a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Komar A.A., Hatzoglou M.. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011; 10:229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gilbert W.V., Zhou K., Butler T.K., Doudna J.A.. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007; 317:1224–1227. [DOI] [PubMed] [Google Scholar]
- 57. Philippe C., Dubrac A., Quelen C., Desquesnes A., Van Den Berghe L., Segura C., Filleron T., Pyronnet S., Prats H., Brousset P et al.. PERK mediates the IRES-dependent translational activation of mRNAs encoding angiogenic growth factors after ischemic stress. Sci. Signal. 2016; 9:ra44. [DOI] [PubMed] [Google Scholar]
- 58. Dobbyn H.C., Hill K., Hamilton T.L., Spriggs K.A., Pickering B.M., Coldwell M.J., de Moor C.H., Bushell M., Willis A.E.. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene. 2008; 27:1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee S.C., Zhang J., Strom J., Yang D., Dinh T.N., Kappeler K., Chen Q.M.. G-Quadruplex in the NRF2 mRNA 5′ Untranslated Region Regulates De Novo NRF2 Protein Translation under Oxidative Stress. Mol. Cell Biol. 2017; 37:e00122–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Morris M.J., Negishi Y., Pazsint C., Schonhoft J.D., Basu S.. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc. 2010; 132:17831–17839. [DOI] [PubMed] [Google Scholar]
- 61. Bonnal S., Schaeffer C., Creancier L., Clamens S., Moine H., Prats A.C., Vagner S.. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003; 278:39330–39336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Koukouraki P., Doxakis E.. Constitutive translation of human alpha-synuclein is mediated by the 5′-untranslated region. Open Biol. 2016; 6:160022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Freibaum B.D., Taylor J.P.. The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front. Mol. Neurosci. 2017; 10:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.