Abstract
Tumor suppressor genes cooperate with each other in tumors. Three important tumor suppressor proteins, retinoblastoma (Rb), p53, phosphatase, and tensin homolog deleted on chromosome ten (PTEN) are functionally associated and they regulated by post-translational modification (PTMs) as well. PTMs include phosphorylation, SUMOylation, acetylation, and other novel modifications becoming growing appreciated. Because most of PTMs are reversible, normal cells use them as a switch to control the state of cells being the resting or proliferating, and PTMs also involve in cell survival and cell cycle, which may lead to abnormal proliferation and tumorigenesis. Although a lot of studies focus on the importance of each kind of PTM, further discoveries shows that tumor suppressor genes (TSGs) form a complex “network” by the interaction of modification. Recently, there are several promising strategies for TSGs for they change more frequently than carcinogenic genes in cancers. We here review the necessity, characteristics, and mechanisms of each kind of post-translational modification on Rb, p53, PTEN, and its influence on the precise and selective function. We also discuss the current antitumoral therapies of Rb, p53 and PTEN as predictive, prognostic, and therapeutic target in cancer.
Subject terms: Epigenetics, Senescence
Background
It has generally acknowledged that cancer is caused by somatic mutations, which is a concept significantly confirmed by demonstrating that cellular proto-oncogenes contribute to carcinogenesis when mutations deregulated or abnormally overexpressed.1,2 Our understanding is that many of these genes encode proteins that control cell proliferation, differentiation, and development, while mutations that affect their function constitutively deregulate specific signal pathways, providing some of the clearest insights into how and why abnormal behave of cancer cells happen.3 The discovery of dominant “activating” oncogenes has also generated the idea that a unique class of “suppressor genes” may counter their effects and prevent the development of tumors. In fact, experiments about somatic cell fusion or chromosome separation have shown the presence of genes that inhibit tumorigenicity.4 Carcinogenesis is a very complicated process, which can be attributed to either mutation of oncogene function or tumor suppressor gene (TSGs).5 Our understanding of TSGs mostly comes from the preliminary study of retinoblastoma genes, the first discovery of a TSG, and mutation causes retinoblastoma in children.6,7 This is a genetic disease caused by the retinoblastoma susceptibility gene (Rb1) gene inactivation mutation. Compared with the general population, Rb1 gene inactivation mutation increases the risk of retinoblastoma (usually in the eyes) by 10,000 times. These patients also have a high risk of acquiring osteosarcoma and other sarcomas. However, about 60% of retinoblastomas are sporadic (almost in one eye), and these patients have a low risk of other types of cancer.8 Therefore, in 1969, the presence of TSGs based on the developmental dynamics of sporadic and hereditary retinoblastoma, which suggested a carcinogenic “2-hit” model, and was eventually accepted and successfully cloned Rb1 in 1986.9,10 One of the early famous arguments aimed at the being of TSG was because it is irreconcilable Knudsen’s 2-hit model with Nowell’s cancer clonal evolution model, in which reckoned that cancer is the outcome of cell evolution through continuous clonal selection waves.1 It is now supported that for many TSGs, loss of heterozygote function is associated with tumorigenesis by reduced gene dosage and haploinsufficiency.11,12 TSGs could be classified into two categories: the one is “gatekeeper” gene and the other is “caretaker” gene.13 The gatekeeper gene controls the progress of cells in the growth or division cycle, while the caretaker gene maintains the integrity of the genome.14 The difference between these two types of genes is important to the development of therapies. Intuitively, it is likely that inhibiting highly active oncogenes is easier than restoring the function of inactivated TSG. Although they are more difficult to “medicate”, changes in TSG dysfunction are equally important for tumorigenesis. The promising approaches to “medicine” TSG are focus on regulating, inhibiting, or epigenetic silencing of TSG molecules, and closing abnormally activated signaling pathways due to TSG deletion.15 TSGs can inhibit or repress cell cycle or promote cell apoptosis. Over the past 30 years, many of these TSGs have been recognized (Table 1). Because they usually only need one functional gene to prevent cancer, the typical TSGs are recessive, and they need two alleles of “second strike” inactivation.9,16 Previous studies indicate that only a copy of a TSG is enough to manipulate cell proliferation; in this way, two alleles of a TSG must be consistently inactivated or deleted to bring about tumorigenesis.17,18 Therefore, the earliest identification methods relied on genetic methods, biallelic gene inactivation for example, usually in one mutant allele is passed on through the germline and the other is lost somatically. In retrospect, these characteristics define the three basic properties of a “classical” TSGs. First, they are recessive, and then undertake biphasic inactivation in the tumor. Second, the pass on of a single mutated allele benefits the susceptibility of the tumor, since only the other additional mutation is needed for gene function completely lost. Thus, germline mutations may be the root cause of familial cancer syndromes that will inherit. Third, the same gene is often losing activity in sporadic cancers.19 At present, TSG, which does not meet the definition of this standard, includes genes that are inactivated by epigenetic silencing rather than deletion. In addition, ubiquitination of proteasome degradation, mis-localization, and abnormal transcriptional regulation are also engaged in the deactivated of TSGs.20 Various kinds of cancer, including prostate, breast, glioblastoma, stomach, liver, lung, and leukemia, have abnormal patterns of DNA methylation, including hypermethylation and hypomethylation.21 Hypermethylation of CpG islands which are in TSG promoters, such as Braca1, Rb, or p53 promoters, leads to inactivation of each protein, causing cancer.22 Two cytosine analogs have been approved by FDA for the treatment of myelodysplastic syndrome (MDS), they are 5-azacytidine/vidaza (AZA) and 5-aza-2′-deoxycytidine/dacogen (DAC).23 The second generation of simulated guadecitabine (SGI-110), an active metabolite of gemcitabine, is currently undergoing clinical trials in MDS and acute myeloid leukemia (AML).24 Besides, most drugs for cancer are targeted to oncogene, TSGs have difficult to be “drug” treated for they are more likely to change than oncogenes. Nowadays, promising strategies have emerged for TSGs or pathways controlled by these genes.15 TSGs nowadays also generally divided into five types: (1) Genes that control cells to enter specific stages of cell cycle;25 (2) a signal receptor, a signal transduction gene or a hormone that can inhibit cell proliferation;26 (3) Genes that code for checkpoint control proteins trigger cell cycle stagnation when DNA damage or chromosomal defects occur;27 (4) Genes that induce apoptosis;28 (5) Genes associated with DNA repair.29 TSGs have become an important vector response to chemotherapy.30,31 TSGs are often affected by mutation or epigenetic disorder in cancer, therefore occurrence and development of all types of cancer along with an important signal molecule in cells.8,32 Manipulation of cell survival and death is important to development and growth of organisms.33 Activation or inhibition of the cell death is essential for molding and organizing tissues in the process of development organisms.34 Signal balance promotes or damages cell survival by impacting on cell aging and various pathologies. Improper cell loss can result in degenerative and autoimmune diseases, and the mutant cells were not eliminated from the constraints of normal cell growth control causes cancer.35 Therefore, survive and death signals work co-operational to control cell quality viability.36,37
Table 1.
Selected tumor suppressor genes
| Gene | Function | Associated cancer | Others major tumor |
|---|---|---|---|
| p53 | Transcription factor | Li-Fraumeni syndrome | >50% of cancers470,471 |
| Rb | Transcriptional corepression | Retinoblastoma | Many114,119 |
| PTEN | Phosphatase | Cowden syndrome | Glioblastoma, endometrial, thyroid, and prostate cancers472,473 |
| RASSF | Transcription factor | Many | Many474 |
| ARF | MDM2 antagonist p53 activator | Melanoma | Many475–477 |
| APC | Wnt/Wingless signaling | Familial adenomatous polyposis | Colorectal cancer |
| Gastro-intesinal tumors478,479 | |||
| ATM | DNA damage sensor (protein kinase) | Ataxia telangiectasia | Lymphoreticular malignancies480 |
| CHK2 | Protein kinase (G1 checkpoint control) | Li-Fraumeni syndrome | Solid tumors481,482 |
| Carcinomas of the colon, stomach, and endometrium482,483 | |||
| BRCA1, BRCA2 | DNA repair | Familial breast and ovarian cancer | Skin cancer, colorectal cancer484,485 |
| TSC1,2 | GTPase activating protein complex | Tuberous sclerosis | Renal cell carcinoma, angiofibromas486 |
| NF1 | GTPase activating protein for Ras | Neurofibromatosis | Sarcomas, gliomas487,488 |
| LKB1 | Serine/threonine kinase | Peutz-Jeghers syndrome (PJS) | Non-small lung cancer (NSCLC), cervical cancer, ovarian cancer, and breast cancer489 |
| FOXO3a | Transcription factor | Many | Many490 |
RASSF Ras association domain family, APC adenomatous polyposis coli, ARF ADP-ribosylation factor, ATM ataxia telangiectasia mutated, CHK2 checkpoint kinase 2, BRCA1 breast cancer 1 protein, TSC tuberous sclerosis complex, NF1 neurofibromatosis type 1, LKB1 the liver kinase B1, FOXO3a forkhead box class O3a
Post-translational modifications (PTMs) are key steps in signal transduction of phosphoric acid, acetyl, and glycosyl groups from one protein to another. Because most PTMs are reversible, normal cells use PTMs as a “switch” to decide the cell’s static and proliferative state, which can quickly and strictly regulate cell proliferation. In cancer cells, the oncogene activation and/or inactivation of TSGs supply with ongoing proliferation signals by regulating the diversity of PTMs states of effector proteins involved in cell survival, cell cycle, and proliferation regulation, resulting in abnormal proliferation of cancer cells.38,39 PTMs are the core of many cellular signaling events. In addition to a single regulatory PTM, there are some PTMs that work in a coordinated manner. This PTM crosstalk is usually a fine-tuning mechanism that adjusts the cell’s response to small changes in the environment.40 Specific protein modification manages almost all cellular physiological processes, such as immune function, as well as the precise location, duration, and intensity of physiological processes to ensure rapid and dynamic cellular responses to extracellular and intracellular stimuli.41 Further, PTMs can play as a tight junction (TJ) protein and regulate the function of epithelial barrier.42 Compared with transcription or translation regulation, PTMs are fast and dynamic processes, and engaged in the context of barrier maintenance, therefore, PTMs may be essential to work with the altar of environment or external impact. PTMs can regulate formation of membrane-free organelles and supply a potential new treatment strategy for neurodegenerative diseases that cannot be treated at present.43
So far, more than 450 unique protein modifications have been found, including phosphorylation, acetylation, ubiquitination, and SUMOylation. These modifications can change the activity, intracellular distribution, protein interaction, and protein life span of the target protein.44 Phosphorylation mainly takes place in serine, threonine, and tyrosine residues of the targeted protein.45 According to different substrates and phosphorylation sites, protein stability, protein interaction, protein location, and enzyme activity were determined.46 Ubiquitination is a well-known post-translational protein modification that manages biological processes, immune responses, apoptosis, and cancer, for example.47 As a post-translational protein modification, SUMOylation has attracted more and more attention, for this pathway is necessary to maintain genome integrity, transcriptional regulation, gene expression, and signal transduction in cells.47
TSGs work cooperativity in cancers and their function is largely influenced by the posttranslational modification.15,17 Ten genes in the human genome are collectively referred to as Ras related domain family (RASSF). RASSF consists of two subclasses: C-RASSF and N-RASSF. N-RASSF and C-RASSF encode Ras related proteins, which are often inhibited by DNA hypermethylation in human cancer. But C-RASSF and N-RASSF are very different. Six C-RASSF proteins are reckoned by a C-terminal coiled-coil motif called the Salvador/RASSF/Hippo domain, while N-RASSF proteins interact with the mammalian Ste20 like kinase, which is the core kinase of the tumor suppressor Hippo pathway.48
ADP-ribosylation factor (ARF) plays a crucial role in preventing the development of cancer by regulating cell proliferation, aging, and apoptosis. As a factor inducing aging, the role of ARF as an antitumor factor is closely related to the p53-MDM2 axis, which is an important process to inhibit tumor formation. Although it is generally believed that ARF expression is majorly modulated at the transcriptional level, studies on post-translational regulation of ARF have shown that ARF proteins can be degraded through ubiquitination.49
Adenomatous polyposis coli (APC) is considered to be a tumor suppressor gene for colorectal cancer (CRC) and is dysregulated at the germ line and somatic level.50 APC activity is related to phosphorylation mediated by CK1 and GSK3β kinase,51 which dramatically enhances its affinity for β-catenin to inhibition of Wnt signaling.52
MKRN1 plays as an activator of the Wnt/β-catenin signaling pathway by inhibiting APC for MKRN1 is an E3 ligase which can be ubiquitinated APC.53
Serine threonine kinase checkpoint kinase 2 (CHK2) is an important DNA damage checkpoint protein for the ATM-p53 signaling pathway. Phosphorylation and ubiquitination are both important post-translational modifications for its function.54
Two key factors of TSGs engaged in the homologous recombination (HR) pathway in humans: breast cancer type 1 susceptibility protein (BRCA1) and its obligatory partner BRCA1-associated RING domain protein 1 (BARD1). Mutations in BRCA1 bring about not only familial breast and ovarian cancers but are also the promoters of different kinds of sporadic cancers. BRCA1–BARD1 heterodimers, through their ability of E3 ubiquitin ligase and interact with DNA and DNA damage response factors, benefit to import DNA double-strand breaks, into the HR pathway for repair.55 Partner and locator of BRCA2 (PALB2) has become a crucial and versatile participant in genome integrity maintenance. The double allele mutation in PALB2 results in Fanconi anemia (FA) subtype FA-N, while monoallelic mutation is prone to breast and pancreatic cancer.56 Regulation of PALB2 involves different post translational modifications of protein, such as phosphorylation and ubiquitination.57
Tuberous sclerosis complex (TSC) is an autosomal dominant disease, which is caused by the loss of function mutation of TSC1 or TSC2. It is characterized by a wide range of clinical characteristics in multiple organs such as skin, brain, eyes, lungs, heart, and kidney.58 TSC-1 and TSC-2 are tumor suppressors that inhibit cell growth. Mutations in both genes can lead to multiple benign tumors. The products of TSC1 and TSC2 gene form a functional complex with GTP enzyme activating protein (GAP) activity, which has the effect of inhibiting the target of mammalian rapamycin complex 1 (mTORC1), while mTORC1 is constitutively activated in TSC mutant tumor.
Neurofibromatosis type 1 (NF1) is an autosomal dominant genetic disease with an estimated prevalence of 1 in 3000–4000 person. NF1 is characterized by the development of benign tumors in the peripheral nervous system and an enhanced risk of malignancy. The phenotype of NF1 is variable and several organ systems are affected, including bone, skin, iris, and central, and peripheral nervous systems.59
The liver kinase B1 (LKB1, encoded by STK11) is a tumor suppressor function as a highly conserved serine/threonine kinase. Phosphorylation is the most common post-translational modification of LBK1 that affects the conformation of LBK1 and creates new surfaces that interact with other proteins. Ubiquitination of proteins is a post-translational modification that, in addition to its well-known functions in protein degradation, is engaged in many other cellular processes, such as activation of the LKB1–AMPK axis.60 The location of LKB1 is not limited to plasma membrane, but occurs in nucleus and cytoplasm, which depends on cell type and state, but on C-terminal conserved cysteine 430, LKB1 is farnesylated. Farnesylation is another kind of post-translational modification that mediates a transient membrane connection.61 LKB1 is also a target for endogenous neddylation and its endogenous neddylation level is increased in hepatocellular carcinoma (HCC). Neddylation is a post-translational modification that relies on NEDD8 binding to target proteins. Similar to ubiquitination and SUMOylation, neddylation needs E1, E2, and E3 enzymes62
The forkhead box class O (FOXO) family is a widely expressed transcription factor that woks in higher organisms. FOXO3a, or FOXO3 or forehead in rhabdomyosarcoma like 1 (FKHRL1), is a member of FOXO3 subfamily, which was first found in human placenta. FOXO3a activity can be modulated by many PTMs, such as phosphorylation, ubiquitination, acetylation, and methylation. Translocation of FOXO3a can be altered by those reversible PTMs, affected its capability of DNA binding affinity, and transcriptional activity patterns at stated gene sites63 (Table 1).
Among TSGs, we focus on three important tumor suppressor proteins, Rb, p53, and PTEN, for they are tightly functionally connected and more closely related to post-translational modification. In triple-negative breast cancer (TNBC), Rb and PTEN are often deactivated with p53.64 p53, PTEN, and Rb are the most frequently altered TSGs in primary prostate cancer, with abnormal PI3K/AKT, RAS/RAF, and cell cycle signals.65 The genomic changes of p53, PTEN, and Rb in early and late prostate cancer (as well as the combined loss of these genes) indicate a poor prognosis.66 The changes of p53, Rb, and PTEN have been discovered that they are enriched in drug-resistant diseases, by the genome analysis of metastatic castration resistant tumors.67 The formation of glioblastoma requires the disorder of three core pathways: Rb controlled cell cycle progression, p53 signaling pathway, and receptor tyrosine kinase (RTK)/phosphatidylinositol 3′-kinase (PI3K)/AKT axis,68 and PTEN negatively mediates the PI3K–AKT–MDM2 pathway that downregulates p53. In addition, p53 also activates PTEN, therefore protecting itself from overly powerful survival signals.35 These relationships indicate that proteins induce or inhibit the function of cell death are interconnected.69,70 Genetic aberrations influencing the intermediates of these three pathways have been found in almost all glioblastomas.68 Rb, p53, or PTEN are TSGs that are found to be inactivated in the tumor matrix of oropharyngeal, breast, and other human cancers.66 The mouse model verified the tumor promoting effect of Rb, p53, and PTEN deletion on fibroblasts, which can transform normal fibroblasts into cancer-related fibroblasts (CAFs).71 Thus, TSGs are networked to promote normal cell function and eliminate abnormal cells, and this paper attempts to pay more attention to these three tumor suppressor genes.
Moreover, these three tumor suppressor genes, Rb, p53, and PTEN are also deeply influenced by post-translational modifications. In sum, we here explore the influence of those three TSGs, on their functions, as well as new drug targets and strategies for cancer treatment.
The Rb gene, the first tumor suppressor gene and inactivation by multisite phosphorylation
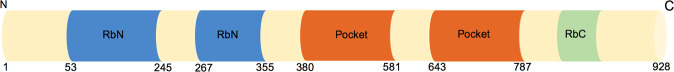
Rb recognition was initially associated with the formation of a rare retinal neoplasm in children, called retinoblastoma.10,72,73 Further research shows that changes in the Rb gene or inactivation of Rb protein appeared in many kinds of human cancers, and it is widely believed that Rb inactivation could be one of the most common events in cancer.74,75 The functional regulation of Rb includes inhibition of phosphorylation and activation of dephosphorylation events.76,77 The Rb phosphorylated by cyclin-dependent kinase (CDK) and checkpoint kinase 2 (CHK2),78 while the activation of Rb by dephosphorylation is still rare.79 Except a few cases, phosphorylation of Rb brings about inactivation, transcriptional inhibition, and cell cycle progression.80 Phosphorylation of Rb regulates the interaction between Rb and other proteins, and this modification usually promotes conformational transition from disordered structure to ordered structure, thus concealing the protein binding surface.81–83 Therefore, understanding how Rb is phosphorylated and inactivated requires studying how Rb structure promotes protein–protein interactions and how phosphorylation regulates these interactions.84 Rb consists of two independently folded domains and a substantial number of inherently disordered first-order sequences (approximately 33% of the 928 amino acids). The structure of N terminal domain (RbN) and central pocket domain are composed of two helical subdomains (Fig. 1).85,86
Fig. 1.
Rb structural domains. Rb structured domains include the N-terminal domain (RbN), the pocket domain, and parts of the C-terminal domain (RbC)
Rb deletion allows cancer cells to bypass two different barriers in the progression of tumors.87,88 Firstly, Rb loss decreased the requirement of amplification of p38 mitogen-activated protein kinase (MAPK) signal when malignant progression. Rb phosphorylated by CDK2 is an effector of p38 mitogen-activated protein kinase (MAPK) signal and a regulator of resisting CDK4 and CDK6 suppression.89 Secondly, Rb inactivation relieves the expression of cell state determinants, promotes lineage infidelity, and increases the acquisition of metastasis ability.90 The high phosphorylation level of Rb controls its association with early region 2 binding factor (E2F) and depresses its tumor suppressive properties. However, activated Rb can be mono-phosphorylated at any of the 14 CDK phosphorylation sites during G1, and the 14 sites coordinate the interaction of Rb, which endow it with functional specificity.91 The mono-phosphorylation of Rb at serine 811 (S811) alters the transcriptional activity of Rb by promoting its binding with nucleosome remodeling and histone deacetylation (NuRD) complex. Mono-phosphorylation of Rb at S811 or threonine (T826) activates the expression of oxidative phosphorylation genes, which increases cell oxygen consumption. The activation signal of Rb might be integrated into a phosphorylation code that controls the different activities of Rb.91 The interaction between Rb and nuclear factor-kappa B (NF-kappa B) protein p65 is mainly dependent on the phosphorylation of S249/T252 mediated by CDK4/6 of Rb, and S249/T252 phosphorylated Rb was negatively correlated with programmed death ligand-1 (PD-L1) expression in patient samples, which indicates that hyperphosphorylated Rb-NF-kappa B axis can be used to overcome cancer immune evasion induced by traditional or targeted therapies.92 Phosphorylated proteomics data suggest that Rb phosphorylation is associated with reduced proliferation and inhibited apoptosis in colon cancer cells, explaining why this classical tumor suppressor is enrichment in colon cancer and provides a theoretical basis for the application of targeted Rb phosphorylation.93 Those results reveal that Rb activation signals can be integrated in a phosphorylation code that will control the diversity of Rb activity,91 indicating that phosphorylation of Rb manages interaction with different proteome, chooses different targets, and controls different aspects of Rb function.
Effects of other post-translation modifications on Rb
Rb is also controlled by other types of post-translation modifications, which may affect Rb in different ways. Oncoproteins binded Rb are often targeted at Rb and degraded by proteasomes during carcinogenesis.94 In proteasome, Rb protein is degraded by ubiquitin dependent and non-ubiquitin dependent pathways. Human U3 protein 14a (hUTP14a) interacts with Rb and promotes poly-ubiquitination and turnover of Rb, indicating that nucleolar proteins can be used as nucleolar sensors to directly send nucleolar interruption signals to p53 and Rb, which protect cells from nucleolar damage.94 TRIM71, protein kinase A (PKA)-mediated phosphorylation of the E3 ubiquitin ligase, degrades Rb, p53, and antigen peptide-loading complex (PLC) by catalyzing K48 linked polyubiquitination, thus reducing immune monitoring.95 HAUSP increases in glioma and regulates Rb, which is by stabilizing effect of MDM2 leading to a decrease in Rb levels in cancer cells.96 However, CMV PP71 promotes Rb degradation through non-ubiquitin dependent pathway.97 The oncoprotein MDM2, a p53 ubiquitin-E3 ligase that mediates Rb degradation through the ubiquitin-dependent and non-ubiquitin dependent pathways.98,99
In the whole cell cycle, Rb is by small ubiquitin like modifier (SUMO)ylated at the early G1 phase,8,100,101 which activates Rb phosphorylated in the early G1 phase. The SUMOylation of Rb stimulates its phosphorylation level by recruiting a kinase CDK2 containing SUMO-interaction motif (SIM), resulting in over phosphorylation of Rb and release of E2F-1. On the contrary, the lack of SUMO in Rb led to the decrease of Rb phosphorylation, the CDK2 binding, and E2F-1 isolation.101 This suggests that in addition to phosphorylation, SUMOylation is also involved in the regulation of Rb during the cell cycle. SUMO protease SENP1 regulates SUMO1 binding of Rb and lamin A/C. SUMOylation is required for the interaction of these two proteins. Importantly, this SUMO1 dependent complex shelters Rb and Lamin A/C from proteasome degradation. SENP1 regulated Rb desumoylation in cell cycle regulation further deepens understanding of Rb proteasome-dependent degradation.102 Therefore, those results present that SUMOylation is a molecular switch controlling phosphorylation and cell cycle regulator function.
