Abstract
Computer-aided diagnosis (CAD) has been a major field of research for the past few decades. CAD uses machine learning methods to analyze imaging and/or non-imaging patient data and make assessment of the patient’s condition, which can then be used to assist clinicians in their decision making process. The recent success of the deep learning technology in machine learning spurs new research and development efforts to improve CAD performance and to develop CAD for many other complex clinical tasks. In this paper, we discuss the potential and challenges in developing CAD tools using deep learning technology or artificial intelligence (AI) in general, the pitfalls and lessons learned from CAD in screening mammography and considerations needed for future implementation of CAD or AI in clinical use. It is hoped that the past experiences and the deep learning technology will lead to successful advancement and lasting growth in this new era of CAD, thereby enabling CAD to deliver intelligent aids to improve health care.
Keywords: Computer-aided diagnosis, deep learning, artificial intelligence
I. Introduction
In computer-aided diagnosis (CAD), machine learning methods are utilized to analyze imaging and non-imaging data from past case samples of a patient population and develop a model to associate the extracted information with certain disease outcome. The developed model is expected to predict the outcome of a new unknown case when data from a new case are input. If properly trained and validated, the CAD prediction may be used as a second opinion or supporting information in a clinician’s decision making process. The approach of using machine learning technology to analyze patient data for decision support is applicable to any patient care process, such as disease or lesion detection, characterization, cancer staging, treatment planning, treatment response assessment, recurrence monitoring, and prognosis prediction. More often than not, imaging data play a major role in each of these stages and thus image analysis is a main component in CAD.
Prior to the 1980’s, a few studies had attempted to develop computerized methods for automated lesion detection in radiologic images; these studies did not attract strong interest, probably due to the limitations in computational power and in accessing high-quality digitized or digital images. Systematic research and development of CAD methods for various diseases started in the early 1980’s in the Kurt Rossmann Laboratory at the University of Chicago1. Chan et al. developed a computer-aided detection (CADe) system for clustered microcalcifications on digitized mammograms2 and conducted the first observer study3 to demonstrate the potential that CADe as a second opinion to radiologists can improve their performance. In 1998, the U. S. Food and Drug Administration (FDA) approved the first commercial CADe system for use as a second reader to assist in the detection of breast cancer in screening mammography. Research in various areas of CAD has been increasing over the years4–8. Although the majority of the work is directed at detection and characterization of various types of diseases on images, there are increasing interests and efforts in applying CAD methods to the quantitative image analysis of tumor heterogeneity, correlation of image phenotypes with underlying genetic and biological processes, differentiation of cancer subtypes, cancer staging, treatment planning and response assessment. The CAD area of quantitative analysis of image features in these applications has been called radiomics in recent years.
II. Machine Learning/Artificial Intelligence in Computer-Aided Diagnosis (CAD)
Machine learning is a broad field in computer science with applications to many areas such as face recognition, text and speech recognition, robotics, satellite imagery analysis, and target detection and characterization in military or civilian use. Machine learning makes use of knowledge and techniques from multidisciplinary fields to analyze the input imaging or non-imaging data or a combination of both and extract relevant information to interpret the data or predict the outcome for a given task. For example, mathematics and statistics are important tools to develop new machine learning methods and build predictive models from the data, understanding of biological pathways and genetics are critical to guide the analysis of radiomics and genomics associations, and domain knowledge of a specific type of diseases and how they manifest in a given medical imaging modality is needed to guide feature design and extraction. With deep learning that does not require hand-engineered features, domain knowledge is even more important for understanding whether the machine has learned relevant features, interpreting the output and correlating it with the clinical condition of the patients.
To develop a robust machine learning system for a given task, one has to collect a sufficiently large and representative set of sample data of each class from the population of interest so that the machine learning algorithm can correctly model the statistical properties of the population and assess any new unknown case from the same population. For robust training, the proportion of the classes ideally should be balanced, and thus classes of rare events will require even more effort to collect. Machine learning technology has been evolving over time from so-called conventional methods to the recent deep learning methods. In a conventional approach of supervised machine learning, the features to be extracted from the input data are usually designed by human developers based on expert domain knowledge, and the best features and their relationships (or predictive model) are chosen statistically with the guidance by the predictive performance in a training set of labeled case samples. In unsupervised learning the case samples are not labeled and the machine analysis is expected to discover the underlying characteristics and the relationships among the case samples, which generally requires a much larger set of training samples.
Deep learning has emerged as the state-of-the-art machine learning method9. Deep learning learns multiple levels of representations from the training data by iteratively adjusting the layers of weights in a deep neural network architecture. It has found success in many fields such as speech and text recognition, natural language understanding and translation, object detection and classification. At present, convolutional neural networks (CNN) are most commonly used in deep learning for computer vision and pattern recognition tasks in images. CNN is one type of artificial neural networks that could find its origin from the neocognitron proposed by Fukushima et al in the early 1980’s10. LeCun11 adapted the method and demonstrated its application in recognition of handwritten digits. CNN is different from other pattern classification methods in that it is a type of representation learning that discovers useful features from the input data without the need of manually designed features. To achieve high discriminative power for complex patterns, relatively large number of training samples is required. In 1993, Lo et al.12,13 first introduced CNN into medical image analysis and applied it to lung cancer detection in chest radiographs. Chan et al.14–16 applied CNN to the classification of true and false microcalcifications in a CADe system for mammography in 1993 and trained another CNN for classification of true and false masses in 199417–21. Zhang et al.22 applied a similar shift-invariant neural network for detection of clustered microcalcifications in 1994. Due to the limited computational power of computers, the limited training data available, and the vanishing gradient problem23, the early CNNs contained very few convolutional layers and very few kernels in each layer, which limit the learning capacity of the CNN. Nonetheless, these studies demonstrated the potential of applying CNN to pattern recognition in medical imaging.
A number of factors spur the advancement of machine learning techniques in the past decade. The popularity of social media and personal devices drives the Information Technology industry to develop automated and interactive functionalities for various applications. The need to reduce manual costs in various industries also stimulates the growth of automation and computer-assisted technologies. In addition, the availability of low-cost graphical processing units (GPUs) and memory from the video gaming industry makes it possible to use CNN with large number of layers and kernels. The fast internet and cloud facilitate the collection of large data samples for training. More importantly, effective network training strategies for deep architectures have been developed over time,9,24 such as layer-wise unsupervised pretraining followed by supervised fine-tuning25–27, replacing sigmoid-type activation functions with rectified linear unit (ReLU)28,29, and regularization with dropout30. These new techniques reduce the risk of vanishing gradient and overfitting and increase training convergence speed, thus enabling deep neural networks containing millions of weights to be trained. In 2012, Krizhevsky et al31 showed that a “deep” CNN (DCNN) with five convolutional layers for feature extraction and 3 fully connected layers (known as the AlexNet) could outperform other methods in an ImageNet Large Scale Visual Recognition Challenge (ILSVRC)32 that required the classification of over 1000 classes of objects. Since the AlexNet, another important technique, batch normalization33, was proposed as a regularizer for network training, which reduces the internal covariate shift, allows higher learning rate, and reduces overfitting, thus facilitating training deeper and deeper CNN structures. It has been shown that the errors for complex classification tasks decreased with the depth of CNN34. The ImageNet data set provided by the ILSVRC contains over 1.2 million images but studies indicated that the classification accuracy can further increase by using DCNNs with deeper architecture and greater learning capacity if even larger training set is available35.
