Abstract
“SARS-CoV2”, a previously unknown strain of coronaviruses caused a severe respiratory disease called Coronavirus disease (COVID-19) which emerged from Wuhan city of China on 30 December 2019, and declared as Global health problem by World Health Organisation within a month. In less than two and half months (11 March, 2020) it was declared as a pandemic disease due to its rapid spreading ability, it covered more than 211 countries infecting around 1.7 million persons and claiming around 1.1 lakhs lives within merely 100 days of its emergence. Containment of the infection of this virus is the only available measure to control the disease as no vaccine or specific antiviral treatment is available. Confirmed detection of the virus followed by isolation of the infected person at the earliest possible is the only measure to prevent this disease. Although there are number of methods available for detection of virus and to combat this disease in the present pandemic situation, but these available diagnostic methods have their own limitations. The speedy and exponential global spread of this disease strongly urges the fast and economic diagnostics tools. Additional to the available diagnostic methods, there is a sudden surge for development of various of methods and platforms to diagnose the COVID-19. The review summarized the advantage and disadvantage of various diagnostic approaches being used presently for COVID-19, newer detection methods in developmental stage and the feasibility of advanced platforms like newer nano-sensor based on-the-spot detection technologies.
Keywords: COVID-19, SARS-CoV2, RT-PCR, Diagnostic method for COVID-19, Biosensor
Introduction
During the end of December 2019, some patients having symptoms of flu like illness were admitted in Wuhan, China, the infecting organism remained unknown as preliminary etiological agents suspected like influenza, other respiratory viruses, Chlamydia pneumoniae and Mycoplasma pneumoniae were not found in the laboratory investigations. So, to identify the pathogen responsible, metagenomic RNA sequencing of this patient’s sample was done. The complete viral genome data suggested, this is a new RNA virus related to the family Coronaviridae which was later on designated as ‘2019-nCoV’ or Novel CoV-19. This analysis revealed that this virus has more than 89% genomic similarity with a SARS-like bat coronaviruses which belongs to Sarbecovirus subgenus and Betacoronavirusgenus. On 11 February 2020, International Virus Classification Commission renamed this Novel CoV-19 as “severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)” on the basis of its genetic composition and similarity with other coronaviruses and the disease caused by the virus was renamed as COVID-19 [18].
Coronaviruses are a group of large sized (100–160 nm), spherical, positively sense, non-segmented, single-stranded RNA with genome sized 26–32 kb (the largest among known RNA viruses), and known to infect both animals and humans [2, 44, 45, 47]. Coronavirus has been classified into four genera (α-alpha, β-beta, γ-gamma and δ-Delta), out of which only two genera–alpha which contains CoV-NL63 & CoV-229E, and -beta contains CoV-OC43, CoV-HKU1, Middle East respiratory syndrome coronavirus (MERS-CoV) and SARS-CoV, found to be infectious for human [27, 44]. The genome of COVID-19 virus constitutes 29,903 nucleotides which upon fresh reannotation and mapping of the RNA-sequences obtained, presented the 123,613 reads assembly, and was very similar to SL-CoVZC45—an already known bat strain and SARS-CoV [17, 18, 42, 46]. This group of viruses can easily undergo mutation and recombination to adapt any environment and thus survive by altering wide host range [40] causing constant and long-term health threats, therefore it is necessary to understand its virology to prevent its rapid spreading and safety of mankind.
Coronavirus are among top ten deadliest viruses known for human beings with a high fatality rate of up to 36% by MERS-CoV during 2012 and 10% by SARS-CoV in 2002–2003 [40, 42, 46]. The current SARS-CoV-2 has already infected around 1.3 million persons and killing 72,774 persons from over 211 countries, and all it happened within 100 days of emergence of this new virus (WHO, as on 8 April 2020). International air travel facility and asymptomatic carriers state of the patients has been mainly responsible for rapid and exponential increase of the incidences of COVID-19 infections over the globe. To slow down or curtain the COVID-19 spread in very first step many countries including India have followed the complete “Lockdown” of their countries.
The second step to slow down the spread of disease is to identify the infected persons as early as possible for early prevention and cure but patient infected with this virus shows no symptoms or only mild symptoms of infection matching or confusing with common cold/flu. The potential of a pathogen is estimated by its reproduction number (R0) which is the average secondary cases can be infected by an individual. R0 of SARS-CoV-2 has been told much higher (i.e. > 2.5) than that earlier human coronaviruses, SARS (< 2) and MERS (< 1), hence potential of COVID-19 have more potential to causing pandemic, which has been turned true within short time span [10, 28, 49]. Even in some reported studies, the R0 for SARS-CoV2 was estimated to be around 4, thus indicating even bigger pandemic situation than the present status [7, 30]. Asymptomatic carriers can increase the disease transmission to an uncontrollable manner if they will not be identified and quarantine in early stage. In such situation, mass screening for the disease becomes necessary and hence the fast testing devices are strongly advised to prevent the spread of the virus.