Rb can be acetylated and methylated in addition to being phosphorylated, SUMOylated, and ubiquitinated. Rb at Lys873 and Lys874 can be acetylated, resulting in increased their affinity for MDM2, and then reduced phosphorylation of Rb.103 DNA damage may lead to Rb acetylation, which engaged in cell differentiation.104 Methyltransferase Set7/9 methylate Rb at K810, which has negative effects on Rb phosphorylation and growth of cells.105
Given the loss or inactivation of Rb function in most human malignancies, further research is necessary to explore whether PTMs affect the molecular interactions of Rb and mediate Rb’s cell cycle function, as well as the immune function that mediates Rb overlap, or whether it is possible to target various aspects of Rb.
Targeting the CDK–Rb–E2F axis for cancer treatment
In cancer, cell cycles are frequently activated by interfering with the CDK–Rb–E2F pathway, leading to drug efforts to block the pathway.75,106 Kinase inhibitors are the most advanced in drug development, although some compounds that target this pathway are also in different stages of development.107 The most promising option among CDK inhibitors is undoubtedly inhibitors of CDK4 and CDK6 (called CDK4/6 inhibitors) and compounds are intended to target the ATP binding sites of the CDK complexes.108 More than a decade after Pfizer first synthesized palbociclib in 2001, which is the most advanced component of its kind nowadays.109–111 Hypo-phosphorylation of Rb is related to G0/G1 stagnation by inhibiting the activity of E2F transcription factors, while hyper-phosphorylation of Rb promotes E2F release and cell cycle to progress from G0/G1 to S phase,and CDK regulates the hyper-phosphorylation of Rb in the cell cycle.101 Therefore, CDK–Rb–E2F axis constructs the core transcriptional mechanism that promotes cell cycle progression, determines the time and fidelity of genome replication, and ensures that genetic material precisely goes through each cell division cycle.75 Evaluations of a few small molecules that are highly specific CDK4/6 are under way, besides palbociclib (PD332991) there are ribociclib and abemaciclib, which induces pocket protein hypo-phosphorylation and reactivation, bring about cell cycle arrest in G1.112–115 Many clinical trials are under way, with the result being reviewed by several groups, PALOMA-2 is in clinical phase III trial and two other CDK4/6 inhibitors, ribociclib (Novartis) and the other abemaciclib (Eli Lilly) are in clinical trials for breast and other cancers as well116–119 (Table 2). In addition, PALOMA-3 was in a randomized, double-blind, placebo-controlled phase III trial, compared the efficacy of palbociclib and fulvestrant (an ER antagonist) for ER+HER2− breast cancer that recurred or progressed during hormone therapy.120–123
Table 2.
Components that are being explored to target Rb, p53 family, and PTEN
| Drug target | Drug | Indication |
|---|---|---|
| CDK kinase | Palbociclib | Perturbations in CDK4, CDK6113 |
| CDK kinase | Ribociclib | Perturbations in CDK4, CDK6114 |
| CDK kinase | Abemaciclib | Perturbations in CDK4, CDK6115 |
| CDK kinase | PALOMA-1 | Perturbations in CDK4, CDK6118 |
| CDK kinase | LEE 001 | Perturbations in CDK4, CDK6119,491 |
| CDK kinase | LY 2835219 | Perturbations in CDK4, CDK6492 |
| MDM2 | Nutlins | Inhibitors of the MDM2–p53 interaction278,493 |
| MDM2 | RG7112 | Inhibitors of the MDM2–p53 interaction494 |
| MDM2 | RG7388 | Inhibitors of the MDM2–p53 interaction495 |
| MDM2 | SAR405838 | Inhibitors of the MDM2–p53 interaction273 |
| Mutant p53 | PRIMA-1 | Conversion mutant p53 to wild-type496 |
| Mutant p53 | NSC319726 | Conversion mutant p53 to wild-type497 |
| Mutant p53 | STIMA-1 | Conversion mutant p53 to wild-type498 |
| Mutant p53 | SCH529074 | Conversion mutant p53 to wild-type367 |
| Mutant p53 | CP31398 | Conversion mutant p53 to wild-type499,500 |
| Mutant p53 | Zinc | Conversion mutant p53 to wild-type501 |
| Mutant p73 | RETRA | Inhibition mutant p73 interaction with other protein502,503 |
| PI3K | Wortmannin | PIK3CA related excessive growth423 |
| AKT | ARQ 092 | AKT1-associated Proteus syndrome422 |
Since the Rb gene was isolated in 1986 and the first E2Fs gene was cloned in 1992, we have a deep understanding of the role of CDK–Rb–E2F pathway in cancer. In fact, in almost all human malignant tumors, this pathway is out of control in one way or another, leading this pathway an extremely attractive target for cancer treatment.
Post translational modification in the non-canonical Rb pathway facilitates histone modification and modulates chromosome structure
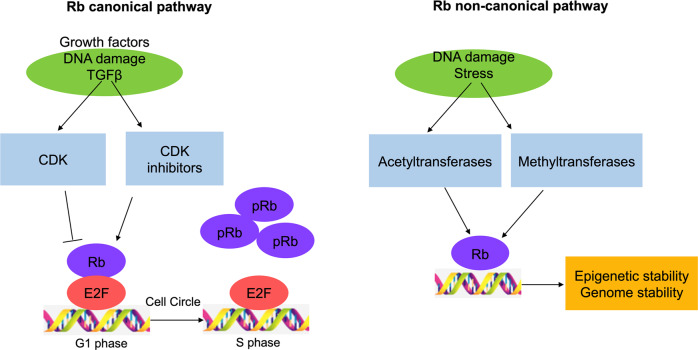
The canonical model of Rb as a TSG developed in the past 30 years is based on the modulation of E2F transcription factors to limit cell cycle progression.124–126 In mechanism, non-canonical Rb pathway regulates histone modification and modulates chromosome structure in a way different from cell cycle modulation127,128 (Fig. 2).
Fig. 2.
The Rb canonical and non-canonical pathways. Signals of growth factors, DNA damage, and transforming growth factor-β (TGFβ) activate CDKs to phosphorylate and inactivate Rb, whereas CDK inhibitors activates Rb. Inactivation of Rb in the canonical pathway results in transcription of E2F target genes; Stresses and DNA damage stimulates acetylation and methylation of Rb, which can maintain genomic stability by locating DNA break sites and stimulating non-homologous end connections or homologous recombination repair. Rb also recruits enhancers from EZH2 to H3K27me3 to ensure the fidelity of DNA replication and chromosome aggregation
Chromatin replication maintains Rb-dependent epigenetic markers. An important and indirect mechanism explains the preserve Rb function in the S phase is intrinsic to the chromatin replication. Rb is known to recruit histone methyltransferase enhancer of zeste homolog 2 (EZH2) to guide trimethylhistone H3 lysine 27 (H3K27me3) to deposit and promote octamer-binding protein 4 (OCT4) and Sox2 expressed,129–131 which is preserved in the S-phase of cell circle during DNA replication.132,133 In turn, the location of H3K27me3 is enhanced in daughter cells via another EZH2 recruitment by Rb in subsequent G1. Thus, mechanisms of maintain epigenetic memory during DNA replication can keep Rb-dependent characteristics without the need for the persistence of the Rb protein presence.127 In summary, Rb can be protected by high levels of CDK activity (thus maintaining low phosphorylation levels of Rb), high phosphorylation of Rb preserves function, and Rb relies on histone modification characteristics through the cell cycle. These properties allow Rb to play a role in proliferating cells.134
Although the repetitive sequences and sites of DNA damage that is the target of Rb do not appear to require a consistent E2F element (TTTCGCGC), studies have demonstrated that Rb is often engaged in these genomic locations in a E2F1 dependent manner.129,135 A mutation in Rb invalidates the interaction between its labeled box domain and E2F1, which has been shown to break the binding of Rb and E2F1 with different types of repetitive sequences.129,136
The acetylation and methylation of Rb are caused by DNA damage.103,105,137,138 These modifications decrease phosphorylation of Rb by CDK, which further implies that Rb–E2F1 complexes may have a protective effect on CDK activity when it participates in the function of non-homologous end joining (NHEJ) and homologous recombination (HR), for E2F1 is recruited to the sites of DNA double strand break, which is very important for NHEJ, HR135,139 (Fig. 2). Hyperphosphorylated Rb also interacts with E2F1 when DNA breaks.140,141 These results further suggest that Rb–E2F1 interaction is not sensitive to CDK activity and therefore, these mechanisms of epigenetic and genomic stability depending on Rb–E2F1 are not related to CDK and belong to the non-canonical functions of Rb.
Finally, the multifunctional nature of Rb makes it a key target in many cancer-associated environments. Further, the trans-differentiation phenotype about recurrent cancer from a series of molecular targeted therapies shows that Rb loss is related to acquired treatment resistance, and its pathway is beyond the control of cell cycle. Understanding how Rb loss leads to drug resistance is critical to realizing the function of these targeted molecules.127 The loss of Rb in both regulatory pathways in cancer may produce a powerful synergistic cancer promotion combination. These functions of Rb are significant for chemotherapy response and drug resistance of targeted anticancer drugs. This view provides a framework for Rb research in future basic and clinical research.
Tumor suppressor p53: determinants of its post-translational modifications
Transcription factors (TFs) are always activated through two main mechanisms: (i) the TF levels are increased in the nucleus, or (ii) via post-translational modifications (PTMs).142 Tumor protein p53, a TF, is encoded by homologous genes in different organisms, and it is crucial in multiple organisms.143–145 p53 is a short-lived protein because of its rapid proteasomal degradation, and it controls the cellular response to different stress signals;146,147 therefore, p53 undergoes a variety post-translational modifications following genotoxic stress, leading to enhanced protein stability and translocation to the nucleus.148–151 It is well accepted that protein modifications play a significant role in p53 regulation, whose functions vary from regulating p53 stability and localization, to controlling cell proliferation, and cell death.145 Post-transcriptional modifications of p53 occur at approximately 50 sites on the peptide, and include phosphorylation, acetylation, mono- and dimethylation, glycosylation, ubiquitination, neddylation, sumoylation, and poly-ribosylation.152 Many post-translational modifications occur with or without genotoxic pressure and are relatively independent of each other. Less is known about a possible direct connection between chromatin modification and post-translational modifications. p53 also plays a crucial role in regulating the epigenetic changes that occur in cells due to cross-talk between p53 associated with its modifications.153,154 In addition to the role of chromatin remodeling proteins in metabolism and ferroptosis,155–157 we have suggested that these proteins may also have post-translational modification functions.151
Phosphorylation of p53 is a critical modification guiding its regulation of apoptotic cell death
Human p53 contains serine (S) and threonine (T) phosphorylation sites across the entire protein, but they are enriched in the transcriptional activation area of the N-terminal domain and the regulatory region of the C-terminal domain.158 Some stimuli, including genotoxic stress (DNA damage-inducing agents) or glucose deprivation, induce many reversible PTMs of p53.159,160
The phosphorylation of p53 two transactivation domains (TAD) at serine 15 is the initially activated phosphorylation site, and it is phosphorylated by both the ataxia telangiectasia mutated gene (ATM) and ataxia-telangiectasia-mutated-and-Rad3-related kinase (ATR) protein kinases,161–163 phosphorylation also can stimulate the association between p53 and histone/lysine acetyltransferase (HATS),164 which is quite crucial for the stability and activation of p53. The activation of ATM leads to the phosphorylation of a number of substrates, such as casein kinase (CK1), checkpoint kinase 1 (Chk2), and p53, mediating the effects of ATM on DNA repair, cell-cycle arrest, apoptosis, and other downstream processes. In addition, ATM depleted and p53 mutation are usually mutually exclusive, which shows that these proteins are the same in promoting the survival of cancer cells.165 The phosphorylation of Ser15 also triggers a series of other p53 phosphorylation events that contribute to p53 induction and activation, suggesting that Ser15 phosphorylation is a key point in p53 activation.162,166 It was reported that phosphorylation of Ser15 led to the dissociation of MDM2 from p53, which increases the stability of p53.167 Ser15 can also be phosphorylated via the AMP-activated protein kinase (AMPK) pathway, which is mediated by glucose-dependent cell cycle arrest at G1/S.168,169 Further, both IR and UV light can induce phosphorylation of p53 on Ser-20, for ATM and ATR can phosphorylate p53 on Ser-20, which mediates stabilization of human p53 in response to DNA damage.170
In addition, p53 function altars from “arrestor” and “repairer” to “killer” depending on many post-translational amino-terminal phosphorylation of p53. The function of Ser46 phosphorylation in p53 is closely related to the killer function of p53 bringing about apoptosis and can be phosphorylated by a number of candidate kinases, such as homeodomain-interacting protein kinase 2 (HIPK2), p38 and dual specificity tyrosine-phosphorylation-regulated kinase 2 (DYRK2).171–173
The interactions between p53 and MDM2 or p300/CBP are regulated by various phosphorylation events in the amino terminus of p53, which leads to the simultaneous binding of one monomer of p300/CBP to tetrameric p53 to mediate p53-dependent transactivation in response to genotoxic stress.174,175 p53 cooperates with the apoptosis stimulating proteins of p53 (ASPP) proteins being able to bind and work p300 together, selectively regulating the apoptotic function of p53.176,177 The Ser 6 and 9 sites were initially thought to be phosphorylation sites of the protein kinase CK1 family members, CK1d and CK11.178 The function of Ser 6 and Ser 9 phosphorylation in p53 is to integrate TGF-beta and FGF-signaling by inducing the interaction between p53 and Smad, which may be important in tumourigenesis and metastatic progression.179 Smad plays as crucial platforms in mutant-p53/p63 protein complex, and when Ras signaling accelerates mutant-p53 phosphorylation, mutant p53 and Smad interrupt p63 to form a ternary complex, in doing so, the p63 transcriptional functions are antagonized.179 The role of amino-terminal phosphorylation is to regulate the interaction between p53 and its inhibitor, MDM2, or its coactivators p300/CBP, and growth factor-mediated phosphorylation coordinates physiological and developmental signals.152,180 Those results suggest that transcriptional coactivator p300/CBP is an important player in activating p53.
Acetylation of p53 engaged in the fine tuning of cellular responses to DNA damage and genotoxic stress
Acetylation of p53 is an important form of post-translational modification that is essential for its activation, and the acetylation occurs via a reversible enzymatic process.181–183 Both acetylation and ubiquitination can modify the same lysine residues at the C terminus of p53 (similar to neddylation and methylation), and these modifications are mutually exclusive and have different effects on p53 regulation.184–188
Six p53 lysine (K) residues within the C-terminal regulatory domain (K370, K372, K373, K381, K382, and K386) can be targeted by MDM2.158 These modifications lead to activation of the transcriptional activation activity of p53 and increase its stability. CBP/p300 are transcriptional coactivator proteins that play a dual role in regulating p53 function. For one thing, an interaction between p300 and either p53 or E2F1 has a significant impact on early cell cycle progression, suggesting that a critical role for p300 in cooperation with the pathways of growth arrest regulated by E2F and p53.189 For another, they facilitate the ubiquitination of p53 by MDM2, which decreases p53 levels in the presence of genotoxic stress.190 They also protect p53 from degradation by acetylating the p53 carboxyl terminus, which contains targets for ubiquitination. K320, present in the tetramerization domain, can be acetylated by PCAF after DNA damage, and this acetylation is beneficial for cell survival as it boosts the expression of p53-controlled cell cycle arrest target genes, such as cyclin-dependent kinase inhibitor 1A (CDKN1A, commonly known as p21).191,192
Unique to these residues, K120-acetylated p53 accumulates at mitochondria, which is thought to negatively regulate apoptosis by affecting the Bak/Mcl-1 interaction.193 In the p53 DNA-binding domain, K120 also can be acetylated by human males absent on the first (hMOF) and Tip60, which is quite essential for the activation of target genes connected to apoptosis but not to those involved in cell cycle arrest.194 In addition, K120 and K164 are present in the p53 DNA-binding domain, which is the most common region for p53 mutations in malignant solid tumors, indicating that they might be connected with p53 function in cancer. A K120 mutation was found in Ewing’s Sarcoma and esophageal SCC cells, while a mutation in K164 was discovered in glioblastoma and bladder carcinoma.188,195 These data indicate the key role of p53 acetylation in tumor suppressive activity.
p53 methylation contributes to its tumor suppressor activity
Lysine (K) and arginine (R) residues in p53 can be methylated, and a growing number of studies in recent years have shown that p53 methylation takes place during the DNA damage response.196–198 Methylation of lysine and arginine residues in histones has long been known to impact chromatin structure and gene expression.199 In recent years, the methylation of p53 has emerged as an important modification that affects its function in various processes, such as cell cycle arrest, DNA repair, senescence, apoptosis, and tumourigenesis.199 Whether p53 is activated or depressed depends on the location of the modification and the number of methyl groups attached.200 Protein arginine N-methyl transferase 5 (PRMT5) was first shown to methylate p53 at several arginine residues (R333, R335, and R337) in the tetramerization domain,196 which specifically controls the functions of p53 in cell cycle arrest and is suggested to inactivate p53 during lymphomagenesis.201,202 There are three different lysine methyl transferases (KMTs) that could mono-methylate p53, and there are at least two KMTs could di-methylate p53.203
Monomethylation of p53 by SET and MYND domain-containing protein 2 (SMYD2) at K370, which was shown to repress p53-mediated transactivation, decreases the binding of p53 to the promoters of its target genes, such as p21.204 Monomethylation at K372 by SET7/9 boosts the activation of p53 downstream target genes, but monomethylation of K370 by SET8 inhibits p53 transcriptional activity.205,206 In addition, a second methyl group can be conjugated to p53 to form K370me2, which then promotes p53 function via stimulating its binding to the Tudor-domain-containing reader, p53 binding protein 1(p53BP1). Like K370Me2, K382Me2 has also been shown to be related to the stabilization and activation of p53. Interestingly, lysine-specific demethylase 1 (LSD1) selectively wipes off this second methyl group, thus inhibiting p53 function by interrupting the association of p53 with 53BP1, which contributes to these effects.207,208 Thus, p53 contributes to keep DNA methylation homeostasis and clone homogeneity, which may benefit to its anti-cancer activity.
p53 SUMOylation regulates p53 localization
The tumor suppressor p53 experience dynamic nuclear output, because its tetramer domain contains a nuclear export signal (NES) domain full of leucine.209 The N-terminal transactivation domain of p53 seems containing another NES, in which phosphorylation blocks the nuclear output of p53, bring about its nuclear accumulation.210 SUMOylation occurs at K386 of p53 and SUMO-1, SUMO-2, or SUMO-3 that accelerates the output of the p53 from nucleus.211–213 p53 in the nucleus not only promotes the expression of pro-apoptotic genes but also prevents cell death by increasing p21 expression.214 Most p53 anti-apoptotic functions happen in the nucleus, especially under resting conditions.214,215 p53 is normally SUMOylated at a single site, K386, by the protein inhibitor of activated stat (PIAS) family members and Topors.216,217 SUMO E3 ligase PIASy and lysine acetyltransferase Tip60 involved in p53-mediated autophagy. The combination of PAISy to p53 and then PAISy activated Tip60 resulted K386 sumoylation and K120 acetylation of p53, respectively. Although these two modifications are not interdependent, they together act as “binary death signals” and promote the accumulation of p53 cytoplasm and the execution of PUMA mediated autophagy.218 When the COOH-terminal nuclear export signal of p53 is masked by its unmodified C-terminal region, it remains in the nucleus. Moreover, the SUMOylation of p53 releases it from the chromosomal region maintenance 1 (CRM1) Huntington-EF3-PP2A subunit-HEAT9 loop to disassemble the transporting complex and promote the translocation of p53 to the cytoplasm.219 Thus, the nuclear export of p53 can facilitate cellular proliferation through the loss of its antigrowth function. Cytosolic p53 performs a non-transcriptional function by interacting with, and then counteracting, the anti-apoptotic function of Bcl (B cell lymphoma/leukemia)-2.220 In addition, p53-Bcl-2 binding depends on p53 SUMOylation,221 and a lot of cytoplasmic p53 localization is clinically associated with poor prognosis and disease progression to hormone-resistance status.222
Ubiquitination/proteasome-dependent protein degradation is important for rapid signal transduction
Ubiquitin is a highly conserved, stable, small molecule protein with 76 amino acid residues.223 The ubiquitin-proteasome system (USP) depends on the small polypeptide ubiquitin and is a delicate process requiring of three classes enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2) and a unique ubiquitin ligase (E3).224 Consequently, ubiquitination includes three main steps: activation, conjugation, and ligation by E1s, E2s, and E3s, respectively.225,226 Ubiquitin conjugation to proteins can control various biochemical reactions, such as precursor protein maturation, degradation of unneeded proteins, and protein turnover.227 Ubiquitination begins with the attachment of a ubiquitin molecule to Lys residue.228The key characteristic of ubiquitin is its seven Lys residues can be ubiquitinated to produce ubiquitin chains linked to isopeptides. When a ubiquitin is connected to the N-terminal of the second ubiquitin, the eighth chain type, MET1 chain or “linear” chain, is generated.229–232 Consecutively assembled ubiquitin molecules generate a poly-ubiquitin chain that is formed on the target proteins and is the degradation signal recognized by the 26S proteasome subunit (Fig. 3).233,234 Subsequently, the protein substrate would be degraded into shorter peptides, resulting in the release and reuse of ubiquitin.235 In addition to ubiquitin, Small Ubiquitin like MOdifier (SUMO), NEDD8 (downregulated protein 8 of neural precursor cell expression), ISG15 (interferon stimulation gene 15) or FAT10 (HLA-F adjacent transcript 10) can also be coupled to the target proteins. These peptides are classified into the ubiquitin like protein (UBL) family and have similar structure with ubiquitin.236
Fig. 3.
