Abstract
Purpose
The challenges of understanding how interventions influence follow-up medical care are magnified during genomic testing because few patients have received it to date and because the scope of information it provides is complex and often unexpected. We tested a novel strategy for quantifying downstream health care utilization following genomic testing to more comprehensively and efficiently identify related services. We also evaluated the effectiveness of different methods for collecting these data.
Methods
We developed a “risk-based” approach for a trial of newborn genomic sequencing where we defined primary conditions based on existing diagnoses and family histories of disease and defined secondary conditions based on unexpected findings. We then created patient-specific lists of services associated with managing primary and secondary conditions. Services were quantified based on medical record reviews, surveys and telephone check-ins with parents.
Results
By focusing on services that genomic testing would most likely influence in the short-term, we reduced the number of services in our analyses by over 90% compared to analyses of all observed services. We also identified the same services that were ordered in response to unexpected findings as were identified during expert review and by confirming whether recommendations were completed. Data also showed that quantifying health care utilization with surveys and telephone check-ins alone would have missed the majority of attributable services.
Conclusions
Our risk-based strategy provides an improved approach for assessing the short-term impact of genomic testing and other interventions on health care utilization while conforming as much as possible to existing best-practice recommendations.
Keywords: Whole Exome Sequencing, Health Care Utilization, Genetic Testing, Genomics, Health Services, Humans, Infant, newborn, Risk Factors, Surveys and Questionnaires, Medical Records
Concise Summary
We summarize a novel approach for quantifying health care utilization that comprehensively assesses services that may be influenced by genomic testing while omitting unrelated services.
INTRODUCTION
Best practices for assessing the value of any intervention are to include how it impacts ongoing medical care [1–3]. Quantifying downstream health care utilization can be difficult, though, especially in the context of genomic testing. Methods that consider all observed services can be uninformative when they require large sample sizes [4], and relatively few patients have received genomic testing to date. Alternative strategies that focus on services to manage specific conditions can miss important procedures when the conditions are complex [5, 6], and the scope of information provided by genomic testing can be especially vast and complicated [7]. Moreover, findings are often unanticipated: laboratories frequently disclose secondary findings that are unrelated to test indications, such as monogenic disease risks, carrier status, and pharmacogenomic information [8–10]. Thus, best practices for assessing health care utilization, to track services that may be influenced by the intervention of interest, independent of cause [2, 11], are difficult to apply.
The challenges of assessing health care utilization downstream of genomic testing and its associated costs have been addressed previously [12, 13]. In 2018, we addressed this methodological challenge by proposing a solution in which health care utilization is assessed through multiple strategies [14]. A pilot study implemented these approaches, and generated a series of analyses that included (a) all health care identified, (b) only services that were immediately attributable to interventions, and (c) all services that could have been ordered during a full genetics workup [14, 15]. While this strategy provided unique insight into the impact of genomic testing on health care utilization, it also had important conceptual and analytic limitations. Analyses of immediately attributable services, for instance, may have omitted services initiated in response to the initial workup, while the overwhelming majority of health care utilization included in analyses of all observed services were clearly unrelated to the interventions. The latter approach also generated tremendously large confidence intervals for mean group differences in health sector costs.
The work presented here addresses the above-mentioned concerns through use of a novel approach for identifying downstream health care utilization associated with genomic testing. We describe a “risk-based” approach that narrows the scope of health care services we expect genomic testing would influence. We then compared the services we identified using this approach against the services we identified using standard approaches. We also compare the effectiveness of different strategies for identifying and quantifying health care utilization. The goal of this manuscript is to provide an alternative approach for researchers who use real world data to assess the short-term impact of genomic testing on health care utilization.
METHODS
Design of the Case Study
We developed the approach described here for the BabySeq Project, the first randomized clinical trial of newborn genomic sequencing (nGS) [16]. Newborn genomic sequencing in the BabySeq Project included the use of exome sequencing to screen nearly 1,000 genes for variants associated with monogenic childhood-onset conditions and highly actionable adult-onset conditions [17]. nGS also identified carrier status for childhood-onset disease and pharmacogenomic variants for medications that may be prescribed during childhood. Providers had the option to order additional indication-based analyses based on symptoms that suggested a possible genetic disorder.
