Abstract
Survival has increased in early stage breast cancer (BC), and the late effects of treatment persist for decades. Molecular mechanisms underlying the acceleration of age-related diseases after chemotherapy and radiotherapy are poorly understood. We examined epigenetic changes in peripheral whole blood cells in early stage BC patients undergoing surgery followed by adjuvant radiotherapy, or surgery followed by adjuvant chemotherapy and radiotherapy. DNA methylation experiments were performed on whole blood samples collected before and after adjuvant therapy. Methylation profiles were used to estimate four measures of epigenetic age acceleration—intrinsic, extrinsic, phenotypic, and Grim—and cell counts. We found significant increases in extrinsic, phenotypic, and Grim epigenetic age acceleration and in estimated proportions of senescent T lymphocytes from pre- to post-treatment. When examining differential effects by treatment category, most of these increases were significant only in women undergoing radiation alone. Further studies are needed to examine whether these effects are related to the risk of cognitive and functional decline in BC survivors.
Subject terms: Breast cancer, Epigenetics, Senescence
While multimodality cancer treatments prolong life in early stage breast cancer (BC) patients, they increase the risk for age-related health problems1–6, functional decline, and fatigue that profoundly impact the quality of life7,8. These secondary problems may be a consequence of accelerated biological aging by exposure to cancer therapies. However, although animal studies demonstrate that chemotherapy and radiation therapy induce cellular senescence9–13, relatively few studies have examined these effects in clinical cohorts14–18, and none have examined effects on epigenetic aging markers.
A promising biomarker of organism aging, the epigenetic clock, measures tissue age based on methylation levels of CpGs co-locating with genes underlying pathways associated with cell survival and self-renewal19,20. Estimated epigenetic age is tightly correlated with chronologic age, accelerated in disease states, and predictive of frailty and mortality21–30. Here we examine whether markers of epigenetic age acceleration increase following two common treatment regimens for women with early-stage BC: radiation and chemotherapy.
Patients were recruited from oncology practices in Los Angeles to participate in a longitudinal study of cancer-related fatigue31. Women were eligible for the parent study if they had been recently diagnosed with Stage 0–IIIA BC and had not yet started adjuvant or neoadjuvant therapy with radiation, chemotherapy, or endocrine therapy. Assessments were conducted before the onset of adjuvant therapy, after completion of radiation and/or chemotherapy, and over an 18-month follow-up (see Supplementary Fig. 1). The current analysis focuses on a subset of women (n = 72) who had blood samples available for epigenetic analyses at baseline and post-treatment. We selected women treated with radiation alone (n = 37) and women treated with chemotherapy followed by radiation (n = 35) to evaluate individual and combined effects of those treatment exposures. All women had completed surgery prior to the baseline assessment.
We examined four measures of epigenetic age acceleration: intrinsic (IEAA), extrinsic (EEAA), phenotypic (PEAA), and Grim (GEAA), based on weighted averages of methylation levels at 353, 71, 513, and 1030 CpGs, respectively, with adjustment for chronologic age. Details of the epigenetic clock, DNA extraction/methylation experiments, and statistical analyses are provided in refs. 19,20,25,26,32 and Supplementary Methods. Briefly, while IEAA captures epigenetic age acceleration independent of changes in cell distribution that occur with advancing age, both EEAA and PEAA capture the functional decline of the immune system and exhibit negative and positive correlations with naive and late differentiated/senescent cytotoxic T lymphocytes, respectively. PEAA is also highly correlated with age-related phenotypes32. GEAA is strongly predictive of lifespan33. We estimated blood cell proportions using the advanced analysis option of the epigenetic clock software19 available online (http://dnamage.genetics.ucla.edu), which estimates the percentage of late differentiated CD8+ T cells (CD8+CD28−CD45RA−) and the number (count) of naive T cells (CD8+CD45RA+CCR7+). We examine changes in each of these measures from pre- to post-treatment, adjusting for ethnicity, body mass index (BMI), and tumor characteristics (ER, PR, and HER2 status, and stage), given links with epigenetic aging markers21,34.
