Abstract
Background
DNA-binding one zinc finger (Dof) proteins are plant-specific transcription factors important for seed development, hormone regulation, and defense against abiotic stress. Although drought stress is a key determinant of plant physiology and metabolic homeostasis, the role of Dof genes in different degrees of PEG6000-induced drought stress has received little attention.
Methods
Tea plants (Camellia sinensis) were exposed to mild, moderate and severe drought stress. The Tea Genome and Plant TFDB databases were used to identify Dof gene family members in the tea plant. Clustal W2.1, MEGA6.0, ScanProsite, SMART, ExPASy, GSDS, MEME and STRING were used to build a phylogenetic tree, predict the molecular masses and isoelectric points of the Dof proteins, and construct a predicted protein-protein interaction network between the CsDof TFs and proteins in the A. thaliana database. The expression patterns of Dof genes in different tissues were analyzed, and qRT-PCR was used to measure the expression of Dof genes under different degrees of drought stress in tea.
Results
We identified 16 Dof genes in tea (C. sinensis cv. Huangjinya) using whole-genome analysis. Through comparative analysis of tea and Arabidopsis thaliana, we divided the Dof genes into four families (A, B, C, and D). We identified 15 motifs in the amino acid sequences of the CsDof proteins. Gene sequences and motif structures were highly conserved among families, especially in the B1 and C2 subfamilies. The protein-protein interaction network indicated that multiple CsDof proteins may be involved in the response to drought stress. Real-time PCR was used to examine the tissue-specific expression patterns of the CsDof genes and to measure their responses to different levels of PEG6000-induced drought stress in mature leaves. Most CsDof genes responded to drought stress. These results provide information on the Dof gene family in tea, offer new insights into the function of CsDof genes in a perennial species, and lay the foundation for further analysis of their functions.
Keywords: Drought stress, Dof transcription factors, Gene expression, Tea
Introduction
Drought stress refers to the phenomenon of water imbalance that occurs when the plant’s water transpiration is greater than its water absorption (Wang et al., 2016). Drought seriously endangers the growth and development of crops and is one of the main environmental factors that limit plant growth and reduce crop yields (Sharma et al., 2017; Zhu et al., 2018). Under severe drought stress, the stomatal conductance, net photosynthetic rate, and transpiration rate of Camellia sinensis cv. Tieguanyin decreased significantly (Lin, 1998). Studies have also shown that the superoxide dismutase activity of tea increases under short-term or mild drought stress but decreases under long-term or severe drought stress, causing tea plants to age more rapidly (Liu, 2006; Fu, 2018). It is therefore important to study and improve plant drought resistance.
Transcription factors play a particularly important role in plant growth and development (Lu et al., 2009). DNA-binding one zinc finger (Dof) proteins are a subfamily of the zinc finger protein family (Wei et al., 2018a; Wei et al., 2018b). This family is unique to plants and has not been found in yeast or nematodes (Gupta et al., 2015). The common ancestor of the Dof transcription factor family was discovered in Chlamydomonas reinhardtii, which has only one Dof transcription factor (Moreno-Risueno et al., 2007). Dof proteins consist of a conserved N-terminal single zinc finger DNA-binding domain (the Dof domain) and a C-terminal domain (Kushwaha et al., 2011; Rueda-Romero et al., 2012; Ma et al., 2015; Chen et al., 2017). Studies have found that Dof proteins are approximately 200–400 amino acids in length (Yanagisawa & Schmidt, 1999). The Dof domain consists of 50–52 amino acids and has a classical four-cysteine zinc finger that specifically binds to the core sequence (A/T)AAAG of target gene promoters (Yanagisawa & Schmidt, 1999). This bifunctional domain mediates both DNA–protein and protein–protein interactions (Yanagisawa, 1997; Krohn, 2002). For example, the Dof transcription factor OBP1 (OBF binding protein) interacts with the bZIP transcription factors OBF4 and OBF5 (two ocs element binding factors) (Zhang et al., 1996). The Dof domain is thought to interact with different regulatory proteins, leading to a diversity of Dof protein functions (Noguero et al., 2013), including plant defense (Chen, Chao & Singh, 1996; Ward et al., 2005; Rueda-Romero et al., 2012), abiotic stress response (Iwamoto, Higo & Takano, 2009), auxin response (Chen & Cao, 2015), and photoperiod response (Gualberti et al., 2002; Wang et al., 2007).
Corrales et al. (2014) found that the overexpression of SlCDF1 and SlCDF3 (two tomato Cycling Dof Factors) significantly enhanced the drought resistance and salt tolerance of Arabidopsis and activated other stress-responsive genes such as COR15, RD29A, and ERD10. Complementing the work of Li et al. (2016), we have identified 16 new CsDof genes, thereby expanding our understanding of drought stress response in tea.
Materials & Methods
Plant materials
Annual tea cuttings (C. sinensis cv. Huangjinya) were bought from a tea plantation located at the tea plantation base of the Chaxi Valley Co., Ltd. in Tai’an, Shandong Province (36.19°N, 117.11°E). Here, the planting area is 200 m above sea level, the soil fertility is moderate, and tea plants grow well. In October 2019, we bought and transplanted the cuttings to a natural light greenhouse at Shandong Agricultural University and carried out a one-week-long seedling treatment under standard horticultural conditions.
After the one-week treatment, we selected at least 30 tea plants (C. sinensis cv. Huangjinya) and collected their flower buds (FBs), stems, terminal buds (TBs), first leaves under new shoots (FLs), second leaves under new shoots (SLs), third leaves under new shoots (TLs), and fourth leaves under new shoots (mature leaves, MLs). The harvested samples were immediately snap frozen in liquid nitrogen and stored at −80 °C for later use.
We also selected another 30 tea plants after the one-week-long seedling treatment, cleaned the soil from the roots, and fixed each plant in a hydroponic box with a foam board. Plants were fully aerated daily with an oxygen pump, and Hoagland’s nutrient solution (Xia, 2010) (Table S1) was replaced every 3.5 days. After one week of pre-culture, a PEG-induced, simulated drought treatment was initiated. PEG6000 concentrations ranged from 10% (mild drought stress) to 30% (moderate drought stress) to 50% (severe drought stress). At 2, 4 and 6 h after treatment, 2–3 mature leaves were removed at the same height from at least three tea seedlings and stored at −80 °C for later use. Mature leaves from plants that had not been subjected to drought treatment (PEG6000 treatment for 0 h) were used as the control; all other processing conditions were the same as in the PEG6000 treatment. Leaf samples were quickly frozen in liquid nitrogen and stored in a −80 °C freezer for later use.