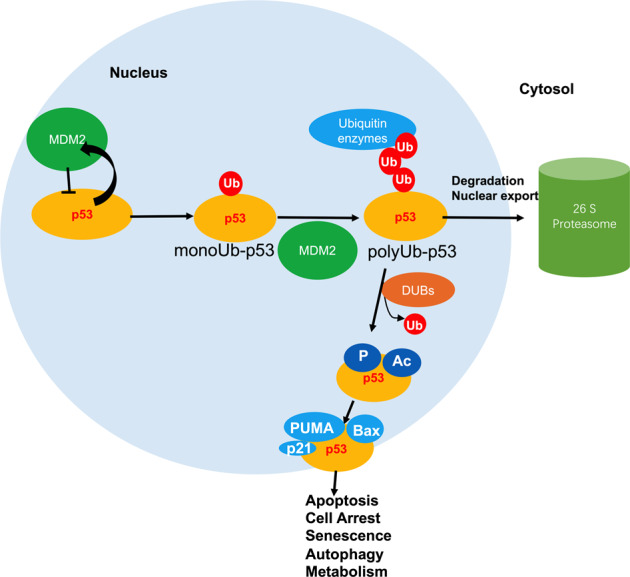
Ubiquitinated and de-ubiquitinated p53 functions and pathways. p53 is degraded after MDM2-mediated ubiquitination, and other DUBs stabilize p53 by eliminating ubiquitin from p53. Under normal conditions, MDM2, a target of p53, inhibits p53 activity by forming a p53/MDM2 auto-regulatory feedback loop. Furthermore, MDM2 can bind to p53 and control p53 monoubiquitination, leading to the nuclear export of p53. Other E3 ligases further promote p53 poly-ubiquitination and 26S proteasomal degradation in the cytoplasm. Upon DNA damage, DUBs localize to the nucleus and de-ubiquitinate p53 to alter its stability, thus boosting p53 activation. Consequently, p53 is activated through various kinase or acetyltransferases, after which it binds to its transcriptional targets, including p21, p53 upregulated modulator of apoptosis (PUMA), Bax and Noxa, for example. Ub ubiquitin, DUBs de-ubiquitinating enzymes
Protein modifications can be achieved by either a ubiquitin molecule (mono-ubiquitination) or by a chain of ubiquitin (poly-ubiquitination).237–239 Polyubiquitination, in which four or more ubiquitin monomers are bound to a substrate, occurs mostly on K48 and K29 and is regarded as a “molecular kiss of death” as it is associated with proteasome-dependent degradation.240–242 K63-linked ubiquitination is associated with aggregate formation, lysosomal degradation, and protein interactions.243–246 Monoubiquitination and multiple monoubiquitinations are involved in various processes, including trafficking, inflammation, DNA repair, and histone regulation.247,248 Therefore, ubiquitination regulates proteins in several ways: it can alter their location in cells, impact their activity, control their degradation by the proteasome, and stimulate or prevent protein interactions.249,250 Recently, more and more attention has been paid to the regulation of transcription factor function by ubiquitination. The primary sites for p53 ubiquitination are located at its C terminus, where acetylation takes place during times of cell stress and functions to block protein degradation, maintaining p53 stability.235
MDM2 is a key negative regulator of p53
Mouse two-minute two (MDM2) is an oncogene that accelerates cell growth, survival, invasion, and contributes to therapeutic resistance, and the most well-known function of MDM2 is that it works as an E3 ubiquitin ligase. Physiologically, MDM2 antagonizes tumor suppressor p53.251 MDM2 inhibits the stability of p53 by ubiquitination. In addition, p53 inactivation was managed by MDM2 and in turn, MDM2 affected the subcellular localization of p53. MDM2 is often overexpressed in some human and mouse malignant tumors.252
MDM2, first recognized E3 ligase to regulate p53 stability, contains a RING finger domain and interacts with Ubc5 (E2 ubiquitin-conjugation enzyme), which can ubiquitinate p53 both in vitro and in vivo and, via the proteasome system, is a crucial negative regulator of p53.253–255 The RING finger domain of MDM2 includes a sequence that prevents the activity of E3 ubiquitin-protein ligase;256,257 therefore, MDM2 can regulate its own levels via auto-ubiquitination.258,259 CBP/p300 and MDM2 target six lysine residues (K370, K372, K373, K381, K382, and K386) in the C-terminal regulatory domain respectively for acetylation and ubiquitination,260 which are essential for the nuclear export of p53. MDM2 is a negative regulator of p53 and can effectively inhibit p53 acetylation mediated by p300/CBP in vivo and in vitro. The suppress activity of MDM2 on p53 acetylation was also eliminated by the tumor suppressor p19 (ARF), suggesting that the regulation of acetylation is an important part of the feedback loop of p53-MDM2-p19 (ARF).261 The MDM2 oncoprotein is overexpressed in many types of human cancers and is a critical component of the p53 pathway.262,263 MDM2 targets p53 for ubiquitination, and for proteasome-mediated degradation, and it maintains an appropriately low level of p53 under unstressed cell conditions.264 MDM2 directly decreases the transcriptional activity of p53 by binding to its N-terminal transactivation domain (TAD).265 When MDM2 is overexpressed, there is a loss of p53 activity, and cells acquire limitless replicative potential. Further, MDM2 mediates the nuclear export of p53.266 Moreover, when p53 is ubiquitinated by MDM2, it cannot be acetylated by p300/CBP, and, therefore, rapid proteasome-mediated degradation takes place.261 As MDM2 is transcriptionally induced in a p53-dependent manner, the two proteins make an elegant feedback loop (Fig. 4).267 When modifications occur on MDM2, the direct interaction between p53 and MDM2 is broken, such as during a DNA damage event in which MDM2 is phosphorylated at serine 395.268
Fig. 4.
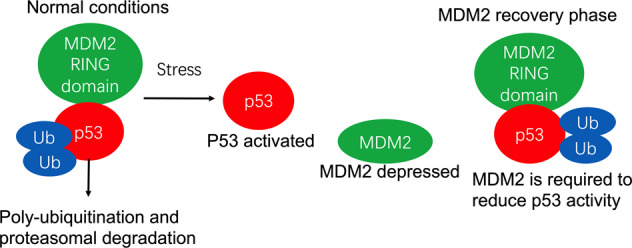
Autoregulatory loop of p53 and MDM2. The RING finger domain of MDM2 is involved in p53 ubiquitination and proteasome-mediated degradation, and, therefore, MDM2 maintains appropriately low p53 levels under unstressed conditions. Activated p53 transcribes MDM2 mRNA and increases the level of MDM2 protein, which in turn reduces p53 activity
Notably, low level of MDM2 activity induces p53 mono-ubiquitination and nuclear output, while high level of MDM2 activity promotes p53 polyubiquitination and nuclear degradation. In summary, MDM2 suppresses p53 in two ways: (i) MDM2 works as an E3 ligase to ubiquitinate p53, thus stimulating its degradation by the proteasomal pathway, and (ii) MDM2 inhibits p53 transcriptional activation by binding to it (Fig. 4).269 Therefore, there are two ways to increase the stability of p53: one is to downregulate the expression of MDM2, the other is to regulate the intracellular localization of MDM2 or p53.270 Activation of p53 results in its uncoupling from MDM2 and the related proteins, such as MDM4, which principally inhibits p53-dependent transactivation.271 Post-translational modification of p53 plays an important role in this process, at least in the DNA damage response. MDM2 is a key mediator of the different stress pathways that impact p53.272
It has been suggested a new cancer treatment strategy is that the small molecular inhibitors intended to block the interaction of MDM2-p53 may effectively treat human cancers that remain wild-type p53 through reactivating the anticancer function of p53.273–277 After two decades of efforts, many nonpeptide, small molecule inhibitors (MDM2 inhibitors) with unique structure and strong efficacy of MDM2–p53 interaction interrupted have been developed, and Nutlins is the first effective and specific small molecule inhibitor which interrupts MDM2–p53 interaction.278 At present, at least seven of these compounds have entered human clinical trials as novel anticancer drugs (Table 2).273
Although MDM2 plays a key role in regulating p53 levels and functions, p53 activity is controlled and fine-managed across a wider range of regulators by multiple mechanisms-monitored PTMs.
Others factors that ubiquitinate p53
p53-induced RING-H2 (Pirh2) is an E3 ligase that has been reported to target p53 for polyubiquitylation and degradation.279 Similar to MDM2, Pirh2 is also a transcriptional target of p53, and its transcription is increased in response to DNA damage.280 Thus, Pirh2 takes part in an autoregulatory feedback loop that mediates p53 function. Interestingly, there are several differences between MDM2 and Pirh2. For example, phosphorylation of Pirh2 can bring about its own inactivation. In addition, MDM2 mainly degrades p53 in unstressed cells, but Pirh2 is capable of degrading p53 after DNA damage.268,281 Furthermore, Pirh2 can regulate the stability of p73, a p53 family member, but MDM2 cannot.282 Thus, it is possible that MDM2 specifically polyubiquitinates and degrades p53, whereas Pirh2 can control the protein stability of other p53 family members. Moreover, Pirh2 interacts with p53 and regulates its polyubiquitylation in association with the E2 ligase ubcH5b, independent of MDM2. Further, Pirh2 preferentially binds to and degrades p53 in its tetrameric active form, but not its monomeric form.283 These data confirm that Pirh2 is a novel tumor suppressor associated with regulation of p53 and MDM2.
Constitutively photomorphogenic 1 (COP1), an E3 ubiquitin ligase, has been regarded as a direct ubiquitin ligase for p53.284,285 COP1 is also a p53-inducible gene (a p53-responsive element exists in the COP1 gene promoter region), and it can ubiquitinate and degrade p53 independently of MDM2, which is necessary for p53 turnover in normal and cancer cells.284 Furthermore, in cancers that involve wild-type p53, the expression of COP1 is associated with a significant reduction in the steady state p53 protein levels and with attenuation of the downstream p53 target gene285–287; therefore, COP1 inhibits p53-mediated G1 arrest, which is important in cell survival, development, and cell growth. In addition, degradation of p53 by COP1 is impaired upon DNA damage, resulting in p53 stabilization and activation.288 The results showed that COP1 was an important negative regulator of p53 and a new pathway for keeping low p53 protein levels in non-stressed cells.
ARF-binding protein 1 (ARF-BP1, HUWE1) is a HECT domain-containing E3 ligase that regulates p53 levels to induce tumor suppression via the stabilization of p53 and the activation of apoptosis.289–291 ARF-BP1 contains a ubiquitin-associated domain (UBA, 1318-54), a WWE domain (1612-92), and a HECT domain in the C-terminal sequences (4036-4734).289 The UBA domain is a small motif shown in various proteins to be related to the ubiquitination pathway.292 ARF-BP1 is a primary binding partner of ARF in cells without p53. Interestingly, ARF effectively represses ARF-BP1-regulated p53 ubiquitination, and it also contributes to the neutralization of ARF-BP1’s p53-independent anti-proliferative effect. In addition, the N-terminal region of ARF showed the strongest inhibition of ARF-BP1-mediated p53 ubiquitination; however, the C-terminal region displays little effect. Therefore, ARF-BP1 plays a crucial role in ARF-mediated p53 stabilization in an MDM2-independent manner.289
Trim24 was identified as a member of family of TRIM/RBCC family of proteins, which contain a conserved amino-terminal tripartite motif consisting of a RING domain, B-box zinc fingers, a coiled-coil region, and carboxy-terminal domains.293,294 Therefore, Trim24 is an E3-ubiquitin ligase that negatively regulates p53 via ubiquitination through its RING domain to promote proteasome-mediated degradation.295,296 Trim24 interacts with phosphorylated p53 to stimulate its degradation. Furthermore, Trim24 is phosphorylated at S768 in response to DNA damage by ATM, which destabilizes Trim24 and interrupts the Trim24–p53 interaction.296 However, DNA-damage-activated p53 induces Trim24 transcription via an interaction with p53 response elements. Similar to MDM2, Trim24 controls p53 levels in an autoregulatory feedback loop.297 However, unlike MDM2, Trim24 also terminates the activated p53-regulated response upon DNA damage.296 p53 is ubiquitinated and negative regulated by Trim24, which indicated that Trim24 is a therapeutic target for p53 to restore tumor inhibition.
Synoviolin, a component of the ER-associated degradation (ERAD) complex, is an E3 ubiquitin ligase that targets p53, and it is engaged in endoplasmic reticulum related degradation, an ATP-dependent ubiquitin-proteasome degradation process that reduces the burden on the ER.298,299 Synoviolin sets p53 apart in the cytoplasm and negatively regulates, for example, its protein level and functions, transcription, and cell cycle regulation.300 Interesting, the regulation of p53 by synoviolin is irrelevant to the other E3 ubiquitin ligase-formed autoregulatory feedback loops, such as those involving MDM2, Pirh2, and Cop1.300 Combined with the antiapoptotic properties of synoviolin previously elucidated in vivo and in vitro studies, its new role in p53 regulation may supply new ideas for studying the pathogenesis of proliferative diseases.
Topoisomerase I-binding protein (Topors) contains an N-terminal C3H4-type RING domain that is similar to the RING domains in E3 ligases, and it contains both ubiquitin-E3 and SUMO-E3 ligase activity.301,302 Human Topors, which was originally regarded as a p53-binding protein and functions as an E3 ubiquitin ligase for p53, leads to the degradation of p53.303
The caspase 8/10-associated RING proteins (CARPs), CARP1 and CARP2, act as RING-domain E3 ligases that target apical caspases for proteasome-mediated degradation.304 In addition to apical caspases, CARPs, which are overexpressed in cancer, physically interact with and target p53 or phospho-p53 for ubiquitination and degradation with or without MDM2. Unlike other E3 ligases, CARPs can ubiquitinate DNA damaged-mediated phospho-p53 at serine 15 or 30.305,306
Human ubiquitination factor E4B (UBE4B) is a human mammalian homolog of the Ufd2 protein found in S. cerevisiae. Yeast Ufd2 is engaged in the Ufd pathway, which is a proteolytic pathway that recognizes ubiquitin as a degradation signal.307 Yeast Ufd2 belongs to a new class of ubiquitination enzyme, E4 (a novel ubiquitin chain assembly factor) and is required for ubiquitin chain assembly.307 Mouse UBE4B regulates ubiquitination as a companion to E1 and E2, and independent of the E3 components. UBE4B physically associates with p53 and MDM2,and then promotes p53 polyubiquitination, which results in p53 degradation, thus inhibiting p53-mediated transactivation and apoptosis.308
p300 and CREB-binding (CBP) were regarded as multifunctional modulators of p53 through their acetylase and poly-ubiquitin ligase (E4) activities.309 p300 and CBP were revealed to be required for endogenous p53 polyubiquitination and rapid turnover in normal cells.310 Interestingly, the ubiquitin ligase activity of p300/CBP is present only in nuclear extracts and not cytoplasmic extracts. In accordance to its E3/E4 activity, CBP specifically destabilizes cytoplasmic, but not nuclear p53.311 In addition, p53 turnover is observed in p300-deficient or CBP-deficient cells via polyubiquitination of mono-p53. Furthermore, p300 exhibits its E3/E4 activity within its N terminus.190 Similar to p300, CBP contains an E3 activity in its N terminus and shows E4 activity towards p53 in vitro.312 Therefore, the E4 activity of cytoplasmic p300/CBP destabilizes p53 by ubiquitinating it, while physically distinct p300/CBP activities in the nucleus, such as p53 acetylation, activates p53.311
E4F transcription factor 1 (E4F1) is a zinc-finger-containing protein identified as an atypical ubiquitin E3 ligase for p53 by activation oligo-ubiquitylation on p53 lysine residues that are different from the targets of MDM2.313 E4F1 physically interacts with p53,314 and then conjugates Ub to p53 that is bound to chromatin, a modification that coincides with the stimulation of a p53 transcriptional program that is engaged only to control cell cycle arrest and not apoptosis. E4F1-mediated modification p53 plays a crucial role in the cellular life-or-death decision.313
Ubc13 is an E2 ubiquitin-conjugating enzyme, but it increases p53 stability by interrupting K63-dependent ubiquitination of p53, which decreases MDM2-dependent polyubiquitination of p53.315 However, Ubc13 increases p53 stability but prevents its tetramerization and increases its location to cytoplasm, which attenuates p53 transcriptional activity.315,316 Like MDM2, p53 activation induces the expression of Ubc13 in response to DNA damage, suggesting a feedback loop between Ubc13 and p53. Ubc13 interaction with p53 requires an intact p53 C-terminal domain, and this interaction negatively effects the tetramerization of p53. However, Ubc13 is not capable of contributing to p53 monomerization in response to DNA damage.316
LINK-A expression increased the degradation of K48 polyubiquitination-mediated endogenous tumor suppressors Rb and p53, which inhibits immune sensitization of breast tumors.95
Thus, p53 are modulated at the level of gene expression and post-translation modification, and at the level of protein stability through ubiquitin proteasome pathway. In the past 20 years, many ubiquitin E3 ligases have been found to promote the degradation of p53 directly or indirectly in vitro and in vivo.
De-ubiquitinating enzymes (DUBs) eliminate ubiquitin from p53
Ubiquitination governs the division, differentiation, and survival of eukaryotic cells. Ubiquitin system is a powerful signal network by consist with multiple E3 ligases (Writers), ubiquitin binding moleculars (Readers) and de-ubiquitylases (erasers) with different functions. From yeast to human, ubiquitin system is used in a similar way.317 De-ubiquitinating enzymes (DUBs) are a group of proteins engaged in the ubiquitin-proteasome system.289 The major function of DUBs is to process and recycle ubiquitin; therefore, DUBs reverse ubiquitination of specific substrate proteins, similar to the reversal of protein phosphorylation by phosphatases.149,284,318 There are several possible reasons why multiple DUBs are needed to regulate p53 stability and activity. First, different DUBs regulate the p53 pathway when confronted with different cellular stresses; second, different DUBs function in different cellular compartments; and last, since p53 is ubiquitinated by many E3 ligases, DUBs are needed to counteract p53 ubiquitination.150,318,319 After p53 is targeted for ubiquitination, de-ubiquitinating enzymes remove ubiquitin from p53 (Fig. 3). It is well known that p53 is a short-lived protein whose levels are low in normal cells and whose stability is tightly regulated through MDM2-mediated ubiquitination.320,321
Abundant evidence suggests that the de-ubiquitinase herpesvirus-associated ubiquitin-specific protease (HAUSP, also known as USP7) plays a critical role in stabilizing p53, even in the presence of excess MDM2, and that it activates p53-dependent cell arrest and apoptosis.322,323 HAUSP was also shown to form a complex with MDM2 and p53. The TRAF-like domain of HAUSP is regarded as the necessary region to bind to p53, and HAUSP interacts with MDM2 both in vivo and in vitro.324,325
In addition to de-ubiquitinating p53, HAUSP also controls MDM2 de-ubiquitination. Thus, HAUSP-mediated de-ubiquitination can bring about increased levels of MDM2 that then accelerate p53 degradation to directly reduce the level of p53. In normal cells, MDM2 is the preferential HAUSP substrate; thus, p53 accumulates due to MDM2 destabilization.326 In stressed cells, ATM is activated by DNA damage, and it then phosphorylates MDM2, which leads to a lowered affinity for HAUSP.248 It is an interesting finding that the effects of HAUSP on the p53 pathway depend on its concentration. Partial reductions in HAUSP levels lead to destabilization of p53, whereas more complete reductions may cause MDM2 destabilization and p53 accumulation.327
USP10 (ubiquitin-specific protease 10) is another de-ubiquitinase enzyme that regulates the levels of p53 by controlling p53 ubiquitination and stability.328,329 Unlike HAUSP, USP10 can interact only with p53, and not with MDM2. Moreover, USP10 is mainly localized in the cytosol, where its function is to maintain the levels of p53 and to counteract MDM2-mediated p53 nuclear export under normal conditions.330 Upon DNA damage, USP10 is phosphorylated by ATM, after which it is re-localized to the nucleus where p53 de-ubiquitination occurs, which is the reverse of the function of residual MDM2, which ubiquitinates p53.329,330 As USP10 plays an anti-cancer role by regulating the nuclear output and degradation of p53 induced by MDM2, down regulating DUBs may have an impact on cancer and other hypoxia related diseases.331
Ovarian tumor domain-containing Ub aldehyde-binding protein 1 (Otub1), DUB from the OTU-domain containing protease family, directly suppresses MDM2-mediated p53 ubiquitination in cells and in vitro.332 However, Otub1 decreases p53 ubiquitination, stabilizing and activating p53 in cells via inhibition of UbcH5, a cognate ubiquitin-conjugating enzyme of MDM2.333 Thus, Otub1 mediates p53 ubiquitination in cells independently of its de-ubiquitinating enzymatic activity.194,332,334,335 Furthermore, Otub1 plays a crucial role in the stability and activity of p53 after DNA damage, because Otub1 can inhibit DNA damage-induced chromatin ubiquitination and slow down DNA repair.336 In conclusion, Otub1 regulates the p53-MDM2 loop as a potential inhibitor of the E2 enzyme.
The ubiquitin-specific protease 2 (USP2) has two isoforms formed by alternative splicing, USP2a and USP2b.337 USP2a is a de-ubiquitinating enzyme that regulates the p53 pathway by interacting with and ubiquitinating MDM2 in vivo.338 USP2a can directly de-ubiquitinate MDM2, but not reverse MDM2-mediated ubiquitination of p53. Overexpression of USP2a causes an increase in the MDM2 protein level and accelerates the degradation of p53. Knock down of USP2a results in increased p53 protein accumulation and activation of its target genes.338 Thus, USP2a was identified as an important regulator of the MDM2/p53 pathway, which is important for repressing p53 activity in vivo.
The DUB ubiquitin-specific protease 24, USP24, is a 2620-amino-acid ubiquitin-specific protease, containing several conserved domains: a UBA domain, a UBL domain and a USP domain.339 USP24 is a DUB that increases p53 stability and activity. USP24 directly de-ubiquitinates p53 in response to DNA damage, as well as in unstressed cells.339 Therefore, USP24 plays a crucial role in the apoptosis pathway by maintaining p53 activation after DNA damaged.150 Furthermore, the USP24 level is increased by DNA damaging agents, and it plays a crucial role in maintaining genome stability.340
Ubiquitin-specific peptidase 29 (USP29) deconjugates ubiquitin from p53 and stabilizes p53.341 USP29 is activated by the far upstream element binding protein (FBP) and reverses MDM2-directed p53 ubiquitination to protect p53 from degradation. Furthermore, USP29 could stabilize p53 in an alternative mechanism via recognizing p38/AIMP2 (JTV1) pro-apoptotic potential.341 As a pro-apoptotic stabilizer of p53, USP29 expression is restricted in most tissues and cells through DNA methylation or repressive chromatin compaction.342
USP22 was initially regarded as part of an 11 gene “death from cancer signature”, which referred to tumors with a cancer stem cell phenotype.343,344 USP22 is a positive regulator of the NAD-dependent histone deacetylase Sirt1. USP22 mediates stabilization of Sirt1 by interacting and removing poly-ubiquitin chains previously conjugated to Sirt1. Sirt1 negatively regulates p53 transcriptional activity to inhibit cell apoptosis. Therefore, USP22 stabilizes Sirt1, leading to suppression of p53-meditated functions.345
In the past decade, DUBs have become an attractive target for cancer treatment for their actions are involved in many diseases such as cancer. The knowledge in the field of DUB and E3 ligase demands further exploration which may benefit to future therapies.331 To summarize, ubiquitination and degradation processes have a profound effect on the activity of p53. Similarly, a series of molecules are involved in de-ubiquitination, which ensures that p53 activity is strictly controlled (summarized in Table 3) (Fig. 3).
Table 3.
Deubiquitinases and ubiquitin-like proteins that impact on the p53 pathway
| De-ubiquitinase /Ubiquitinase | Target | Function |
|---|---|---|
|
De-ubiquitinase HAUSP/USP7 USP2a USP10 Otub1 USP24 USP29 USP22 Ubiquitinase Pirh2 COP1 ARF-BP1 Ubc13 Synoviolin CARP1 Trim24 Topors UBE4B p300/CBP |
p53/MDM2/MdmX p53/MDM2 p53 MDM2 p53 p53 Sirt1 p53 p53 p53 p53 p53 p53 p53 p53 p53/MDM2 p53 |
Stabilization341 Stabilization345 Proteasome degradation Proteasome degradation315 Proteasome degradation509 Proteasome degradation216,308,511 Transactivation216 |
In addition, DUBs is engaged in ubiquitin precursors processing, ubiquitin recycling, and ubiquitin chains editing.346 Thus, it is not surprising that inappropriate activity of DUBs directly or indirectly causes many diseases, including cancer, and affects many signaling pathways. Therefore, the study of p53 related DUB inhibitors and drug modification has become an important study focus in the world, such as USP10 inhibitor Spautin.331
Cross talk between post-translational modifications on p53 following DNA damage
p53 is a key mediator of cellular responses to numerous types of cellular stresses, such as DNA damage. The C terminus of p53 (positions K370, K372, K373, K381, K382, and K386) can be modified by both acetylation and ubiquitination (Fig. 5). Acetylation of p53 interrupts the interaction between p53 and MDM2 by inhibiting the recruitment of MDM2 to the p53 promoter resulting in p53 activation independent of its phosphorylation status.186 After DNA damage, N-terminal phosphorylation of p53 promotes the interaction of p53 with p300/CBP or PCAF and, subsequently, leads to the acetylation of the C-terminal K382 or K320 residues to active the DNA-binding activity of p53. However, repressive K382 methylation prevents acetylation by CBP/p300 at this same site, and the level of methylation at K382 decreased upon DNA damage, counteracting its inhibitory effect and promoting CBP/p300-dependent acetylation of K382.206 Thus, the interplay between p53 methylation, and phosphorylation, as well as acetylation, demonstrates a mechanism for modulating p53 transcriptional activity upon stress. Notably, phosphorylation at S46 and acetylation at K120 are crucial modifications for switching on p53’s pro-apoptotic function, which enables tumor cells to be removed.347 In short, methylation occurs at the C-terminal K370. K372 and K382 residues can also be ubiquitinated and acetylated, and p53 activity can be increased or inhibited depending on the modification site and modification mode. Normally, lysine methylation occurs upon DNA damage and then accelerates or prevents the successive acetylation of other residues158 (Fig. 5). Moreover, ubiquitination and deacetylation quickly weaken p53 expression and function. Therefore, cells can re-enter the cell cycle by escaping from p53-mediated cell cycle arrest.347–349 Collectively, these data suggest that the post-translational of p53 at different sites has different regulatory effects on the transcriptional activity of p53 through different mechanisms.