Table 1 shows patient demographic, tumor, and treatment characteristics. A large percentage of women undergoing radiotherapy alone had ER+ and PR+ tumors and underwent lumpectomy rather than mastectomy. Women undergoing chemotherapy and radiotherapy had higher stage disease. There were no significant group differences in age, ethnicity, or BMI.
Table 1.
Characteristics of the study sample.
| Radiation therapy alone (N = 37) | Chemotherapy and radiotherapy (N = 35) | Total (N = 72) | p-Valuea | |
|---|---|---|---|---|
| Demographic characteristics | ||||
| Age (years), mean ± SD | 57.3 ± 9.3 | 56.1 ± 11.2 | 56.7 ± 10.2 | 0.44 |
| Ethnicity, N (%) | ||||
| Hispanic | 5 (14) | 2 (6) | 7 (10) | 0.47 |
| Non-Hispanic | 32 (86) | 33 (94) | 65 (90) | |
| Education, N (%) | ||||
| HS degree | 8 (22) | 8 (23) | 16 (22) | 0.41 |
| College degree | 12 (43) | 16 (46) | 28 (39) | |
| Postgraduate degree | 17 (46) | 11 (31) | 28 (39) | |
| Body mass index, mean ± SD | 25.6 ± 5.3 | 25.7 ± 6.7 | 25.7 (6.0) | 0.96 |
| Tobacco smoking, N (%) | ||||
| Current | 1 (3) | 1 (3) | 2 (3) | 0.59 |
| Former | 11 (30) | 7 (20) | 18 (25) | |
| Never | 24 (65) | 27 (77) | 51 (71) | |
| Menopausal status, N (%) | ||||
| Pre-menopausal | 8 (22) | 9 (26) | 17 (24) | 0.64 |
| Peri-menopausal | 1 (3) | 3 (8) | 4 (5) | |
| Post-menopausal | 26 (70) | 22 (63) | 48 (67) | |
| Hysterectomy | 2 (5) | 1 (3) | 3 (4) | |
| Tumor characteristics | ||||
| ER positive, N (%) | 36 (97) | 26 (74) | 62 (86) | 0.013 |
| PR positive, N (%) | 32 (86) | 21 (60) | 53 (7) | 0.023 |
| HER2 amplified, N (%) | 2 (5) | 2 (6) | 4 (5) | 0.95 |
| Stage, N (%) | ||||
| 0 | 5 (13) | 0 (0) | 5 (7) | 0.00015 |
| 1 | 25 (68) | 11 (31) | 36 (50) | |
| 2 | 7 (19) | 18 (51) | 25 (35) | |
| 3 | 0 (0) | 5 (14) | 5 (7) | |
| Type of surgery | ||||
| Lumpectomy | 35 (94) | 27 (77) | 62 (86) | 0.032 |
| Mastectomy | 2 (5) | 8 (23) | 10 (14) | |
ap-Value comparing treatment groups.
p-values were in bold if < 0.05.
Predicted epigenetic age (DNAm age) was significantly correlated with chronologic age for IEAA (r = 0.85, p < 0.00001), EEAA (r = 0.8, p < 0.00001), PEAA (r = 0.8, p < 0.00001), and GEAA (r = 0.89, p < 0.00001) in pre-treatment samples from the full cohort. Corresponding age acceleration measures for each of these epigenetic biomarkers are defined as the residuals from regressing DNAm age on chronologic age, and are measured in years.
