Abstract
Purpose
Our research aimed to illuminate the role of miR-100-5p in chordoma and potential mechanism.
Materials and Methods
We used microRNA array analysis to explore differentially expressed miRNAs in chordoma tissue and then verified by qRT-PCR. Cell proliferation and transwell assay were used to evaluate the function of miR-100-5p. Cell apoptosis was analyzed by flow cytometry, and using biological software, we predicted that the insulin-like growth factor 1 receptor (IGF1R) could be the target gene of miR-100-5p, which was then validated by dual luciferase assays and Western blot.
Results
miR-100-5p was downregulated in chordoma tissues. Overexpression of miR-100-5p could suppress the growth of chordoma both in vitro and in vivo, and miR-100-5p could inhibit the migration and invasion of chordoma cells partially by suppressing epithelial–mesenchymal transition (EMT). Furthermore, IGF1R was validated as the target gene of miR-100-5p and expressed in most chordoma tissues.
Conclusion
In conclusion, our results showed that miR-100-5p was lowly expressed in chordoma and inhibited tumor malignant progression by targeting IGF1R.
Keywords: chordoma, miRNA, miR-100-5p, IGF1R
Introduction
Chordoma is a mesenchymal tissue malignant bone tumor that arises from notochord remnants, which is most prevalent in the 50 to 60-year age group and the axial skeleton is the most common position.1 Chordoma is unresponsive to radiotherapy and conventional chemotherapy drugs, and characterized by a high recurrence rate. Duo to the complex anatomical structure of chordoma, patients who underwent surgical resection often cannot receive a good prognosis.2 Therefore, it is urgent to investigate novel molecular markers of chordoma.
miRNAs are 17–25 nt non-coding RNAs that can suppress mRNAs at the post-transcriptional level by complementary binding to 3′-UTRs. Highly conserved miRNAs are believed to play important roles in regulating tumor progressions such as migration, invasion, and apoptosis.3 Furthermore, increasing researches showed that miRNAs dysregulation was significant in the progression of chordoma.4 Thereby, we want to detect the expression of miRNAs in chordoma tissues and illuminate the potential function and mechanism of dysregulated miRNAs in chordoma.
In our study, we found that miR-100-5p was downregulated in chordoma tissues. And current studies showed a controversial role of miR-100 in tumor progressions, for example, miR-100 was found to function as an oncogene in renal cell carcinoma,5 but a tumor suppressor in epithelial ovarian cancer.6 Therefore, in this research, we aim to clarify the real role of miR-100-5p in chordoma and further illuminate the potential molecular mechanism. And finally, we demonstrated that in chordoma, miR-100-5p played a crucial role as a tumor suppressor gene and inhibited tumor malignant progression by targeting IGF1R.
Materials and Methods
Clinical Tissue
All tissues in this study were gathered from the Musculoskeletal Tumor Center, under the protocols approved by the Ethics Committee of Peking University People’s Hospital. Fresh tissues were collected and frozen in liquid nitrogen immediately. Written informed consents were obtained from all patients following the Declaration of Helsinki.
Cell Lines and Cell Transfection
The MUG and U-CH1 were purchased from American Type Culture Collection (ATCC) and were maintained in Iscove’s Modified Dulbecco’s Medium (IMDM; Gibco, Grand Island, NY, USA): RPMI-1640 medium (Gibco) (4:1) with 10% FBS (Gibco) and 1% penicillin/streptomycin (Gibco) at 37°C with 5% CO2. For transfection, cells were cultured in a 6‑well plate, and miR-100-5p mimics and negative control (NC) (GenePharma, Suzhou, China) were transfected into cells by Lipofectamine3000 (Invitrogen, Carlsbad, CA, USA). The final concentration of miRNA mimics was 100 nM, the sense sequence of miR-100-5p mimic was 5ʹ-AACCCGUAGAUCCGAACUUGUG-3ʹ, the antisense was 5ʹ-CACAAGUUCGGAUCUACGGGUU-3ʹ), the sense sequence of mimic NC was 5ʹ-UUUGUACUACACAAAAGUACUG-3ʹ, and the antisense was 5ʹ-CAGUACUUUUGUGUAGUACAAA-3ʹ.
qRT-PCR
The total RNA was isolated using TRIzol reagent (Invitrogen) and the miRNAs were extracted with RNeasy/miRNeasy Mini Kit (Qiagen, Limburg, The Netherlands). The reverse transcription was carried out with Super Script First Strand cDNA System (Invitrogen, Grand Island, NY, USA). All primer sequences are provided in Table 1.