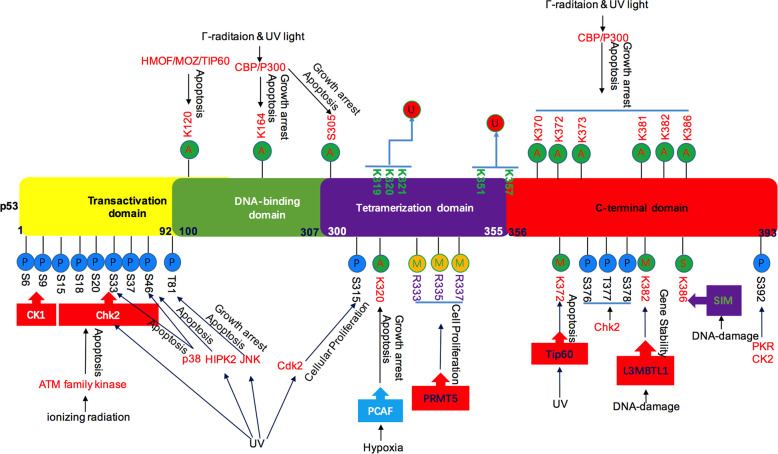
Fig. 5.
p53 structural domains and sites for post-transcriptional modifications. More than 36 amino acids of p53 are reported to be modified. The major sites of p53 post-transcriptional modification are shown with the corresponding main modifying enzymes. The modifications directly responsible for the listed effects are shown. M methylation, A acetylation, U ubiquitination, S sumoylation, P phosphorylation. CK1 casein kinase 1, Chk2 checkpoint kinase 2, ATM ataxia telangiectasia, mutated, hMOF human males absent on the first, MOZ monocytic leukemia zinc-finger protein, TIP60 HIV1-TAT interactive protein, HIPK2 homeodomain interacting protein kinase-2, JNK c-Jun amino terminal kinase, CDK2 cell cycle control regulator cyclin-dependent kinase-2, PCAF p300/CBP-associated factor, PRMT protein arginine methyltransferase, L3MBTL1 lethal 3 malignant brain tumor-like 1, SIM mortality information system, PKR double-stranded RNA-dependent protein kinase
Complex post-translational modifications on p53 in tumor tissues
Furthermore, as many as 150 different PTMs have been identified on p53, suggesting that the mechanisms of p53 post-transcriptional regulation are highly complex in normal and tumor tissues.350 Methylation of lysine and arginine were normally regarded as a reversible mechanism that modulate p53 function. The C terminus of p53 might function as a major site where single modifications occur, and where the K-to-R mutations occur. The variety of modifications and the many modification sites make it very complicated to elucidate the mechanisms by which p53 function is fine-tuned.351 Therefore, extremely careful research using mouse models is needed to study tissue-specific and cell-type-specific changes in p53 function that result from changes in post-translational modifications. Currently, it is not completely clear whether there are other sites, new functions, or new mechanisms that take part in the post-transcriptional modification of p53. Moreover, it is unclear how the modification of p53 influences cells and tissue in a tumor-specific manner. Further studies of specific tumors may help to identify additional attractive targets for radiotherapy and chemotherapy.34,352
Post-translational modifications—modifying the p53 function in mice model
p53S18A knock-in mice, in which serine 18 was mutated to a non-phosphorylatable alanine.353 Phosphorylation of p53 serine 18 does not affect the stability of p53 protein, but contributes to the activation of p53 target genes, thus participating in p53-dependent apoptosis and delayed tumor suppression.354 p53S23A knock-in mice, in which serine 23 was mutated to a non-phosphorylatable alanine. There are data indicate that serine 23 phosphorylation response to DNA damage contributes to the stabilization of p53 protein and cell type dependence of p53-dependent apoptosis, as well as to inhibit the occurrence of B-cell lymphoma.355 p53HupKIS46A, a HupKI mouse strain with serine 46 mutated to non-phosphorylatable alanine, was established to study the role of serine 46 phosphorylation in vivo.356,357 This residue plays a major role in p53-mediated apoptosis. p53S389A knock-in mice was produced and studies have shown that serine 389 phosphorylation selectively promotes apoptosis and tumor suppression under ultraviolet irradiation.358 p53S312A knock-in mice was generated and at this site, ES cells play a key role in the Nanog inhibition and ES cell differentiation, suggesting that serine 315 phosphorylation also plays a role in stem cells.359 Mouse p53 C-terminal contains many lysine residues (K367, K369, K370, K378, K379, K383, and K384), which can be modified by ubiquitination, acetylation, diacylation, sumoyation, or methylation. Two knock-in mouse strains address the importance of these residues by mutating all C-terminal lysine into arginine to block any modification of these residues. The “p536KR” knock-in mouse strain carries six C-terminal lysine mutations (K367R, K369R, K370R, K378R, K379R, and K383R), while the second “p537KR” mouse strain has seven mutations, including the above mutation and one mutation at lysine 384 (K384R), which is a non-conservative sequence in human genes.359,360 To clarify the role of a single lysine, some studies have examined the effects of altering a single lysine, such as a murine strain, p53K317R in lysine knock-in mice, causing acetylation loss on the residue, and acetylation at lysine 317 negatively regulates p53 transcriptional activity.361 The Asn-to-Ser substitution p53 (p53N236S) knock in mice model promotes female embryos neural tube defects.362
The mouse models mentioned above are summarized in Table 4 to provide insight into how post-translational modifications of p53 is linked to its function. PTM mutant mice may exhibit positive or negative regulation of p53 activity.363,364 Thus, future research will further understand the specific role of each PTM and how modification can be used as a therapeutic target for cancer. Thus, PTM site mutant mice may exhibit positive or negative regulation of p53 activity. Future research will understand the specific role of each PTM and show how modifications can be used as a therapeutic target for cancer.
Table 4.
p53 modifications in vivo for p53 as a tumor suppressor protein
| Mouse model | Function | p53 modifications in vivo |
|---|---|---|
|
p53S18A knock-in mice p53S23A knock-in mice p53HupKIS46Aknock-in mice p53S389A knock-in mice p53S312A knock-in mice p536KR knock-in mice |
p53-dependent apoptosis and tumor suppression Stabilization of p53 protein and cell type dependence of p53-dependent apoptosis p53-mediated apoptosis Selectively promotes apoptosis and tumor suppression under ultraviolet irradiation Stem cells DNA damage |
Serine18 mutated to non-phosphorylatable alanine353,512,513 Serine18 mutated to non-phosphorylatable alanine355,513,514 A HupKI mouse strain with serine 46 mutated to non-phosphorylatable alanine356,357 Serine389 mutated to non-phosphorylatable alanine358,515 Serine312 mutated to non-phosphorylatable alanine359,364 Six C-terminal lysine mutations (K367R, K369R, K370R, K378R, K379R, and K383R)516 |
| p537KR knock-in mice | DNA damage | Seven C-terminal lysine mutations (K367R, K369R, K370R, K378R, K379R, K383R, and K384R)359,360 |
| p53K317R knock-in mice | Negatively regulates p53 transcriptional activity | Lysine317 mutated to non-acetylated arginine361 |
| p53N236S knock-in mice | Female embryos neural tube defects | Asparagine236 substitute to serine362,517 |
| p53K120R knock-in mice | mRNA decay | Lysine120 substitute to arginine518 |
Therapeutic strategies to restore wild-type activity of mutant p53
A variety of strategies for tumor expressing p53 mutant, for p53 having many different mutations. Wild-type p53 in tumor cell is an effective activator of apoptosis and senescence, making the reactivation of certain wild-type functions of mutant p53 (usually overexpressed in cancer) a promising therapeutic pathway. Interestingly, the wild-type loss of function caused by some unstable tumor-derived mutations can be remedied by another point mutations that help stabilize the integration of the p53 protein, suggesting that the change of structure is reversible.365 Small molecules such as PhiKan083 and PK7088 bind to a site of p53 and form the Y220C mutant, which will stabilize this mutant and increase the level of wild-type p53.366 Further, other molecules bind to a variety of mutant p53 proteins and interact with DNA binding domains to promote the correct folding of the mutant protein and the recovery of p53 function, PRIMA-1, PRIMA-met/APR-246, CP-31398, and SCH29074 for example.367–369 Wild-type p53 needs to bind to Zn (2+) to fold correctly, while R175H p53 mutant is damaged in zinc binding.370,371 While the addition of zinc to the conformational mutants G245C and G245D p53 partially recover the wild-type constellation.372,373 Therefore, the potential of zinc to restore wild-type folding has been discovered, and this method has been proved to recover chemosensitivity to anticancer drugs in cells which express mutant p53 protein.374 In addition, it was found that NSC31926, a thiourea metal chelator, can restore p53 wild-type function in many different cell lines expressing p53 mutants, possibly by enhancing the bioavailability of zinc to p53 mutants.375
Although there are components targeted to mutant p53, many of them also interact and inhibit p53 family proteins, p63 and p73. A small component called RETRA, discovered by chance in a screening of a drug used to determine stable wild-type p53, is thought to disrupt the p73 mutant with p53 interaction. RETRA induced p73 release led to activation the targeted gene for p73, suppressed tumor cell survival and inhibited xenograft tumor growth376 (Table 2).
Complexity of p53 regulation: post-translational modifications and cross talk with each other
The scope of the post-translational modifications of p53 is deeper and more complex than previously reported. These modifications engaged in p53 level, activity, protein–protein interaction, subcellular localization, and crosstalk from other signaling pathways. The extensive list of p53 post-translational modifications suggest that there is a dazzling arrangement that may exist in p53, therefore, for its functional status at any given time and in any particular biological context. Due to the complexity of those PTMs, future analysis will focus on some certain amino acid sites of p53 and cross talk of PTMs with good characteristics.
PTEN: multiple roles in human cancers
Tumor suppressor, PTEN, a phosphatidylinositol 3,4,5-triphosphate (PIP3) lipid phosphatase, is frequently inactivated in cancer by mutation, epigenetic silencing, or PTMs.377,378 PTEN plays an important role in regulating cell growth, apoptosis, mobility, proliferation, signal transduction and other key cell processes.379 PTEN is affected by phosphorylation, ubiquitination, acetylation, SUMOylation, and oxidation of active sites.380,381 Some post-translational modifications can lead to the deactivation of PTEN function rather than the goal of PTEN gene integrity.382–384 Post-translational modification can dynamically change activity and function of PTEN and abnormal in the post-translational modulation of PTEN brings about cell proliferation, migration, and adhesion, which are related to the occurrence, development and metastasis of cancer.385,386
PTEN phosphorylation is a new mechanism of PTEN inactivation that plays an important role in tumorigenesis
PTEN is a double lipid and protein phosphatase that works as a tumor suppressor through several AKT-dependent and independent pathways.387 PTEN protein has 403 amino acids and contains five crystal domains. One N-terminal (PIP2) binding domain, one N-terminal phosphatase domain, one C2 domain, one C-terminal tail domain rich in proline (P), glutamic acid (E), serine (S), and threonine (T) and various phosphorylation sites and one PDZ interaction region (Fig. 6).388 PTEN has six sites of phosphorylation, which are related to the regulation of tumor suppressive function, stability, and subcellular regionalization.389 Phosphorylation of Ser380, Thr382, Thr383, and Ser385 which are sites of PTEN in its C-tail region results in the intramolecular binding of C-terminal tail of PTEN with the rest of the PTEN body, which leads to the blocking/inactive conformation of PTEN, thus reducing the catalytic activity and membrane binding.390 Each of the four sites helps to stabilize the closed conformation of PTEN, and at least three sites are needed to make up with the full effect of tetraphosphate PTEN, which imply that the dynamic step-by-step closure of PTEN conformation may occur by modifying only one subset of Ser/Thr residues, which in turn may lead to the sliding scale of cell signaling effects.391
Fig. 6.
PTEN structural domains and sites for phosphorylation. PTEN structured domains include the PIP2 binding domain, phosphatase domain, two C-terminal domains, and PDZ domain. PIP2 phosphatidylinositol diphosphate, PDZ post-synaptic95, disks large, zonula occludens, CK2 casein kinase 2, GSK3β glycogen synthase kinase-3β, LKB1 liver kinase B1, PICT1 protein interacting with carboxyl terminus 1, PLK1 polo-like kinase 1, PTEN phosphatase and tensin homolog, ROCK rhoA-associated protein kinase, Ser serine, Thr threonine, Tyr tyrosine
Under the treatment of ionizing radiation (IR), the phosphorylation of PTEN at 240 sites facilitates the interaction between pY240-PTEN and Ki-67, which promotes the recruitment of RAD51 to accelerate DNA repair.392 In glioblastoma (GBM) preclinical model, blocking Y240 phosphorylation can enhance radio sensitivity and prolong survival and Y240F-PTEN knock in mice showed radio sensitivity. FGFR-regulated pY240-PTEN is the key mechanism of anti-radiation therapy and an effective target to improve the efficacy of radiotherapy.392,393 E3 ubiquitin ligase Parkin mediates ubiquitination of many substrate proteins, leading to proteasome degradation. Parkin directly binds with epidermal growth factor receptor (EGFR) and promotes the ubiquitination of EGFR, leading to the decrease of activation of PI3K/AKT signal induced by EGFR, and in turn Parkin depletion promoted the inhibition of PTEN by S-nitrosylation and ubiquitination, which imply that PTEN involved in Parkin depleted PI3K/AKT-mediated cellular survival.394
Casein kinase 2 (CK2) interacts with PTEN physically,395 can phosphorylate PTEN on Thr366, Ser370, Ser380, Thr382, Thr383, and Ser385 (Fig. 6).395–397 The phosphorylation of PTEN by protein kinase CK2 promotes the stabilization of PTEN protein and the associated inactivation of PTEN function.398 Post-translational inactivation of PTEN mediated by CK2 is related to the over-activation of PI3K/AKT pathway, which is a common event in adult B-cell acute lymphoblastic leukemia, suggesting that inhibition of CK2-regulated PTEN may be an effective and novel therapeutic tool for this malignant tumor.399
Ser370, Ser380, Thr382, Thr383, and Ser385 of PTEN can be phosphorylated by liver kinase (LKB1), resulting in its inactivation.400–402 Using the conditional gene knockout alleles of LKB1 and PTEN, the inactivation of the dual alleles of the two tumor suppressor factors in the lung resulted in the pure squamous cell phenotype of lung tumors.403 Glycogen synthase kinase 3β (GSK3β) also play a synergistic role in PTEN phosphorylation with CK2.396 R280T mutation of p53 mediates the proliferation of human glioma cells associated with GSK-3β/PTEN pathway.404 Moreover, rhoA-associated protein kinase (ROCK) can inhibit PTEN after phosphorylation of Ser229, Thr232, Thr319, and Thr321, and then transfer it to the membrane. ROCK1 is a physiological regulator of PTEN. Its function is to inhibit excessive recruitment of macrophages and neutrophils in response to acute inflammation.405 Rak is a tyrosine kinase that interacts with PTEN and phosphorylates it on Tyr336 and plays a real role of tumor suppressor gene by regulating the stability and function of PTEN protein in Breast cancer.406 Furthermore, polo-like kinase 1 (PLK1) phosphorylated Ser-380, Thr-382, and Thr-383 of PTEN which are a cluster of residues regulating the stability of PTEN and the phosphorylation of PTEN was associated with the accumulation of it on chromatin and regulated cell cycle.407 Protein interacting with the carboxy terminus-1 (PICT1) was able to bind to PTEN and phosphorylated Ser-380 which is required for stability of PTEN and its mediated cervical carcinoma.408
In conclusion, phosphorylation of PTEN have potential to restore or enhance PTEN activity, thereby inhibiting cancer cell proliferation and resistance to chemotherapy drugs.
Monoubiquitination of PTEN promotes nuclear localization, and polyubiquitination leads to proteasome degradation in cytosol, resulting in loss of tumor suppressive activity of PTEN
Neuronal precursor cell-expressed developmentally downregulated-4-1 (Nedd4-1), is the first considered E3 ligase for PTEN ubiquitination. NEDD4-1 can mono-ubiquitinate PTEN, which is related to nuclear shuttle, genomic stability and cell cycle arrest.409 NEDD4-1 also can promote polyubiquitylation of PTEN, which accelerate proteasome degradation of PTEN.410
In addition, X-linked apoptotic inhibitors and E3 ubiquitin ligase WW domain (WWP2) is also thought to interacts with and ubiquitinate PTEN, which regulates PTEN degradation via ubiquitination pathway.411,412 It is reported that WWP2 mediates cellular apoptosis, which is a necessary condition for tumorigenesis.413 Therefore, WWP2 might play a crucial role in the survival of cancer cells in a PTEN dependent way.414 Both gene ablation and drug inhibition of WWP1 can activate PTEN and release tumor suppressive activity. WWP1 is a direct MYC (MYC proto-oncogene) target gene that is key for MYC-promoted tumorigenesis. Indole-3-methanol, a compound discovered in cruciferous vegetables, as a natural and effective WWP1 inhibitor.415 Further, linc02023 specifically interacts PTEN, and inhibits its interaction and ubiquitination with PTEN through WWP2, making it stable and inhibiting its downstream expression, suggests that linc02023 may be a new therapeutic target by restoring the antitumor activity of PTEN.416 Therefore, it is a potential therapeutic strategy for the prevention and treatment of cancer through via activation of PTEN.
PTEN-opathies: molecular targeted therapy
The changes of PI3K/AKT/mTOR signaling pathway in PTEN mutant cancer patiens indicated that PI3K, AKT or mTOR are target for molecular therapy.417 PTEN hamartoma tumor syndrome (PHTS) is caused by pathogenic PTEN mutation in germline. mTORC1 inhibitor rapamycin reduces symptoms and excessive growth in PHTS patients.418–420 In fact, rapamycin has been tested in patients with PHTS in phase II open clinical trials.421 In addition, the upstream proteins of PTEN signaling pathway, PI3K and AKT for example, can also be used as drug inhibition candidates for PTEN mutant patients. Drug Wortmannin and AQR 092 are target PI3K and AKT, respectively422,423 (Table 2). Therefore, inhibitors of AKT and PIK3CA are used in Poteus and Proteus-like syndromes and PIK3CA related over growth disorders.422,424,425 In addition, constitutional PTEN pathway dysfunction theoretically requires some kind of chronic treatment program. However, lifelong suppression of mTOR and PIK3CA may not be executive due to immunosuppression, the destruction of systemic glucose homeostasis and the important role of PTEN pathway in normal tissue and organ development.425–427 Another important warning for molecular targeting of PI3K/AKT/mTOR pathway is that feedback induction of collateral carcinogenic signaling pathway leads to drug resistance. This has led to the study of combination therapies that, in theory, can effectively target excessive growth signals without losing control of feedback. In fact, mTORC1 inhibition has been demonstrated effectively bring about feedback activation of upstream signaling components.428 Although most treatment strategies aim to reduce the downstream carcinogenic signal caused by PTEN dysfunction, strategies to improve PTEN level and/or activity also demonstrate promising treatment models. This is especially relevant for cell infiltration of PTEN-L, the first found isoform delegates a long PTEN protein called PTEN long (PTEN-L), which will theoretically allow the recovery of PTEN levels in the context of insufficient PTEN haplotypes.429 Another possible way is to restore or even enhance the function of PTEN by editing the mutated PTEN allele. Although gene editing has brought many challenges, including miss target effect and induction of adaptive immune response, recent progress shows hope in reducing these results.188,430 There is no doubt that gene editing will be very challenging in the reproductive environment of the whole organism.430–432
PI3K/AKT/mTOR pathway is also a crucial pathway of immune regulation.433,434 Since PTEN is the main controller of this pathway, it is not surprising that the destruction of PTEN leads to immune disorders. The latter is closely related to the occurrence of cancer. Immune surveillance, immune recognition evasion and the microenvironment of chronic inflammation are the main immune characteristics of cancer.435 In addition, activation of PI3K/AKT/mTOR pathway has been discovered to regulate the response of immunotherapy. The loss of PTEN in sporadic environment has always been related to the drug resistance of anti PD-1 in the treatment of melanoma. Recently, PTEN has been used in the case study of metastatic uterine leiomyosarcoma.436 Interestingly, the PI3K/AKT/mTOR pathway activated has been demonstrated to drive the expression of PD-1/PD-1L in some solid tumors, leading to immune tolerance.437–439
Significance of the Rb–p53–PTEN network to cancer
Rb is the most common mutation gene in childhood cancer retinoblastoma, and its deletion leads to E2F transcription factor induced proliferation related genes.440,441 However, the increase of E2F level after pRb loss can also activate apoptosis associated genes, as a protective mechanism against sudden tumor. Further, the accumulation and apoptosis induced p53 are considered to be the main mechanism to reduce the abnormal high level of E2F activity.442 Thus, PTEN/PI3K/AKT pathway on Rb/E2F apoptosis suppression may supply a potential therapy for retinoblastoma.
PTEN encodes a lipid phosphatase which antagonizes PI3K, and these two genes are often lost in many human cancers.35,443 Further, mutated PTEN are discovered in rare autosomal dominant cancer susceptibility syndromes, such as Cowden’s disease.444 The gene p53 deleted, point mutated and allele lost are common in most human cancers.445,446 p53 mutation is also related to Li Fraumeni syndrome which is susceptible to hereditary cancer.253 Therefore, Rb, E2F, PTEN, PI3K, AKT, and p53 are all involved in the function of cell growth, and gain or loss function of TSGs and oncogene. The abnormal network of those genes can bring about unregulated growth. PTEN can inhibit PI3K-AKT pathway that can promote the nuclear localization of MDM2 and the downregulation of p53, which may reveal the mechanism of cancer chemotherapy resistance to a certain extent.447,448 Cancer produces growth and survival factors that activate PI3K through autocrine or paracrine mechanisms. PI3K-kinase is a component that can be detected in many human cancers and it is associated with cell cycle arrest, inhibited apoptosis, increased tumor cells resistance to chemotherapy.449–453 Chemotherapy resistance stems from the following facts:
Treatment drugs could damage DNA, which promotes p53 activation. Lack of functional PTEN, or inappropriate activation of PI3K–AKT will ring from downstream target of PTEN, which will decrease p53 activity and disable cancer cells make a proper response to DNA damage. Restoration of PTEN, the development of small molecule inhibitors of PI3K and its targets, including MDM2, or elevation of p53 expression in tumor cells through gene therapy could inhibit tumor growth and sensitize refractory cancers to chemotherapy. The recovery of PTEN function and investigation of small molecule inhibitors to PI3K and its targets, covering MDM2, or the enhancement of p53 expression in tumor cells through gene therapy, can stop tumor growth and make refractory tumors sensitive to chemotherapy35 (Fig. 7).