The success of deep learning in pattern recognition and its adaptation to various applications such as self-driving vehicles, face recognition, voice recognition, chess and Go games, and personal assistants, etc. bring strong interests in applying deep learning to the CAD field in medicine. DCNN has been applied to medical image analysis for various CAD tasks despite the lack of sufficiently large medical data set compared to non-medical imaging data21,36,37. Most of the DCNNs in CAD to-date are trained for differentiation of abnormal images with disease patterns from images with normal or benign patterns for a given imaging examination. Application of DCNN to other CAD tasks such as segmentation of organs and tumors, detection of changes in tumor size or texture patterns in response to treatment, classification of image patterns associated with the risk of recurrence or prognosis, and differentiation of image patterns that may be predictive of high risk or low risk of developing a certain disease or evolving into invasive disease in the future, are also being explored. Similar CAD tasks are also applicable to optical coherence tomography image analysis for eye diseases38 or histopathological image analysis at the cellular level39. The potential of DCNN in improving the accuracy and performance of computer-assisted decision support systems has created a lot of excitement in the medical imaging community. Even though most of the current deep learning applications are still far from exhibiting the characteristics of “intelligence” that are expected of humans, both the developers and users are contented with labeling the computer-assisted technologies as “Artificial Intelligence (AI)”.
III. Deep learning approach to CAD
III.A. Data collection
DCNN certainly brings strong promise in advancing CAD as routine clinical decision support tools in health care and there is even prediction that AI will replace radiologists in the near future40. However, unrealistic expectations may not sustain long-term growth. Extensive research effort and resources are needed to overcome many of the hurdles in developing and integrating CAD tools into clinical workflow. One of the major challenges in developing an accurate and generalizable DCNN for a given task is a large well-curated data set for training. The data set has to cover the variabilities in patient population, imaging devices and acquisition protocols in real world clinical settings for which the DCNN is intended to be used. Collecting such a data set is costly, especially that the labeling and annotation often require the effort of more than a single expert clinician due to the inter- and intra-reader variabilities in image interpretation and disease assessment. Researchers have attempted to use data mining and natural language processing of the electronic health record (EHR) and the picture archiving and communication system (PACS) for extracting clinical data and diagnosis from the physicians’ and pathology reports41. The accuracy of the retrieved labels depends on the methods used42. It has been shown that automatically mined disease labels or annotations can contain substantial noise43. The challenge from mining the EHR may be attributed to many factors, such as the non-standardized reporting and formatting in the clinical reports to date, the errors by the data mining tools in the correlation and interpretation of the various stages of diagnosis and patient management in complicated cases, and incomplete prior or follow-up information due to patient referral and transfer between different health systems. It has also been reported that information from the DICOM header of images can also be inaccurate with as much as over 15% error in labeling body parts44, thereby introducing noise into automatically retrieved DICOM data for DCNN training or testing. Collaborative efforts by the vendors and users to standardize the reporting among the various data archiving systems are needed to facilitate mining big data for CAD development. Furthermore, if secure electronic communication of patient records can be established among different health systems, it may not only improve patient care by transferring clinical data more efficiently and accurately during patient referral, but also increase the accuracy of data mining of these cases.
Training with mislabeled data reduces the accuracy and generalizability of the trained DCNN. Samala et al.45 conducted a simulation study of training a DCNN for the classification of malignant and benign breast masses on mammograms using a training set with corrupted labels over a range of 0% to 50% of the training samples. It was shown that the classification performance could reach 100% on the training set but decreased on unknown test cases as the amount of training label corruption increased. Methods have been proposed for training DCNN with noisy labels46,47. In case a small training set with clean labels can be constructed or is available, one approach is to first train the DCNN using the large training set with noisy labels and then fine-tune the DCNN using the data set with clean labels. A recent study48 proposed a multi-task network that jointly learned to clean noisy labels in the large data set and fine-tuned the network using both the small clean data set and the large data set with reduced label noise. Another study49 proposed a loss correction technique that used a small data set with trusted labels to estimate the noise distribution of the label noise and showed that the method could improve the robustness of the deep networks for several vision and natural language processing tasks. Whether these methods can reduce the effort in labeling large data set of medical images remains to be studied.
III.B. Transfer learning
To alleviate the problems of limited data available for training of DCNN in medical imaging, a common approach is to use transfer learning. In transfer learning, a DCNN that has been trained with data for a task in a source domain, is adapted to a new target task by further training it with data from the target domain. Since a DCNN works as an automatic feature extractor and many image features are composed of common basic elements, a DCNN having its weights pre-trained to extract features for an imaging domain will make it easier to be re-trained for a new imaging domain than to train from randomly initialized weights. If the available training data in the source domain is abundant while the training data in the target domain is scarce, transfer learning will enable a DCNN to learn the target task with the limited data set which may be impossible otherwise. Most of the DCNN models in medical imaging to-date were trained by transfer learning using models pre-trained with the large ImageNet data set32.
Although transfer learning may reduce the requirement of training sample size in the target domain to a certain extent, the performance of the transfer-trained DCNN for the target task still depends on the training sample size. Samala et al.50 studied the effect of training sample size on transfer learning for the task of classifying breast masses as malignant and benign in digital breast tomosynthesis (DBT). Because DBT data was limited, they collected a relatively large data set of mammograms in addition to the small set of DBTs from different patients. The classification of masses on mammograms is a similar but still different task than that in DBT. From the mammography set, 2242 unique views with 2454 regions of interest (ROIs) containing breast masses were extracted. From the DBT set, 324 unique views with 1585 ROIs were extracted and partitioned into a training set of 1140 ROIs and an independent test set of 445 ROIs. Several transfer learning strategies were compared. The AlexNet that was pre-trained with the ImageNet data was modified by adding two fully connected layers to adapt it to a two-class classification task. The AlexNet was then transfer-trained either in a single stage with the mammography or DBT data alone, or in two stages with mammography data followed by DBT data. A range of training sample size ranging from 1% to 100% of the original set was simulated by randomly sampling a subset from the entire mammography data or the DBT training set. In addition, the effectiveness of transfer learning was also studied by freezing either the first convolution layer (C1) alone or the C1-to-4th fully connected layers (C1-to-F4) of the AlexNet. The transfer learning strategies were compared in terms of the area under the receiver operating characteristic curve (AUC) on the independent DBT test set. Fig. 1 and Fig. 2 summarize the results: (1) the classification performance increases steadily as the training sample size in either stage 1 or stage 2 increases, indicating that the training sample size has a strong impact on the robustness of the transfer-trained DCNN even if the DCNN has been pre-trained with millions of samples from the source domain, i.e., the non-medical image data from ImageNet (see Fig. 1 and Fig. 2), (2) when the available data set in the target domain (DBT) is small, another stage of pre-training using data from a similar domain (mammography) can improve the robustness of the trained DCNN, in comparison to transfer learning with the DBT training set alone (see A vs B in Fig. 1 and B vs D in Fig. 2), and (3) if too many layers of the pre-trained DCNN are frozen during transfer learning, the learning capacity of the DCNN is restricted and not able to fully learn the information available in the training set (see B vs C in Fig. 1). On the other hand, if the training sample size of the target domain is too small, allowing too many layers to be re-trained can degrade the performance compared to re-training with fewer layers (see B vs C in Fig. 2). This study demonstrates that the performance of a transfer-trained DCNN depends on the training sample size of the target task and the potential usefulness of multi-stage transfer learning.
