Abstract
Purpose
Lung cancer remains the leading cancer-associated deaths worldwide. Cisplatin (CDDP) was used in combination with curcumin (CUR) for the treatment of non-small cell lung cancer. The aim of this study was to prepare and characterize CDDP prodrug and CUR co-encapsulated layer-by-layer nanoparticles (CDDP-PLGA/CUR LBL NPs) to induce cooperative response, maximize the therapeutic effect, overcome drug resistance, and reduce adverse side effects.
Methods
CDDP prodrug (CDDP-PLGA) was synthesized. CDDP-PLGA/CUR LBL NPs were constructed and their physicochemical properties were investigated by particle-size analysis, zeta potential measurement, drug loading, drug entrapment efficiency, and in vitro drug release behavior. In vitro cytotoxicity against human lung adenocarcinoma cell line (A549 cells) was investigated, and in vivo anti-tumor efficiency of CDDP-PLGA/CUR LBL NPs was evaluated on mice bearing A549 cell xenografts.
Results
CDDP-PLGA/CUR LBL NPs have a size of 179.6 ± 6.7 nm, a zeta potential value of −29.9 ± 3.2 mV, high drug entrapment efficiency of 85.6 ± 3.9% (CDDP) and 82.1 ± 2.8% (CUR). The drug release of LBL NPs exhibited a sustained behavior, which made it an ideal vehicle for drug delivery. Furthermore, CDDP-PLGA/CUR LBL NPs could significantly enhance in vitro cytotoxicity and in vivo antitumor effect against A549 cells and lung cancer animal model compared to the single drug-loaded LBL NPs and free drug groups.
Conclusion
CDDP-PLGA/CUR LBL NPs were reported for the first time in the combination therapy of lung cancer. The results demonstrated that the CDDP-PLGA/CUR LBL NPs might be a novel promising system for the synergetic treatment of lung carcinoma.
Keywords: lung cancer, combination therapy, layer-by-layer, cisplatin prodrug, curcumin
Introduction
Lung cancer is the leading cause of cancer-related death worldwide.1 Non-small cell lung cancer (NSCLC) is the most frequent lung cancer of all types, which accounts for about 85% of all cases of lung cancer.2,3 Although some progresses have been made in radiotherapy, targeted therapy and immunotherapy of NSCLC, the overall survival rate has only slightly improved.4,5 Currently, cisplatin (CDDP) based chemotherapy has become a first-line adjuvant therapy strategy for NSCLC patients after surgical resection.6 However, drug resistance has become a major obstacle to cancer treatment. Polymeric conjugates of conventional drugs (polymeric drug conjugates) have several advantages over their low molecular weight precursors.7 Because of the advantages over free-form drugs, polymer-drug conjugates have led to a new era of polymer drug delivery systems.8,9 One of the most widely used polymers is the biodegradable and biocompatible poly(D,L-lactide-co-glycolide) (PLGA), which has been approved by the FDA for certain human clinical uses.10 In the present study, PLGA was applied to conjugate with CDDP to form a prodrug (CDDP-PLGA).
Combination chemotherapy is preferred over treatment with single agents to combat most cancers as it targets multiple cell-survival pathways at the same time and delays the onset of resistance.11 Combined chemotherapy can regulate different signaling pathways in cancer cells, induce cooperative response, maximize the therapeutic effect, overcome drug resistance, and become increasingly important for achieving long-term prognosis and reducing adverse side effects.12 Curcumin (CUR) is a hydrophobic polyphenol, derived from the plant curcuma longa (turmeric), with low intrinsic toxicity.13 CUR has been widely studied for its anti-inflammatory, anti-angiogenic, antioxidant, wound healing and anti-cancer effects.14 Due to its water-insolubility and instability, CUR has been loaded into liposomes, polymers or nanoparticles to improve its water-solubility, stability and thus bioavailability.15,16 Furthermore, CUR has been co-delivered with doxorubicin, paclitaxel and docetaxel for combination therapy of cancer.17–19 So in this research, CUR was combined with CDDP prodrug for the NSCLC treatment.
Layer-by-layer (LBL) technology is a versatile method to develop multilayer films by the electrostatic attraction of oppositely charged polyelectrolytes.20 This method of alternative deposition of polyelectrolytes has become a new method to functionalize the surface of nanoparticles or to form a core-shell nanoparticle.21 The diversity of the interactions including electrostatic attraction, hydrogen bonding, and chemical reactions been used to produce multilayer films allows a broad range of materials to be used to fabricate an array of functional LBL materials for various applications, such as drug delivery and tissue engineering.22 In particular, drug delivery systems prepared by LBL deposition of polyelectrolytes can significantly promote the delivery of therapeutic proteins by increasing tolerance to extended shelf storage and drug loading.23 LBL nanoparticle platforms have been applied for cancer active targeting,24 including the delivery of CDDP alone or together with other drugs to the tumor site.25,26
In our previous study, doxorubicin and curcumin were co-delivered by polymeric nanocarriers.27 In this paper, we developed a CDDP prodrug (CDDP-PLGA) and CUR co-encapsulated, LBL, lipid-polymer hybrid nanoparticles (CDDP-PLGA/CUR LBL NPs) for the combination therapy of lung cancer to induce cooperative response, maximize the therapeutic effect, overcome drug resistance, and reduce adverse side effects. The in vitro and in vivo anticancer effects of CDDP-PLGA/CUR LBL NPs were evaluated in comparison with non-LBL polymeric nanoparticles. The combination efficiency of CDDP-PLGA/CUR LBL NPs was also investigated compared with single drug-loaded LBL nanoparticles. CDDP-PLGA/CUR LBL NPs were reported for the first time in the combination therapy of lung cancer and was expected to be a novel promising system for the synergetic treatment of lung carcinoma.
Materials and Methods
Materials
Poly (D,L-lactic-co-glycolic) (PLGA, 75:25, MW 17000) was purchased from Shanghai ZZBIO Co., Ltd. (Shanghai, China). CDDP, glyceryl monostearate (GMS), poly(vinyl alcohol) (PVA), dimethyl sulfoxide (DMSO), coumarin-6 (Cou-6), and 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) were obtained from Sigma–Aldrich Co. (St. Louis, MO, USA). Fetal Bovine Serum (FBS) and Roswell Park Memorial Institute medium (RPMI 1640) were purchased from Invitrogen Corporation (Carlsbad, CA). Human NSCLC cell line: A549 and NCI-H1299 cells; human lung epithelial cell line: BEAS-2B cells were obtained from the American Type Culture Collection. Female BALB/c nude mice (15–20 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). All animal experiments should comply with the ARRIVE guidelines and should be carried out in accordance with the U. K. Animals (Scientific Procedures) Act, 1986 and associated guidelines, EU Directive 2010/63/EU for animal experiments and were approved by the Animal Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University.