RNA extraction and quantitative real-time PCR analysis of the Dof genes
Total RNA was extracted with the RNAprep Pure Polysaccharide Polyphenol Plant Total RNA Extraction Kit (Tiangen, Cat No. DP441), and first-strand cDNA was synthesized using the Evo M-MLV RT Kit with gDNA Clean for qPCR (Accurate Biotechnology (Hunan) Co., Ltd, China) according to the manufacturer’s instructions. Real-time quantitative reverse transcription PCR (qRT-PCR) was used to detect the expression level of each gene using a cDNA template. Sixteen quantitative primers were designed using BD software (Tables S2). The internal reference was glyceraldehyde 3-phosphate dehydrogenase (GAPDH), which was synthesized by Sangon Biotech (Shanghai) Co., Ltd. GAPDH is considered to be the best reference gene under drought stress (Hao, 2012; Fang et al., 2017), and its expression does not differ among different developmental stages. We selected 16 Dof genes for qRT-PCR analysis (Table S3 and S4) using the SYBR® Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology (Hunan) Co., Ltd, China). The 20 µL qRT-PCR reaction system contained 10.0 µL 2 ×ChamQ Universal SYBR qPCR Master Mix, 0.4 µL (10 µmol L−1) upstream and downstream primers, 1.0 µL template, and 8.2 µL ddH2O. Three technical replicates were performed for each sample. The reaction conditions were pre-denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. A dissociation curve was drawn using 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s. The experimental data were quantitatively analyzed using the 2−ΔΔCT method (Chen et al., 2017). We measured the expression of Dof genes in various tissues and in mature leaves under different simulated drought conditions. Each reaction was repeated three times, and the results are an average of three independent biological replicates.
Database searches and identification of Dof family members in tea
The tea genome was downloaded from the Tea Genome Database (http://itak.feilab.net/cgi-bin/itak/index.cgi) (Xia et al., 2017). To identify all Dof genes in tea, the Dof domain (Pfam PF02701) was obtained from Pfam (http://pfam.xfam.org) (Finn et al., 2015). To verify the authenticity of candidate sequences, an HMM (hidden Markov model) profile of the Dof domain (PF02701) was used as a query to identify Dofs using the HMMER3.0 program (http://hmmer.janelia.org) (Finn et al., 2015). SMART (http://smart.embl.de) (Letunic, Doerks & Bork, 2015) and ScanProsite (http://www.expasy.ch/tools/scanprosite/) (Castro et al., 2006) were used to examine the CsDof domains of the deduced amino acid sequences (Table S4). The isoelectric points, molecular weights, instability indices, aliphatic indices, and grand average of hydropathicity (GRAVY) scores of the proteins were predicted using the ExPASy Proteomics Server (http://expasy.org/). If more than one allele was present in the genome file, we selected the longest allele for analysis. The Dof transcription factor sequences of Arabidopsis (Table S5) were downloaded from the Plant Transcription Factor Database (http://planttfdb.cbi.edu.cn/), and redundant genes were removed (Jin et al., 2014a). The CsDof proteins were used as BLASTP query sequences against the Arabidopsis thaliana (TAIR10) protein sequence file with default parameters (E-value < 1e−5) (Wang et al., 2019). Homologous Arabidopsis Dof proteins with the highest bit scores were used to construct a protein-protein interaction (PPI) network with STRING (version 10.0) (http://string-db.org/) (Damian et al., 2015), using the Arabidopsis database as the selected organism.
Phylogenetic analysis, gene structure and motif identification
The Dof family protein sequences of Arabidopsis and tea were obtained as described above. All sequences were aligned using the default settings of ClustalX 2.1 (Larkin et al., 2007), and a phylogenetic tree was constructed with the neighbor-joining algorithm in MEGA 6.0 (http://www.megasoftware.net/mega6/). The reliability of the resulting tree was assessed using 1,000 bootstrap replicates. The structures of the Dof genes were analyzed online using the Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/). The Dof family transcription factor database was downloaded from the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/) (Jin et al., 2014a). MEME Suite was used to identify motifs in the CsDof protein sequences (Bailey et al., 2009) using a motif width of 6–50 and a maximum of 15 motifs (Ma et al., 2015).
Results
Identification of the CsDof genes
We identified 16 non-redundant putative CsDof genes in the tea genome (Tables S3 and S4). Their lengths and predicted molecular weights varied widely, but there were fewer differences in their theoretical isoelectric points (Table 1). The predicted Dof transcription factors were 486 to 1,428 amino acids in length; their molecular weights ranged from 39.3 to 120.2 kDa, and their theoretical isoelectric points were close to 5 (4.99 to 5.22).
Table 1. Physiochemical properties of the tea CsDof genes and their corresponding proteins.
| Gene name | Gene ID | Size (aa) | Molecular weight (kDa) | PI | Instability index | Aliphatic index | Grand average of hydropathicity (GRAVY) | |
|---|---|---|---|---|---|---|---|---|
| A | CsDof1 | CsA005492 | 864 | 71.81608 | 5.05 | 60.65 unstable | 25.93 | 0.948 |
| B 1 | CsDof2 | CsA012146 | 1,047 | 87.01927 | 5.03 | 51.35 unstable | 24.45 | 0.813 |
| CsDof3 | CsA012347 | 870 | 72.83445 | 5.07 | 59.00 unstable | 27.82 | 0.870 | |
| B 2 | CsDof4 | CsA009371 | 1,428 | 120.21870 | 5.00 | 44.41 unstable | 26.47 | 0.745 |
| CsDof5 | CsA032220 | 1,122 | 95.09702 | 5.04 | 45.53 unstable | 28.34 | 0.804 | |
| C 1 | CsDof6 | CsA002607 | 1,047 | 86.47761 | 5.07 | 49.66 unstable | 29.23 | 0.779 |
| CsDof7 | CsA002685 | 966 | 79.51451 | 5.09 | 49.79 unstable | 26.71 | 0.717 | |
| C 2.1 | CsDof8 | CsA020146 | 804 | 65.50705 | 5.13 | 51.25 unstable | 30.35 | 0.794 |
| CsDof9 | CsA028787 | 807 | 66.88200 | 5.12 | 51.32 unstable | 30.98 | 0.832 | |
| C 2.2 | CsDof10 | CsA027884 | 744 | 61.30654 | 5.13 | 33.36 stable | 26.61 | 0.756 |
| C 3 | CsDof11 | CsA002683 | 765 | 64.08197 | 5.09 | 60.62 unstable | 28.76 | 0.940 |
| CsDof12 | CsA007538 | 930 | 78.49438 | 5.02 | 63.63 unstable | 27.31 | 1.022 | |
| D 1 | CsDof13 | CsA013235 | 486 | 39.32758 | 5.22 | 32.61 stable | 26.75 | 0.700 |
| CsDof14 | CsA013544 | 1,407 | 116.56649 | 4.99 | 46.46 unstable | 31.56 | 0.896 | |
| CsDof15 | CsA021984 | 1,389 | 113.63680 | 5.00 | 45.31 unstable | 31.61 | 0.884 | |
| CsDof16 | CsA027886 | 1,311 | 107.98698 | 5.00 | 46.87 unstable | 30.28 | 0.900 |
The instability index, the aliphatic index, and the GRAVY score were similar within each subfamily, but there were large differences among different subfamilies (Table 1). For example, the two members of the C2.1 subfamily had instability indices of 51.25 and 51.32 (unstable), aliphatic indices of 30.35 and 30.98, and GRAVY scores of 0.794 and 0.832. However, the two members of the C3 subfamily had instability indices of 60.62 and 63.63 (unstable), aliphatic indices of 28.76 and 27.31, and GRAVY scores of 0.94 and 1.022.
