Abstract
The self-renewal ability is a unique property of fetal-derived innate-like B-1a lymphocytes, which survive and function without being replenished by bone marrow (BM) progenitors. However, the mechanism by which IgM-secreting mature B-1a lymphocytes self-renew is poorly understood. Here, we showed that Bmi1 was critically involved in this process. Although Bmi1 is considered essential for lymphopoiesis, the number of mature conventional B cells was not altered when Bmi1 was deleted in the B-cell lineage. In contrast, the number of peritoneal B-1a cells was significantly reduced. Peritoneal cell transfer assays revealed diminished self-renewal ability of Bmi1-deleted B-1a cells, which was restored by additional deletion of Ink4-Arf, the well-known target of Bmi1. Fetal liver cells with B-cell-specific Bmi1 deletion failed to repopulate peritoneal B-1a cells, but not other B-2 lymphocytes after transplantation assays, suggesting that Bmi1 may be involved in the developmental process of B-1 progenitors to mature B-1a cells. While Bmi1 deletion has also been shown to alter the microenvironment for hematopoietic stem cells, fat-associated lymphoid clusters, the reported niche for B-1a cells, were not impaired in Bmi1−/− mice. RNA expression profiling suggested lysine demethylase 5B (Kdm5b) as another possible target of Bmi1, which was elevated in Bmi1−/− B-1a cells in a stress setting and might repress B-1a cell proliferation. Our work has indicated that Bmi1 plays pivotal roles in self-renewal and maintenance of fetal-derived B-1a cells.
Introduction
Murine B-1 cells are innate-like mature B-lymphocytes distinct from conventional adoptive immune B-lymphocytes (B-2 cells). B-1 cells harbor unique characteristics, including specific surface markers (IgMhighIgDlowCD19hiB220lo), production of natural IgM antibodies, and their primal localization in the pleural and peritoneal cavities (1). One of the most striking characteristics of CD5+ B-1 cells (referred to as B-1a cells) is their unique origin and self-replenishing ability. Cell transfer studies using fetal liver (FL) and bone marrow (BM) progenitors demonstrated that only FL progenitors, not BM cells, efficiently reconstitute peritoneal B-1a cells upon transplantation (2). Lin−AA4.1+CD19+B220lo-neg B-1-specific progenitors have been found in the FL and neonatal BM and were shown to decline in number during aging, suggesting that B-1 cells are mostly derived from fetal and neonatal progenitors (3, 4) and represent a distinct lineage from conventional B-2 cells. Additionally, long-term hematopoietic stem cells (LT-HSCs) in the FL and adult BM reportedly failed to reconstitute peritoneal B-1a cells in transplantation assays (5, 6), indicating that some (if not most) B-1a cells develop independently of HSCs (7, 8) and are maintained throughout life without being replenished by adult HSC-derived progenitors (9). Finally, adoptive transfer of mature B-1a cells into congenic recipient mice was demonstrated to be sufficient to repopulate and maintain the B-1a cell compartment in the long-term, supporting the notion that B-1a cells are maintained in vivo by self-renewal mechanisms.
The self-renewal ability is one of the most important features of stem cells. Both HSCs and neural stem cells rely on the Bmi1 polycomb ring finger proto-oncogene (Bmi1) for their long-term self-renewal (10-12). BMI1 consists of the polycomb repressor complex 1 (PRC1), which represses gene expression. In Bmi1−/− mice, the number of HSCs and all lymphoid cell subsets were shown to be significantly reduced, and the self-renewal ability of HSCs upon transplantation was lost (11). The Ink4-Arf locus is a known target of Bmi1 in both HSCs and neural stem cells, and deletion of Ink4-Arf in Bmi1−/− mice was demonstrated to dramatically correct the number and self-renewal ability of HSCs (13). Moreover, Bmi1 has also been shown to regulate the regeneration of skeletal muscles (14) and the proliferation and self-renewal of intestinal stem cells (15). Additionally, it has been reported that overexpression of Bmi1 induced an extensive capacity for self-renewal in adult BM erythroblasts to similar levels as those seen in embryo-derived self-renewing erythroblasts (16). We recently reported that overexpressing Bmi1 in mouse embryonic stem cell-derived B-1 cells enhanced long-term engraftment in recipient mice upon transplantation (17). Therefore, we hypothesized that Bmi1 might play an important role in the homeostatic ability of self-renewal in B-1a cells.
Here we report the critical role of Bmi1 in the maintenance of B-1a cells in vivo. We found that, compared with levels in other lymphoid cell subsets, Bmi1 is highly expressed in B-1a cells. We confirmed that the total number of T- and B-lymphocytes were reduced in the spleen and BM of Bmi1−/− mice, as previously reported (18). However, the percentage and number of total peritoneal B-1a cells were specifically decreased at a higher rate than those of B-2 and B-1b cells. Importantly, in B-cell-specific Bmi1 knockout mice, only B-1a cells, not other mature B cell subsets (follicular, marginal zone, and B-1b B cells), showed a reduction in their number and frequency. Accordingly, Bmi1-deficient peritoneal B-1a cells lost their self-renewal ability upon transplantation, which was restored by overexpression of Bmi1 or deletion of a well-known target of Bmi1, Ink4-Arf. Although this restoration by Ink4-Arf deletion is consistent with previous reports on HSCs (13), microarray analysis suggested lysine demethylase 5B (Kdm5b) as another novel candidate target gene for Bmi1. Our results indicated that mature B-1a lymphocytes utilize Bmi1 for their maintenance of self-renewal ability.
Materials and Methods
Mice
All mice were bred and maintained under specific pathogen–free conditions in the Indiana University School of Medicine Laboratory Animal Resource Center (LARC) and the Center for Laboratory Animal Medicine and Care (CLAMC) at University of Texas Health Science Center at Houston (UTHealth). C57BL/6J, Boy/J, NOD/SCID/IL2Rγc−/− (NSG), Bmi1-flox/flox (Bmi1F/F) and CD19-Cre knock-in mice (CD19Cre/Cre) were purchased from Jackson laboratory. Bmi1F/F mice were also provided by Dr. Xicheng Liu (National Institute of Biological Science, Beijing). Bmi1−/− mice were provided by Dr. Maarten van Lohuizen (The Netherlands Cancer Institute, The Netherlands). Ink4a-Arf−/− mice were provided by Dr. Ronald A. DePinho (MD Anderson Cancer Center, Houston). Bmi1−/−and Bmi1−/−Ink4-Arf−/− mice had CD45.1+CD45.2+ or CD45.2+ (depending on the experiment). The experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Indiana University and the Animal Welfare Committee (AWC) in UTHealth.
Flow cytometry
FL, spleen, BM, and peritoneal cells were prepared as single cell suspension. For analysis and sorting of hematopoietic subsets (Table S1), the following antibodies were used at different fluorescent color combinations: anti-mouse AA4.1 (AA4.1), CD19 (1D3), B220 (RA3–6B2), IgM (II/41), CD21 (8D9), CD23 (B3B4), CD11b (M1/70), Gr1 (RB6–8C5), CD5 (53–7.3), CD3e (145–2C11), Ter119 (TER-119), c-kit (2B8), Sca-1 (D7), CD150 (TC15–12F12.1), CD48 (HM48–1), IL- 7Ra (A7R34) and annexin V (all purchased from eBioscience or Biolegend). Cells were analyzed on LSRII or sorted on FACS Aria (Becton Dickinson).
Real-time PCR
Briefly, total RNA was extracted with RNeasy micro (QIAGEN), followed by reverse-transcription with SuperScript III (Invitrogen). The input cDNA was standardized and then amplified an ABI Prism 7500HT (Applied Biosystems) with SYBR Green Master Mix (Applied Biosystems), the following primer sets were used: β-actin F, 5’-CCTAAGGCCAACCGTGAAAAG and R, 5’- CAGAGGCATACAGGGACAGCA, HPRT F, 5’- TCCTCCTCAGACCGCTTTT and R, 5’-CCTGGTTCATCATCGCTAATC, Bmi1 F, 5’- CAGGTTCACAAAACCAGACCAC and R, 5’- TGACGGGTGAGCTGCATAAA, Arf F, 5’- CATGTTGTTGAGGCTAGAGAGGA and R, 5’-CTGCACCGTAGTTGAGCAGA, Kdm5b F, 5’- TCAGGGGACATTATGAGCGAA and R, 5’- TCGGAGGTCAGGTTAGGCTTC, p21 F, 5’-AACATCTCAGGGCCGAAAAC R, 5’- TCCTGACCCACAGCAGAAGA, Bcl-2 F, 5’-TGAGTACCTGAACCGGCATCT and R, 5’- TCAAACAGAGGTCGCATGCTG.