Synthesis of PLGA Conjugated CDDP Prodrug
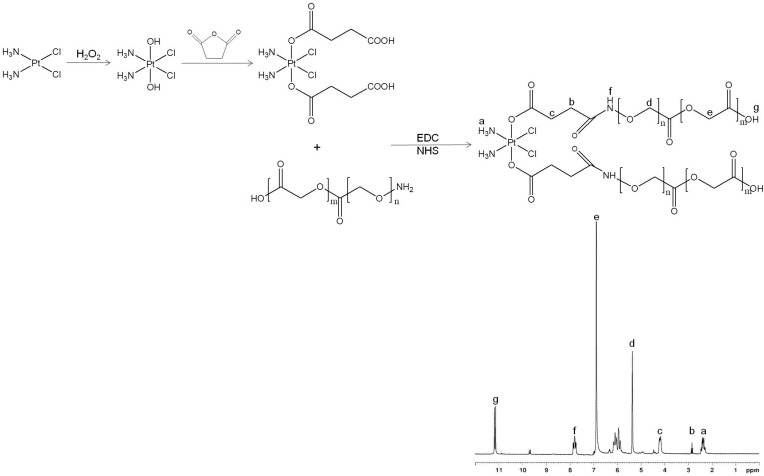
PLGA conjugated CDDP prodrug (CDDP-PLGA) was synthesized by amidation of the carboxyl groups of CDDP with the amine groups of PLGA (Figure 1). CDDP-(COOH)2 was firstly prepared by adding H2O2 to CDDP (dissolved in DMSO) to get CDDP-(OH)2, then succinic anhydride was added to CDDP-(OH)2 and stirred for 10 h.28 EDC·HCl and NHS were added into CDDP-(COOH)2 solution, followed by adding PLGA-NH2 into the solution and stirred for 24 h to form CDDP-PLGA.29 CDDP-PLGA was then dialyzed against water for 20 h and lyophilized. 1H-NMR and FTIR analysis were applied to determine the structure of CDDP-PLGA.
Figure 1.
Synthesis route and 1H NMR spectrum of PLGA conjugated CDDP prodrug (CDDP-PLGA). CDDP-PLGA was synthesized by amidation of the carboxyl groups of CDDP with the amine groups of PLGA.
Abbreviations: PLGA, poly(D,L-lactic-co-glycolic); CDDP, cisplatin.
Preparation of CDDP-PLGA/CUR LBL NPs
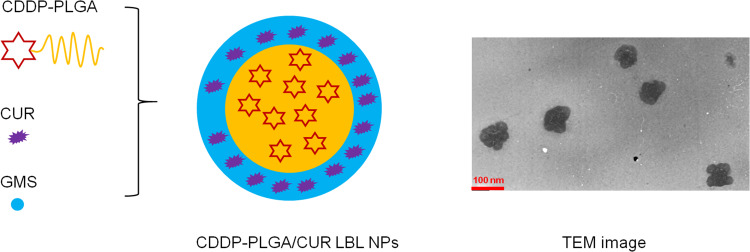
CDDP-PLGA/CUR LBL NPs were prepared by a solvent diffusion technique (Figure 2).26 Briefly, CDDP-PLGA (200 mg) was dissolved in acetone and added into PVA (2%, w/v) solution under gentle stirring. CUR (30 mg) and GMS (100 mg) were dissolved in acetone solution and added drop-by-drop into the CDDP-PLGA solution. To remove the organic solvent, the mixture was stirred overnight at room temperature.30 The final solution was centrifuged at 15,000 rpm for 20 min and washed three times with water for injection. To prepare CDDP (not prodrug) and CUR co-encapsulated LBL, lipid-polymer hybrid nanoparticles (CDDP/CUR LBL NPs), CDDP (30 mg) and PLGA (170 mg) was used to replace CDDP-PLGA, and CUR was excluded from the preparation process. To prepare drug-free LBL, lipid-polymer hybrid nanoparticles (LBL NPs), PLGA was used to replace CDDP-PLGA and CUR was excluded from the preparation process. To prepare single drug (CDDP or CUR) encapsulated LBL NPs (CDDP LBL NPs or CUR LBL NPs), another drug (CUR or CDDP) was excluded from the preparation process.
Figure 2.
Scheme and TEM image of CDDP-PLGA/CUR LBL NPs. CDDP-PLGA/CUR LBL NPs were prepared by a solvent diffusion technique.
Abbreviations: TEM, transmission electronic microscopy; CDDP, cisplatin; PLGA, poly(D,L-lactic-co-glycolic); CUR, curcumin; LBL, layer-by-layer; NPs, lipid-polymer hybrid nanoparticles.
Characterization of CDDP-PLGA/CUR LBL NPs
The prepared CDDP-PLGA/CUR LBL NPs were dispersed air dried, stained with sodium phosphotungstate, and observed on a JEM transmission electronic microscopy (TEM) microscope (JEOL Ltd., Tokyo, Japan).31 The particle size, polydispersity index (PDI) and zeta potential of LBL NPs were determined with dynamic light scattering (DLS; Malvern Instruments, Malvern, UK).32 To measure the drug encapsulation and loading efficiency (EE and LE), LBL NPs samples were added to 1 mL deionized water (pH 7.4), sonicated (10 min) and centrifuged (15,000 rpm, 10 min, 4°C). CDDP concentration was measured by UV–visible spectrophotometric method.33 LBL NPs samples were heated at 90°C for 30 min with o-phenylenediamine (OPDA) solution in dimethylformamide (DMF) and estimated at 705 nm on the UV–visible spectrophotometer (UV-1800, Shimadzu, Japan). CUR concentration was measured with an HPLC method at 420 nm using Agilent 1260 Infinity LC (Agilent Technologies, Santa Clara, CA).34 EE and LE were calculated using formulations: EE (%) = weight of loading drugs/weight of feeding drugs ×100; LE (%) = weight of loading drugs/weight of LBL-NPs ×100.
In vitro Drug Release from CDDP-PLGA/CUR LBL NPs
In vitro release of CDDP or CUR from LBL NPs was conducted by dialysis bag diffusion method.35 Briefly, CDDP-PLGA/CUR LBL NPs, CDDP/CUR LBL NPs, CDDP LBL NPs, or CUR LBL NPs suspensions (1 mL) were sealed in dialysis bags (Molecular weight cut-off 10 KDa), respectively. Then, the bays were placed in degassed PBS solution (pH 7.4, 10 mL) which was degassing with nitrogen in the presence of NADPH (100 mM) for the entire period of the release experiment to maintain the hypoxic condition under constantly shaken (100 rpm).36 The release medium (0.5 mL) was taken out at determined time intervals and the amount of CDDP or CUR released was evaluated as the method in “Characterization of CDDP-PLGA/CUR LBL NPs” section. The same amount of fresh release medium (0.5 mL) was added to maintain sink conditions. The normal condition (non-hypoxic condition) was prepared using PBS (pH 7.4) containing 100 mM NADPH without degassing as control.
In vitro Cytotoxicity of CDDP-PLGA/CUR LBL NPs
In vitro cytotoxicity of LBL NPs on A549 cells was assessed by the MTT assay.37 Briefly, A549 cells, NCI-H1299 cells, or BEAS-2B cells (5×104 cells/mL, 200 µL) were seeded into 96-well plates and incubated with Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% FBS and Penicillin-Streptomycin (100 μg/mL) for 24 h. Then, CDDP-PLGA/CUR LBL NPs, CDDP/CUR LBL NPs, CDDP LBL NPs, CUR LBL NPs, LBL NPs, free CDDP/CUR, free CDDP and free CUR in different concentrations were added and incubated for 72 h. MTT solution (5 mg/mL, 200 µL/well) was added and incubated at 37°C for another 4 h. DMSO (150 µL) was added into each well after the removal of medium and shaken for 10 min. A microplate reader was utilized to record the absorbance at the wavelength of 570 nm.