Phylogenetic analysis and classification of Dof genes in tea and Arabidopsis
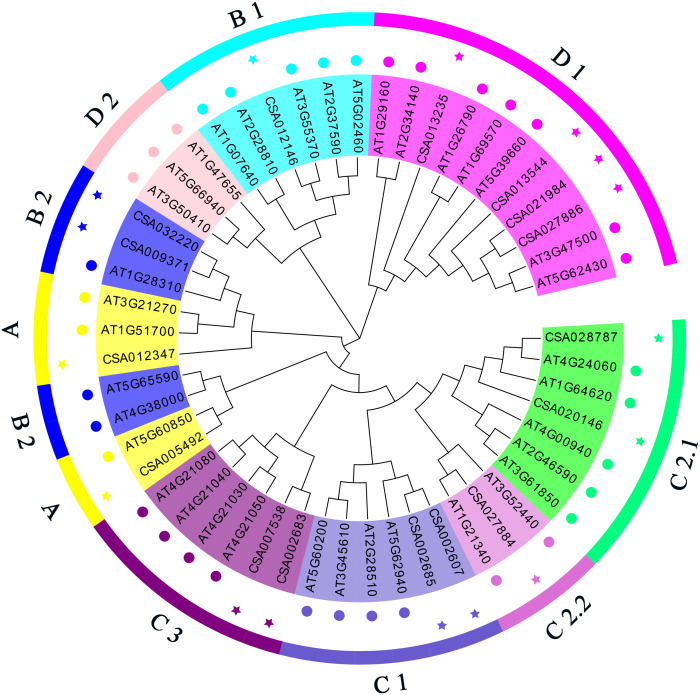
To study the molecular evolution of the tea CsDof genes and predict their functions, tea and Arabidopsis Dof proteins (Tables S4 and S5) were used to construct a phylogenetic tree (Fig. 1). Based on the phylogenetic tree and previous reports, the predicted tea CsDof genes were divided into four major families (A, B, C, and D) and seven subfamilies (A, B1, B2, C1, C2, D1, and D2). Together, the C and D families constituted the largest group, including 11 members and accounting for 68.75% of the total number of predicted genes. Family B contained four members, accounting for 25% of the predicted genes, and Family A contained only one member, accounting for 6.25% of the predicted genes. By comparing tea and Arabidopsis Dof proteins, we found that Dof transcription factors from different species within the same family (any of the four major families A, B, C, and D) were more similar to one another than were Dof transcription factors from the same species in different families. For example, Arabidopsis and tea Dof genes classified into Family A were more similar to one another than were tea Dof genes classified into Families A and B.
Figure 1. Phylogenetic relationships among C. sinensis and A. thaliana Dof proteins.
The neighbor-joining tree was created using the MEGA6.0 program (bootstrap value set at 1,000). Thirty-six AtDof proteins marked with various colors pentacle and 19 CsDof proteins marked with various colors pentacle. The resulting phylogenetic tree was clustered into four major groups (A, B, C and D). The different colors of the pentacles represent different subfamilies.
Gene structures and protein motifs of the CsDof gene family
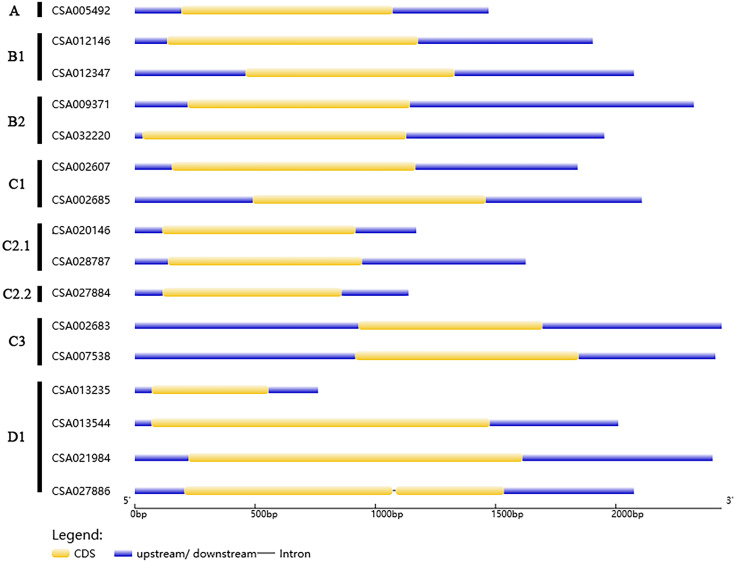
We analyzed the structures of the CsDof genes based on their coding sequences and genomic sequences (Fig. 2). The number of introns per gene ranged from zero to one. Only one CsDof gene (CsDof16) had an intron, and the others had no introns. In general, CsDof genes from the same subfamily had the same gene structure, indicating that tea Dof gene evolution is conserved. To reveal the diversity of Dof proteins in tea, we used the MEME Suite to identify motifs in the CsDof protein sequences. A total of 15 motifs were identified in the predicted CsDof proteins (Figs. 3 and 4). All CsDof proteins contained motif 1, which represents the conserved Dof domain. Furthermore, motif 13 was another conserved motif found in seven CsDofs. Basically, each subfamily had a specific motif, such as motif 14 in subfamily B2, motif 12 in subfamily C1, and motifs 7 and 10 in subfamily C2.1. Motifs 2, 3, and 11 were found only in subfamily D1. Several closely related CsDofs in the phylogenetic tree contained common motifs, suggesting that CsDofs from the same subfamily have similar functions. Analysis of gene structure and protein motif locations in the CsDofs indicated that most members were conserved in individual subfamilies.
Figure 2. Phylogenetic analysis and structure of Dof genes in tea.
In the gene structure diagram, yellow box, blue and black lines represent exons, upstream/downstream regions of the gene and introns, respectively.
Figure 3. Common motifs of CsDof family proteins.
Dof domains are represented by boxes of different colours.
Figure 4. Sequence logos of tea Dof domains.