Table 1.
Primers for Real-Time PCR
| Primers | Sequences | |
|---|---|---|
| E-cadherin | F | 5′-TGCTCACATTTCCCAACTC-3' |
| R | 5′-TCTGTCACCTTCAGCCATC-3' | |
| N-cadherin | F | 5′-CTGACAATGACCCCACAGC-3' |
| R | 5′-TCCTGCTCACCACACTACTT-3' | |
| Vimentin | F | 5′-CTGGATTTCCTCTTCGTGGA-3' |
| R | 5′-CGAAAACACCCTGCAATCTT-3' | |
| GAPDH | F | 5′-GCACCGTCAAGGCTGAGAAC-3' |
| R | 5′-ATGGTGGTGAAGACGCCAGT-3' | |
| miR-100-5p | F | 5′-GTGTTCAAGCCTAGATGCCCAA-3' |
| R | 5′-GCATCTAGGCTTGAACACGCC-3' | |
| U6 | F | 5′-CTCGCTTCGGCAGCACA-3' |
| R | 5′-AACGCTTCACGAATTTGCGT-3' |
Western Blot
Proteins were collected from cells lysates and loaded to 10% SDS-PAGE gels, and then transferred to PVDF membranes. After blocking, membranes were incubated with primary antibodies and then secondary antibodies. The primary antibodies used in this study include anti-Vimentin (Cell Signaling Technology (CST), Rabbit, 1:1000, MA, USA), anti-N-cadherin (CST, Rabbit, 1:1000), anti- IGF1R (CST, Rabbit, 1:1000), anti-GAPDH (Abcam, Mouse, 1:1000), and anti-E-cadherin (Abcam, Rabbit, 1:1000).
CCK-8 Assay and Colony Formation Assay
After transfection, 5,000 cells were plated in 96-well plates, and 10μL CCK-8 (Dojindo Laboratories, Kumamoto, Japan) was added to examining cell viability. For colony formation assay, 1,000 cells were plated in 6-well plates. After 10 days, cells were fixed and stained, then colonies were counted for analysis.
Wound-Healing and Transwell Assay
After transfection, cells were plated in 6-well plates and cultured to a confluent state. P-200 pipette tip was used to scratch, then data was recorded at different time points. For transwell assay, 3 × 104 cells in 200ul serum-free medium were seeded in the upper chambers (BD Biosciences) and there was 600ul medium with 20% FBS in the lower chamber. After two days, the cells were fixed and stained for analyzing under microscope.
Flow Cytometry for Apoptosis Analysis
After transfection, chordoma cells were stained with the Annexin V/FITC Kit (BD Biosciences) according to the manufacturer’s instructions and analyzed by flow cytometry.
Luciferase Reporter Assay
Wild-type/mutant IGF1R luciferase reporter vector was synthesized by Genscript (Nanjing, Jiangsu, China), and U-CH1 cells were co-transfected with the wild-type/mutant IGF1R luciferase reporter vector and miR-100-5p mimic/NC using Lipofectamine 3000. After 48h, Luciferase activity was measured, and the results were normalized to Renilla luciferase activity.
Tumorigenicity Assay
Briefly, 5 × 106 U-CH1 cells were injected subcutaneously to the right flank of 8 BALB/c athymic nude mice (Vitalriver, Beijing, China). Then, 8 mice were randomly divided into two groups and intratumor injection of 10 nmol mir-100-5p agomir (Ribobio Co. Guangzhou, China) or negative control (scramble) was performed once every seven days for seventy days. The tumor volume (length × width2/2) was measured every week.7 Finally, the mice were sacrificed and the tumor was removed for further research.
All animal experiments were performed with written confirmation authorized by the Animal Care and Use Committee of Peking University People’s Hospital, and followed the guidelines of Animal Care and Institutional Ethical in China.
Immunohistochemistry
Immunohistochemistry (IHC) was performed as reported before.4 Simply, 8 xenograft tumors were used to detect the expression of EMT markers and 45 primary chordoma samples were used to analyze the relationship between the expression of IGF1R and clinical characteristics.
MicroRNA Array Analysis and Statistical Analyses
MicroRNA array analysis was performed as reported before.4 Simply, 12 chordoma tissues and 12 nucleus pulposus tissues (non-tumor tissues) were collected for microRNA array analysis, and additional 16 chordoma tissues and 16 nucleus pulposus tissues were used as the validation set.