Cellular Uptake of CDDP-PLGA/CUR LBL NPs
Cou-6 was used as a fluorescent probe to determine the cellular uptake efficiency of LBL-NPs.38 Cou-6 containing LBL NPs were prepared using the method in section 2.3 using Cou-6 (0.5 mg) dissolved in acetone along with PLGA or CDDP-PLGA. A549 cells, NCI-H1299 cells, or BEAS-2B cells (1×106 cells/well) were seeded in 6-well culture plate and incubated at 37°C for 24 h.39 CDDP-PLGA/CUR LBL NPs, CDDP/CUR LBL NPs, CDDP LBL NPs, and CUR LBL NPs (200 μg/mL) were added to the wells and incubated for 1 h. The cellular uptake efficiency of LBL NPs was visualized using a fluorescent-inverted microscope. A flow cytometer (BD Biosciences, Franklin Lakes, NJ) was applied to quantitatively analyze cellular uptake of LBL NPs after the cells were washed three times with D-Hank’s solution, collected and centrifuged at 1500 rpm for 5 min.40
In vivo Anti-Tumor Efficacy of CDDP-PLGA/CUR LBL NPs
A549 cells (2×106 cells/mouse) were subcutaneous implanted into the right flank region of the BALB/c nude mice and let the tumor volumes grow to about 50 mm3.41 The tumor volumes were calculated using formulation: V (mm3) = 1/2×(length)×(width)2.42 Then, the mice were randomized into 8 groups (n = 6) and (1) 0.9% saline as a control, (2) CDDP-PLGA/CUR LBL NPs, (3) CDDP/CUR LBL, (4) CDDP LBL NPs, (5) CUR LBL NPs, (6) free CDDP/CUR, (7) free CDDP, (8) free CUR (10 mg CDDP and/or CUR per kg body weight) were intravenously injected on day 0, 4, 8, 12 and 16. The behaviors of mice were monitored every 12 h along with the tumor growth and tumor volumes were measured with a vernier caliper every 3 days. After 21 days of injection, mice were sacrificed by cervical decapitation and the final tumor weight and body weight of mice were recorded to calculate the anti-tumor activity.
In vivo Tissue Distribution of CDDP-PLGA/CUR LBL NPs
Tumor-bearing BALB/c nude mice were randomly divided into 3 groups (n = 6) and (1) CDDP-PLGA/CUR LBL NPs, (2) CDDP/CUR LBL, (3) free CDDP/CUR (10 mg CDDP and/or CUR per kg body weight) were intravenously injected. Mice were sacrificed at 24 h after intravenous injection.43 The tumor tissue, lung, heart, kidney, liver, and spleen were harvested, decomposed on heating in nitric acid, evaporated to dryness, and redissolved in acetonitrile and water (50/50, v/v) solution.44 The distribution of CDDP and CUR was evaluated as the method in “Characterization of CDDP-PLGA/CUR LBL NPs” section.
Statistical Analysis
The data of the studies were expressed as the mean ± standard deviation (mean ± SD) and statistical analysis was performed using a post hoc test following ANOVA. P < 0.05 was considered statistically significant (*).
Results
Synthesis of CDDP-PLGA
Figure 1 illustrates the 1H NMR spectrum of CDDP-PLGA in accordance with the chemical structure. The peak (a) 2.35 ppm belongs to CDDP; and the peaks at (d) 5.31, (e) 6.85, and (g) 11.18 ppm are marked on the PLGA structure. Peaks (b) 2.83 and (f) 7.81 are the protons of CH2 next to the amide linkage and the NH in the amide linkage. These peaks could prove the existence of the CDDP, PLGA and amide linkage. IR spectrum (ν/cm−1): peaks at 3282, 2933, and 1306 belong to CDDP; PLGA have the peaks of 1763, 1427, and 1093. The peak of 1661 is the evidence of the amide linkage.
Characterization of CDDP-PLGA/CUR LBL NPs
TEM image of CDDP-PLGA/CUR LBL NPs exhibited a core-shell morphology (Figure 2). The particle size, zeta potential, EE and LE are summarized in Table 1. CDDP-PLGA/CUR LBL NPs and LBL NPs showed similar sizes (about 180 nm), which means the encapsulation of CDDP and CUR within LBL NPs did not change the size of the system. The PDIs of LBL NPs ranged from 0.116 to 0.157. Zeta potential of LBL NPs was negative, −29.9 ± 3.2 mV for CDDP-PLGA/CUR LBL NPs. EE of CDDP and CUR in LBL NPs were over 80%.
Table 1.
Characterization of LBL NPs (Mean ± SD, n=6)
| Formulation | CDDP-PLGA/CUR LBL NPs | CDDP/CUR LBL NPs | CDDP LBL NPs | CUR LBL NPs | LBL NPs |
|---|---|---|---|---|---|
| Particle size (nm) | 179.6 ± 6.7 | 172.3 ± 5.3 | 177.9 ± 4.4 | 176.3 ± 4.2 | 175.1 ± 3.9 |
| PDI | 0.157 ± 0.031 | 0.143 ± 0.028 | 0.125 ± 0.021 | 0.131 ± 0.023 | 0.116 ± 0.018 |
| Zeta potential (mV) | −29.9 ± 3.2 | −28.7 ± 3.6 | −31.4 ± 3.5 | −30.2 ± 3.1 | −27.6 ± 2.5 |
| EE of CDDP (%) | 85.6 ± 3.9 | 89.1 ± 3.6 | 87.9 ± 3.2 | N/A | N/A |
| EE of CUR (%) | 82.1 ± 2.8 | 81.6 ± 2.7 | N/A | 80.8 ± 2.3 | N/A |
| LE of CDDP (%) | 10.2 ± 0.9 | 11.3 ± 1.1 | 10.8 ± 0.8 | N/A | N/A |
| LE of CUR (%) | 10.8 ± 1.1 | 11.8 ± 1.3 | N/A | 12.1 ± 1.4 | N/A |
Notes: This table summarized the particle size, zeta potential, EE and LE of LBL NPs.