Fig. 1.
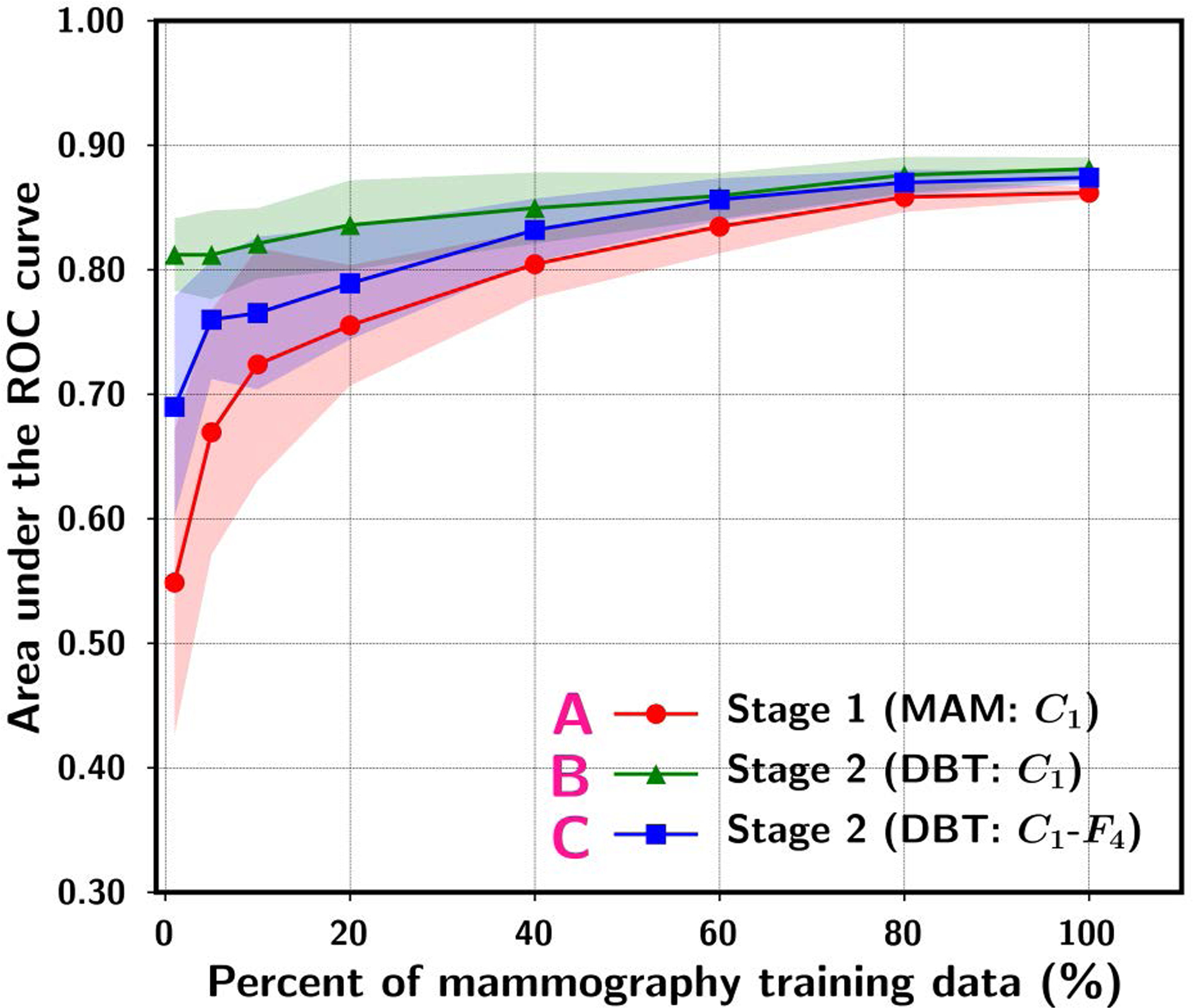
ROI-based area under the receiver operating characteristic curve (AUC) performance on the DBT test set while varying the simulated mammography sample size available for training. The data point and the upper and lower range show the mean and standard deviation of the test AUC resulting from ten random samplings of the training set of a given size from the original set. “A. Stage 1 (MAM:C1)” denotes single-stage training using mammography data and the C1-layer of the ImageNet pre-trained AlexNet frozen during transfer learning without stage 2. “B. Stage 2 (DBT:C1)” denotes stage 2 C1-frozen transfer learning at a fixed (100%) DBT training set size after stage 1 transfer learning (curve A). “C. Stage 2 (DBT:C1-F4)” denotes stage 2 C1-to-F4-frozen transfer learning at a fixed (100%) DBT training set size after stage 1 transfer learning (curve A). [reprint with permission50]
Fig. 2.