Therefore, rapid, on-the-spot and accurate screening of potential virus carriers along with critical observation of patients without any indicative symptoms is very much essential for controlling the spread of COVID-19. Thus, the present review paper summarized the various detection methods available for the detection of COVID-19, their advantages, disadvantages and urgent need for a rapid POC detection method, on the spot biosensor their feasibly and importance in presence ever increasing countdown of COVID- 19 infected patients.
Diagnosis of COVID-19
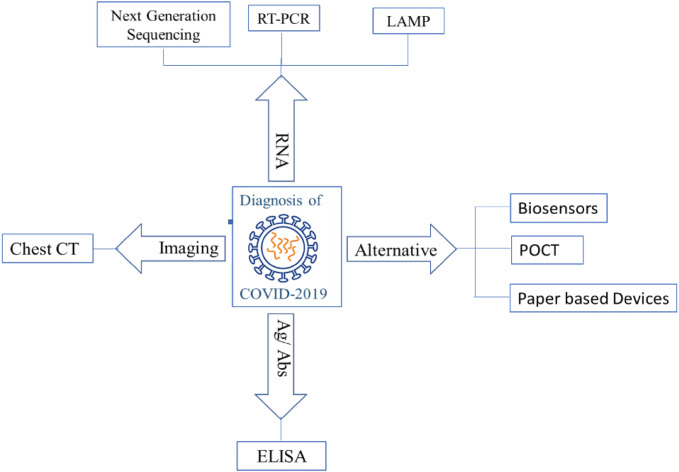
Coronavirus detection approaches are generally based on the travel history of the person from the affected areas as well as the analysis of their clinical symptoms along with some auxiliary examinations. Clinical symptoms like pneumonia, due to COVID-19 are highly atypical, and quite similar to diseases due to other respiratory viral pneumonia. A rapid and sensitive diagnosis of COVID-19 is still unavailable, although some diagnosis methods are available presently (Fig. 1) for virus detection, each having different degree of specificity and based on single or multiple target molecule from the SARS-CoV2. These methods use the pathological changes in the patient’s organ by imaging like CT, or viral nucleic acid like RT PCR using one or more gene, or Next Generation Sequencing whole genome, immunological molecules produced by the patient or by the virus in the patient’s body- Antigen–antibody reaction based tests like ELISA and utilizing each of these diagnosis approach has its their own advantages and shortcoming in present scenario (Table 1). Out of these, some methods were already established and considered as Gold Standard methods which could be replicated for this novel virus also while others are being developed and evaluated for the diagnosis of this virus. On other hands, there are other methods, technologies/devices also which has been developed but pending for regulatory approval and are intended for the use in COVID-19 methods are described here.
Fig. 1.
Diagnosis Approaches for COVID- 19
Table 1.
Current Diagnosis method available for COVID-19
| Method available | Working principle | Advantage | Time required | Disadvantage |
|---|---|---|---|---|
| Next generation sequencing (NGS) | Whole genome sequencing |
Highly sensitive and specific, Provide all related information; Can identify novel strain |
1–2 day |
High expertise Equipment dependency and high cost Highly sophisticated Lab required |
| RT-PCR | Specific primer-probe based detection |
Fast results Higher sensitivity Needs small amount of DNA Can be performed in a single step Well established methodology in viral diagnostics |
3–4 h |
Higher costs due to the use of expensive consumables Expensive lab equipment Detection is also complex and time consuming |
| LAMP | More than two sets of specific primers pair based detection |
Highly repeatable and accurate Single working temperature |
1 h | Too sensitive, highly prone to false positives due to carry-over or cross-contamination |
| Serological (traditional) | Antigen/Antibodies IgG/IgM | Sensitive and specific | 4–6 h |
Testing come after 3-4 days of infection False positive |
| Rapid serological | Antigen/Antibodies IgG/IgM | POCT | 15–30 min |
Testing come after 3-4 days of infection False positive |
| CT scan | Chest images | Enhance sensitivity of detection if findings combined with RT-PCR results | 1 h | Indistinguishability from other viral pneumonia and the hysteresis of abnormal CT |
| Virus isolation | In vitro live virus isolation and propagation |
Highly (100%) specific Gold standard |
5–15 days | Low sensitivity as isolation is not 100% |
Nucleic acid based method
Nucleic Acid based technologies are utilizes the genetic material such as DNA/RNA and are based on the principle of their highly specific base paring with homologous strands. Genetic materials-based detection and diagnostics are comparatively faster than traditional culture-based methods and very useful for high-throughput testing and also provide clinical useful information like drug resistance, virulence factors or strain sub-types within some hours, but are relatively expensive. These technologies such as polymerase chain reaction (PCR), DNA microarrays, and high throughput automated sequencing methods have tremendous role in the routine clinical diagnosis and discovering novel strains of bacteria and viruses and other pathogens. Here, the current state of the art in the nucleic acid-based technologies for diagnostics of COVID-19, and their future advancements along with pro and cons of using these technologies are being described.