Retrovirus production and Infection
The knockdown plasmid pMKO was purchased from addgene (#10676) and pMKO-shLuc or -shKdm5b (sh1–3) were prepared with standard cloning strategy. For retrovirus production, pMigR1-Mock, -Bmi1 or knockdown plasmids were transfected into Phoenix-Eco producer cells respectively, followed by collection and concentration of the virus supernatant with the standard method(19). The established spin-infection was used for infection into FL cells. Target sequences are followings; Control (luciferase) TAAGGCTATGAAGAGATAC, Ms_Kdm5b-sh1 GCAGAGGCTATGAATATTAA, Ms_Kdm5b-sh3 CGCCAGTGTGTGGAGCATTA, and Ms_Kdm5b-shB GAGATGCACTCCGATACAT.
Transplantation
For peritoneal cell transplantation, donor peritoneal cells (CD45.2 or CD45.1+CD45.2+) were injected into the peritoneal cavity of sublethally irradiated (250rad) NSG mice (CD45.1). FL lineage− cells with or without Bmi1 expressing vector were injected into the tail vein of lethally irradiated (950 rad) Boy/J mice. Fetal liver B-1 progenitor cells were injected into the peritoneal cavity of sublethally irradiated (150 rad) NSG neonates.
Microarray analysis
Peritoneal B-1a cells were collected from Bmi1−/− and wild type mice and RNA was extracted using RNeasy micro kit (QIAGEN) and submitted to Miltenyi Biotech for gene expression analysis using Agilent Whole Mouse Genome Oligo Microarrays. Each 7 samples were examined. Complete raw data set and normalized data were produced by Miltenyi. The micro array data were submitted to GEO (Accession code: GSE97202), https://www.ncbi.nlm.nih.gov/geo/.
Bioinformatics analysis of microarray
The normalized expression data from Miltenyi was first filtered by the expression level with a cut-off value 5. If a gene is expressed higher than this cut-off level in four or more WT/KO samples, it is kept for the downstream differential expression (DE) analysis. The DE analysis was performed on Partek Genomic Suite (Partek, St. Charles, MO). We then calculated Benjamini-Hochberg multi-test adjusted FDR based on the p-values from the DE analysis. Genes with an FDR<0.2 were used for the Ingenuity Pathway Analysis (IPA).
For hierarchical clustering, filtered data were imported into GenePattern (v. 3.9.9, Broad Institute) for hierarchical clustering. Clustering and distance measurements were calculated using pairwise complete linkage and Pearson’s correlation. Output was used to render heat map data.
Chromatin IP (ChIP)
ChIP was done with standard protocols. DNA from Baf-3 cell (1 × 107) was crosslinked with 1% formaldehyde and was sheared into fragments approximately 200–500 base pairs in length with a sonicator (505 Sonic Dismembrator, Fisher Scientific). All immunoprecipitation used 5μg control IgG, anti-Bmi1 (AF27, Active Motif) or Ring-1b (D139–3, MBL International) Ab with protein A Dynabeads (Invitrogen). Quantification of precipitated genomic DNA relative to input was done in triplicate after real-time PCR with SYBR Green Master Mix (Applied Biosystems). The following primer sets were used: Ms_Kdm5b/ChIP/+700 F, 5’- GTCTGGAGCGGCTGGTTGAG and R, 5’- CCCACCATCCTCAAAGTGTCG, Ms_Kdm5b/ChIP/−200 F, 5’-GTCTGTCCTTGCTGCTCCTTG and R, 5’-AAACCCGAGAAGCAGAGTACT, Ms_Kdm5b/ChIP/−2000 F, 5’- TCAGGCTCCAAATCCCTGTAGA and R, 5’-GCTCTATCGAAGTACCTGGCC, Ms_Kdm5b/ChIP/−3300 F, 5’-ACTGAGATGGTGCATTGCTGA and R, 5’-CCTGGCCTTACTGTTAGTGCG, Ms_Arf/ChIP F, 5’- AAAACCCTCTCTTGGAGTGGG and R, 5’-GCAGGTTCTTGGTCACTGTGAG, Ms_Ink4a/ChIP F, 5’- GCCCGAGAAATCCTAGAGAATCC and R, 5’-GGATTCTCTAGGATTTCTCGGGC, Ms_GAPDH/ChIP F, 5’- CCCACTTGCCTCTGTATTGG and R, 5’- CTGTGGGGAGTCCTTTTCAG.
FALCs staining
Fat tissues were removed from the peritoneal cavity and fixed with 4% paraformaldehyde for 1 hour at 4°C. The samples were stained with 1:200 Alexa Fluor 555 conjugated anti-mouse IgM antibody (SouthernBiotech) and FITC conjugated anti-CD31 antibody or FITC-conjugated anti-CD11b antibody in 0.1% triton X100-PBS for 1 hour at room temperature. Then Alexa Fluor 555+ (red) areas were dissected out under a Leica™ mz9.5 fluorescent stereomicroscope and then mounted on Superfrost™ Plus Gold Slides (Thermo Fisher Scientific) with ProLong® Gold Antifade solution with DAPI (Thermo Fisher Scientific). Confocal images were taken using an Olympus II microscope with UApoN340 20x/0.7W objectives (Olympus). ImageJ software was used to adjust and output the images.
Ag stimulation assays
The sorted B-1a cells (20–10×103) from each genogroup were plated in the 96 well plate in IMDM with 10% FBS, 10ng/ml IL-5, 5ug/ml R848 (TLR7/8 ligand), and 5ug/ml LPS. Fourty-eight hours later, cells were collected and cell numbers were counted.
Statistical Analysis
Student t-test was used for all statistical analysis.
Results
B-1a cells show higher dependency on Bmi1 than do other lymphoid subsets
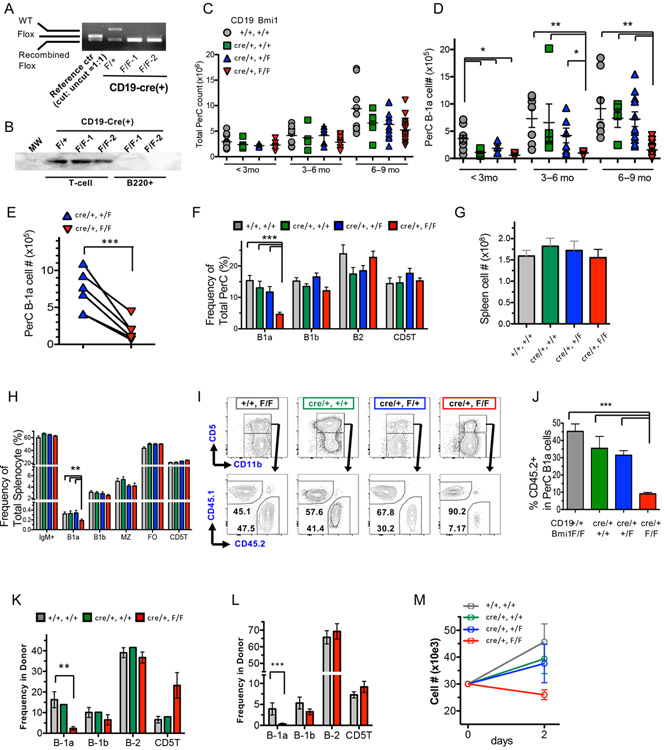
We examined the expression of the Bmi1 gene by quantitative PCR (qPCR) in various FACS-sorted lymphocyte subsets as shown in Table S1. Notably, Bmi1 was highly expressed in peritoneal B-1a and B-1b cells compared with other lymphocyte subsets (Fig. 1A). In the FL, Bmi1 was highly expressed in B-1-specific progenitors (lin−AA4.1+CD19+B220lo/−) compared with other progenitor subsets (Fig. 1B). Although previous reports have indicated that the lymphoid compartment is globally reduced in the lymphoid organs of Bmi1−/− mice, the effect of Bmi1 deletion on peritoneal B-1 cells has not been reported. Therefore, we examined the peritoneal cavity and found that the total number of peritoneal cells was markedly reduced in Bmi1−/− mice (Fig. 1C). While the frequency of CD5+ T-cells was not altered, the frequency of IgM+ B-cells was significantly reduced (Fig. 1D). Interestingly, among the peritoneal IgM+ cells, the percentage of B-1a cells was specifically reduced, whereas B-2 and B-1b subsets showed similar percentages to those in wild-type (WT) mice (Fig. 1E, F). The absolute number of B-1a cells was also dramatically reduced. Both the frequency and absolute number of B-1 progenitors in the Bmi1−/− FL were modestly increased compared with those of WT FL (Fig. 1G-J). Thus, loss of Bmi1 resulted in a profound reduction in both the frequency and absolute number of peritoneal B-1a cells.