Fig. 7.
Response of cell stress on regulation of p53 function by Rb and PTEN. a The response of cells to stress is phosphorylation of Rb, and the stabilization of the p53 protein. Such stabilization readies the cell for an apoptosis. b One of the target genes activated by p53 is MDM2. Nuclear entry of MDM2 blocks p53 transactivation and promotes p53 degradation. c p53 activates the gene encoding PTEN. PTEN protein inhibits PI3K signaling and increases cellular levels of p53. Induction of PTEN by p53 could enhance p53 function and activate apoptotic response of cells
Conclusions
Under physiological conditions, tumor suppressor genes are finely regulated. These genes act as a role in the normal survival of cells by modulating the cell cycle and activating other genes engaged in the cell’s response to DNA damage, as well as inhibiting carcinogenesis, and mutation or deletion of these tumor suppressor genes may result in the deactivation of tumor suppressor, and then lead to the occurrence of malignant tumors. However, Rb deletions are almost universal in neuroendocrine prostate cancer, characterized by frequent concurrent changes in PTEN and p53. p53 mutation may also lead to poor response to androgen receptor targeted therapy of castration-resistant prostate cancer.454 Absent of PTEN is linked to the enhanced risk of cancer recurrence and metastasis after treatment.455 The loss of PTEN accelerated the medullary thyroid carcinoma induced by the loss of p53 and Rb.456 In high grade serous ovarian cancer, there is signaling between p53, PTEN, and Rb which contributes to tubal epithelial stem cell maintenance and the main drivers of cell transformation.457 In adult brain, the synergistic effect of PTEN, p53, and Rb pathway can produce high-grade astrocytoma.68 Inactivation of these three tumor suppressor genes was also detected in the stroma of oropharyngeal, breast, and other tumors. The mouse model demonstrated the tumor promoting effect of deletion of Rb, Pten, or p53 in fibroblasts, which transformed normal fibroblasts into cancer-related fibroblasts.71 The above suggests the interaction of signaling pathways managed through tumor suppressors, and those three major tumor suppressor genes interact with each other in the development and progression of these tumors, and PTMs play an important role in it.
In addition, PTM can improve the stability of complex signaling pathways through a variety of regulatory mechanisms. PTM is closely related to the occurrence, spread and metastasis of tumors; however, the underlying molecular mechanisms are still poorly understood.47,449 In most cancers, PTM is significantly changed, so it may become a potential target of cancer treatment. PTMs can be used as a biomarker of disease status, and its application in the assessment and monitoring of cancer disorders is a new clinical focus.458,459 p53 gene is now thought to encode as many as 12 different isoforms, some of which may experience PTM, suggesting that there is a great number of structural permutations possible for p53 and its function can change based on a profoundly complex variety of PTMs.153
Dysfunctional of TSG is part of signal pathway, and the carcinogenesis is regulated by over activation of the pathway. In this case, inactivated TSG can be a therapeutic target by inhibiting the downstream associated pathways. One example is PTEN, one of the most common TSG changes in human malignancies. PTEN is inactivated with a significant proportion of mutations or deletions in a variety of cancer types, such as glioblastoma, endometrial, prostate, uterine and breast cancers, and melanoma.15,426,460 Post-translational modifications of TSG impact downstream targets of TSG, and can influence their functions involving in cancer, ageing, heart failure, autoimmune disease and so on (Fig. 8).461 The reversible processes of post-translational modification provide a complex regulatory net in the TSG pathway, including the maintenance of low p53 protein levels via ubiquitination, and p53 localization, which is related to ubiquitination, de-ubiquitination and SUMOylation. The TSG post-translational modification network may be different in different species. For example, the p53-responsive binding sites guiding apoptosis in mice do not appear to be functional in primates.462,463 Ubiquitination and de-ubiquitination have received much more attention.235,464–466 Nevertheless, many questions remain about how E3 ligases mediate p53 ubiquitination or what controls the activity of de-ubiquitinating enzymes. Future studies will most likely focus on in vivo experiments to elucidate the complexity and functions of post-translational modifications in the modulation of TSG activity. Clinical strategies may be intended to overcome chemo-resistance by inhibiting TSG degradation or other modifications.467 The design of TSG molecular inhibitors that target the ubiquitination pathway might be an intriguing anticancer strategy in the future.468,469
Fig. 8.
Interplay among post-translational modifications (PTMs) in the regulation of disease. Five main PTMs (phosphorylation, ubiquitination, acetylation, sumoylation and glycosylation) as well as their relative reverse processes (dephosphorylation, deubiquitination, and deacetylation) are involved in the regulation of cancer. In conclusion, the balance of cell (Yin–Yang; Yin, black; Yang, white) is crucial for maintaining cell fundamental functions, whereas dysfunction is associated with normal aging as well as with many human diseases including premature aging diseases, cancer, heart failure, autoimmune disease, and neurodegenerative disease
There are several questions to be launched. Do any other kinds of PTMs exist? Are there any other PTM enzymes not related to what have already been found? Are PTMs genuinely associated with tumor suppression or progression? If PTMs enzymes do not directly play a key role in tumor suppression or progression, then is it possible that they control one or new homeostatic mechanisms? Furthermore, given that TSG wild-type or mutant forms inhibit or promote the expression of many target genes, what role do PTMs enzymes play in these processes? Future research shows that absolute modifying factors of disease performance and related signal networks will be the most important factors to define more accurate and effective prevention and treatment strategies for individuals at risk.
Of note, few other studies have reported the role of PTMs in crosstalk of tumor suppressor genes, especially in Rb, p53, and PTEN which are more obviously affected by PTMs. Future research will be necessary to pay attention to the proteomics so that we can fully understand the role of different PTMs in regulating TSGs in cancer.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [81672787 (Y.Tao), 81874139 and 81672991 (S.Liu)], and the Overseas Expertise Introduction Project for Discipline Innovation (111 Project, No. 111-2-12).
Author contributions
L.C. and Y.T. wrote the manuscript, and S.L. and Y.T. contributed to revise the content of the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Bourguignon LYW. Matrix hyaluronan-CD44 interaction activates microRNA and LncRNA signaling associated with chemoresistance, invasion, and tumor progression. Front. Oncol. 2019;9:492. doi: 10.3389/fonc.2019.00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop JM, et al. Origin and function of avian retrovirus transforming genes. Cold Spring Harb. Symp. Quant. Biol. 1980;44:919–930. doi: 10.1101/sqb.1980.044.01.099. [DOI] [PubMed] [Google Scholar]
- 4.Parris GE. The cell clone ecology hypothesis and the cell fusion model of cancer progression and metastasis: history and experimental support. Med. Hypotheses. 2006;66:76–83. doi: 10.1016/j.mehy.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Bashyam MD, Animireddy S, Bala P, Naz A, George SA. The Yin and Yang of cancer genes. Gene. 2019;704:121–133. doi: 10.1016/j.gene.2019.04.025. [DOI] [PubMed] [Google Scholar]
- 6.Dyson NJ. RB1: a prototype tumor suppressor and an enigma. Genes Dev. 2016;30:1492–1502. doi: 10.1101/gad.282145.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalsoom S, et al. Alterations in the RB1 gene in Pakistani patients with retinoblastoma using direct sequencing analysis. Mol. Vis. 2015;21:1085–1092. [PMC free article] [PubMed] [Google Scholar]
- 8.Joyce, C. & Kasi, A. Cancer, tumor-suppressor genes. (StatPearls, 2019). [PubMed]
- 9.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc. Natl Acad. Sci. USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friend SH, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 11.Quon KC, Berns A. Haplo-insufficiency? Let me count the ways. Genes Dev. 2001;15:2917–2921. doi: 10.1101/gad.949001. [DOI] [PubMed] [Google Scholar]
- 12.Cook WD, McCaw BJ. Accommodating haploinsufficient tumor suppressor genes in Knudson’s model. Oncogene. 2000;19:3434–3438. doi: 10.1038/sj.onc.1203653. [DOI] [PubMed] [Google Scholar]
- 13.Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 14.Epstein RJ. A periodic table for cancer. Future Oncol. 2015;11:785–800. doi: 10.2217/fon.14.315. [DOI] [PubMed] [Google Scholar]
- 15.Morris LG, Chan TA. Therapeutic targeting of tumor suppressor genes. Cancer. 2015;121:1357–1368. doi: 10.1002/cncr.29140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comings DE. A general theory of carcinogenesis. Proc. Natl Acad. Sci. USA. 1973;70:3324–3328. doi: 10.1073/pnas.70.12.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, et al. Targeting tumor suppressor genes for cancer therapy. Bioessays. 2015;37:1277–1286. doi: 10.1002/bies.201500093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGarvey TW, Malkowicz SB. The role of the cell cycle in genitourinary carcinoma. World J. Urol. 1996;14:310–317. doi: 10.1007/BF00184603. [DOI] [PubMed] [Google Scholar]
- 19.Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 20.Wang LH, Wu CF, Rajasekaran N, Shin YK. Loss of tumor suppressor gene function in human cancer: an overview. Cell Physiol. Biochem. 2018;51:2647–2693. doi: 10.1159/000495956. [DOI] [PubMed] [Google Scholar]
- 21.Klughammer J, et al. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat. Med. 2018;24:1611–1624. doi: 10.1038/s41591-018-0156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JW, Han JW. Targeting epigenetics for cancer therapy. Arch. Pharm. Res. 2019;42:159–170. doi: 10.1007/s12272-019-01126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santos FP, Kantarjian H, Garcia-Manero G, Issa JP, Ravandi F. Decitabine in the treatment of myelodysplastic syndromes. Expert Rev. Anticancer Ther. 2010;10:9–22. doi: 10.1586/era.09.164. [DOI] [PubMed] [Google Scholar]
- 24.Kantarjian HM, et al. Guadecitabine (SGI-110) in treatment-naive patients with acute myeloid leukaemia: phase 2 results from a multicentre, randomised, phase 1/2 trial. Lancet Oncol. 2017;18:1317–1326. doi: 10.1016/S1470-2045(17)30576-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leiderman YI, Kiss S, Mukai S. Molecular genetics of RB1—the retinoblastoma gene. Semin. Ophthalmol. 2007;22:247–254. doi: 10.1080/08820530701745165. [DOI] [PubMed] [Google Scholar]
- 26.Smith AL, Robin TP, Ford HL. Molecular pathways: targeting the TGF-beta pathway for cancer therapy. Clin. Cancer Res. 2012;18:4514–4521. doi: 10.1158/1078-0432.CCR-11-3224. [DOI] [PubMed] [Google Scholar]
- 27.Savage KI, Harkin DP. BRCA1, a ‘complex’ protein involved in the maintenance of genomic stability. FEBS J. 2015;282:630–646. doi: 10.1111/febs.13150. [DOI] [PubMed] [Google Scholar]
- 28.Nayak SK, Panesar PS, Kumar H. p53-Induced apoptosis and inhibitors of p53. Curr. Med. Chem. 2009;16:2627–2640. doi: 10.2174/092986709788681976. [DOI] [PubMed] [Google Scholar]
- 29.Rahman N, Scott RH. Cancer genes associated with phenotypes in monoallelic and biallelic mutation carriers: new lessons from old players. Hum. Mol. Genet. 2007;16:R60–R66. doi: 10.1093/hmg/ddm026. [DOI] [PubMed] [Google Scholar]
- 30.Xu JH, Hu SL, Shen GD, Shen G. Tumor suppressor genes and their underlying interactions in paclitaxel resistance in cancer therapy. Cancer Cell Int. 2016;16:13. doi: 10.1186/s12935-016-0290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu K, et al. Oncogenes and tumor suppressor genes: comparative genomics and network perspectives. BMC Genomics. 2015;16:S8. doi: 10.1186/1471-2164-16-S7-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai D, Visser-Grieve S, Yang X. Tumour suppressor genes in chemotherapeutic drug response. Biosci. Rep. 2012;32:361–374. doi: 10.1042/BSR20110125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tulipano, G. How treatments with endocrine and metabolic drugs influence pituitary cell function. Endocr. Connect. 10.1530/EC-19-0482 (2020). [DOI] [PMC free article] [PubMed]
- 35.Mayo LD, Donner DB. The PTEN, Mdm2, p53 tumor suppressor-oncoprotein network. Trends Biochem. Sci. 2002;27:462–467. doi: 10.1016/s0968-0004(02)02166-7. [DOI] [PubMed] [Google Scholar]
- 36.Gudipaty SA, Conner CM, Rosenblatt J, Montell DJ. Unconventional ways to live and die: cell death and survival in development, homeostasis, and disease. Annu Rev. Cell Dev. Biol. 2018;34:311–332. doi: 10.1146/annurev-cellbio-100616-060748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong YN, Crawford JC, Heckmann BL, Green DR. To the edge of cell death and back. FEBS J. 2019;286:430–440. doi: 10.1111/febs.14714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horita, H., Law, A. & Middleton, K. Utilizing optimized tools to investigate PTM crosstalk: identifying potential PTM crosstalk of acetylated Mitochondrial proteins. Proteomes10.3390/proteomes6020024 (2018). [DOI] [PMC free article] [PubMed]
- 39.Stram AR, Payne RM. Post-translational modifications in mitochondria: protein signaling in the powerhouse. Cell Mol. Life Sci. 2016;73:4063–4073. doi: 10.1007/s00018-016-2280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vu LD, Gevaert K, De Smet I. Protein language: post-translational modifications talking to each other. Trends Plant Sci. 2018;23:1068–1080. doi: 10.1016/j.tplants.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 41.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 42.Reiche, J. & Huber, O. Post-translational modifications of tight junction transmembrane proteins and their direct effect on barrier function. Biochim. Biophys. Acta Biomembr.10.1016/j.bbamem.2020.183330 (2020). [DOI] [PubMed]
- 43.Ryan VH, Fawzi NL. Physiological, pathological, and targetable membraneless organelles in neurons. Trends Neurosci. 2019;42:693–708. doi: 10.1016/j.tins.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venne AS, Kollipara L, Zahedi RP. The next level of complexity: crosstalk of posttranslational modifications. Proteomics. 2014;14:513–524. doi: 10.1002/pmic.201300344. [DOI] [PubMed] [Google Scholar]
- 45.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 46.Johnson LN. The regulation of protein phosphorylation. Biochem. Soc. Trans. 2009;37:627–641. doi: 10.1042/BST0370627. [DOI] [PubMed] [Google Scholar]
- 47.Han ZJ, Feng YH, Gu BH, Li YM, Chen H. The post-translational modification, SUMOylation, and cancer (Review) Int. J. Oncol. 2018;52:1081–1094. doi: 10.3892/ijo.2018.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iwasa H, Hossain S, Hata Y. Tumor suppressor C-RASSF proteins. Cell Mol. Life Sci. 2018;75:1773–1787. doi: 10.1007/s00018-018-2756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 50.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 51.Ikeda S, Kishida M, Matsuura Y, Usui H, Kikuchi A. GSK-3beta-dependent phosphorylation of adenomatous polyposis coli gene product can be modulated by beta-catenin and protein phosphatase 2A complexed with Axin. Oncogene. 2000;19:537–545. doi: 10.1038/sj.onc.1203359. [DOI] [PubMed] [Google Scholar]
- 52.Ha NC, Tonozuka T, Stamos JL, Choi HJ, Weis WI. Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation. Mol. Cell. 2004;15:511–521. doi: 10.1016/j.molcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 53.Lee HK, et al. Ubiquitylation and degradation of adenomatous polyposis coli by MKRN1 enhances Wnt/beta-catenin signaling. Oncogene. 2018;37:4273–4286. doi: 10.1038/s41388-018-0267-3. [DOI] [PubMed] [Google Scholar]
- 54.Bohgaki M, et al. The E3 ligase PIRH2 polyubiquitylates CHK2 and regulates its turnover. Cell Death Differ. 2013;20:812–822. doi: 10.1038/cdd.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarsounas, M. & Sung, P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 10.1038/s41580-020-0218-z (2020). [DOI] [PMC free article] [PubMed]
- 56.Ducy M, et al. The tumor suppressor PALB2: inside out. Trends Biochem. Sci. 2019;44:226–240. doi: 10.1016/j.tibs.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 57.Luijsterburg, M. S. et al. A PALB2-interacting domain in RNF168 couples homologous recombination to DNA break-induced chromatin ubiquitylation. Elife10.7554/eLife.20922 (2017). [DOI] [PMC free article] [PubMed]
- 58.Martin KR, et al. The genomic landscape of tuberous sclerosis complex. Nat. Commun. 2017;8:15816. doi: 10.1038/ncomms15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abramowicz A, Gos M. Neurofibromin—protein structure and cellular functions in the context of neurofibromatosis type I pathogenesis. Postepy Hig. Med. Dosw. 2015;69:1331–1348. doi: 10.5604/17322693.1185213. [DOI] [PubMed] [Google Scholar]
- 60.Lee SW, et al. Skp2-dependent ubiquitination and activation of LKB1 is essential for cancer cell survival under energy stress. Mol. Cell. 2015;57:1022–1033. doi: 10.1016/j.molcel.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dogliotti G, et al. Membrane-binding and activation of LKB1 by phosphatidic acid is essential for development and tumour suppression. Nat. Commun. 2017;8:15747. doi: 10.1038/ncomms15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barbier-Torres L, et al. Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget. 2015;6:2509–2523. doi: 10.18632/oncotarget.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, et al. Critical role of FOXO3a in carcinogenesis. Mol. Cancer. 2018;17:104. doi: 10.1186/s12943-018-0856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu JC, et al. Identification of CDC25 as a common therapeutic target for triple-negative breast cancer. Cell Rep. 2018;23:112–126. doi: 10.1016/j.celrep.2018.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamid AA, et al. Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. Eur. Urol. 2019;76:89–97. doi: 10.1016/j.eururo.2018.11.045. [DOI] [PubMed] [Google Scholar]
- 67.Armenia J, et al. The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 2018;50:645–651. doi: 10.1038/s41588-018-0078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chow LM, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19:305–316. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drake LE, Macleod KF. Tumour suppressor gene function in carcinoma-associated fibroblasts: from tumour cells via EMT and back again? J. Pathol. 2014;232:283–288. doi: 10.1002/path.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benavente CA, Dyer MA. Genetics and epigenetics of human retinoblastoma. Annu. Rev. Pathol. 2015;10:547–562. doi: 10.1146/annurev-pathol-012414-040259. [DOI] [PubMed] [Google Scholar]
- 73.Fabian ID, et al. The management of retinoblastoma. Oncogene. 2018;37:1551–1560. doi: 10.1038/s41388-017-0050-x. [DOI] [PubMed] [Google Scholar]
- 74.Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat. Rev. Mol. Cell Biol. 2013;14:297–306. doi: 10.1038/nrm3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer. 2019;19:326–338. doi: 10.1038/s41568-019-0143-7. [DOI] [PubMed] [Google Scholar]
- 76.Egger JV, Lane MV, Antonucci LA, Dedi B, Krucher NA. Dephosphorylation of the retinoblastoma protein (Rb) inhibits cancer cell EMT via Zeb. Cancer Biol. Ther. 2016;17:1197–1205. doi: 10.1080/15384047.2016.1235668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Antonucci LA, Egger JV, Krucher NA. Phosphorylation of the Retinoblastoma protein (Rb) on serine-807 is required for association with Bax. Cell Cycle. 2014;13:3611–3617. doi: 10.4161/15384101.2014.964093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Munro S, Carr SM, La Thangue NB. Diversity within the pRb pathway: is there a code of conduct? Oncogene. 2012;31:4343–4352. doi: 10.1038/onc.2011.603. [DOI] [PubMed] [Google Scholar]
- 79.Kolupaeva V, Janssens V. PP1 and PP2A phosphatases-cooperating partners in modulating retinoblastoma protein activation. FEBS J. 2013;280:627–643. doi: 10.1111/j.1742-4658.2012.08511.x. [DOI] [PubMed] [Google Scholar]
- 80.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 81.Rubin SM, Gall AL, Zheng N, Pavletich NP. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell. 2005;123:1093–1106. doi: 10.1016/j.cell.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 82.Burke JR, Deshong AJ, Pelton JG, Rubin SM. Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J. Biol. Chem. 2010;285:16286–16293. doi: 10.1074/jbc.M110.108167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burke JR, Hura GL, Rubin SM. Structures of inactive retinoblastoma protein reveal multiple mechanisms for cell cycle control. Genes Dev. 2012;26:1156–1166. doi: 10.1101/gad.189837.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rubin SM. Deciphering the retinoblastoma protein phosphorylation code. Trends Biochem. Sci. 2013;38:12–19. doi: 10.1016/j.tibs.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hassler M, et al. Crystal structure of the retinoblastoma protein N domain provides insight into tumor suppression, ligand interaction, and holoprotein architecture. Mol. Cell. 2007;28:371–385. doi: 10.1016/j.molcel.2007.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee JO, Russo AA, Pavletich NP. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 87.Nozaki M, et al. Roles of the functional loss of p53 and other genes in astrocytoma tumorigenesis and progression. NeuroOncology. 1999;1:124–137. doi: 10.1093/neuonc/1.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marshall, A. E. et al. RB1 deletion in retinoblastoma protein pathway-disrupted cells results in DNA damage and cancer progression. Mol. Cell Biol.10.1128/MCB.00105-19 (2019). [DOI] [PMC free article] [PubMed]
- 89.Gubern A, et al. The N-Terminal phosphorylation of RB by p38 bypasses its inactivation by CDKs and prevents proliferation in cancer cells. Mol. Cell. 2016;64:25–36. doi: 10.1016/j.molcel.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 90.Walter DM, et al. RB constrains lineage fidelity and multiple stages of tumour progression and metastasis. Nature. 2019;569:423–427. doi: 10.1038/s41586-019-1172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sanidas I, et al. A code of mono-phosphorylation modulates the function of RB. Mol. Cell. 2019;73:985–1000 e1006. doi: 10.1016/j.molcel.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jin X, et al. Phosphorylated RB promotes cancer immunity by inhibiting NF-kappaB activation and PD-L1 expression. Mol. Cell. 2019;73:22–35 e26. doi: 10.1016/j.molcel.2018.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vasaikar S, et al. Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell. 2019;177:1035–1049. doi: 10.1016/j.cell.2019.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu H, et al. Human U3 protein14a is a novel type ubiquitin ligase that binds RB and promotes RB degradation depending on a leucine-rich region. Biochim. Biophys. Acta. 2018;1865:1611–1620. doi: 10.1016/j.bbamcr.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 95.Hu Q, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat. Immunol. 2019;20:835–851. doi: 10.1038/s41590-019-0400-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bhattacharya S, Ghosh MK. HAUSP, a novel deubiquitinase for Rb - MDM2 the critical regulator. FEBS J. 2014;281:3061–3078. doi: 10.1111/febs.12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kalejta RF, Shenk T. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc. Natl Acad. Sci. USA. 2003;100:3263–3268. doi: 10.1073/pnas.0538058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Uchida C, et al. Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBO J. 2005;24:160–169. doi: 10.1038/sj.emboj.7600486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Higashitsuji H, Liu Y, Mayer RJ, Fujita J. The oncoprotein gankyrin negatively regulates both p53 and RB by enhancing proteasomal degradation. Cell Cycle. 2005;4:1335–1337. doi: 10.4161/cc.4.10.2107. [DOI] [PubMed] [Google Scholar]