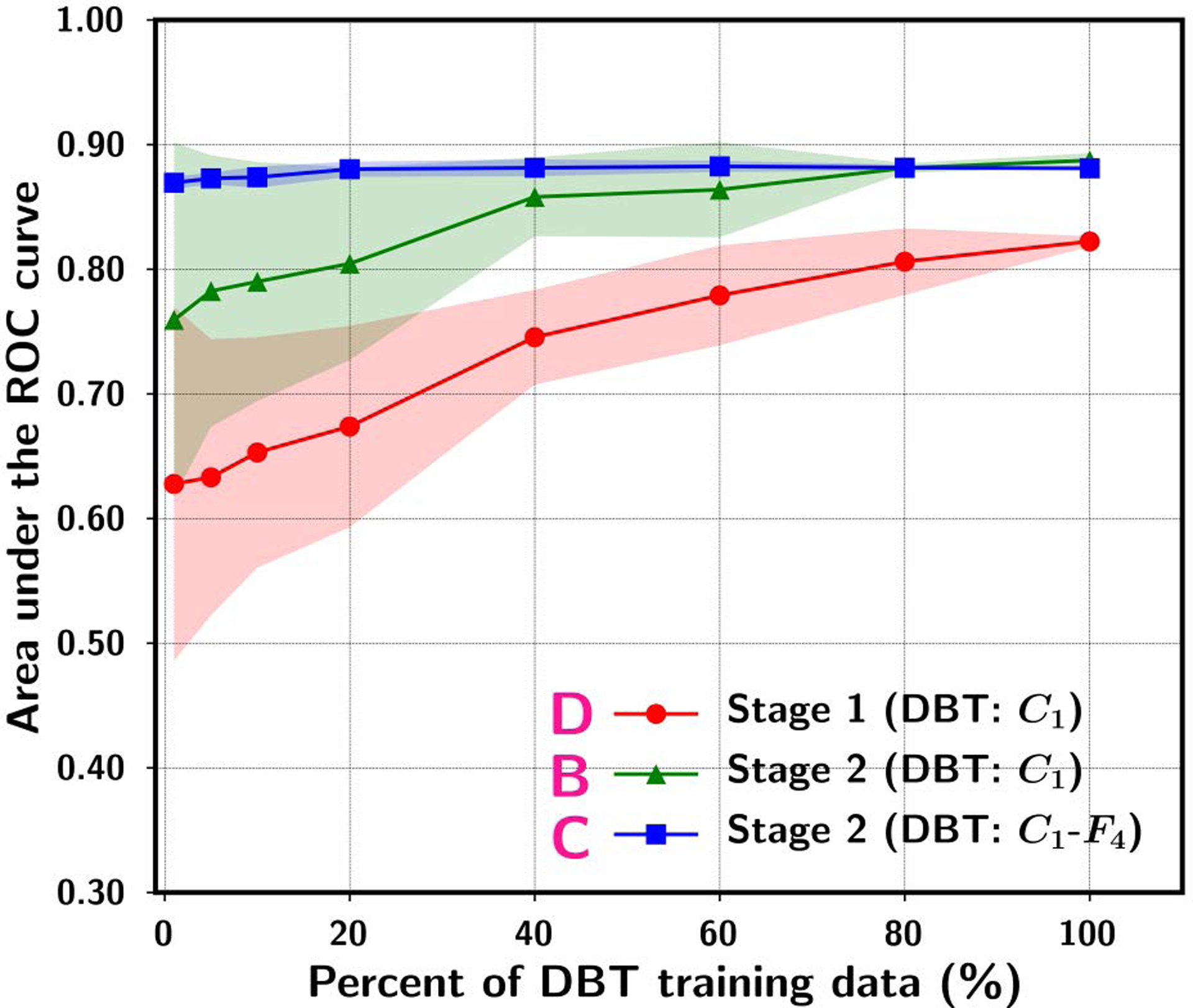
ROI-based AUC performance on the DBT test set while varying the simulated DBT sample size available for training. The data point and the upper and lower range show the mean and standard deviation of the test AUC resulting from ten random samplings of the DBT training set of a given size from the original set. “D. Stage 1 (DBT:C1)” denotes single-stage training using DBT training set with the C1-layer of the ImageNet pre-trained AlexNet frozen during transfer learning without stage 2. “B. Stage 2 (DBT:C1)” denotes stage 2 C1-frozen transfer learning after stage 1 transfer learning with a fixed (100%) mammography training set. “C. Stage 2 (DBT:C1-F4)” denotes stage 2 C1-to-F4-frozen transfer learning after stage 1 transfer learning with a fixed (100%) mammography training set. [reprint with permission50]
III.C. Data augmentation
For DCNN training in medical imaging, a commonly used method to alleviate the limited data problem is to increase training sample size via data augmentation, i.e., to generate multiple slightly different versions of images from each image in the original set. Data augmentation can be implemented off-line or on-line, and each of which may be implemented in many different ways. For off-line data augmentation, for example, all augmented versions of each image are usually pre-generated and mixed with the original data set before being input to the DCNN, which then uses the data set in randomized mini-batches for training. The augmentation techniques may include rotating the image within a range of angles (Na), scaling the image size over a range of factors (Ns), translating and flipping the image in various directions (Nt), cropping the image (Nc), and generating shape- and/or intensity-transformed images with different methods (Nd). If all techniques are applied in combinations, the original sample size of No can be apparently increased to N = No × Na × Ns × Nt × Nd. For on-line augmentation, a common approach is to implement the operations (e.g., rotating, scaling, translating, flipping, cropping, transformation) as a part of the DCNN pipeline and the user selects the range and the probability of each type of augmentation as input parameters. The original training set is used as input and each image in a mini-batch is randomly altered according to the probabilities. By properly choosing the parameters and the number of epochs for training, the augmented training set can be made statistically similar between on-line and off-line augmentation. The major difference is that in on-line augmentation an augmented image is unlikely to repeat itself because each type of operation (except for flipping) is usually set up to randomly select a value within a continuous range, whereas in off-line augmentation the augmented training set is repeatedly used except that the mini-batches are randomly regrouped for each epoch. Off-line augmentation requires more memory space and on-line augmentation costs more computation time. Typically, the choice between off-line and on-line augmentation depends on the size of the data set; off-line augmentation is preferred for small data sets and on-line augmentation is preferred for large data sets especially if the augmentation can be implemented on the GPU. Data augmentation has been shown to reduce the risk of overfitting to a small training set and improve generalizability by introducing some variations or jittering to the original data31,51,52. Thus, data augmentation is a type of regularization-by-data approach, which in general also includes other types such as dropout and data normalization53. However, the augmented images are highly correlated and the CNN learning is invariant to many of these small differences so that there is only limited knowledge the DCNN can learn from the augmented images. Furthermore, if the original training set lacks the representation of certain characteristics of the target lesion and the surrounding tissue in the population due to its limited size, these augmentation methods cannot create lesions with characteristics that do not exist in the original samples. For example, if the original set does not contain spiculated lesions, the augmentation techniques will not be able to generate realistic spiculated lesions. Investigators are also developing more complex augmentation methods that use DCNN such as generative adversarial networks (GANs) to generate altered images with mixed features learned from different images after training on the available sample images54, and methods to digitally generate artificial lesions for various purposes55,56. These methods require more computation time to generate each image and may not be practical to be used in on-line augmentation. Further investigation is needed, especially in medical imaging applications, to study issues such as how effective the augmented lesions or artificial lesions are compared to real independent sample of a similar size in training DCNN, whether they provide useful new features or knowledge for the DCNN to learn, whether the tissue texture in the artificially generated images will improve or impede DCNN learning if texture is an important feature for a given CAD task, how effective the augmentation methods are compared to one another, and whether the effectiveness depends on the classification task.
IV. CAD in retrospect and looking ahead
IV.A. Pitfalls and lessons learned from CAD to-date
Although research and development of CAD in medicine encompasses a wide range of applications in the patient care process such as risk assessment, disease detection, treatment, prognosis prediction and recurrence monitoring, large scale clinical studies on the effect of CAD are mostly focused on CAD in screening mammography, probably because it was the first FDA-approved CAD system for use as a second reader and screening mammography was widely used. Nevertheless, the experience of CAD in screening mammography may provide some useful information to guide future CAD development and clinical implementation in general.