A more detailed summary of the BabySeq Project, molecular findings, and parental attitudes at enrollment has been published previously [16–23]. The study included healthy newborns born at Brigham and Women’s Hospital (BWH) and newborns in intensive care units at BWH, Boston Children’s Hospital (BCH), and Massachusetts General Hospital (MGH). Newborns were randomized to a modified standard of care (standard newborn screening and an in-depth family history analyses), or standard of care plus nGS. Results were disclosed to parents by a genetic counselor and physician. Reports were uploaded into newborns’ medical records and sent to clinicians involved in the newborns’ care. Primary endpoints included health care utilization over a 10-month period following results disclosure. The trial was registered with ClinicalTrials.gov (Identifier: NCT02422511) and was approved by the Institutional Review Boards of BWH, BCH, MGH, and the Baylor College of Medicine.
Defining Relevant Health Care Services
An overview of the approach we tested is summarized in Table 1. First, we defined “primary conditions” and “secondary conditions.” Primary conditions are akin to indications in diagnostic testing, and are conditions known prior to randomization for which we would expect nGS to influence care immediately. In the BabySeq Project, these conditions included existing clinical diagnoses and family history reviews that raised the possibility of a genetic disorder. We omitted acute transient conditions that resolved by the time the newborn was discharged from the hospital. We defined secondary conditions as those associated with unexpected monogenic findings and carrier status known to cause disease. To improve the feasibility of implementation, we omitted conditions associated with carrier status that could not manifest as disease. We also omitted conditions associated with pharmacogenomic information because they could not be well-defined.
Table 1.
Key terms. Definitions were adapted from the National Human Genome Research Institute’s Talking Glossary of Genetic Terms [43], the National Cancer Institute’s Dictionary of Genetics Terms [44], the Genetics Home Reference [45], the World Health Organization’s web site [46], and the CSER TOOLKIT [47].
| Genomic testing | A laboratory process that can identify changes in multiple genes. Genomic testing can include exome and genome sequencing. |
| Genome sequencing | A laboratory process that determines nearly all of the approximately 3 billion nucleotides of an individual’s complete DNA sequence, including noncoding sequences. |
| Exome sequencing | A laboratory process that is used to determine the nucleotide sequence primarily exonic (or protein-coding) regions of an individual’s genome and related sequences, representing approximately 1% of the complete DNA sequence. |
| Newborn genomic sequencing | A laboratory approach, currently under study, that uses genomic testing to collect and analyze large amounts of DNA sequence data in the newborn period. https://ghr.nlm.nih.gov/primer/newbornscreening/newborngenomicsequencing |
| Monogenic disease | Disease that result from modifications in a single gene occurring in all cells of the body. |
| Carrier status | In order to have some diseases, an individual must usually have two mutated copies of a gene. “Carrier status” is positive when an individual has one normal copy of a gene and one mutated copy of a gene. With rare exceptions, individuals with positive carrier status not expected to develop the disease. |
| Pharmacogenomics | Pharmacogenomics is a branch of pharmacology concerned with using DNA and amino acid sequence data to inform drug development and testing. An important application of pharmacogenomics is correlating individual genetic variation with drug responses. |
Based on the primary and secondary conditions identified, we developed lists for each newborn that included specialist encounters, tests, procedures, prescriptions and devices that may be ordered to diagnose, screen for, or treat each condition. Services were identified based on condition summaries in GeneReviews [24], Online Mendelian Inheritance in Man [25], the National Comprehensive Cancer Network [26]; and UpToDate [27]. To blind reviewers of medical records and survey data from each newborn’s randomization status and nGS findings, lists of services omitted the names of the associated conditions. Study personnel then used these patient-specific lists to review the newborns’ medical records and family-reported data to determine how often the services occurred within the trial period. The purpose of distinguishing primary conditions and secondary conditions was to improve the statistical precision of analyses by limiting the scope of considered services to those we may expect nGS to influence in the short-term.
Approaches to Quantifying Health Care Utilization
After developing patient-specific lists of services to analyze, we documented how often the services occurred by reviewing medical records, surveying and interviewing families, and surveying physicians who provided care to the newborn. Approaches are summarized in Table 2. To identify health care associated with primary conditions, we reviewed the medical records of the health care systems in which the newborns had been enrolled (i.e., for newborns enrolled at BWH or MGH, we reviewed each newborn’s medical records at Partners HealthCare; and for newborns enrolled at BCH, we reviewed newborns medical records at BCH). To identify health care associated with secondary conditions, we reviewed the medical records at both Partners HealthCare and BCH for all newborns with unexpected monogenic findings, and included services that parents reported during follow-up calls from study staff that were conducted periodically post-disclosure.