Figure 1. Peritoneal B-1a cells have higher Bmi1 dependency than other lymphoid subsets.
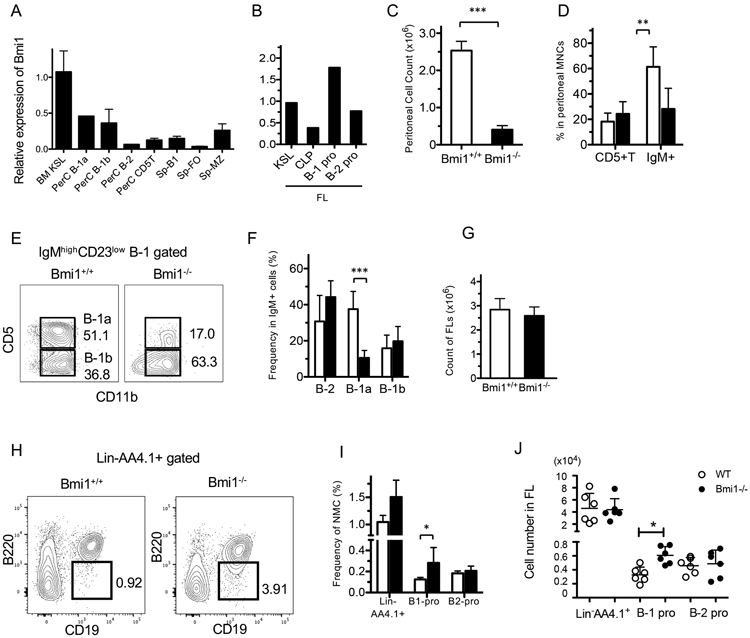
A. Bmi1 mRNA expression measured by qPCR in various sorted populations from adult BM, peritoneal cells (PerC), or spleen (Sp). KLS: kit+Sca-1+lin− HSC population, FO: follicular B-cells, MZ: marginal zone B-cells. B. Bmi1 expressions in various fetal liver (FL) progenitor populations. CLP: common lymphoid progenitors. C. Marked reduction of peritoneal cell count in Bmi1−/− mice (n=6, p<0.001). D. The frequency of IgM+ B-cell and CD5+ T-cells in the WT and Bmi1−/− peritoneal cavity is depicted (n=5, ** p<0.01). E. Representative FACS plots for peritoneal B-1a cells in WT and Bmi1−/− mice are depicted. F. The frequency of B-cell subsets among the peritoneal IgM+ cells is depicted. The percentage of B-1a cells is reduced (n=5, ***p<0.001). (G-J) Total cell number of FLs (G), representative FACS plots for lin−AA4.1+CD19+B220− B-1 progenitor population in the FL (H), the frequency (I) and absolute number (J) of FL B-1 and B-2 progenitor cells in WT and Bmi1−/− embryos are depicted. B-1 progenitor cell number is increased in the Bmi1−/− FL (n=6, *p<0.03). White bar & circle: WT, black bar & circle: Bmi1−/−. All data were obtained from experiments more than 3 times.
Loss of Bmi1 impairs self-renewal ability of peritoneal B-1a cells
B-1a cells have a self-renewal ability; their numbers are maintained without being replenished by BM progenitors. Since Bmi1 is crucial for the self-renewal capacity of various stem cells (10-15), we hypothesized that Bmi1 might also play a critical role in self-renewal ability of B-1a cells. Accordingly, we measured the self-renewal ability of peritoneal B-1 cells using transplantation assays in which peritoneal cells containing the same number of B-1a cells (4,000–80,000 cells) from either WT or Bmi1−/− mice were transplanted into sublethally irradiated (250 rad) NSG mice. Our results showed that 3 weeks after transplantation, WT donor peritoneal B-1 cells repopulated both B-1a and B-1b subsets, while Bmi1−/− donor B-1a cells showed poor engraftment (Fig. S1A). Furthermore, 12 weeks after transplantation, WT B-1a cells were fully reconstituted in the recipient peritoneal cavity, while Bmi1−/− B-1a cells were diminished (Fig. 2A, B). Taken together, these results indicate that B-1a cells lost the self-renewal ability in the absence of Bmi1.
Figure 2. Loss of B-1a self-renewal by Bmi1 disruption.
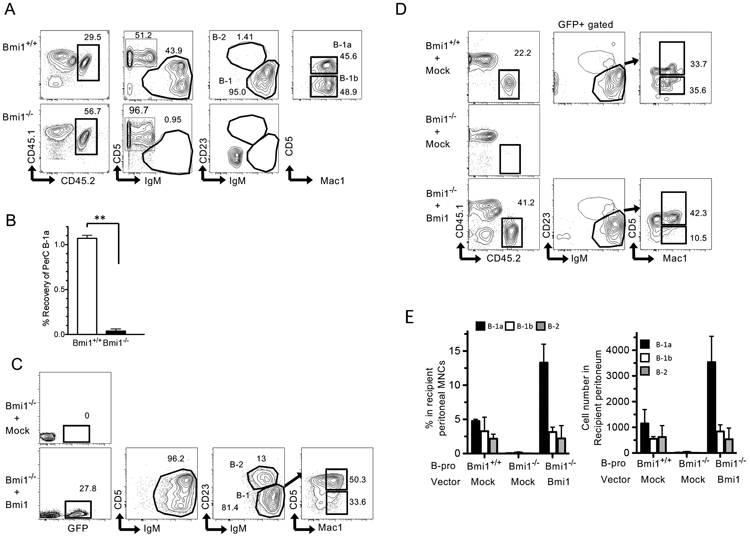
A: Representative FACS plots of recipient peritoneal cells at 12 weeks post-transplant. The peritoneal cells of the recipient NSG mice transplanted with Bmi1+/+ (upper panel) or Bmi1−/− (lower panel) peritoneal cells are depicted (n=3). B: Recovery ratio of WT and Bmi1−/− B-1a cells after transplantation, calculated by (number of recovered B-1a cells)/ (number of injected B-1a cells) 12 weeks after transplantation (n=3 for each group). Around 4000-80000 B-1a cells were injected. C: Retrovirus with mock or Bmi1 vector was infected into Bmi1−/− FL Lin− cells. Subsequently, infected cells were transplanted into sub-lethally irradiated (250 rad) NSG mice. Representative FACS plots at 4 months post-transplant are depicted (n=3). D: WT and Bmi1−/− FL B-1 progenitor cells with a mock or Bmi1-overexpressing retrovirus were injected into sub-lethally irradiated NSG mice. The recipient peritoneal cell analysis 4 months after transplantation is depicted (n=3). E: The percentage (left) and cell number (right) of donor-derived B-cell population in the peritoneal cavity of NSG mice transplanted with Bmi1−/− FL B-1 progenitor cells with or without Bmi1-overexpressing vector are depicted (n=3). All data were obtained from experiments more than 3 times.