CAD was initially developed as an “aid”, and not as a pre-screener or primary decision maker. Given the limitations of the machine learning technology in the early days of CAD, CAD algorithms can achieve a sensitivity comparable to radiologists but at the expense of relatively high false positive rates. However, CAD may make different types of errors than human experts; the complementary use of CAD by clinicians can improve the overall accuracy as demonstrated in many observer studies1. CAD in screening mammography was therefore approved by FDA only as a second reader. As such, the radiologist is expected to read as vigilant as they should without CAD, and only uses CAD as a “spell checker” after their own reading. They also should not dismiss their own findings if there is no CAD mark at the suspected lesion that they have found in their own first reading. If CAD is used as it is intended and approved for, the disease detection sensitivity should increase or at least cannot be worse than radiologists reading alone. Since the sensitivity can be gained only if radiologists would review the CAD marks and recall some suspected lesions, the users should expect an increase in reading time and also an increase in recalls. The amount of increases would depend on a radiologist’s ability in distinguishing true from false positives on screening mammograms and experience in using CAD.
A number of prospective and retrospective studies have been conducted to compare breast cancer detection in screening mammography with and without CAD or to compare single reader with CAD and double reading57. Most prospective clinical studies use historical statistics of performance measures as controls such as cancer detection rate and recall rate collected over a period of time before CAD was implemented in the clinic, and compared similar data collected after CAD was implemented. These study designs involve a number of confounding factors such as changes in patient populations and radiologists’ experiences between the two periods of time that may contribute to differences in the performance statistics in addition to the use of CAD. Some studies used a matched design in which radiologists’ decision before and after seeing the CAD output were recorded, which would be more consistent with using CAD as a second opinion and eliminate the differences in the patient cohorts and radiologists’ experiences, but there were concerns that the reader could be influenced and become either less vigilant or overly competitive against the computer aid. Gilbert et al.58 conducted a relatively well-controlled three-center prospective randomized clinical trial in the United Kingdom (CADET II) to compare single reading using CAD with double reading. Each of the three centers enrolled a comparable number of patients at over 9,000 with a total of over 28,000. Each patient’s screening mammogram was independently read in the two arms and the first readers in the double reading arm had experience matched to those of the single readers in the CAD arm. The results showed that the sensitivity of the two reading methods were comparable at 87.2% and 87.7%, respectively. The recall rates in two centers were similar in the two arms (3.7% vs 3.6% and 2.7% vs 2.7%) but the third center had significantly higher recall rates in single reading with CAD than in double reading (5.2% vs 3.8%), resulting in an overall recall rate averaged over all centers at 3.9% and 3.4%, respectively, for the two arms. Gromet et al.59 conducted a retrospective review in a single center to compare double reading before CAD implementation to single reading with CAD by the same group of high-volume radiologists. In their double reading setting, the result from the first reading, which could be considered a single reading, was also recorded as a reference. The additional positive by the second reader would be read by a third reader for a final decision on recall. They reported that the sensitivity and recall rate from the first reading were 81.4% and 10.2%, double reading were 88% and 11.9%, and single reading with CAD were 90.4% and 10.6%, respectively. Single reading with CAD therefore achieved 11% higher sensitivity than the first reading and comparable sensitivity with double reading, and 3.9% higher recall rate than the first reading but 12% lower than double reading. These studies indicate that single reading with CAD has the potential to improve cancer detection sensitivity to the level achieved by double reading but at the cost of a moderately increase in the recall rate compared to single reading without CAD if properly used as a second reader.
The review of studies in the literature by Taylor et al.57 shows that the outcomes of radiologists using CAD in screening mammography varied over a wide range. The change in cancer detection ranged from 0% to 19% with a weighted average of 4% and the increase in recall rate from 0% to 37% with a weighted average of 10%. In addition to the differences in the study designs, the clinical environments and the experience of the radiologists, the variations may be attributed partly to how the radiologists used CAD. Some users and promoters might have misunderstood the limitations and capabilities of CAD and ignore its intended use. Many users appeared to over-rely on using the CAD marks for recall decisions while others used CAD as pre-screener to reduce reading time and improve workflow during their readings. There have not been systematic studies to investigate these issues but the reported results and the discussions in some of these studies revealed that the problems may be prevalent. Fenton et al.60 observed a 30% increase in the recall rate and 4.5% gain in cancer detection sensitivity, although they found in a follow-up study61 that the increase in recall rate decreased to 6% after some time post CAD implementation but the gain in sensitivity also decreased to 1.8%. Fenton et al.61 noted that “radiologists with variable experience and expertise may use CAD in a nonstandardized idiosyncratic fashion”, and “Some community radiologists, for example, may decide not to recall women because of the absence of CAD marks on otherwise suspicious lesions”. Lehman et al.62 compared reading digital mammograms with and without CAD by 271 radiologists in 66 facilities of the Breast Cancer Surveillance Consortium (BCSC). They reported that the average sensitivity decreased by 2.3% and the recall rate increased by 4.5% with the use of CAD. They acknowledged that “Prior reports have confirmed that not all cancers are marked by CAD and that cancers are overlooked more often if CAD fails to mark a visible lesion” and that “CAD might improve mammography performance when appropriate training is provided on how to use it to enhance performance”. Unfortunately, Lehman et al. simply concluded that insurers pay more for CAD with no established benefit to women instead of addressing the problems. These studies showed that the lack of understanding of the intended use and the limitations of CAD by users as well as the lack of post-market monitoring and regulation by FDA on the misleading promotion and off-label use of computer aids are significant factors that lead to improper use and CAD “failure” to-date. Furthermore, these experiences indicate that a mismatch of the performance levels of the available CAD systems with the expectation and the need of the clinicians will increase the risk of misuse and negative outcomes.
IV.B. Challenges and opportunities of CAD with deep learning
The success of deep learning in many machine learning tasks revives interests in research and development of various types of CAD. In the recent challenges of developing CAD methods for various classification tasks in medical imaging, all winning teams used deep learning approach63,64. Numerous studies have reported promising results and many showed significantly higher accuracy than radiologists or clinicians. Although the enthusiasm drives a positive change for the CAD field, the excessive optimism and high expectations should be viewed with cautions.
While many studies have shown that deep learning can be more accurate and robust than conventional machine learning approach in many CAD applications, these algorithms have not been extensively tested in routine clinical settings, where many seemingly ideal hardware and software tools could fail when factors such as real-life variabilities in patient population, data quality, user experiences, human-machine interaction, and workflow efficiency play crucial roles. Although some deep learning algorithms claimed to achieve near perfect AUC or better performance than expert clinicians in laboratory testing38,65,66, whether the performance can be reproduced in clinical practice is yet to be proven. In reality, no machine learning techniques can guarantee to be free of false negatives and false positives, which is true also even for the most experienced clinicians. Zech et al.67 reported large variabilities in the generalization performance of DCNNs when different combinations of training and test data collected from three clinical sites were used. They also demonstrated that DCNNs could learn information irrelevant to the patient’s medical conditions and used it effectively for disease classification. In their study, when a DCNN was trained with case cohorts of varying disease prevalence, it would learn to exploit the prevalence to make prediction, and thus generalized poorly to test cohorts that had very different prevalence than the training cohort. A DCNN could also learn subtle differences in the images, such as acquisition equipment and techniques, image processing, and data compression protocols, to distinguish images from different departments within a hospital or from different hospitals, and apparently associate the differences with disease prevalence. Other studies also reported that the DCNNs would learn features irrelevant to the specific abnormalities of interest but correlate them with the presence of the disease43. These studies highlight the importance to train and test the DCNNs properly with internal and external data sets as well as to analyze and understand what information the DCNN has learned for a given classification task. Researchers have developed methods to visualize the feature maps at each convolutional layer inside the deep learning structure68,69 and to highlight the target objects recognized by the DCNN with a class activation map70. Initial efforts have been made to use these tools to visualize the detected location of abnormalities67,71 or to visualize the characteristics of the deep features72 on medical images. These efforts are the first steps towards understanding the inner-workings of deep learning but they are still far from being able to present the network response to clinicians with more insightful medical interpretation, especially for more complex applications than detection. Unlocking the blackbox-like prediction from deep learning and discovering the correlation or causal relationship of the machine findings with other clinical data of the patient will be crucial areas of investigation to enable CAD to deliver interpretable diagnosis and reasoning to clinicians and advance CAD towards AI in medicine.