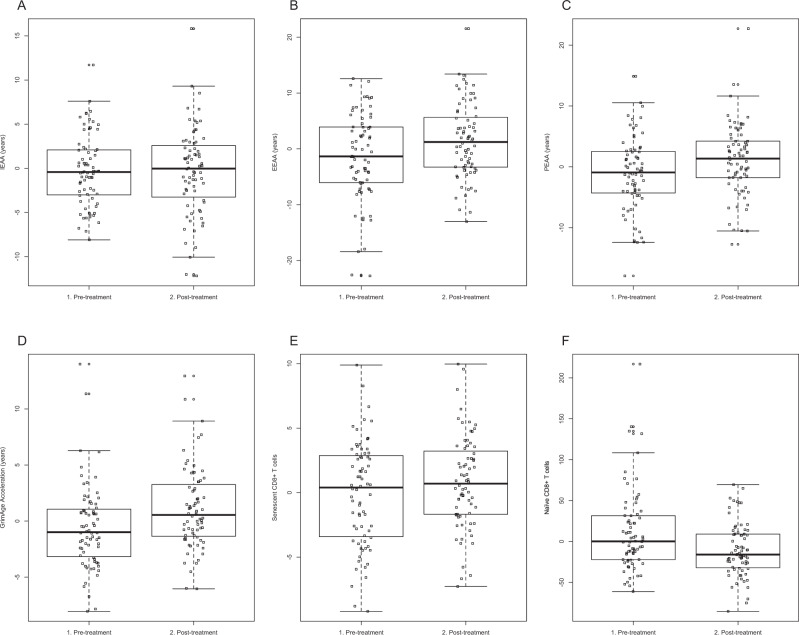
Figure 1 shows box pots of age acceleration measures from pre- to post-treatment for the full sample; plots for the two treatment groups are shown in Supplementary Fig. 2.
Fig. 1. Full distributions for measures of age acceleration in peripheral blood from pre- to post-treatment.
In addition to intrinsic (IEAA, a), extrinsic (EEAA, b), phenotypic (PEAA, c), and Grim (GEAA, d) measures of age-acceleration, age-adjusted estimates of senescent cytotoxic T lymphocytes (e) and naive T lymphocytes (f) are shown for the full sample. Our repeated measures ANOVA analysis revealed significant increases in EEAA (p = 0.0021), PEAA (p = 0.015), GEAA (P = 3.2 × 10−6), and age-adjusted estimates of senescent cytotoxic T lymphocytes (p = 0.038). Age-adjusted estimates of naive T lymphocytes decreased after treatment (p = 2.2 × 10−5). There was no significant change in IEAA with treatment (p = 0.83). Supplementary Fig. 2 reveals boxplots for each of these age acceleration measures and estimated cell counts, separated by treatment type.
In the full sample receiving adjuvant treatment, repeated measures analysis of variance showed a significant increase in EEAA (F(1,71) = 10.2, p = 0.0021), PEAA (F(1,71) = 6.22, p = 0.015), and GEAA (F(1,71) = 25.6, p = 3.2 × 10−6) from pre- to post-treatment, adjusting for ethnicity, BMI, stage, and ER/PR/HER2 status. Amongst patients receiving radiotherapy alone, EEAA (F(1,36) = 16.0, p = 3.0 × 10−4), PEAA (F(1,36) = 5.94, p = 0.020), and GEAA (F(1,36) = 11.7, p = 0.0015) were significantly increased, but not IEAA. Patients receiving both chemotherapy and radiation exhibited a significant increase in GEAA (F(1,34) = 13.6, p = 8.0 × 10−4), and non-significant increases in EEAA (F(1,34) = 0.66, p = 0.42) and PEAA (F(1,34) = 0.87, p = 0.36). There was a notable decrease (of borderline significance) from pre- to post-treatment in IEAA in the group receiving both chemotherapy and radiotherapy (F = 3.68, p = 0.064), and this finding is consistent with a recent report of decreased DNAm age with G-CSF administration35.