Table 2.
Strategy for analyzing the impact of population newborn genomic sequencing (nGS) on downstream health care costs.
| Step | BabySeq Project Implementation |
|---|---|
| 1. Define relevant conditions | |
| Primary conditions | Conditions identified prior to disclosure for which care may be influenced by nGS • Existing diagnoses • Family histories suggestive of possible genetic disease |
| Secondary conditions | Conditions identified during disclosure for which care may be influenced by nGS • Unexpected monogenic disease risks • Manifesting carrier status |
| 2. Identify relevant services | For each primary and secondary condition, develop patient-specific lists of services that may be used for • Diagnosis • Screening • Treatment Services of interest are identified based on review of • GeneReviews [28], • Online Mendelian Inheritance in Man [29] • National Comprehensive Cancer Network guidelines [30] • UpToDate [31] |
| 3. Quantify health care utilization | Record how often the relevant services were received by each patient based on • Medical records reviews • Family self-report ○ Surveys ○ Periodic calls with families ○ 10-month check-in • Provider surveys |
Given the potential for subjects to receive care associated with nGS outside of the Partners HealthCare (BWH and MGH) or BCH systems over the study period, we also collected data from subjects’ families. All families were asked to complete surveys administered 3 months and 10 months after disclosure sessions that asked whether anyone in their families, including the newborn, received genetic counseling outside of the study. Surveys also asked parents to report the number of specialist visits their newborn received. In addition, parents were called after ten months had passed from their newborns’ results disclosure sessions as part of a “10-month check-in” where they were asked to report any follow-up their newborns received regarding their results. Parents of newborns identified with unexpected monogenic disease risks were also called by study staff periodically and asked about the services their newborn received.
As a final strategy for quantifying health care utilization, we invited all physicians who provided care to newborns in the BabySeq Project to complete surveys that asked them to describe any referrals or additional tests they initiated based on BabySeq reports.
Data Analysis
To provide insight into how much our approach reduced the scope of analyzed services, we descriptively compared the total number of services it identified against the total number of outpatient services observed in medical records data over the 10-month trial period. These analyses were restricted to comparisons of newborns enrolled at Partners HealthCare sites, given challenges in distinguishing inpatient and outpatient care received at BCH.
To provide insight into the effectiveness of our risk-based approach for identifying services associated with secondary conditions, we descriptively compared the number of services it identified against the number of services identified by expert review of newborns’ medical records and data from periodic follow-up calls. We also compared the number of services identified using our risk-based approach against the number of confirmed services that newborns received in response to recommendations for clinical follow-up issued during results disclosure sessions. Given the heterogeneity of conditions associated with unexpected genomic findings and the potential for families to pursue follow-up outside the systems of enrollment, analyses of health care utilization associated with secondary conditions and unexpected monogenic disease risks included medical records data from both Partners HealthCare and BCH for all newborns and services that were identified through family self-report and provider report. Concordance analyses were conducted by calculating Kappa statistics that compared specific approaches. Analyses were conducted using R version 3.6.1.
RESULTS
Participant Characteristics and Data Availability
Results from family history analyses and nGS were disclosed to families of 237 well newborns and 61 ill newborns (newborn characteristics are summarized in the Supplementary Materials). Newborns were 4.6 months old, on average (sd=55 days), at the time of results disclosure sessions. No genetic variants were identified that conclusively explained existing clinical features, but unexpected findings about monogenic disease risks were identified in 17 of 158 infants (6.6%) randomized to nGS [23], excluding one ill newborn identified with a monogenic disease risk variant who passed away prior to the results disclosure session.
Our analyses provide insight about how non-response to surveys can compromise the ability to identify health care utilization. Thirty-one families (10.4%) reported health care utilization at 3 months post-disclosure but not 10 months post-disclosure, and 68 families (22.8%) did not report health care utilization data at either time point. Among the 17 families of newborns with unexpected monogenic disease risks, one (5.9%) did not complete survey items about health care utilization at 10-months post-disclosure only, and five others (29.4%) did not complete survey items about health care utilization at 3- or 10-months post-disclosure. Data from provider surveys were also limited, with no data available for 167 newborns overall (56.0%) and no data available for 10 newborns with unexpected monogenic disease risks (58.8%).