The Interaction Network of Dof TFs between C. sinensis and Arabidopsis
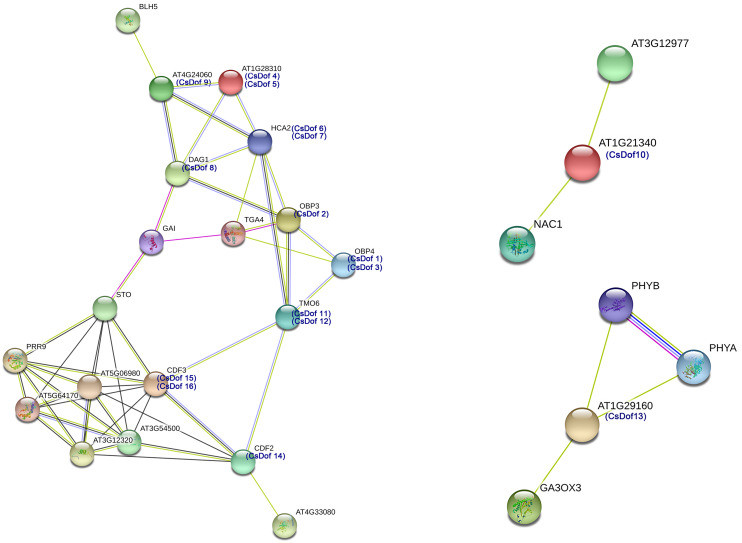
We predicted protein-protein interactions between Arabidopsis homologs of the CsDof TFs and other Arabidopsis proteins (Fig. 5). Different line colors represent the types of evidence for the association. The amino acid sequence of CDF2 was highly similar to that of CsDof14, and the sequence of CDF3 was highly similar to those of CsDof15 and CsDof16. There was a predicted interaction between AT1G21340, which is highly similar to CsDof10, and the drought resistance protein NAC1 (Li et al., 2014). Moreover, AT1G29160, which was highly similar to CsDof13, was predicted to have complex interactions with the drought resistance-related protein PHYB (Yoo et al., 2017) and the abiotic stress protein GA3ox3 (Pan et al., 2017). In addition, HCA2, which was highly similar to CsDof6 and CsDof7, was predicted to have complex interactions with seven CsDofs (CsDof2, CsDof4, CsDof5, CsDof8, CsDof9, CsDof11 and CsDof12). Similarly, OBP3, which was highly similar to CsDof2, was also predicted to have complex interactions with seven CsDofs (CsDof1, CsDof3, CsDof6, CsDof7, CsDof8, CsDof11 and CsDof12).
Figure 5. The interaction networks of Dofs in C. sinensis according to the orthologs in Arabidopsis.
Tissue expression patterns of CsDof genes
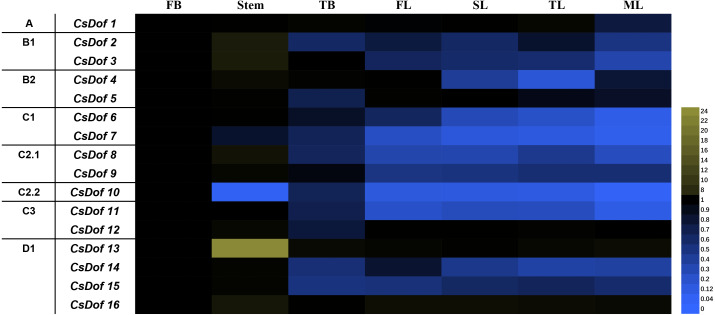
Sixteen CsDof genes were expressed in the flower buds (FBs), stems, terminal buds (TBs), first leaves under new shoots (FLs), second leaves under new shoots (SLs), third leaves under new shoots (TLs), and fourth leaves under new shoots (mature leaves, MLs). There were differences in expression patterns among different tissues (Fig. 6). For example, the expression levels of CsDof7 and CsDof10 were higher in FBs than in other tissues, and CsDof2, CsDof3, CsDof8, CsDof13, and CsDof16 had the highest expression in stems. Approximately 56.25% of the CsDof genes were significantly downregulated in TBs compared to FBs. The expression levels of CsDof4 and CsDof14 in TLs were significantly lower than those in other tissues. CsDof 8 and CsDof9 were significantly downregulated in FLs. Eleven (68.75%) of the CsDof genes were significantly downregulated in SLs compared to FBs. CsDof1, CsDof2, CsDof3, and CsDof6 were significantly downregulated in MLs. The expression levels of CsDof16 were similar among all tissues.
Figure 6. Relative expression profiles of Dof genes in different tissues of tea plants.
The gene expression of different tissues of tea plants was analyzed by qRT-PCR. Expression levels were normalized against that of GAPDH. FB denotes the flower bud, TB means the terminal bud, FL denotes the first leaf of new sprouting shoots, SL means the second leaf, TL denotes the third leaf and ML means the mature fourth leaf.
Expression patterns of CsDof genes under PEG6000-induced drought stress
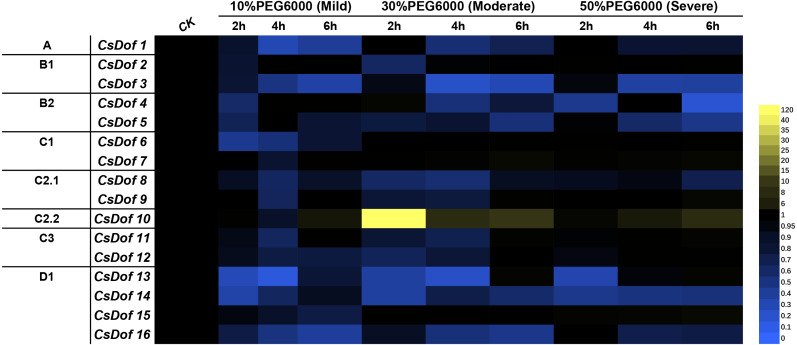
We analyzed the expression of 16 CsDof genes at 0, 2, 4 and 6 h after exposure to different degrees of PEG6000-induced drought stress (mild, moderate and severe drought stress) (Fig. 7) and found that most CsDof genes responded to drought stress.
Figure 7. Expression patterns of CsDof genes in response to drought stress in tea plant cultivar ‘Huangjinya’.
Mild means mild drought stress, Moderate means moderate drought stress, Severe means severe drought stress.
The expression of CsDof10 after 6 h of mild drought stress was significantly higher than its expression under the control treatment. CsDof1, CsDof3, CsDof6, CsDof8, CsDof12 and four genes in the D1 subfamily were significantly downregulated after 2, 4 and 6 h of mild drought stress. CsDof2 and CsDof4 were significantly downregulated only after 2 h of mild drought stress. The genes whose expression levels decreased significantly after 4 h of mild drought stress were CsDof7, CsDof9 and CsDof10.
CsDof10 was significantly upregulated after 2 h of moderate drought stress; its expression declined gradually at 4 and 6 h but remained significantly higher than that of control plants. In addition, 75% (12) of the CsDof genes showed significantly lower expression under moderate drought stress than under control conditions, and the expression of CsDof5, CsDof8, CsDof14 and CsDof16 continued to decrease after 2, 4, and 6 h of drought treatment.
Under severe drought stress, the expression of 43.75% (7) genes was significantly lower than under the control treatment, and the expression of CsDof1, CsDof3, CsDof5, CsDof14, and CsDof16 continued to decrease after 2–6 h of treatment. CsDof13 was significantly downregulated only when treated with severe drought stress for 2 h. Interestingly, the expression of CsDof10 showed a gradual increase with time, and its expression was the highest after 6 h of severe drought stress.