Next generation sequencing (NGS)
The, next-generation sequencing (NGS) is also called as high-throughput sequencing (HTS). By this method we can determine the genomic sequence, even more than 1 million base pairs in a single experiment. By this technique, we can diagnose the inheritable diseases, cancer, and infectious diseases. Earlier, also the same technology was used for in UK for tracking an outbreak due to the Methicillin Resistant Staphylococcus aureus (MRSA) [8, 22] with high precision and traceability even in a single patient, while other routine surveillance techniques could not do it with that much precision.
NGS helps not only in discovery of novel viral strains on large scale but also provides very rapid detection of these viruses which link with human diseases. The NGS technology along with bioinformatics tools have largely influenced the modern viral parthenogenesis studies and viral diagnostics. This technology also played great application in the present COVID-19 outbreak. At initiation of current outbreak of SARS-CoV2, the samples from the patients admitted acute respiratory distress syndrome were negative for the all suspected already known pathogens, the etiological pathogen was identified by only NGS by doing metagenomic RNA sequencing and the phylogenetic analysis of its complete genome generated could conclude that it is a new strain of an RNA virus which belonged to the Coronaviridae family and was designated as SARS-CoV2 after nucleotide similarity and genome matching with the existing pathogen’s genome [33].Therefore, this technology has great importance for identifying unknown pathogens, and mutation or recombination in the genome of the pathogen in a short span of time, but the huge cost of the equipment and chemicals required in this technique restricts its utilization in routine laboratory diagnosis of the diseases.
RT-PCR
Presently, quantitative reverse transcription-polymerase chain reaction (rRT-PCR) is being used for diagnosis of COVID-19 and is a gold standard molecular diagnostic technique for many viruses as well. Single step quantitative RT-PCR with TaqMan chemistry is more sensitive and specific. As this technology is well established, and so can be used easily, only needs specific primer- probe designed and synthesized, remaining components of the reaction remain same as used for other viruses without or with a little change. Once the first sequence results of the SARS-CoV-2 virus from China were out, candidate diagnostic rRT-PCR assays were designed and made available in the public domain for researchers. Various agencies or manufacturers have opted different set of genes out of many genes of SARS-CoV2 (ORF-1a gene, ORF-1b gene, RdRp gene, N gene, E gene etc.), so every assay has varied degree of sensitivity.
As per the standard protocol one patient is confirmed of infection when both the selected target genes come to be positive (http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html) [13]. While in some reported studies two individual single-step RT-PCR assays (Based upon TaqMan-chemistry) were performed for identification, and amplification of two segment of any two genes, mostly N or ORF1b from viral genome separately, others have used multiplex assays using more than one genes amplification in single reaction (https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer–probes.html (2020). Although, these methods are very sensitive (almost 100%), but take longer time for confirmation as the test has to be done in a well sophisticated laboratory. The testing has to be done in two steps; first step for screening assay, using the SARS-CoV-2-specific E gene and second step for confirmatory assays targeted the ‘RdRp gene’, ‘N gene’ and ‘ORF-1b’. The “Positive control” material used for these assays was in vitro transcribed RNA of known copy numbers. RNAse P gene detection as used in other most respiratory viruses, is being used as an internal control give the information of the quality of sample collection, RNA extraction process. The standard testing protocol as per WHO (https://apps.who.int/iris/bitstream/handle/10665/331509/WHO-COVID-19-lab_testing-2020.1-eng.pdf) involves 5 steps, (1) sample collection from patient; (2) Proper transportation of collected samples to the laboratory; (3) Providing demographic and clinical information to the laboratory; (4) Sample testing by the laboratory; (5) preparing and reporting the correct and appropriate test results. Testing is carried out at specific centers which further delays the diagnosis and make condition of patient severe. These PCR assays provide good results but on the other hand they are laborious and expensive as well.