Health Care Services Associated with Primary Conditions
Analyses of services potentially associated with follow-up care of primary conditions identified 316 services ordered, including 154 services for care of newborns in the nGS arm and 162 services for care of newborns in the control arm (p=0.25). Analyses of services received by the 266 newborns enrolled in the Partners HealthCare system showed that restricting analyses to services that may be associated with the care of primary conditions reduced the number of services overall by over 90%, from 521 total services to 48.
Health Care Services Associated with Secondary Conditions
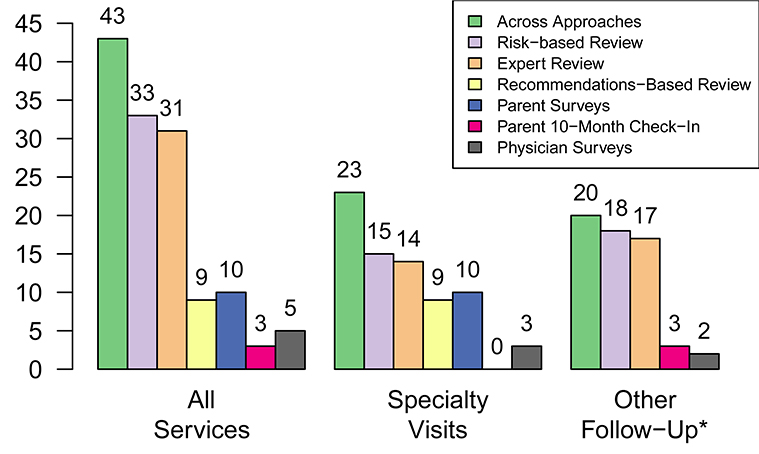
Among the 17 newborns who were identified with unexpected genomic findings during nGS findings and were followed for the trial period, we identified 43 services associated with follow-up care. See Figure 1. The most services were identified using our proposed risk-based strategy (33 of 43; 76.7%), followed by expert review (31 of 43; 72.1%). All other approaches identified less than 25% of the total number. The concordance between services identified using our risk-based approach and expert review was high (kappa = 0.92; p<0.001). The only discrepancy was a hematology encounter for a newborn with a likely-pathogenic variant in the GLMN gene that was omitted during expert review. Concordance between our risk-based approach and all other approaches was slight (kappa = 0.22; p=0.012) in comparison with recommendations-based reviews and poor (kappa<0.07, all p>0.08) in comparisons with parent surveys, parent interviews, and physician surveys.
Figure 1.
Number of health care services associated with follow-up of secondary findings from newborn genomic sequencing using different approaches.
* Parent surveys were omitted because they included no items to assess health care utilization other than specialist visits.
Figure 1 also summarizes differences between the number of follow-up services identified using a risk-based approach and other approaches specific to follow-up visits and other services. The additional visits identified using a risk-based approach rather than a recommendations-based approach were encounters that occurred subsequent to an initial follow-up visit for four newborns. No specialist visits were reported at the 10-month check-in, and only four of the 17 total specialist visits (23.5%) were observed in both our risk-based medical approach and family surveys. All three referrals to specialists reported by providers were for a single patient, and could not be corroborated by data from medical record reviews, surveys of families, or interviews of families. Risk-based reviews and expert reviews also identified the majority of other types of follow-up services. All services reported by parents at the 10-month check-in were also observed during the risk-based and expert reviews. Physician surveys included recommendations for an echocardiogram and electrocardiogram for the aforementioned newborn in the ill newborn cohort with the GLMN variant that were not observed in other approaches.
DISCUSSION
Even if genomic testing provides clinical benefits, it will need to demonstrate that it provides value. By distinguishing between primary and secondary conditions and defining relevant services on a per-patient basis, we narrowed the scope of services included in our analyses to only those that we would anticipate genomic testing to influence in the short-term and reduced the number of services that were included in our analyses by over 90%. At the same time, our risk-based approach identified the same services that were ordered in response to unexpected genomic findings as an expert review and by confirming whether recommended services occurred. Our successful proof-of-concept work provides a starting point for other studies of the impact of genomic testing on health care utilization which comprehensively identifies attributable services while improving the precision of statistical analyses.