In vitro Drug Release from CDDP-PLGA/CUR LBL NPs
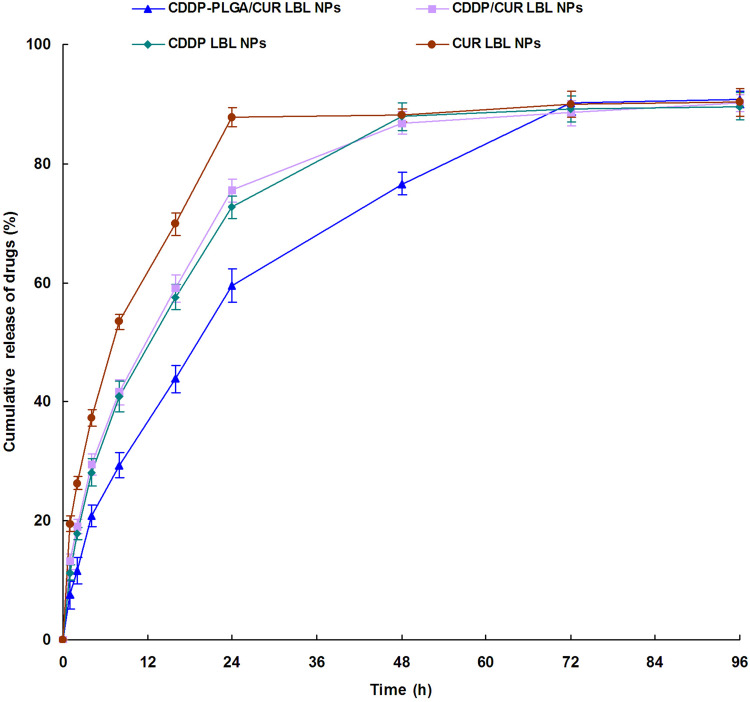
Different release profiles were observed in CDDP-PLGA/CUR LBL NPs and CDDP/CUR LBL NPs systems (Figure 3). Although both of them appeared sustained drug release patterns, while CDDP released from CDDP-PLGA/CUR LBL NPs in a more sustained manner. Nearly complete release of CDDP-PLGA/CUR LBL NPs and CDDP/CUR LBL NPs was observed at 72 h and 48 h, respectively. This could be the evidence that CDDP-PLGA prodrug hindered the CDDP release and produced a more sustained release behavior. The release of CUR from LBL NPs was faster than that of CDDP, which may be explained by the location of CUR is in the outer lipid layer and may release easier than CDDP.
Figure 3.
In vitro drug release profiles of CDDP or CUR from LBL NPs. In vitro release of CDDP or CUR from LBL NPs was conducted by dialysis bag diffusion method. Data presented as mean ± SD, n = 3.
Abbreviations: CDDP, cisplatin; CUR, curcumin; LBL, layer-by-layer; NPs, lipid-polymer hybrid nanoparticles.
In vitro Cytotoxicity of CDDP-PLGA/CUR LBL NPs
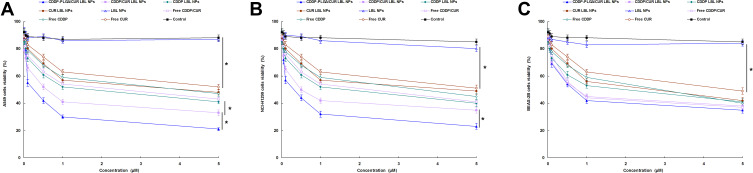
Figure 4 shows the viability of A549 cells, NCI-H1299 cells, and BEAS-2B cells when incubated with CDDP-PLGA/CUR LBL NPs and other formulas. Firstly, blank LBL NPs did not exhibit significant cytotoxicity on these cells, which means the materials used for the preparation of NPs have good biocompatibility. On A549 and NCI-H1299 cells, free drugs showed obvious cytotoxicity than the control group, but not as good as their LBL NPs loaded counterparts, the latter illustrated more notable inhibition effect on the cancer cells (P < 0.05). CDDP-PLGA/CUR LBL NPs displayed the most remarkable cytotoxicity, which is better than CDDP/CUR LBL NPs and others (P < 0.05). In contrast, free drugs and their LBL NPs loaded counterparts showed similar cytotoxicity on BEAS-2B cells.
Figure 4.
The viability of A549 cells (A), NCI-H1299 cells (B), and BEAS-2B cells (C) when incubated with CDDP-PLGA/CUR LBL NPs, CDDP/CUR LBL NPs, CDDP LBL NPs, CUR LBL NPs, LBL NPs, free CDDP/CUR, free CDDP and free CUR in different concentrations for 72 h. In vitro cytotoxicity of LBL NPs on A549 cells was assessed by the MTT assay. Data presented as mean ± SD, n = 6. *means P < 0.05.
Abbreviations: NSCLC, non-small cell lung cancer; CDDP, cisplatin; CUR, curcumin; LBL, layer-by-layer; NPs, lipid-polymer hybrid nanoparticles; MTT, 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide.
Cellular Uptake of CDDP-PLGA/CUR LBL NPs
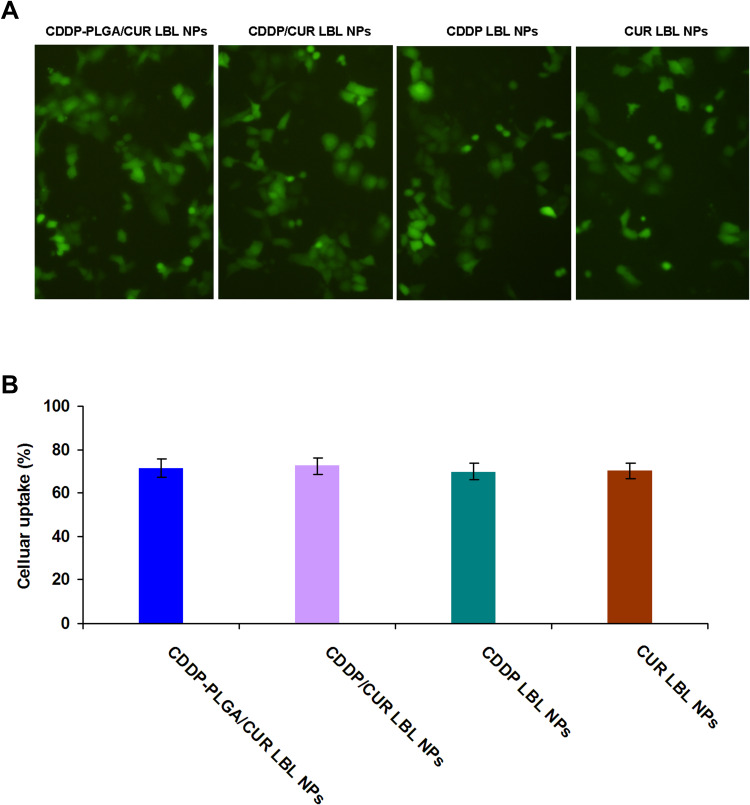
Cellular uptake efficiency of LBL NPs on A549 cells was presented by the fluorescent images (Figure 5A) and flow cytometry (Figure 5B). Images captured by fluorescence microscopy and the quantitative results indicated that the cellular uptake of LBL NPs was about 70%, indicating the high cellular uptake of the carriers. The uptake efficiency of LBL NPs on NCI-H1299 cells, and BEAS-2B cells were summarized and compared with A549 cells in Table 2. The A549 cells and NCI-H1299 cells exhibited similar uptake efficiencies, which are higher than that of BEAS-2B cells.
Figure 5.
Cellular uptake efficiency of LBL NPs presented by the fluorescent images (A) and flow cytometry (B). Cou-6 was used as a fluorescent probe to determine the cellular uptake efficiency of LBL NPs. Scale bar: 20 μm. Data presented as mean ± SD, n = 6.
Abbreviations: LBL, layer-by-layer; NPs, lipid-polymer hybrid nanoparticles.
Table 2.
The Uptake Efficiency of LBL NPs (Mean ± SD, n=6)
| Formulation | Uptake Efficiency (%) | ||
|---|---|---|---|
| A549 Cells | NCI-H1299 Cells | BEAS-2B Cells | |
| CDDP-PLGA/CUR LBL NPs | 71.3 ± 4.3 | 68.9 ± 4.1 | 50.3 ± 3.1 |
| CDDP/CUR LBL NPs | 72.5 ± 3.8 | 70.6 ± 3.8 | 52.7 ± 2.9 |
| CDDP LBL NPs | 69.7 ± 3.9 | 72.1 ± 3.9 | 51.6 ± 3.3 |
| CUR LBL NPs | 70.2 ± 3.5 | 71.7 ± 4.2 | 52.5 ± 3.5 |
Notes: This table summarized the tumor inhibition rates of CDDP-PLGA/CUR LBL NPs, CDDP/CUR LBL NPs, and free CDDP/CUR groups.