Besides proper training and testing to ensure the generalizability of a CAD tool, whether it can be successful will still be determined by how clinicians use the CAD tool and the overall value of implementing the tool in the clinic. Misunderstanding the limitations and capabilities of a CAD tool and lack of proper training for the users can lead to unrealistic expectation, misuse and disappointment. Similar to the use of medical devices or some medical procedures, it will be prudent to implement quality assurance monitoring of the performance of CAD over time and establish appropriate metrics to track the effectiveness and efficiency of CAD in clinical use. These outcome measures can provide useful evidence to encourage wider adoption of the CAD tool or, even if negative, can provide important data to guide further improvement. The FDA recently proposed to reclassify medical image analysis devices, including computer-aided detection devices, that are intended to direct the clinician’s attention to portions of an image that may reveal abnormalities during interpretation of patient’s radiology images by the clinician from class III (premarket approval) to class II (special controls), and proposed special controls that the Agency believes are necessary to provide a reasonable assurance of safety and effectiveness of the device. However, the special controls require the manufacturers to label the intended use of the device and user training, but no post-market monitoring and regulations are proposed to enforce that the specified requirements are followed during clinical use. The overhype on AI could incite misuse of the deep-learning-based CAD devices, sending these new generation of CAD devices down the same path as CAD in screening mammography. We have seen early warning signs from the sensational news on accidents by self-driving cars, whose drivers might have ignored the warning that they should be the hands-on drivers, or on machine recommending incorrect or unsafe cancer treatment after the initial excitement about its helpfulness. The AAPM CAD Subcommittee has published two opinion papers on the proper training and evaluation of CAD devices73, and the quality assurance and user training on CAD devices in clinical use74. The discussions have not attracted much attention previously but it will be timely to revisit these issues in view of the renewed interests in deep-learning-based CAD and computer-assisted quantitative image analysis, or AI in medical imaging, under the leadership of organizations such as the AAPM, the American College of Radiology (ACR) and the Radiological Society of North America (RSNA).
In current clinical practice, workflow efficiency and costs are major considerations. Clinicians will not be receptive to a supplemental tool that requires additional time and/or costs without obvious clinical benefits. It is important for CAD researchers and developers to understand the preferred mode of assistance by clinicians for each type of clinical tasks, and design CAD tools and user interface appropriately by taking into consideration the practical issues in clinical settings. In radiology, clinicians may prefer to have CAD as a concurrent or first reader that can help identify abnormality more efficiently or reduce workload, or AI tools that can help manage workflow by automatically triaging cases to prioritize reading or treatment. A good example is the application of CAD in digital breast tomosynthesis (DBT) to generating synthetic mammograms (SM). DBT as an adjunct to mammogram has been shown to be effective in increasing sensitivity and reducing recalls in breast cancer screening but a major concern is that it adds significant reading time to each case. A synthetic mammogram is generated from each DBT volume in replace of the 2D mammogram to reduce radiation dose and to provide an overview of the volume. Due to the limited depth resolution of DBT, direct generation of a projected 2D mammogram from DBT cannot recover all information of a true 2D mammogram, especially the subtle lesions. With CAD technology, an SM can be reconstructed with the CAD-detected suspected lesions enhanced on the SM but no CAD marks are explicitly shown. The CAD-detected lesions again include both true and false positives; however, the false positives on an SM without artificial markers seem to be less disturbing to radiologists. A recent study75 showed that, in comparison to reading DM alone, combo DM+DBT reading reduced recall rate without increasing sensitivity, but DM+SM significantly increased the sensitivity and further reduced recalls. Another recent observer study76 compared detection of breast cancer in DBT with and without deep-learning-based CAD as a concurrent reader that marked suspected lesions and the confidence of malignancy on the DBT slices. The results demonstrated that reading with the CAD tool could significantly reduce the average reading time by more than 50% for a DBT case, increase sensitivity and specificity, as well as reduce recall rate. The concurrent CAD had a case-based sensitivity of over 90% and a specificity of over 40%, which are higher than all of the CAD tools currently used in screening DM. These and other studies indicate that, in addition to improving the performance of CAD tools, designing smart interfaces to deliver CAD assistance or utilizing CAD to enhance visualization and navigation that can improve reading efficiency will also be areas of research interest to facilitate integration of CAD into clinical workflow.
V. Summary
CAD as a second reader has been shown to improve the detection of early stage breast cancer, but the accompanied increases in recall rate and reading time cause criticism. The use of CAD as a concurrent reader before it is validated results in no or little gain in cancer detection but can still increase the recall rate. These experiences are useful lessons to guide the evolution of CAD into practical clinical tools in the future. The deep learning technology has demonstrated strong potential to bring CAD to high performance levels, opening the opportunities of adapting CAD as concurrent reader or even first screener to improve both accuracy and workflow, and more importantly, developing CAD for other complex clinical tasks in the patient care process. However, among the excessive hype and high expectations, CAD developers and users should be mindful of the importance of rigorous training, validation, and independent testing, as well as user training in clinical settings to ensure not only the generalizability of the standalone performance to the real world environment but also the effectiveness of clinicians using CAD in practice. With proper user training and understanding of the capability and limitations the deep learning technology, together with proper monitoring, objective assessments and constructive feedback to enable further research and development, it can be expected that CAD technology will continue to progress and reach the goal of providing truly intelligent aids to improve health care.
Acknowledgment
This work is supported by National Institutes of Health award number R01 CA214981.
Footnotes
Disclosures
The authors have no conflicts to disclose.