Our approach balances best practices and pragmatism. For primary conditions, our strategy is akin to standard analysis of health care utilization for indication-based testing. Rather than focusing on services associated with managing a single condition, our approach encompasses an expansive set of conditions that genomic testing may influence over shorter periods of time. In this initial work, we focused on patient diagnoses and family histories of disease suggestive of possible genetic disorders. An alternative approach that could narrow the scope of relevant services further would be to define primary conditions as those with validated gene-disease associations. Another improvement could be to consider services associated with a diagnostic workup and treatment of unexplained phenotypes. Regardless, the approach we currently tested would improve the precision of statistical analyses over traditional approaches while conforming as much as possible to best practices recommendations [11]. The improved methods we have proposed for short-term analyses may help address the needs of policymakers and payers who are making decisions about genomic testing today.
Our work also provides an improved strategy for assessing health care associated with secondary conditions and unexpected genomic findings. Our strategy was effective not only for confirming whether recommendations made at disclosure were completed, but also for identifying services ordered after the initial workup. The concordance between the services identified through our risk-based approach and the services identified by confirming recommendations as well as expert review was not surprising, given that the informational resources we used to determine relevant services are frequently used by genetic counselors and medical geneticists [28]. Investigators may interpret the concordance in two ways. On the one hand, our findings suggest that expert review of services can be as effective at identifying attributable services as our risk-based approach but requires less effort to implement. On the other hand, our risk-based approach identified the same services as the expert review, even though it was executed by study team members with limited expertise in genomics. Moreover, developing lists of relevant services for managing secondary conditions and unanticipated findings provides an approach for assessing health care utilization that is potentially more scalable. The genes that are frequently queried for unexpected findings, such as the incidental findings list developed by ACMG for secondary findings disclosure [28] or Geisinger Health System’s MyCode list [29], typically have major overlap. Developing consensus lists of services that include associated diagnostic and billing codes could help standardize analyses across studies and facilitate automated data abstraction from electronic medical records. These strategies are integral to cost of illness studies that use encounter-based approaches [30] or the development of condition-specific resource use questionnaires [31]. One approach to developing these lists of procedures and diagnostic codes may be to capitalize on NIH-based efforts such as ClinVar [32] and ClinGen [33]. These efforts have already honed collaborative processes for developing standards and for disseminating data to researchers.
In addition to providing a strategy for reducing the number of services to consider in analyses, our work provides insight into the effectiveness of different strategies for determining how often services occurred. The data we summarize demonstrate that limiting the approach to confirming whether families followed through on clinical recommendations can miss many of the follow-up services that occurred. Similarly, relying on surveys administered to families may miss many specialty visits, although the surveys we administered did identify visits that medical record reviews and periodic phone calls did not. It is likely that structuring our surveys to query parents about specific provider visits helped families recall some visits that may have occurred at medical systems for which we had no access to medical records [34, 35].
Surveys of families provided little insight about other types of health care utilization because they did not ask about services aside from additional genetic testing (which no parents of newborns with unexpected monogenic disease risks reported). Similarly, few parents of newborns with unexpected monogenic disease risks reported services during their end-of-study phone interview. Given that study staff had been calling these families periodically to collect information about the medical follow-up their newborns were receiving, it is possible that many families considered the 10-month check in to be redundant because they had already shared relevant information.
Admittedly, our approach is suited primarily for studies with shorter time horizons. Over longer time horizons, the approach is likely to overstate the impact of unexpected genomic findings on health care utilization because the approach would not account for potential reductions in resource use enabled by early identification of genetic risk factors and the initiation of preventive care. Greater standardization about what genomic testing results should be disclosed as unexpected findings could also better allow our approach to be suitable for longer-term analyses because it would allow lists of the most common secondary conditions to be created a priori. Such lists could then be used to assess downstream health care utilization in all patients included in analyses, regardless of whether they received genomic testing or whether they were identified with relevant pathogenic variants.