Some studies using RT-PCR SYBER green dye based assay found to be less specificity than TaqMan probe based assays. Similar results were reported recently in China by patients who self-collected saliva and showed 91.7% (11/12) positive SARS-CoV-2 while diagnosing by SYBR based RT-qPCR [43]. RT-qPCR assays have been reported highly sensitive and specific for SARS-CoV, MERS-CoV detection and also same for COVID-19, but this technology is prone to its false negative rates which could result in severe consequences due to missed diagnosis of COVID-19 [48]. The real example is present from the current outbreak of SARS-CoV2 where five patients were reported as negative by RT-qPCR, but found positive when CT scan examination of their chest was done and recollected samples repeated RT-qPCR, all patients were confirmed positive for SARS-CoV-2 [50]. The sensitivity of RT-qPCR for detection of SARS-CoV was reported between 50% and 79%, that too depends on their adopted protocol, quality of sample (time of collection, amount, maintenance of cold chain) and total number of samples [9, 51], and needs further improvement using synergistic approaches.
Besides sensitivity problem, RT-qPCR has some other drawbacks such as possible biological safety hazards occurred during transport and sample processing, nucleic acid extraction, and requirement of sophisticated laboratory equipment like biosafety cabinets that is often available only in few main central laboratory [14, 16]. Technical expertise along with sample transportation which is inevitable makes the overall process time consuming. All these drawbacks could make the process less useful in case of health emergency or present global outbreak situation. Moreover, in PCR we are able to detect not only target virus, but it can also perform co-detection of several other respiratory viruses which leads increase in false positive or negative results [12, 21].
Loop-mediated isothermal amplification (LAMP)
LAMP, comparatively novel technique which in process of approval for COVID-19 diagnosis is molecular amplification technique that can amplify any genomic material with high efficiency and in shorter time. The technique is based on synthesis of target DNA at constant temperature of 60–65 °C using specially designed primer sand enzyme (DNA polymerase) having strand displacement activity instead of heat denaturation as in other PCR techniques [35] and in an hour or lesser time can amplify the target sequence up to more than 109 copies forming a cauliflower shaped structure as a final product consisting a stem and a loop form of DNA with many inverted repeats.
LAMP is a user-friendly technique which can provides reliable, sensitive and specific results in lesser time as compared to other conventional techniques, and therefore become quite popular just after its development focusing its applications in microbial detection [19, 20]. This technique has the advantage of requiring only single constant temperature, and thus eliminating the need of thermocycler and so as power consumption.
Computed tomography (CT) scan
CT Scan is also one of the diagnosis techniques having high sensitivity due to which many researchers recommend its use as one of the necessary auxiliary diagnostic method for COVID-19, moreover its results come even before clinical symptoms appear. Typical features by CT of COVID-19 patient include bilateral multi-lobar ground-glass opacificities with differently distributions in posteriors and also in peripheral [39], along with sub-pleura ascendance, thickened lobular septa with variable alveolar filling, and amalgation [41].
According to a recent report from Wuhan, the CT is significantly more sensitive than PCR for iSARS-CoV-2 suspected persons. The results concluded that in patients having negative RT-qPCR reports, more sensitive and accurate conclusion can be achieved using a combination of CT-Scan and other standard techniques like RT-qPCR or other sensitive diagnostic tests.
Moreover, the high-resolution CT of the chest is also proved as an essential tool for detection of SARS-CoV-2, at early stage and to take rapid and necessary intervention [15]. Therefore, various studies recently utilizing chest CT images to diagnose the COVID-19 [1, 15, 38]. Earlier also the typical CT images in patients infected with SARS-CoV and MERS-CoV showed similar symptoms as in COVID-19 (38, 39). As per these findings, CT scans found to be a great diagnostic tool for screening of COVID-19 patients especially in the high prevalence or pandemic areas.
As the CT scans are indicative and not confirmatory tool for pathogen detection in the COVID-19 diagnosis and associated with few shortcomings also such as inability to separate the cases of other pneumonia (viral or non-viral) and the hysteresis of the abnormal CT imaging.