Overexpression of Bmi1 rescues the self-renewal ability of B-1a cells
To confirm that Bmi1 deficiency is the primary cause of the defect in B-1a cell self-renewal in Bmi1−/− mice, we retrovirally overexpressed Bmi1 in Bmi1−/− FL lineage-negative cells (Fig. S1B) and transplanted them into sublethally irradiated NSG mice. While Bmi1−/− FL cells transduced with mock vector failed to reconstitute in recipient mice, Bmi1−/− FL cells overexpressing Bmi1 efficiently repopulated all lineages in the BM, including HSCs (Fig. S1C). Donor-derived B-1a, B-1b, and B-2 cells were all reconstituted in the peritoneal cavity of the recipient (Fig. 2C).
Next, Bmi1−/− FL B-1-specific progenitors retrovirally transduced with Bmi1 were injected into sublethally irradiated NSG mice to determine the requirement for Bmi1 in B-1 progenitors. We found that 4 months after transplantation, Bmi1−/− B-1 progenitors transduced with mock vector failed to repopulate B-1a cells, as expected (Fig. 2D). However, Bmi1-overexpressing Bmi1−/− B-1 progenitors exhibited efficient B-1a cell repopulation in recipient NSG mice (Fig. 2E). Interestingly, donor-derived B-1a cells were more frequent in mice transplanted with Bmi1-expressing Bmi1−/− B-1 progenitors relative to those transplanted with WT B-1 progenitors (Fig. 2D). Thus, Bmi1 overexpression not only restored, but also enhanced the self-renewal ability of Bmi1−/− B-1a cells.
B-cell restricted deletion of Bmi1 leads to B-1a-specific reduction and loss of self-renewal ability
To exclude the potential influence of defects in either HSCs or the microenvironment on the reduction of B-1a cells in Bmi1−/− mice, we used a mouse model in which Bmi1 was conditionally deleted in the B-cell lineage by crossing CD19Cre/+ and Bmi1-floxed (Bmi1F/F) mice. Notably, since homozygous CD19Cre/Cre knock-in mice show defects in B-1a cells (20-22), we utilized CD19Cre/+ heterozygous mice to examine B-cell-specific depletion of Bmi1 to avoid any confusing results. Efficient deletion of Bmi1 alleles was confirmed in sorted peritoneal B-1 cells from CD19Cre/+: Bmi1F/F mice by genomic PCR, as well as by western blotting of sorted spleen B220+ cells, including B-1, marginal zone, and follicular B cells (Fig. 3A, B).
Figure 3. B-cell-specific deletion of Bmi1 leads to reduction in the B-1a cell pool and loss of self-renewal ability.
The efficiency of CD19-Cre recombinase was confirmed by genomic PCR (A) in FACS-sorted B-1 cells or bead-selected spleen B-cells (B). C: Total peritoneal cell numbers in WT, CD19Cre/+, CD19Cre/+: Bmi1F/+, and CD19Cre/+: Bmi1F/F mice are shown. No obvious difference was found among the 4 groups. D: The numbers of peritoneal B-1a cells from 4 groups at different ages are shown. (n=7-12 for each genotype for each age point, *p<0.05, **p<0.01). E: The number of B-1a cells in CD19Cre/+: Bmi1F/+ and CD19Cre/+: Bmi1F/F mice at 3-9 months of age in the same litter is depicted. The line connects the mice in the same litter. The frequency of lymphoid subsets in the peritoneal cavities (F) and the spleens (G, H) of WT, CD19Cre/+, CD19Cre/+: Bmi1F/+, and CD19Cre/+: Bmi1F/F mice are shown (n=6). I, J: Competitive peritoneal cell transplantation assays. The FACS plots of peritoneal B-1 cells of competitive peritoneal cell-transplanted mice (I) and the donor percentage (J) are shown. K, L: Fetal liver MNC transplantation assays. The percentage of donor-derived lymphoid subsets in the peritoneal cavities of transplanted mice with WT or CD19Cre/+: Bmi1F/F FL MNCs at 6 weeks (K) and 9 months (J) post-transplant (n=3 for each genotyping, at each time point). M: B-1a cell proliferation upon stimulation with TRL7 agonist. Cell number was counted 48 h after stimulation (n=3). MZ: marginal zone B cell, FO: follicular B cell, CD5T: CD5+ T cell. All data were obtained from experiments more than 3 times.
Because CD19 is known to play a key role in B-cell receptor (BCR) signaling, we investigated any adverse effects of CD19 heterozygosity on B-1a cells by comparing the frequency and absolute numbers of B-1a cells and other lymphoid subsets in the peritoneal cavity and spleen of WT, CD19Cre/+, CD19Cre/+: Bmi1F/+, and CD19Cre/+: Bmi1F/F mice at different developmental ages (Fig. 3C). Although the numbers of peritoneal B-1a cells were significantly lower in any of the CD19Cre/+ groups at < 3 months old compared with those in WT mice, after 3 months, only CD19Cre/+: Bmi1F/F knockout mice showed a significant reduction in the numbers of peritoneal B-1a cells among the 4 groups (Fig. 3C, D). When we compared the numbers of B-1a cells in CD19Cre/+: Bmi1F/+control and CD19Cre/+: Bmi1F/F mice in the same litter, the reduction of B-1a cells was clear (Fig. 3E). Therefore, we evaluated the effect of B-cell-specific deletion of Bmi1 in mice older than 3 months. The frequency and absolute number of peritoneal and splenic B-1a cells were significantly reduced in CD19Cre/+: Bmi1F/F mice among the 4 groups, whereas no significant change was seen in the other B-cell subsets (B-1b, B-2 cells) (Fig. 3F-H, Fig. S2A). In addition, no significant change was observed in FL B-1 progenitors (Fig. S2B).
To compare the self-renewal ability of B-1a cells from CD19Cre/+: Bmi1F/F and WT mice in the same microenvironment, we performed competitive transplantation assays. To this end, 1 × 105 peritoneal B-1a cells from Bmi1F/F or CD19Cre/+: Bmi1F/F mice (CD45.2) were injected into the peritoneal cavities of NSG mice along with an equal number of congenic BoyJ (B/J) WT B-1a cells (CD45.1, competitor). We noted that 12 weeks after the injection, CD19Cre/+: Bmi1F/F B-1a cells were able to reconstitute only approximately 8% of total B-1a cells in recipient mice, whereas both the control Bmi1F/F and WT B/J B-1a cells achieved comparable levels of reconstitution (Fig. 3I, J). Thus, the self-renewal ability of B-1a cells was severely impaired in the absence of Bmi1.
The in vivo environment in Bmi1−/− mice permits development and maintenance of B-1a cells
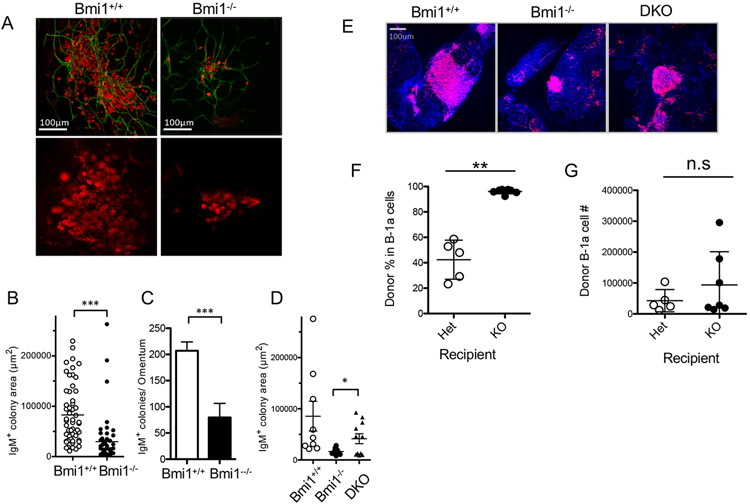
The difference between the severities of the reductions in B-1a cells of Bmi1−/− and CD19Cre/+: Bmi1F/F mice led us to hypothesize that the microenvironment supporting maintenance of B-1a cells might be altered in Bmi1−/− mice, similar to the impaired Bmi1−/− BM environment that reportedly failed to support HSC self-renewal(13, 23, 24). Fat-associated lymphoid clusters (FALCs) have been reported as a niche for B-1 cells (25, 26), with IgM+ cell colonies surrounded by CD31+ endothelial cells (Fig. 4A). In Bmi1−/− mice, the IgM+ lymphoid clusters/colonies were smaller in size and number than were those in WT mice (Fig. 4B, C, p < 0.01). Interestingly, FALCs in the omentum of Bmi1−/−Ink4a-Arf−/− mice were partially rescued in size compared with those in Bmi1−/− mice (Fig. 4D, E). However, it was not clear whether the reduced number and size of FALCs in Bmi1−/− mice was the result of environmental defects or of the reduction of B-1a cells itself. To investigate this, we examined the peritoneal environment of Bmi1−/− mice using reciprocal transplantation assays. Sorted WT B-1a cells were injected into the peritoneal cavity of sublethally irradiated Bmi1−/− and Bmi1+/− mice. Against our expectations, the percentage of donor-derived B-1a cells was greater in Bmi1−/− than in Bmi1+/− recipient mice, although the actual cell numbers were similar (Fig. 4F, G). These results indicated that there were no environmental defects related to the engraftment/maintenance of B-1a cells in the peritoneal cavities of Bmi1−/− mice.