Discussion
Li et al. (2016) identified 29 tea tree Dof genes and predicted that CsDof-22 interacted with ABA1 and participated in drought stress. However, Li et al. (2016) only studied the expression of 8 of the 29 Dof family members at different time points under single levels of high temperature, low temperature, drought stress, and salt stress. They found that only CsDof-8 and CsDof-13 responded to drought stress at the transcriptional level and that CsDof-22 did not change significantly at the transcriptional level compared to the control. Here, we identified 16 new members of the Dof gene family in tea and focused on their role in the mechanism of drought response under different degrees of drought stress. We specifically studied the response of the 16 new members to light, moderate and severe drought stress over time, expanding our understanding of the role of tea tree Dofs in the response to drought stress.
Dof gene numbers in multiple plant species
With advances in genome sequencing technology, members of the Dof gene family have been identified in many species, including Arabidopsis (Kushwaha et al., 2011), tomato (Cai et al., 2013), rice (Lijavetzky, Carbonero & Vicente-Carbajosa, 2003), castor bean (Jin, Chandrasekaran & Liu, 2014b), peach (Chen et al., 2017), eggplant (Wei et al., 2018b), physic nut (Zou & Zhang, 2019), and others. The tea genome has been sequenced (Wei et al., 2018a; Xia et al., 2017), and Li et al. (2016) identified 29 putative Dof TFs. In this work we identified 16 new CsDof genes (Fig. S2).
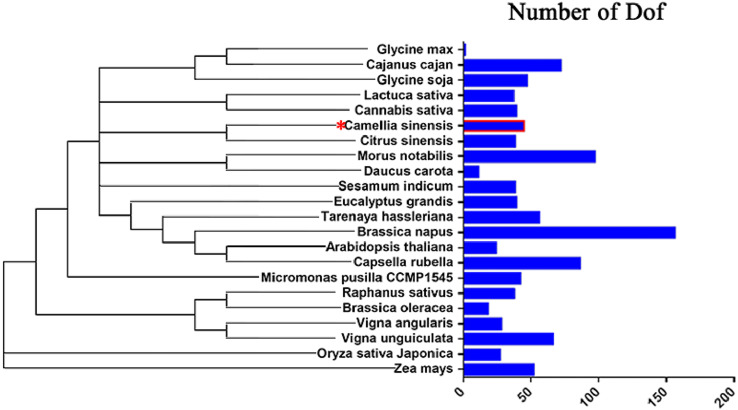
To study the evolution of Dof genes in plants, we compared 22 different algal and plant species, including species from the Chlorophyta and the Embryophyta subkingdoms, and determined how many Dof genes were present in each species (Chen et al., 2017; Letunic & Bork, 2007; Letunic & Bork, 2011). The number of Dof genes in different species ranged from 1 to 156. Embryophyte species had more Dof genes than chlorophyte algae (Fig. 8), suggesting that Dof genes have played an important role in the evolutionary process.
Figure 8. Distribution of Dof transcription factors in different species.
Duplication of the CsDof genes
Dof transcription factors are found not only in angiosperms and gymnosperms but across all plant lineages, presumably because a longer breeding time has led to a greater diversity of the Dof family and the Dof transcription factors participate in more biological processes (Yanagisawa, 2004; Yin et al., 2017). The Dof family has been found in Arabidopsis (Kushwaha et al., 2011), tomato (Cai et al., 2013), rice (Lijavetzky, Carbonero & Vicente-Carbajosa, 2003) and peach (Chen et al., 2017), which have 36, 34, 30, and 25 Dof transcription factors, respectively. To date, a total of 45 CsDof genes have been reported in tea (Li et al., 2016), but the number of Dof family members is independent of genome size. For example, the number of Dof family members in peach (25) (Chen et al., 2017) is less than that in Arabidopsis (36) (Kushwaha et al., 2011), although the genome size of peach (224.6 Mb) is almost twice that of Arabidopsis (125 Mb). We found that the number of Arabidopsis Dofs was four-fifths that of tea (45), despite the fact that the tea genome (3.1 Gb) is 25 times bigger than that of Arabidopsis (Xia et al., 2017). Studies have shown that more than half of the bases (67%) in the tea genome are retrotransposon sequences, with numerous copies and insertions into different sites, leading to a dramatic expansion of genome size (Xia et al., 2017). Therefore, although the size of the tea genome is much larger than that of Arabidopsis, the number of Dof family members is only slightly greater, perhaps due to gene duplication during evolution.
Homology analysis of the tea and Arabidopsis Dof genes
Dof proteins in Arabidopsis are usually divided into four families (A–D) (Kushwaha et al., 2011). We also divided the tea Dof genes into four families based on the positions of their proteins on a phylogenetic tree. Moreover, we found that gene structures were consistent within families, suggesting that the genes within a family may have similar functions (Chen et al., 2017).
Papi et al. (2000) demonstrated that AtDAG1 (Dof affecting germination) is expressed in flowers and mature pericarp tissue, mainly in the seed coat and phloem. We found that tea genes assigned to the C2.1 subfamily with AtDAG1 were mainly expressed in flower buds, stems, and terminal buds. Their expression levels in leaves were lower, consistent with previous studies (Papi et al., 2000). Therefore, we speculate that genes of the C2.1 subfamily in tea are similar to those in Arabidopsis and that most of them are involved in the plant vascular system and seed development (Gabriele et al., 2009). The results of gene expression analysis provide a basis for the functional characterization of CsDof genes, and the phylogenetic analysis of the Dof family provides a theoretical basis for further functional genomics studies in tea.
Transcript profiles of CsDof paralogs
We found that the expression patterns of the two C1 subfamily members differed: CsDof7 (CsA002685) was mainly expressed in flower buds, whereas CsDof6 (CsA002607) was primarily expressed in flower buds and stems. Some genes and their paralogs play redundant roles (Fornara et al., 2009), but other paralogs, such as AtDof3.4 (OBP1) and AtDof5.8 (SCAP1), have different functions. Although both are OG-2b orthologs (Zou & Zhang, 2019), AtDof3.4 participates in defensive response (Zhang et al., 1995) and cell cycle regulation (Skirycz et al., 2008), whereas AtDof5.8 (SCAP1) participates in vascular development (Konishi & Yanagisawa, 2007), stomatal function and morphogenesis (Negi et al., 2013). Therefore, we speculate that genes that are expressed differently in some subfamilies may be involved in different growth and developmental processes. Moreover, differences in the expression of Dof genes from the same subfamily may be related to sequences other than conserved motifs.