The proportion of late differentiated/senescent T lymphocytes increased after treatment in the full sample (Fig. 1e) (F(1,71) = 4.5, p = 0.038), and in the group treated with radiotherapy alone (Supplementary Fig. 2e) (F(1,36) = 7.7, p = 0.0077). There was an accompanying decrease in naive T lymphocytes in the full sample (F(1,71) = 20.7, p = 2.2 × 10−5) (Fig. 1f), and in both treatment groups (F(1,36) = 14.8, p = 4.5 × 10−4) for radiotherapy alone; (F(1,34) = 6.1, p = 0.019 for chemotherapy plus radiotherapy) (Supplementary Fig. 2f).
Because epigenetic biomarkers are not independent, we adjusted for multiple testing using a Bonferroni correction of 0.05/2. Using this criterion we find that increases in EEAA, PEAA, and GEAA, and the decrease in estimated naive T lymphocytes remained significant in the full sample and in the group receiving radiotherapy alone, while the increase in estimated senescent T lymphocytes remained significant only in the group receiving radiotherapy alone, and GEAA and the decrease in estimated naive T lymphocytes remained significant in the group receiving chemotherapy and radiotherapy.
Using a biomarker of biologic aging in whole blood, we found evidence of accelerating aging and immunosenescence after adjuvant therapy in women with early stage BC. To contrast these changes with “normal” aging, we examined age acceleration patterns in women of similar age but without cancer using published longitudinal data on DNA methylation studies of peripheral blood mononuclear cells of healthy women36. All women in the healthy cohort had at least two assessments, and the length of time between first and second visits ranged from 2 to 7 years. In this group, we found no significant changes from baseline to follow up in EEAA (mean 0.37 to −0.091, p = 0.35), IEAA (mean 0.0124–0.0115, p = 0.99), PEAA (0.90 to −0.30, p = 0.051), or GEAA (−0.063 to 0.018, p = 0.29). We also considered the possibility that changes in aging markers in the women undergoing cancer therapy might be driven by stress associated with a cancer diagnosis and treatment, but controlling for scores on the Perceived Stress Scale (PSS) yielded comparable results.
Examination of specific treatment exposures in our sample suggests a specific role of radiation in accelerating aging, although additional work is needed to confirm differential treatment effects and investigate mechanisms underlying these effects. Our findings are consistent with reports showing that adjuvant treatment for early stage BC accelerates biologic aging, as measured by p16INK4A expression in T lymphocytes14 and lower telomerase activity15. Because epigenetic aging biomarkers predict frailty and mortality23,32, our results raise concerns about treatment as a potential accelerator of these processes and highlight the importance of identifying strategies to prevent accelerated aging in cancer survivors.
Limitations of our study include the small sample size, and the inability to directly examine the effects of chemotherapy alone on epigenetic age. Further studies are needed to examine the acute effects of chemotherapy alone and to examine whether accelerations in DNAm age persist in the years following recovery from surgery, chemotherapy, and radiotherapy. Another important limitation of our study is our inability to disentangle the relative contributions of cancer diagnosis and surgery from the presumed effects of adjuvant therapies. Women undergoing surgery alone after early stage breast cancer diagnosis do not provide an adequate comparison group, as these patients tend to carry a Stage 0 (ductal carcinoma in situ) diagnosis, and undergo mastectomy more frequently. Future work will examine global methylation changes associated with radiation and/or chemotherapy, particularly within pathways related to DNA repair. However, we would not expect global hypo- or hyper-methylation to influence our finding of age-related methylation patterns, as these are based on weighted averages of a specific subset of CpGs. Our results raise questions about whether the observed acceleration in epigenetic age is associated with adverse outcomes and toxicities associated with combined treatment modalities in early stage breast cancer, and further work should test the predictive utility of the epigenetic clock for adverse late effects in longitudinal research designs. Ultimately this information could be used to estimate risks of late effects due to accelerated aging after treatment of early stage breast cancer.
Methods
Patients recruited to this longitudinal study provided informed written consent. This study was approved by the UCLA Institutional Review Board.