Figure 4. Fat-associated lymphoid clusters (FALCs) in Bmi1−/− peritoneum and reciprocal transplantation assays.
A: Representative pictures of FALCs marked by IgM staining (red) in WT or Bmi1−/− peritoneum. CD31+ endothelial cells are also stained (green). The size (B) and number (C) of FALCs in the omentum are depicted. D: IgM+ colony areas in the omentum among WT, Bmi1−/−, and DKO mice. E: Representative pictures of FALCs in WT, Bmi1−/−, or DKO mice are shown. The percentage (F) and number (G) of donor-derived B-1a cells in recipient peritoneal cavity 6 weeks after reciprocal transplantation assays. Bar=100μm. The data were obtained from 3-4 animals for each genotype from 3 experiments.
Development of B-1a cells from FL progenitors is impaired in the absence of Bmi1
Next, to understand the effect of Bmi1 deletion during development of B-1a cells, we transplanted E14.5 FL mononuclear cells (MNCs) containing B-1 progenitors from Bmi1F/F, CD19Cre/+, or CD19Cre/+: Bmi1F/F embryos into sublethally irradiated adult NSG mice. Accordingly, 9 months after transplantation, CD19Cre/+: Bmi1F/F FL MNCs failed to repopulate the peritoneal B-1a cells, but successfully repopulated the B-2 and B-1b cells to the same levels as did Bmi1F/F FL MNCs (Fig. 3L). Strikingly, defects in the reconstitution of B-1a cells in both the peritoneum and spleen of recipient mice could be observed as early as 6 weeks after transplantation (Fig. 3K, S2C), suggesting that Bmi1 might also be important for the development of B-1a cells from B-1 progenitors in the FL.
TLR-dependent proliferation of B-1a is impaired in the absence of Bmi1
Based on our findings that Bmi1 was essential to B-1a cell self-renewal, we hypothesized that Bmi1-deficient B-1a cells might lose their proliferation ability upon antigen stimulation. To this end, we examined the ability of B-1 cells to proliferate upon stimulation with Toll-like receptors (TLRs). FACS-sorted B-1a cells from each genotype were stimulated in vitro using 5 μg/mL R848 (TRL7/8 agonist) (27). Our results showed that 48 h after stimulation, Bmi1-deficient B-1a cells failed to proliferate, and instead decreased in number, whereas B-1a cells from all control groups showed robust proliferation (Fig. 3M). Taken together, these data suggested that, in addition to its role in self-renewal, Bmi1 was essential for the proliferation of B-1a cells upon stimulation.
Ink4-Arf locus is a target of Bmi1 in B-1a cells
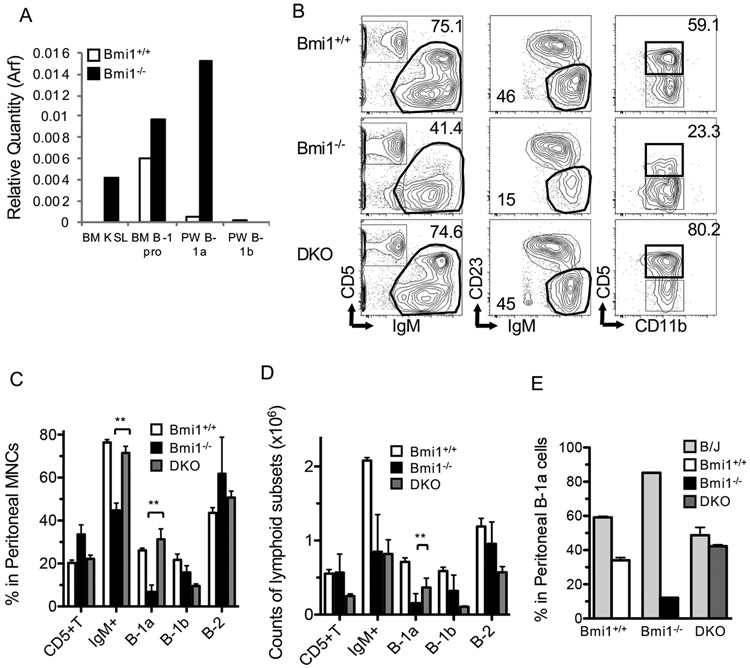
Because the Ink4a-Arf locus is a well-known target of Bmi1 in HSCs, and loss of Ink4-Arf in Bmi1−/− mice has been shown to rescue the numbers of HSCs and their ability to self-renew (13), we hypothesized that Ink4-Arf might also be a target of Bmi1 in B-1a cells. Correspondingly, qPCR analysis showed a marked elevation of the expression of Arf in Bmi1−/− peritoneal B-1a cells (Fig. 5A). In Bmi1−/−Ink4-Arf−/− (DKO) mice, the absolute number of peritoneal cells was significantly higher than that in Bmi1−/− mice, but still lower than that in WT mice, probably due to their smaller body sizes (WT: 2.7± 0.1×106, DKO: 1.2 ± 0.2 × 106, Bmi1−/−: 0.5 ± 0.2 × 106, p < 0.01 between DKO and Bmi1−/−). The percentage of B-1a cells in DKO mice was similar to that in WT mice (Fig. 5B, C), suggesting that deletion of Ink4-Arf in Bmi1−/− mice was able to rescue the maintenance of B-1a cells, although the recovery of absolute numbers of B-1a cells in DKO was only partial (Fig. 5D). When we performed a competitive assay by transplanting DKO along with B/J WT peritoneal B-1a cells into sublethally irradiated NSG mice, DKO cells achieved similar chimerism to that in WT cells, whereas Bmi1−/− B-1a cells failed to repopulate, indicating that deletion of Ink4-Arf in Bmi1−/− B-1a cells completely restored the self-renewal ability of B-1a cells (Fig. 5E). These results demonstrated that the Ink4-Arf locus appears to be a critical target of Bmi1 in B-1a cells and might play a key role in the self-renewal ability of peritoneal B-1a cells.
Figure 5. Arf-Ink4a is a major target for self-renewal of Bmi1−/− B-1a cells.
A: Arf expression in WT and Bmi1−/− B-cells was measured by qPCR. PW: peritoneal wash. B: Representative FACS plots of peritoneal cells from WT, Bmi1−/−, and Bmi1−/−Arf-Ink4−/− (DKO) mice are depicted. C: Frequencies of CD5+ T-cell and B-cell subsets in the peritoneal cavity of WT, Bmi1−/−, and DKO mice are shown (n=3). D: Absolute numbers of the peritoneal subpopulation from the same 3 groups (C) were measured. E: Equal numbers of sorted B-1a cells from WT, Bmi1−/−, and DKO mice were injected into sublethally irradiated NSG mice in a competitive manner against B/J B-1a cells, followed by analysis at 6 weeks. The results of donor contribution are shown. The Bmi1−/− B-1a cells failed to repopulate, while the DKO cells achieved repopulation similar to that of WT (n=3 for each genotyping). The data were obtained from 3 experiments.