Antigen–antibody based methods
Serological based testing methods normally use blood samples for detection of virus instead of nasopharyngeal swab samples used in PCR test. The blood samples contain either a significant and measurable concentration of antibodies or virus specific antigens. The two main type of antibodies in the blood which the test looks for are the immunoglobin G (IgG) and immunoglobulin M (IgM). The body’s way of remembering the prior infection, and how it responded to infection in previous encounter is very crucial, so that the body is able to attack the same pathogen again is through antibodies. IgM appears within few days and act as first line of active defense, followed by production of IgG to start clearing the infection. All kind of infections are fought through IgM and IgG. The body’s immune response mechanism can be utilized to detect the particular pathogen. The blood test for COVID-19 detect the protein (signature antigen/biomarkers) or antibodies particular to the virus so as for SARS-CoV2 with the confirmed SARS-CoV2 specific antibodies in case of antigen detection or confirmed SARS-CoV2 antigen in case of antibodies detection, and not produced for the seasonal flu or other virus. Currently, two types of COVID-19 tests have been reported one direct utilizing antigen based on detection of viral component present during the time of infection and the second indirect using antibodies that appears in patient’s serum later due to development of immune response against the virus (https://www.ecdc.europa.eu/en/publications-data/overview-rapid-test-situation-covid-19-diagnosis-eueea).
Antigen Detection tests: FIND, non-governmental organization (https://www.finddx.org/covid-19/pipeline/) lists ten rapid antigen detection tests for COVID-19 with EU approval under IVDs directives (98/79/EC), but yet to come in the market due to non-availability of distributors for these devices. However, reports from competent authorities indicates the availability of three such CE-marked devices very soon.
Antibody Detection tests: There are nearly 60 antibody tests marked rapid SARS-CoV-2 that are expected to come soon in market along various other in-house validated tests for SARS-CoV-2 by many researchers, which can help in early diagnostic at commercial scale [37].
Also, a number of point of care (POCT) kits based on IgM or IgG, and ELISA for COVID-19 showing higher detection rates compared to nucleic acid based detection methods, have been developed and pre-tested by many companies but still not in commercial stage [3]. ELISA based detection kits developed or being developed using antibodies against spike, nucleocapsid or membrane and envelope proteins are considered as the one of the most sensitive method for COVID-19 diagnosis. Earlier also this method using N-based IgG ELISA and S-based IgG ELISA showed good sensitivity for SARS-CoV i.e. 94.7% and 58.9% respectively [9]. The sensitivity of ELISA kit for SARS-CoV-2 is still under study. Also, antibodies-based diagnostic assays are not useful for early or active diagnosis of COVID-19, due to their longer time requirement (7 days or more) to be developed by the host to provides positive results, and so, the detectable antibodies are produced late after appearance of symptoms [3, 37] and once developed can persist long after the infection has been cleared. There is great urgency to develop an auxiliary method for accurate diagnosis of COVID-19 which should be enough sensitive, specific and cost effective.
Rapid test
Rapid tests are the one which involve non-automated, mostly qualitative but in some cases quantitative also, are used for in vitro diagnostics (IVDs) of many diseases already, and now also being tried for COVID-19 diagnosis. These tests can provide results within 10–30 min, so their results are considered as instant as compared to the molecular tests which generally takes 4–6 h. Moreover, these tests are user friendly, thus won’t require any extensive training or expertise to operate and can be used either in hospital environment, in the laboratories or at patient bedside without any difficulty.
Advanced/alternative (POCT) approaches
Point of care testing (POCT), as the name indicates can be used at the patient’s bedside with ease without any experts or trained person to operate. These devices are useful for detecting various diseases including infectious viral like HIV, influenza, Hepatitis etc. and bacterial disease in cost effective and user-friendly way, and help in finding the source of any health outbreaks quickly and providing the enough time to the authorities for taking necessary preventive or therapeutic measures. Out of many types of POC devices, the handheld POCTs are of great importance in medical diagnostics which includes various type of biosensors.
Biosensor
Biosensor is a self-contained integrated analytical device consisting of the bioreceptor, transducer and a signal detector. The interaction of bioreceptor with the target analyte produces an electronic signal and through transduces which can then be further amplified by a detector circuit, processed, and displayed.
Biosensors helps in development of point of care, portable devices for sensitive, specific and rapid diagnosis of disease in cost effective way. They use various diagnostics principles, such as PCR involving RNA or DNA sequences, gel electrophoresis, enzyme-linked immunosorbent assay (ELISA) also called sandwich assay involving interaction of antigen antibodies, and other detection procedures coupled with fluorescent and or radioactive labeling [5].