- 100.Meng, F., Li, X., Ren, H. & Qian, J. In vivo detection and analysis of Rb protein SUMOylation in human cells. J. Vis. Exp. 10.3791/56096 (2017). [DOI] [PMC free article] [PubMed]
- 101.Meng F, Qian J, Yue H, Li X, Xue K. SUMOylation of Rb enhances its binding with CDK2 and phosphorylation at early G1 phase. Cell Cycle. 2016;15:1724–1732. doi: 10.1080/15384101.2016.1182267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sharma P, Kuehn MR. SENP1-modulated sumoylation regulates retinoblastoma protein (RB) and Lamin A/C interaction and stabilization. Oncogene. 2016;35:6429–6438. doi: 10.1038/onc.2016.177. [DOI] [PubMed] [Google Scholar]
- 103.Chan HM, Krstic-Demonacos M, Smith L, Demonacos C, La Thangue NB. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat. Cell Biol. 2001;3:667–674. doi: 10.1038/35083062. [DOI] [PubMed] [Google Scholar]
- 104.Markham D, Munro S, Soloway J, O’Connor DP, La Thangue NB. DNA-damage-responsive acetylation of pRb regulates binding to E2F-1. EMBO Rep. 2006;7:192–198. doi: 10.1038/sj.embor.7400591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carr SM, Munro S, Kessler B, Oppermann U, La Thangue NB. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. EMBO J. 2011;30:317–327. doi: 10.1038/emboj.2010.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Michaloglou C, et al. Combined inhibition of mTOR and CDK4/6 is required for optimal blockade of E2F function and long-term growth inhibition in estrogen receptor-positive breast cancer. Mol. Cancer Ther. 2018;17:908–920. doi: 10.1158/1535-7163.MCT-17-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Singh S, Johnson J, Chellappan S. Small molecule regulators of Rb-E2F pathway as modulators of transcription. Biochim. Biophys. Acta. 2010;1799:788–794. doi: 10.1016/j.bbagrm.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goel S, DeCristo MJ, McAllister SS, Zhao JJ. CDK4/6 inhibition in cancer: beyond cell cycle arrest. Trends Cell Biol. 2018;28:911–925. doi: 10.1016/j.tcb.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Finn RS, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16:25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 110.Fry DW, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 111.Toogood PL, et al. Discovery of a potent and selective inhibitor of cyclin-dependent kinase 4/6. J. Med. Chem. 2005;48:2388–2406. doi: 10.1021/jm049354h. [DOI] [PubMed] [Google Scholar]
- 112.Whittaker SR, Mallinger A, Workman P, Clarke PA. Inhibitors of cyclin-dependent kinases as cancer therapeutics. Pharm. Ther. 2017;173:83–105. doi: 10.1016/j.pharmthera.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McCartney A, et al. Mechanisms of resistance to CDK4/6 inhibitors: potential implications and biomarkers for clinical practice. Front. Oncol. 2019;9:666. doi: 10.3389/fonc.2019.00666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tripathy D, Bardia A, Sellers WR. Ribociclib (LEE011): mechanism of action and clinical impact of this selective cyclin-dependent kinase 4/6 inhibitor in various solid tumors. Clin. Cancer Res. 2017;23:3251–3262. doi: 10.1158/1078-0432.CCR-16-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Naz S, et al. Abemaciclib, a selective CDK4/6 inhibitor, enhances the radiosensitivity of non-small cell lung cancer in vitro and in vivo. Clin. Cancer Res. 2018;24:3994–4005. doi: 10.1158/1078-0432.CCR-17-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mukai H, et al. Palbociclib in combination with letrozole in patients with estrogen receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: PALOMA-2 subgroup analysis of Japanese patients. Int. J. Clin. Oncol. 2019;24:274–287. doi: 10.1007/s10147-018-1353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dickson MA. Molecular pathways: CDK4 inhibitors for cancer therapy. Clin. Cancer Res. 2014;20:3379–3383. doi: 10.1158/1078-0432.CCR-13-1551. [DOI] [PubMed] [Google Scholar]
- 118.Rugo HS, et al. Palbociclib plus endocrine therapy in older women with HR+/HER2- advanced breast cancer: a pooled analysis of randomised PALOMA clinical studies. Eur. J. Cancer. 2018;101:123–133. doi: 10.1016/j.ejca.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 119.Johnson J, et al. Targeting the RB-E2F pathway in breast cancer. Oncogene. 2016;35:4829–4835. doi: 10.1038/onc.2016.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: from discovery to therapy. Cancer Discov. 2016;6:353–367. doi: 10.1158/2159-8290.CD-15-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Klein ME, Kovatcheva M, Davis LE, Tap WD, Koff A. CDK4/6 inhibitors: the mechanism of action may not be as simple as once thought. Cancer Cell. 2018;34:9–20. doi: 10.1016/j.ccell.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lynce F, Shajahan-Haq AN, Swain SM. CDK4/6 inhibitors in breast cancer therapy: current practice and future opportunities. Pharm. Ther. 2018;191:65–73. doi: 10.1016/j.pharmthera.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 125.Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 126.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat. Rev. Cancer. 2008;8:714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dick FA, Goodrich DW, Sage J, Dyson NJ. Non-canonical functions of the RB protein in cancer. Nat. Rev. Cancer. 2018;18:442–451. doi: 10.1038/s41568-018-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Burkhart DL, Morel KL, Sheahan AV, Richards ZA, Ellis L. The role of RB in prostate cancer progression. Adv. Exp. Med. Biol. 2019;1210:301–318. doi: 10.1007/978-3-030-32656-2_13. [DOI] [PubMed] [Google Scholar]
- 129.Ishak CA, et al. An RB-EZH2 complex mediates silencing of repetitive DNA sequences. Mol. Cell. 2016;64:1074–1087. doi: 10.1016/j.molcel.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kareta MS, et al. Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell. 2015;16:39–50. doi: 10.1016/j.stem.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Blais A, Dynlacht BD. E2F-associated chromatin modifiers and cell cycle control. Curr. Opin. Cell Biol. 2007;19:658–662. doi: 10.1016/j.ceb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 2012;13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- 133.Alabert C, et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015;29:585–590. doi: 10.1101/gad.256354.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Calo E, et al. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010;466:1110–1114. doi: 10.1038/nature09264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Velez-Cruz R, et al. RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Genes Dev. 2016;30:2500–2512. doi: 10.1101/gad.288282.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Coschi CH, et al. Haploinsufficiency of an RB-E2F1-Condensin II complex leads to aberrant replication and aneuploidy. Cancer Discov. 2014;4:840–853. doi: 10.1158/2159-8290.CD-14-0215. [DOI] [PubMed] [Google Scholar]
- 137.Munro S, Khaire N, Inche A, Carr S, La Thangue NB. Lysine methylation regulates the pRb tumour suppressor protein. Oncogene. 2010;29:2357–2367. doi: 10.1038/onc.2009.511. [DOI] [PubMed] [Google Scholar]
- 138.Saddic LA, et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem. 2010;285:37733–37740. doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen J, et al. E2F1 promotes the recruitment of DNA repair factors to sites of DNA double-strand breaks. Cell Cycle. 2011;10:1287–1294. doi: 10.4161/cc.10.8.15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ianari A, et al. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell. 2009;15:184–194. doi: 10.1016/j.ccr.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carnevale J, Palander O, Seifried LA, Dick FA. DNA damage signals through differentially modified E2F1 molecules to induce apoptosis. Mol. Cell Biol. 2012;32:900–912. doi: 10.1128/MCB.06286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Loffreda A, et al. Live-cell p53 single-molecule binding is modulated by C-terminal acetylation and correlates with transcriptional activity. Nat. Commun. 2017;8:313. doi: 10.1038/s41467-017-00398-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang M, et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019;26:2329–2343. doi: 10.1038/s41418-019-0304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mao C, et al. A G3BP1-interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res. 2018;78:3484–3496. doi: 10.1158/0008-5472.CAN-17-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu S, et al. As a novel p53 direct target, bidirectional gene HspB2/alphaB-crystallin regulates the ROS level and Warburg effect. Biochim. Biophys. Acta. 2014;1839:592–603. doi: 10.1016/j.bbagrm.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 146.Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 147.Tavana O, Gu W. Modulation of the p53/MDM2 interplay by HAUSP inhibitors. J. Mol. Cell Biol. 2017;9:45–52. doi: 10.1093/jmcb/mjw049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fukuda T, et al. CACUL1/CAC1 attenuates p53 activity through PML post-translational modification. Biochem. Biophys. Res. Commun. 2017;482:863–869. doi: 10.1016/j.bbrc.2016.11.125. [DOI] [PubMed] [Google Scholar]
- 149.Rodriguez J, et al. PHD3 regulates p53 protein stability by hydroxylating proline 359. Cell Rep. 2018;24:1316–1329. doi: 10.1016/j.celrep.2018.06.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kwon SK, Saindane M, Baek KH. p53 stability is regulated by diverse deubiquitinating enzymes. Biochim. Biophys. Acta. 2017;1868:404–411. doi: 10.1016/j.bbcan.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 151.Chen L, et al. DNA methylation modifier LSH inhibits p53 ubiquitination and transactivates p53 to promote lipid metabolism. Epigenet. Chromatin. 2019;12:59. doi: 10.1186/s13072-019-0302-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Meek DW, Anderson CW. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 2009;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Levine AJ, Berger SL. The interplay between epigenetic changes and the p53 protein in stem cells. Genes Dev. 2017;31:1195–1201. doi: 10.1101/gad.298984.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mishra A, Brat DJ, Verma M. P53 tumor suppression network in cancer epigenetics. Methods Mol. Biol. 2015;1238:597–605. doi: 10.1007/978-1-4939-1804-1_31. [DOI] [PubMed] [Google Scholar]
- 155.Xiao D, et al. Chromatin remodeling factor LSH is upregulated by the LRP6-GSK3beta-E2F1 axis linking reversely with survival in gliomas. Theranostics. 2017;7:132–143. doi: 10.7150/thno.17032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jiang Y, He Y, Liu S, Tao Y. Chromatin remodeling factor lymphoid-specific helicase inhibits ferroptosis through lipid metabolic genes in lung cancer progression. Chin. J. Cancer. 2017;36:82. doi: 10.1186/s40880-017-0248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.He X, et al. Chromatin remodeling factor LSH drives cancer progression by suppressing the activity of fumarate hydratase. Cancer Res. 2016;76:5743–5755. doi: 10.1158/0008-5472.CAN-16-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol. Med. 2010;16:528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chao CC. Mechanisms of p53 degradation. Clin. Chim. Acta. 2015;438:139–147. doi: 10.1016/j.cca.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 160.Zhang J, Biggar KK, Storey KB. Regulation of p53 by reversible post-transcriptional and post-translational mechanisms in liver and skeletal muscle of an anoxia tolerant turtle, Trachemys scripta elegans. Gene. 2013;513:147–155. doi: 10.1016/j.gene.2012.10.049. [DOI] [PubMed] [Google Scholar]
- 161.Lezina L, et al. CD40L/IL-4-stimulated CLL demonstrates variation in translational regulation of DNA damage response genes including ATM. Blood Adv. 2018;2:1869–1881. doi: 10.1182/bloodadvances.2017015560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Loughery J, Cox M, Smith LM, Meek DW. Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res. 2014;42:7666–7680. doi: 10.1093/nar/gku501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rao F, et al. Inositol pyrophosphates mediate the DNA-PK/ATM-p53 cell death pathway by regulating CK2 phosphorylation of Tti1/Tel2. Mol. Cell. 2014;54:119–132. doi: 10.1016/j.molcel.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sakaguchi K, et al. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dahl ES, Aird KM. Ataxia-telangiectasia mutated modulation of carbon metabolism in cancer. Front. Oncol. 2017;7:291. doi: 10.3389/fonc.2017.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 167.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 168.Jones RG, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 169.Yang T, et al. Phosphorylation of p53 serine 15 is a predictor of survival for patients with hepatocellular carcinoma. Can. J. Gastroenterol. Hepatol. 2019;2019:9015453. doi: 10.1155/2019/9015453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc. Natl Acad. Sci. USA. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer. 2016;16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 172.Liebl MC, Hofmann TG. Cell fate regulation upon DNA damage: p53 serine 46 kinases pave the cell death road. Bioessays. 2019;41:e1900127. doi: 10.1002/bies.201900127. [DOI] [PubMed] [Google Scholar]
- 173.Yoshida S, Yoshida K. Multiple functions of DYRK2 in cancer and tissue development. FEBS Lett. 2019;593:2953–2965. doi: 10.1002/1873-3468.13601. [DOI] [PubMed] [Google Scholar]
- 174.Avantaggiati ML, et al. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 175.Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 176.Trigiante G, Lu X. ASPP [corrected] and cancer. Nat. Rev. Cancer. 2006;6:217–226. doi: 10.1038/nrc1818. [DOI] [PubMed] [Google Scholar]
- 177.Gillotin S, Lu X. The ASPP proteins complex and cooperate with p300 to modulate the transcriptional activity of p53. FEBS Lett. 2011;585:1778–1782. doi: 10.1016/j.febslet.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 178.Higashimoto Y, et al. Human p53 is phosphorylated on serines 6 and 9 in response to DNA damage-inducing agents. J. Biol. Chem. 2000;275:23199–23203. doi: 10.1074/jbc.M002674200. [DOI] [PubMed] [Google Scholar]
- 179.Adorno M, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 180.Ghosh R, et al. Tumor suppressor p53-mediated structural reorganization of the transcriptional coactivator p300. Biochemistry. 2019;58:3434–3443. doi: 10.1021/acs.biochem.9b00333. [DOI] [PubMed] [Google Scholar]
- 181.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 182.Chiarugi V, Cinelli M, Magnelli L. Acetylation and phosphorylation of the carboxy-terminal domain of p53: regulative significance. Oncol. Res. 1998;10:55–57. [PubMed] [Google Scholar]
- 183.Wang Y, et al. The role of acetylation sites in the regulation of p53 activity. Mol. Biol. Rep. 2020;47:381–391. doi: 10.1007/s11033-019-05141-7. [DOI] [PubMed] [Google Scholar]
- 184.Liang L, et al. A designed peptide targets two types of modifications of p53 with anti-cancer activity. Cell Chem. Biol. 2018;25:761–774 e765. doi: 10.1016/j.chembiol.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 185.Wang B, et al. A dual role of miR-22 modulated by RelA/p65 in resensitizing fulvestrant-resistant breast cancer cells to fulvestrant by targeting FOXP1 and HDAC4 and constitutive acetylation of p53 at Lys382. Oncogenesis. 2018;7:54. doi: 10.1038/s41389-018-0063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- 187.Wang D, et al. Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature. 2016;538:118–122. doi: 10.1038/nature19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang J, Shen L, Sun LQ. The regulation of radiosensitivity by p53 and its acetylation. Cancer Lett. 2015;363:108–118. doi: 10.1016/j.canlet.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 189.Lee CW, Sorensen TS, Shikama N, La Thangue NB. Functional interplay between p53 and E2F through co-activator p300. Oncogene. 1998;16:2695–2710. doi: 10.1038/sj.onc.1201818. [DOI] [PubMed] [Google Scholar]
- 190.Grossman SR, et al. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 191.Kruse JP, Gu W. SnapShot: p53 posttranslational modifications. Cell. 2008;133:930–930 e931. doi: 10.1016/j.cell.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Knights CD, et al. Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J. Cell Biol. 2006;173:533–544. doi: 10.1083/jcb.200512059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sykes SM, Stanek TJ, Frank A, Murphy ME, McMahon SB. Acetylation of the DNA binding domain regulates transcription-independent apoptosis by p53. J. Biol. Chem. 2009;284:20197–20205. doi: 10.1074/jbc.M109.026096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Juang YC, et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol. Cell. 2012;45:384–397. doi: 10.1016/j.molcel.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.He Y, et al. Nuclear localization of metabolic enzymes in immunity and metastasis. Biochim. Biophys. Acta. 2017;1868:359–371. doi: 10.1016/j.bbcan.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 196.Jansson M, et al. Arginine methylation regulates the p53 response. Nat. Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 197.Campaner S, et al. The methyltransferase Set7/9 (Setd7) is dispensable for the p53-mediated DNA damage response in vivo. Mol. Cell. 2011;43:681–688. doi: 10.1016/j.molcel.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 198.Abaev-Schneiderman E, Admoni-Elisha L, Levy D. SETD3 is a positive regulator of DNA-damage-induced apoptosis. Cell Death Dis. 2019;10:74. doi: 10.1038/s41419-019-1328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Raposo AE, Piller SC. Protein arginine methylation: an emerging regulator of the cell cycle. Cell Div. 2018;13:3. doi: 10.1186/s13008-018-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hill SY, Rompala G, Homanics GE, Zezza N. Cross-generational effects of alcohol dependence in humans on HRAS and TP53 methylation in offspring. Epigenomics. 2017;9:1189–1203. doi: 10.2217/epi-2017-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Li Y, et al. PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov. 2015;5:288–303. doi: 10.1158/2159-8290.CD-14-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Berger SL. Out of the jaws of death: PRMT5 steers p53. Nat. Cell Biol. 2008;10:1389–1390. doi: 10.1038/ncb1208-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sims RJ, III, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat. Rev. Mol. Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- 204.Huang J, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 205.Chuikov S, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 206.Shi X, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol. Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Huang J, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 208.Carr SM, Poppy Roworth A, Chan C, La Thangue NB. Post-translational control of transcription factors: methylation ranks highly. FEBS J. 2015;282:4450–4465. doi: 10.1111/febs.13524. [DOI] [PubMed] [Google Scholar]
- 209.Stommel JM, et al. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zhang Y, Xiong Y. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science. 2001;292:1910–1915. doi: 10.1126/science.1058637. [DOI] [PubMed] [Google Scholar]
- 211.Gostissa M, et al. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Rodriguez MS, et al. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chen L, Chen J. MDM2-ARF complex regulates p53 sumoylation. Oncogene. 2003;22:5348–5357. doi: 10.1038/sj.onc.1206851. [DOI] [PubMed] [Google Scholar]
- 214.Garner E, Raj K. Protective mechanisms of p53-p21-pRb proteins against DNA damage-induced cell death. Cell Cycle. 2008;7:277–282. doi: 10.4161/cc.7.3.5328. [DOI] [PubMed] [Google Scholar]
- 215.Kung CP, Khaku S, Jennis M, Zhou Y, Murphy ME. Identification of TRIML2, a novel p53 target, that enhances p53 SUMOylation and regulates the transactivation of proapoptotic genes. Mol. Cancer Res. 2015;13:250–262. doi: 10.1158/1541-7786.MCR-14-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Stehmeier P, Muller S. Regulation of p53 family members by the ubiquitin-like SUMO system. DNA Repair. 2009;8:491–498. doi: 10.1016/j.dnarep.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 217.Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 218.Naidu SR, Lakhter AJ, Androphy EJ. PIASy-mediated Tip60 sumoylation regulates p53-induced autophagy. Cell Cycle. 2012;11:2717–2728. doi: 10.4161/cc.21091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Santiago A, Li D, Zhao LY, Godsey A, Liao D. p53 SUMOylation promotes its nuclear export by facilitating its release from the nuclear export receptor CRM1. Mol. Biol. Cell. 2013;24:2739–2752. doi: 10.1091/mbc.E12-10-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Mihara M, et al. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 221.Heo KS, Berk BC, Abe J. Disturbed flow-induced endothelial proatherogenic signaling via regulating post-translational modifications and epigenetic events. Antioxid. Redox Signal. 2016;25:435–450. doi: 10.1089/ars.2015.6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ashikari D, et al. Androgen induces G3BP2 and SUMO-mediated p53 nuclear export in prostate cancer. Oncogene. 2017;36:6272–6281. doi: 10.1038/onc.2017.225. [DOI] [PubMed] [Google Scholar]
- 223.Goldstein G, et al. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl Acad. Sci. USA. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Caldeira MV, Salazar IL, Curcio M, Canzoniero LM, Duarte CB. Role of the ubiquitin-proteasome system in brain ischemia: friend or foe? Prog. Neurobiol. 2014;112:50–69. doi: 10.1016/j.pneurobio.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 225.Bulatov, E., Valiullina, A., Sayarova, R. & Rizvanov, A. Promising new therapeutic targets for regulation of inflammation and immunity: RING-type E3 ubiquitin ligases. Immunol. Lett. 10.1016/j.imlet.2018.08.001 (2018). [DOI] [PubMed]
- 226.Bozi LHM, Campos JC. Targeting the ubiquitin proteasome system in diabetic cardiomyopathy. J. Mol. Cell Cardiol. 2017;109:61–63. doi: 10.1016/j.yjmcc.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 227.Mendes, M. L., Fougeras, M. R. & Dittmar, G. Analysis of ubiquitin signaling and chain topology cross-talk. J. Proteomics10.1016/j.jprot.2020.103634 (2020). [DOI] [PubMed]
- 228.Kaiser SE, et al. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat. Methods. 2011;8:691–696. doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wagner SA, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell Proteom. 2011;10:M111 013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ziv I, et al. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol. Cell Proteom. 2011;10:M111 009753. doi: 10.1074/mcp.M111.009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Thomson, S. M., Pulido, P. & Jarvis, R. P. Protein import into chloroplasts and its regulation by the ubiquitin-proteasome system. Biochem. Soc. Trans. 10.1042/BST20190274 (2020). [DOI] [PMC free article] [PubMed]
- 234.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 235.Bang, S., Kaur, S. & Kurokawa, M. Regulation of the p53 family proteins by the ubiquitin proteasomal pathway. Int. J. Mol. Sci.10.3390/ijms21010261 (2019). [DOI] [PMC free article] [PubMed]
- 236.Ribet D, Cossart P. Ubiquitin, SUMO, and NEDD8: key targets of bacterial pathogens. Trends Cell Biol. 2018;28:926–940. doi: 10.1016/j.tcb.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Akutsu M, Dikic I, Bremm A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016;129:875–880. doi: 10.1242/jcs.183954. [DOI] [PubMed] [Google Scholar]
- 238.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mevissen TET, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 240.You J, Pickart CM. A HECT domain E3 enzyme assembles novel polyubiquitin chains. J. Biol. Chem. 2001;276:19871–19878. doi: 10.1074/jbc.M100034200. [DOI] [PubMed] [Google Scholar]