Dysregulation of epigenetic gene modifiers in Bmi1−/− B-1a cells suggested Kdm5b as a target of Bmi1
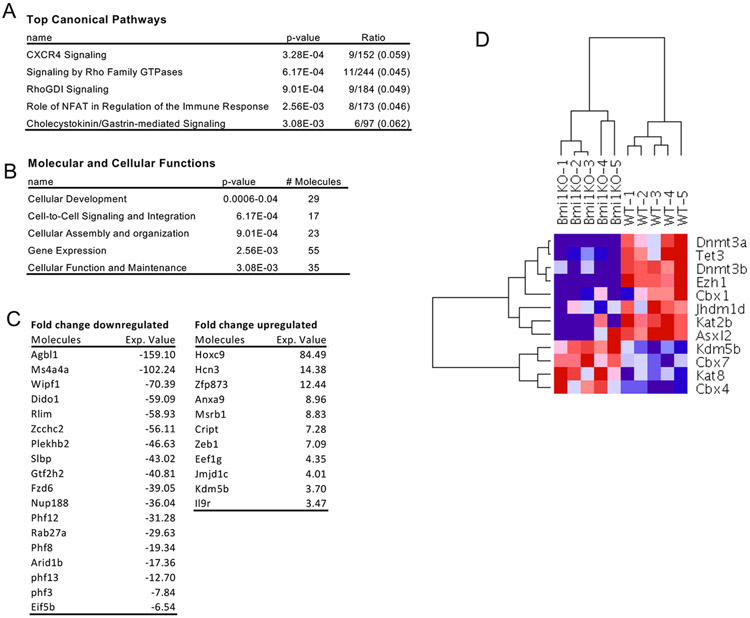
To investigate the Bmi1-related genes involved in the maintenance of B-1a cells, we performed microarray analysis of sorted peritoneal B-1a cells from WT and Bmi1−/− mice. We identified 364 genes that were shown to be significantly changed (> 2-fold) between the 2 groups. Significantly upregulated and downregulated genes are shown in Figure 6. Multiple genes mediating genome integrity and translational control, including AT-rich interaction domain 1B (Arid1b) and PHD finger protein 12 (Phf12), were downregulated (Fig. 6C). Interestingly, various epigenetic modifiers, including DNA methyltransferase 3α (Dnmt3a), 3b, enhancer of zeste 1 polycomb repressive complex 2 subunit (Ezh1), and chromobox 1 (Cbx1), were also downregulated (Fig. 6D).
Figure 6. Gene expression profiling of Bmi1−/− reveals dysregulation of epigenetic genes.
B-1a cells were sorted from WT and Bmi1−/− peritoneal cavity and subjected to microarray analysis. Affected canonical signaling pathways (A) and affected biofunctions (B) analyzed by Ingenuine Pathway Analysis are listed. C. Differentially upregulated and downregulated transcripts are listed. D. Hierarchical clustering of epigenetic genes is shown. n=5 for each group.
Since Bmi1 represents a PRC1 gene repressor, many transcription factors were elevated in Bmi1−/− B-1a cells. Among them, we were interested in Kdm5b because it has been reported to play a role in the self-renewal and proliferation of cells (28). The expression of Kdm5b was significantly increased in the peritoneal B-1a cells, adult LT-HSCs, FL B-1 progenitors, and FL LT-HSCs in the Bmi1−/− mice (Fig. 7AB). However, the expression of Kdm5b was not elevated in CD19Cre/+: Bmi1F/F B-1a cells in steady states (Fig. 7C, pre). Forty-eight hours after transplantation into sublethally irradiated NSG mice, Kdm5b expression significantly increased, and Arf expression in CD19Cre/+: Bmi1F/F peritoneal B-1a cells markedly increased (Fig. 7C). In addition, the expression of p21 (cell cycle repressor) at 48 h was not repressed in CD19Cre/+: Bmi1F/F peritoneal B-1a cells as it was in the control cells (Fig. 7C). Moreover, a higher percentage of dead cells was observed among CD19Cre/+: Bmi1F/F B-1a cells (Fig. 7D). These results suggested that Bmi1-deficient B-1a cells could not proliferate after transplantation, presumably due to overexpression of Arf and p21.
Figure 7. Kdm5b is a possible Bmi1 target and crucial for post-transplant B-1a cell expansion.
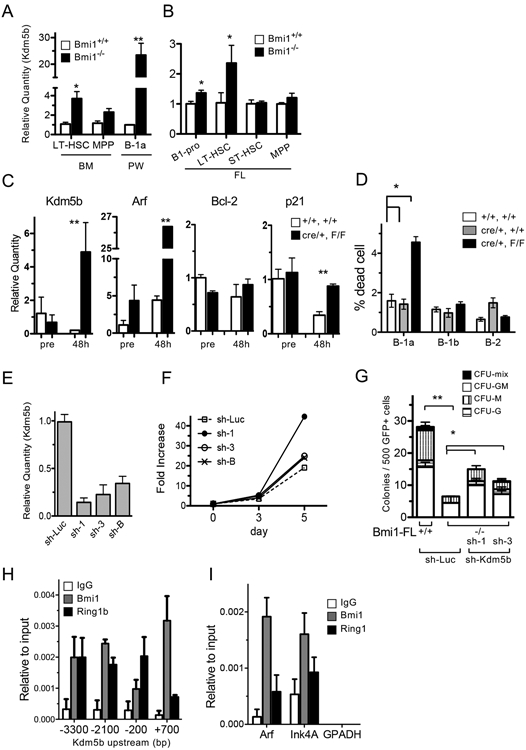
Expression of Kdm5b in various cell fractions from WT and Bmi1−/− BM and peritoneum (A) or FL (B). (n=3, *p<0.03, **p<0.01) C: Sorted B-1a cells from CD19Cre/+: Bmi1F/F or control mice were injected into sub-lethally irradiated NSG mice, followed by harvesting of B-1a cells 48 h after transplantation by FACS sorting. Changes in the expression of various mRNAs before and after transplantation (day 2) were measured (n=3, **p<0.01). D: Cell death in experiment (C) was measured by Annexin-V and DAPI staining. n=3. E: Confirmation of knockdown of Kdm5b. Sh-RNAs targeting kdm5b or control (sh-Luc) were introduced retrovirally into Baf/3 pro-B cells, and GFP+ cells were sorted. The quantity of Kdm5b mRNA in each clone is shown. F: Cell proliferation of each Baf/3 subline targeting Kdm5 is shown. G: Colony forming assays of Kdm5 knockdown Bmi1−/− FL cells. Bmi1−/− FLs with sh-Kdm5b formed more colonies than did the sh-control (n=3, **p<0.01, *p<0.05). H, I: ChIP assays were performed in Baf3 cells to confirm Bmi1 binding to the Kdm5b or Arf-Ink4 regions. Binding to upstream regions of Kdm5b (H) and Arf-Ink4a (I) is shown. The data were obtained from 3 experiments.
To understand the molecular role of Kdm5b in cell proliferation, we knocked down the expression of Kdm5 in the Baf3 pro-B cell line using a sh-Kdm5b retroviral construct (Fig. 7E). Sh-Kdm5b-transduced cells showed more proliferation than did control cells (sh-Luc) (Fig. 7F). Next, we introduced the viral sh construct into Bmi1−/− FL cells and performed colony forming assays. The number of colonies that formed from Bmi1−/− FL cells was significantly lower than that formed from WT cells, as previously reported (12) (Fig. 7G). However, the knockdown of Kdm5b in Bmi1−/− FLs induced an increase in the number of colonies formed (Fig. 7G). Thus, knockdown of Kdm5b led to enhanced cell proliferation in Baf3 and FL cells. In addition, a chromatin immunoprecipitation assay showed that Bmi1 bound to the upstream region of Kdm5b (Fig. 7H), as well as Arf, a known target of Bmi1 (Fig.7I). We also confirmed that ring finger protein 1b (Ring1b), a main component of the PRC1 complex protein, bound to the same regions. Collectively, our data suggested that Kdm5b appears to be a downstream target of Bmi1, and an optimal low expression level of Kdm5b seems to be important for cell proliferation.