Tissue acquisition and processing
Genomic DNA was extracted from buffy coats of peripheral blood samples using the MIDI DNAEasy Blood and Tissue Kit for the QIASymphony automated extractor (Qiagen). Purified DNA was placed into 96-well plates and concentrated using a SpedVac, and suspended in AE buffer to a minimum of 100 ng/μL. DNA was quantified using the Invitrogen Quant-iT dsDNA Assay Kit, high sensitivity (Invitrogen).
DNA methylation data pre-processing
Bisulfite conversion using the Zymo EZ DNA Methylation Kit (ZymoResearch, Orange, CA, USA) as well as subsequent hybridization of the Human Methylation 850K EPIC chip (Illumina, San Diego, CA), and scanning (iScan, Illumina) was performed by the UCLA Neuroscience Genomics Core facilities according to the manufacturer’s protocols by applying standard settings. DNA methylation levels (β values) were determined by calculating the ratio of intensities between methylated (signal A) and un-methylated (signal B) sites. Specifically, the β value was calculated from the intensity of the methylated (M corresponding to signal A) and un-methylated (U corresponding to signal B) sites, as the ratio of fluorescent signals β = max(M,0)/[max(M,0) + max(U,0) + 100]. Thus, β values range from 0 (completely un-methylated) to 1 (completely methylated). To impute missing β values, we used a Euclidean metric to find k-nearest neighbors and impute the missing elements by averaging non-missing elements of its neighbors, using the impute.knn function in R37. Quantile normalization was applied to the raw data, in order to detect and remove outliers, and with the goal of making data comparable to the training data of the epigenetic clock.
Measures
Survey data were available from the RISE study on patients’ age, ethnicity, and education. BMI was determined through measurement of height and weight at the baseline assessment. Tumor characteristics and type of treatments received were determined by medical record review.
Statistical methods and analysis
We used four well-established measures to estimate epigenetic age based on weighted averages of CpGs: intrinsic (353 CpGs19,25), extrinsic (71 CpGs20,26), phenotypic (513 CpGs32), and grim (1030 CpGs33). Residuals from linear regression of these measures on chronologic age are used to define the age-adjusted age acceleration measures: IEAA, EEAA, PEAA, and GEAA. Details of the epigenetic clock methodology are provided in the online clock software and tutorial19,25. Briefly, IEAA adjusts for imputed measures of blood cell counts and captures epigenetic age acceleration independent of cell distribution, while EEAA employs a weighted adjustment for the estimated blood cell counts from three blood cell types that change with age: naive (CD45RA+CCR7+) cytotoxic T lymphocytes (reflecting stem cell self-renewal), late differentiated/senescent (CD28−CD45RA−) cytotoxic T lymphocytes, and plasma B lymphocytes. PEAA is highly correlated with age-related phenotypes. Unlike IEAA, both EEAA and PEAA correlate with markers of immunosenescence, and both exhibit negative and positive correlations with naive and late differentiated/senescent cytotoxic T lymphocytes, respectively. Grim age is calculated using DNA methylation-based surrogate biomarkers of smoking pack-years and serum biomarkers known to be predictive of morbidity or mortality33. After regressing time-to-death on these DNAm-based biomarkers, the mortality risk estimate is transformed into an age estimate (Grim Age), and this age estimate is strongly predictive of lifespan33. We further estimated proportions of cytotoxic T lymphocytes in naive and senescent states using global methylation data, using the methods of Horvath25.