References
- 1.Doi K Chapter 1. Historical overview In: Computer-Aided Detection and Diagnosis in Medical Imaging. eds. Li Q, Nishikawa RM, Boca Raton,FL: Taylor & Francis Group, LLC, CRC Press; 2015:1–17. [Google Scholar]
- 2.Chan H-P, Doi K, Galhotra S, Vyborny CJ, MacMahon H, Jokich PM. Image feature analysis and computer-aided diagnosis in digital radiography. 1. Automated detection of microcalcifications in mammography. Medical Physics. 1987;14:538–548. [DOI] [PubMed] [Google Scholar]
- 3.Chan H-P, Doi K, Vyborny CJ, et al. Improvement in radiologists’ detection of clustered microcalcifications on mammograms. The potential of computer-aided diagnosis. Investigative Radiology. 1990;25:1102–1110. [DOI] [PubMed] [Google Scholar]
- 4.Computer aided Detection and Diagnosis in Medical Imaging. First ed eds. Li Q, Nishikawa RM, Boca Raton, FL: Taylor & Francis Group, LLC. CRC Press; 2015. [Google Scholar]
- 5.Shiraishi J, Li Q, Appelbaum D, Doi K. Computer-Aided Diagnosis and Artificial Intelligence in Clinical Imaging. Seminars in Nuclear Medicine. 2011;41(6):449–462. [DOI] [PubMed] [Google Scholar]
- 6.van Ginneken B, Schaefer-Prokop CM, Prokop M. Computer-aided Diagnosis: How to Move from the Laboratory to the Clinic. Radiology. 2011;261(3):719–732. [DOI] [PubMed] [Google Scholar]
- 7.Chan H-P, Hadjiiski LM, Zhou C, Sahiner B. Computer-Aided Diagnosis of Lung Cancer and Pulmonary Embolism in Computed Tomography—A Review. Acad Radiol. 2008;15(5):535–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giger ML, Chan H-P, Boone J. Anniversary Paper: History and status of CAD and quantitative image analysis: The role of Medical Physics and AAPM. Medical Physics. 2008;35(12):5799–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. [DOI] [PubMed] [Google Scholar]
- 10.Fukushima K, Miyake S. Neocognitron: a new algorithm for pattern recognition tolerant of deformations and shifts in position. Pattern Recognition. 1982;15:455–469 [Google Scholar]
- 11.LeCun Y, Boser B, Denker JS, et al. Handwritten digit recognition with a back-propagation network. Proc Advances in Neural Information Processing Systems. 1990. 396–404. [Google Scholar]
- 12.Lo SCB, Lin JS, Freedman MT, Mun SK. Computer-assisted diagnosis of lung nodule detection using artificial convolution neural network. Proc SPIE. 1993;1898:859–869. [Google Scholar]
- 13.Lo SCB, Chan H-P, Lin JS, Li H, Freedman M, Mun SK. Artificial Convolution neural network for medical image pattern recognition. Neural Networks. 1995;8:1201–1214. [Google Scholar]
- 14.Chan H-P, Lo SCB, Helvie MA, Goodsitt MM, Cheng SNC, Adler DD. Recognition of mammographic microcalcifications with artificial neural network. Radiology. 1993;189(P):318. [Google Scholar]
- 15.Chan H-P, Lo SCB, Sahiner B, Lam KL, Helvie MA. Computer-aided detection of mammographic microcalcifications: Pattern recognition with an artificial neural network. Medical Physics. 1995;22:1555–1567. [DOI] [PubMed] [Google Scholar]
- 16.Ge J, Sahiner B, Hadjiiski LM, et al. Computer aided detection of clusters of microcalcifications on full field digital mammograms. Medical Physics. 2006;33(8):2975–2988. [DOI] [PubMed] [Google Scholar]
- 17.Chan H-P, Sahiner B, Lo SCB, et al. Computer-aided diagnosis in mammography: Detection of masses by artificial neural network. Medical Physics 1994;21:875–876. [Google Scholar]
- 18.Petrick N, Chan H-P, Sahiner B, et al. Automated detection of breast masses on digital mammograms using adaptive density-weighted contrast-enhancement filtering. Proc SPIE. 1995;2434:590–597. [Google Scholar]
- 19.Sahiner B, Chan H-P, Petrick N, et al. Image classification using artificial neural networks. Proc SPIE. 1995;2434:838–845. [Google Scholar]
- 20.Sahiner B, Chan H-P, Petrick N, et al. Classification of mass and normal breast tissue: A convolution neural network classifier with spatial domain and texture images. IEEE Transactions on Medical Imaging. 1996;15:598–610. [DOI] [PubMed] [Google Scholar]
- 21.Litjens G, Kooi T, Bejnordi BE, et al. A Survey on Deep Learning in Medical Image Analysis. Medical Image Analysis. 2017;42:60–88. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Doi K, Giger ML, Wu Y, Nishikawa RM, Schmidt RA. Computerized detection of clustered microcalcifications in digital mammograms using a shift-invariant artificial neural network. Med Phys. 1994;21:517–524. [DOI] [PubMed] [Google Scholar]
- 23.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Computation. 1997;9(8):1735–1780. [DOI] [PubMed] [Google Scholar]
- 24.Sutskever I, Martens J, Dahl G, Hinton G. On the importance of initialization and momentum in deep learning. Proc International Conference on Machine Learning. 20131139–1147. [Google Scholar]
- 25.Hinton GE, Osindero S, Teh Y-W. A Fast Learning Algorithm for Deep Belief Nets. Neural computation. 2006;18(7):1527–1554. [DOI] [PubMed] [Google Scholar]
- 26.Bengio Y, Lamblin P, Popovici D, Larochelle H. Greedy layer-wise training of deep networks. Proc Advances in Neural Information Processing Systems. 2006;19:153–160. [Google Scholar]
- 27.Erhan D, Bengio Y, Courville A, Manzagol P-A, Vincent P, Bengio S. Why Does Unsupervised Pre-training Help Deep Learning? Journal of Machine Learning Research. 2010;11:625–660. [Google Scholar]
- 28.Nair V, Hinton GE. Rectified linear units improve restricted boltzmann machines. Proc 27th International Conference on Machine Learning. 2010807–814. [Google Scholar]
- 29.Glorot X, Bordes A, Y. B. Deep sparse rectifier neural networks. Proc 14th International Conference on Artificial Intelligence and Statistics 2011315–323. [Google Scholar]
- 30.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Machine Learning Res. 2014;15:1929–1958 [Google Scholar]
- 31.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems. 20121097–1105. [Google Scholar]
- 32.Russakovsky O, Deng J, Su H, et al. Imagenet large scale visual recognition challenge. International Journal of Computer Vision. 2015;115(3):211–252. [Google Scholar]
- 33.Ioffe S, Szegedy C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. Proc 32nd International Conference on Machine Learning (ICML’15). 2015;37:448–456, arXiv:1502.03167. [Google Scholar]
- 34.He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2015770–778, arXiv:1512.03385. [Google Scholar]
- 35.Sun C, Shrivastava A, Singh S, Gupta A. Revisiting unreasonable effectiveness of data in deep learning era. 2017. arXiv:1707.02968.
- 36.Sahiner B, Pezeshk A, Hadjiiski LM, et al. Deep learning in medical imaging and radiation therapy. Medical Physics. 2019;46(1):e1–e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazurowski MA, Buda M, Saha A, Bashir MR. Deep learning in radiology: An overview of the concepts and a survey of the state of the art with focus on MRI. Journal of Magnetic Resonance Imaging 2019;49(4):939–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fauw JD, Ledsam JR, Romera-Paredes B, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nature Medicine 2018;24(9):1342–1350. [DOI] [PubMed] [Google Scholar]
- 39.Janowczyk A, Madahushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J Pathol Informatics. 2016;7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chockley K, Emanuel E. The End of Radiology? Three Threats to the Future Practice of Radiology. J Am Coll Radiol. 2016;13:1415–1420. [DOI] [PubMed] [Google Scholar]
- 41.Shin H, Le L, Kim L, Seff A, Yao J, Summers RM. Interleaved text/image Deep Mining on a large-scale radiology database. Proc IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 20151090–1099. [Google Scholar]
- 42.Zech J, Pain M, Titano J, et al. Natural Language-based Machine Learning Models for the Annotation of Clinical Radiology Reports. Radiology. 2018;287(2):570–580. [DOI] [PubMed] [Google Scholar]
- 43.Oakden-Rayner L Exploring the ChestXray14 dataset: problems. 2017;https://lukeoakdenrayner.wordpress.com/2017/12/18/the-chestxray14-dataset-problems/.