Discussion
B-1a lymphocytes are derived from embryonic precursors and maintained in the long-term through their self-renewal ability independently of HSCs (5-9). However, despite extensive studies on the development and function of B-1 cells, the molecular mechanism of B-1a cell self-renewal remains largely unknown. Traditionally, the spleen is known as the organ important for the maintenance of B-1a cells, in addition to their specific BCR and CD19 signaling, all of which continuously stimulate B-1a cells (22, 29-31). A recent study showed that deletion of the basic helix-loop-helix family member E41 (Bhlhe41) transcription factor significantly reduced the number of peritoneal B-1a cells, likely by altering BCR signaling and preventing the maturation of transitional B-1 progenitors in the spleen (32). Another interesting report has demonstrated that the autophagy-related 7 (Atg7) gene was required for the self-renewal of B-1a cells, because of their specific metabolic status that is different from that of B-2 cells (33). These findings have gradually started to elucidate the mechanisms that support B-1a cell self-renewal and suggest its complexity. In this study, we demonstrated that expression of Bmi1 was indispensable for the self-renewal ability of peritoneal B-1a cells. The importance of this finding is that mature IgM-secreting B-1a lymphocytes utilize BMI1 for their self-renewal ability in a similar way to HSCs.
Bmi1, a component of PRC1, is a widely known critical factor in the self-renewal of various stem cells. Importantly, Bmi1 is also considered to be involved in lymphopoiesis, because a large reduction has been observed in lymphocyte counts in the spleen and thymus of Bmi1−/− mice (18). However, the frequency and number of B-2 lymphocytes were not altered when Bmi1 was deleted in the B-cell lineage (using CD19-Cre mice). Therefore, it seems that a defect in stem cells (differentiation to B-progenitors) mainly contributed to the reduction of B-2 cells in Bmi1−/− mice, and that among the mature B cell subsets, only B-1a cells depend on Bmi1 expression. In the field of HSC biology, self-renewal ability is confirmed only by transplantation assays. Therefore, we carried out peritoneal cell transfer assays in which wild-type peritoneal B-1a cells repopulate the recipient peritoneal cavity in the long-term, and we further demonstrated a defect in the self-renewal ability of Bmi1-deficient B-1a cells, which was restored by additional deletion of Ink4-Arf. Proliferation of Bmi1-deficient B-1a cells was also impaired after exposure to a TLR7/8 agonist, as well as after transplantation. Bmi1-deficient B-1a cells showed upregulation of Arf and failure to downregulate p21 after stimulation, which suggested that Bmi1 maintains the self-renewal ability of B-1a cells through the Arf-p21 axis. Although Bmi1 seems to play a role in the self-renewal of B-1a cells similar to its role in HSCs, we have further identified Kdm5b as a possible target of Bmi1 in B-1a cells. The KDM5B histone modifier specifically demethylates H3 at lysine 4 and is known to be required for self-renewal of embryonic stem cells, HSC function, and leukemia/cancer progression, and plays different roles depending on cell type and physiological context (28, 34-36). Our results showed that Kdm5b was upregulated in Bmi1-deficient B-1a cells in a stress setting, but not in steady state, suggesting that Kdm5b suppresses proliferation of B-1a cells in the absence of Bmi1. Indeed, knockdown of Kdm5b enhanced proliferation of Baf3 cells and rescued the colony-forming ability of Bmi1−/− FL cells (Fig. 7F, G). Therefore, it can be speculated that B-1a cells fail to proliferate and undergo apoptosis upon antigen exposure in the absence of Bmi1 followed by upregulated Kdm5b. As such, further investigation on the precise roles of Kdm5b in B-1a cells will be required.
Furthermore, FL MNCs that contain Bmi1-deleted B-progenitors failed to repopulate only B-1a cells and not other mature B-cell subsets, even at an early time point after transplantation. This result suggests a possibility that Bmi1 may also be required for the developmental process of fetal B-1 progenitor cells into mature B-1a cells. In addition, it is notable that overexpressing Bmi1 in Bmi1−/− FL MNCs not only rescued but further enhanced B-1a cell repopulation ability (Fig. 2E). This result is consistent with previous reports showing enhanced self-renewal after overexpressing Bmi1 in BM HSCs and erythroblasts (12, 16). Thus, Bmi1 seems to enhance both B-1a cell differentiation from FL progenitors and self-renewal of mature B-1a cells. Since the developmental pathway from FL B-1 progenitors to mature peritoneal B-1a cells has not been fully characterized, further study to determine the stage at which Bmi1 might be involved in the differentiation into B-1a cells is required.
Recent reports have shown the important role of Bmi1 in controlling the fat volume in the HSC niche(23, 24). We also sought to investigate the effect of the lack of Bmi1 on FALCs, a niche for B-1a cells, but found that the microenvironment of B-1a cells was not altered. Therefore, Bmi1 appears to maintain the self-renewal ability of B-1a cells in a cell-intrinsic manner.
In summary, our studies here showed that the self-renewal ability of B-1a lymphocytes depends heavily on Bmi1 expression, and this dependency is not observed in other mature lymphocyte subsets. Since B-1a cells are not replenished by adult BM progenitors, the loss of B-1a cells after bone marrow transplantation might thus result in acquired immunodeficiency (37). The knowledge presented here could pave the way to fully elucidate the mechanisms of the in vivo self-renewal of B-1a cells, an important step toward producing pluripotent stem cell-derived B-1 cells for immune cell therapy (17).
Supplementary Material
Key points.
Bmi1 regulates self-renewal ability of B-1a cells via Ink4-Arf and possibly Kdm5b.
Bmi1 may be involved in the developmental process of B-1 progenitors to B-1a cells.
FALCs, the niche for B-1a cells, are not altered in Bmi1−/− mice.
Acknowledgments
This work was supported by NIAID R56AI110831 and NIAID R01AI121197 (MY).
Abbreviations used in this article:
- BM
bone marrow
- DKO
double knockout
- FALCs
fat-associated lymphoid clusters
- FL
fetal liver
- LT
long-term
- HSC
hematopoietic stem cell
- Kdm5b
lysine demethylase 5B
- NSG
NOD/SCID/IL2Rγc−/−
- WT
wild-type
- MNC
mononuclear cell
- PRC1
polycomb repressor complex 1
Footnotes
Disclosures: The authors have no financial conflicts of interest.