Limitations to our work also include the post-hoc identification of primary and secondary conditions and related services. We classified services as having occurred based on family report even though respondent recall is often biased or incomplete [36, 37], particularly for minor services received over longer time horizons [38, 39]. Patient- and provider-reported services were not always verified, and the scope of care received by newborns outside of the BCH and Partners HealthCare systems was unclear. Data were collected from a single trial in metro Boston and may not generalize well to other studies and settings.
CONCLUSIONS
A risk-based approach that defines primary and secondary conditions and relevant health care services for individual patients can provide a more comprehensive assessment of services that are impacted by genomic testing in the short-term and omit unrelated services from analyses. Approaches such as these can help address the dearth of evidence about the impact of genomic testing [7, 40–42] and provide more convincing evidence about its value. Our approach can also be used to assess health care utilization for other interventions where current usage is limited and the downstream impact on care may be complicated and unexpected.
Supplementary Material
Table 3.
Summary of approaches that were implemented to quantify health care utilization.
| Approach | Description |
|---|---|
| Medical Records Data | |
| Review of medical records | Investigators and study staff reviewed electronic medical records of newborns at their system of enrollment. Medical records at both BCH and Partners HealthCare were reviewed if newborns were identified with an unexpected monogenic disease risk. |
| Family Self-Report | |
| Phone calls with families | Study staff called families of newborns with unexpected monogenic disease risks periodically to ask about health care that were received |
| Surveys | Surveys administered 3 and 10-months after disclosure sessions asked parents to report how often their newborn received genetic counseling or specialist visits |
| 10-month check-in | Parents of newborns randomized to nGS were called after 10 months had passed from disclosure sessions and asked to report follow-up regarding their newborns’ results. |
| Provider Report | |
| Surveys | Providers of all enrolled newborns were sent surveys asking whether they made referrals or ordered additional tests |
HIGHLIGHTS.
What is already known about the topic?
Challenges to quantifying downstream health care utilization are common, but magnified by genomic testing because of the limited number of patients who have received it to date and the complexity and diversity of information it can provide.
What does the paper add to existing knowledge?
We summarize a novel approach for quantifying health care utilization related to genomic testing that balances the need to comprehensively define the scope of services that may be influenced by results while avoiding consideration of unrelated services. We also summarize the effectiveness of different approaches for determining how often patients received genomic result-related services.
What insights does the paper provide for informing health care-related decision making?
The approach we propose provides methods for researchers to use in analyses of real world data about the short-term impact of genomic testing on health care utilization that more effectively identifies related services and excludes unrelated services that could also be applied in other contexts. The paper also provides insight about how effectively health care services are identified using different strategies for reviewing medical records, survey data, and family-reported data.
Acknowledgments
This study was supported by National Institutes of Health grants U19HD077671, K01HG009173, U01HG009599, and R01CA221870 and the American College of Medical Genetics and Genomics Summer Genetics Scholars Program. Members of the BabySeq Project Team also include Pankaj B. Agrawal, Ozge Ceyhan-Birsoy, Shawn Fayer, Leslie A. Frankel, Casie A. Genetti, Amanda M. Gutierrez, Maegan Harden, Ingrid A. Holm, Joel B. Krier, Matthew S. Lebo, Kalotina Machini, Amy L. McGuire, Medha Naik, Tiffany T. Nguyen, Stacey Pereira, Vivek Ramanathan, Heidi L. Rehm, Amy Roberts, Jill O. Robinson, Sergei Roumiantsev, Talia S. Schwartz, Tina K. Truong, Grace E. VanNoy, Susan E. Waisbren, and Timothy W. Yu.