Nowadays several advanced biosensors-based diagnosis approaches has been utilized for fabrication of innovated and novel handheld devices which can overcome the drawbacks of lengthy gold standard detection protocol. These biosensors use the nanomaterials with tunneling and quantum properties leading to enhancement in signal amplification [34]. Further, the nanomaterials are having high surface-to-volume ratio which enhanced their high sensitivity many fold [29], moreover the viruses (target analytes) are also in nanoscale, these all features make the nano-sensors a potential diagnostics tool [4].
Nano-biosensors using aptamers are one of such potent analytical tools for rapid diagnosis of diseases with high sensitivity and specificity in a cost effective and user-friendly manner compared to conventional methods [6]. Such nano-sensor will have great potential for detection of SARS-CoV-2 even in person without any symptoms with high sensitivity, specificity and selectivity only for COVID-19.
Aptamer based nano-biosensor
Aptamers originated from a word ‘aptus’ (a Latin word) means “fit” in 1990 [23, 24], and consist of oligonucleotides of nucleic acids or even small peptide molecules having high specific binding affinity for certain target molecules leading to increase in sensitive and accurate detection. These molecules can be any membrane protein, amino acids, toxins, immunoglobulins, cytokines, growth factors, coupling agents, ionic metals, intact cells or other small molecules. Apta-sensors can be easily converted to any specific design through surface activation or modification by chemical treatment to induce linkers and coupling sites. Due to their high reproducibility and purity, stability and reversibility under harsh environmental conditions with vast availability of target specific linkers, aptamers are being used as novel diagnostics tools [25, 26, 36].
Aptamers, can specially designed and synthesized for the of SARS-CoV-2, using its nucleocapsid protein to obtain fast test results within few seconds only, and it won’t require any sample preparation step. Pinpoint’s aptamer based POC for detection of SARS-CoV2 is in developmental stage, for which the developers are claiming that will provide the SARS-CoV2 test result within 1 min and to be precise in only 30 s (https://www.rapidmicrobiology.com/news/pinpoint39s-low-cost-handheld-covid-19-aptamer-based-diagnostic-device-in-development).
Paper based detection
An alternative paper-based technology using waste water as samples has been suggested by Kang Mao et al. [32].
Paper based device based on integration of different functional area like for extraction, elution, purification, amplification and detection all in a small inexpensive, disposable paper and printed with wax on its surface in the form of zones. It is very much possible to complete the whole testing process without any power source or energy, just by simply folding the paper in various modes, thus it is more beneficial than expensive and complicated multistep techniques. These analytical devices provide high-quality, fast still very precise method for pathogen’s detection, and additionally low manufacturing cost and user-friendly nature [11, 31].
This technology can act as an alternative detection tool for rapid tracing for the source or presence of causative agents like COVID-19 in any pandemic area. Faeces and urine from disease carriers in the community, entering in the sewer system could contain many biomarkers of the virus, and same has been confirmed in a recent study which showed that these infectious agents can remain active for several days even after has been disseminated from the patients, if found suitable environment [11, 31]. There is strong potential in this paper-based device to trace the COVID-19 transmission in community wastewater by analyzing SARS-CoV-2 in faeces, urine and other excreted output of human.
Present status of rapid test for COVID-19
Presently, various WHO referral laboratories along European Commission and Member States especially working for validation of various commercial testing assays developed for COVID-19, and also trying to find rapid diagnostic tests for COVID-19. Researches are regularly doing clinical trials of rapid diagnostic tests for finally getting approval from regulatory bodies for their use in public health with safety. All regulatory authorities, like European Commission, Member State authorities, FIND and WHO are working in close association, and updating each and every significant research outcome in the form of product or protocol as earlier as it is validated, and being approved at earliest so that these can further be upscaled for device production and distribution to meet the pace of present demand of testing and screening.
Concluding remarks
The rapid spread of Covid-19 across the world has become an intense concern for health officials globally, and urgent need for developing better methods for mass screening to prevent the spread of the virus has emerged. Today, only scanning of foreheads for fever using thermal scanner is widely used, but this test cannot detect asymptomatic or pre symptomatic infections, nor it distinguish the novel coronavirus from other respiratory illnesses either. Other than RT-PCR, SARS-CoV2 specific other diagnostic tests like rapid antibodies-based kits being used or under development seems not worthy for mass screening. It is essential to diagnose suspected cases at the clinic or hospital, but results take time anywhere from few hours to some days which is too slow for front-line screening. Thus, the requirement of rapid diagnostic tool like nano-biosensor based technology which can provide the diagnosis result within few seconds is quite high for mass screening and need to be developed at commercial scale as soon as possible. There are many methodology and devices aiming rapid diagnosis of COVID19, are in pipeline for development and are at different stages. These POCT, Biosensors and other alternative devices have the potential to become the technology of future with high sensitivity, specificity and reproducibility.