References
- 1.Herzenberg LA, Stall AM, Lalor PA, Sidman C, Moore WA, and Parks DR. 1986. The Ly-1 B cell lineage. Immunological reviews 93: 81–102. [DOI] [PubMed] [Google Scholar]
- 2.Hardy RR, and Hayakawa K. 1991. A developmental switch in B lymphopoiesis. Proc Natl Acad Sci U S A 88: 11550–11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montecino-Rodriguez E, Leathers H, and Dorshkind K. 2006. Identification of a B-1 B cell-specified progenitor. Nat Immunol 7: 293–301. [DOI] [PubMed] [Google Scholar]
- 4.Barber CL, Montecino-Rodriguez E, and Dorshkind K. 2011. Reduced production of B-1-specified common lymphoid progenitors results in diminished potential of adult marrow to generate B-1 cells. Proceedings of the National Academy of Sciences of the United States of America 108: 13700–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosn EE, Yamamoto R, Hamanaka S, Yang Y, Herzenberg LA, Nakauchi H, and Herzenberg LA. 2012. Distinct B-cell lineage commitment distinguishes adult bone marrow hematopoietic stem cells. Proc Natl Acad Sci U S A 109: 5394–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosn EE, Waters J, Phillips M, Yamamoto R, Long BR, Yang Y, Gerstein R, Stoddart CA, Nakauchi H, and Herzenberg LA. 2016. Fetal Hematopoietic Stem Cell Transplantation Fails to Fully Regenerate the B-Lymphocyte Compartment. Stem Cell Reports 6: 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi M, Shelley WC, Seo W, Vemula S, Lin Y, Liu Y, Kapur R, Taniuchi I, and Yoshimoto M. 2014. Functional B-1 progenitor cells are present in the hematopoietic stem cell-deficient embryo and depend on Cbfbeta for their development. Proc Natl Acad Sci U S A 111: 12151–12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshimoto M, Montecino-Rodriguez E, Ferkowicz MJ, Porayette P, Shelley WC, Conway SJ, Dorshkind K, and Yoder MC. 2011. Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc Natl Acad Sci U S A 108: 1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawai CM, Babovic S, Upadhaya S, Knapp DJ, Lavin Y, Lau CM, Goloborodko A, Feng J, Fujisaki J, Ding L, Mirny LA, Merad M, Eaves CJ, and Reizis B. 2016. Hematopoietic Stem Cells Are the Major Source of Multilineage Hematopoiesis in Adult Animals. Immunity 45: 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, and Morrison SJ. 2003. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425: 962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, and Clarke MF. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302–305. [DOI] [PubMed] [Google Scholar]
- 12.Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, Tsukui H, Ema H, Kamijo T, Katoh-Fukui Y, Koseki H, van Lohuizen M, and Nakauchi H. 2004. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity 21: 843–851. [DOI] [PubMed] [Google Scholar]
- 13.Oguro H, Iwama A, Morita Y, Kamijo T, van Lohuizen M, and Nakauchi H. 2006. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. The Journal of experimental medicine 203: 2247–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Foggia V, Zhang X, Licastro D, Gerli MF, Phadke R, Muntoni F, Mourikis P, Tajbakhsh S, Ellis M, Greaves LC, Taylor RW, Cossu G, Robson LG, and Marino S. 2014. Bmi1 enhances skeletal muscle regeneration through MT1-mediated oxidative stress protection in a mouse model of dystrophinopathy. J Exp Med 211: 2617–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, Sangiorgi E, Capecchi MR, and Kuo CJ. 2012. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A 109: 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim AR, Olsen JL, England SJ, Huang YS, Fegan KH, Delgadillo LF, McGrath KE, Kingsley PD, Waugh RE, and Palis J. 2015. Bmi-1 Regulates Extensive Erythroid Self-Renewal. Stem Cell Reports 4: 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y, Kobayashi M, Azevedo Portilho N, Mishra A, Gao H, Liu Y, Wenzel P, Davis B, Yoder MC, and Yoshimoto M. 2019. Long-Term Engraftment of ESC-Derived B-1 Progenitor Cells Supports HSC-Independent Lymphopoiesis. Stem Cell Reports 12: 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, Hulsman D, Leung C, Arsenijevic Y, Marino S, and van Lohuizen M. 2005. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev 19: 1438–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao R, Chen S, Kobayashi M, Yu H, Zhang Y, Wan Y, Young SK, Soltis A, Yu M, Vemula S, Fraenkel E, Cantor A, Antipin Y, Xu Y, Yoder MC, Wek RC, Ellis SR, Kapur R, Zhu X, and Liu Y. 2015. Bmi1 promotes erythroid development through regulating ribosome biogenesis. Stem Cells 33: 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rickert RC, Roes J, and Rajewsky K. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res 25: 1317–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rickert RC, Rajewsky K, and Roes J. 1995. Impairment of T-cell-dependent B-cell responses and B-1 cell development in CD19-deficient mice. Nature 376: 352–355. [DOI] [PubMed] [Google Scholar]
- 22.Sato S, Ono N, Steeber DA, Pisetsky DS, and Tedder TF. 1996. CD19 regulates B lymphocyte signaling thresholds critical for the development of B-1 lineage cells and autoimmunity. J Immunol 157: 4371–4378. [PubMed] [Google Scholar]
- 23.Hu T, Kitano A, Luu V, Dawson B, Hoegenauer KA, Lee BH, and Nakada D. 2019. Bmi1 Suppresses Adipogenesis in the Hematopoietic Stem Cell Niche. Stem Cell Reports 13: 545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato Y, Hou LB, Miyagi S, Nitta E, Aoyama K, Shinoda D, Yamazaki S, Kuribayashi W, Isshiki Y, Koide S, Si S, Saraya A, Matsuzaki Y, van Lohuizen M, and Iwama A. 2019. Bmi1 restricts the adipogenic differentiation of bone marrow stromal cells to maintain the integrity of the hematopoietic stem cell niche. Exp Hematol 76: 24–37. [DOI] [PubMed] [Google Scholar]
- 25.Benezech C, Luu NT, Walker JA, Kruglov AA, Loo Y, Nakamura K, Zhang Y, Nayar S, Jones LH, Flores-Langarica A, McIntosh A, Marshall J, Barone F, Besra G, Miles K, Allen JE, Gray M, Kollias G, Cunningham AF, Withers DR, Toellner KM, Jones ND, Veldhoen M, Nedospasov SA, McKenzie AN, and Caamano JH. 2015. Inflammation-induced formation of fat-associated lymphoid clusters. Nat Immunol 16: 819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J, Ohtani M, Fujii H, and Koyasu S. 2010. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature 463: 540–544. [DOI] [PubMed] [Google Scholar]
- 27.Genestier L, Taillardet M, Mondiere P, Gheit H, Bella C, and Defrance T. 2007. TLR agonists selectively promote terminal plasma cell differentiation of B cell subsets specialized in thymus-independent responses. J Immunol 178: 7779–7786. [DOI] [PubMed] [Google Scholar]
- 28.Cellot S, Hope KJ, Chagraoui J, Sauvageau M, Deneault E, MacRae T, Mayotte N, Wilhelm BT, Landry JR, Ting SB, Krosl J, Humphries K, Thompson A, and Sauvageau G. 2013. RNAi screen identifies Jarid1b as a major regulator of mouse HSC activity. Blood 122: 1545–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kretschmer K, Stopkowicz J, Scheffer S, Greten TF, and Weiss S. 2004. Maintenance of peritoneal B-1a lymphocytes in the absence of the spleen. J Immunol 173: 197–204. [DOI] [PubMed] [Google Scholar]
- 30.Pedersen GK, Li X, Khoenkhoen S, Adori M, Beutler B, and Karlsson Hedestam GB. 2018. B-1a Cell Development in Splenectomized Neonatal Mice. Front Immunol 9: 1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wardemann H, Boehm T, Dear N, and Carsetti R. 2002. B-1a B cells that link the innate and adaptive immune responses are lacking in the absence of the spleen. J Exp Med 195: 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kreslavsky T, Vilagos B, Tagoh H, Poliakova DK, Schwickert TA, Wohner M, Jaritz M, Weiss S, Taneja R, Rossner MJ, and Busslinger M. 2017. Essential role for the transcription factor Bhlhe41 in regulating the development, self-renewal and BCR repertoire of B-1a cells. Nat Immunol 18: 442–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke AJ, Riffelmacher T, Braas D, Cornall RJ, and Simon AK. 2018. B1a B cells require autophagy for metabolic homeostasis and self-renewal. J Exp Med 215: 399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kidder BL, Hu G, Yu ZX, Liu C, and Zhao K. 2013. Extended self-renewal and accelerated reprogramming in the absence of Kdm5b. Mol Cell Biol 33: 4793–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Song C, Ding Y, Pan X, Ge Z, Tan BH, Gowda C, Sachdev M, Muthusami S, Ouyang H, Lai L, Francis OL, Morris CL, Abdel-Azim H, Dorsam G, Xiang M, Payne KJ, and Dovat S. 2016. Transcriptional Regulation of JARID1B/KDM5B Histone Demethylase by Ikaros, Histone Deacetylase 1 (HDAC1), and Casein Kinase 2 (CK2) in B-cell Acute Lymphoblastic Leukemia. J Biol Chem 291: 4004–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart MH, Albert M, Sroczynska P, Cruickshank VA, Guo Y, Rossi DJ, Helin K, and Enver T. 2015. The histone demethylase Jarid1b is required for hematopoietic stem cell self-renewal in mice. Blood 125: 2075–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moins-Teisserenc H, Busson M, Herda A, Apete S, Peffault de Latour R, Robin M, Xhaard A, Toubert A, and Socie G. 2013. CD19(+)CD5(+) B Cells and B1-Like Cells Following Allogeneic Hematopoietic Stem Cell Transplantation. Biol Blood Marrow Transplant 19: 988–991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.