Funding/Support: This study was supported by National Institutes of Health grants U19HD077671, K01HG009173, U01HG009599 and R01CA221870, and by the American College of Medical Genetics and Genomics Summer Genetics Scholars Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Sanders GD, Neumann PJ, Basu A, et al. Recommendations for conduct, methodological practices, and reporting of cost-effectiveness analyses: Second panel on cost-effectiveness in health and medicine. JAMA. 2016;316:1093–103. [DOI] [PubMed] [Google Scholar]
- [2].Glick HA. Economic Evaluation in Clinical Trials. 2nd ed. New York: Oxford University Press, 2015. [Google Scholar]
- [3].Caro JJ, Briggs AH, Siebert U, et al. Modeling good research practices--overview: a report of the ISPOR-SMDM Modeling Good Research Practices Task Force-1. Med Decis Making. 2012;32:667–77. [DOI] [PubMed] [Google Scholar]
- [4].Diehr P, Yanez D, Ash A, et al. Methods for analyzing health care utilization and costs. Annu Rev Public Health. 1999;20:125–44. [DOI] [PubMed] [Google Scholar]
- [5].Robertson C, Archibald D, Avenell A, et al. Systematic reviews of and integrated report on the quantitative, qualitative and economic evidence base for the management of obesity in men. Health Technol Assess. 2014;18:v-vi, xxiii-xxix, 1–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nestler-Parr S, Korchagina D, Toumi M, et al. Challenges in research and health technology assessment of rare disease technologies: report of the ISPOR Rare Disease Special Interest Group. Value Health. 2018;21:493–500. [DOI] [PubMed] [Google Scholar]
- [7].Phillips KA, Deverka PA, Marshall DA, et al. Methodological issues in assessing the economic value of next-generation sequencing tests: many challenges and not enough solutions. Value Health. 2018;21:1033–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ackerman SL, Koenig BA. Understanding variations in secondary findings reporting practices across U.S. genome sequencing laboratories. AJOB Empir Bioeth. 2018;9:48–57. [DOI] [PubMed] [Google Scholar]
- [9].Ormond KE, O’Daniel JM, Kalia SS. Secondary findings: how did we get here, and where are we going? J Genet Couns. 2019;28:326–33. [DOI] [PubMed] [Google Scholar]
- [10].Delanne J, Nambot S, Chassagne A, et al. Secondary findings from whole-exome/genome sequencing evaluating stakeholder perspectives. A review of the literature. Eur J Med Genet. 2019;62:103529. [DOI] [PubMed] [Google Scholar]
- [11].Ramsey SD, Willke RJ, Glick H, et al. Cost-effectiveness analysis alongside clinical trials II—an ISPOR Good Research Practices Task Force report. Value Health. 2015;18:161–72. [DOI] [PubMed] [Google Scholar]
- [12].Reeves P, Doran C, Carey M, et al. Costs and cost-effectiveness of targeted, personalized risk information to increase appropriate screening by first-degree relatives of people with colorectal cancer. Health Educ Behav. 2019:1090198119835294. [DOI] [PubMed] [Google Scholar]
- [13].Fahr P, Buchanan J, Wordsworth S. A review of the challenges of using biomedical big data for economic evaluations of precision medicine. Appl Health Econ Health Policy. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Christensen KD, Phillips KA, Green RC, et al. Cost analyses of genomic sequencing: lessons learned from the MedSeq Project. Value Health. 2018;21:1054–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Christensen KD, Vassy JL, Phillips KA, et al. Short-term costs of integrating whole-genome sequencing into primary care and cardiology settings: a pilot randomized trial. Genet Med. 2018;20:1544–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Holm IA, Agrawal PB, Ceyhan-Birsoy O, et al. The BabySeq project: implementing genomic sequencing in newborns. BMC Pediatr. 2018;18:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ceyhan-Birsoy O, Machini K, Lebo MS, et al. A curated gene list for reporting results of newborn genomic sequencing. Genet Med. 2017;19:809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].VanNoy GE, Genetti CA, McGuire AL, et al. Challenging the current recommendations for carrier testing in children. Pediatrics. 2019;143:S27–s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Holm IA, McGuire A, Pereira S, et al. Returning a genomic result for an adult-onset condition to the parents of a newborn: insights from the BabySeq Project. Pediatrics. 2019;143:S37–s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pereira S, Robinson JO, Gutierrez AM, et al. Perceived benefits, risks, and utility of newborn genomic sequencing in the BabySeq Project. Pediatrics. 2019;143:S6–s13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Murry JB, Machini K, Ceyhan-Birsoy O, et al. Reconciling newborn screening and a novel splice variant in BTD associated with partial biotinidase deficiency: a BabySeq Project case report. Cold Spring Harbor molecular case studies. 2018;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Genetti CA, Schwartz TS, Robinson JO, et al. Parental interest in genomic sequencing of newborns: enrollment experience from the BabySeq Project. Genet Med. 2019;21:622–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ceyhan-Birsoy O, Murry JB, Machini K, et al. Interpretation of genomic sequencing results in healthy and ill newborns: results from the BabySeq Project. Am J Hum Genet. 2019;104:76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pagon RA, Tarczy-Hornoch P, Baskin PK, et al. GeneTests-GeneClinics: Genetic testing information for a growing audience. Hum Mutat. 2002;19:501–09. [DOI] [PubMed] [Google Scholar]