- 241.Kristariyanto YA, et al. K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin. Mol. Cell. 2015;58:83–94. doi: 10.1016/j.molcel.2015.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Michel MA, et al. Assembly and specific recognition of k29- and k33-linked polyubiquitin. Mol. Cell. 2015;58:95–109. doi: 10.1016/j.molcel.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.van Wijk SJ, et al. Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol. Cell. 2012;47:797–809. doi: 10.1016/j.molcel.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Tran H, Hamada F, Schwarz-Romond T, Bienz M. Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev. 2008;22:528–542. doi: 10.1101/gad.463208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Emmerich CH, et al. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl Acad. Sci. USA. 2013;110:15247–15252. doi: 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Laplantine E, et al. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 2009;28:2885–2895. doi: 10.1038/emboj.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Miranda M, Sorkin A. Regulation of receptors and transporters by ubiquitination: new insights into surprisingly similar mechanisms. Mol. Interv. 2007;7:157–167. doi: 10.1124/mi.7.3.7. [DOI] [PubMed] [Google Scholar]
- 248.Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 250.Yau RG, et al. Assembly and function of heterotypic ubiquitin chains in cell-cycle and protein quality control. Cell. 2017;171:918–933. doi: 10.1016/j.cell.2017.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Finlay CA. The mdm-2 oncogene can overcome wild-type p53 suppression of transformed cell growth. Mol. Cell Biol. 1993;13:301–306. doi: 10.1128/mcb.13.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Cao Z, et al. MDM2 promotes genome instability by ubiquitinating the transcription factor HBP1. Oncogene. 2019;38:4835–4855. doi: 10.1038/s41388-019-0761-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 254.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 255.Todoric J, et al. Stress-activated NRF2-MDM2 cascade controls neoplastic progression in pancreas. Cancer Cell. 2017;32:824–839. doi: 10.1016/j.ccell.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Yang, L., Song, T., Cheng, Q., Chen, L. & Chen, J. Mutant p53 sequestration of the MDM2 acidic domain inhibits E3 ligase activity. Mol. Cell Biol.10.1128/MCB.00375-18 (2019). [DOI] [PMC free article] [PubMed]
- 257.Tian H, Tackmann NR, Jin A, Zheng J, Zhang Y. Inactivation of the MDM2 RING domain enhances p53 transcriptional activity in mice. J. Biol. Chem. 2017;292:21614–21622. doi: 10.1074/jbc.RA117.000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ranaweera RS, Yang X. Auto-ubiquitination of Mdm2 enhances its substrate ubiquitin ligase activity. J. Biol. Chem. 2013;288:18939–18946. doi: 10.1074/jbc.M113.454470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wei S, et al. Lenalidomide promotes p53 degradation by inhibiting MDM2 auto-ubiquitination in myelodysplastic syndrome with chromosome 5q deletion. Oncogene. 2013;32:1110–1120. doi: 10.1038/onc.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ito A, et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20:1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Spiegelberg D, et al. The MDM2/MDMX-p53 antagonist PM2 radiosensitizes wild-type p53 tumors. Cancer Res. 2018;78:5084–5093. doi: 10.1158/0008-5472.CAN-18-0440. [DOI] [PubMed] [Google Scholar]
- 264.Zhu D, et al. BAI1 suppresses medulloblastoma formation by protecting p53 from Mdm2-mediated degradation. Cancer Cell. 2018;33:1004–1016 e1005. doi: 10.1016/j.ccell.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Oliner JD, et al. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 266.Li M, et al. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 267.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 268.Maya R, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol. Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ashcroft M, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell Biol. 2000;20:3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J. Cell Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 272.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat. Rev. Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 273.Wang, S., Zhao, Y., Aguilar, A., Bernard, D. & Yang, C. Y. Targeting the MDM2-p53 protein–protein interaction for new cancer Therapy: progress and challenges. Cold. Spring Harb. Perspect. Med.10.1101/cshperspect.a026245 (2017). [DOI] [PMC free article] [PubMed]
- 274.Capoulade C, et al. Overexpression of MDM2, due to enhanced translation, results in inactivation of wild-type p53 in Burkitt’s lymphoma cells. Oncogene. 1998;16:1603–1610. doi: 10.1038/sj.onc.1201702. [DOI] [PubMed] [Google Scholar]
- 275.Momand J, Wu HH, Dasgupta G. MDM2–master regulator of the p53 tumor suppressor protein. Gene. 2000;242:15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 276.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 277.Nayak SK, Khatik GL, Narang R, Monga V, Chopra HK. p53-Mdm2 interaction inhibitors as novel nongenotoxic anticancer agents. Curr. Cancer Drug Targets. 2018;18:749–772. doi: 10.2174/1568009617666170623111953. [DOI] [PubMed] [Google Scholar]
- 278.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 279.Yang G, Gong Y, Wang Q, Wang L, Zhang X. miR-100 antagonism triggers apoptosis by inhibiting ubiquitination-mediated p53 degradation. Oncogene. 2017;36:1023–1037. doi: 10.1038/onc.2016.270. [DOI] [PubMed] [Google Scholar]
- 280.Leng RP, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 281.Prives C, Hall PA. The p53 pathway. J. Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 282.Jung YS, Qian Y, Chen X. The p73 tumor suppressor is targeted by Pirh2 RING finger E3 ubiquitin ligase for the proteasome-dependent degradation. J. Biol. Chem. 2011;286:35388–35395. doi: 10.1074/jbc.M111.261537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Shloush J, et al. Structural and functional comparison of the RING domains of two p53 E3 ligases, Mdm2 and Pirh2. J. Biol. Chem. 2011;286:4796–4808. doi: 10.1074/jbc.M110.157669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Dornan D, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 285.Ka WH, Cho SK, Chun BN, Byun SY, Ahn JC. The ubiquitin ligase COP1 regulates cell cycle and apoptosis by affecting p53 function in human breast cancer cell lines. Breast Cancer. 2018;25:529–538. doi: 10.1007/s12282-018-0849-5. [DOI] [PubMed] [Google Scholar]
- 286.Moscetti I, Bizzarri AR, Cannistraro S. Imaging and kinetics of the bimolecular complex formed by the tumor suppressor p53 with ubiquitin ligase COP1 as studied by atomic force microscopy and surface plasmon resonance. Int. J. Nanomed. 2018;13:251–259. doi: 10.2147/IJN.S152214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Zou S, et al. The ubiquitin ligase COP1 promotes glioma cell proliferation by preferentially downregulating tumor suppressor p53. Mol. Neurobiol. 2017;54:5008–5016. doi: 10.1007/s12035-016-0033-x. [DOI] [PubMed] [Google Scholar]
- 288.Dornan D, et al. ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science. 2006;313:1122–1126. doi: 10.1126/science.1127335. [DOI] [PubMed] [Google Scholar]
- 289.Chen D, et al. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 290.Canfield K, et al. Inverse association between MDM2 and HUWE1 protein expression levels in human breast cancer and liposarcoma. Int. J. Clin. Exp. Pathol. 2016;9:6342–6349. [PMC free article] [PubMed] [Google Scholar]
- 291.Wei J, et al. Bacterial CagA protein induces degradation of p53 protein in a p14ARF-dependent manner. Gut. 2015;64:1040–1048. doi: 10.1136/gutjnl-2014-307295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Wang L, et al. Dichotomous role of pancreatic HUWE1/MULE/ARF-BP1 in modulating beta cell apoptosis in mice under physiological and genotoxic conditions. Diabetologia. 2014;57:1889–1898. doi: 10.1007/s00125-014-3295-8. [DOI] [PubMed] [Google Scholar]
- 293.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 295.Allton K, et al. Trim24 targets endogenous p53 for degradation. Proc. Natl Acad. Sci. USA. 2009;106:11612–11616. doi: 10.1073/pnas.0813177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Jain AK, Allton K, Duncan AD, Barton MC. TRIM24 is a p53-induced E3-ubiquitin ligase that undergoes ATM-mediated phosphorylation and autodegradation during DNA damage. Mol. Cell Biol. 2014;34:2695–2709. doi: 10.1128/MCB.01705-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Jain AK, Barton MC. Regulation of p53: TRIM24 enters the RING. Cell Cycle. 2009;8:3668–3674. doi: 10.4161/cc.8.22.9979. [DOI] [PubMed] [Google Scholar]
- 298.Yagishita N, Yamasaki S, Nishioka K, Nakajima T. Synoviolin, protein folding and the maintenance of joint homeostasis. Nat. Clin. Pract. Rheumatol. 2008;4:91–97. doi: 10.1038/ncprheum0699. [DOI] [PubMed] [Google Scholar]
- 299.Wu ZZ, Sun NK, Chien KY, Chao CC. Silencing of the SNARE protein NAPA sensitizes cancer cells to cisplatin by inducing ERK1/2 signaling, synoviolin ubiquitination and p53 accumulation. Biochem. Pharm. 2011;82:1630–1640. doi: 10.1016/j.bcp.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 300.Yamasaki S, et al. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ‘Synoviolin’. EMBO J. 2007;26:113–122. doi: 10.1038/sj.emboj.7601490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Lin L, et al. topors, a p53 and topoisomerase I-binding RING finger protein, is a coactivator of p53 in growth suppression induced by DNA damage. Oncogene. 2005;24:3385–3396. doi: 10.1038/sj.onc.1208554. [DOI] [PubMed] [Google Scholar]
- 302.Haluska P, Jr., et al. Interaction between human topoisomerase I and a novel RING finger/arginine-serine protein. Nucleic Acids Res. 1999;27:2538–2544. doi: 10.1093/nar/27.12.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Yang X, et al. Plk1-mediated phosphorylation of Topors regulates p53 stability. J. Biol. Chem. 2009;284:18588–18592. doi: 10.1074/jbc.C109.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.McDonald ER, III, El-Deiry WS. Suppression of caspase-8- and -10-associated RING proteins results in sensitization to death ligands and inhibition of tumor cell growth. Proc. Natl Acad. Sci. USA. 2004;101:6170–6175. doi: 10.1073/pnas.0307459101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Yang W, et al. CARPs are ubiquitin ligases that promote MDM2-independent p53 and phospho-p53ser20 degradation. J. Biol. Chem. 2007;282:3273–3281. doi: 10.1074/jbc.M610793200. [DOI] [PubMed] [Google Scholar]
- 306.Yang W, Dicker DT, Chen J, El-Deiry WS. CARPs enhance p53 turnover by degrading 14-3-3sigma and stabilizing MDM2. Cell Cycle. 2008;7:670–682. doi: 10.4161/cc.7.5.5701. [DOI] [PubMed] [Google Scholar]
- 307.Koegl M, et al. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 308.Wu H, et al. UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat. Med. 2011;17:347–355. doi: 10.1038/nm.2283. [DOI] [PubMed] [Google Scholar]
- 309.Dutto I, Scalera C, Prosperi E. CREBBP and p300 lysine acetyl transferases in the DNA damage response. Cell Mol. Life Sci. 2018;75:1325–1338. doi: 10.1007/s00018-017-2717-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Schuldner M, et al. Exosome-dependent immune surveillance at the metastatic niche requires BAG6 and CBP/p300-dependent acetylation of p53. Theranostics. 2019;9:6047–6062. doi: 10.7150/thno.36378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Shi D, et al. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc. Natl Acad. Sci. USA. 2009;106:16275–16280. doi: 10.1073/pnas.0904305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Lundblad JR, Kwok RP, Laurance ME, Harter ML, Goodman RH. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 313.Le Cam L, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127:775–788. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 314.Sandy P, et al. p53 is involved in the p120E4F-mediated growth arrest. Oncogene. 2000;19:188–199. doi: 10.1038/sj.onc.1203250. [DOI] [PubMed] [Google Scholar]
- 315.Laine A, et al. Regulation of p53 localization and activity by Ubc13. Mol. Cell Biol. 2006;26:8901–8913. doi: 10.1128/MCB.01156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Topisirovic I, et al. Control of p53 multimerization by Ubc13 is JNK-regulated. Proc. Natl Acad. Sci. USA. 2009;106:12676–12681. doi: 10.1073/pnas.0900596106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Oh E, Akopian D, Rape M. Principles of ubiquitin-dependent signaling. Annu. Rev. Cell Dev. Biol. 2018;34:137–162. doi: 10.1146/annurev-cellbio-100617-062802. [DOI] [PubMed] [Google Scholar]
- 318.Sharma A, et al. USP14 regulates DNA damage repair by targeting RNF168-dependent ubiquitination. Autophagy. 2018;14:1–15. doi: 10.1080/15548627.2018.1496877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Tu R, et al. USP49 participates in the DNA damage response by forming a positive feedback loop with p53. Cell Death Dis. 2018;9:553. doi: 10.1038/s41419-018-0475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Wang, W. et al. Targeting MDM2 for novel molecular therapy: Beyond oncology. Med. Res. Rev. 10.1002/med.21637 (2019). [DOI] [PMC free article] [PubMed]
- 321.Jovanovic KK, et al. Deregulation and targeting of TP53 pathway in multiple myeloma. Front. Oncol. 2018;8:665. doi: 10.3389/fonc.2018.00665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Li M, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 323.Liu X, et al. Trip12 is an E3 ubiquitin ligase for USP7/HAUSP involved in the DNA damage response. FEBS Lett. 2016;590:4213–4222. doi: 10.1002/1873-3468.12471. [DOI] [PubMed] [Google Scholar]
- 324.Hu M, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 325.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 326.Kon N, et al. Roles of HAUSP-mediated p53 regulation in central nervous system development. Cell Death Differ. 2011;18:1366–1375. doi: 10.1038/cdd.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Meulmeester E, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol. Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 328.Ko A, et al. Oncogene-induced senescence mediated by c-Myc requires USP10 dependent deubiquitination and stabilization of p14ARF. Cell Death Differ. 2018;25:1050–1062. doi: 10.1038/s41418-018-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Jochemsen AG, Shiloh Y. USP10: friend and foe. Cell. 2010;140:308–310. doi: 10.1016/j.cell.2010.01.034. [DOI] [PubMed] [Google Scholar]
- 330.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Mennerich D, Kubaichuk K, Kietzmann T. DUBs, hypoxia, and cancer. Trends Cancer. 2019;5:632–653. doi: 10.1016/j.trecan.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 332.Sun XX, Challagundla KB, Dai MS. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 2012;31:576–592. doi: 10.1038/emboj.2011.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Sun XX, Dai MS. Deubiquitinating enzyme regulation of the p53 pathway: a lesson from Otub1. World J. Biol. Chem. 2014;5:75–84. doi: 10.4331/wjbc.v5.i2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Li Y, et al. Monoubiquitination is critical for ovarian tumor domain-containing ubiquitin aldehyde binding protein 1 (Otub1) to suppress UbcH5 enzyme and stabilize p53 protein. J. Biol. Chem. 2014;289:5097–5108. doi: 10.1074/jbc.M113.533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Wiener R, Zhang X, Wang T, Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature. 2012;483:618–622. doi: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Nakada S, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010;466:941–946. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 337.Tong X, Buelow K, Guha A, Rausch R, Yin L. USP2a protein deubiquitinates and stabilizes the circadian protein CRY1 in response to inflammatory signals. J. Biol. Chem. 2012;287:25280–25291. doi: 10.1074/jbc.M112.340786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Stevenson LF, et al. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26:976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Zhang L, et al. The deubiquitinating enzyme USP24 is a regulator of the UV damage response. Cell Rep. 2015;10:140–147. doi: 10.1016/j.celrep.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Zhang L, Gong F. Involvement of USP24 in the DNA damage response. Mol. Cell Oncol. 2016;3:e1011888. doi: 10.1080/23723556.2015.1011888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Liu J, et al. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 2011;30:846–858. doi: 10.1038/emboj.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Huang JM, Kim J. DNA methylation analysis of the mammalian PEG3 imprinted domain. Gene. 2009;442:18–25. doi: 10.1016/j.gene.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Glinsky GV. Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle. 2006;5:1208–1216. doi: 10.4161/cc.5.11.2796. [DOI] [PubMed] [Google Scholar]
- 344.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Lin Z, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell. 2012;46:484–494. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 346.Haq, S. & Ramakrishna, S. Deubiquitylation of deubiquitylases. Open Biol. 10.1098/rsob.170016 (2017). [DOI] [PMC free article] [PubMed]
- 347.Gu B, Zhu WG. Surf the post-translational modification network of p53 regulation. Int. J. Biol. Sci. 2012;8:672–684. doi: 10.7150/ijbs.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Olsson A, Manzl C, Strasser A, Villunger A. How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ. 2007;14:1561–1575. doi: 10.1038/sj.cdd.4402196. [DOI] [PubMed] [Google Scholar]
- 349.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 350.Meek DW. Regulation of the p53 response and its relationship to cancer. Biochem. J. 2015;469:325–346. doi: 10.1042/BJ20150517. [DOI] [PubMed] [Google Scholar]
- 351.Baugh EH, Ke H, Levine AJ, Bonneau RA, Chan CS. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018;25:154–160. doi: 10.1038/cdd.2017.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Sabapathy K, Lane DP. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol. 2018;15:13–30. doi: 10.1038/nrclinonc.2017.151. [DOI] [PubMed] [Google Scholar]
- 353.Armata HL, Garlick DS, Sluss HK. The ataxia telangiectasia-mutated target site Ser18 is required for p53-mediated tumor suppression. Cancer Res. 2007;67:11696–11703. doi: 10.1158/0008-5472.CAN-07-1610. [DOI] [PubMed] [Google Scholar]
- 354.Garcia PB, Attardi LD. Illuminating p53 function in cancer with genetically engineered mouse models. Semin. Cell Dev. Biol. 2014;27:74–85. doi: 10.1016/j.semcdb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.MacPherson D, et al. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J. 2004;23:3689–3699. doi: 10.1038/sj.emboj.7600363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Feng L, Hollstein M, Xu Y. Ser46 phosphorylation regulates p53-dependent apoptosis and replicative senescence. Cell Cycle. 2006;5:2812–2819. doi: 10.4161/cc.5.23.3526. [DOI] [PubMed] [Google Scholar]
- 357.Kenzelmann Broz D, Attardi LD. In vivo analysis of p53 tumor suppressor function using genetically engineered mouse models. Carcinogenesis. 2010;31:1311–1318. doi: 10.1093/carcin/bgp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Bruins W, et al. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol. Cell Biol. 2004;24:8884–8894. doi: 10.1128/MCB.24.20.8884-8894.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Lin T, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 360.Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc. Natl Acad. Sci. USA. 2005;102:10188–10193. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Chao C, et al. Acetylation of mouse p53 at lysine 317 negatively regulates p53 apoptotic activities after DNA damage. Mol. Cell Biol. 2006;26:6859–6869. doi: 10.1128/MCB.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Zhao J, et al. p53 Mutant p53(N236S) induces neural tube defects in female embryos. Int. J. Biol. Sci. 2019;15:2006–2015. doi: 10.7150/ijbs.31451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Hamard PJ, et al. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev. 2013;27:1868–1885. doi: 10.1101/gad.224386.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Lee MK, Tong WM, Wang ZQ, Sabapathy K. Serine 312 phosphorylation is dispensable for wild-type p53 functions in vivo. Cell Death Differ. 2011;18:214–221. doi: 10.1038/cdd.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 366.Bossi G, et al. Conditional RNA interference in vivo to study mutant p53 oncogenic gain of function on tumor malignancy. Cell Cycle. 2008;7:1870–1879. doi: 10.4161/cc.7.12.6161. [DOI] [PubMed] [Google Scholar]
- 367.Demma M, et al. SCH529074, a small molecule activator of mutant p53, which binds p53 DNA binding domain (DBD), restores growth-suppressive function to mutant p53 and interrupts HDM2-mediated ubiquitination of wild type p53. J. Biol. Chem. 2010;285:10198–10212. doi: 10.1074/jbc.M109.083469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Bykov VJ, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 369.Foster BA, Coffey HA, Morin MJ, Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286:2507–2510. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- 370.Loh SN. The missing zinc: p53 misfolding and cancer. Metallomics. 2010;2:442–449. doi: 10.1039/c003915b. [DOI] [PubMed] [Google Scholar]
- 371.Butler JS, Loh SN. Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry. 2003;42:2396–2403. doi: 10.1021/bi026635n. [DOI] [PubMed] [Google Scholar]
- 372.Puca R, et al. Restoring wtp53 activity in HIPK2 depleted MCF7 cells by modulating metallothionein and zinc. Exp. Cell Res. 2009;315:67–75. doi: 10.1016/j.yexcr.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 373.Pintus SS, et al. The substitutions G245C and G245D in the Zn(2+)-binding pocket of the p53 protein result in differences of conformational flexibility of the DNA-binding domain. J. Biomol. Struct. Dyn. 2013;31:78–86. doi: 10.1080/07391102.2012.691364. [DOI] [PubMed] [Google Scholar]
- 374.Puca R, et al. Restoring p53 active conformation by zinc increases the response of mutant p53 tumor cells to anticancer drugs. Cell Cycle. 2011;10:1679–1689. doi: 10.4161/cc.10.10.15642. [DOI] [PubMed] [Google Scholar]
- 375.Yu X, Vazquez A, Levine AJ, Carpizo DR. Allele-specific p53 mutant reactivation. Cancer Cell. 2012;21:614–625. doi: 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Kravchenko JE, et al. Small-molecule RETRA suppresses mutant p53-bearing cancer cells through a p73-dependent salvage pathway. Proc. Natl Acad. Sci. USA. 2008;105:6302–6307. doi: 10.1073/pnas.0802091105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Hillmann, P. & Fabbro, D. PI3K/mTOR pathway inhibition: opportunities in oncology and rare genetic diseases. Int. J. Mol. Sci.10.3390/ijms20225792 (2019). [DOI] [PMC free article] [PubMed]
- 378.Salvatore, L. et al. PTEN in colorectal cancer: shedding light on its role as predictor and target. Cancers10.3390/cancers11111765 (2019). [DOI] [PMC free article] [PubMed]
- 379.Chen CY, Chen J, He L, Stiles BL. PTEN: tumor suppressor and metabolic regulator. Front. Endocrinol. 2018;9:338. doi: 10.3389/fendo.2018.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Leslie NR, Kriplani N, Hermida MA, Alvarez-Garcia V, Wise HM. The PTEN protein: cellular localization and post-translational regulation. Biochem. Soc. Trans. 2016;44:273–278. doi: 10.1042/BST20150224. [DOI] [PubMed] [Google Scholar]
- 381.Allison SJ. Ubiquitylation of PTEN drives fibrosis in diabetic kidney disease. Nat. Rev. Nephrol. 2019;15:254. doi: 10.1038/s41581-019-0130-y. [DOI] [PubMed] [Google Scholar]
- 382.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 383.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 384.Tamguney T, Stokoe D. New insights into PTEN. J. Cell Sci. 2007;120:4071–4079. doi: 10.1242/jcs.015230. [DOI] [PubMed] [Google Scholar]
- 385.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 386.Ho J, Bassi C, Stambolic V. Characterization of nuclear PTEN and its post translational modifications. Methods. 2015;77-78:104–111. doi: 10.1016/j.ymeth.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 387.Blanco-Aparicio C, Renner O, Leal JF, Carnero A. PTEN, more than the AKT pathway. Carcinogenesis. 2007;28:1379–1386. doi: 10.1093/carcin/bgm052. [DOI] [PubMed] [Google Scholar]