In vivo Anti-Tumor Efficacy of CDDP-PLGA/CUR LBL NPs
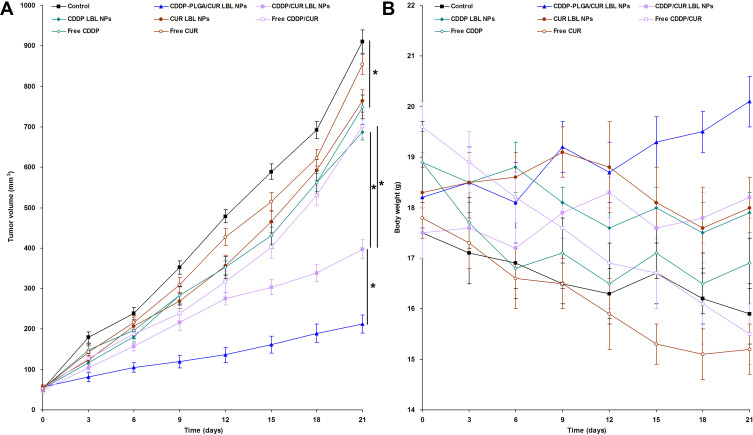
In vivo anti-tumor efficacy of LBL NPs was evaluated on NSCLC bearing mice, and tumor growth and body weight curves are presented in Figure 6. Figure 6A shows that CDDP and CUR encapsulated LBL NPs and free drugs groups significantly suspend the tumor growth (P < 0.05). The most remarkable anti-tumor efficiency was observed in CDDP-PLGA/CUR LBL NPs group, which is obviously higher than CDDP/CUR LBL NPs group and other groups (P < 0.05). Among these groups, CDDP/CUR LBL NPs group exhibited higher tumor inhibition efficacy than free CDDP/CUR group and CDDP LBL NPs group (P < 0.05). The tumor inhibition rates of CDDP-PLGA/CUR LBL NPs, CDDP/CUR LBL NPs, and free CDDP/CUR groups were 76.7 ± 3.1, 56.4 ± 2.2 and 23.1 ± 1.5% (Table 3). Figure 6B exhibits that the body weight of mice was not significantly changed when administrated with drugs loaded LBL NPs but slightly decreased with free drugs administration.
Figure 6.
In vivo anti-tumor efficacy of CDDP-PLGA/CUR LBL NPs evaluated on NSCLC bearing mice, tumor growth (A) and body weight (B) were presented. The tumor volumes were calculated using formulation: V (mm3) = 1/2×(length)×(width)2. After 21 days of injection, mice were sacrificed by cervical decapitation and the final tumor weight and body weight of mice was recorded. Data presented as mean ± SD, n = 6. * means P < 0.05.
Abbreviations: CDDP, cisplatin; CUR, curcumin; LBL, layer-by-layer; NPs, lipid-polymer hybrid nanoparticles; NSCLC, non-small cell lung cancer.
Table 3.
Tumor Inhibition Rates (Mean ± SD, n=6)
| Formulation | Tumor Inhibition Rates (%) |
|---|---|
| CDDP-PLGA/CUR LBL NPs | 76.7 ± 3.1 |
| CDDP/CUR LBL NPs | 56.4 ± 2.2 |
| CDDP LBL NPs | 24.6 ± 1.9 |
| CUR LBL NPs | 16.1 ± 1.2 |
| Free CDDP/CUR | 23.1 ± 1.5 |
| Free CDDP | 17.8 ± 1.1 |
| Free CUR | 6.3 ± 0.8 |
Notes: This table summarized the tumor inhibition rates of CDDP-PLGA/CUR LBL NPs, CDDP/CUR LBL NPs, and free CDDP/CUR groups.
In vivo Tissue Distribution of CDDP-PLGA/CUR LBL NPs
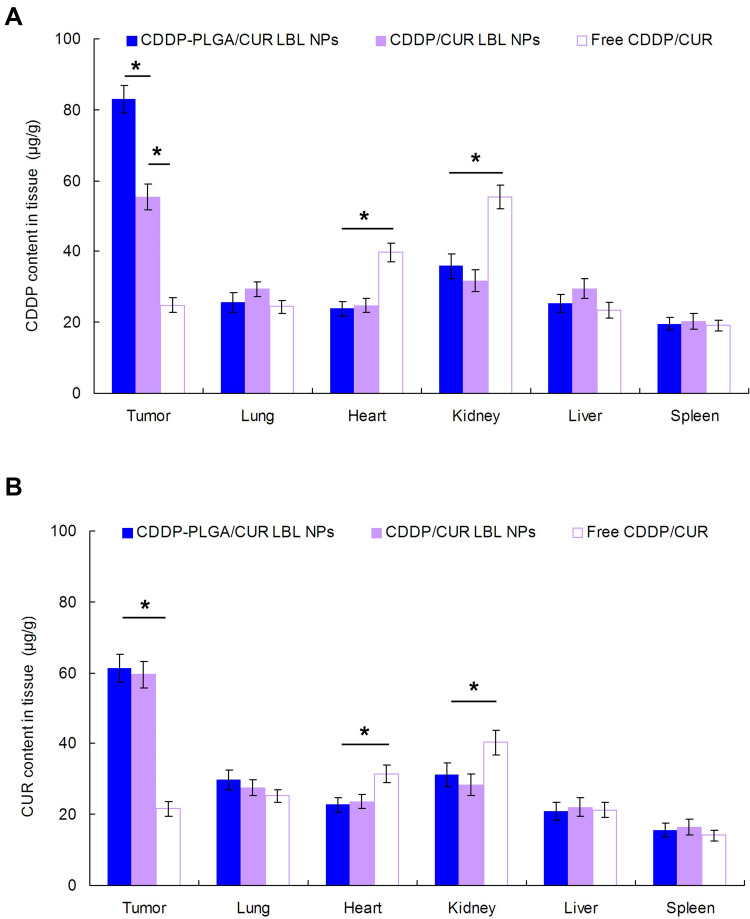
In vivo tissue CDDP or CUR distribution in tissues is different (Figure 7). CDDP distribution in tumor of CDDP-PLGA/CUR LBL NPs was higher than that of CDDP/CUR LBL NPs (P < 0.05); the latter was higher than free CDDP/CUR (P < 0.05) (Figure 7A). However, CUR distribution of CDDP-PLGA/CUR LBL NPs and CDDP/CUR LBL NPs showed no obvious different (Figure 7B). On the other hand, CDDP and CUR distribution in the hearts and kidneys of mice were remarkably reduced by drugs encapsulated LBL NPs than the free drugs group (P < 0.05).
Figure 7.
In vivo tissue CDDP (A) and CUR (B) distribution in tissues at 24 h. Mice were sacrificed at 24 h after intravenous injection. The tumor tissue, lung, heart, kidney, liver, and spleen were harvest, decomposed on heating in nitric acid, evaporated to dryness, and redissolved in acetonitrile and water (50/50, v/v) solution. Data presented as mean ± SD, n = 6. * means P < 0.05.