Compliance with ethical standards
Conflict of interest
There is no conflict of interest in the present work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ajlan AM, Ahyad RA, Jamjoom LG, et al. Middle east respiratory syndrome coronavirus (MERS-CoV) infection: chest CT findings. AJR Am J Roentgenol. 2014;203:782–787. doi: 10.2214/AJR.14.13021. [DOI] [PubMed] [Google Scholar]
- 2.Almeida JD, Tyrrell DA. The morphology of three previously uncharacterized human respiratory viruses that grow in organ culture. J Gen Virol. 1967;1:175–178. doi: 10.1099/0022-1317-1-2-175. [DOI] [PubMed] [Google Scholar]
- 3.Amanat F, Nguyen T, Chromikova V, 8. Strohmeier S, Stadlbauer D, Javier A et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. MedRxiv 2020. Available here: 10.1101/2020.03.17.20037713. [DOI] [PMC free article] [PubMed]
- 4.Arugula MA, Simonian A. Novel trends in affinity biosensors: current challenges and perspectives. Meas Sci Technol. 2014 doi: 10.1088/0957-0233/25/3/032001. [DOI] [Google Scholar]
- 5.Azab SM, Fekry AM. Electrochemical design of a new nanosensor based on cobalt nanoparticles, chitosan and MWCNT for the determination of daclatasvir: a hepatitis C antiviral drug. RSC Adv. 2017;7(2):1118–1126. doi: 10.1039/C6RA25826C. [DOI] [Google Scholar]
- 6.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC. Quantum dot_aptamer conjugates for synchronous cancer imaging, therapy, and sensing of drug delivery based on bi-fluorescence resonance energy transfer. Nano Lett. 2007;7:3065–3070. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 7.Bauch CT, Lloyd-Smith JO, Coffee MP, Galvani AP. Dynamically modeling SARS and other 19 newly emerging respiratory illnesses. Epidemiology. 2005;16(6):791–801. doi: 10.1097/01.ede.0000181633.80269.4c. [DOI] [PubMed] [Google Scholar]
- 8.Brown JR, Bharucha T. Breuer J Encephalitis diagnosis using metagenomics: application of next generation sequencing for undiagnosed cases. J Infect. 2018;76:225–240. doi: 10.1016/j.jinf.2017.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chantal B.F. Vogels, Nathan D. Grubaugh et.al. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR assays.MedRxiv. 2020.10.1101/2020.03.30.20048108.
- 10.Chen J. Pathogenicity and transmissibility of 2019-nCoV-a quick overview and comparison with other emerging viruses. Microbes Infect. 2020 doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin A, Chu J, Perera M, Hui K, Yen HL, Chan M, Peiris M, Poon L. Stability of in different environmental conditions. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho CH, Lee CK, Nam MH, Yoon SY, Lim CS, Cho Y, Kim YK. Evaluation of the Advan Sure. real-time RT-PCR compared with culture and Seeplex RV15 for simultaneous detection of respiratory viruses. Diagn Microbiol Infect Dis. 2014;79:14–18. doi: 10.1016/j.diagmicrobio.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu DKW, Pan Y, Cheng SMS, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020 doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu DKW, Pan Y, Cheng SMS, Hui KPY, Krishnan P, Liu Y, Ng DYM, Wan CKC, Yang P, Wang Q, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;7:1–7. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung M, Bernheim A, Mei X, et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2019;2020:200230. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drosten C, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 18.Fan Wu, Zhao Su, Bin Yu, Chen Yan-Mei, Wang Wen, Song Zhi-Gang, Yi Hu, Tao Zhao-Wu, Tian Jun-Hua, Pei Yuan-Yuan, Yuan Ming-Li, Zhang Yu-Ling, Dai Fa-Hui, Liu Yi, Wang Qi-Min, Zheng Jiao-Jiao, Lin Xu, Holmes Edward C, Zhang Yong-Zhen. A new coronavirus associated with human respiratory disease in China. Nature. 2020;12(579):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francois P, Tangomo M, Hibbs J, Bonetti E-J, Boehme CC, Notomi T, Perkins MD, Schrenzel J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol Med Microbiol. 2011;62:41–48. doi: 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- 20.Galvez LC, Barbosa CFC, Koh RBL, Aquino VM. Loop-mediated isothermal amplification (LAMP) assays for the detection of abaca bunchy top virus and banana bunchy top virus in abaca. Crop Prot. 2020;131:105101. doi: 10.1016/j.cropro.2020.105101. [DOI] [Google Scholar]
- 21.Gaunt ER, Hardie A, Claas ECJ, Simmonds P, Templeton KE. Epidemiology and Clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris SR, Cartwright EJ, Török ME, et al. Whole-genome sequencing for analysis of an outbreak of methicillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis. 2013;13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–1650. doi: 10.1093/clinchem/45.9.1628. [DOI] [PubMed] [Google Scholar]
- 24.Jellinek D, Green LS, Bell C, Lynott CK, Gill N, Ellington AD, Szostak JW. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;1992(355):850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 25.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;1994(263):1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 26.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Fang. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liebel M, Hugall JT, van Hulst NF. Ultrasensitive label-free nanosensing and high-speed tracking of single proteins. Nano Lett. 2017;17(2):1277–1281. doi: 10.1021/acs.nanolett.6b05040. [DOI] [PubMed] [Google Scholar]
- 30.Liu T, Hu J, Kang M, et al. Transmission dynamics of 2019 novel coronavirus (2019-nCoV). bioRxiv. January 2020:2020.01.25.919787. 10.1101/2020.01.25.919787.