The data were analyzed by GraphPad Prism 8 using a Student’s t-test or one-way ANOVA. All results were shown as the mean ± SD and P<0.05 means statistical significance. P < 0.05 (*), P < 0.01 (**).
Results
miR-100-5p Is Downregulated in Chordoma Samples
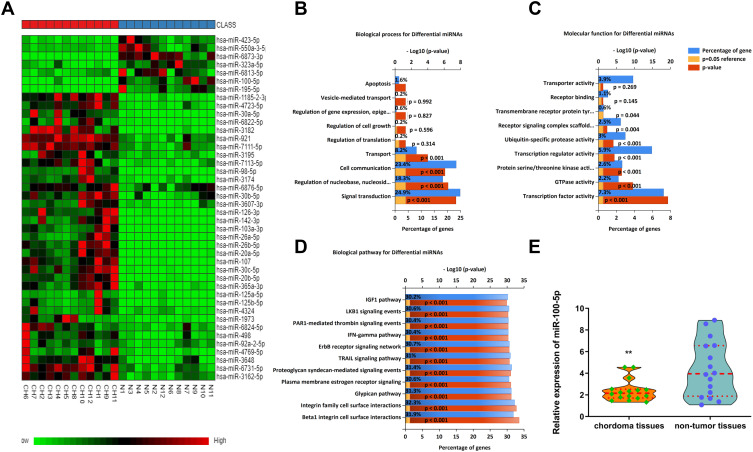
Using miRNA microarray assay, we found that some miRNAs were dysregulated in chordoma samples. Partial dysregulated miRNAs were shown in Heat-map, among which, miR-100-5p was downregulated (Figure 1A). Bioinformatics analysis results showed that dysregulated miRNAs might associate with biological processes such as signal transduction, cell communication, transport et al (Figure 1B). And these miRNAs might have functions like transcription factor activity, GTPase activity, and transcription regulator activity (Figure 1C). Pathway analysis showed that several different pathways corresponded to the significantly dysregulated miRNAs. In which, we could see TRAIL signaling pathway, IFN-γ signaling pathway, IGF1 pathway, and so on (Figure 1D). All these findings indicated that dysregulated miRNAs played a significant role in chordoma. Further validation by qRT-PCR showed that miR-100-5p was significantly downregulated in chordoma (Figure 1E).
Figure 1.
The miR-100-5p expression level was downregulated in chordoma tissues. (A) Heat-map representation of the differentially expressed miRNA patterns in chordoma (CH) versus nucleus pulposus tissues (N). Relative expression is presented as a color gram (red: high expression). (B–D) Bioinformatics analysis of biological processes, molecular functions, and biological pathways about dysregulated miRNAs. (E) Expression of miR-100-5p as determined by real-time RT-PCR analysis. A comparison of the average relative expression levels of miR-100-5p in chordoma and nucleus pulposus samples is shown in the Violin plot. **P < 0.01.
miR-100-5p Can Suppress Chordoma Cell Proliferation and Induce Apoptosis
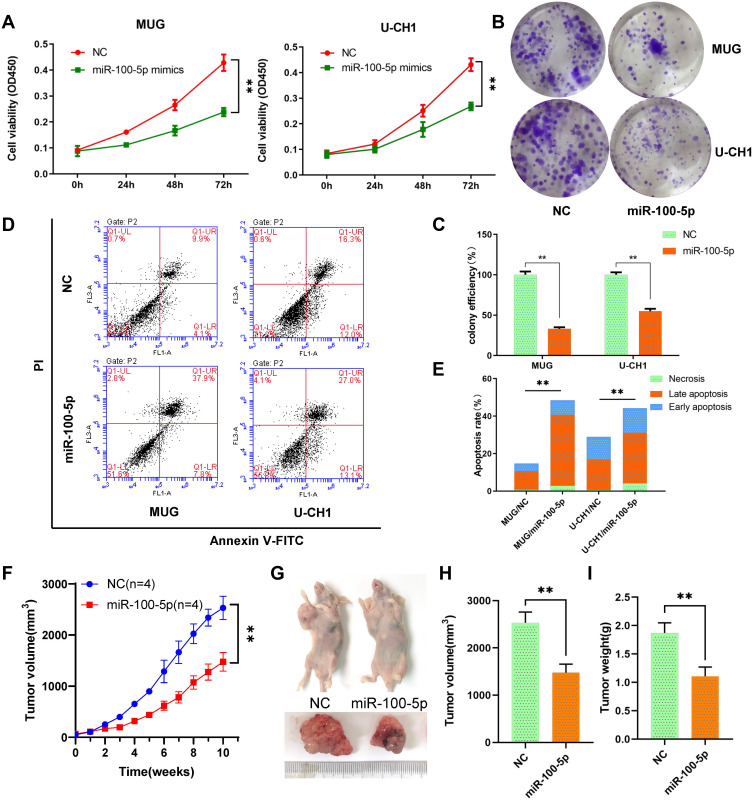
Cck-8 assays results indicated that the viability of chordoma cells MUG and U-CH1 was significantly suppressed by miR-100-5p mimics compared with NC (Figure 2A). And, miR-100-5p mimics decreased both MUG and U-CH1 cell colony formation compared with negative control (Figure 2B and C). To determine whether miR-100-5p could inhibit cell proliferation by inducing apoptosis, flow cytometry was used and results showed that miR-100-5p mimics could induce apoptosis significantly compared with NC (Figure 2D and E). These data collectively showed that miR-100-5p could inhibit cell proliferation and partially by inducing apoptosis.