Abbreviations: NSCLC, non-small cell lung cancer; CDDP, cisplatin; CUR, curcumin.
Discussion
In this paper, prodrug of CDDP (CDDP-PLGA) was synthesized. Amino acid was selected as the connection and it is a relatively weak linkage, under certain conditions the chemical bond would easily rupture and release CDDP.45 Using amino acid as spacer could adjust the drug release behavior and achieve better therapeutic efficiency.46 The LBL NPs drug delivery platform enables ratiometric, synergistic, and modular delivery of combinations of drugs, leading to synthetic lethality and a level of effectiveness not achievable by conventional anticancer drug delivery approaches.47 CDDP-PLGA/CUR LBL NPs showed the size of about 180 nm. Particle sizes smaller than 200 nm are conducive to drug accumulation at the tumor site by the enhanced permeation and retention (EPR) effect.48
The drug release behaviors of CDDP and CUR were summarized. CDDP released from CDDP-PLGA/CUR LBL NPs in a more sustained manner than CDDP/CUR LBL NPs, which could be the evidence that CDDP-PLGA prodrug hindered the CDDP release and produced a more sustained release behavior. Longer time was needed to release CDDP from CDDP-PLGA/CUR LBL NPs may slightly reduce the systemic toxicity. In addition, the time-dependent drug release could lead to stronger cytotoxicity to the cancer cells.49
The internalization and retention ability of nanoparticles in cancer cells has a strong impact on the therapeutic effects of the system.27 Cellular uptake research could provide the evidence of the percentage of LBL NPs that entered the cells. In this study, Cou-6 loaded LBL NPs were applied to observe the uptake efficiency. The quantitative results revealed that about 70% of NPs were uptake by A549 cells and NCI-H1299 cells, which revealed that the LBL NPs had excellent ability to enter cancer cells. This characteristic could improve the therapeutic efficacy of this system due to the high cancer cells uptake ability.
Anti-tumor efficiency of CDDP-PLGA/CUR LBL NPs was evaluated in vitro and in vivo.29 Firstly, the viabilities of A549, NCI-H1299, and BEAS-2B cells were applied to determine the tumor cell inhibition ability of the LBL NPs and the influence on normal cells. CDDP-PLGA/CUR LBL NPs displayed the most remarkable cytotoxicity on cancer cells, which is better than CDDP/CUR LBL NPs group and other groups. This phenomenon may be explained by the better efficiency of the CDDP prodrug that enhanced the antitumor ability. CDDP/CUR LBL NPs illustrated better cell inhibition efficiency than free CDDP/CUR group, which may be explained by the protective effect of LBL NPs that could reduce the degradation of CDDP and CUR in blood circulation and increase the amount of drugs accumulated within the tumor cells. In vivo antitumor study showed that the tumor growth was more prominently inhibited by CDDP-PLGA/CUR LBL NPs than the CDDP/CUR LBL NPs, and free CDDP/CUR groups. These results are in accordance with the results of the in vitro cytotoxicity and could be the evidence that CDDP-PLGA/CUR LBL NPs have better tumor inhibition efficiency. Higher tumor inhibition efficacy achieved by CDDP/CUR LBL NPs group than CDDP LBL NPs group can be the proof of the combined efficiency of CDDP and CUR.50
In vivo drug distribution of CDDP-PLGA/CUR LBL NPs was evaluated and considered along with the body weight changes of the mice.51 Higher CDDP concentration in tumor tissue was observed in CDDP-PLGA/CUR LBL NPs, which is higher than that of CDDP/CUR LBL NPs, and free CDDP/CUR groups. This phenomenon could be explained by the EPR effect at the tumor site.52 Due to little or no lymphatic drainage in tumor tissues, NPs are accumulation more in the tumor, termed the EPR effect.53 The size range (172.6–479.65 nm) reported by Mahmood et al achieve EPR effect, which is in accordance with the sizes in our study.54 This would better prove the better ability of the prodrug-encapsulated system to the tumor tissue. Lower CDDP and CUR distribution in the hearts and kidneys were found in CDDP-PLGA/CUR LBL NPs and CDDP/CUR LBL NPs groups in heart and kidney, which could decrease the systemic toxicity during the tumor therapy. Thus, it is safe and necessary for the application of nanoparticles in the drug delivery system.55 At day 21, mice treated with drugs loaded LBL NPs showed no obvious changes in body weight, which illustrated the well tolerance of the systems. Considering the results along with the antitumor and tissue distribution experiments, the resulting CDDP-PLGA/CUR LBL NPs could be utilized as a safe and effective system for the treatment of NSCLC.
Conclusion
CDDP-PLGA was synthesized and encapsulated along with CUR into the LBL NPs. CDDP-PLGA/CUR LBL NPs showed high uptake and remarkable tumor inhibition efficiency with no obvious changes in body weight. CDDP-PLGA/CUR LBL NPs were reported for the first time in the combination therapy of lung cancer and was expected to be a novel promising system for the synergetic treatment of lung carcinoma. This LBL NPs system may also be used as delivery vectors for the co-encapsulation of drugs and utilized for various cancer therapies.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Guo S, Zhang L, Zhang Y, et al. Long non-coding RNA TUG1 enhances chemosensitivity in non-small cell lung cancer by impairing microRNA-221-dependent PTEN inhibition. Aging (Albany NY). 2019. 11. doi: 10.18632/aging.102271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu B, Ren C, Du K, et al. Olean-28,13b-olide 2 plays a role in cisplatin-mediated apoptosis and reverses cisplatin resistance in human lung cancer through multiple signaling pathways. Biochem Pharmacol. 2019;170:113642. doi: 10.1016/j.bcp.2019.113642 [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Zhang D, Huang L, et al. Discovery of novel biomarkers of therapeutic responses in Han Chinese pemetrexed-based treated advanced NSCLC patients. Front Pharmacol. 2019;10:944. doi: 10.3389/fphar.2019.00944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu Z, Ye B, Zhao S, et al. Non-canonical Raf-1/p70S6K signalling in non-small-cell lung cancer. J Cell Mol Med. 2019;23(11):7632–7640. doi: 10.1111/jcmm.14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta A, Tun A, Ticona K, Baqui A, Guevara E. Invasive aspergillosis in a patient with stage III (or 3a or 3b) non-small-cell lung cancer treated with durvalumab. Case Rep Oncol Med. 2019;2019:2178925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge P, Cao L, Yao YJ, Jing RJ, Wang W, Li HJ. lncRNA FOXD2-AS1 confers cisplatin resistance of non-small-cell lung cancer via regulation of miR185-5p-SIX1 axis. Onco Targets Ther. 2019;12:6105–6117. doi: 10.2147/OTT.S197454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song H, Wang R, Xiao H, et al. A cross-linked polymeric micellar delivery system for cisplatin(IV) complex. Eur J Pharm Biopharm. 