CsDof proteins may interact with proteins that respond to drought stress
We predicted protein-protein interactions between Arabidopsis homologs of the CsDof TFs and other Arabidopsis proteins. In Arabidopsis and tomato, CDFs are involved in the response to drought stress (Corrales et al., 2014; Hoekstra, Golovina & Buitink, 2001; Rizhsky, 2004). We predicted by protein-protein interaction network analysis that the CDF2 amino acid sequence was highly similar to that of CsDof14, and the CDF3 sequence was highly similar to those of CsDof15 and CsDof16. Moreover, CDF3 was predicted to have complex interactions with CDF2, STO (salt tolerance protein), CsDof11, CsDof12, and five other proteins (AT5G06980, PRR9, AT5G64170, AT3G54500, AT3G12320). Similarly, CsDof10 was predicted to interact with NAC1 (drought resistance protein) (Li et al., 2014), and CsDof13 was predicted to interact with PHYB, which is involved in drought resistance (Yoo et al., 2017). Therefore, we speculate that CsDof10, CsDof13, CsDof14, CsDof15, CsDof16 may play important roles in the drought stress response. Moreover, CsDof15 and CsDof16 may participate in the drought stress response through interaction with CsDof11, CsDof12 and CsDof14.
The Dof gene family may be involved in drought stress response
In this study, we investigated the responses of CsDof genes to varying degrees of PEG6000-induced drought stress. Most CsDofs responded to different degrees of drought stress, although the details of their responses differed. This suggests that the CsDof genes may play various roles in drought stress.
Corrales et al. (2014) found that all SlCDF genes, which are members of the Dof gene family in tomato, are regulated by drought and that members of this gene family may be upstream activators of drought stress response pathways, directly or indirectly acting on different stress-regulated target genes (Corrales et al., 2014). In Arabidopsis, the overexpression of SlCDF3 promoted the accumulation of compounds such as proline, glutamine, GABA and sucrose (Hoekstra, Golovina & Buitink, 2001; Rizhsky, 2004). The levels of these compounds usually change significantly under drought stress (Kerepesi & Galiba, 2000; Farrant & Moore, 2011; Pinheiro & Chaves, 2011), increasing stress tolerance through osmotic adjustment, detoxification of ROS, and intracellular pH regulation (Munns & Tester, 2008; Bressan, Bohnert & Zhu, 2009; Chaves, Flexas & Pinheiro, 2008), Here, we found that CsDof15 (CsA021984) and CsDof16 (CsA027886) exhibited up to 100% identity with SlCDF1 and SlCDF3 (Fig. S1) and were responsive to drought stress. In particular, the expression of CsDof16 under various levels of drought stress gradually decreased through time. By contrast, CsDof15 expression showed a gradual downward trend only under mild drought stress as the treatment time increased from 2 to 6 h. We found only one C2.2 subfamily member (CsDof10) whose expression was significantly upregulated after 2 h of moderate drought stress in comparison to the control condition. Thus, CsDof10 may play an important role in moderate drought stress.
Conclusions
In summary, 16 new CsDof genes were identified. Analysis of their physiochemical properties, phylogeny, gene structure and PPI network provided more complete information for this gene family in tea. Gene expression profiles after drought stress indicated that some of the CsDof s may play a role in drought resistance. The results of this study provide a basis for future functional characterization of the role of Dof genes in drought stress in eukaryotes.
Supplemental Information
Funding Statement
This research was supported by Funds of Shandong ‘Double Tops’ Program (SYL2017YY03).
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Qian Yu and Chen Li conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Jiucheng Zhang performed the experiments, prepared figures and/or tables, and approved the final draft.
Yueyue Tian and Hanyue Wang performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Yue Zhang, Qinzeng Xiang and Xiaoyang Han analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Zhengqun Zhang analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Lixia Zhang conceived and designed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
The 16 CsDofs sequences listed in Table 1 are available at http://itak.feilab.net/cgi-bin/itak/db_family_gene_list.cgi?acc=C2C2-Dof&plant=4442.
Data Availability
The following information was supplied regarding data availability:
The raw gene data of tea and Arabidopsis are available in Tables S4 and S5. The alignment analysis of the amino acid sequences of Dof (CsDof1-29 amino acid sequence was already studied (Li et al., 2016) and 16 distinct CsDofs we identified) is available in Fig. S2.
References
- Bailey et al. (2009).Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren JY, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan, Bohnert & Zhu (2009).Bressan R, Bohnert H, Zhu J-K. Abiotic stress tolerance: from gene discovery in model organisms to crop improvement. Molecular Plant. 2009;2(1):1–2. doi: 10.1093/mp/ssn097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai et al. (2013).Cai XF, Zhang YY, Zhang C, Zhang T, Hu T, Ye J. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. Journal of Integrative Plant Biology. 2013;55:552–566. doi: 10.1111/jipb.12043. [DOI] [PubMed] [Google Scholar]
- Castro et al. (2006).Castro ED, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. Scanprosite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Research. 2006;34:362–365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves, Flexas & Pinheiro (2008).Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Annals of Botany. 2008;103(4):551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen & Cao (2015).Chen Y, Cao J. Comparative analysis of Dof transcription factor family in maize. Plant Molecular Biology Reporter. 2015;33(5):1245–1258. doi: 10.1007/s11105-014-0835-9. [DOI] [Google Scholar]
- Chen, Chao & Singh (1996).Chen W, Chao G, Singh KB. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. The Plant Journal. 1996;10:955–966. doi: 10.1046/j.1365-313X.1996.10060955.x. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2017).Chen M, Liu X, Huan L, Sun M, Liu L, Chen X, Gao D, Li L. Genome-wide analysis of Dof family genes and their expression during bud dormancy in peach (Prunus persica) Scientia Horticulturae. 2017;214:18–26. doi: 10.1016/j.scienta.2016.11.014. [DOI] [Google Scholar]
- Corrales et al. (2014).Corrales AR, Nebauer SG, Carrillo L, Fernández-Nohales P, Marqués J, Renau-Morata B, Granell A, Pollmann S, Vicente-Carbajosa J, Molina RV, Medina J. Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. Journal of Experimental Botany. 2014;65(4):995–1012. doi: 10.1093/jxb/ert451. [DOI] [PubMed] [Google Scholar]