Figure 2.
miR-100-5p suppressed cell proliferation in vitro and vivo. (A) Cell proliferation was evaluated by the CCK-8 assay at 24, 48, and 72 h. (B and C) miR-100-5p decreased both MUG and U-CH1 cell colony formation compared with control. (D and E) miR-100-5p induced apoptosis in chordoma cells. (F) The growth curves of the tumors. (G–I) Both tumor volume and weight were significantly decreased in the miR-100-5p agomir treatment group compared with the negative control group. All experiments were repeated three times. Data are presented as the mean ± S.D. **P < 0.01.
miR-100-5p Can Retard the Growth of Chordoma in vivo
To further evaluate the effect of miR-100-5p on chordoma, we established an in vivo xenograft model. Results indicated that overexpression of miR-100-5p significantly retarded the growth of chordoma (Figure 2F), and both tumor volume and tumor weight were significantly decreased after miR-100-5p agomir treatment (Figure 2G–I).
miR-100-5p Can Attenuate Chordoma Cell Migration and Invasion by Regulating EMT
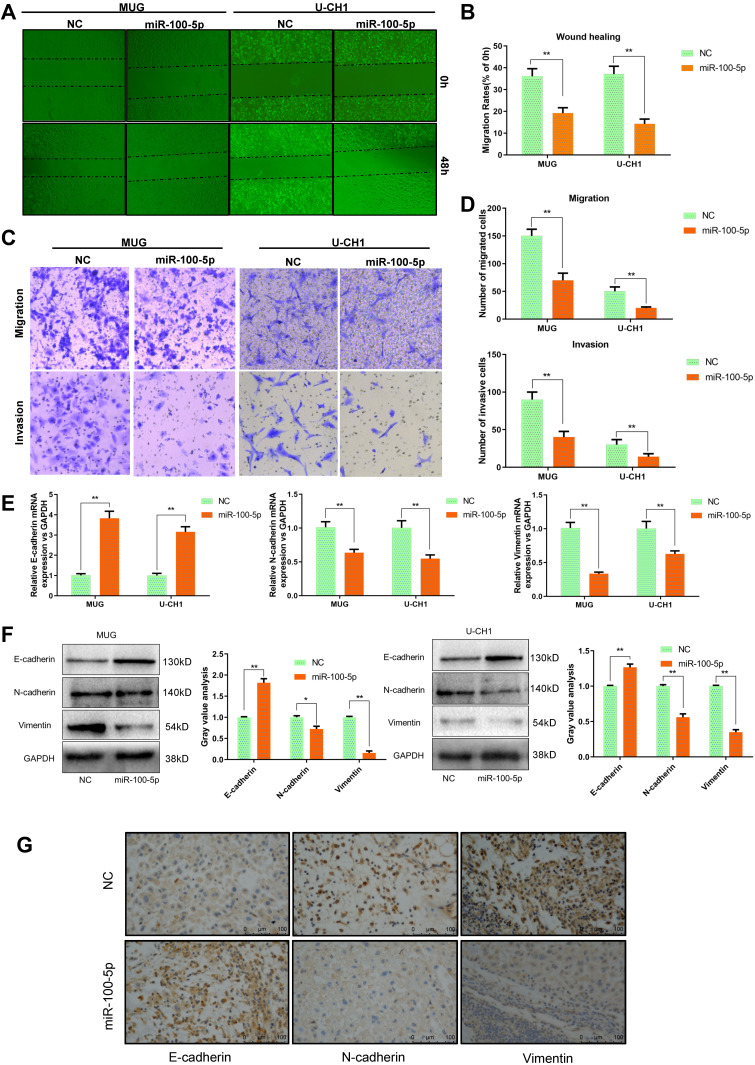
As shown in Figure 3A and B, wound healing assay indicated that overexpression of miR-100-5p could significantly inhibit the migration capacity of chordoma cells. And transwell assays showed that both migration and invasion ability of chordoma cells were attenuated significantly by miR-100-5p mimics (Figure 3C and D). Considering that EMT plays a crucial role in tumor metastasis. We further explored whether miR-100-5p could regulate EMT process and qRT-PCR and Western blotting results showed that miR-100-5p could significantly reduce the expression of N-cadherin and vimentin but increase the expression of E-cadherin in chordoma cells (Figure 3E and F). Furthermore, in vivo experiments showed the same results (Figure 3G).