2013;83(1):63–75. doi: 10.1016/j.ejpb.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 8.Kesharwani SS, Ahmad R, Bakkari MA, et al. Site-directed non-covalent polymer-drug complexes for inflammatory bowel disease (IBD): formulation development, characterization and pharmacological evaluation. J Control Release. 2018;290:165–179. doi: 10.1016/j.jconrel.2018.08.004 [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Kesharwani SS, Mathur H, Tyagi M, Bhat GJ, Tummala H. Molecular complexation of curcumin with pH sensitive cationic copolymer enhances the aqueous solubility, stability and bioavailability of curcumin. Eur J Pharm Sci. 2016;82:86–96. doi: 10.1016/j.ejps.2015.11.010 [DOI] [PubMed] [Google Scholar]
- 10.Zou W, Liu C, Chen Z, Zhang N. Studies on bioadhesive PLGA nanoparticles: a promising gene delivery system for efficient gene therapy to lung cancer. Int J Pharm. 2009;370(1–2):187–195. doi: 10.1016/j.ijpharm.2008.11.016 [DOI] [PubMed] [Google Scholar]
- 11.Desale SS, Soni KS, Romanova S, Cohen SM, Bronich TK. Targeted delivery of platinum-taxane combination therapy in ovarian cancer. J Control Release. 2015;220(Pt B):651–659. doi: 10.1016/j.jconrel.2015.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Q, Sun W, Wang C, Gu Z. Recent advances of cocktail chemotherapy by combination drug delivery systems. Adv Drug Deliv Rev. 2016;98:19–34. doi: 10.1016/j.addr.2015.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Chen T, Xu H, et al. Curcumin-loaded solid lipid nanoparticles enhanced anticancer efficiency in breast cancer. Molecules. 2018;23(7):1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lv ZD, Liu XP, Zhao WJ, et al. Curcumin induces apoptosis in breast cancer cells and inhibits tumor growth in vitro and in vivo. Int J Clin Exp Pathol. 2014;7(6):2818–2824. [PMC free article] [PubMed] [Google Scholar]
- 15.Tang H, Murphy CJ, Zhang B, et al. Curcumin polymers as anticancer conjugates. Biomaterials. 2010;31(27):7139–7149. doi: 10.1016/j.biomaterials.2010.06.007 [DOI] [PubMed] [Google Scholar]
- 16.Jelezova I, Drakalska E, Momekova D, et al. Curcumin loaded pH-sensitive hybrid lipid/block copolymer nanosized drug delivery systems. Eur J Pharm Sci. 2015;78:67–78. doi: 10.1016/j.ejps.2015.07.005 [DOI] [PubMed] [Google Scholar]
- 17.Misra R, Sahoo SK. Coformulation of doxorubicin and curcumin in poly(D,L-lactide-co-glycolide) nanoparticles suppresses the development of multidrug resistance in K562 cells. Mol Pharm. 2011;8(3):852–866. doi: 10.1021/mp100455h [DOI] [PubMed] [Google Scholar]
- 18.Baek JS, Cho CW. A multifunctional lipid nanoparticle for co-delivery of paclitaxel and curcumin for targeted delivery and enhanced cytotoxicity in multidrug resistant breast cancer cells. Oncotarget. 2017;8(18):30369–30382. doi: 10.18632/oncotarget.16153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan J, Wang Y, Jia Y, et al. Co-delivery of docetaxel and curcumin prodrug via dual-targeted nanoparticles with synergistic antitumor activity against prostate cancer. Biomed Pharmacother. 2017;88:374–383. doi: 10.1016/j.biopha.2016.12.138 [DOI] [PubMed] [Google Scholar]
- 20.Gao Z, Li Z, Yan J, Wang P. Irinotecan and 5-fluorouracil-co-loaded, hyaluronic acid-modified layer-by-layer nanoparticles for targeted gastric carcinoma therapy. Drug Des Devel Ther. 2017;11:2595–2604. doi: 10.2147/DDDT.S140797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramasamy T, Tran TH, Choi JY, et al. Layer-by-layer coated lipid-polymer hybrid nanoparticles designed for use in anticancer drug delivery. Carbohydr Polym. 2014;102:653–661. doi: 10.1016/j.carbpol.2013.11.009 [DOI] [PubMed] [Google Scholar]
- 22.Go DP, Hung A, Gras SL, O’Connor AJ. Use of a short peptide as a building block in the layer-by-layer assembly of biomolecules on polymeric surfaces. J Phys Chem B. 2012;116(3):1120–1133. doi: 10.1021/jp208898m [DOI] [PubMed] [Google Scholar]
- 23.Jeon S, Yoo CY, Park SN. Improved stability and skin permeability of sodium hyaluronate-chitosan multilayered liposomes by Layer-by-Layer electrostatic deposition for quercetin delivery. Colloids Surf B Biointerfaces. 2015;129:7–14. doi: 10.1016/j.colsurfb.2015.03.018 [DOI] [PubMed] [Google Scholar]
- 24.Suh MS, Shen J, Kuhn LT, Burgess DJ. Layer-by-layer nanoparticle platform for cancer active targeting. Int J Pharm. 2017;517(1–2):58–66. doi: 10.1016/j.ijpharm.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 25.Zhou D, Xiao H, Meng F, et al. Layer-by-layer assembled polypeptide capsules for platinum-based pro-drug delivery. Bioconjug Chem. 2012;23(12):2335–2343. doi: 10.1021/bc300144e [DOI] [PubMed] [Google Scholar]
- 26.Zhang R, Ru Y, Gao Y, Li J, Mao S. Layer-by-layer nanoparticles co-loading gemcitabine and platinum (IV) prodrugs for synergistic combination therapy of lung cancer. Drug Des Devel Ther. 2017;11:2631–2642. doi: 10.2147/DDDT.S143047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong Y, Che S, Hui B, et al. Lung cancer therapy using doxorubicin and curcumin combination: targeted prodrug based, pH sensitive nanomedicine. Biomed Pharmacother. 2019;112:108614. doi: 10.1016/j.biopha.2019.108614 [DOI] [PubMed] [Google Scholar]
- 28.Tan S, Wang G. Lung cancer targeted therapy: folate and transferrin dual targeted, glutathione responsive nanocarriers for the delivery of cisplatin. Biomed Pharmacother. 2018;102:55–63. doi: 10.1016/j.biopha.2018.03.046 [DOI] [PubMed] [Google Scholar]
- 29.Yang F, Li A, Liu H, Zhang H. Gastric cancer combination therapy: synthesis of a hyaluronic acid and cisplatin containing lipid prodrug coloaded with sorafenib in a nanoparticulate system to exhibit enhanced anticancer efficacy and reduced toxicity. Drug Des Devel Ther. 2018;12:3321–3333. doi: 10.2147/DDDT.S176879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alibolandi M, Abnous K, Anvari S, Mohammadi M, Ramezani M, Taghdisi SM. CD133-targeted delivery of self-assembled PEGylated carboxymethylcellulose-SN38 nanoparticles to colorectal cancer. Artif Cells Nanomed Biotechnol. 2018;46(sup1):1159–1169. doi: 10.1080/21691401.2018.1446969 [DOI] [PubMed] [Google Scholar]