- Damian et al. (2015).Damian S, Andrea F, Stefan W, Kristoffer F, Davide H, Jaime H-C, Milan S, Alexander R, Alberto S, Kalliopi PK, Michael K, Peer B, Lars JJ, Christian von M. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;2015(43):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang et al. (2017).Fang J, Li C, Ma C, Chen L. Molecular cloning, bioinformatics and expression analysis of GGPS gene family in tea plant. Journal of Tea Science. 2017;37(2):130–138. doi: 10.13305/j.cnki.jts.2017.02.002. [DOI] [Google Scholar]
- Farrant & Moore (2011).Farrant JM, Moore JP. Programming desiccation-tolerance: from plants to seeds to resurrection plants. Current Opinion in Plant Biology. 2011;14(3):340–345. doi: 10.1016/j.pbi.2011.03.018. [DOI] [PubMed] [Google Scholar]
- Finn et al. (2015).Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research. 2015;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara et al. (2009).Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupl G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Developmental Cell. 2009;17(1):75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Fu (2018).Fu Z. Master thesis. 2018. Effects of exogenous hydrogen sulfide on seeds germination and seedlings physiological characteristics of tea (Camellia sinensis) under drought stress. [Google Scholar]
- Gabriele et al. (2009).Gabriele S, Rizza A, Martone J, Circelli P, Costantino P, Vittorioso P. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. The Plant Journal. 2009;61(2):312–323. doi: 10.1111/j.1365-313X.2009.04055.x. [DOI] [PubMed] [Google Scholar]
- Gualberti et al. (2002).Gualberti G, Papi M, Bellucci L, Ricci I, Bouchez D, Camilleri C, Costantino P, Vittorioso P. Mutations in the dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of arabidopsis seeds. The Plant Cell. 2002;14:1253–1263. doi: 10.1105/tpc.010491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta et al. (2015).Gupta S, Malviya N, Kushwaha H, Nasim J, Bisht NC, Singh VK, Yadav D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta. 2015;241(3):549–562. doi: 10.1007/s00425-014-2239-3. [DOI] [PubMed] [Google Scholar]
- Hao (2012).Hao S. Master thesis. 2012. Selection of appropriate reference genes for expression studies in Camellia sinensis by real-time polymerase chain reaction. [Google Scholar]
- Hoekstra, Golovina & Buitink (2001).Hoekstra FA, Golovina EA, Buitink J. Mechanisms of plant desiccation tolerance. Trends in Plant Science. 2001;6(9):0–438. doi: 10.1016/s1360-1385(01)02052-0. [DOI] [PubMed] [Google Scholar]
- Iwamoto, Higo & Takano (2009).Iwamoto M, Higo K, Takano M. Circadian clock- and phytochrome-regulated Dof-like gene, Rdd1, is associated with grain size in rice. Plant, Cell and Environment. 2009;32:592–603. doi: 10.1111/j.1365-3040.2009.01954.x. [DOI] [PubMed] [Google Scholar]
- Jin, Chandrasekaran & Liu (2014b).Jin Z, Chandrasekaran U, Liu A. Genome-wide analysis of the Dof transcription factors in castor bean (Ricinus communisL.) Genes & Genomics. 2014b;36(4):527–537. doi: 10.1007/s13258-014-0189-6. [DOI] [Google Scholar]
- Jin et al. (2014a).Jin J, Zhang H, Kong L, Gao G, Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Research. 2014a;42(D1):1182–1187. doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerepesi & Galiba (2000).Kerepesi I, Galiba G. Osmotic and salt stress-induced alteration in soluble carbohydrate content in wheat seedlings. Crop Science. 2000;40(2):482–487. doi: 10.2135/cropsci2000.402482x. [DOI] [Google Scholar]
- Konishi & Yanagisawa (2007).Konishi M, Yanagisawa S. Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana. Plant Physiology and Biochemistry. 2007;45(8):623–629. doi: 10.1016/j.plaphy.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Krohn (2002).Krohn NM. Specificity of the stimulatory interaction between chromosomal HMGB proteins and the transcription factor Dof2 and its negative regulation by protein kinase CK2-mediated phosphorylation. Journal of Biological Chemistry. 2002;277(36):32438–32444. doi: 10.1074/jbc.M203814200. [DOI] [PubMed] [Google Scholar]
- Kushwaha et al. (2011).Kushwaha H, Gupta S, Singh VK, Rastogi S, adav DY. Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Molecular Biology Reports. 2011;38:5037–5053. doi: 10.1007/s11033-010-0650-9. [DOI] [PubMed] [Google Scholar]
- Larkin et al. (2007).Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Letunic & Bork (2007).Letunic I, Bork P. Interactive tree of life (itol): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Letunic & Bork (2011).Letunic I, Bork P. Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Research. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, Doerks & Bork (2015).Letunic I, Doerks T, Bork P. Smart: recent updates, new developments and status in 2015. Nucleic Acids Research. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2016).Li H, Huang W, Liu Z, Wang Y, Zhuang J. Transcriptome-based analysis of Dof family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis) International Journal of Genomics. 2016;2016(21):1–15. doi: 10.1155/2016/5614142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2014).Li XL, Yang X, Hu YX, Yu XD, Li QL. A novel NAC transcription factor from Suaeda liaotungensis K. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance. Plant Cell Reports. 2014;33(5):767–778. doi: 10.1007/s00299-014-1602-y. [DOI] [PubMed] [Google Scholar]
- Lijavetzky, Carbonero & Vicente-Carbajosa (2003).Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evolutionary Biology. 2003;3:17. doi: 10.1186/1471-2148-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin (1998).Lin J. Effect of water stress on the photosynthesis of tea. Journal of Fujian Agricultural University. 1998;27(04):40–44. [Google Scholar]
- Liu (2006).Liu Y. Master thesis. 2006. Study on the physiological and biochemical mechanism of drought-resistance in tea [Camellia sinensis (L.) O. Kuntze] [Google Scholar]
- Lu et al. (2009).Lu G, Gao C, Zheng X, Han B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta. 2009;229(3):605–615. doi: 10.1007/s00425-008-0857-3. [DOI] [PubMed] [Google Scholar]
- Ma et al. (2015).Ma J, Li MY, Wang F, Tang J, Xiong AS. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics. 2015;16:1–15. doi: 10.1186/s12864-015-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno et al. (2007).Moreno-Risueno MA, Martinez M, Vicente-Carbajosa J, Carbonero P. The family of DOF transcription factors: from green unicellular algae to vascular plants. Molecular Genetics and Genomics. 2007;277:379–390. doi: 10.1007/s00438-006-0186-9. [DOI] [PubMed] [Google Scholar]
- Munns & Tester (2008).Munns R, Tester M. Mechanisms of salinity tolerance. Annual Review of Plant Biology. 2008;59(1):651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Negi et al. (2013).Negi J, Moriwaki K, Konishi M, Yokoyama R, Nakano T, Kusumi K, Hashimoto-Sugimoto M, Schroeder JI, Nishitani K, Yanagisawa S, Iba K. A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Current Biology. 2013;23(6):479–484. doi: 10.1016/j.cub.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguero et al. (2013).Noguero M, Atif RM, Ochatt S, Thompson RD. The role of the DNA-binding one zinc finger (DOF) transcription factor family in plants. Plant Science. 2013;209:32–45. doi: 10.1016/j.plantsci.2013.03.016. [DOI] [PubMed] [Google Scholar]