Figure 3.
miR-100-5p suppressed the invasion and migration of chordoma cells. (A and B) Wound-healing assays were performed in chordoma cells transfected with miR-100-5p mimics or negative controls. (C and D) Transwell assays showed that migration and invasion ability of chordoma cells can be attenuated significantly by miR-100-5p mimics. (magnification 100X). (E and F) The mRNA and protein expression levels of E-cadherin, N-cadherin, and vimentin in chordoma cells transfected with miR-100-5p mimics or negative control were measured by qRT-PCR and Western blotting separately. (G) IHC results showed that miR-100-5p increased the expression of E-cadherin and decreased the expression of vimentin and N-cadherin in tumors formed by chordoma cells. All experiments were repeated three times. Data are presented as the mean ± S.D. *P < 0.05, **P < 0.01.
IGF1R Is the Target of miR-100-5p and Highly Expressed in Chordoma
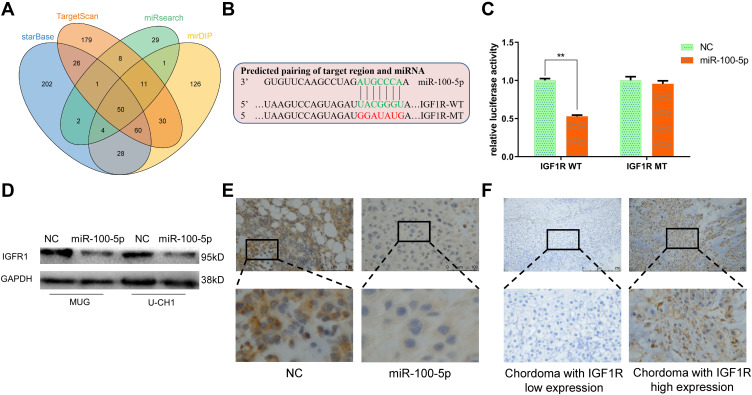
TargetScan, miRsearch, starBase, and mirDIP were used to predict target genes of miR-100-5p and by which we found IGF1R (Figure 4A). Dual-luciferase reporter assay and Western blotting results confirmed that IGF1R was the target gene of miR-100-5p (Figure 4B–D). And the immunohistochemistry results of xenograft models showed that IGF1R was downregulated in the miR-100-5p group when compared with the NC group. Furthermore, immunohistochemistry results showed that IGF1R was highly expressed in chordoma tissues (31 of 45) (Figure 4E), and highly expressed IGF1R was correlated with surrounding invasion (p < 0.05, Table 2).
Figure 4.
Identification of IGF1R as a target of miR-100-5p. (A) Venn diagram showed the comparison of targets of miR-100-5p predicted by TargetScan, miRsearch, starBase, and mirDIP. (B) Schematic representation of IGF1R 3ʹ-UTR containing the wild-type or mutant binding site for miR-100-5p. (C) Luciferase reporter activity following expression after transfection (negative control and miR-100-5p mimics) in U-CH1 cells. (D) Western blot analysis of IGF1R expression in miR-100-5p-overexpressed cells. (E) Representative IHC staining images of IGF1R expression in xenograft models. (F) Representative IHC staining images of IGF1R expression in chordoma tissues. All experiments were repeated three times. Data are presented as the mean ± S.D. **P < 0.01.