- 31.Guo S, Zhang Y, Wu Z, et al. Synergistic combination therapy of lung cancer: cetuximab functionalized nanostructured lipid carriers for the co-delivery of paclitaxel and 5-Demethylnobiletin. Biomed Pharmacother. 2019;118:109225. doi: 10.1016/j.biopha.2019.109225 [DOI] [PubMed] [Google Scholar]
- 32.Zhang D, Kong YY, Sun JH, et al. Co-delivery nanoparticles with characteristics of intracellular precision release drugs for overcoming multidrug resistance. Int J Nanomedicine. 2017;12:2081–2108. doi: 10.2147/IJN.S128790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang G, Liu F, Jia E, Jia L, Zhang Y. Folate-modified, cisplatin-loaded lipid carriers for cervical cancer chemotherapy. Drug Deliv. 2016;23(4):1393–1397. doi: 10.3109/10717544.2015.1054052 [DOI] [PubMed] [Google Scholar]
- 34.Cui T, Zhang S, Sun H. Co-delivery of doxorubicin and pH-sensitive curcumin prodrug by transferrin-targeted nanoparticles for breast cancer treatment. Oncol Rep. 2017;37(2):1253–1260. doi: 10.3892/or.2017.5345 [DOI] [PubMed] [Google Scholar]
- 35.Song X, Zhao Y, Hou S, et al. Dual agents loaded PLGA nanoparticles: systematic study of particle size and drug entrapment efficiency. Eur J Pharm Biopharm. 2008;69(2):445–453. doi: 10.1016/j.ejpb.2008.01.013 [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Angelidaki I. Submersible microbial fuel cell sensor for monitoring microbial activity and BOD in groundwater: focusing on impact of anodic biofilm on sensor applicability. Biotechnol Bioeng. 2011;108(10):2339e47. doi: 10.1002/bit.23204 [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Zhang P, Zhu T. Ovarian carcinoma biological nanotherapy: comparison of the advantages and drawbacks of lipid, polymeric, and hybrid nanoparticles for cisplatin delivery. Biomed Pharmacother. 2019;109:475–483. doi: 10.1016/j.biopha.2018.10.158 [DOI] [PubMed] [Google Scholar]
- 38.Ruan C, Liu L, Lu Y, et al. Substance P-modified human serum albumin nanoparticles loaded with paclitaxel for targeted therapy of glioma. Acta Pharm Sin B. 2018;8(1):85–96. doi: 10.1016/j.apsb.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srinivasan AR, Lakshmikuttyamma A, Shoyele SA. Investigation of the stability and cellular uptake of self-associated monoclonal antibody (MAb) nanoparticles by non-small lung cancer cells. Mol Pharm. 2013;10(9):3275–3284. doi: 10.1021/mp3005935 [DOI] [PubMed] [Google Scholar]
- 40.Rajput MKS, Kesharwani SS, Kumar S, Muley P, Narisetty S, Tummala H. Dendritic cell-targeted nanovaccine delivery system prepared with an immune-active polymer. ACS Appl Mater Interfaces. 2018;10(33):27589–27602. doi: 10.1021/acsami.8b02019 [DOI] [PubMed] [Google Scholar]
- 41.Zhu D, Tao W, Zhang H, et al. (DTX)-loaded polydopamine-modified TPGS-PLA nanoparticles as a targeted drug delivery system for the treatment of liver cancer. Acta Biomater. 2016;30:144–154. doi: 10.1016/j.actbio.2015.11.031 [DOI] [PubMed] [Google Scholar]
- 42.Xiong Y, Zhao Y, Miao L, Lin CM, Huang L. Co-delivery of polymeric metformin and cisplatin by self-assembled core-membrane nanoparticles to treat non-small cell lung cancer. J Control Release. 2016;244(Pt A):63–73. doi: 10.1016/j.jconrel.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding Q, Li Z, Yang Y, et al. Preparation and therapeutic application of docetaxel-loaded poly(d,l-lactide) nanofibers in preventing breast cancer recurrence. Drug Deliv. 2016;23(8):2677–2685. doi: 10.3109/10717544.2015.1048490 [DOI] [PubMed] [Google Scholar]
- 44.Nishiyama N, Okazaki S, Cabral H, et al. Novel cisplatin-incorporated polymeric micelles can eradicate solid tumors in mice. Cancer Res. 2003;63(24):8977–8983. [PubMed] [Google Scholar]
- 45.Lu Z, Su J, Li Z, Zhan Y, Ye D. Hyaluronic acid-coated, prodrug-based nanostructured lipid carriers for enhanced pancreatic cancer therapy. Drug Dev Ind Pharm. 2017;43(1):160–170. doi: 10.1080/03639045.2016.1226337 [DOI] [PubMed] [Google Scholar]
- 46.Feng X, Yuan YJ, Wu JC. Synthesis and evaluation of water-soluble paclitaxel prodrugs. Bioorg Med Chem Lett. 2002;12(22):3301–3303. doi: 10.1016/S0960-894X(02)00694-7 [DOI] [PubMed] [Google Scholar]
- 47.Mensah LB, Morton SW, Li J, et al. Layer-by-layer nanoparticles for novel delivery of cisplatin and PARP inhibitors for platinum-based drug resistance therapy in ovarian cancer. Bioeng Transl Med. 2019;4(2):e10131. doi: 10.1002/btm2.10131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yugui F, Wang H, Sun D, Zhang X. Nasopharyngeal cancer combination chemoradiation therapy based on folic acid modified, gefitinib and yttrium 90 co-loaded, core-shell structured lipid-polymer hybrid nanoparticles. Biomed Pharmacother. 2019;114:108820. doi: 10.1016/j.biopha.2019.108820 [DOI] [PubMed] [Google Scholar]
- 49.Li M, Zhao L, Zhang T, et al. Redox-sensitive prodrug nanoassemblies based on linoleic acid-modified docetaxel to resist breast cancers. Acta Pharm Sin B. 2019;9(2):421–432. doi: 10.1016/j.apsb.2018.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang H, Geng D, Liu H, Li Z, Cao J. Co-delivery of etoposide and curcumin by lipid nanoparticulate drug delivery system for the treatment of gastric tumors. Drug Deliv. 2016;23(9):3665–3673. doi: 10.1080/10717544.2016.1217954 [DOI] [PubMed] [Google Scholar]
- 51.Duan W, Liu Y. Targeted and synergistic therapy for hepatocellular carcinoma: monosaccharide modified lipid nanoparticles for the co-delivery of doxorubicin and sorafenib. Drug Des Devel Ther. 2018;12:2149–2161. doi: 10.2147/DDDT.S166402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baeza A. Tumor targeted nanocarriers for immunotherapy. Molecules. 2020;25(7):1508. doi: 10.3390/molecules25071508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maruyama K. Intracellular targeting delivery of liposomal drugs to solid tumors based on EPR effects. Adv Drug Deliv Rev. 2011;63(3):161–169. doi: 10.1016/j.addr.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 54.Mahmood MA, Madni A, Rehman M, Rahim MA, Jabar A. Ionically cross-linked chitosan nanoparticles for sustained delivery of docetaxel: fabrication, post-formulation and acute oral toxicity evaluation. Int J Nanomedicine. 2019;20(14):10035–10046. doi: 10.2147/IJN.S232350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen D, Jiang X, Liu J, Jin X, Zhang C, Ping Q. In vivo evaluation of novel pH-sensitive mPEG-Hz-Chol conjugate in liposomes: pharmacokinetics, tissue distribution, efficacy assessment. Artif Cells Blood Substit Immobil Biotechnol. 2010;38(3):136–142. doi: 10.3109/10731191003685481 [DOI] [PubMed] [Google Scholar]