- 388.Xu W, Yang Z, Zhou SF, Lu N. Posttranslational regulation of phosphatase and tensin homolog (PTEN) and its functional impact on cancer behaviors. Drug Des. Dev. Ther. 2014;8:1745–1751. doi: 10.2147/DDDT.S71061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Milella M, et al. PTEN: multiple functions in human malignant tumors. Front. Oncol. 2015;5:24. doi: 10.3389/fonc.2015.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Rahdar M, et al. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc. Natl Acad. Sci. USA. 2009;106:480–485. doi: 10.1073/pnas.0811212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Chen Z, et al. Molecular features of phosphatase and tensin homolog (PTEN) regulation by C-terminal phosphorylation. J. Biol. Chem. 2016;291:14160–14169. doi: 10.1074/jbc.M116.728980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Ma J, et al. Inhibition of nuclear PTEN tyrosine phosphorylation enhances glioma radiation sensitivity through attenuated DNA repair. Cancer Cell. 2019;35:504–518 e507. doi: 10.1016/j.ccell.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Yan Y, et al. FGFR2-mediated phosphorylation of PTEN at tyrosine 240 contributes to the radioresistance of glioma. J. Cell Commun. Signal. 2019;13:279–280. doi: 10.1007/s12079-019-00518-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Gupta A, et al. PARK2 depletion connects energy and oxidative stress to PI3K/Akt activation via PTEN S-nitrosylation. Mol. Cell. 2017;65:999–1013 e1017. doi: 10.1016/j.molcel.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Miller SJ, Lou DY, Seldin DC, Lane WS, Neel BG. Direct identification of PTEN phosphorylation sites. FEBS Lett. 2002;528:145–153. doi: 10.1016/s0014-5793(02)03274-x. [DOI] [PubMed] [Google Scholar]
- 396.Al-Khouri AM, Ma Y, Togo SH, Williams S, Mustelin T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. J. Biol. Chem. 2005;280:35195–35202. doi: 10.1074/jbc.M503045200. [DOI] [PubMed] [Google Scholar]
- 397.Torres J, Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J. Biol. Chem. 2001;276:993–998. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- 398.Tolkacheva T, et al. Regulation of PTEN binding to MAGI-2 by two putative phosphorylation sites at threonine 382 and 383. Cancer Res. 2001;61:4985–4989. [PubMed] [Google Scholar]
- 399.Gomes AM, et al. Adult B-cell acute lymphoblastic leukemia cells display decreased PTEN activity and constitutive hyperactivation of PI3K/Akt pathway despite high PTEN protein levels. Haematologica. 2014;99:1062–1068. doi: 10.3324/haematol.2013.096438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Mehenni H, et al. LKB1 interacts with and phosphorylates PTEN: a functional link between two proteins involved in cancer predisposing syndromes. Hum. Mol. Genet. 2005;14:2209–2219. doi: 10.1093/hmg/ddi225. [DOI] [PubMed] [Google Scholar]
- 401.Song P, et al. Reactive nitrogen species induced by hyperglycemia suppresses Akt signaling and triggers apoptosis by upregulating phosphatase PTEN (phosphatase and tensin homologue deleted on chromosome 10) in an LKB1-dependent manner. Circulation. 2007;116:1585–1595. doi: 10.1161/CIRCULATIONAHA.107.716498. [DOI] [PubMed] [Google Scholar]
- 402.Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Xu C, et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell. 2014;25:590–604. doi: 10.1016/j.ccr.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Lin C, Liang Y, Zhu H, Zhang J, Zhong X. R280T mutation of p53 gene promotes proliferation of human glioma cells through GSK-3beta/PTEN pathway. Neurosci. Lett. 2012;529:60–65. doi: 10.1016/j.neulet.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 405.Vemula S, Shi J, Hanneman P, Wei L, Kapur R. ROCK1 functions as a suppressor of inflammatory cell migration by regulating PTEN phosphorylation and stability. Blood. 2010;115:1785–1796. doi: 10.1182/blood-2009-08-237222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Yim EK, et al. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell. 2009;15:304–314. doi: 10.1016/j.ccr.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Choi BH, Pagano M, Dai W. Plk1 protein phosphorylates phosphatase and tensin homolog (PTEN) and regulates its mitotic activity during the cell cycle. J. Biol. Chem. 2014;289:14066–14074. doi: 10.1074/jbc.M114.558155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Okahara F, et al. Critical role of PICT-1, a tumor suppressor candidate, in phosphatidylinositol 3,4,5-trisphosphate signals and tumorigenic transformation. Mol. Biol. Cell. 2006;17:4888–4895. doi: 10.1091/mbc.E06-04-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Wang X, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Li H, et al. WWP2 is a physiological ubiquitin ligase for phosphatase and tensin homolog (PTEN) in mice. J. Biol. Chem. 2018;293:8886–8899. doi: 10.1074/jbc.RA117.001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Chen C, Matesic LE. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 2007;26:587–604. doi: 10.1007/s10555-007-9091-x. [DOI] [PubMed] [Google Scholar]
- 412.Chen C, et al. Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene. 2007;26:2386–2394. doi: 10.1038/sj.onc.1210021. [DOI] [PubMed] [Google Scholar]
- 413.Chen C, Zhou Z, Ross JS, Zhou W, Dong JT. The amplified WWP1 gene is a potential molecular target in breast cancer. Int. J. Cancer. 2007;121:80–87. doi: 10.1002/ijc.22653. [DOI] [PubMed] [Google Scholar]
- 414.Maddika S, et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 2011;13:728–733. doi: 10.1038/ncb2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Lee, Y. R. et al. Reactivation of PTEN tumor suppressor for cancer treatment through inhibition of a MYC-WWP1 inhibitory pathway. Science10.1126/science.aau0159 (2019). [DOI] [PMC free article] [PubMed]
- 416.Wang Q, et al. Long noncoding RNA Linc02023 regulates PTEN stability and suppresses tumorigenesis of colorectal cancer in a PTEN-dependent pathway. Cancer Lett. 2019;451:68–78. doi: 10.1016/j.canlet.2019.02.041. [DOI] [PubMed] [Google Scholar]
- 417.Yehia L, Ngeow J, Eng C. PTEN-opathies: from biological insights to evidence-based precision medicine. J. Clin. Invest. 2019;129:452–464. doi: 10.1172/JCI121277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Marsh DJ, et al. Rapamycin treatment for a child with germline PTEN mutation. Nat. Clin. Pract. Oncol. 2008;5:357–361. doi: 10.1038/ncponc1112. [DOI] [PubMed] [Google Scholar]
- 419.Schmid GL, et al. Sirolimus treatment of severe PTEN hamartoma tumor syndrome: case report and in vitro studies. Pediatr. Res. 2014;75:527–534. doi: 10.1038/pr.2013.246. [DOI] [PubMed] [Google Scholar]
- 420.Zak M, Ledbetter M, Maertens P. Infantile Lhermitte-Duclos disease treated successfully with rapamycin. J. Child Neurol. 2017;32:322–326. doi: 10.1177/0883073816681340. [DOI] [PubMed] [Google Scholar]
- 421.Mester J, Charis E. PTEN hamartoma tumor syndrome. Handb. Clin. Neurol. 2015;132:129–137. doi: 10.1016/B978-0-444-62702-5.00009-3. [DOI] [PubMed] [Google Scholar]
- 422.Lindhurst MJ, et al. Repression of AKT signaling by ARQ 092 in cells and tissues from patients with Proteus syndrome. Sci. Rep. 2015;5:17162. doi: 10.1038/srep17162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Loconte DC, et al. Molecular and functional characterization of three different postzygotic mutations in PIK3CA-related overgrowth spectrum (PROS) patients: effects on PI3K/AKT/mTOR signaling and sensitivity to PIK3 inhibitors. PLoS ONE. 2015;10:e0123092. doi: 10.1371/journal.pone.0123092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Ranieri C, et al. In vitro efficacy of ARQ 092, an allosteric AKT inhibitor, on primary fibroblast cells derived from patients with PIK3CA-related overgrowth spectrum (PROS) Neurogenetics. 2018;19:77–91. doi: 10.1007/s10048-018-0540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Venot Q, et al. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature. 2018;558:540–546. doi: 10.1038/s41586-018-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27:5477–5485. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 427.Dumont FJ, Su Q. Mechanism of action of the immunosuppressant rapamycin. Life Sci. 1996;58:373–395. doi: 10.1016/0024-3205(95)02233-3. [DOI] [PubMed] [Google Scholar]
- 428.Mahalingam D, Sankhala K, Mita A, Giles FJ, Mita MM. Targeting the mTOR pathway using deforolimus in cancer therapy. Future Oncol. 2009;5:291–303. doi: 10.2217/fon.09.9. [DOI] [PubMed] [Google Scholar]
- 429.Hopkins BD, et al. A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science. 2013;341:399–402. doi: 10.1126/science.1234907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Ma H, et al. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548:413–419. doi: 10.1038/nature23305. [DOI] [PubMed] [Google Scholar]
- 431.Ricciardi AS, et al. In utero nanoparticle delivery for site-specific genome editing. Nat. Commun. 2018;9:2481. doi: 10.1038/s41467-018-04894-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Collins FS, Gottlieb S. The next phase of human gene-therapy oversight. N. Engl. J. Med. 2018;379:1393–1395. doi: 10.1056/NEJMp1810628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annu. Rev. Immunol. 2013;31:675–704. doi: 10.1146/annurev-immunol-032712-095946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Chen HH, et al. Immune dysregulation in patients with PTEN hamartoma tumor syndrome: analysis of FOXP3 regulatory T cells. J. Allergy Clin. Immunol. 2017;139:607–620 e615. doi: 10.1016/j.jaci.2016.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Cavallo F, De Giovanni C, Nanni P, Forni G, Lollini PL. 2011: the immune hallmarks of cancer. Cancer Immunol. Immunother. 2011;60:319–326. doi: 10.1007/s00262-010-0968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.George S, et al. Loss of PTEN is associated with resistance to anti-PD-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017;46:197–204. doi: 10.1016/j.immuni.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Parsa AT, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat. Med. 2007;13:84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 438.Crane CA, et al. PI(3) kinase is associated with a mechanism of immunoresistance in breast and prostate cancer. Oncogene. 2009;28:306–312. doi: 10.1038/onc.2008.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Lastwika KJ, et al. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76:227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 440.Kamihara J, et al. Retinoblastoma and neuroblastoma predisposition and surveillance. Clin. Cancer Res. 2017;23:e98–e106. doi: 10.1158/1078-0432.CCR-17-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Mendoza PR, Grossniklaus HE. The biology of retinoblastoma. Prog. Mol. Biol. Transl. Sci. 2015;134:503–516. doi: 10.1016/bs.pmbts.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 442.Xie C, et al. Co-deleting Pten with Rb in retinal progenitor cells in mice results in fully penetrant bilateral retinoblastomas. Mol. Cancer. 2015;14:93. doi: 10.1186/s12943-015-0360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 444.Liaw D, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 445.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 446.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 447.Zhou BP, et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 448.Mayo LD, Dixon JE, Durden DL, Tonks NK, Donner DB. PTEN protects p53 from Mdm2 and sensitizes cancer cells to chemotherapy. J. Biol. Chem. 2002;277:5484–5489. doi: 10.1074/jbc.M108302200. [DOI] [PubMed] [Google Scholar]
- 449.Asselin E, Mills GB, Tsang BK. XIAP regulates Akt activity and caspase-3-dependent cleavage during cisplatin-induced apoptosis in human ovarian epithelial cancer cells. Cancer Res. 2001;61:1862–1868. [PubMed] [Google Scholar]
- 450.Ghoneum, A. & Said, N. PI3K-AKT-mTOR and NFkappaB pathways in ovarian cancer: implications for targeted therapeutics. Cancers10.3390/cancers11070949 (2019). [DOI] [PMC free article] [PubMed]
- 451.Oh, T. I. et al. Fascaplysin sensitizes anti-cancer effects of drugs targeting AKT and AMPK. Molecules10.3390/molecules23010042 (2017). [DOI] [PMC free article] [PubMed]
- 452.Razzini G, et al. Novel functional PI 3-kinase antagonists inhibit cell growth and tumorigenicity in human cancer cell lines. FASEB J. 2000;14:1179–1187. doi: 10.1096/fasebj.14.9.1179. [DOI] [PubMed] [Google Scholar]
- 453.Mills GB, et al. The role of genetic abnormalities of PTEN and the phosphatidylinositol 3-kinase pathway in breast and ovarian tumorigenesis, prognosis, and therapy. Semin. Oncol. 2001;28:125–141. doi: 10.1016/s0093-7754(01)90290-8. [DOI] [PubMed] [Google Scholar]
- 454.Maughan BL, et al. p53 status in the primary tumor predicts efficacy of subsequent abiraterone and enzalutamide in castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. 2018;21:260–268. doi: 10.1038/s41391-017-0027-4. [DOI] [PubMed] [Google Scholar]
- 455.Tosoian JJ, et al. Prevalence and prognostic significance of PTEN loss in African-American and European-American men undergoing radical prostatectomy. Eur. Urol. 2017;71:697–700. doi: 10.1016/j.eururo.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Song H, et al. Selective ablation of tumor suppressors in parafollicular C cells elicits medullary thyroid carcinoma. J. Biol. Chem. 2017;292:3888–3899. doi: 10.1074/jbc.M116.765727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Hoffmann K, et al. Stable expansion of high-grade serous ovarian cancer organoids requires a low-Wnt environment. EMBO J. 2020;39:e104013. doi: 10.15252/embj.2019104013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Diaz-Fernandez A, Miranda-Castro R, de-Los-Santos-Alvarez N, Lobo-Castanon MJ. Post-translational modifications in tumor biomarkers: the next challenge for aptamers? Anal. Bioanal. Chem. 2018;410:2059–2065. doi: 10.1007/s00216-018-0861-9. [DOI] [PubMed] [Google Scholar]
- 459.Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer. 2016;15:18. doi: 10.1186/s12943-016-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Wang, L., Qi, H., Tang, Y. & Shen, H. M. Post-translational modifications of key machinery in the control of mitophagy. Trends Biochem. Sci. 10.1016/j.tibs.2019.08.002 (2019). [DOI] [PubMed]
- 462.Jegga AG, Inga A, Menendez D, Aronow BJ, Resnick MA. Functional evolution of the p53 regulatory network through its target response elements. Proc. Natl Acad. Sci. USA. 2008;105:944–949. doi: 10.1073/pnas.0704694105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Warnock LJ, Knox A, Mee TR, Raines SA, Milner J. Influence of tetramerisation on site-specific post-translational modifications of p53: comparison of human and murine p53 tumor suppressor protein. Cancer Biol. Ther. 2008;7:1481–1489. doi: 10.4161/cbt.7.9.6473. [DOI] [PubMed] [Google Scholar]
- 464.Lehmann AR. Ubiquitin-family modifications in the replication of DNA damage. FEBS Lett. 2011;585:2772–2779. doi: 10.1016/j.febslet.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 465.Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014;20:1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 466.Cai J, Culley MK, Zhao Y, Zhao J. The role of ubiquitination and deubiquitination in the regulation of cell junctions. Protein Cell. 2018;9:754–769. doi: 10.1007/s13238-017-0486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.M JR, S V. BMI1 and PTEN are key determinants of breast cancer therapy: a plausible therapeutic target in breast cancer. Gene. 2018;678:302–311. doi: 10.1016/j.gene.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 468.Jiang A, et al. Curcumin reactivates silenced tumor suppressor gene RARbeta by reducing DNA methylation. Phytother. Res. 2015;29:1237–1245. doi: 10.1002/ptr.5373. [DOI] [PubMed] [Google Scholar]
- 469.Gupta A, Shah K, Oza MJ, Behl T. Reactivation of p53 gene by MDM2 inhibitors: a novel therapy for cancer treatment. Biomed. Pharmacother. 2019;109:484–492. doi: 10.1016/j.biopha.2018.10.155. [DOI] [PubMed] [Google Scholar]
- 470.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Lane DP, Benchimol S. p53: oncogene or anti-oncogene? Genes Dev. 1990;4:1–8. doi: 10.1101/gad.4.1.1. [DOI] [PubMed] [Google Scholar]
- 472.Schultz KAP, et al. PTEN, DICER1, FH, and their associated tumor susceptibility syndromes: clinical features, genetics, and surveillance recommendations in childhood. Clin. Cancer Res. 2017;23:e76–e82. doi: 10.1158/1078-0432.CCR-17-0629. [DOI] [PubMed] [Google Scholar]
- 473.Que WC, Qiu HQ, Cheng Y, Liu MB, Wu CY. PTEN in kidney cancer: a review and meta-analysis. Clin. Chim. Acta. 2018;480:92–98. doi: 10.1016/j.cca.2018.01.031. [DOI] [PubMed] [Google Scholar]
- 474.Volodko N, Gordon M, Salla M, Ghazaleh HA, Baksh S. RASSF tumor suppressor gene family: biological functions and regulation. FEBS Lett. 2014;588:2671–2684. doi: 10.1016/j.febslet.2014.02.041. [DOI] [PubMed] [Google Scholar]
- 475.Ko A, Han SY, Song J. Regulatory network of ARF in cancer development. Mol. Cells. 2018;41:381–389. doi: 10.14348/molcells.2018.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Gallagher SJ, Kefford RF, Rizos H. The ARF tumour suppressor. Int. J. Biochem. Cell Biol. 2006;38:1637–1641. doi: 10.1016/j.biocel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 477.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Minde DP, Anvarian Z, Rudiger SG, Maurice MM. Messing up disorder: how do missense mutations in the tumor suppressor protein APC lead to cancer? Mol. Cancer. 2011;10:101. doi: 10.1186/1476-4598-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Morin PJ. Colorectal cancer: the APC-lncRNA link. J. Clin. Invest. 2019;129:503–505. doi: 10.1172/JCI125985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 481.Choi M, Kipps T, Kurzrock R. ATM mutations in cancer: therapeutic implications. Mol. Cancer Ther. 2016;15:1781–1791. doi: 10.1158/1535-7163.MCT-15-0945. [DOI] [PubMed] [Google Scholar]
- 482.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 483.Antoni L, Sodha N, Collins I, Garrett MD. CHK2 kinase: cancer susceptibility and cancer therapy—two sides of the same coin? Nat. Rev. Cancer. 2007;7:925–936. doi: 10.1038/nrc2251. [DOI] [PubMed] [Google Scholar]
- 484.Sopik V, Phelan C, Cybulski C, Narod SA. BRCA1 and BRCA2 mutations and the risk for colorectal cancer. Clin. Genet. 2015;87:411–418. doi: 10.1111/cge.12497. [DOI] [PubMed] [Google Scholar]
- 485.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 486.Lam HC, Nijmeh J, Henske EP. New developments in the genetics and pathogenesis of tumours in tuberous sclerosis complex. J. Pathol. 2017;241:219–225. doi: 10.1002/path.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Evans DGR, et al. Cancer and central nervous system tumor surveillance in pediatric neurofibromatosis 1. Clin. Cancer Res. 2017;23:e46–e53. doi: 10.1158/1078-0432.CCR-17-0589. [DOI] [PubMed] [Google Scholar]
- 488.Brosseau JP, et al. NF1 heterozygosity fosters de novo tumorigenesis but impairs malignant transformation. Nat. Commun. 2018;9:5014. doi: 10.1038/s41467-018-07452-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Kullmann L, Krahn MP. Controlling the master-upstream regulation of the tumor suppressor LKB1. Oncogene. 2018;37:3045–3057. doi: 10.1038/s41388-018-0145-z. [DOI] [PubMed] [Google Scholar]
- 490.Wang X, Hu S, Liu L. Phosphorylation and acetylation modifications of FOXO3a: Independently or synergistically? Oncol. Lett. 2017;13:2867–2872. doi: 10.3892/ol.2017.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Ma CX, et al. NeoPalAna: neoadjuvant palbociclib, a cyclin-dependent kinase 4/6 inhibitor, and anastrozole for clinical stage 2 or 3 estrogen receptor-positive breast cancer. Clin. Cancer Res. 2017;23:4055–4065. doi: 10.1158/1078-0432.CCR-16-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Abemaciclib, In: Drugs and Lactation Database (LactMed). (National Library of Medicine (US), Bethesda, MD, 2006).
- 493.Carvajal D, et al. Activation of p53 by MDM2 antagonists can protect proliferating cells from mitotic inhibitors. Cancer Res. 2005;65:1918–1924. doi: 10.1158/0008-5472.CAN-04-3576. [DOI] [PubMed] [Google Scholar]
- 494.Vu B, et al. Discovery of RG7112: a small-molecule MDM2 inhibitor in clinical development. ACS Med. Chem. Lett. 2013;4:466–469. doi: 10.1021/ml4000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Khurana A, Shafer DA. MDM2 antagonists as a novel treatment option for acute myeloid leukemia: perspectives on the therapeutic potential of idasanutlin (RG7388) OncoTargets Ther. 2019;12:2903–2910. doi: 10.2147/OTT.S172315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Ramraj, S. K. et al. Novel ovarian cancer maintenance therapy targeted at mortalin and mutant p53. Int. J. Cancer10.1002/ijc.32830 (2019). [DOI] [PMC free article] [PubMed]
- 497.Yu X, et al. Thiosemicarbazones functioning as zinc metallochaperones to reactivate mutant p53. Mol. Pharm. 2017;91:567–575. doi: 10.1124/mol.116.107409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Zache N, et al. Mutant p53 targeting by the low molecular weight compound STIMA-1. Mol. Oncol. 2008;2:70–80. doi: 10.1016/j.molonc.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Liu L, et al. CP31398 attenuates endometrial cancer cell invasion, metastasis and resistance to apoptosis by downregulating MDM2 expression. Int. J. Oncol. 2019;54:942–954. doi: 10.3892/ijo.2019.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Poulain S, et al. TP53 mutation and its prognostic significance in Waldenstrom’s macroglobulinemia. Clin. Cancer Res. 2017;23:6325–6335. doi: 10.1158/1078-0432.CCR-17-0007. [DOI] [PubMed] [Google Scholar]
- 501.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Santag S, et al. Recruitment of the tumour suppressor protein p73 by Kaposi’s Sarcoma Herpesvirus latent nuclear antigen contributes to the survival of primary effusion lymphoma cells. Oncogene. 2013;32:3676–3685. doi: 10.1038/onc.2012.385. [DOI] [PubMed] [Google Scholar]
- 503.Sonnemann J, et al. RETRA exerts anticancer activity in Ewing’s sarcoma cells independent of their TP53 status. Eur. J. Cancer. 2015;51:841–851. doi: 10.1016/j.ejca.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 504.Hu M, et al. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Kim J, Keay SK, You S, Loda M, Freeman MR. A synthetic form of frizzled 8-associated antiproliferative factor enhances p53 stability through USP2a and MDM2. PLoS ONE. 2012;7:e50392. doi: 10.1371/journal.pone.0050392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Reece KM, Figg WD. A novel regulator (USP10) of p53: implications for tumor suppression and therapeutic targeting. Cancer Biol. Ther. 2010;9:583–584. doi: 10.4161/cbt.9.8.11690. [DOI] [PubMed] [Google Scholar]
- 507.Iglesias-Gato D, et al. OTUB1 de-ubiquitinating enzyme promotes prostate cancer cell invasion in vitro and tumorigenesis in vivo. Mol. Cancer. 2015;14:8. doi: 10.1186/s12943-014-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Kon N, Zhong J, Qiang L, Accili D, Gu W. Inactivation of arf-bp1 induces p53 activation and diabetic phenotypes in mice. J. Biol. Chem. 2012;287:5102–5111. doi: 10.1074/jbc.M111.322867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Yang W, El-Deiry WS. CARPs are E3 ligases that target apical caspases and p53. Cancer Biol. Ther. 2007;6:1676–1683. doi: 10.4161/cbt.6.11.4939. [DOI] [PubMed] [Google Scholar]
- 510.Rajendra R, et al. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J. Biol. Chem. 2004;279:36440–36444. doi: 10.1074/jbc.C400300200. [DOI] [PubMed] [Google Scholar]
- 511.Wu H, Leng RP. UBE4B, a ubiquitin chain assembly factor, is required for MDM2-mediated p53 polyubiquitination and degradation. Cell Cycle. 2011;10:1912–1915. doi: 10.4161/cc.10.12.15882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Borges HL, Chao C, Xu Y, Linden R, Wang JY. Radiation-induced apoptosis in developing mouse retina exhibits dose-dependent requirement for ATM phosphorylation of p53. Cell Death Differ. 2004;11:494–502. doi: 10.1038/sj.cdd.4401366. [DOI] [PubMed] [Google Scholar]
- 513.Chao C, Herr D, Chun J, Xu Y. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J. 2006;25:2615–2622. doi: 10.1038/sj.emboj.7601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.El-Dahr SS, Aboudehen K, Dipp S. Bradykinin B2 receptor null mice harboring a Ser23-to-Ala substitution in the p53 gene are protected from renal dysgenesis. Am. J. Physiol. Ren. Physiol. 2008;295:F1404–F1413. doi: 10.1152/ajprenal.90378.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Iwakuma T, et al. Mutation at p53 serine 389 does not rescue the embryonic lethality in mdm2 or mdm4 null mice. Oncogene. 2004;23:7644–7650. doi: 10.1038/sj.onc.1207793. [DOI] [PubMed] [Google Scholar]
- 516.Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol. Cell Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Wang B, et al. The transcription and expression profile of p53(N236S) mutant reveals new aspects of gain of function for mutant p53. FEBS Lett. 2018;592:3183–3197. doi: 10.1002/1873-3468.13223. [DOI] [PubMed] [Google Scholar]
- 518.Lee SY, et al. K120R mutation inactivates p53 by creating an aberrant splice site leading to nonsense-mediated mRNA decay. Oncogene. 2019;38:1597–1610. doi: 10.1038/s41388-018-0542-3. [DOI] [PubMed] [Google Scholar]