Table 2.
The Expression of IGF1R in Chordoma Tissues
| Biological Characteristics | n | Lower Expression | Higher Expression | χ2 | p | ||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| Age (years) | |||||||
| >60 | 24 | 8 | 57.14 | 16 | 51.61 | 0.119 | 0.731 |
| ≤60 | 21 | 6 | 42.86 | 15 | 48.39 | ||
| Gender | |||||||
| Male | 32 | 10 | 71.43 | 22 | 70.97 | 0.001 | 0.975 |
| Female | 13 | 4 | 28.57 | 9 | 29.03 | ||
| Relapse | |||||||
| Yes | 16 | 6 | 42.86 | 10 | 32.26 | 0.473 | 0.492 |
| No | 29 | 8 | 57.14 | 21 | 67.74 | ||
| Surrounding invasion | |||||||
| Yes | 27 | 4 | 28.57 | 23 | 74.19 | 8.364 | 0.004 |
| No | 18 | 10 | 71.43 | 8 | 25.81 | ||
Discussion
Chordoma is a malignant tumor with the ability of metastasis and recurrence. However, there are few researches about the molecular and functional mechanism of chordoma. Our study focused on the aberrantly expressed miRNAs in chordoma. Currently, there are some researches have revealed that several miRNAs might play crucial roles in the progression of chordoma. For example, Osaka et al revealed that upregulated miR-155 could regulate cell proliferation, apoptosis, and EMT in chordoma.8 Malgulwar et al found that miR-193a-5p and miR-671-5p could influence the tumor by targeting TGF-β signaling in chordoma.9 Zou et al found that downregulated miR-1237-3p in chordoma was related to poor prognosis.10 Zhang et al found that miR-34a and miR-608 were participated in the progression of chordoma by regulating EGFR and MET.11 Furthermore, our previous study indicated that downregulated miR-16-5p could regulate the malignant development of chordoma by targeting Smad3.4
miR-100-5p has been found aberrantly expressed in some type of cancers, such as oral cancer, acute myeloid leukemia, nasopharyngeal carcinoma and endometrioid endometrial carcinoma.12–15 However, there has no research about the expression and function of miR-100-5p in chordoma. In our study, we found that downregulated miR-100-5p could be a tumor suppressor gene and further research confirmed that miR-100-5p could suppress the progression of chordoma, and IGF1R was the target gene.
IGF1R is a widely expressed transmembrane tyrosine kinase receptor and involved in many types of solid tumors, such as osteosarcoma, colorectal cancer, breast cancer, and prostate cancer.16–19 Previous studies have demonstrated that IGF-1R participated in various biological processes of tumorigenesis and development, including proliferation, apoptosis, migration, invasion, vascularization, and distant metastasis.20 Our results demonstrated that IGF1R was highly expressed in most of the chordoma tissues and highly expressed IGF1R was correlated with surrounding invasion. All these findings indicated that miR-100-5p could inhibit the malignant behavior of chordoma cells by targeting IGF1R.
Conclusion
In summary, we identified that miR-100-5p was lowly expressed in chordoma tissues, and miR-100-5p could inhibit the malignant behavior of chordoma by targeting IGF1R which provided a new insight into the molecular mechanism of chordoma and highlight the potential of miR-100-5p and IGF1R as new therapeutic targets.