We used analysis of variance (ANOVA) with repeated measures to examine changes in four measures of age acceleration (IEAA, EEAA, PEAA, and GEAA) and estimated cell counts (naive and senescent cytotoxic T lymphocytes) from pre- to post-treatment, first combining across the two treatment groups, and subsequently examining each treatment group individually in order to assess whether the addition of chemotherapy further contributes to the effects of radiotherapy. All analyses were adjusted for ethnicity and BMI, given their known associations with epigenetic clock accelerations34,38. All analyses were also adjusted for tumor characteristics, including stage, and ER, PR, and HER2 status. Furthermore in order to examine for the effects of stress, we performed additional analyses adjusting for the PSS.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors thank the participants in the RISE study for their generous participation in both surveys and sample donations. The authors would like to thank Laura Petersen for her help with data management and variable selection, and Deborah Garet for study coordination. This work was supported by the National Institutes of Health (R01 5R01CA160427), by a Career Catalyst Award for Basic and Translational Research from the Susan G. Komen Foundation, by a pilot award from the UCLA Older Americans Independence Center, and by a seed grant from the Jonsson Comprehensive Cancer Center. Support also provided from the Cousins Center for Psychoneuroimmunology to JEC and JEB, from the American Cancer Society to JEC, and from the Breast Cancer Research Foundation to JEB.
Author contributions
All authors contributed to the design of this work, analysis and interpretation of data, and writing of the manuscript. All authors have approved the completed version of the manuscript and are accountable for all aspects of the work.
Data availability
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.1184736939. The methylation data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus40 and are accessible through GEO Series accession https://identifiers.org/geo:GSE140038 41.
Competing interests
S.H. is listed as inventor of several patent applications surrounding epigenetic biomarkers of aging. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41523-020-0161-3.
References
- 1.Agrawal S. Late effects of cancer treatment in breast cancer survivors. South Asian J. Cancer. 2014;3:112–115. doi: 10.4103/2278-330X.130445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfano CM, et al. Inflammatory cytokines and comorbidity development in breast cancer survivors versus noncancer controls: evidence for accelerated aging? J. Clin. Oncol. 2017;35:149–156. doi: 10.1200/JCO.2016.67.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordan JH, et al. Incident comorbidities and all-cause mortality among 5-year survivors of Stage I and II breast cancer diagnosed at age 65 or older: a prospective-matched cohort study. Breast Cancer Res. Treat. 2014;146:401–409. doi: 10.1007/s10549-014-3021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bluethmann SM, Mariotto AB, Rowland JH. Anticipating the “Silver Tsunami”: prevalence trajectories and comorbidity burden among older cancer survivors in the United States. Cancer Epidemiol. Biomark. Prev. 2016;25:1029–1036. doi: 10.1158/1055-9965.EPI-16-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett JA, Winters-Stone KM, Dobek J, Nail LM. Frailty in older breast cancer survivors: age, prevalence, and associated factors. Oncol. Nurs. Forum. 2013;40:E126–E134. doi: 10.1188/13.ONF.E126-E134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowland JH, Bellizzi KM. Cancer survivorship issues: life after treatment and implications for an aging population. J. Clin. Oncol. 2014;32:2662–2668. doi: 10.1200/JCO.2014.55.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bower JE. Behavioral symptoms in patients with breast cancer and survivors. J. Clin. Oncol. 2008;26:768–777. doi: 10.1200/JCO.2007.14.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bower JE. Cancer-related fatigue—mechanisms, risk factors, and treatments. Nat. Rev. Clin. Oncol. 2014;11:597–609. doi: 10.1038/nrclinonc.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang J, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demaria M, et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 2017;7:165–176. doi: 10.1158/2159-8290.CD-16-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Schulte BA, LaRue AC, Ogawa M, Zhou D. Total body irradiation selectively induces murine hematopoietic stem cell senescence. Blood. 2006;107:358–366. doi: 10.1182/blood-2005-04-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 13.Carbonneau CL, et al. Ionizing radiation-induced expression of INK4a/ARF in murine bone marrow-derived stromal cell populations interferes with bone marrow homeostasis. Blood. 2012;119:717–726. doi: 10.1182/blood-2011-06-361626. [DOI] [PubMed] [Google Scholar]