- 31.Magro L, Escadafal C, Garneret P, Jacquelin B, Kwasiborski A, Manuguerra JC, Monti F, Sakuntabhai A, Vanhomwegen J, Lafaye P, Tabeling P. Paper microfluidics for nucleic acid amplification testing (NAAT) of infectious diseases. Lab Chip. 2017;17(14):2347–2371. doi: 10.1039/C7LC00013H. [DOI] [PubMed] [Google Scholar]
- 32.Mao Kang, Zhang Hua, Yang Zhugen. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Envir Sci Technol. 2020 doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- 33.Massart S, et al. Virus detection by high-throughput sequencing of small RNAs: large scale performance testing of sequence analysis strategies strategies. Phytopathology. 2019;109(3):488–497. doi: 10.1094/PHYTO-02-18-0067-R. [DOI] [PubMed] [Google Scholar]
- 34.Mokhtarzadeh A, Eivazzadeh-Keihan R, Pashazadeh P, Hejazi M, Gharaatifar N, Hasanzadeh M, et al. Nanomaterial-based biosensors for detection of pathogenic virus. TrAC Trends Anal Chem. 2017;97:445–457. doi: 10.1016/j.trac.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes. 2002;16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 36.O’Sullivan CK. Aptasensors—the future of biosensing? Anal Bioanal Chem. 2002;372:44–48. doi: 10.1007/s00216-001-1189-3. [DOI] [PubMed] [Google Scholar]
- 37.Okba N, Muller M, Li W, Wang C, Geurts vanKessel C, Corman V et al medRxiv 2020. SARS-CoV-2 specific antibody responses in COVID-19 patients. Available here 10.1101/2020.03.18.20038059.
- 38.Ooi GC, Khong PL, Muller NL, et al. Severe acute respiratory syndrome: temporal lung changes at thin-section CT in 30 patients. Radiology. 2004;230:836–844. doi: 10.1148/radiol.2303030853. [DOI] [PubMed] [Google Scholar]
- 39.Pan Y, Guan H, Zhou S, et al. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur Radiol. 2020 doi: 10.1007/s00330-020-06731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paules CI, Marston HD, Fauci AS. Coronavirus infections more than just the common cold. JAMA. 2020 doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 41.Shi H, Han X, Zheng C. Evolution of CT manifestations in a patient recovered from 2019 novel coronavirus (2019-nCoV) pneumonia in Wuhan, China. Radiology. 2019;2020:200269. doi: 10.1148/radiol.2020200269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su S, Wong G, Shi W, Liu J, Lai AC, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.To KK, Tsang OT, Chik-Yan Yip C, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tok TT, Tatar G. Structures and functions of coronavirus proteins: molecular modeling of viral nucleoprotein. Int J Virol Infect Dis. 2017;2(1):001–007. [Google Scholar]
- 45.Van der H, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, et al. Identification of a new human coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woo PC, Lau SK, Wong BH, et al. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J Clin Microbiol. 2005;43:3054–3058. doi: 10.1128/JCM.43.7.3054-3058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie X, Zhong Z, Zhao W, et al. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yam WC, Chan KH, Poon LL, et al. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J Clin Microbiol. 2003;41:4521–4524. doi: 10.1128/jcm.41.10.4521-4524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]