- 44.Gueld MO, Kohnen M, Keysers D, et al. Quality of DICOM header information for image categorization. Proc SPIE. 2002;4685:280–287. [Google Scholar]
- 45.Richter CD, Samala RK, Chan H-P, Hadjiiski L, Cha KH. Generalization Error Analysis: Deep Convolutional Neural Network in Mammography. Proc SPIE. 2018;10575:1057520. [Google Scholar]
- 46.Reed SE, Lee H, Anguelov D, Szegedy C, Erhan D, Rabinovich A. Training deep neural networks on noisy labels with bootstrapping. 2015arXiv:1412.6596.
- 47.Natarajan N, Dhillon IS, Ravikumar PK, Tewari A. Learning with noisy labels. Proc Advances in Neural Information Processing Systems 26 (NIPS 2013). 20131196–1204 [Google Scholar]
- 48.Veit A, Alldrin N, Chechik G, Krasin I, Gupta A, Belongie S. Learning From Noisy Large-Scale DatasetsWith Minimal Supervision. 2017arXiv:1701.01619. [Google Scholar]
- 49.Hendrycks D, Mazeika M, Wilson D, Gimpel K. Using Trusted Data to Train Deep Networks on Labels Corrupted by Severe Noise. 2018. arXiv:1802.05300v05304.
- 50.Samala RK, Chan H-P, Hadjiiski LM, Helvie MA, Richter CD, Cha K. Breast Cancer Diagnosis in Digital Breast Tomosynthesis: Effects of Training Sample Size on Multi-Stage Transfer Learning using Deep Neural Nets. IEEE Trans Medical Imaging. 2019;38(3):686–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor L, Nitschke G. Improving Deep Learning using Generic Data Augmentation. 2017. arXiv:1708.06020.
- 52.Wang J, Perez L. The Effectiveness of Data Augmentation in Image Classification using Deep Learning. 2017arXiv:1712.04621.
- 53.Kukačka J, Golkov V, Cremers D. Regularization for deep learning: A taxonomy. 2017arXiv:1710.10686.
- 54.Goodfellow IJ, Pouget-Abadie J, Mirza M, et al. Generative Adversarial Nets. 2014arXiv:1406.2661v1401. [Google Scholar]
- 55.Badano A, Graff CG, Badal A, et al. Evaluation of digital breast tomosynthesis as replacement of full-field digital mammography using an in silico imaging trial. JAMA Network Open. 2018;1(7):e185474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Graff CG. A new, open-source, multi-modality digital breast phantom. Proc SPIE. 2016;9783:978309. [Google Scholar]
- 57.Taylor P, Potts HWW. Computer aids and human second reading as interventions in screening mammography: Two systematic reviews to compare effects on cancer detection and recall rate. European Journal of Cancer. 2008;44:798–807. [DOI] [PubMed] [Google Scholar]
- 58.Gilbert FJ, Astley SM, Gillan MGC, et al. Single reading with computer-aided detection for screening mammography. The New England Joural of Medicine. 2008;359(16):1675–1684. [DOI] [PubMed] [Google Scholar]
- 59.Gromet M Comparison of Computer-Aided Detection to Double Reading of Screening Mammograms: Review of 231,221 Mammograms. Am J Roentgenol. 2008;190(4):854–859. [DOI] [PubMed] [Google Scholar]
- 60.Fenton JJ, Taplin SH, Carney PA, et al. Influence of computer-aided detection on performance of screening mammography. New England Journal of Medicine. 2007;356:1399–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fenton JJ, Abraham L, Taplin SH, et al. Effectiveness of Computer-Aided Detection in Community Mammography Practice. Journal of the National Cancer Institute. 2011;103(15):1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lehman CD, Wellman RD, Buist DSM, Kerlikowske K, Tosteson ANA, Miglioretti DL. Diagnostic Accuracy of Digital Screening Mammography With and Without Computer-Aided Detection. JAMA Intern Med. 2015;175(11):1828–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Data Science Bowl. https://www.kaggle.com/competitions.
- 64.Dream Challenges. http://dreamchallenges.org/.
- 65.Lakhani P, Sundaram B. Deep Learning at Chest Radiography: Automated Classification of Pulmonary Tuberculosis by Using Convolutional Neural Networks. Radiology. 2017;284:574–582. [DOI] [PubMed] [Google Scholar]
- 66.Rajpurkar P, Irvin J, Zhu K, et al. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. 2017arXiv:1711.05225.
- 67.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLOS Medicine. 2018;15(11):e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeiler MD, Fergus R. Visualizing and Understanding Convolutional Networks. Lecture Notes in Computer Science Computer Vision – European Conference on Computer Vision (ECCV) 2014. 2014;8689:818–833. [Google Scholar]
- 69.Yosinski J, Clune J, Nguyen A, Fuchs T, Lipson H. Understanding Neural Networks Through Deep Visualization. 2015arXiv:1506.06579v06571.
- 70.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning Deep Features for Discriminative Localization. Proc IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 20162921–2929. [Google Scholar]
- 71.Dunnmon JA, Yi D, Langlotz CP, Ré C, Rubin DL, Lungren MP. Assessment of Convolutional Neural Networks for Automated Classification of Chest Radiographs. Radiology. 2019;290(2):537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Samala RK, Chan H-P, Hadjiiski LM, Helvie MA, Cha KH, Richter CD. Multi-task transfer learning deep convolutional neural network: Application to computer-aided diagnosis of breast cancer on mammograms. Physics in Medicine and Biology. 2017;62:8894–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petrick N, Sahiner B, Armato SG, et al. Evaluation of computer-aided detection and diagnosis systems. Medical Physics. 2013;40(8):087001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huo ZM, Summers RM, Paquerault S, et al. Quality assurance and training procedures for computer-aided detection and diagnosis systems in clinical use. Medical Physics. 2013;40(7):077001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aujero MP, Gavenonis SC, Benjamin R, Zhang Z, Holt JS. Clinical Performance of Synthesized Two-dimensional Mammography Combined with Tomosynthesis in a Large Screening Population. Radiology. 2017;283:70–76. [DOI] [PubMed] [Google Scholar]
- 76.Conant EF, Toledano AY, Periaswamy S, et al. Improving Accuracy and Efficiency with Concurrent Use of Artificial Intelligence for Digital Breast Tomosynthesis Screening. Radiological Society of North America Scientific Assembly and Annual Meeting. 2018RC215–214. [Google Scholar]