- 14.Sanoff HK, et al. Effect of cytotoxic chemotherapy on markers of molecular age in patients with breast cancer. J. Natl. Cancer Inst. 2014;106:dju057. doi: 10.1093/jnci/dju057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scuric Z, et al. Biomarkers of aging associated with past treatments in breast cancer survivors. npj Breast Cancer. 2017;3:50. doi: 10.1038/s41523-017-0050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diker-Cohen T, et al. The effect of chemotherapy on telomere dynamics: clinical results and possible mechanisms. Leuk. Lymphoma. 2013;54:2023–2029. doi: 10.3109/10428194.2012.757765. [DOI] [PubMed] [Google Scholar]
- 17.Lee JJ, et al. Telomere length shortening in non-Hodgkin’s lymphoma patients undergoing chemotherapy. Ann. Hematol. 2003;82:492–495. doi: 10.1007/s00277-003-0691-4. [DOI] [PubMed] [Google Scholar]
- 18.M’Kacher R, et al. Telomere shortening: a new prognostic factor for cardiovascular disease post-radiation exposure. Radiat. Prot. Dosim. 2015;164:134–137. doi: 10.1093/rpd/ncu296. [DOI] [PubMed] [Google Scholar]
- 19.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvath S, et al. Obesity accelerates epigenetic aging of human liver. Proc. Natl Acad. Sci. USA. 2014;111:15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvath S, et al. Accelerated epigenetic aging in Down syndrome. Aging Cell. 2015;14:491–495. doi: 10.1111/acel.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marioni RE, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rickabaugh TM, et al. Acceleration of age-associated methylation patterns in HIV-1-infected adults. PLoS ONE. 2015;10:e0119201. doi: 10.1371/journal.pone.0119201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horvath S, Levine AJ. HIV-1 infection accelerates age according to the epigenetic clock. J. Infect. Dis. 2015;212:1563–1573. doi: 10.1093/infdis/jiv277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen BH, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging. 2016;8:1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christiansen L, et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15:149–154. doi: 10.1111/acel.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson’s disease patients. Aging. 2015;7:1130–1142. doi: 10.18632/aging.100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breitling LP, et al. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin. Epigenet. 2016;8:21. doi: 10.1186/s13148-016-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, et al. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat. Commun. 2017;8:14617. doi: 10.1038/ncomms14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bower JE, et al. Testing a biobehavioral model of fatigue before adjuvant therapy in women with breast cancer. Cancer. 2019;125:633–641. doi: 10.1002/cncr.31827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine ME, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10:573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu AT, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. 2019;11:303–327. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horvath S, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17:171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soraas A, et al. Epigenetic age is a cell-intrinsic property in transplanted human hematopoietic cells. Aging Cell. 2019;18:e12897. doi: 10.1111/acel.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sehl ME, Henry JE, Storniolo AM, Ganz PA, Horvath S. DNA methylation age is elevated in breast tissue of healthy women. Breast Cancer Res. Treat. 2017;164:209–219. doi: 10.1007/s10549-017-4218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Troyanskaya O, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 38.Dugue PA, et al. Association of DNA methylation-based biological age with health risk factors and overall and cause-specific mortality. Am. J. Epidemiol. 2018;187:529–538. doi: 10.1093/aje/kwx291. [DOI] [PubMed] [Google Scholar]
- 39.Sehl, M. E., Carroll, J. E., Horvath, S. & Bower, J. E. Metadata supporting data files in the published article: The acute effects of adjuvant radiation and chemotherapy on peripheral blood epigenetic age in early stage breast cancer patients. figshare.10.6084/m9.figshare.11847369 (2020). [DOI] [PMC free article] [PubMed]
- 40.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sehl, M. E., Horvath, S., Bower, J. & Carroll, J. The acute effects of chemotherapy and radiotherapy on peripheral blood epigenetic age in early stage breast cancer patients. Gene Expression Omnibus.https://identifiers.org/geo:GSE140038 (2020). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and analyzed during this study are described in the following data record: 10.6084/m9.figshare.1184736939. The methylation data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus40 and are accessible through GEO Series accession https://identifiers.org/geo:GSE140038 41.