Acknowledgments
This work was supported by the Beijing Municipal Natural Science Foundation (No. 7172227) and the National Nature Science Foundation of China (No. 81972509).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Yamaguchi T, Yamato M, Saotome K. First histologically confirmed case of a classic chordoma arising in a precursor benign notochordal lesion: differential diagnosis of benign and malignant notochordal lesions. Skeletal Radiol. 2002;31(7):413–418. doi: 10.1007/s00256-002-0514-z [DOI] [PubMed] [Google Scholar]
- 2.Smoll NR, Gautschi OP, Radovanovic I, Schaller K, Weber DC. Incidence and relative survival of chordomas: the standardized mortality ratio and the impact of chordomas on a population. Cancer. 2013;119(11):2029–2037. doi: 10.1002/cncr.28032 [DOI] [PubMed] [Google Scholar]
- 3.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13(1):48–57. doi: 10.1016/j.ccr.2007.12.008 [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Yang K, Ren T, Huang Y, Tang X, Guo W. miR-16-5p inhibits chordoma cell proliferation, invasion and metastasis by targeting Smad3. Cell Death Dis. 2018;9(6):680. doi: 10.1038/s41419-018-0738-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen P, Lin C, Quan J, et al. Oncogenic miR-100-5p is associated with cellular viability, migration, and apoptosis in renal cell carcinoma. Mol Med Rep. 2017;16(4):5023–5030. doi: 10.3892/mmr.2017.7139 [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Yu M, Guan S, Zhang G, Wang J, Cheng Y. Prognostic significance of microRNA-100 in solid tumors: an updated meta-analysis. Onco Targets Ther. 2017;10:493–502. doi: 10.2147/OTT.S122774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faustino-Rocha A, Oliveira PA, Pinho-Oliveira J, et al. Estimation of rat mammary tumor volume using caliper and ultrasonography measurements. Lab Anim. 2013;42(6):217–224. doi: 10.1038/laban.254 [DOI] [PubMed] [Google Scholar]
- 8.Osaka E, Kelly AD, Spentzos D, et al. MicroRNA-155 expression is independently predictive of outcome in chordoma. Oncotarget. 2015;6(11):9125–9139. doi: 10.18632/oncotarget.3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malgulwar PB, Pathak P, Singh M, et al. Downregulation of SMARCB1/INI1 expression in pediatric chordomas correlates with upregulation of miR-671-5p and miR-193a-5p expressions. Brain Tumor Pathol. 2017;34(4):155–159. doi: 10.1007/s10014-017-0295-7 [DOI] [PubMed] [Google Scholar]
- 10.Zou MX, Huang W, Wang XB, et al. Reduced expression of miRNA-1237-3p associated with poor survival of spinal chordoma patients. Eur Spine J. 2015;24(8):1738–1746. doi: 10.1007/s00586-015-3927-9 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Schiff D, Park D, Abounader R. MicroRNA-608 and microRNA-34a regulate chordoma malignancy by targeting EGFR, Bcl-xL and MET. PLoS One. 2014;9(3):e91546. doi: 10.1371/journal.pone.0091546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henson BJ, Bhattacharjee S, O’Dee DM, Feingold E, Gollin SM. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer. 2009;48(7):569–582. doi: 10.1002/gcc.20666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rucker FG, Russ AC, Cocciardi S, et al. Altered miRNA and gene expression in acute myeloid leukemia with complex karyotype identify networks of prognostic relevance. Leukemia. 2013;27(2):353–361. doi: 10.1038/leu.2012.208 [DOI] [PubMed] [Google Scholar]
- 14.Sun X, Liu X, Wang Y, Yang S, Chen Y, Yuan T. miR-100 inhibits the migration and invasion of nasopharyngeal carcinoma by targeting IGF1R. Oncol Lett. 2018;15(6):8333–8338. doi: 10.3892/ol.2018.8420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres A, Torres K, Pesci A, et al. Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinoma. BMC Cancer. 2012;12:369. doi: 10.1186/1471-2407-12-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YH, Wang ZX, Qiu Y, et al. Lentivirus-mediated RNAi knockdown of insulin-like growth factor-1 receptor inhibits growth, reduces invasion, and enhances radiosensitivity in human osteosarcoma cells. Mol Cell Biochem. 2009;327(1–2):257–266. doi: 10.1007/s11010-009-0064-y [DOI] [PubMed] [Google Scholar]
- 17.Codony-Servat J, Cuatrecasas M, Asensio E, et al. Nuclear IGF-1R predicts chemotherapy and targeted therapy resistance in metastatic colorectal cancer. Br J Cancer. 2017;117(12):1777–1786. doi: 10.1038/bjc.2017.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pian L, Wen X, Kang L, et al. Targeting the IGF1R pathway in breast cancer using antisense lncRNA-mediated promoter cis competition. Mol Ther Nucleic Acids. 2018;12:105–117. doi: 10.1016/j.omtn.2018.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahearn TU, Peisch S, Pettersson A, et al. Expression of IGF/insulin receptor in prostate cancer tissue and progression to lethal disease. Carcinogenesis. 2018;39(12):1431–1437. doi: 10.1093/carcin/bgy112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jentzsch T, Robl B, Husmann M, Bode-Lesniewska B, Fuchs B. Worse prognosis of osteosarcoma patients expressing IGF-1 on a tissue microarray. Anticancer Res. 2014;34(8):3881–3889. [PubMed] [Google Scholar]