- [25].Hamosh A, Scott AF, Amberger J, et al. Online Mendelian Inheritance in Man (OMIM). Hum Mutat. 2000;15:57–61. [DOI] [PubMed] [Google Scholar]
- [26].Winn RJ, Botnick W, Dozier N. The NCCN guidelines development program. Oncology (Williston Park, NY). 1996;10:23–8. [PubMed] [Google Scholar]
- [27].Fox GN, Moawad N. UpToDate: a comprehensive clinical database. J Fam Pract. 2003;52:706–10. [PubMed] [Google Scholar]
- [28].Christensen KD, Bernhardt BA, Jarvik GP, et al. Anticipated responses of early adopter genetic specialists and nongenetic specialists to unsolicited genomic secondary findings. Genet Med. 2018;20:1186–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Carey DJ, Fetterolf SN, Davis FD, et al. The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research. Genet Med. 2016;18:906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rosen AB, Cutler DM. Challenges in building disease-based national health accounts. Med Care. 2009;47:S7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Leggett LE, Khadaroo RG, Holroyd-Leduc J, et al. Measuring resource utilization: a systematic review of validated self-reported questionnaires. Medicine. 2016;95:e2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rehm HL, Berg JS, Brooks LD, et al. ClinGen — The Clinical Genome Resource. N Engl J Med. 2015;372:2235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bhandari A, Wagner T. Self-reported utilization of health care services: improving measurement and accuracy. Medical care research and review : MCRR. 2006;63:217–35. [DOI] [PubMed] [Google Scholar]
- [35].Gama H, Correia S, Lunet N. Effect of questionnaire structure on recall of drug utilization in a population of university students. BMC Med Res Methodol. 2009;9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ansah EK, Powell-Jackson T. Can we trust measures of healthcare utilization from household surveys? BMC Public Health. 2013;13:853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hunger M, Schwarzkopf L, Heier M, et al. Official statistics and claims data records indicate non-response and recall bias within survey-based estimates of health care utilization in the older population. BMC Health Serv Res. 2013;13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mauldin PD, Guimaraes P, Albin RL, et al. Optimal frequency for measuring health care resource utilization in Parkinson’s disease using participant recall: the FS-TOO resource utilization substudy. Clinical therapeutics. 2008;30:1553–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kjellsson G, Clarke P, Gerdtham UG. Forgetting to remember or remembering to forget: a study of the recall period length in health care survey questions. J Health Econ. 2014;35:34–46. [DOI] [PubMed] [Google Scholar]
- [40].Buchanan J, Wordsworth S, Schuh A. Issues surrounding the health economic evaluation of genomic technologies. Pharmacogenomics. 2013;14:1833–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Prosser LA, Grosse SD, Kemper AR, et al. Decision analysis, economic evaluation, and newborn screening: challenges and opportunities. Genet Med. 2013;14:703–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Phillips KA, Trosman JR, Kelley RK, et al. Genomic sequencing: assessing the health care system, policy, and big-data implications. Health Aff (Millwood). 2014;33:1246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].National Human Genome Research Institute. Talking glossary of genetic terms. Available at: https://www.genome.gov/genetics-glossary. [Accessed November 15, 2019].
- [44].National Cancer Institute. NCI Dictionary of Genetics Terms. Available at: https://www.cancer.gov/publications/dictionaries/genetics-dictionary. [Accessed November 15, 2019].
- [45].U. S. National Library of Medicine. What is newborn genomic sequencing? Available at: https://ghr.nlm.nih.gov/primer/newbornscreening/newborngenomicsequencing. [Accessed November 15, 2019].
- [46].World Health Organization. Human Genomics in Global Health. Available at: https://www.who.int/genomics/public/geneticdiseases/en/index2.html. [Accessed November 15, 2019].
- [47].Practitioner Education Working Group of the Clinical Sequencing Exploratory Research (CSER) Consortium. Introduction and overview to genomic test reports. Available at: https://www.ashg.org/education/csertoolkit/intro.html. [Accessed November 15, 2019].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.