- Pan et al. (2017).Pan C, Tian KH, Ban QY, Wang LG, Sun QL, He Y, Yang YF, Pan YT, Li YY, Jiang JY, Jiang CJ. Genome-wide analysis of the biosynthesis and deactivation of gibberellin-dioxygenases gene family in Camellia sinensis (L.) O. Kuntze. Gene. 2017;8(9):235. doi: 10.3390/genes8090235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi et al. (2000).Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, Vittorioso P. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes & Development. 2000;14(1):28–33. [PMC free article] [PubMed] [Google Scholar]
- Pinheiro & Chaves (2011).Pinheiro C, Chaves MM. Photosynthesis and drought: can we make metabolic connections from available data? Journal of Experimental Botany. 2011;62(3):869–882. doi: 10.1093/jxb/erq340. [DOI] [PubMed] [Google Scholar]
- Rizhsky (2004).Rizhsky L. When defense pathways collide. The Response of arabidopsis to a combination of drought and heat stress. Plant Physiology. 2004;134(4):1683–1696. doi: 10.1104/pp.103.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda-Romero et al. (2012).Rueda-Romero P, Barrero-Sicilia C, Gomez-Cadenas A, Carbonero P, Onate-Sanchez L. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. Journal of Experimental Botany. 2012;63(5):1937–1949. doi: 10.1093/jxb/err388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma et al. (2017).Sharma M, Gupta SK, Majumder B, Maurya VK, Deeba F, Alam A, Pandey V. Salicylic acid mediated growth, physiological and proteomic responses in two wheat varieties under drought stress. Journal of Proteomics. 2017;163(23):28–57. doi: 10.1016/j.jprot.2017.05.011. [DOI] [PubMed] [Google Scholar]
- Skirycz et al. (2008).Skirycz A, Radziejwoski A, Busch W, Hannah MA, Czeszejko J, Kwaśniewski M, Zanor MI, Lohmann JU, De Veylder L, Witt I, Mueller-Roeber B. The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant Journal. 2008;56(5):779–792. doi: 10.1111/j.1365-313X.2008.03641.x. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2019).Wang L, Liu X, Wang X, Pan Z, Geng X, Chen B, Liu B, Du X, Song X. Identification and characterization analysis of sulfotransferases (SOTs) gene family in cotton (Gossypium) and its involvement in fiber development. BMC Plant Biology. 2019;19(1):595. doi: 10.1186/s12870-019-2190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2007).Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plant. The Plant Journal. 2007;52:716–729. doi: 10.1111/j.1365-313X.2007.03268.x. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2016).Wang X, Wang Y, Tang X, Li C, Wang Y. Research progress of drought resistance mechanisms and breeding of Camellia sinensis. Chinese Agricultural Science Bulletin. 2016;32(13):12–17. [Google Scholar]
- Ward et al. (2005).Ward JM, Cufr CA, Denzel MA, Neff MM. The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. The Plant Cell. 2005;17(2):475–85. doi: 10.1105/tpc.104.027722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei et al. (2018b).Wei Q, Wang W, Hu T, Hu H, Mao W, Zhu Q, Bao C. Genome-wide identification and characterization of Dof transcription factors in eggplant (Solanum melongena L) PeerJ. 2018b;6(4):e4481. doi: 10.7717/peerj.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei et al. (2018a).Wei C, Yang H, Wang S, Zhao J, Liu C, Gao L, Xia E, Lu Y, Tai Y, She G, Sun J, Cao H, Tong W, Gao Q, Li Y, Deng W, Jiang X, Wang W, Chen Q, Zhang S, Li H, Wu J, Wang P, Li P, Shi C, Zheng F, Jian J, Huang B, Shan D, Shi M, Fang C, Yue Y, Li F, Li D, Wei S, Han B, Jiang C, Yin Y, Xia T, Zhang Z, Bennetzen JL, Zhao S, Wan X. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proceedings of the National Academy of Sciences of the United States of America. 2018a;115(18):E4151–E4158. doi: 10.1073/pnas.1719622115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia (2010).Xia JB. Master thesis. 2010. Young tea plants gene expression analysis under drought stress. [Google Scholar]
- Xia et al. (2017).Xia EH, Zhang HB, Sheng J, Li K, Zhang QJ, Kim C, Zhang Y, Liu Y, Zhu T, Li W, Huang H, Tong Y, Nan H, Shi C, Shi C, Jiang JJ, Mao SY, Jiao JY, Zhang D, Zhao Y, Zhao Y, Zhang LP, Liu YL, Liu BY, Yu Y, Shao SF, Ni DJ, Eichler EE, Gao LZ. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Molecular Plant. 2017;10(6):866–877. doi: 10.1016/j.molp.2017.04.002X. [DOI] [PubMed] [Google Scholar]
- Yanagisawa (1997).Yanagisawa S. Dof DNA binding domains of plant transcription factors contribute to multiple protein-protein interactions. European Journal of Biochemistry. 1997;250(2):403–410. doi: 10.1111/j.1432-1033.1997.0403a.x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa (2004).Yanagisawa S. Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant and Cell Physiology. 2004;45:386–391. doi: 10.1093/pcp/pch055. [DOI] [PubMed] [Google Scholar]
- Yanagisawa & Schmidt (1999).Yanagisawa S, Schmidt RJ. Diversity and similarity among recognition sequences of Dof transcription factors. The Plant Journal. 1999;17(2):209–214. doi: 10.1046/j.1365-313X.1999.00363.x. [DOI] [PubMed] [Google Scholar]
- Yin et al. (2017).Yin M, Wang Y, Zhang L, Li J, Quan W, Yang L, Wang Q, Chan Z. The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. Journal of Experimental Botany. 2017;68(11):2991–3005. doi: 10.1093/jxb/erx157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo et al. (2017).Yoo YH, Anil K. Nalini Chandran Park, JC, Gho YS, Lee SW, A Gynheung, Jung KH. OsPhyB-mediating novel regulatory pathway for drought tolerance in rice root identified by a global RNA-Seq transcriptome analysis of rice genes in response to water deficiencies. Frontiers in Plant Science. 2017;8:580. doi: 10.3389/fpls.2017.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (1995).Zhang B, Chen W, Foley RC, Büttner M, Singh KB. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. The Plant Cell. 1995;7(12):2241–2252. doi: 10.1105/tpc.7.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (1996).Zhang B, Chen W, Foley RC, Buttner M, Singh KB. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. The Plant Cell. 1996;7(12):2241–2252. doi: 10.1105/tpc.7.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu et al. (2018).Zhu M, Meng X, Cai J, Li G, Dong T, Li Z. Basic leucine zipper transcription factor SlbZIP1 mediates salt and drought stress tolerance in tomato. BMC Plant Biology. 2018;18(1):83. doi: 10.1186/s12870-018-1299-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou & Zhang (2019).Zou Z, Zhang X. Genome-wide identification and comparative evolutionary analysis of the Dof transcription factor family in physic nut and castor bean. PeerJ. 2019;7:e6354. doi: 10.7717/peerj.6354/table-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw gene data of tea and Arabidopsis are available in Tables S4 and S5. The alignment analysis of the amino acid sequences of Dof (CsDof1-29 amino acid sequence was already studied (Li et al., 2016) and 16 distinct CsDofs we identified) is available in Fig. S2.